Abstract
Usher syndrome (USH) is the most common form of deaf-blindness in humans. Molecular characterization revealed that the USH gene products form a macromolecular protein network in hair cells of the inner ear and in photoreceptor cells of the retina via binding to PDZ domains in the scaffold protein harmonin encoded by the Ush1c gene in mice and humans. Although several mouse mutants for the Ush1c gene have been described, we generated a targeted null mutation Ush1c mouse model in which the first four exons of the Ush1c gene were replaced with a reporter gene. Here, we assessed the expression pattern of the reporter gene under control of Ush1c regulatory elements and characterized the phenotype of mice defective for Ush1c. These Ush1 knockout mice are deaf but do not recapitulate vision defects before 10 months of age. Our data show LacZ expression in multiple layers of the retina but in neither outer nor inner segments of the photoreceptor layers in mice bearing the knockout construct at 1–5 months of age. The fact that Ush1c expression is much higher in the ear than in the eye suggests a different role for Ush1c in ear function than in the eye and may explain why Ush1c mutant mice do not recapitulate vision defects.
Keywords: Usher syndrome, Ush1c, knockout mouse, deafness, gene, mutation, ear
1. INTRODUCTION
Usher syndrome (USH) is the most frequent cause of deaf-blindness in humans (Rosenberg et al., 1997). USH is divided into three clinical subtypes (USH1, USH2 and USH3) on the basis of the severity of hearing loss and vestibular dysfunction (Petit, 2001). All subtypes are both clinically and genetically heterogeneous. USH1 is the most severe form with a prevalence of approximately 3 to 6 per 100,000. It is characterized by severe to profound congenital sensorineural hearing loss, constant vestibular dysfunction and a pre-pubertal onset of retinitis pigmentosa (RP). In the most frequently occurring type, USH2, the congenital hearing loss is milder, vestibular function is normal and the onset of RP is after puberty. USH3 is distinguished from USH1 and USH2 by the later initiation of progressive deafness combined with variable RP and vestibular dysfunction (Ahmed et al., 2003; Davenport et al., 1978; Petit, 2001). To date, seven genetic loci (USH1B-H) associated with USH1 have been mapped, the newest addition being USH1H (Ahmed et al., 2008). Five of the corresponding genes have been cloned: the actin-based motor protein myosin VIIa (Myo7a, USH1B) (Gibson et al., 1995; Weil et al., 1995); two cadherin-related proteins, otocadherin or cadherin 23 (Cdh23, USH1D) (Bolz et al., 2001; Bork et al., 2001) and protocadherin 15 (Pcdh15, USH1F) (Ahmed et al., 2001; Alagramam et al., 2001a and b); and two scaffold proteins, harmonin (USH1C) (Bitner-Glindzicz et al., 2000; Verpy et al., 2000) and sans (USH1G) (Kikkawa et al., 2003; Weil et al., 2003).
At least one mouse mutant has been reported for each of the known Ush1 genes; shaker-1 (sh1) for Myo7a (Gibson et al., 1995), waltzer (v) for Cdh23 (Di Palma et al., 2001; Wilson et al., 2001), Ames waltzer (av) for Pcdh15 (Alagramam et al., 2001a), deaf circler (dfcr) for Ush1c (Johnson et al., 2003), and Jackson shaker (js) for Ush1g (Kikkawa et al., 2003). All of these mice are deaf, exhibit vestibular dysfunction and display similar morphological abnormalities in hair bundle development. In all of these models, the hair cell stereocilia vary irregularly in height and splay out from one another indicating defective lateral interactions. Given the very similar severe ear phenotype of these mutations, we may surmise that either products of all of these genes are required simultaneously for proper functioning of the USH1 interactome, or that these gene products each play a critical role in a single pathway in the mouse ear as well as in the human ear. Notably, none of the USH 1 mouse models recapitulate the severe retinal/vision defects of USH1 patients. This may be explained by some alternative USH1 gene transcripts the expression of which is either not required, or is substantially lower in the mouse eye than in the ear. The data presented here in our new Ush1c−/− model provide new evidence in support of the latter explanation.
Analysis of Ush1c transcripts predicts the existence of at least 10 isoforms (Reiners et al., 2005b). These alternative transcripts form three subclasses (a, b, and c) according to their protein domain composition (Verpy et al., 2000). The isoform “a” transcript subclass is expressed in many tissues whereas the longest “b” transcript is expressed in just a few tissues including the inner ear. The short isoform “c” subclass lacks both the second coiled-coil domain (CC2) and the third PDZ domain. We have previously shown that mouse mutant dfcr is defective in harmonin a, b and c isoforms and dfcr-2 Jackson (dfcr-2J) is defective only in the harmonin b isoform subclass (Johnson et al., 2003). However, partially functional harmonin isoforms might be translated from dfcr mutant transcripts because the reading frames are unaffected in the shortened dfcr transcripts of either isoform a or isoform b, and transcription proceeds to the normal stop codons. Furthermore, dfcr mutant transcripts retain all three PDZ-encoding domains. Neither gene expression nor localization was analyzed in the previous mutant mice. Additionally, previously published knockout (KO) mouse models of the Ush1c gene have not shown a clear gene expression pattern in the ear and eye (Lentz et al., 2007; Lefevre et al., 2008).
To establish an animal model and investigate the gene expression profiles in situ in the ear and eye, we have generated and characterized a null-mutant Ush1c KO mouse by targeted deletion of the first four Ush1c exons that are used in all known harmonin isoforms. The fourth exon encodes part of the first PDZ domain. The first four exons were replaced with β-galactosidase (lacZ) reporter and neomycin resistance (neo) genes. Our data show LacZ expression in multiple layers of the retina (but not in outer segments nor in most inner segments of the photoreceptor layers) in mice bearing the KO construct at 1–5 months of age. The apparent high level of Ush1c expression in the ear implies that there is a significant role for this protein in the ear. In contrast, the levels in the eye, based on mRNA expression and LacZ detection, appear to be quite low, which may indicate a minor or cryptic role for harmonin protein in eye function that could explain why Ush1c-deficient mice have not exhibited an obvious vision phenotype.
2. RESULTS
2.1. Generation of Ush1c knockout mice by homologous recombination
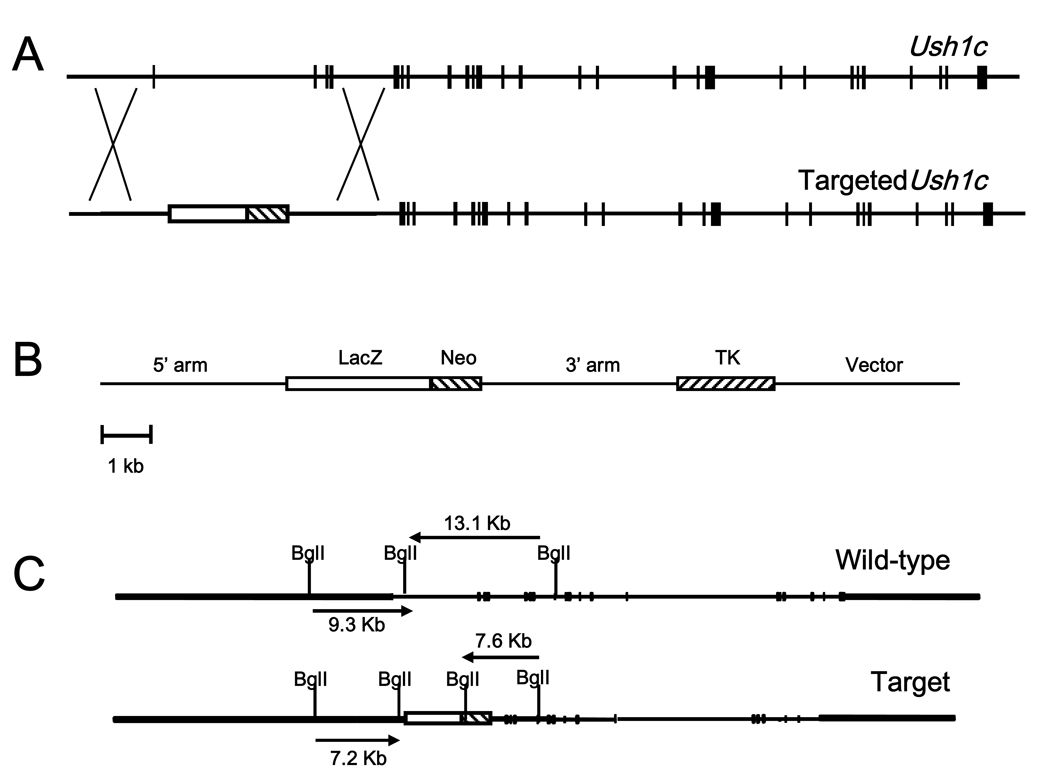
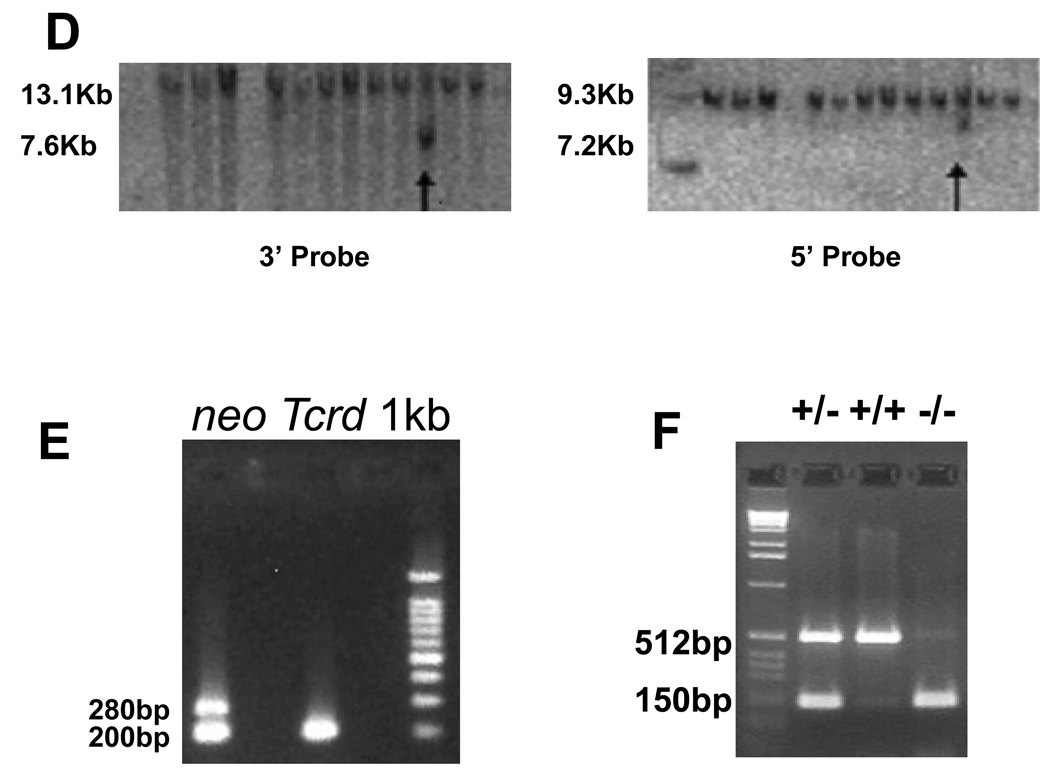
To generate an Ush1c null mutant, we designed a targeting vector that replaced the first four exons of the gene with β-galactosidase (lacZ) reporter and neomycin resistance (neo) genes. This design prevents any and all Ush1c expression by replacing the initial transcription start site with the β-gal reporter, which was placed under control of the endogenous Ush1c promoters and enhancers. The β-gal/neo replacement cassette was flanked by short and long arms of homology, identified and subcloned from a bacterial artificial chromosome (BAC) SV129 mouse genomic clone of the Ush1c gene. The herpes simplex virus thymidine kinase gene cassette (TK) was added downstream of the long arm of homology (Figs. 1A and B). The linearized construct was transfected into ES cells cultured in neomycin to select for presence of the targeting construct and ganciclovir to select for loss of the TK cassette coincident with homologous construct integration. The targeting vector included a BglI site in the β-gal/neo cassette of the targeting vector for genotyping (Fig. 1C). Correct targeting events were identified by Southern blot of BglI-digested genomic DNA from selected ES cells (Figs. 1C and D). Of 172 different ES colonies, two clones were found to have Ush1c properly disrupted on both the 5’ and 3’ ends, yielding a homologous recombination rate of approximately 1.16%. Appropriately targeted clones were injected into blastocysts and the resulting chimeric mice were used to breed heterozygous and homozygous mice, which were genotyped by neo-PCR and allele-specific primers to confirm the Ush1c targeted deletion (Figs. 1E and F).
Figure 1.
Targeted disruption of Ush1c. The native Ush1c gene (A), the properly targeted deletion locus (A) and the targeting vector (B) are shown. The first four exons of the native gene were replaced with a β-gal/neo cassette downstream of the putative promoter region to drive β-gal reporter expression and positive selection by neomycin resistance. A herpes simplex virus thymidine kinase gene (TK) cassette was added downstream of the 3′ sequence to permit negative selection of ES cells. (C) Restriction maps of the wild-type and the properly targeted deletion locus for Southern-blot analysis of embryonic stem (ES) cells and mutant mice. (D) Southern-blot analysis was used for screening of embryonic stem (ES) cells. A probe to the 3’ end of the targeting vector yielded a 13.1 Kb fragment for the wild type allele and a 7.6 Kb fragment for the allele targeted for disruption. Likewise, a probe to the 5’ end yielded fragments of 9.3 Kb and 7.2 Kb for the wild type allele and the targeted allele respectively. Arrows indicate targeted clones on each blot. (E) The presence of the neomycin gene is detected in heterozygous KO mice (lane 1) as a 280-bp band in PCR from tail DNA. Wild-type mice (lane 2) lack the neo gene, but give a 200-bp band for the Tcrd gene. (F) Ush1c PCR yields a 512-bp band for the endogenous Ush1c gene and a 150-bp band for the KO allele from tail DNA. Lane 2, heterozygous knockout; lane 3, wild-type, lane 4, homozygous Ush1c knockout.
2.2. Loss of Ush1c expression confirmed at mRNA and protein levels
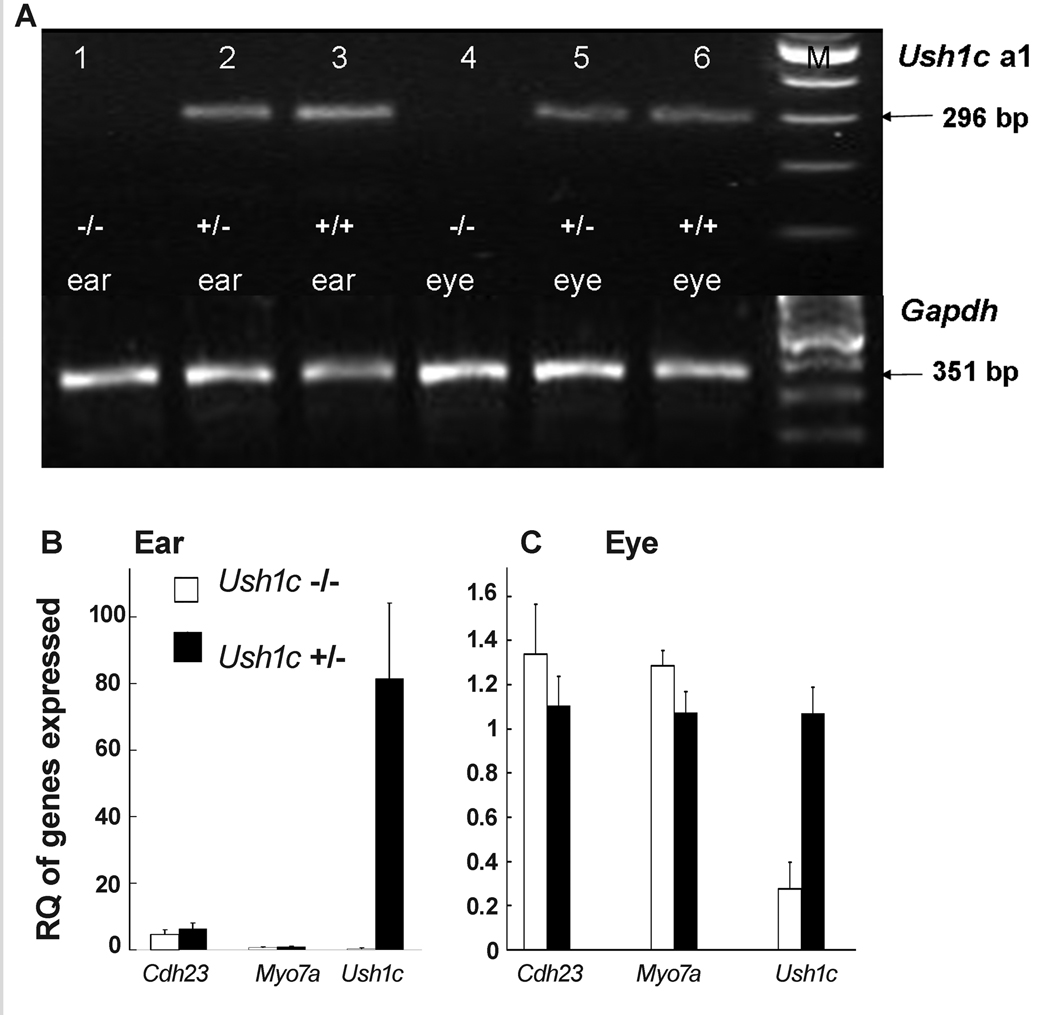
RT-PCR results at P25 revealed that Ush1c isoform a1 was expressed in neither the inner ears nor eyes of Ush1c−/− knockout (KO) mice (Fig. 2A). The putative 296-bp-specific band (spanning exons 13–17) was absent from the inner ears (lane 1) and eyes (lane 4) of the Ush1c−/− mice, but was detected in the inner ears and eyes of the Ush1c+/− mice and Ush1c+/+ mice. By sequencing, we determined that the 296-bp fragment was amplified from mouse Ush1c isoform a1 (data not shown). Agarose gel electrophoresis of RT-PCR products from inner ear RNA of mice at P6 showed the expected bands in Ush1c+/− mice for the a1 isoform (primers spanning exons 13–17), b isoform (exons 16–18), and c isoform (exons 10–22), as well as exons 1–4, 8–11, 12–14, 23–25, and 10–27 of the Ush1c gene, but only the control GAPDH band appeared in Ush1c−/− mice, confirming the elimination of all Ush1c gene expression in the Ush1c−/− mice (Supplementary Fig. S1).
Figure 2.
RT-PCR analysis of gene expression in KO mice. (A) 2% agarose gel electrophoresis of RT-PCR products to detect the Ush1c a1 isoform (296 bp) in mice at P25. Lane M: 100-bp DNA ladders; lanes 1, 2 and 3, RT-PCR products from inner ears of Ush1c−/−, Ush1c+/− and Ush1c+/+ mice, respectively; lanes 4, 5 and 6, RT-PCR products from eye tissue of Ush1c−/−, Ush1c+/− and Ush1c+/+mice, respectively. Lower panel, expression level of GAPDH as the control; Upper panel, expression level of Ush1c a1. (B) Gene expression in the inner ears of the of Ush1c−/− and Ush1c+/− mice. The gene being assayed (by real-time RT-PCR, N=5) is indicated on the X axis and RQ is relative quantitation. (C) Gene expression in the eye tissue of Ush1c−/− (white bars) and Ush1c+/−(black bars) mice. The error bars in (B) and (C) refer to one standard deviation (SD). The conditions for the real time RT-PCR experiment (N=5) performed for gene expression in the eye were identical to the assay conditions in the inner ears, but the expression level of Ush1c was too high to be presented in one histogram panel. N=5 for both (B) and (C).
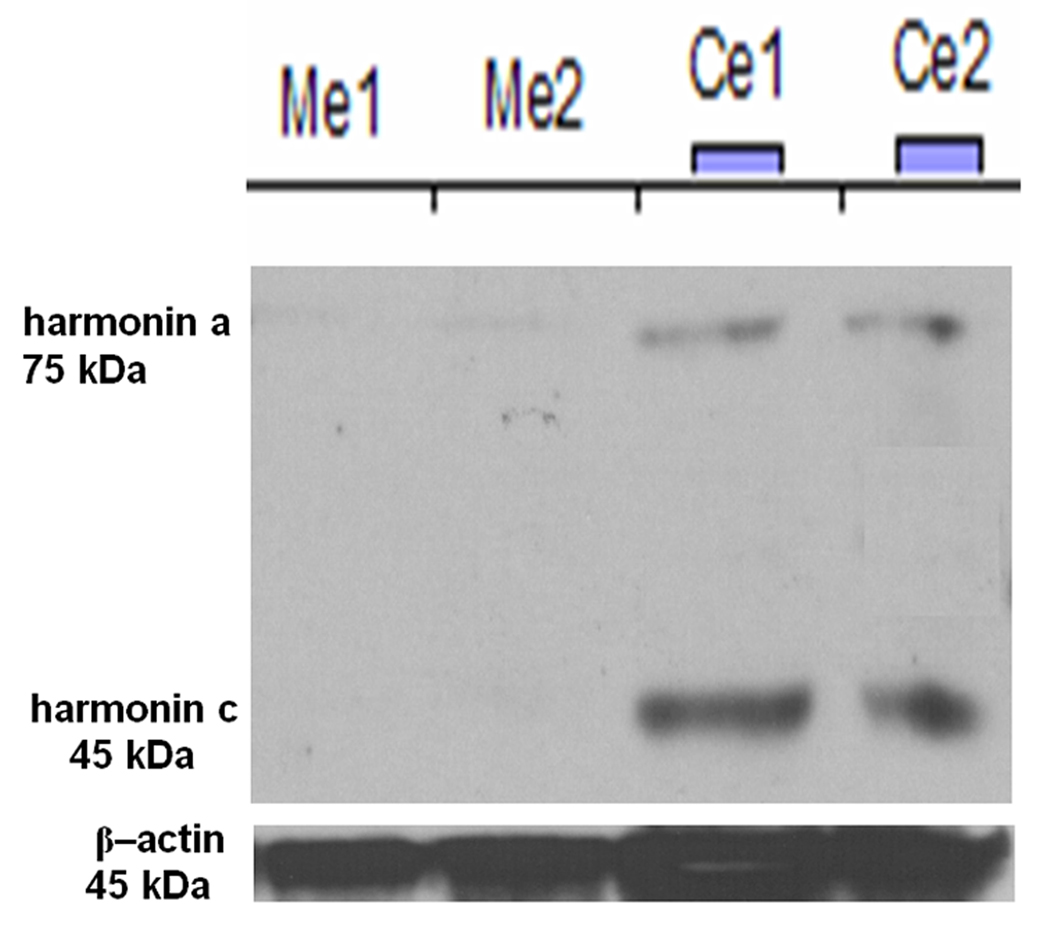
Gene expression levels of Ush1c, Cdh23 and Myo7a in the inner ears and eyes of the Ush1c−/− and Ush1c+/− mice at P25 were measured by real-time PCR. Samples from five mice of each group were assayed. The GAPDH gene was used as an endogenous control. In the inner ears, the Ush1c and Cdh23 expression levels were significantly lower (P<0.001) in Ush1c−/− than that in Ush1c+/− mice. However, there was no significant difference (P=0.154) between the two groups for Myo7a gene expression in the inner ears (Fig. 2B). In eye tissue, the Ush1c expression level was significantly lower (P<0.001) in Ush1c−/− mice than in Ush1c+/− mice. Furthermore, there was a significant difference (P=0.022) for Myo7a gene expression between the two groups. No difference (P=0.063) existed between the two groups for Cdh23 gene expression in eye tissue (Fig. 2C). Real-time RT-PCR experiments were performed on all samples using identical conditions simultaneously by including all samples on a single real-time RT-PCR plate for both ears (Fig. 2B) and eyes (Fig. 2C). However, because the Ush1c expression level in the inner ears of Ush1c+/− mice is more than 80 times greater than in the eyes, there would be no visible bars for gene expression levels from the eye if we displayed them all in one panel. On the other hand, the Ush1c expression level in the eye of Ush1c−/− mice at 0.27 RQ is no higher than the background noise level for this technique. Similar results were obtained when harmonin protein levels were assayed by Western blot analysis with a harmonin peptide-antibody and quantitated against its control β-actin band. Harmonin/Ush1c isoforms a and c (harmonin a=75 kDa and harmonin c=45 kDa) were absent in the inner ears of two Ush1c−/− mice but were clearly detectable in the ear samples from Ush1c+/− mice (Fig. 3). Harmonin isoform b was undetectable even in wild-type inner ears, presumably because expression levels are below the level of detection in this assay. Probably for similar reasons, none of the a, b or c harmonin isoforms were detected in the eyes of the same sets of both Ush1c−/− and Ush1c+/+ mice (n=10) (data not shown).
Figure 3.
Western blot analysis with an anti-harmonin antibody to show that there is no protein production in the inner ears of Ush1c−/− (knockout) mice. Ears from two mice of each genotype were assayed for harmonin protein. Me1: Ush1c−/− ear 1, Ce1: Ush1c+/− ear 1. Bands for two isoforms of harmonin were detected and quantitated against an actin control band using ImageQuant software.
We then used a specific antibody to test in situ whether the harmonin protein is absent as predicted in homozygous mutant mice (Ush1c−/−). As described previously, harmonin localizes to the tips of hair bundles of both the outer and inner hair cells in the cochleae of wild-type mice (Lefevre, et al., 2008). We examined harmonin expression in the cochlea of a P0 Ush1c−/− mouse and a heterozygous littermate control. In the control mouse, harmonin localized to tips of hair cell stereocilia as expected (Supplementary Fig. S2); however, harmonin expression was not detected in any part of the inner ear of the Ush1c−/− KO mouse (Fig. S2). The lack of immunofluorescence in cochleae of mutant mice is explained by the absence of harmonin protein and is not the result of missing or dysmorphic hair bundles as evidenced by phalloidin-stained organ of Corti surface preparations, which clearly show the presence of normal-appearing hair bundles in mutant mice at P0 (Fig. S2). This is further confirmed by SEM images of hair bundles in Fig. 6.
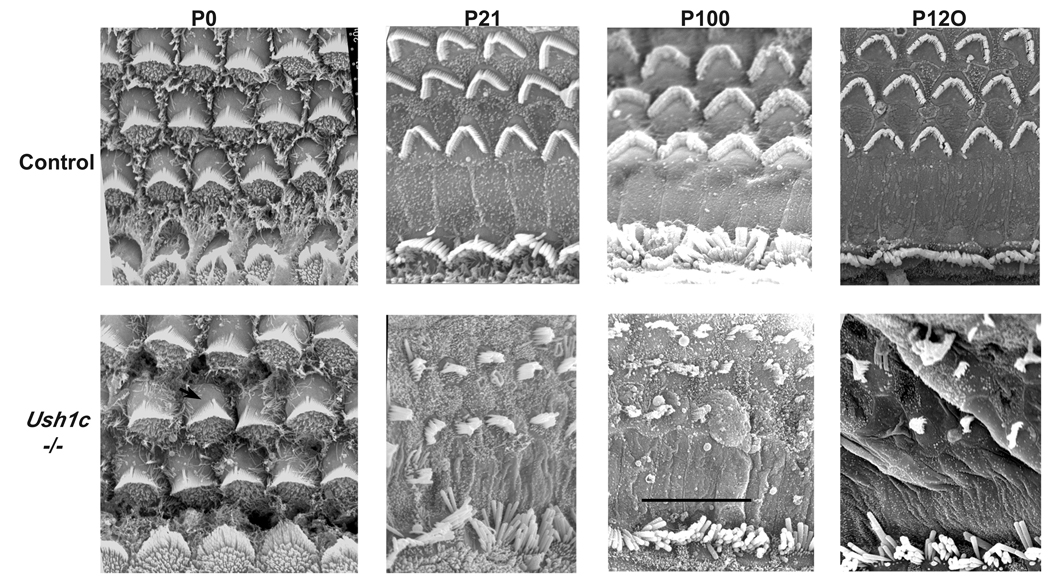
Figure 6.
Scanning Electron Microscopy of organ of Corti surface preparations to examine hair cell morphology. At birth (P0), Ush1c−/− mice (lower panels) exhibit damage predominantly in the OHCs, with kinociliar rotation deviating roughly 20° from normal. Severe damage at P21, P100, and P120 exhibits progressive degeneration of the hair cells and stereocilia of the cochlea in Ush1c−/− mice compared to the rigid organization of the age-matched wild type controls (upper panels). Some of the stereocilia of the outer hair cell bundles are missing in the KO mice, and the inner hair cells are all severely splayed. Bar = 10 µm for all panels.
2.3. β-Gal is appropriately expressed from the integrated deletion construct
To disrupt the Ush1c gene, we replaced the portion corresponding to Ush1c with the bacterial lacZ gene (Fig. 1), with the aim of studying β-galactosidase expression in KO animals. In embryonic and postnatal mice (both heterozygous Ush1c+/− and homozygous Ush1c−/−), lacZ expression was detected in the outer pillar cells and Deiter’s cells of mice that possessed the KO construct (Fig. 4D).
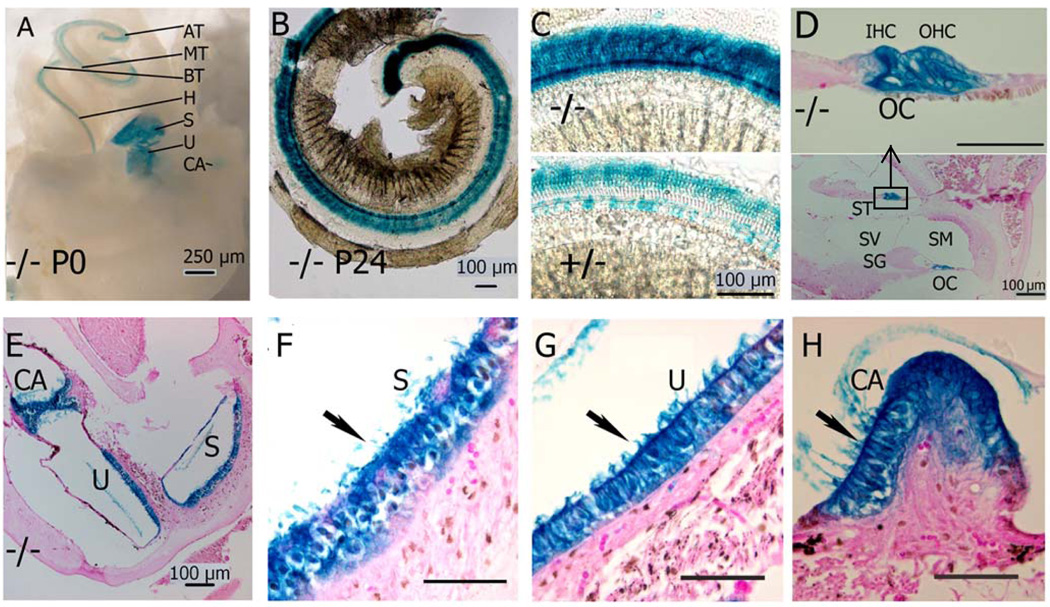
Figure 4.
Ush1c reporter gene expression in the inner ear. The lacZ reporter gene for Ush1c expression was visualized by histochemical reaction of its product, β-gal. Whole mount preparations of inner ears from P0 mice A show Ush1c expression is limited to the neurosensory epithelia: apical turn (AT), middle (MT), basal (BT) and hook (H) of the organ of Corti (OC) in the cochlea, the maculae of the saccule (S) and utricle (U), cristae ampullares (CA) of the semicircular canals. B Surface preparations of the dissected organ of Corti of P24 mice show expression in the three rows of outer hair cells (OHC) and one row of inner hair cells (IHC) on the apical turn of the cochlear duct. A uniform expression pattern appears along the entire length of the cochlear duct in Ush1c−/− mice. C Enlarged views of corresponding middle turn of (−/−, heavier stained) and (+/− lighter stained). D Midmodiolar cross section through the cochlea of a P24 mouse shows β-gal expression in the inner and outer hair cells (IHC and OHC) and supporting cells (pillar cells and Deiters cells) of the basal and middle turns. Upper panel is an enlargement of the rectangle box. ST: Scala tympani, SM: Scala media, SV: Scala vestibule, SG: spiral ganglion, E–H Cross section through saccule (S) and utricle (U), cristae ampullares (CA) show only neurosensory epithelia are stained. Wildtype (+/+) is not stained (data not shown). Scale bars D, F–H are 50 µm unless otherwise labeled.
We performed expression analysis on the inner ears of mice of the following ages: embryonic day (E) 16.5, E18.5 and postnatal day (P) 0, P2, P7, P17 and P24. At each of these time points, β-galactosidase expression was detected in outer pillar cells and Deiter’s cells, the hair cells of the saccular and utricular maculae and the cristae. Expression of β-gal in the P0 hair cells was punctate (data not shown) but became more diffuse through the cell body of the hair cells by P24. Fig. 4A shows the macroscopic and microscopic examination of β-galactosidase expression in Ush1c−/− mice, specifically at P0 (A) and P24 (Figs. 4B–H), confirming the effective functioning of the targeting construct and revealing the expression pattern of Ush1c in the inner ear and eye (see below).
2.4. Inner ear phenotype and pathology
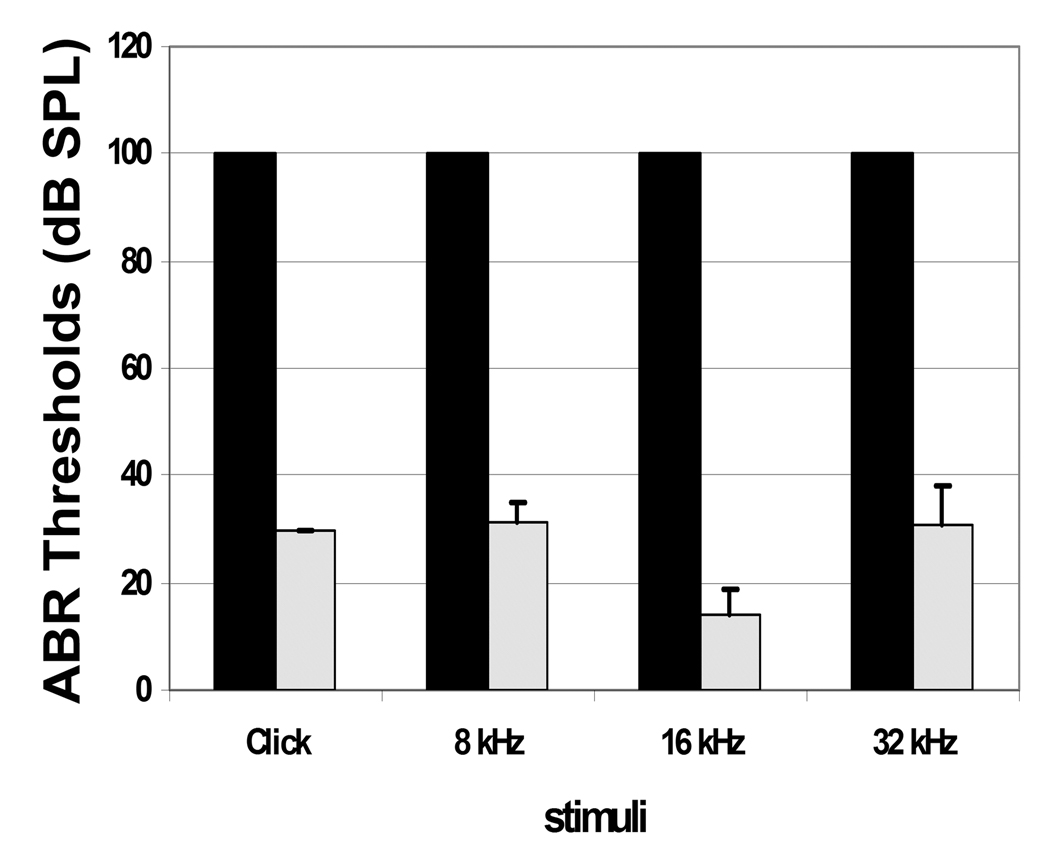
Homozygous mutant mice (Ush1c−/−) exhibit the typical behavior associated with inner ear defects: deafness, hyperactivity, head-tossing and circling, indicators of both auditory and vestibular defects. To quantitatively assess hearing loss in Ush1c−/− mice, we measured auditory-evoked brainstem response (ABR) thresholds to click stimuli and to 8, 16 and 32 kHz pure-tone stimuli on mutant and control mice at P22 (Fig. 5 and supplementary Fig. S3), P15, and P100 (data not shown). Ush1c−/− mice were completely deaf at all the frequencies tested, as there was no detectable ABR with 100 dB SPL stimuli; whereas, age-matched Ush1c+/− controls showed ABR thresholds in the normal hearing-range at all ages. To assay vestibular function, we performed a swimming test. None of 5 adult mutant mice were able to keep their noses above the water’s surface while swimming in a water bath, but all of the littermate Ush1c+/− controls passed the test (see Supplementary Video 1 online, please download all three files together to watch the video clips).
Figure 5.
ABR thresholds in Ush1c−/− and Ush1c+/− littermate mice. Mean ABR thresholds are plotted in response to click or tone-burst stimuli at frequencies shown on the X axis at P22 (n=5) in Ush1c−/− mice (black bars). In Ush1c−/− mice, no ABR was detectable at any frequency up to 100 dB SPL (decibels sound pressure level) at all timepoints tested. Mean ABR thresholds at the same frequencies are shown for Ush1c+/− mice (light bars) at P22. Standard deviations from the mean are shown by vertical error bars. The dark columns without bars indicate all mice have the same 100 value. All the STDEVs were 0 and could not be shown on the chart.
The deafness and balance impairment of mutant mice suggested an inner ear dysfunction. We therefore examined hair cell surface preparations by scanning electron microscopy (SEM, Fig. 6). At P0, some minor cell polarity changes of outer hair cells were present in Ush1c−/− mice. SEM of Ush1c−/− from birth (P0) to P120 showed progressively disorganized outer hair cell stereocilia compared with the well-organized pattern and rigid structure typical of normal stereocilia. Stereocilia of inner hair cells of mutant mice also showed a disorganized appearance, but to a lesser degree than did the outer hair cells (Fig. 6). Examinations of cross sections by light microscopy through apical regions of the cochleae of Ush1c−/− at P2 revealed no apparent hair cell degeneration (data not shown).
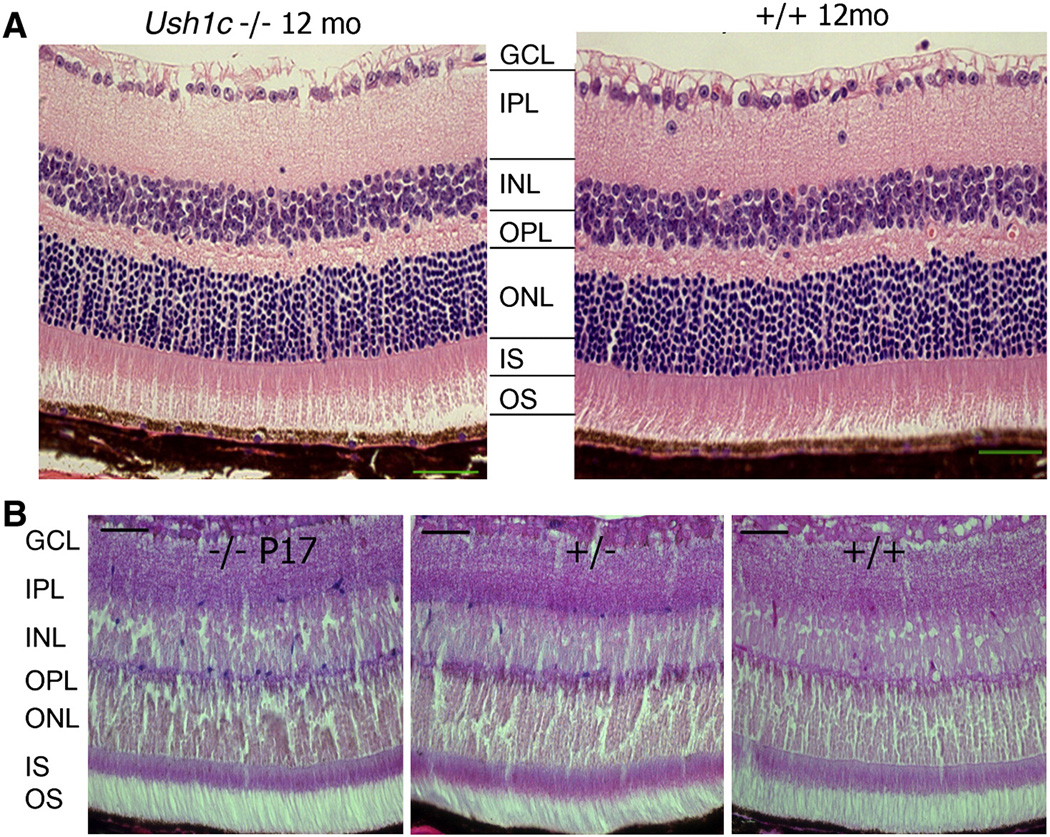
2.5. Eye phenotype, pathology and reporter gene expression profile
We examined the eyes of three Ush1c−/− mice at each age because mutation of the homologous gene in humans causes progressive RP. The eyes of the mutant mice appeared clinically normal upon gross examination, having a normal fundus with thin retinal vessels at 1, 3, and 12 months of age, the same as in the littermate controls. At 12 months of age, histological examinations of eyes from 6 Ush1c−/− and 6 littermate control mice were normal (Fig. 7A). The amplitude and implicit time of both the rod- and cone-mediated electroretinograms (ERGs) appeared normal compared with those of heterozygous littermate controls up to 10 months of age (data not shown); however, by 11 months there was a decline in ERG for both rods and cones (data not shown). Expression of lacZ was detectable in eyes of Ush1c+/− and Ush1c−/− mice at P17 (Fig. 7B) and P150 (Supplementary Fig. S4B, 5 months). The scattered blue dots were distributed with rare frequency in the epithelial layer of the vitreous body (Supplementary Fig. S4C) and in each layer from the outer plexiform layer (OPL) to the ganglion cell layer (GCL) but not in most portions of the photoreceptor layer (Fig. 7B). The level of lacZ staining was much lower in the eye than in the ear (Fig. 4) using similar experimental conditions and tissue processing except that the eye tissue did not need decalcification.
Figure 7.
Retinal histology from KO and control mice shows normal phenotype and lacZ expression in eyes of Ush1c knockout. (A) Eyes from Ush1c−/− mice (left panel, n=3) compared with Ush1c+/+ littermate controls (right panel, n=3) at 12 months of age. Each layer is indicated as follows: ganglion cell layer (GCL), inner plexiform layer (IPL), inner nuclear layer (INL), outer plexiform layer (OPL), outer nuclear layer (ONL), inner segments (IS) and outer segments (OS). (B) LacZ expression in the multiple layers of retina (but not the IS nor OS layers) in the littermate mice bearing the KO construct and higher lacZ expression for −/− (n=3) than +/− (n=4) at P17, No expression of lacZ was observed in the wild-type littermate +/+ (n=2). Scale bar = 50 µm.
3. DISCUSSION
Animal models that reproduce human disease phenotypes are important in revealing disease mechanisms and aiding development of therapies. To date, seven genetic loci (USH1B-H) associated with USH1 have been mapped (Ahmed et al., 2008). Five of the corresponding genes have been cloned and many corresponding genetic mouse models have being generated. According to the JAX database (http://www.informatics.jax.org/), the following 54 mouse mutations have been created for the five USH 1 genes: 13 alleles for Ush1b/Myo7a: Spontaneous (4), Chemically induced (9); 19 alleles for Ush1d/Cdh23: Spontaneous (12), Chemically induced (5), QTL (2); 11 alleles for Ush1f/Pcdh15: Gene trapped (2), Transgenic (1), Spontaneous (6), Chemically induced (2); 4 alleles for Ush1g: Targeted, knockout (1), Spontaneous (3); 7 alleles for Ush1c: Targeted, knock-out (1), Targeted, other (3) Spontaneous (3). Among the above 54 published mouse mutations, there are only three targeted mutations, among which only two exist as live mice (one Ush1g KO exists as a cell line) prior to the KO mice presented here. The two are Usher syndrome 1C homolog targeted mutation 1.1, Unite de Genetique des Deficits Sensoriels (Symbol Ush1ctm1.1Ugds) and Usher syndrome 1C homolog targeted mutation 1, Bronya Keats (Symbol Ush1ctm1Bkts) (Lentz et al., 2007; Lefevre, et al., 2008). Our model is unique because we have generated a targeted null mutation Ush1c mouse model in which the first four exons of the Ush1c gene have been replaced by a reporter gene. None of the previous mutations of the Ush1 genes include a reporter assay to facilitate expression pattern profiles in the ear and eye or other tissues. For comparison, we have summarized the five molecularly defined alleles for Ush1c in Table 1.
Table 1.
molecularly defined Ush1c mouse models
| Allele Symbol Name |
References | Category | Phenotypes/pathology | Genetic background | Mutation | Affected isoforms/ transcripts |
In situ gene expression assay |
|---|---|---|---|---|---|---|---|
| Ush1c-LacZ | this paper | Targeted (Reporter) |
circling or head tossing; congenital deafness |
B6;129S4 Mostly B6 (Backcrossed to B6 more than 10 generations) |
The first four exons replaced with lacZ and neo genes. |
Affects all 3 isoforms. Confirmed by RT-PCR (no band spanning exons 3–8 in the ear). |
LacZ- Reporter |
| Ush1cdfcr deaf circler |
Johnson et al., 2003 | Spontaneous | circling or head tossing; congenital deafness; a slight peripheral retinal degeneration at 9 mo due to genetic background |
BALB/cByJ | 12.7 kb deletion Involves exons 12- 15, A, B, C and D. |
Affects transcripts of both isoforms a and b. |
none |
| Ush1cdfcr-2J deaf circler 2 Jackson |
Johnson et al., 2003 | Spontaneous | circling or head tossing; congenital deafness |
B6;129S4 | 1 bp deletion of the 4th nucleotide in the inner ear-specific exon C |
Affects only isoform b transcripts. |
none |
| Ush1ctm1.1Ugds targeted mutation 1.1, Unite de Genetique des Deficits Sensoriels |
Lefevre, et al., 2008 | Targeted (knock-out) |
circling or head tossing; congenital deafness (most likely) |
129S2/SvPas | Cre was used to remove exon 1. |
Affects all 3 isoforms. Confirmed by RT-PCR (no band spanning exons 3–8 in the ear). |
none |
| Ush1ctm1Bkts targeted mutation 1, Bronya Keats |
Lentz et al., 2007 | Targeted (knock-in) |
behavior?, circling or head tossing; congenital deafness (most likely) |
129S6/SvEvTac C57BL/6J * FVB/N |
216G to A substitution in exon 3 |
A deletion of 35 bases. Confirmed by RT-PCR. |
none |
We have validated our model by confirming that Ush1c gene expression is completely knocked out in the ears and eyes of our Ush1c−/− mice. We can draw this conclusion from the results shown in Fig. 1F; Supplementary Figs. S1 and S2; Figs. 2A, B, and C; Fig. 3; Fig. 1F lane 4; and because the homozygous Ush1c KO genotype correlates with circling behavior and a deafness phenotype. Fig. 2A, Supplementary Figs. S1 and S2, show the absence of mRNA in both ears (lane 1) and eyes (lane 4) of the Ush1c−/− mice when primer pairs were used (296 bp, exons 13–17) that detect the Ush1c a1 variant. Figs. 2B and 2C also show the absence of Ush1c mRNA in ears and eyes of the mutant mice. It should be noted that although Fig. 2c includes a bar graph reading for Ush1c mRNA expression in the Ush1c−/− eye sample, the actual RQ value is considered negative as it is so small as to be indistinguishable from the background reading obtained with negative control samples (the real-time PCR machine never reads exactly zero even for negative controls). Figs. 2B and 2C use different units on the vertical axis in the two coordinate systems in order to clearly display the positive readings in Fig. 2C. Compared to the mean RQ value of 81.52 in the control mice, the mean RQ of 0.30 in the ears of Ush1c−/− mice can be considered the background signal of the real-time PCR, and thus interpreted as negative. Similarly, the mean RQ of 0.27 in the eyes of Ush1c−/− mice would also be equivalent to background noise, as the real-time PCR for Ush1c mRNA in the ears and eyes was done simultaneously in one PCR plate. Furthermore, with realtime PCR, we do not expect the mean RQ value of one group to be zero, as the number of PCR cycles was 40. We further confirmed that this was true by additional RT-PCR reactions in Supplementary Fig. S1. RT-PCR products collectively spanning nearly all exons of all transcripts of the Ush1c gene were absent in ear tissue from Ush1c−/− mice and present in Ush1c+/− ear tissue. We used Ush1c+/−and Ush1c+/+ as normal controls because Ush1c+/− mice have a normal phenotype, the same as Ush1c+/+ mice, which indicates that this KO is a completely recessive allele. As we used 32 PCR cycles for our RT-PCR reaction (Fig. 2A), we indeed see a semi-quantitative measure: the 296-bp band for Ush1c+/+ in lane 3 is brighter than that for Ush1c+/− in lane 2 for inner ears. This semi-quantitative differentiation is in agreement with the reporter gene assay shown in Fig. 4C: the homozygote for LacZ gene (−/−) stained heavier than that for heterozygote (+/−). In Fig. 4A, the much more intense bands (lanes 2 and 3) for the inner ears than for the eyes (lanes 5 and 6) are in good agreement with real-time PCR results in Fig. 4B and 4C showing that Ush1c is expressed more than 80 times higher in normal ears than in normal eyes. At the same time, gene expression levels of other genes were slightly higher (Cdh23) or similar (Myo7a) in the inner ears than in eyes of the Ush1c+/− mice (Fig. 2B and 2C), suggesting that their roles are not as substantial as the Ush1c gene in ear function, although all loss-of-function mutations in the three genes cause similar deafness and loss of balance. However, we do see some clear differences for swimming patterns among them (unpublished observations). In the inner ears (Fig. 2B), the observation that Ush1c and Cdh23 expression levels were significantly lower (P<0.001) in Ush1c−/− than in Ush1c+/− mice may be due to lost hair cells. Furthermore, there was little (Myo7a) or no (Cdh23) mutant effect on the expression levels, which may be because there was no cell loss in the eyes of the Ush1c−/−mice.
We further confirmed the validity of our KO model by showing that USH1C is absent at the level of protein, in addition to mRNA. Fig. 3 shows the presence of Ush1c splicing products (harmonin a and c) in the ears of control mice (Ce1 and Ce2) and absence of products in the ears of mutant mice (Me1 and Me2). Supplementary Fig. S2 shows that N-17 antibodies revealed an absence of harmonin expression at the tips of hair bundle stereocilia but labeled harmonin very clearly on the littermate Ush1c+/+. Although the expected isoform b band did not appear in the Ush1c+/+ mice, the RT-PCR results above confirm its absence. The inability to probe for isoform b by Western may be a sensitivity issue, explained by the antibody not binding strongly enough to the b isoform and/or that the b isoform protein level is low even in wildtype mice. Additionally, the manufacturer’s specifications (www.abcam.com) do not show the b isoform as being detected by the particular harmonin antibody they produced. This antibody is different from the one previously used for their publication (Verpy et al., 2000; Boeda et al., 2002; Michalski et al., 2009; Grillet et al., 2009). Nevertheless, our aim here is not to detect the presence of the b isoform. We intended to prove that all isoforms of harmonin are absent in the Ush1c−/− mice while using any of the isoforms we could detect as control in the normal littermate mice. None of the a, b or c harmonin isoforms were detected in the eyes of the same sets of both Ush1c−/− and Ush1c+/+ mice (n=10) (data not shown), which may also suggest that harmonin protein is expressed at a much lower and undetectable level in the eye than in the ears. This does not suggest this KO is an ear tissue-specific KO because we have provided evidence that the first four exons were replaced with the LacZ and Neo genes and it is well-established that the first four exons are shared by all transcript variants of the Ush1c gene (Verpy et al., 2000; Boeda et al., 2002).
We assessed the expression pattern of the reporter gene under control of Ush1c regulatory elements and characterized the phenotype of mice defective for Ush1c. Harmonin is expressed as several splice variants, including harmonin-a, -b, and -c (Verpy et al., 2000). The longest variant, harmonin-b which contains three PDZ domains and two coiled-coil domains, is expressed in hair bundles of developing cochlear hair cells (Lefevre et al., 2008, Boeda et al., 2002 and Verpy et al., 2000). Harmonin a and harmonin c were present in the eyecups. Harmonin a was concentrated in the photoreceptor synapse. Those studies revealed localization of harmonin, but did not quantitatively compare the in situ expression levels between ear and eye as we have here. Our in situ expression data indeed found that the LacZ/Ush1c expression was significantly higher in the inner ear, including the vestibular organs and the organ of Corti, but was detected at a much lower level in the eye. This is the first detailed in situ expression report in the vestibular organs. This also explains well the severe cycling/balance, deafness phenotype and limited vision problems in our model.
We examined the auditory phenotype of our Ush1c−/− mice. The abnormal stereocilia morphology observed in our Ush1c KO mice is similar to that reported in mouse models for other forms of human USH1, except that the hair cell bundles at P0 are less disorganized in our model than in previously published ones (Lefevre et al 2008). The lesser degree of HC disorganization in our Ush1c−/− mice (Fig. 6, P0) may be explained by a difference in mouse background (ours on C57BL/6 vs. Lefevre’s on 129S2/SvPas- Ush1ctm1.1Ugds). Another model, BALB/cBy-Ush1cdfcr/dfcrJ mice, was confirmed to have a similar lesser degree of HC disorganization around P0 (per personal communication with Dr. Ping Chen from Emory University who studies hair cell polarity). We also have sent some ear tissue from our Ush1c−/− KO mice to her, and she found the same result of a lesser degree of HC disorganization in these mice around P0.
Because the USH complex may play a role in ears and eyes, we also examined the vision phenotype in our model and found no abnormality of retinal cell organization. However, we have detected reduced ERG responses in Ush1c−/− mice at 11 months, which is consistent with findings reported for some of the alleles of shaker-1, waltzer and Ames waltzer mice (Haywood-Watson et al., 2006; Libby and Steel, 2001) as well as Gpr98-mutant mice (McGee et al., 2006) and knockin mice expressing the Acadian USH1C mutant gene (Lentz et al., 2007). In the shaker-1, waltzer and Ames waltzer mice, the a- and b-wave amplitudes were reduced by a similar proportion, pointing out that the defect resides within the photoreceptor cell response. It has been suggested that all the USH1 proteins form a large complex in the photoreceptor synapse (Kremer et al., 2006;Reiners et al., 2006; Reiners and Wolfrum, 2006; van Wijk et al., 2006). However, this localization is not consistent with that reported by other groups who found the connecting cilium region of the photoreceptor cells to be a focus for USH protein interaction and function in the retina (Gibbs and Williams, 2004; Lillo et al., 2005; Liu et al., 2007). Nor is it supported by retinal phenotypes of mutant mice, as none of the Ush1 mouse models presents a mutant phenotype that could be attributed to photoreceptor synaptic function (Haywood-Watson et al., 2006; Libby and Steel, 2001; Libby et al., 2003; Liu et al., 2007; Sun et al., 2006).
The absence of an overt retinal phenotype in mouse mutants defective for USH1 proteins has been difficult to explain, despite USH1 protein expression in the mouse retina. However, here we demonstrate the power of the LacZ reporter gene expression assay in this unique Ush1c KO mouse model. Our findings via the LacZ reporter assay show that the Ush1c gene is expressed at a much higher level in ear than in eye at P24, specifically in the retina (Fig. 7B). The Ush1c gene is expressed at a similar or slightly higher level at five months of age than at one month of age in the retina (Supplementary Fig. S4). LacZ was detectable in each layer from the outer plexiform layer (OPL) to the ganglion cell layer (GCL) but not the photoreceptor layer. This result is much clearer and more definitive than previous controversial results using antibody staining for harmonin localization in the retina (Bork et al., 2002; Gibbs and Williams, 2004; Kremer et al., 2006; Lillo et al., 2005; Liu et al., 2007; Reiners et al., 2005a; Reiners et al., 2006; Reiners and Wolfrum, 2006; van Wijk et al., 2006). However, our retinal LacZ expression pattern is in agreement with the data of Williams et al (2009) which showed that outer segments were not labeled by H1 antibodies (generated against the N-terminal region that is common among all isoforms) or with H3 antibodies (generated against a region common to the a and b isoforms). The reporter expression level in the retina appears to be lower than in the ear at birth, based on the apparent intensity of the LacZ signal (Figs. 4 and 7B) which may point toward an explanation for the lack of a vision defect in the Ush1c mutant mice. Some compensation mechanism may replace the function of the Ush1c gene or its protein product. Further quantitative study may be warranted and will be facilitated by the LacZ system that has been incorporated into our new mouse model.
Real-time RT-PCR revealed that the expression level of Ush1c mRNA in the ear is more than 80 times greater than in the eye (Fig 2B and C), in agreement with our reporter data, supporting this new explanation for the lack of an eye phenotype in the mutant mice. It would be revealing to examine whether the same expression pattern for Ush1c is observed in humans. Our findings are consistent with previous studies showing that mice lacking orthologs of USH1 proteins do not suffer from RP (Alagramam et al., 2001b; Gibson et al., 1995; Johnson et al., 2003; Kikkawa et al., 2003), although the recently generated mouse model for USH2A has been reported to exhibit overt retinal degeneration (Liu et al., 2007).
Other USH-related mouse models may be relevant to the question of auditory and vision phenotypes, for example, deaf circler (dfcr) mice and shaker-1/waltzer double-mutant mice have been reported to display a slight retinal degeneration notable in 9- or 12-month-old mice, respectively (Johnson et al., 2003; Lillo et al., 2003), and recently this was demonstrated to be a BALB/cByJ genetic background effect (Williams et al., 2009). Shaker-1 mice (bearing a mutated Myo7a allele) do exhibit a vision phenotype including abnormal transport of opsin molecules through the connecting cilia of photoreceptor cells, abnormal outer-segment phagocytosis and abnormal melanosome motility in the retinal pigmented epithelium, yet retinal degeneration does not ensue (Gibbs et al., 2003). In addition, electrophysiologic studies on USH1 mouse models (e.g. shaker-1, waltzer and Ames waltzer mice) revealed a slight reduction of electroretinograms that is consistent with the dysfunction of synapses in neuronal retina, without histologic changes at the light microscopy level (Haywood-Watson et al., 2006; Libby et al., 2003; Libby and Steel, 2001). A Ush1c216A mouse model recently generated by knocking in the human 216G→A mutation showed the behavioral characteristics of deaf mice. The mice were hyperactive, displayed circling and head-tossing behavior and mimicked the phenotype of human patients (Lentz et al., 2007). A study on the mouse mutant for protocadherin 15 showed that most of the mutations involve exons that are alternatively spliced, suggesting that loss of the affected exons may preserve some functional protein isoforms in the retina (Haywood-Watson et al., 2006). However, it remains unclear to what extent alternative splicing might explain the absence of retinal phenotype in mouse models with mutations in Ush1 genes. The only USH mouse model that has been reported to exhibit any convincing retinal degeneration is the Ush2a KO mouse. Photoreceptor morphology and numbers were found to be normal in these USH2A null mice at 10 months of age, but by 20 months over half the photoreceptor cells were lost (Liu et al., 2007).
In conclusion, we have generated a targeted null-mutant Ush1c mouse model in which the first four exons of the Ush1c gene have been replaced by a reporter gene. This mutant has a similar ear and eye phenotype as previously reported Ush1c models. The expression pattern of the reporter gene under control of Ush1c regulatory elements suggests that Ush1c is expressed to a lesser extent in the retina than in the ear. Most importantly, Ush1c is not significantly expressed in photoreceptor layers. Real-time RT-PCR showed a dramatically reduced expression of Ush1c in the eye compared to the ear. These data explain why the mouse model does not recapitulate vision defects of the human Usher 1 syndrome before 10 months of age. The decline in ERG amplitude at 11 months of age for both rods and cones warrants further study as a possible late-onset vision dysfunction. Nevertheless, this reporter gene assay and the mouse model will be very valuable in Ush1c gene expression study in other tissues such as kidney and intestine, as Ush1c expression has been demonstrated in many tissues throughout the body.
4. EXPERIMENTAL PROCEDURES
4.1. Gene targeting and generation of Ush1c knockout mice
Mouse (C57BL/6) and human genomic sequences from public databases were compared and aligned to identify the mouse Ush1c exons. Primers were designed within the known sequence to amplify genomic regions at the 5' and 3' ends of the Ush1c gene to screen a bacterial artificial chromosome (BAC) SV129 mouse genomic library (from Research Genetics, Huntsville, AL, USA) to obtain the mouse Ush1c clone. Restriction analysis and sequencing were used to identify suitable regions for subcloning into the targeting vector. The method of Thomas et al. (1992) was used to generate the targeting construct and flanking probes. A targeting vector (Figs. 1A and B) was constructed to replace the first four exons of the Ush1c gene with a β-gal reporter and a neomycin resistance gene (neo). Initially, we obtained the plasmid p13 that contained a neo cassette downstream of the β-gal gene, both in forward transcriptional orientation on a pSAβ-gal plasmid backbone (kind gift of the Soriano lab) (Friedrich and Soriano, 1991). The herpes simplex virus thymidine kinase gene (TK) cassette from pKOSelectTK (Lexicon Genetics Inc., The Woodlands, TX, USA) was cloned into a unique KpnI site in the p13 plasmid immediately downstream of the neo cassette to permit selection for homologously integrated clones. Next, a 3.7-kb fragment (amplified from R1 to 129 genomic DNA) homologous to the intron upstream of the first Ush1c exon was cloned into a unique NotI site immediately upstream of the β-gal gene. Then, a similarly amplified 4-kb fragment homologous to the intron downstream of the fourth Ush1c exon was cloned into a unique SalI site between neo and the TK gene. A BglI site in the β-gal/Neo cassette of the targeting vector facilitates genotyping (Fig. 1C). The integrity of the targeting construct was confirmed by restriction digest analysis and by sequence analysis across the junctions of each fragment that had been cloned into the final construct. The targeting construct was linearized with SacII and electroporated into R1 (129×1/SvJ and 129S1/SV) embryonic stem (ES) cells (a gift from Janet Rossant and Andras Nagy, Mount Sinai Hospital, Toronto, ON, Canada). Targeted ES cells were selected by G418 and ganciclovir resistance. Two clones of 172 ES colonies contained a properly targeted deletion as evidenced by Southern blot (Fig. 1C–D). ES cell clones with targeted integrations were injected into C57BL/6 blastocysts, and the injected embryos were implanted into surrogate mothers. Highly chimeric mice were identified by coat color and were backcrossed to C57BL/6 mice for six more generations. The resultant offspring were genotyped to identify germline transmission. Mice that inherited the mutation were then interbred to produce homozygous mutant animals that lack the normal gene product.
The convention for Ush1c nomenclature when referring to genes, transcripts, and proteins is somewhat unclear and inconsistent throughout the published literature. Because we refer to both human and mouse genes, transcripts, and proteins at various points in this manuscript, consistent and correct nomenclature can still be confusing. In effort to be as correct and consistent as possible, we have used the following convention regarding all of the genes and proteins in this manuscript, using Ush1c as an example: mouse genes/DNA and transcripts/RNA/cDNA and alternate splice variant transcripts (Ush1c); human genes and transcripts (USH1C); and protein and protein isoforms in any species (harmonin). It may also be correct to name the protein as USH1C, but for the sake of clarity, in this manuscript we will use harmonin throughout to refer to the protein. Upon this publication, this KO mouse will be made available for investigators.
4.2. Genotyping of ES Cells and Mice
Genomic DNA was isolated from either ES cell clones or from mouse tail biopsies. ES cells were lysed overnight at 37°C in lysis buffer containing 0.2 M NaCl, 5 mM EDTA, 50 mM Tris-HCl, pH 7.5, 0.2% SDS, and 400 µg/ml proteinase K. Genomic DNA was precipitated with ethanol, washed with 70% ethanol, air-dried, and resuspended in H20. For mouse tail biopsies, approximately 1 cm of tissue was digested overnight at 55°C in lysis buffer containing 200 µg/ml proteinase K, extracted with phenol/chloroform, and ethanol-precipitated. To identify gene-targeting events in ES cells and homozygous KO mice, genomic DNA was digested with BglI and analyzed by Southern blot analysis with a 402-bp 5’ probe generated by PCR using oligos 5’-TGCCCTTTCACCTTTCCACT-3’ and 5’-TCCGTTTTCTCCGTTGGC-3’ and a 411-bp 3’ probe using primers 5’-CCCCAGAACTTCCTCTCCCT-3’ and 5’-TCTGAGGCAGACTGGCAGG-3’. The following amplification protocol was used: 94° for 2 min and 35 cycles of 94°C for 10 s, 58°C for 10 s, and 72°C for 30 s. The probes were labeled with [32P]dCTP by random priming and hybridized overnight at 55°C in ExpressHyb buffer (BD Biosciences Clontech, Palo Alto, CA, USA). The blots were washed twice in 2x SSC and 0.1% SDS, once in 0.2x SSC and 0.1% SDS, and once in 0.1x SSC and 0.1% SDS, each for 15 min at 60°C.
For this study, a total of approximately 500 mice were used. Colonies were maintained either at The Jackson Laboratory (TJL) animal research facility or at Case Western Reserve University (CWRU) Wolstein Animal Facility. All experimental animal protocols were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of either TJL or CWRU. Heterozygous and homozygous KO mice were genotyped by allele-specific PCR. The KO construct was detected by a universal neomycin PCR protocol that detects the neo gene using a forward Neo primer (5’-CTTGGGTGGAGAGGCTATTC-3’) and a reverse primer (5’-AGGTGAGATGACAGGAGATC-3’) to yield a 280-bp band (Fig. 1E). Wild-type mice lacking the KO construct were identified by amplification of a 200-bp band from Tcrd gene primers (5’-CAAATGTTGCTTGTCTGGTG-3’) and (5’-GTCAGTCGAGTGCACAGTTT-3’) in combination with a negative universal Neo PCR result (Fig. 1E). The Tcrd/Neo primer cocktail was used in amplification conditions of 12 cycles of 94°C for 20 s, 64°C for 30 s (-0.5°C per cycle) and 72°C for 35 s followed by 25 cycles of 94°C for 20 s, 58°C for 30 s, and 72°C for 35 s. Proper insertion at the 3’ end of the KO construct (Fig. 1F) was confirmed by a 150-bp band amplified from a primer within the neomycin gene (Ush1c Neo; 5’-GGAAATTGCATCGCATTGT-3’) and a primer in the 3’ homologous region of Ush1c (Ush1c R; 5’-TGGATCAGAAACAGATAGGCAT-3’). The wild-type, endogenous Ush1c allele (Fig. 1F) was detected as a 512-bp band amplified from primers Ush1c F (5’-GGGCAAAGGTCATAACCAG-3’) and Ush1c R. The bands in Fig. 1F were amplified in a cocktail using the following conditions: 9 cycles of 94°C for 15 s, 55°C for 30 s and 72°C for 45 s followed by 19 cycles of 94°C for 15 s, 55°C for 30 s and 72°C for 45 s plus 5 s each cycle.
4.3. Detection of gene expression by qRT-PCR in the inner ears and eyes of Ush1c−/− and Ush1c+/− mice
The Ush1c−/− and Ush1c+/−mice were sacrificed at 25 days of age utilizing Avertin anesthesia. The inner ears and eyes were quickly removed. Total RNA (DNA-free) was prepared using the pure-LinkTM Micro-to-Midi Total RNA Purification System (Invitrogen, Rockville, MD, USA). RNA was quantified using an ND-1000 spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA). cDNA was prepared using the SuperScript TM First-Strand Synthesis System (Invitrogen, Catalog No. 11904-018). To determine the expression of Ush1c isoform a1 in the inner ears and eyes, PCR reactions were carried out using the forward primer 5´–GAAGGCTGCCGAGGAGAATGAG–3´ in exon 13 and reverse primer 5´–CTGCGATCTGCTCTGGCGAGAA–3´ in exon 17 ( Verpy et al., 2000). As an internal control, glyceraldehyde-3-phosphatedehydrogenase (GAPDH) was amplified using primer sequences 5′-AACTTTGGCATTGTGGAAGG-3′ (forward) and 5′-GGAGACAACCTGGTCCTCAG-3′ (reverse) (Liu et al., 2007). We subjected one-tenth of the reaction product to PCR amplification. For the qRT-PCR, archival cDNA was made from ~3 µg of total RNA template per sample using an ABI high-capacity cDNA archive kit in a 100-µl reverse transcription reaction in an ABI 9700 PCR unit (Applied Biosystems Inc., Foster City, CA, USA). All assays were performed in triplicate in 384-well plates using GAPDH as an endogenous control. Assay kits were purchased from ABI and performed according to manufacturer’s instructions for a 40-cycle run. The Assay ID (Applied Biosystems TaqMan® Assay ID) is Mm00458359_m1 for the Ush1c gene. The amplicon is a 60-bp sequence across exons 15 and 16 boundary (GenBank accession no. AF228924.1); Assay ID is Mm00485371_m1for the Myo7a gene. The amplicon is a 62-bp sequence across exons 14 and 15 boundary (GenBank accession no. U81453.1); Assay ID is Mm00465755_m1 for the Cdh23 gene. The amplicon is a 75-bp sequence across the exons 9 and 10 boundary (GenBank accession no. NM_023370.2). Results were generated using ABI SDS 2.0 proprietary software and are presented as relative fold-changes versus a designated calibrator sample (GAPDH). The delta-delta Ct method was used to calculate RQ (relative quantitation) values and 95% confidence intervals (CI) are given.
4.4. Western Blots
Goat polyclonal antibody was purchased from Abcam (Cambridge, UK) against the peptide corresponding to N-terminal amino acids 2–14 of human harmonin (C-DRKVAREFRHKVD-common to all of the isoforms: a, b and c) (ab19045, GenBank accession nos. NP_005700.2; NP_710142.1). Samples were denatured by boiling 5 min and electrophoretically separated on pre-poured SDS-PAGE gels at 100–150 V (Bio-Rad Ready Gel, Tris-HCl Gel, 4–20%, Bio-Rad, Hercules, CA, USA). The protein was electrophoretically transferred (100V for 90 min) to a PDVF membrane that was blocked for 1 h in 5% non-fat milk, TBST. The membrane was incubated with primary antibody (Ab), diluted 1:500 in 5% non-fat-milk/TBST, for 16 h at 4°C. The membrane was then washed 3x with TBST and incubated for 1 h at room temp with secondary Ab, diluted 1:500 in 5% non-fat-milk/TBST. After three washes with TBST, the membrane was developed with chemiluminescent reagents for about 30 seconds according to the Bio-Rad protocol. The harmonin bands were quantitated with respect to actin controls for each lane, using the ImageQuant TL software (Amersham/GE Healthcare, Piscataway, NJ, USA).
4.5. Hearing Assessment by ABR
A computer-aided evoked potential system (Intelligent Hearing System, IHS; Miami, FL, USA) was used to test mouse ABR thresholds as previously described (Zheng et al., 1999). Briefly, mice were anesthetized and their body temperature was maintained at 37–38°C using a heating pad in a sound-attenuating chamber. Subdermal needle electrodes were inserted at the vertex of (active) and ventrolaterally to (reference) the right ear and the left ear (ground). Clicks, and 8, 16 and 32 kHz tone bursts were respectively channeled through plastic tubes into the animal's ear canals. Amplified brainstem responses were averaged by a computer and displayed on the computer screen. Auditory thresholds were obtained for each stimulus by reducing the sound pressure level (SPL) at 10-dB steps and finally at 5-dB steps up and down to identify the lowest level generating a recognizable ABR pattern.
4.6. LacZ staining
LacZ detection was performed as described (Oberdick, 1994) with a few modifications. Mice were decapitated and heads were dissected. Heads were fixed overnight in 2% paraformaldehyde (PFA) in 0.1M modified PIPES buffer and then stained in X-gal buffer overnight at 37°C followed by three washes in PBS. For mice older than one week of age, ears were dissected from the skull and decalcified in 7% EDTA/PBS after staining. Ears were dehydrated through a series of immersion in ethanol, then xylene, followed by paraffin embedding. Five-micrometer sections were cut using an American Optical model 820 rotary microtome (American Optical, Buffalo, New York, USA) and mounted on Superfrost plus slides (Fisher Scientific, Suwanee, GA, USA). Eye slides were prepared in a similar way but without EDTA decalcification. Slides were then counterstained with Eosin Y for 3 minutes. Slides were coverslipped and photographed with a Leica DM4500B light microscope and Leica DFC 500 camera (Leica Microsystems (Switzerland) Ltd., Heerbrugg, Switzerland). Whole-mount preparations of dissected ears and sensory epithelia from X-gal-stained half heads of mice were placed in glycerol and viewed with a Leica DM4500B light microscope.
4.7. Phenotypic evaluation of eyes
Mouse pupils were dilated with 1% atropine ophthalmic drops and evaluated by indirect ophthalmoscopy using a 78-diopter lens. Signs of retinal degeneration were noted, such as vessel attenuation, retinal pigment epithelium alteration, and retinal dots present or absent. ERG and histological microscopy of eyes were performed as described previously (Johnson et al., 2003).
4.8. Immunofluorescence and SEM analyses of inner ears
Isolated mouse inner ears were opened by piercing the apex and rupturing both the oval and round windows in 4% paraformaldehyde (pH 7.4) fixative. The inner ears were then immersed in the same fixative for 2 h at 4°C. The organ of Corti was dissected from the cochlear spiral in PBS using a fine needle, and then was permeabilized in 5% Triton X-100 for 30 min, then washed in PBS. Non-specific binding sites were blocked using 5% normal rabbit serum (Life Technologies, Gaithersburg, MD, USA) and 2% bovine serum albumin (ICN, Aurora, OH, USA) in PBS for 1 h. Samples were incubated with Harmonin (N-17) antibody (sc-26285, Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA), which recognizes all of the (a, b and c) isoforms (antigen peptide from aa position no.25–75), at 1:50 in blocking solution overnight at 44°C. After several rinses in PBS, samples were incubated in Alexa Fluor 488-conjugated rabbit anti-goat IgG at 1:300 (Molecular Probes, Eugene, OR, USA) for 1 h. After several rinses in PBS, samples were incubated with Alexa Fluor 568 labeled phalloidin at 1:200 (A12380, Molecular Probes, Inc., Eugene, OR, USA) to reveal hair cell bundles. Samples were mounted using Vectashield hard set H-1000 mounting medium (Vector Laboratories, Inc., Burlingame, CA, USA) and analyzed and photographed with a Leica DM4500B light microscope and Leica DFC 500 camera (Leica Microsystems (Switzerland) Ltd., Heerbrugg, Switzerland).
For SEM analysis, inner ears were dissected and fixed in 2.5% glutaraldehyde, 0.25% tannic acid, in 0.1 M phosphate buffer (pH 7.2) for 5 h at 4°C. After three washes in phosphate buffer, the temporal bone surrounding the cochlea and the tectorial membrane was removed to expose the organ of Corti. An osmium tetroxide–thiocarbohydrazide (OTOTO) procedure was used to stain prior to dehydrating and critical-point drying. Specimens were sputter-coated with gold and examined at 15 kV under a Hitachi 3000N scanning electron microscope (Hitachi High Technologies America, Inc., Pleasanton, CA, USA).
4.9. Statistical methods
The student’s t-test was used to compare the mean values for relative gene expression. A value of P < 0.05 was considered significant, unless stated specifically otherwise.
Supplementary Material
Supplementary Figure S1:
RT-PCR analysis of Ush1c expression in the inner ear of Ush1c−/− mice and heterozygous littermates Ush1c+/− (n=4) at P6. The RT-PCR primers were designed according to the cDNA sequence of the 3 transcriptional variants of the Ush1c gene from the ensemble data base: (http://www.ensembl.org/, gene: Ush1cMus_musculus/Gene/Summary?g=ENSMUSG00000030838) by using the Primer 3 online software. Three pairs of primers, harmonin-a1F (GAAGGCTGCCGAGGAGAATGAG, in exon 13) and harmonin-a1R (CTGCGATCTGCTCTGGCGAGAA, in exon 17); Ush1c1F (ATGCTGGAGAAGGAGAAGCA, in exon 16) and Us1c1R (GCCAGAGGAGACACAGAAGG, in exon 18); and Ush1c2F (ACTTCACCAACCTGGACCAC, in exon 10) and Ush1c2R (TGTGGGTCAGCTTCCAGGGAGACTTT, in exon 22) (Johnson et al., 2003) were used to identify harmonin isoforms a1, b and c, respectively. The exon numbers refer to the sequential order in each transcript variant. The other 4 pairs of primers, Ush1c3F (CGAGAATTCCGACACAAGGT, in exon 1) and Ush1c3R (GTCCAAGCGTACCTCCTTCA, in exon 4), Ush1c4F (CGGACAACCAAGGAGAAGAA, in exon 8) and Ush1c4R (GGCTGCTCTTCAGGACATTC, in exon 11) (Johnson et al., 2003), Ush1c5F (GCCATGGAGTCCAACAAGAT, in exon 12) and Ush1c5R (CCCAGTCTTCTTTCCACTGC, in exon 14), and Ush1c6F (TGTTTATGAAGGGGGAGCTG, in exon 23) and Ush1c6R (TTAGGGGGACAGACAGCAAC, in exon 25) are at different positions of the Ush1c gene to determine the complete absence of Ush1c gene expression in the mutant mice. The exon numbers refer to the sequential order in the longest b transcript variant. mRNA extraction and cDNA preparation were performed as described above. The 20-µl reaction mixture contained 50 mM KCl, 10 mM Tris-HCl, pH 9.0 (at 25°C), 0.01% Triton X-100, 2 mM MgCl2, 250 nM of each primer (forward and reverse), 200 µM dNTPs, 1 µl of cDNA and 0.5 U of Taq DNA polymerase (New England BioLabs). PCR was performed in a BioRad PTC-200 Peltier Thermal Cycler. Amplification conditions were 94°C for 2 min; followed by 32 cycles of 94°C for 30 s, 60°C for 40 s, and 72°C for 50 s; followed by 5 min at 72°C. Ten microliters of the PCR products was subject to agarose gel electrophoresis (1.8 %). The mRNA level Gapdh was used as internal control as in the qRT-PCR. The sizes of the PCR products of were as follows: Gapdh, 351 bp; a1-isoform, 294 bp; b-isoform, 212 bp; c-isoform, 1081 bp; exons 1–4, 255 bp; exons 8–11, 250 bp; exons 12–14, 166 bp; and exons 23–25, 180 bp. All the cDNA fragments were amplified in the heterozygous mice (Ush1c+/−) as the predicted sizes, but failed to be amplified in the homozygous KO mice (Ush1c−/−), suggesting the deletion of all Ush1c gene expression in the Ush1c−/− KO mice. The 2069-bp PCR products in the Ush1c−/− mice amplified by Ush1c5F (in exon 12) and Ush1c5R (in exon 14) match the predicted size of the fragment amplified from residual genomic DNA of the Ush1c gene. Similar expression levels of the Gapdh gene were detected in −/− and +/− samples, suggesting that total RNA and cDNA were comparable. M, 100 bp DNA ladders.
Supplementary Figure S2: Cochlear whole-mounts of P0 wild-type and Ush1c−/− mice were stained with phalloidin (red) and with the harmonin- N-17 antibody, which recognizes all of the a, b, and c isoforms (green) (Santa Cruz Biotechnology, Inc. Harmonin N-17, sc-26285). An intense harmonin- N-17 staining was detected at the stereocilia tips of Ush1c+/+ hair cells, but was completely lost in Ush1c−/− hair cells. Scale bars: 10 µm.
Supplementary Figure S3: Auditory Brain response (ABR) recordings show normal wave patterns and thresholds in Ush1c+/− heterozygous KO control mice, but there were no evoked responses at all in homozygous Ush1c−/− mutants (n=5) tested at 3 weeks of age. ABRs for broad-band click are shown here, but similar results were obtained for 8, 16, and 32 kHz pure-tone stimuli. Scale Bar=8µV.
Supplementary Figure S4: LacZ expression in the eyes of 3 Ush1c+/− mice. There is a similar expression level for both ages (1 and 5 months). A and B have similar labeling as Fig 7. (C) The scattered blue dots were distributed with rare frequency in the epithelial layer (black arrow) of the vitreous body (C) and in each layer from the outer plexiform layer (OPL) to the ganglion cell layer (GCL) but not in most portions of the photoreceptor layer (A, B). No expression of lacZ was observed in the wild-type littermate (+/+) even at 5 months of age (D). Scale bar = 50 µm.
Supplementary Video S1: Swimming test: C57BL/6J-Ush1c+/− mice (left, n=2) can keep nose above the surface of bathing water while littermate C57BL/6J-Ush1c−/− mice (right, n=5) failed to do so and kept diving and circling in the deep water.
Acknowledgements
The research was supported by NIH DC05575, DC007392, DC009246, DC004301 and by the Gene Expression Core Facility of the Case Comprehensive Cancer Center (P30 CA43703). We wish to thank Heather Carlisle for design and assembly of the gene-targeting construct, and the personnel of the Cell Biology and Microinjection Services for generating the Ush1c-LacZ mice. We also thank Chunling Gu for technical assistance, Ken Johnson for critical review of this manuscript, and Ronald E. Hurd for the ERG recording. We also thank Cindy Benedict-Alderfer for her great effort in editing the manuscript.
Abbreviations
- ABR
auditory-evoked brainstem response
- ERG
electroretinogram
- IHC
inner hair cells
- OHC
outer hair cells
- RP
retinitis pigmentosa
- SEM
scanning electron microscopy
- USH
Usher syndrome
- USH1C
harmonin
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Authors' contributions:
CT performed the immunoassays. XZL contributed to initial research design. FC and CT performed the molecular studies including real-time PCR and sequence analyses. HY performed ABR tests, the immunoassays, contributed to generation of backcross and Ush1c−/− mice. CT and CLG contributed the lacZ reporter gene expression and hair bundle assay, CLG performed the SEM assay. BY and CL performed genotyping, DY performed literature research. QYZ conceived and designed the study, supervised all the experimental data analyses and the manuscript preparation. All authors read and approved the final manuscript.
References
- Ahmed Z, Riazuddin S, Khan S, Friedman P, Riazuddin S, Friedman T. Ush1h, a novel locus for type I Usher syndrome, maps to chromosome 15q22-23. Clin Genet. 2008 doi: 10.1111/j.1399-0004.2008.01038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed ZM, Riazuddin S, Bernstein SL, Ahmed Z, Khan S, Griffith AJ, Morell RJ, Friedman TB, Riazuddin S, Wilcox ER. Mutations of the protocadherin gene pcdh15 cause usher syndrome type 1f. Am J Hum Genet. 2001;69:25–34. doi: 10.1086/321277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed ZM, Riazuddin S, Riazuddin S, Wilcox ER. The molecular genetics of usher syndrome. Clin Genet. 2003;63:431–444. doi: 10.1034/j.1399-0004.2003.00109.x. [DOI] [PubMed] [Google Scholar]
- Alagramam KN, Murcia CL, Kwon HY, Pawlowski KS, Wright CG, Woychik RP. The mouse ames waltzer hearing-loss mutant is caused by mutation of pcdh15, a novel protocadherin gene. Nat Genet. 2001a;27:9–102. doi: 10.1038/83837. [DOI] [PubMed] [Google Scholar]
- Alagramam KN, Yuan H, Kuehn MH, Murcia CL, Wayne S, Srisailpathy CR, Lowry RB, Knaus R, Van Laer L, Bernier FP, Schwartz S, Lee C, Morton CC, Mullins RF, Ramesh A, Van Camp G, Hageman GS, Woychik RP, Smith RJ. Mutations in the novel protocadherin pcdh15 cause usher syndrome type 1f. Hum Mol Genet. 2001b;10:1709–1718. doi: 10.1093/hmg/10.16.1709. [DOI] [PubMed] [Google Scholar]
- Bitner-Glindzicz M, Lindley KJ, Rutland P, Blaydon D, Smith VV, Milla PJ, Hussain K, Furth-Lavi J, Cosgrove KE, Shepherd RM, Barnes PD, O'Brien RE, Farndon PA, Sowden J, Liu XZ, Scanlan MJ, Malcolm S, Dunne MJ, Aynsley-Green A, Glaser B. A recessive contiguous gene deletion causing infantile hyperinsulinism, enteropathy and deafness identifies the usher type 1c gene. Nat Genet. 2000;26:56–60. doi: 10.1038/79178. [DOI] [PubMed] [Google Scholar]
- Boeda B, El-Amraoui A, Bahloul A, Goodyear R, Daviet L, Blanchard S, Perfettini I, Fath KR, Shorte S, Reiners J, Houdusse A, Legrain P, Wolfrum U, Richardson G, Petit C. Myosin viia, harmonin and cadherin 23, three usher i gene products that cooperate to shape the sensory hair cell bundle. EMBO J. 2002;21:6689–6699. doi: 10.1093/emboj/cdf689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolz H, von Brederlow B, Ramirez A, Bryda EC, Kutsche K, Nothwang HG, Seeliger M, del CSCM, Vila MC, Molina OP, Gal A, Kubisch C. Mutation of cdh23, encoding a new member of the cadherin gene family, causes usher syndrome type 1d. Nat Genet. 2001;27:108–112. doi: 10.1038/83667. [DOI] [PubMed] [Google Scholar]
- Bork JM, Morell RJ, Khan S, Riazuddin S, Wilcox ER, Friedman TB, Griffith AJ. Clinical presentation of dfnb12 and usher syndrome type 1d. Adv Otorhinolaryngol. 2002;61:145–152. doi: 10.1159/000066829. [DOI] [PubMed] [Google Scholar]
- Bork JM, Peters LM, Riazuddin S, Bernstein SL, Ahmed ZM, Ness SL, Polomeno R, Ramesh A, Schloss M, Srisailpathy CR, Wayne S, Bellman S, Desmukh D, Ahmed Z, Khan SN, Kaloustian VM, Li XC, Lalwani A, Bitner-Glindzicz M, Nance WE, Liu XZ, Wistow G, Smith RJ, Griffith AJ, Wilcox ER, Friedman TB, Morell RJ. Usher syndrome 1d and nonsyndromic autosomal recessive deafness dfnb12 are caused by allelic mutations of the novel cadherin-like gene cdh23. Am J Hum Genet. 2001;68:26–37. doi: 10.1086/316954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport SL, O'Nuallain S, Omenn GS, Wilkus RJ. Usher syndrome in four hard-of-hearing siblings. Pediatrics. 1978;62:578–583. [PubMed] [Google Scholar]
- Di Palma F, Holme RH, Bryda EC, Belyantseva IA, Pellegrino R, Kachar B, Steel KP, Noben-Trauth K. Mutations in cdh23, encoding a new type of cadherin, cause stereocilia disorganization in waltzer, the mouse model for usher syndrome type 1d. Nat Genet. 2001;27:103–107. doi: 10.1038/83660. [DOI] [PubMed] [Google Scholar]
- Friedrich G, Soriano P. Promoter traps in embryonic stem cells: A genetic screen to identify and mutate developmental genes in mice. Genes Dev. 1991;5:1513–1523. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- Gibbs D, Kitamoto J, Williams DS. Abnormal phagocytosis by retinal pigmented epithelium that lacks myosin viia, the usher syndrome 1b protein. Proc Natl Acad Sci U S A. 2003;100:6481–6486. doi: 10.1073/pnas.1130432100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs D, Williams DS. Usher 1 protein complexes in the retina. Invest Ophthalmol Vis Sci. 2004;45 e-letter. [Google Scholar]
- Gibson F, Walsh J, Mburu P, Varela A, Brown KA, Antonio M, Beisel KW, Steel KP, Brown SD. A type vii myosin encoded by the mouse deafness gene shaker-1. Nature. 1995;374:62–64. doi: 10.1038/374062a0. [DOI] [PubMed] [Google Scholar]
- Grillet N, Xiong W, Reynolds A, Kazmierczak P, Sato T, Lillo C, Dumont RA, Hintermann E, Sczaniecka A, Schwander M, Williams D, Kachar B, Gillespie PG, Müller U. Harmonin mutations cause mechanotransduction defects in cochlear hair cells. Neuron. 2009;62(3):375–387. doi: 10.1016/j.neuron.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haywood-Watson RJ, 2nd, Ahmed ZM, Kjellstrom S, Bush RA, Takada Y, Hampton LL, Battey JF, Sieving PA, Friedman TB. Ames waltzer deaf mice have reduced electroretinogram amplitudes and complex alternative splicing of pcdh15 transcripts. Invest Ophthalmol Vis Sci. 2006;47:3074–3084. doi: 10.1167/iovs.06-0108. [DOI] [PubMed] [Google Scholar]
- Johnson KR, Gagnon LH, Webb LS, Peters LL, Hawes NL, Chang B, Zheng QY. Mouse models of ush1c and dfnb18: Phenotypic and molecular analyses of two new spontaneous mutations of the ush1c gene. Hum Mol Genet. 2003;12:3075–3086. doi: 10.1093/hmg/ddg332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikkawa Y, Shitara H, Wakana S, Kohara Y, Takada T, Okamoto M, Taya C, Kamiya K, Yoshikawa Y, Tokano H, Kitamura K, Shimizu K, Wakabayashi Y, Shiroishi T, Kominami R, Yonekawa H. Mutations in a new scaffold protein sans cause deafness in jackson shaker mice. Hum Mol Genet. 2003;12:453–461. doi: 10.1093/hmg/ddg042. [DOI] [PubMed] [Google Scholar]
- Kremer H, van Wijk E, Marker T, Wolfrum U, Roepman R. Usher syndrome: Molecular links of pathogenesis, proteins and pathways. Hum Mol Genet 15 Spec No. 2006;2:R262–R270. doi: 10.1093/hmg/ddl205. [DOI] [PubMed] [Google Scholar]
- Lefevre G, Michel V, Weil D, Lepelletier L, Bizard E, Wolfrum U, Hardelin JP, Petit C. A core cochlear phenotype in ush1 mouse mutants implicates fibrous links of the hair bundle in its cohesion, orientation and differential growth. Development. 2008;135:1427–1437. doi: 10.1242/dev.012922. [DOI] [PubMed] [Google Scholar]
- Lentz J, Pan F, Ng SS, Deininger P, Keats B. Ush1c216a knock-in mouse survives katrina. Mutat Res. 2007;616:139–144. doi: 10.1016/j.mrfmmm.2006.11.006. [DOI] [PubMed] [Google Scholar]
- Libby RT, Kitamoto J, Holme RH, Williams DS, Steel KP. Cdh23 mutations in the mouse are associated with retinal dysfunction but not retinal degeneration. Exp Eye Res. 2003;77:731–739. doi: 10.1016/j.exer.2003.07.007. [DOI] [PubMed] [Google Scholar]
- Libby RT, Steel KP. Electroretinographic anomalies in mice with mutations in myo7a, the gene involved in human usher syndrome type 1b. Invest Ophthalmol Vis Sci. 2001;42:770–778. [PubMed] [Google Scholar]
- Lillo C, Kitamoto J, Liu X, Quint E, Steel KP, Williams DS. Mouse models for usher syndrome 1b. Adv Exp Med Biol. 2003;533:143–150. doi: 10.1007/978-1-4615-0067-4_18. [DOI] [PubMed] [Google Scholar]
- Lillo C, Siemens J, Kazmierczak P, Mueller U, Williams DS. Roles and interactions of three ush1 proteins in the retina and inner ear. ARVO Abstracts. 2005 #5176. [Google Scholar]
- Liu X, Bulgakov OV, Darrow KN, Pawlyk B, Adamian M, Liberman MC, Li T. Usherin is required for maintenance of retinal photoreceptors and normal development of cochlear hair cells. Proc Natl Acad Sci U S A. 2007;104:4413–4418. doi: 10.1073/pnas.0610950104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee RJ, Goodyear DR, McMillan DR, Stauffer EA, Holt JR, Locke KG, Birch DG, Legan PK, White PC, Walsh EJ, Richardson GP. The very large G-protein-coupled receptor VLGR1: a component of the ankle link complex required for the normal development of auditory hair bundles. J Neurosci. 2006;26:6543–6553. doi: 10.1523/JNEUROSCI.0693-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalski N, Michel V, Caberlotto E, Lefèvre GM, van Aken AF, Tinevez JY, Bizard E, Houbron C, Weil D, Hardelin JP, Richardson GP, Kros CJ, Martin P, Petit C. Harmonin-b, an actin-binding scaffold protein, is involved in the adaptation of mechanoelectrical transduction by sensory hair cells. Pflugers Arch. 2009;459(1):115–130. doi: 10.1007/s00424-009-0711-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberdick J. Evidence for a genetically encoded map of functional development in the cerebellum. Histochemistry. 1994;102:1–14. doi: 10.1007/BF00271044. [DOI] [PubMed] [Google Scholar]
- Petit C. Usher syndrome: From genetics to pathogenesis. Annual Review of Genomics and Human Genetics. 2001;2:271–297. doi: 10.1146/annurev.genom.2.1.271. [DOI] [PubMed] [Google Scholar]
- Reiners J, Marker T, Jurgens K, Reidel B, Wolfrum U. Photoreceptor expression of the usher syndrome type 1 protein protocadherin 15 (ush1f) and its interaction with the scaffold protein harmonin (ush1c) Mol Vis. 2005a;11:347–355. [PubMed] [Google Scholar]
- Reiners J, Nagel-Wolfrum K, Jurgens K, Marker T, Wolfrum U. Molecular basis of human usher syndrome: Deciphering the meshes of the usher protein network provides insights into the pathomechanisms of the usher disease. Exp Eye Res. 2006;83:97–119. doi: 10.1016/j.exer.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Reiners J, van Wijk E, Marker T, Zimmermann U, Jurgens K, te Brinke H, Overlack N, Roepman R, Knipper M, Kremer H, Wolfrum U. Scaffold protein harmonin (ush1c) provides molecular links between usher syndrome type 1 and type 2. Hum Mol Genet. 2005b;14:3933–3943. doi: 10.1093/hmg/ddi417. [DOI] [PubMed] [Google Scholar]
- Reiners J, Wolfrum U. Molecular analysis of the supramolecular usher protein complex in the retina. Harmonin as the key protein of the usher syndrome. Adv Exp Med Biol. 2006;572:349–353. doi: 10.1007/0-387-32442-9_49. [DOI] [PubMed] [Google Scholar]
- Rosenberg T, Haim M, Hauch AM, Parving A. The prevalence of usher syndrome and other retinal dystrophy-hearing impairment associations. Clin Genet. 1997;51:314–321. doi: 10.1111/j.1399-0004.1997.tb02480.x. [DOI] [PubMed] [Google Scholar]
- Sun X, Pawlyk B, Adamian M, Michaud N, Bulgakov O, Li T. Functional and structural deficits of cone photoreceptors in mice lacking PCDH15, a protein encoded by the Ush1F gene. ARVO abstracts. 2006 #5770. [Google Scholar]
- Thomas KR, Deng C, Capecchi MR. High-fidelity gene targeting in embryonic stem cells by using sequence replacement vectors. Mol Cell Biol. 1992;12:2919–2923. doi: 10.1128/mcb.12.7.2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wijk E, van der Zwaag B, Peters T, Zimmermann U, Te Brinke H, Kersten FF, Marker T, Aller E, Hoefsloot LH, Cremers CW, Cremers FP, Wolfrum U, Knipper M, Roepman R, Kremer H. The dfnb31 gene product whirlin connects to the usher protein network in the cochlea and retina by direct association with ush2a and vlgr1. Hum Mol Genet. 2006;15:751–765. doi: 10.1093/hmg/ddi490. [DOI] [PubMed] [Google Scholar]
- Verpy E, Leibovici M, Zwaenepoel I, Liu XZ, Gal A, Salem N, Mansour A, Blanchard S, Kobayashi I, Keats BJ, Slim R, Petit C. A defect in harmonin, a pdz domain-containing protein expressed in the inner ear sensory hair cells, underlies usher syndrome type 1c. Nat Genet. 2000;26:51–55. doi: 10.1038/79171. [DOI] [PubMed] [Google Scholar]
- Weil D, Blanchard S, Kaplan J, Guilford P, Gibson F, Walsh J, Mburu P, Varela A, Levilliers J, Weston MD, et al. Defective myosin viia gene responsible for usher syndrome type 1b. Nature. 1995;374:60–61. doi: 10.1038/374060a0. [DOI] [PubMed] [Google Scholar]
- Weil D, El-Amraoui A, Masmoudi S, Mustapha M, Kikkawa Y, Laine S, Delmaghani S, Adato A, Nadifi S, Zina ZB, Hamel C, Gal A, Ayadi H, Yonekawa H, Petit C. Usher syndrome type i g (ush1g) is caused by mutations in the gene encoding sans, a protein that associates with the ush1c protein, harmonin. Hum Mol Genet. 2003;12:463–471. doi: 10.1093/hmg/ddg051. [DOI] [PubMed] [Google Scholar]
- Williams DS, Aleman TS, Lillo C, Lopes VS, Hughes LC, Stone EM, Jacobson SG. Harmonin in the murine retina and the retinal phenotypes of Ush1c-mutant mice and human USH1C. Invest Ophthalmol Vis Sci. 2009;50(8):3881–3889. doi: 10.1167/iovs.08-3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson SM, Householder DB, Coppola V, Tessarollo L, Fritzsch B, Lee EC, Goss D, Carlson GA, Copeland NG, Jenkins NA. Mutations in cdh23 cause nonsyndromic hearing loss in waltzer mice. Genomics. 2001;74:228–233. doi: 10.1006/geno.2001.6554. [DOI] [PubMed] [Google Scholar]
- Zheng QY, Johnson KR, Erway LC. Assessment of hearing in 80 inbred strains of mice by abr threshold analyses. Hear. Res. 1999;130:94–107. doi: 10.1016/s0378-5955(99)00003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1:
RT-PCR analysis of Ush1c expression in the inner ear of Ush1c−/− mice and heterozygous littermates Ush1c+/− (n=4) at P6. The RT-PCR primers were designed according to the cDNA sequence of the 3 transcriptional variants of the Ush1c gene from the ensemble data base: (http://www.ensembl.org/, gene: Ush1cMus_musculus/Gene/Summary?g=ENSMUSG00000030838) by using the Primer 3 online software. Three pairs of primers, harmonin-a1F (GAAGGCTGCCGAGGAGAATGAG, in exon 13) and harmonin-a1R (CTGCGATCTGCTCTGGCGAGAA, in exon 17); Ush1c1F (ATGCTGGAGAAGGAGAAGCA, in exon 16) and Us1c1R (GCCAGAGGAGACACAGAAGG, in exon 18); and Ush1c2F (ACTTCACCAACCTGGACCAC, in exon 10) and Ush1c2R (TGTGGGTCAGCTTCCAGGGAGACTTT, in exon 22) (Johnson et al., 2003) were used to identify harmonin isoforms a1, b and c, respectively. The exon numbers refer to the sequential order in each transcript variant. The other 4 pairs of primers, Ush1c3F (CGAGAATTCCGACACAAGGT, in exon 1) and Ush1c3R (GTCCAAGCGTACCTCCTTCA, in exon 4), Ush1c4F (CGGACAACCAAGGAGAAGAA, in exon 8) and Ush1c4R (GGCTGCTCTTCAGGACATTC, in exon 11) (Johnson et al., 2003), Ush1c5F (GCCATGGAGTCCAACAAGAT, in exon 12) and Ush1c5R (CCCAGTCTTCTTTCCACTGC, in exon 14), and Ush1c6F (TGTTTATGAAGGGGGAGCTG, in exon 23) and Ush1c6R (TTAGGGGGACAGACAGCAAC, in exon 25) are at different positions of the Ush1c gene to determine the complete absence of Ush1c gene expression in the mutant mice. The exon numbers refer to the sequential order in the longest b transcript variant. mRNA extraction and cDNA preparation were performed as described above. The 20-µl reaction mixture contained 50 mM KCl, 10 mM Tris-HCl, pH 9.0 (at 25°C), 0.01% Triton X-100, 2 mM MgCl2, 250 nM of each primer (forward and reverse), 200 µM dNTPs, 1 µl of cDNA and 0.5 U of Taq DNA polymerase (New England BioLabs). PCR was performed in a BioRad PTC-200 Peltier Thermal Cycler. Amplification conditions were 94°C for 2 min; followed by 32 cycles of 94°C for 30 s, 60°C for 40 s, and 72°C for 50 s; followed by 5 min at 72°C. Ten microliters of the PCR products was subject to agarose gel electrophoresis (1.8 %). The mRNA level Gapdh was used as internal control as in the qRT-PCR. The sizes of the PCR products of were as follows: Gapdh, 351 bp; a1-isoform, 294 bp; b-isoform, 212 bp; c-isoform, 1081 bp; exons 1–4, 255 bp; exons 8–11, 250 bp; exons 12–14, 166 bp; and exons 23–25, 180 bp. All the cDNA fragments were amplified in the heterozygous mice (Ush1c+/−) as the predicted sizes, but failed to be amplified in the homozygous KO mice (Ush1c−/−), suggesting the deletion of all Ush1c gene expression in the Ush1c−/− KO mice. The 2069-bp PCR products in the Ush1c−/− mice amplified by Ush1c5F (in exon 12) and Ush1c5R (in exon 14) match the predicted size of the fragment amplified from residual genomic DNA of the Ush1c gene. Similar expression levels of the Gapdh gene were detected in −/− and +/− samples, suggesting that total RNA and cDNA were comparable. M, 100 bp DNA ladders.
Supplementary Figure S2: Cochlear whole-mounts of P0 wild-type and Ush1c−/− mice were stained with phalloidin (red) and with the harmonin- N-17 antibody, which recognizes all of the a, b, and c isoforms (green) (Santa Cruz Biotechnology, Inc. Harmonin N-17, sc-26285). An intense harmonin- N-17 staining was detected at the stereocilia tips of Ush1c+/+ hair cells, but was completely lost in Ush1c−/− hair cells. Scale bars: 10 µm.
Supplementary Figure S3: Auditory Brain response (ABR) recordings show normal wave patterns and thresholds in Ush1c+/− heterozygous KO control mice, but there were no evoked responses at all in homozygous Ush1c−/− mutants (n=5) tested at 3 weeks of age. ABRs for broad-band click are shown here, but similar results were obtained for 8, 16, and 32 kHz pure-tone stimuli. Scale Bar=8µV.
Supplementary Figure S4: LacZ expression in the eyes of 3 Ush1c+/− mice. There is a similar expression level for both ages (1 and 5 months). A and B have similar labeling as Fig 7. (C) The scattered blue dots were distributed with rare frequency in the epithelial layer (black arrow) of the vitreous body (C) and in each layer from the outer plexiform layer (OPL) to the ganglion cell layer (GCL) but not in most portions of the photoreceptor layer (A, B). No expression of lacZ was observed in the wild-type littermate (+/+) even at 5 months of age (D). Scale bar = 50 µm.
Supplementary Video S1: Swimming test: C57BL/6J-Ush1c+/− mice (left, n=2) can keep nose above the surface of bathing water while littermate C57BL/6J-Ush1c−/− mice (right, n=5) failed to do so and kept diving and circling in the deep water.