Abstract
Background
The availability of the complete chicken (Gallus gallus) genome sequence as well as a large number of chicken probes for fluorescent in-situ hybridization (FISH) and microarray resources facilitate comparative genomic studies between chicken and other bird species. In a previous study, we provided a comprehensive cytogenetic map for the turkey (Meleagris gallopavo) and the first analysis of copy number variants (CNVs) in birds. Here, we extend this approach to the Pekin duck (Anas platyrhynchos), an obvious target for comparative genomic studies due to its agricultural importance and resistance to avian flu.
Results
We provide a detailed molecular cytogenetic map of the duck genome through FISH assignment of 155 chicken clones. We identified one inter- and six intrachromosomal rearrangements between chicken and duck macrochromosomes and demonstrated conserved synteny among all microchromosomes analysed. Array comparative genomic hybridisation revealed 32 CNVs, of which 5 overlap previously designated "hotspot" regions between chicken and turkey.
Conclusion
Our results suggest extensive conservation of avian genomes across 90 million years of evolution in both macro- and microchromosomes. The data on CNVs between chicken and duck extends previous analyses in chicken and turkey and supports the hypotheses that avian genomes contain fewer CNVs than mammalian genomes and that genomes of evolutionarily distant species share regions of copy number variation ("CNV hotspots"). Our results will expedite duck genomics, assist marker development and highlight areas of interest for future evolutionary and functional studies.
Background
Comparative genomics allows the transfer of genomic information from a well-characterized species to another that is less well described. It can be applied at all levels from that of the chromosome to the genome sequence. However, despite the recent advances in sequencing technologies, the considerable effort involved in producing a genome sequence assembly is reflected by the small number of vertebrate genomes that have been sequenced to date. In birds, there is only one published genome sequence, that of the chicken [1], with the zebra finch genome due to be published soon.
Combining cross-species fluorescent in-situ hybridization (FISH) and microarray analysis using resources developed in the chicken provides a powerful tool for the identification of gross genomic rearrangements, gene gains/losses, copy number variants (CNVs) and gene order in other bird species. These techniques do not require sequence data for any species other than the reference (i.e. chicken). We have previously successfully used this approach for a genome wide comparison of chromosomal rearrangements and CNVs between chicken and turkey[2]. This revealed a strong conservation of genome structure over about 30 million years of evolution between chicken and turkey[3]. In particular, our results suggested that, when compared to mammalian genomes, bird genomes contain a low number of CNVs (i.e. polymorphisms in the number of copies of a DNA fragment 1 kb or larger[4], with the exception of insertions or deletions of transposable elements[5]). The latter finding indicates that patterns of CNVs in bird genomes mirror the low number of chromosomal rearrangements in this phylogenetic group[2,6].
Following on from the turkey, the Pekin duck (Anas platyrhynchos, APL) is the next obvious target among domestic birds for detailed genomic studies due to its agricultural importance, with worldwide duck consumption being between 4 and 5% of the total poultry market[7]. Duck is also an important target for immunological studies because of its resistance to avian influenza[8]. Despite this, genomic information about the duck is limited to a few linkage and physical mapping studies. Huang et al. [9] produced a preliminary genetic map based on 240 microsatellite loci and assigned 11 out of 19 linkage groups to ten duck (APL) chromosomes by FISH mapping of 28 BACs. Cross-species chromosome painting and G-banding studies [10-12] have suggested one interchromosomal difference between the chicken and duck karyotypes – the ancestral chromosomes 4 and 10, fused in the chicken lineage to give GGA4q and GGA4p respectively, remain separate in duck[6]. This interchromosomal rearrangement presumably explains the difference in diploid chromosome number between the two species, which is 2n = 78 in chicken and 2n = 80 in duck. FISH mapping of 57 chicken BACs revealed small intrachromosomal rearrangements in APL2, 7, 8 and Z and confirmed synteny for GGA9, 11, 13–15, 18 and 28 in the duck genome[13]. However, no molecular markers are available for the remaining microchromosomes, which are indistinguishable by conventional cytogenetics. It is also unclear which duck chromosome corresponds to GGA4p (ancestral chromosome 10). Thus, from a molecular cytogenetic standpoint, the duck genome is at present only partially defined, and given the low number of physical markers mapped by FISH, it is possible that hitherto undetected intrachromosomal rearrangements exist.
No data are currently available concerning CNVs in duck or indeed any other bird species than chicken and turkey. CNVs have been found to contribute significantly to normal and disease-related genetic and phenotypic variation in humans and other primates[5,14]. Studies of the evolutionary significance of CNVs have largely focused on primates and revealed numerous lineage-specific gene gains and losses and CNVs (e.g. [15-20]).
Our previous study of CNVs in chicken and turkey revealed a total of 16 CNVs[2]. Five of these CNVs appear to be shared in layer and broiler chickens, and in turkey, at regions dubbed "CNV hotspots" (i.e. genomic regions in which CNVs of approximately equal size were found in both chicken breeds and in turkey). Given that the contribution of CNVs to phenotypic variation is becoming increasingly clear, analyses of this kind of structural variation in organismic groups other than mammals are clearly needed.
Here, we present a detailed molecular cytogenetic map for the duck based on comparative FISH mapping of 155 chicken BACs, which revealed several hitherto undescribed intrachromosomal rearrangements. We also provide an analysis of CNVs in the duck genome by array comparative genomic hybridisation (array CGH) of duck DNA to a commercially available chicken whole-genome oligonucleotide tiling path microarray. The analysis of CNVs supports the hypotheses that bird genomes contain fewer CNVs than mammalian genomes and that some CNVs appear to be consistently shared across species, forming CNV hotspots.
Results
Comparative FISH mapping between chicken and duck
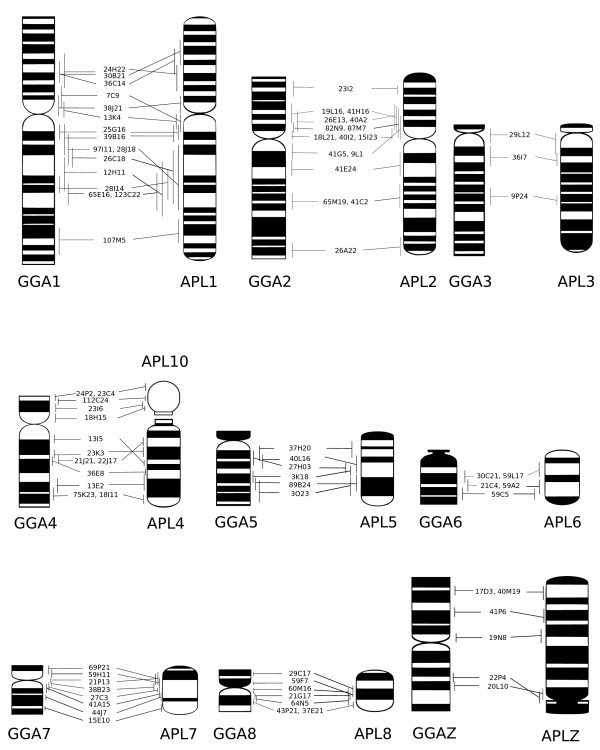
Of 400 BACs that successfully hybridized to chicken metaphases, 155 (39%) could be visualized with confidence on duck chromosomes. These covered the majority of the karyotype i.e. APL1–29 and Z (except 26). Figure 1 shows the G-banded ideograms for GGA1–8 and Z and APL1–8 and Z[13,21], with the positions of the BACs mapped to these chromosomes; the full data are presented in Additional file 1. Figure 2 shows example FISH results. Only one interchromosomal difference was detected among the macrochromosomes, the retention of the ancestral chromosomes 4 and 10 in duck (which are fused in chicken).
Figure 1.
Comparative map of chicken and duck chromosomes 1–8 and Z. The G-banded ideograms of chicken (Gallus gallus; GGA) and duck (Anas platyrhynchos; APL) chromosomes 1–8 and Z are shown with the positions of all BACs successfully hybridized to both species as determined by FLpter measurements. Intrachromosomal rearrangements can be seen on GGA and APL1, 2, 4, 7, 8 and Z. GGA4p corresponds to APL10. Note the orientation of APLZ. Ideograms were prepared from[13,21]. Error bars represent one standard deviation.
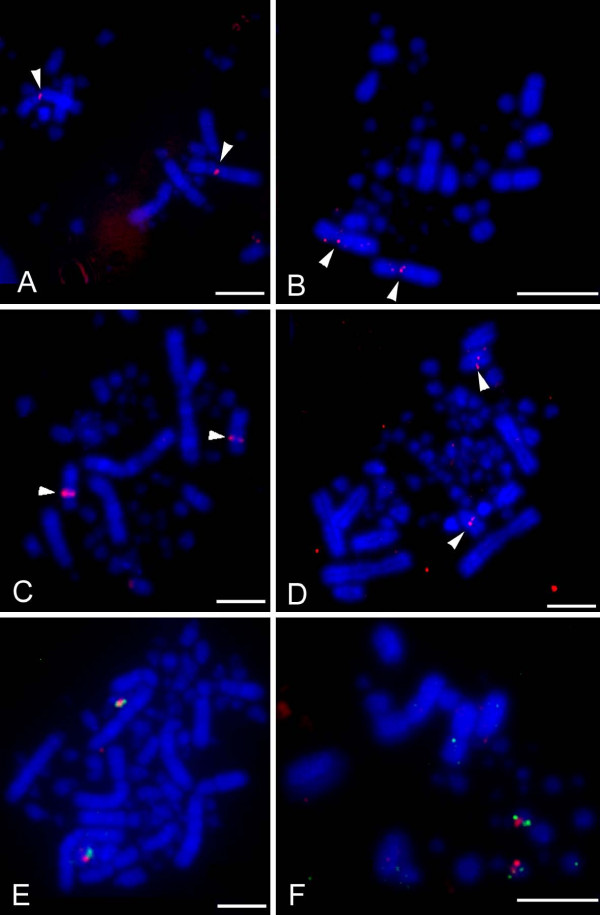
Figure 2.
Example FISH Results. BAC WAG13K4 on A) chicken (Gallus gallus; GGA) chromosome 1p and B) duck (Anas platyrhynchos; APL) chromosome 1q. BAC WAG13I5 on C) chicken chromosome 4q and D) duck chromosome 4 demonstrating part of the evidence that led us to deduce a paracentric inversion. Conserved synteny among the microchromosomes was tested by dual color FISH. An example is shown for E) GGA15 and F) APL16 using BACs WAG10L1 and WAG93I1. Scale bars represent five microns.
FISH mapping suggested intrachromosomal rearrangements on GGA1, 2, 4, 7, 8, Z and APL1, 2, 4, 7, 8 and Z. BACs WAG24H22, WAG30B21 and WAG36C14 clearly showed a rearrangement on GGA1p and APL1p. The order of BACs was not completely inverted, suggesting that the underlying rearrangement may be a translocation rather than a paracentric inversion. BACs WAG7C9 and WAG13K4 (Figure 2A, B) mapped to GGA1p and APL1q, indicating a small pericentric inversion. Besides, some BACs mapping to GGA1q and APL1q suggested possible rearrangements on these chromosome arms; however, the high standard deviations in the FLpter measurements determined for these BACs in duck made it difficult to distinguish artefacts from real changes in marker order.
BACs WAG42G5 and WAG9L1 mapped to GGA2q and APL2p, providing clear evidence of a pericentric inversion. However, BAC WAG18G1, which mapped close to the centromere on GGA2p, also hybridized close to the centromere in APL2p. This demonstrated that the inversion involves only a small fraction of 2p.
BACs WAG13I5, WAG23K3, WAG21J21 AND WAG22J17 clearly demonstrated a paracentric inversion on GGA4q and APL4 (example image of WAG13I5 in Figure 2C, D).
The morphological differences between GGA7 and APL7 were reflected in a change in marker order involving BACs WAG69P21, WAG59H11 and WAG21P13. However, like in the rearrangement on GGA1p and APL1p, marker order was not completely inverted, indicating that this rearrangement may be more complex than a simple pericentric inversion. Similarly, our FISH mapping results did not provide clear evidence for a pericentric inversion causing the morphological differences between GGA8 and APL8.
Marker order on GGAZ and APLZ chromosome was largely conserved, with the possible exception of a small inversion involving BACs WAG22P4 and WAG20L10. Thus, it seems that the morphological differences between the metacentric GGAZ and the subtelocentric APLZ are due to neocentromere formation rather than a pericentric inversion.
Dual-color FISH experiments with BACs mapping to either end of a microchromosome were used to check for conserved synteny among the microchromosomes. This demonstrated conserved synteny for APL9, 11, 14–16, 19, 21, 27–29. The BACs successfully hybridized to the microchromosomes are listed in Additional file 2; example results are shown in Figure 2E, F.
FISH mapping of BACs WAG112C24 and WAG23I06 facilitated the identification of the duck orthologue of GGA4p. Chromosome area measurements suggested that this chromosome should be numbered as APL10. There were no major differences in size order between the remaining microchromosomes and their chicken orthologues.
Copy number variation between chicken and duck
Hybridisation of genomic DNA from two female ducks to the Nimblegen chicken whole genome tiling path microarray revealed 32 CNVs, of which ten were seen in both individuals (CNVs marked with a in Table 1). Both gains and losses relative to chicken were seen (8 gains and 24 losses). The mean and median lengths of the detected CNVs were 281 kb and 50 kb respectively, ranging from 2.8 kb to 4.4 Mb. The CNV locations were compared with those previously found in turkey and chicken broilers and layers[2]. Six CNVs overlapped with CNVs identified in turkey, and of these five CNVs matched the five potential CNV 'hotspots' described by Griffin et al. [2] (bolded in Table 1). Three of the 'hotspot' CNVs were seen in both ducks. Known or predicted genes were found to be associated with 22 of the 32 CNVs (68.75%). In the ten cases where no genes were associated, all were losses, and all except one were found either near to the beginning or end of the chromosome (between 2 bp and 4 Mb of the sequence start or end according to Ensembl, http://www.ensembl.org/Gallus_gallus). The exception, CNV#14, covered a sequence gap, which likely contains centromeric repeats. Two of the CNVs, #7 and #9, were in regions that are potentially involved in rearrangements on GGA7 and 8 respectively.
Table 1.
CNVs detected in duck (Anas platyrhynchos) relative to chicken reference DNA.
| CNV | Chicken chromosome | Start (bp) | Stop (bp) | Size (kb) | Fold change c.f. Red Junglefowl | Comments |
|---|---|---|---|---|---|---|
| 1ab | 1 | 140580435 | 140655038 | 74.603 | -4.23 | Novel genes |
| 2 | 3 | 107012500 | 108262500 | 1250 | -1.43 | Mitochondrial oxidoreductase (β-oxidation cycle) |
| 3ab | 3 | 113237500 | 113651334 | 413.834 | -1.73 | 39S ribosomal protein L19 (ribosomal subunit) |
| 4 | 3 | 113605240 | 113652668 | 47.428 | -3.88 | 39S ribosomal protein L19 (ribosomal subunit) |
| 5b | 4 | 88710224 | 89072982 | 362.758 | -1.61 | Histone demethylation protein |
| 6 | 6 | 37387597 | 37400184 | 12.587 | -1.86 | |
| 7 | 7 | 352 | 5048 | 4.696 | -2.66 | |
| 8 | 7 | 38350336 | 38380092 | 29.756 | -2.85 | N-acetyltransferase 5 (acetylation of amino-terminal methionine residues) |
| 9a | 8 | 102 | 35016 | 34.914 | -3.54 | |
| 10 | 10 | 252 | 27706 | 27.454 | -1.54 | Novel genes |
| 11 | 10 | 22517883 | 22537510 | 19.627 | -2.26 | |
| 12a | 11 | 202 | 15241 | 15.039 | -8.50 | |
| 13 | 11 | 12500 | 62500 | 50 | -1.41 | Novel genes |
| 14a | 11 | 2675046 | 3190227 | 515.181 | -2.76 | Centromere |
| 15a | 12 | 12500 | 62500 | 50 | -2.86 | Senescent cell antigen-like-containing domain protein (Plays a role in modulating cell spreading and migration) |
| 16 | 13 | 4932529 | 4935329 | 2.8 | 3.75 | Teneurin 2 (transcription factor activity) |
| 17b | 13 | 18865302 | 18907544 | 42.242 | -2.28 | |
| 18 | 15 | 10240354 | 10295196 | 54.842 | 2.29 | Novel genes |
| 19b | 16 | 352 | 432851 | 432.499 | -1.43 | Covers all GGA16 sequence available; MHC |
| 20 | 16 | 262500 | 287500 | 25 | 1.50 | Predicted similar to MHC Rfp-Y class I alpha chain |
| 21a | 17 | 802976 | 867945 | 64.969 | 1.99 | Neural proliferation differentiation and control protein |
| 22 | 18 | 1812500 | 1837500 | 25 | -1.46 | Novel genes |
| 23 | 19 | 3665182 | 3845095 | 179.913 | 1.48 | Myosin regulatory light chain, cardiac muscle isoform |
| 24 | 22 | 3912586 | 3935246 | 22.66 | 1.43 | Similar to 2-oxoglutarate dehydrogenase (TCA cycle) |
| 25 | 26 | 481 | 5060 | 4.579 | -1.92 | |
| 26 | 26 | 4577801 | 4750398 | 172.597 | 1.62 | Forkhead box protein; MyoD family inhibitor (transcriptional regulators) |
| 27a | 26 | 2911 | 837508 | 834.597 | -1.79 | Novel proteins |
| 28a | 28 | 2 | 12526 | 12.524 | -7.87 | |
| 29 | 28 | 12500 | 62500 | 50 | -2.48 | Major vault protein (ribonucleoprotein complex) |
| 30 | E64 | 402 | 45047 | 44.645 | -1.43 | Predicted similar to ligase (DNA replication) |
| 31 | Z | 2090459 | 6530135 | 4439.676 | 1.59 | Synaptotagmin-4 (neurotransmitter secretion) |
| 32ab | Z | 71512500 | 71762500 | 250 | -5.09 | Novel genes |
Discussion
Comparative cytogenetic map of the duck genome
Previous studies of avian genome evolution using cross-species FISH have suggested that gross genome structure is remarkably conserved among birds. Duck is no exception from this pattern; in fact it appears that the duck karyotype corresponds very closely to the putative ancestral avian karyotype [6]. The conservation of ancestral chromosomes 4 and 10, as APL4 and APL10 are consistent with both the previous studies on duck and with broader patterns of avian genome evolution[6]. These chromosomes appear intact in almost all birds, and ancestral chromosome 4 is seen intact in human chromosome 4 as well[6,13]. Our finding that only 39% of the BACs provided identifiable signals is attributable to the evolutionary distance between chicken and duck (90 million years); we achieved a 70% success rate mapping chicken BACs in turkey [2] (which diverged from chicken 30 million years ago), and our experience suggests that success rates of cross-species FISH with BACs rapidly decrease with increased evolutionary distance. This is also reflected by increased levels of non-specific background hybridisation from repetitive content, especially on the macrochromosomes, accounting for our wider error bars when assigning positions to BACs on these chromosomes.
In agreement with previous studies, no other interchromosomal rearrangements were detected between chicken and duck. However, our BAC mapping data are consistent with intrachromosomal rearrangements distinguishing chromosomes GGA1, 2, 4, 7, 8, Z and APL1, 2, 4, 7, 8 and Z, which confirms and expands on previous findings[13]. The detection of additional rearrangements on GGA1 and APL1 and GGA4q and APL4 was due to the much higher number of BACs hybridized in this study compared to previous studies. Likewise, higher density mapping demonstrated that the morphological differences between GGAZ and APLZ are probably due to the formation of a neocentromere rather than a pericentric inversion. This type of chromosomal rearrangement was previously reported in birds only for the red-legged partridge [23] and the Japanese quail[24]. However, despite the good coverage of the duck cytogenetic map presented here, it was not possible to determine unequivocally the nature of all chromosomal rearrangements observed between chicken and duck. It seems likely however that, in addition to peri- and paracentric inversions and neocentromere formation, translocations contributed to avian genome evolution. This conclusion is based on the order of BACs associated with rearrangements on GGA1, 7 and 8 and APL1, 7 and 8, which is not entirely consistent with the order expected if the rearrangements were inversions. Thus, it appears that while the available data from comparative FISH mapping suggest a relatively low frequency of intrachromosomal rearrangements in the evolution of bird genomes, the underlying processes may be more diverse than previously appreciated. Undoubtedly, the higher resolution afforded by genome sequencing projects such as that of the zebra finch will help to resolve this question.
The evolutionary direction of intrachromosomal changes could be determined for GGA8 and APL8 only. Comparison with the turkey map [2] suggested that APL8 likely represents the ancestral state; the order of BACs on turkey chromosome 10 (ancestral chromosome 8) and the morphology of this chromosome is the same as in duck, indicating that the rearrangement has occurred in the chicken lineage. Due to a lack of comparative data, it was not possible to determine the polarity of the remaining intrachromosomal rearrangements. If BAC positions are compared between this map and the turkey map (Additional file 1, [2]), the differences fit the patterns expected from the previously described inter- and intra-chromosomal rearrangements.
Among the rearrangements that we detected, the inversion observed in GGA4q and APL4 is of particular interest. Morphological differences in GGA4 have been described between different chicken breeds[25], and the ancestral bird chromosome 4 (corresponding to GGA4q and APL4, respectively) is also one of the chromosomes most prone to convergent independent fusions in birds (with ancestral chromosome 10) [6]. This contrasts with the conserved synteny of the ancestral bird chromosome 4 (corresponding to GGA4q and APL4, respectively) in humans. Together, the data suggest that rearrangements in chromosome 4 may be more common than has been suspected from previous comparative genomic studies, and analyzing them will prove valuable for understanding avian and other vertebrate genome evolution.
The present study extended the data previously available for conserved synteny among the microchromosomes. Fillon et al. [13]demonstrated conserved synteny for seven microchromosome pairs (APL9, 12, 14–16, 19, 29); here we demonstrate conserved synteny for ten microchromosome pairs (APL9, 11, 14–16, 19, 21, 27–29). The lack of detected rearrangements makes it reasonable to suggest that there are very few, if any, rearrangements among the remaining microchromosomes – including the as-yet unexamined smallest microchromosomes for which no markers exist. Indeed, no sequence data from the chicken genome has yet been assigned to these smallest chromosomes, and it is still unclear why, although there is a suggestion that there may be a cloning or sequencing bias against microchromosomal sequences[26]. Data on conserved microchromosomal synteny in other bird species are restricted to the Japanese quail (Coturnix japonica[27]) and the turkey[2]. Despite the paucity of data, the emerging picture is one of remarkable conservation among most avian species, with the exception of a few groups where large-scale interchromosomal rearrangements are common (such as the Falconiformes or Psittaciformes[28,29]).
The detailed cytogenetic map allowed for an improved definition of the duck karyotype. Chromosome banding and macrochromosome painting studies had previously shown orthology of APL1–9 and Z to GGA1–3, 4q, 5–9 and Z [10-12]. However, it was not known which duck chromosome corresponded to chicken chromosome 4p; Fillon et al. [13]suggested that this was approximately APL10–13. In this study we have used a combination of BAC mapping and chromosome area measurements which suggest that this chromosome should be numbered as APL10. Moreover we found no evidence for rearrangements among the microchromosomes. Accordingly, we propose that duck chromosomes be numbered as per chicken for 1–9; that APL10 corresponds to GGA4p; then GGA10 onwards correspond to APL11 onwards. The successful hybridization of at least one BAC from GGA1–28 (except 25) means markers are now available for APL1–29 (except 26) and Z. Taken together, these results enabled us to define unequivocally chromosomes APL1–29.
Copy number variation between chicken and duck
The purpose of our array CGH experiments was to test two of our earlier hypotheses: (i) that birds show a reduced number of CNVs compared to mammals, (ii) that genomic CNV hotspots described previously in chicken and turkey [2] are found in the duck as well (indicating conservation over a large evolutionary distance). The successful hybridizations that we observed, despite approximately 90 million years of divergence between chicken and duck[31], extend avian cross-species microarray experiments from the sole previous study in turkey[2]. The present study revealed a total of 32 CNVs in the duck when compared to the chicken, which is substantially fewer than the 58 CNVs discovered in a comparison of human and chimpanzee[31], which diverged only 6–7 million years ago[32]. While this result supports the hypothesis that bird genomes show fewer CNVs than mammalian ones, it should also be noted that we found twice as many CNVs in duck as were found in the comparison of turkey and chicken (32 versus 16) [2]. Only ten of these CNVs were found in both duck specimens examined, indicating substantial intraspecific variation. These findings highlight the need for further studies with larger sample sizes and call for some caution when comparing the frequency of CNVs in birds and mammals.
The comparison of CNVs between chicken breeds and turkey revealed five tentative CNV hotspots by virtue of the fact that they contained CNVs of similar size in different chicken lines and in turkey[2]. Of these five hotspot regions, three contained CNVs in both ducks and two contained CNVs in one duck. The hotspot regions contain a number of novel genes including most of the available sequence for GGA16, covering the MHC loci. Moreover these findings lend support to our avian "CNV hotspot hypothesis" but, of course, need to be confirmed by analysing a wider number of species.
All copy number gains (in duck compared to chicken) were located in coding regions. Genes in regions of copy number gain relative to chicken included transcription factors, neural proliferation control and neurotransmitter activity, and a predicted MHC class I gene. This is consistent with previously described duplication of the MHC class I locus in the mallard duck, followed by subsequent inactivation of some of the extra gene copies[33].
Where copy number losses relative to chicken were detected, two possible explanations exist: a true copy number loss, or sequence divergence leading to lack of hybridization on the microarray. If such sequence divergence had occurred, however, the loss might be expected in both individuals. This was only seen for 9 CNVs; the remainder are more likely to be true polymorphic copy number differences. However, it is important to note that about two thirds of all apparent copy number losses were observed in coding regions; hence, the observed loss in hybridization efficiency is likely associated with functional consequences, regardless of whether it is due to copy number change or sequence divergence. Thus these results highlight genomic regions that are of particular interest for further functional and evolutionary studies.
It has been suggested that segmental duplications are correlated with CNVs, and facilitate chromosomal rearrangements, the lack of segmental duplications in birds therefore explaining the relative paucity of CNVs[1]. Due to the vastly different levels of resolution afforded by cytogenetic mapping and microarray analysis, it is difficult to directly correlate the results of these two methods. Nevertheless, it is interesting to note that two of the CNVs revealed in this paper appear to coincide with rearrangements detected from the cytogenetic mapping. These are CNVs #7 and #9, on chromosomes GGA7 and 8 respectively. Further studies are necessary to examine this link between chromosomal rearrangements and CNVs in more detail.
Conclusion
The comparative cytogenetic map of the duck presented here highlights the extraordinary conservation seen among the genomes of many bird species, and how little structural genetic variation is readily apparent. The cytogenetic map will allow the transfer of genetic information directly from chicken to duck, expediting mapping studies in the duck and help to target marker development in duck through the prediction of new loci. The combination of area measurements and FISH mapping of chicken BACs allowed the identification of markers for chromosomes APL1–24 and 26–29 which will facilitate further mapping studies in the duck. Moreover, we have extended the analysis of CNVs in birds, providing further evidence that birds have low numbers of CNVs when compared to mammals and that bird genomes contain CNV hotspots. While overall we confirm the evolutionary conservation of bird genomes, the intrachromosomal differences and CNVs found highlight areas of future interest for evolutionary and functional studies.
Methods
Cell culture and chromosome preparation
All chromosome preparations were made from cultured cells derived from fertilized eggs. Chicken eggs were supplied by Hill Top Farm, Cambridgeshire, UK and Friday's Farm, Kent, UK. Duck eggs were provided by Cherry Valley Ltd, Market Rasen, UK. Fibroblast cultures were established from 5- to 7-day-old embryos. Chromosome preparation followed standard protocols[34,35]: mitostatic treatment with colcemid at a final concentration of 0.1 μg/ml for 1 h at 37°C, hypotonic treatment with 75 mM KCl for 15 min at 37°C and fixation with 3:1 methanol:acetic acid.
Selection and preparation of BAC clones
400 BAC clones were selected from the Wageningen chicken BAC library [36] based on the position of markers on the chicken consensus linkage map[37]. The BACs were labeled by nick translation with biotin-16-dUTP or digoxigenin-11-dUTP (Roche) following standard protocols. At least one BAC was available for GGA1–28 (except 25) and Z. The 155 BACs successfully hybridized to both chicken and duck are detailed in Additional files 1 and 2.
Fluorescent in-situ hybridisation (FISH)
Slides with metaphase preparations were aged for one hour at 70°C on a hotplate then treated with 4 mg/ml RNase A for one hour at 37°C. The chromosomes were denatured for 1 minute 30 seconds in 70% formamide in 2×SSC at 70°C. BACs were applied to slides and sealed under coverslips with rubber cement. Hybridization was carried out in a humidified chamber for 72 hours at 37°C. Following post-hybridization washes (40% or 30% formamide in 2×SSC for 20 minutes; 1 minute in 2×SSC/0.1% Igepal at RT; 15 minutes in 4×SSC/0.05% Tween 20 at RT; 25 minutes in 4×SSC/0.05% Tween 20/2% BSA at RT), probes were detected with 1:200 streptavidin-Cy3 (Amersham), plus 1:200 FITC-anti-digoxigenin (Amersham) for dual-color experiments, in 4×SSC, 0.05% Igepal, 1.25% BSA for 35 minutes. Slides were washed in 4×SSC, 0.05% Igepal for 3 × 5 minutes then counterstained using Vectashield with DAPI (Vector Labs).
Dual-color FISH was used to determine whether there was conserved synteny among the microchromosomes, selecting BACs that were as close as possible to the ends of the chromosomes. For GGA11, only one hybridizing BAC in duck was available.
Image capturing and analysis
Slides were analyzed on an Olympus BX-61 epifluorescence microscope equipped with a cooled CCD camera and appropriate filters. Images were captured using SmartCapture 3 (Digital Scientific UK). The signal positions were measured as the fractional length from the p-terminus (FLpter[38]). FLpter and area measurements were carried out in ImageJ[39]. The area of duck chromosomes was determined as a ratio of the easily recognized chromosome 5, as per[40]. For area measurements, 10 metaphases were measured per chromosome; the numbers of metaphases used for FLpter measurements are given in Additional file 1.
Array CGH
The NimbleGen chicken whole-genome tiling array (Catalogue Number/Design Name B3791001-00-01, galGal3 WG CGH – Roche NimbleGen, Milton Keynes, UK) was used for the array CGH experiments. It contains 385,000 50-mer oligonucleotides with an average spacing of 2,586 base pairs (source – UCSC, build – galGal3) and was interrogated with duck whole genomic DNA extracted from blood from two female ducks using a DNeasy Animal Blood and Tissue kit (Qiagen, #69504); the reference (Red Jungle Fowl) DNA, from the same animal used in the chicken genome sequencing project, was kindly provided by Dr Hans Cheng (Michigan State University). Labeling of genomic DNA and hybridization to the Roche NimbleGen array were performed by the company (Roche NimbleGen) and used random priming to incorporate modified nucleotides by either amino-allyl or direct linkage to either of the two dyes used (Cy3 and Cy5). All of the hybridizations in this experiment used two dyes per slide (Cy3 and Cy5). Red Jungle Fowl reference DNA (Cy5) was co-hybridized with duck test DNA (Cy3).
CGH analysis proceeded in three stages, normalization, window averaging and segmentation. After combining the signal intensity and genomic coordinate information, the Cy3 and Cy5 signal intensities were normalized to one another using Qspline normalization[41]. Qspline is a robust non-linear method for normalization using array signal distribution analysis and cubic splines. Once normalized, the data was prepared for DNA segmentation analysis. This included a window averaging step, where the probes that fall into a defined base pair window size (25 kb) are averaged, using the Tukey's biweight mean[42]. The Tukey's biweight method yields a robust weighted mean that is relatively insensitive to outliers, even when extreme. A new position was assigned to this average, which is the midpoint of the window. Segmentation was also performed on unaveraged data to permit smaller segments than the window size to be detected. The circular binary segmentation algorithm [43] was used to segment the averaged log2 ratio data. DNA segments were called by attempting to break the segments into sub-segments by looking at the t-statistic of the means. Permutations (n = 1000) were used to provide the reference distribution. If the resulting p-value was below the threshold (default of p = 0.01), then a breakpoint was called. A pruning step was used to remove spurious segments, rejecting segments where the standard deviation of the means was not sufficiently different. By default, a cut off of 1.5 standard deviations was used. CNVs were called for segments in which the log2 ratio was greater than | ± 0.5|. Where overlapping CNVs were detected in window averaged and unaveraged data, the data were considered to represent a single CNV.
Authors' contributions
BMS performed probe preparation, hybridisations, microscopy and data analysis and prepared the manuscript. LBWR, HGT, EJL, DI and KEF assisted with probe preparation and microscopy. RPMAC provided the BAC clones. ADH and DKG conceived and designed the study. DKG was the PI on the BBSRC grant that funded the project and supervisor of BMS, LBWR, HGT, EJL, DI, KEF and MV. DKG critically revised the manuscript. MV performed hybridisations, data analysis and prepared the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Chicken (Gallus gallus; GGA) BACs successfully hybridized to duck (Anas platyrhynchos; APL) macrochromosomes. BACs with these markers successfully hybridized to duck chromosomes. FLpter represents Fractional Length from the p terminus[38]; SD represents standard deviation. For reference, our previous data on turkey (Meleagris gallopavo, MGA) [2] is also included.
BACs on duck (Anas platyrhynchos; APL) microchromosomes. BACs with the named markers successfully hybridized to duck chromosomes. Duck chromosome number after APL10 was assigned as chicken (GGA) chromosome number plus one. The orthologous turkey (Meleagris gallopavo, MGA) chromosome (from [2]) is also listed.
Contributor Information
Benjamin M Skinner, Email: bs74205@gmail.com.
Lindsay BW Robertson, Email: Lindsay.Robertson@icr.ac.uk.
Helen G Tempest, Email: h.tempest@thebridgecentre.co.uk.
Elizabeth J Langley, Email: E.J.Langley@kent.ac.uk.
Dimitris Ioannou, Email: di9@kent.ac.uk.
Katie E Fowler, Email: kef20@kent.ac.uk.
Richard PMA Crooijmans, Email: Richard.Crooijmans@wur.nl.
Anthony D Hall, Email: D.K.Griffin@kent.ac.uk.
Darren K Griffin, Email: D.K.Griffin@kent.ac.uk.
Martin Völker, Email: mv53121@gmail.com.
Acknowledgements
The authors would like to thank Hans Cheng (MSU) for the Red Junglefowl DNA, and the BBSRC and Genesis-Faraday for the project grant which funded this study.
References
- Hillier LW, Miller W, Birney E, Warren W, Hardison RC, Ponting CP, Bork P, Burt DW, Groenen MA, Delany ME, Dodgson JB, Chinwalla AT, Cliften PF, Clifton SW, Delehaunty KD, Fronick C, Fulton RS, Graves TA, Kremitzki C, Layman D, Magrini V, McPherson JD, Miner TL, Minx P, Nash WE, Nhan MN, Nelson JO, Oddy LG, Pohl CS, Randall-Maher J, Smith SM, Wallis JW, Yang SP, Romanov MN, Rondelli CM, Paton B, Smith J, Morrice D, Daniels L, Tempest HG, Robertson L, Masabanda JS, Griffin DK, Vignal A, Fillon V, Jacobbson L, Kerje S, Andersson L, Crooijmans RP, Aerts J, Poel JJ van der, Ellegren H, Caldwell RB, Hubbard SJ, Grafham DV, Kierzek AM, McLaren SR, Overton IM, Arakawa H, Beattie KJ, Bezzubov Y, Boardman PE, Bonfield JK, Croning MD, Davies RM, Francis MD, Humphray SJ, Scott CE, Taylor RG, Tickle C, Brown WR, Rogers J, Buerstedde JM, Wilson SA, Stubbs L, Ovcharenko I, Gordon L, Lucas S, Miller MM, Inoko H, Shiina T, Kaufman J, Salomonsen J, Skjoedt K, Wong GK, Wang J, Liu B, Yu J, Yang H, Nefedov M, Koriabine M, Dejong PJ, Goodstadt L, Webber C, Dickens NJ, Letunic I, Suyama M, Torrents D, von Mering C, Zdobnov EM. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432(7018):695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- Griffin DK, Robertson LB, Tempest HG, Vignal A, Fillon V, Crooijmans RP, Groenen MA, Deryusheva S, Gaginskaya E, Carre W, Waddington D, Talbot R, Volker M, Masabanda JS, Burt DW. Whole genome comparative studies between chicken and turkey and their implications for avian genome evolution. BMC Genomics. 2008;9:168. doi: 10.1186/1471-2164-9-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira SL, Baker AJ. A molecular timescale for galliform birds accounting for uncertainty in time estimates and heterogeneity of rates of DNA substitutions across lineages and sites. Mol Phylogenet Evol. 2006;38(2):499–509. doi: 10.1016/j.ympev.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Feuk L, Marshall CR, Wintle RF, Scherer SW. Structural variants: changing the landscape of chromosomes and design of disease studies. Human Molecular Genetics. 2006;15(Spec No 1):R57–66. doi: 10.1093/hmg/ddl057. [DOI] [PubMed] [Google Scholar]
- Freeman JL, Perry GH, Feuk L, Redon R, McCarroll SA, Altshuler DM, Aburatani H, Jones KW, Tyler-Smith C, Hurles ME, Carter NP, Scherer SW, Lee C. Copy number variation: new insights in genome diversity. Genome Res. 2006;16(8):949–961. doi: 10.1101/gr.3677206. [DOI] [PubMed] [Google Scholar]
- Griffin DK, Robertson LB, Tempest HG, Skinner BM. The evolution of the avian genome as revealed by comparative molecular cytogenetics. Cytogenet Genome Res. 2007;117(1–4):64–77. doi: 10.1159/000103166. [DOI] [PubMed] [Google Scholar]
- Food and Agriculture Organisation of the United Nations. http://faostat.fao.org/
- Munster VJ, Veen J, Olsen B, Vogel R, Osterhaus AD, Fouchier RA. Towards improved influenza A virus surveillance in migrating birds. Vaccine. 2006;24(44–46):6729–6733. doi: 10.1016/j.vaccine.2006.05.060. [DOI] [PubMed] [Google Scholar]
- Huang Y, Zhao Y, Haley CS, Hu S, Hao J, Wu C, Li N. A Genetic and Cytogenetic Map for the Duck (Anas platyrhynchos) Genetics. 2006;173(1):287–296. doi: 10.1534/genetics.105.053256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denjean B, Ducos A, Darre A, Pinton A, Seguela A, Berland H, Blanc M, Fillon V, Darre R. Caryotype des canards commun (Anas platyrhynchos), Barbarie (Cairina moschata) et de leur hybride. Revue Med Vet. 1997;148(8–9):695–704. [Google Scholar]
- Schmid M, Nanda I, Guttenbach M, Steinlein C, Hoehn M, Schartl M, Haaf T, Weigend S, Fries R, Buerstedde JM, Wimmers K, Burt DW, Smith J, A'Hara S, Law A, Griffin DK, Bumstead N, Kaufman J, Thomson PA, Burke T, Groenen MA, Crooijmans RP, Vignal A, Fillon V, Morisson M, Pitel F, Tixier-Boichard M, Ladjali-Mohammedi K, Hillel J, Maki-Tanila A, Cheng HH, Delany ME, Burnside J, Mizuno S. First report on chicken genes and chromosomes 2000. Cytogenet Cell Genet. 2000;90(3–4):169–218. doi: 10.1159/000056772. [DOI] [PubMed] [Google Scholar]
- Schmid M, Nanda I, Hoehn H, Schartl M, Haaf T, Buerstedde JM, Arakawa H, Caldwell RB, Weigend S, Burt DW, Smith J, Griffin DK, Masabanda JS, Groenen MA, Crooijmans RP, Vignal A, Fillon V, Morisson M, Pitel F, Vignoles M, Garrigues A, Gellin J, Rodionov AV, Galkina SA, Lukina NA, Ben-Ari G, Blum S, Hillel J, Twito T, Lavi U, David L, Feldman MW, Delany ME, Conley CA, Fowler VM, Hedges SB, Godbout R, Katyal S, Smith C, Hudson Q, Sinclair A, Mizuno S. Second report on chicken genes and chromosomes 2005. Cytogenet Genome Res. 2005;109(4):415–479. doi: 10.1159/000084205. [DOI] [PubMed] [Google Scholar]
- Fillon V, Vignoles M, Crooijmans RP, Groenen MA, Zoorob R, Vignal A. FISH mapping of 57 BAC clones reveals strong conservation of synteny between Galliformes and Anseriformes. Anim Genet. 2007;38(3):303–307. doi: 10.1111/j.1365-2052.2007.01578.x. [DOI] [PubMed] [Google Scholar]
- Emanuel BS, Saitta SC. From microscopes to microarrays: dissecting recurrent chromosomal rearrangements. Nat Rev Genet. 2007;8(11):869–883. doi: 10.1038/nrg2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JA, Eichler EE. Primate segmental duplications: crucibles of evolution, diversity and disease. Nat Rev Genet. 2006;7(7):552–564. doi: 10.1038/nrg1895. [DOI] [PubMed] [Google Scholar]
- Dumas L, Kim YH, Karimpour-Fard A, Cox M, Hopkins J, Pollack JR, Sikela JM. Gene copy number variation spanning 60 million years of human and primate evolution. Genome Res. 2007;17(9):1266–1277. doi: 10.1101/gr.6557307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortna A, Kim Y, MacLaren E, Marshall K, Hahn G, Meltesen L, Brenton M, Hink R, Burgers S, Hernandez-Boussard T. et al. Lineage-specific gene duplication and loss in human and great ape evolution. PLoS Biol. 2004;2(7):E207. doi: 10.1371/journal.pbio.0020207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke DP, Segraves R, Carbone L, Archidiacono N, Albertson DG, Pinkel D, Eichler EE. Large-scale variation among human and great ape genomes determined by array comparative genomic hybridization. Genome Res. 2003;13(3):347–357. doi: 10.1101/gr.1003303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman TL, Tuzun E, Morrison VA, Hayden KE, Ventura M, McGrath SD, Rocchi M, Eichler EE. A genome-wide survey of structural variation between human and chimpanzee. Genome Research. 2005;15(10):1344–1356. doi: 10.1101/gr.4338005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samonte RV, Eichler EE. Segmental duplications and the evolution of the primate genome. Nat Rev Genet. 2002;3(1):65–72. doi: 10.1038/nrg705. [DOI] [PubMed] [Google Scholar]
- Ladjali-Mohammedi K, Bitgood JJ, Tixier-Boichard M, Ponce De Leon FA. International system for standardized avian karyotypes (ISSAK): standardized banded karyotypes of the domestic fowl (Gallus domesticus) Cytogenet Cell Genet. 1999;86(3–4):271–276. doi: 10.1159/000015318. [DOI] [PubMed] [Google Scholar]
- Chowdhary BP, Raudsepp T. HSA4 and GGA4: remarkable conservation despite 300-Myr divergence. Genomics. 2000;64(1):102–105. doi: 10.1006/geno.1999.6085. [DOI] [PubMed] [Google Scholar]
- Kasai F, Garcia C, Arruga MV, Ferguson-Smith MA. Chromosome homology between chicken (Gallus gallus domesticus) and the red-legged partridge (Alectoris rufa); evidence of the occurrence of a neocentromere during evolution. Cytogenet Genome Res. 2003;102(1–4):326–330. doi: 10.1159/000075770. [DOI] [PubMed] [Google Scholar]
- Galkina S, Deryusheva S, Fillon V, Vignal A, Crooijmans R, Groenen M, Rodionov A, Gaginskaya E. FISH on avian lampbrush chromosomes produces higher resolution gene mapping. Genetica. 2006;128(1–3):241–251. doi: 10.1007/s10709-005-5776-7. [DOI] [PubMed] [Google Scholar]
- Musa HH, Li BC, Chen GH, Lanyasunya TP, Xu Q, Bao WB. Karyotype and Banding Patterns of Chicken Breeds. International Journal of Poultry Science. 2005;4(10):741–744. doi: 10.3923/ijps.2005.741.744. [DOI] [Google Scholar]
- Douaud M, Feve K, Gerus M, Fillon V, Bardes S, Gourichon D, Dawson DA, Hanotte O, Burke T, Vignoles F. et al. Addition of the microchromosome GGA25 to the chicken genome sequence assembly through radiation hybrid and genetic mapping. BMC Genomics. 2008;9:129. doi: 10.1186/1471-2164-9-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayang BB, Fillon V, Inoue-Murayama M, Miwa M, Leroux S, Feve K, Monvoisin JL, Pitel F, Vignoles M, Mouilhayrat C, Beaumont C, Ito S, Minvielle F, Vignal A. Integrated maps in quail (Coturnix japonica) confirm the high degree of synteny conservation with chicken (Gallus gallus) despite 35 million years of divergence. BMC Genomics. 2006;7(1):101. doi: 10.1186/1471-2164-7-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bed'Hom B, Coullin P, Guillier-Gencik Z, Moulin S, Bernheim A, Volobouev V. Characterization of the atypical karyotype of the black-winged kite Elanus caeruleus (Falconiformes: Accipitridae) by means of classical and molecular cytogenetic techniques. Chromosome Res. 2003;11(4):335–343. doi: 10.1023/A:1024091923939. [DOI] [PubMed] [Google Scholar]
- Nanda I, Karl E, Griffin DK, Schartl M, Schmid M. Chromosome repatterning in three representative parrots (Psittaciformes) inferred from comparative chromosome painting. Cytogenet Genome Res. 2007;117(1–4):43–53. doi: 10.1159/000103164. [DOI] [PubMed] [Google Scholar]
- van Tuinen M, Hedges SB. Calibration of avian molecular clocks. Mol Biol Evol. 2001;18(2):206–213. doi: 10.1093/oxfordjournals.molbev.a003794. [DOI] [PubMed] [Google Scholar]
- Perry GH, Tchinda J, McGrath SD, Zhang J, Picker SR, Caceres AM, Iafrate AJ, Tyler-Smith C, Scherer SW, Eichler EE, Stone AC, Lee C. Hotspots for copy number variation in chimpanzees and humans. Proc Natl Acad Sci USA. 2006;103(21):8006–8011. doi: 10.1073/pnas.0602318103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiper ME, Young NM. Primate molecular divergence dates. Mol Phylogenet Evol. 2006;41(2):384–394. doi: 10.1016/j.ympev.2006.05.021. [DOI] [PubMed] [Google Scholar]
- Moon DA, Veniamin SM, Parks-Dely JA, Magor KE. The MHC of the duck (Anas platyrhynchos) contains five differentially expressed class I genes. J Immunol. 2005;175(10):6702–6712. doi: 10.4049/jimmunol.175.10.6702. [DOI] [PubMed] [Google Scholar]
- Ahlroth MK, Kola EH, Ewald D, Masabanda J, Sazanov A, Fries R, Kulomaa MS. Characterization and chromosomal localization of the chicken avidin gene family. Anim Genet. 2000;31(6):367–375. doi: 10.1046/j.1365-2052.2000.00681.x. [DOI] [PubMed] [Google Scholar]
- Griffin DK, Haberman F, Masabanda J, O'Brien P, Bagga M, Sazanov A, Smith J, Burt DW, Ferguson-Smith M, Wienberg J. Micro- and macrochromosome paints generated by flow cytometry and microdissection: tools for mapping the chicken genome. Cytogenet Cell Genet. 1999;87(3–4):278–281. doi: 10.1159/000015449. [DOI] [PubMed] [Google Scholar]
- Crooijmans RP, Vrebalov J, Dijkhof RJ, Poel JJ van der, Groenen MA. Two-dimensional screening of the Wageningen chicken BAC library. Mamm Genome. 2000;11(5):360–363. doi: 10.1007/s003350010068. [DOI] [PubMed] [Google Scholar]
- Groenen MA, Cheng HH, Bumstead N, Benkel BF, Briles WE, Burke T, Burt DW, Crittenden LB, Dodgson J, Hillel J, Lamont S, de Leon AP, Soller M, Takahashi H, Vignal A. A consensus linkage map of the chicken genome. Genome Res. 2000;10(1):137–147. doi: 10.1101/gr.10.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichter P, Tang CJ, Call K, Hermanson G, Evans GA, Housman D, Ward DC. High-resolution mapping of human chromosome 11 by in situ hybridization with cosmid clones. Science. 1990;247(4938):64–69. doi: 10.1126/science.2294592. [DOI] [PubMed] [Google Scholar]
- Abramoff MD, Magelhaes PJ, Ram SJ. Image Processing with ImageJ. Biophotonics International. 2004;11(7):36–42. [Google Scholar]
- Morris WB, Stephenson JE, Robertson LB, Turner K, Brown H, Ioannou D, Tempest HG, Skinner BM, Griffin DK. Practicable approaches to facilitate rapid and accurate molecular cytogenetic mapping in birds and mammals. Cytogenet Genome Res. 2007;117(1–4):36–42. doi: 10.1159/000103163. [DOI] [PubMed] [Google Scholar]
- Workman C, Jensen LJ, Jarmer H, Berka R, Gautier L, Nielser HB, Saxild HH, Nielsen C, Brunak S, Knudsen S. A new non-linear normalization method for reducing variability in DNA microarray experiments. Genome Biol. 2002;3(9) doi: 10.1186/gb-2002-3-9-research0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tukey J. In: Contributions to Probability and Statistics. Olkin E, editor. Stanford University Press; 1960. A survey of sampling from contaminated distributions; pp. 448–485. [Google Scholar]
- Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5(4):557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Chicken (Gallus gallus; GGA) BACs successfully hybridized to duck (Anas platyrhynchos; APL) macrochromosomes. BACs with these markers successfully hybridized to duck chromosomes. FLpter represents Fractional Length from the p terminus[38]; SD represents standard deviation. For reference, our previous data on turkey (Meleagris gallopavo, MGA) [2] is also included.
BACs on duck (Anas platyrhynchos; APL) microchromosomes. BACs with the named markers successfully hybridized to duck chromosomes. Duck chromosome number after APL10 was assigned as chicken (GGA) chromosome number plus one. The orthologous turkey (Meleagris gallopavo, MGA) chromosome (from [2]) is also listed.