Abstract
Background
Versatile transgenic manipulation of skeletal muscle requires knowledge of the expression profiles of diverse promoter/enhancer elements in the transcriptionally specialized fiber types of which muscle is composed. "Universal" viral promoters/enhancers, e.g., cytomegalovirus IE1 (CMV IE1), are of interest as reagents that may drive broad expression. However, a previous study noted a marked heterogeneity of CMV IE1-driven transgene expression among muscle fibers, raising the possibility of fiber-type-restricted expression. The purpose of the present study was to characterize CMV IE1-driven expression in terms of fiber type.
Results
We produced two lines of transgenic mice carrying the CMV IE1/ β-galactosidase construct CMVLacZ, and analyzed transgene expression and fiber type by histochemical analysis of hindlimb muscle sections. In both lines CMVLacZ was expressed in all four major fiber types: type I (slow) and types IIA, IIB and IIX (fast). There was no unique pattern of fiber-type-preferential expression; fiber-type quantitative differences were observed but details varied between muscle regions and between lines. Both lines showed similar fiber-type-independent regional differences in overall expression levels, and a high level of within-fiber-type variability of expression, even among nearby fibers. The soleus muscle showed strong expression and comparatively little within-fiber-type or between-fiber-type variability.
Conclusions
The CMV IE1 promoter/enhancer is not fiber-type-restricted and can be useful for driving germ-line transgene expression in all four fiber types. However, not all fibers express the gene at high levels due in part to regional differences in overall expression levels, and to a high level of within-fiber-type variability. Given the multinucleate syncitial nature of muscle fibers, it is not likely that this variability is due to variegating heterochromatinization. The soleus muscle would make a suitable subject for near-uniform experimental gene expression driven by CMV IE1 elements.
Background
Versatile genetic manipulation of skeletal muscle through expression of experimental germ-line transgenes requires knowledge of the expression characteristics of diverse promoter/enhancer elements in the various skeletal muscle fiber types.
Rodent limb and axial muscles contain four major fiber types, one slow-contracting (type I) and three fast-contracting (types IIA, IIB, and IIX (also called IID)), that differ in activity patterns [1] and in contractile and metabolic properties [2,3,4,5]. Each of the four principal fiber types expresses a distinct myosin heavy chain gene that largely determines fast or slow contractile speed [6] and many other muscle protein gene families contain members that are differentially expressed in fast and slow fibers [2, 5]. Recent studies have begun to characterize the cis-regulatory elements and trans-acting factors that direct fiber-type-specific gene expression [7,8,9].
"Universal" viral promoters, such as the cytomegalovirus (CMV) IE1 promoter/enhancer [10, 11] are of interest in terms of their potential for broad, high-level expression. However the transcriptional specialization of the fiber types raises the possibility of fiber-type-restricted or differential expression in skeletal muscle. Several transgenic mouse studies have shown CMV IE1-driven expression in skeletal muscle as well as in other tissues [12,13,14,15], and Baskar et al [14], who studied expression at the cellular level, noted a marked differential expression among skeletal muscle fibers. However, the possible contribution of fiber-type heterogeneity to this differential expression has not been addressed. The purpose of the present study was to characterize CMV IE1-driven transgene expression in skeletal muscle in relation to fiber type.
Through production of transgenic mice carrying the CMV IE1/ β-galactosidase (β-gal) construct CMVLacZ, and histochemical characterization of transgene expression in hindlimb skeletal muscle, we show that CMV IE1-driven expression is not fiber-type-restricted. CMVLacZ is expressed effectively, though at varying mean levels, in all the major fiber types. However, its expression is highly variable within each of the fiber types in most muscles/regions, even among nearby fibers of the same type. We discuss the implications of our findings for the potential use of the CMV IE1 promoter/enhancer as a tool for driving experimental germ-line transgene expression in skeletal muscle. We also discuss fiber-to-fiber differences in physiological status as a possible cause of within-fiber-type expression variability, and note that variegating heterochromatinization [16, 17] would not be expected to generate expression variability in multinucleate syncitial cells such as skeletal muscle fibers.
Results
We produced two transgenic mouse lines (lines 9 and 18) carrying the construct CMVLacZ, in which β-gal expression is driven by the CMV IE1 promoter/enhancer. Expression features shared by both lines presumably reflect intrinsic capabilities of the transgene, whereas features unique to each line may reflect chromosomal insertion site-specific effects. Except where specifically indicated we restrict our comments to features common to both lines.
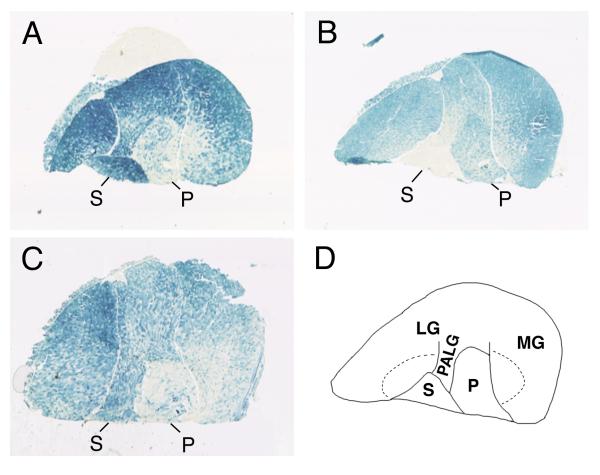
In agreement with previous studies [12,13,14,15] we found detectable β-gal expression in a variety of tissues in CMVLacZ transgenic mice, including skeletal muscle. As shown in Fig 1A, 1C, β-gal histochemistry on cryostat sections of hindlimb crural muscles revealed transgene expression in all muscles and muscle regions. Expression was strong in most regions of the gastrocnemius muscle, including the plantaris-adjacent lateral gastrocnemius (PALG), and also in the soleus muscle, but was weaker in the plantaris muscle and in the complex zone of the medial gastrocnemius (MG) muscle. This pattern does not correspond to the distribution of any particular fiber types.
Figure 1.
Expression patterns of CMVLacZ and TnILacZ1 transgenes in hindlimb crural muscles. Panels A-C show β-gal histochemistry of hindlimb muscle sections from adult transgenic mice of A) CMVLacZ line 18, B) TnILacZ1 line 29, C) CMVLacZ line 9. Panel D outlines muscles and muscle regions: S = soleus, P = plantaris, PALG = plantaris-adjacent lateral gastrocnemius, LG = lateral gastrocnemius, MG = medial gastrocnemius. The "complex zones" (regions containing all four fiber types) of the medial and lateral gastrocnemius are indicated by dashed lines. The superficial gastrocnemius includes LG and MG regions outside of the PALG and complex zones. The tissue block in panel A includes a piece of liver tissue (unstained) placed against the gastrocnemius muscle.
Figure 1B shows, for comparison, the expression pattern of a known fiber-type-specific transgene construct, TnILacZ1, in which β-gal expression is driven by elements from the fast fiber-specific troponin I (TnIfast) gene. The TnILacZ1 transgene is known to be expressed specifically in fast, but not in slow fibers, and among the fast fiber types in a IIB>IIX>IIA pattern [18]. Thus the pattern of TnILacZ1 transgene expression on whole muscle cross-sections largely reflects the distribution of IIB muscle fibers. This is shown in Fig 1B by 1) strong expression in the superficial regions of the gastrocnemius muscle, which consist principally of IIB fibers, 2) distinctly weaker expression in the plantaris and internal regions of the gastrocnemius including the PALG and the complex zones of the MG and lateral gastrocnemius (LG), which contain mixtures of IIB fibers among other fiber types, and 3) extremely weak expression in the soleus muscle, which consists mostly of type I (~ 50%) and type IIA fibers and contains no IIB fibers.
By contrast with TnILacZ1 (Fig 1B), the strong expression of CMVLacZ (Fig 1A,C) both in the soleus muscle, which is devoid of IIB fibers, and in the superficial gastrocnemius, which consists predominantly of IIB fibers, indicated that CMV IE1-driven expression was not principally associated with any one fiber type.
In most muscle regions histochemical analysis at the single-cell level showed considerable fiber-to-fiber variation of CMVLacZ expression as visualized by X-Gal staining (Fig 2A), in agreement with the observations of Baskar et al [14]. The same pattern of fiber-to-fiber staining differences was observed repeatedly on serial cross-sections, consistent with the data of Baskar et al [14] showing uniform staining intensities along the lengths of fibers caught in longitudinal section. Identification of fiber types, by histochemical and immunohistochemical analysis of myosin isoforms on serial sections, showed that the CMVLacZ transgene was expressed in each of the four fiber types, but at variable levels within each fiber type (see examples of within-fiber-type variability in Fig 2).
Figure 2.
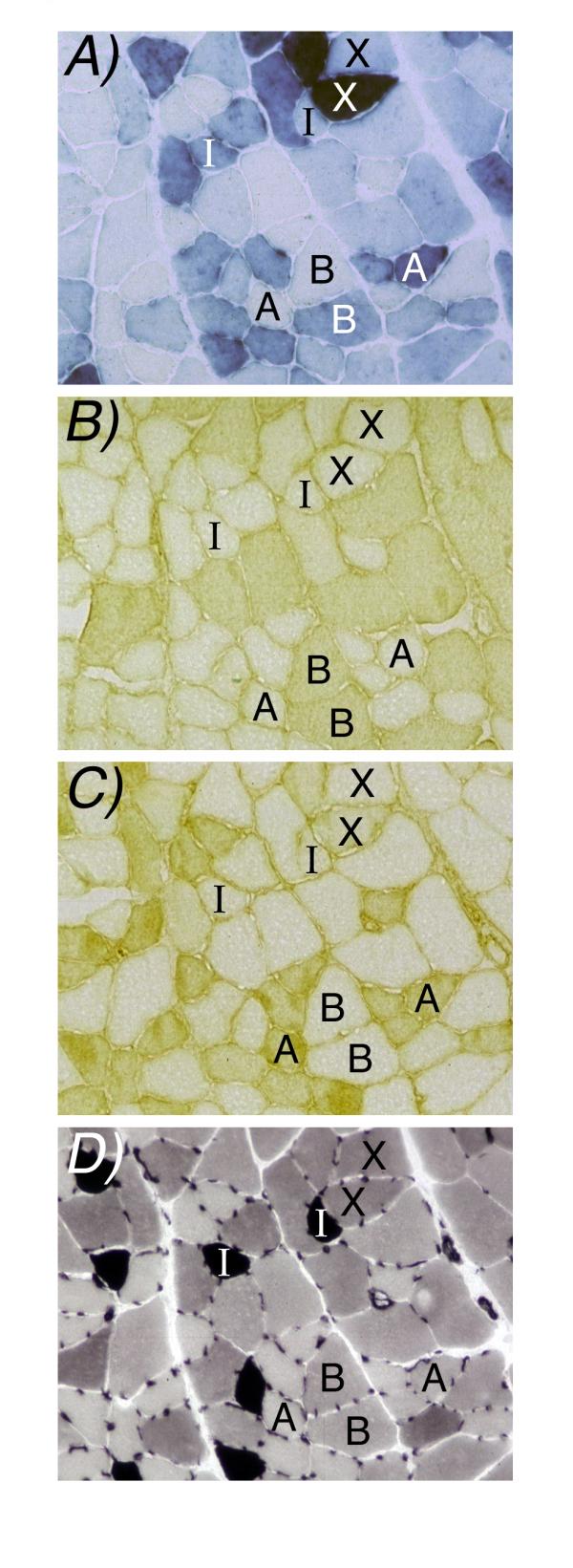
CMVLacZ transgene expression at the level of individual fibers. Panels A-D are near-serial sections of the medial gastrocnemius complex zone of an adult CMVLacZ (line 9) transgenic mouse. Panel A shows β-gal histochemistry and Panels B-D are treated to identify fiber types: Panel B, immunohistochemistry for IIB myosin heavy chain; Panel C immunohistochemistry for IIA myosin heavy chain; Panel D, ATPase histochemistry after preincubation at pH 4.6, with type I (slow) fibers showing strong stain, type IIA fibers showing weak stain, and type IIX and IIB fibers showing intermediate stain. Eight fibers are identified by fiber type where they appear in each panel; A=IIA, B=IIB, X=IIX. Note that nearby fibers of the same type can show very different β-gal expression levels.
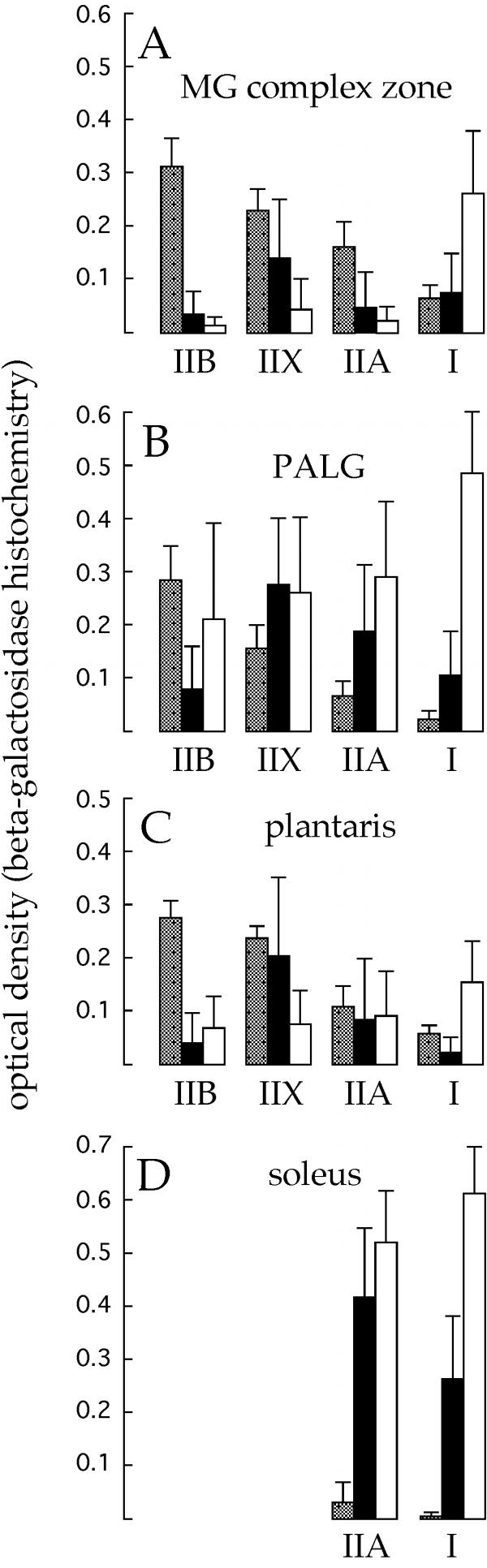
β-gal expression was measured quantitatively in individual typed fibers by microdensitometry of X-Gal-stained cross sections. These data are summarized for several muscle regions in Fig 3 for both CMVLacZ lines and, for comparison, TnILacZ1 transgenic mice. The quantitative data confirm CMVLacZ expression in all four fiber types and also show, by the relatively high standard deviations, that expression levels were highly variable within each fiber type, more so than was the case for TnILacZ1. Coefficients of variation for the CMVLacZ data shown in Fig 3 averaged 1.9- (line 18) to 2.4- (line 9) fold higher than for TnILacZ1. The smallest coefficients of variation for CMVLacZ expression were obtained for type I and type IIA fibers in the soleus muscle. There was also relatively little between-fiber-type difference in mean expression levels in the soleus, so that expression in this muscle was comparatively uniform.
Figure 3.
Quantitative β-gal histochemical analysis of CMVLacZ and TnILacZ1 expression patterns. The histograms show the optical densities of X-Gal histochemical reaction product measured in individual typed muscle fibers. Each panel corresponds to a different muscle/region. In each muscle or region ~ 70 fibers in a contiguous patch were measured. Three histogram bars are shown for each fiber type in each muscle region, representing CMVLacZ line 9 (black), CMVLacZ line 18 (white), and TnILacZ1 line 36 (gray). Error bars show standard deviations. CMVLacZ expression differences among the fiber types were statistically significant (P < 0.01; t test for soleus, ANOVA for other muscle regions) in every case except for CMVLacZ line 18 plantaris muscle.
Although there was active expression in all four fiber types, in most muscles/regions the between-fiber-type differences in CMVLacZ mean expression levels were statistically significant (see legend to Fig 3). However, due to regional and line-to-line variability these differences did not define a unique pattern of fiber-type-preferential expression. For example in both lines the type I:type IIA expression ratio was higher in the MG complex zone than in other muscle regions. Moreover, whereas in line 9 type IIX fibers had the highest mean expression levels in most muscle regions, in line 18 type I fibers did.
Quantitative analysis also revealed regional differences in CMVLacZ overall expression levels, independent of fiber type. The lower expression evident in the plantaris and MG complex zone in Fig 1 reflects lower expression in all fiber types in these regions (Fig 3, compare A,C with B,D).
Discussion
CMV IE1-driven transgene expression in skeletal muscle shows a marked fiber-to-fiber variability [14]. Because skeletal muscle fibers fall into several transcriptionally specialized fiber types, we assessed the possibility that this variable expression reflects fiber-type-specificity. We found that the CMV IE1-driven transgene CMVLacZ does not show marked fiber-type-specificity; it is expressed in all four major fiber types, including slow (type I), and fast (types IIA, IIB, and IIX) fibers. Moreover, although there were statistically significant differences in mean expression levels among the fiber types in most regions, there was no characteristic fiber type pattern of preferential expression shared by both transgenic lines examined. Between-line differences in the details of the fiber-type expression profile imply that although expression in all muscle fiber types is an intrinsic feature of the CMVLacZ construct, the precise quantitative balance of expression among the different fiber types can be influenced by chromosomal insertion site effects. Fiber-type mean expression level differences do contribute to the overall fiber-to-fiber heterogeneity observed, but they cannot account for a large component, i.e., the marked heterogeneity of expression within each fiber type, even among nearby fibers of the same type. Within-fiber-type variability was greater for CMVLacZ than for a contractile protein gene reporter construct TnILacZ1.
Variegating heterochromatinization (also known as position effect variegation) is a stochastic chromosomal process that has been postulated to account for cell-to-cell mosaicism in transgene expression in tissues composed of uninucleate cells [16, 17]. However, this mechanism would not be expected to generate variability in skeletal muscle because muscle fibers are multinucleate syncitia produced by the fusion of hundreds or thousands of individual uninucleate myoblasts, so that stochastic events operating at the level of individual nuclei would be expected to average out, giving uniform expression. Alternatively, the CMV IE1 promoter/enhancer may be responsive to cellular physiological status, which could differ even among nearby fibers of the same type, particularly those in different motor units. The presence of cis-elements for several highly-regulated trans-factors including CREB and NF-κB [13] in the CMV IE1 promoter/enhancer is consistent with the possibility of sensitive physiological regulation. Moreover, because all muscle fibers have a functional requirement for contractile protein gene expression, it might be expected that contractile protein gene constructs, such as TnILacZ1, would be relatively insensitive to physiologically regulated transcription factors whose activity may differ significantly among fibers of the same type. Further studies will be required to clarify the mechanism responsible for within-fiber-type variable expression of CMV IE1-driven transgenes.
The CMV IE1 promoter/enhancer has novel properties that may make it useful in the transgenic manipulation of skeletal muscle. To our knowledge this is the first report documenting expression of a germline transgene construct in all fiber types, including comparable expression in both fast and slow fibers. Quantitative studies of reporter gene expression that have employed direct fiber type analysis have generally revealed marked preferential expression of muscle transgenes in either slow or fast fibers [see e.g. 18, 19,20,21,22]. Immunofluorescence studies of dystrophin/utrophin transgene expression from mouse muscle creatine kinase [23,24,25] and human skeletal muscle actin [26,27,28] promoters have shown detectable expression in most or all fibers in muscle regions that might be expected to contain multiple fiber types. However in these studies fiber types were not determined, moreover immunofluorescence analysis of sarcolemma proteins may not reveal quantitative fiber-to-fiber differences in transgene expression. Indeed, other data suggest that human skeletal actin (29) and mouse muscle creatine kinase (30) promoters have a marked quantitative preference for fast glycolytic (type IIB) fibers, which are abundant in many mouse muscles. With its comparable expression in fast and slow fibers, and comparable expression in both fast glycolytic (IIB) and fast oxidative/glycolytic (IIA) fibers, CMV IE1 may provide a useful addition to the promoters used to drive experimental germ-line transgene expression in muscle. Of course, because it is also active in tissues other than muscle, the CMV IE1 promoter/enhancer would not be applicable in cases where expression of the gene product in nonmuscle cells would be deleterious. In addition, it is important to note that because of within-fiber-type variation, and regional/muscle differences in overall expression levels, not all fibers may express CMV IE1-driven transgenes at high levels. Among hindlimb muscles the soleus shows relatively little fiber-to-fiber variability of expression, and relatively little between-fiber-type difference, and hence would make a suitable subject muscle for near-uniform experimental gene expression driven by CMV IE1 elements.
Our characterization of the fiber type expression pattern of the CMV IE1 promoter/enhancer provides information useful for its assessment as a tool for experimental genetic modification of muscle through germ-line transgene expression. Similar study of additional "universal" promoters would be of interest and would improve our knowledge of the behavior of gene regulatory elements in skeletal muscle.
Conclusion
The CMV IE1 promoter/enhancer is not fiber-type-restricted and can be useful for driving germ-line transgene expression in all four major skeletal muscle fiber types. It is the first promoter for which germ-line transgene expression in all major fiber types, including comparable expression in slow and fast fibers, has been documented. However, because of regional effects on expression levels, and within-fiber-type variability, not all muscle fibers express the transgene at high levels. The cause of the marked within-fiber-type variability is unknown. It does not seem likely that variegating heterochromatinization would generate expression heterogeneity in multinucleate syncitial cells such as skeletal muscle fibers - an alternative hypothesis is that within-fiber-type expression variation may reflect fiber-to-fiber differences in physiological status. Among hindlimb muscles the soleus shows comparatively little between- and within-fiber-type variation and thus would make a suitable subject for near-uniform experimental germ-line transgene expression driven by CMV IE1 elements.
Materials and methods
Transgenic mouse production
CMVLacZ transgenic mice were prepared by microinjection of a 5 kb XhoI/SpeI fragment of pUT535 (Cayla, Toulouse, France) including the human CMV (Towne) immediate early enhancer/promoter region from - 511 to +97 relative to the IE1 transcription start site (nucleotides 622-1241 in ref 31) linked to Sh ble/E. coli LacZ β-gal coding sequences [32]. Transgenic founders were identified by tail DNA Southern blot and were bred with CD1 mates; lines were carried in the hemizygous condition. TnILacZ1 transgenic mice were from lines 29 and 36 described in ref 18. All analyses used adult mice at least 6 weeks old.
Fiber typing
Two methods were used to identify muscle fiber types. Immunohistochemical analysis used isoform-specific anti myosin heavy chain monoclonal antibodies as described in [33]: IIB-specific, BF-F3; IIA specific, SC-71; type I, BA-D5 or 10D10 (34). Fibers that did not react with any of the antibodies were typed as IIX. In addition, myosin ATPase histochemistry was performed after preincubation at pH 4.6 according to the method of Brooke and Kaiser [35] as modified for visualization of IIX fibers in the mouse [36]. This procedure gave a positive signal for IIX fibers, and was also useful for interpreting occasional fibers expressing more than one myosin isoform (0-~ 10% of fibers, varying between muscles and between individual animals). For simplicity, such hybrid fibers were counted among the "pure" fiber types as follows; IIB/IIX with IIB, IIX/IIA with IIA, and IIA/I with I. None of our conclusions are affected by this pooling.
β-gal histochemistry
Cryostat sections (10 μ) of frozen muscle samples were fixed 3 min in 0.25% glutaraldehyde in water at room temperature and were incubated in phosphate-buffered saline containing 5 mM ferricyanide, 5 mM ferrocyanide, 2 mM MgCl2, 0.1 mg/ml X-Gal at room temperature for 2-20 h. Image analysis was by microdensitometric measurement of optical densities of stained muscle fibers, using a reference series of standard neutral density filters for calibration [18]. Statistical analyses were performed at the VassarStats website [http://faculty.vassar.edu/~lowry/VassarStats.html#menu] created and maintained by Richard Lowry, Vassar College, Poughkeepsie, New York.
Acknowledgments
Acknowledgements
We thank Peter Merrifield and Stefano Schiaffino for providing antibodies. This work was supported by grants from the Medical Research Council of Canada. K.E.M.H is a Killam Scholar of the Montreal Neurological Institute.
Contributor Information
Patricia L Hallauer, Email: cxph@musica.mcgill.ca.
Kenneth EM Hastings, Email: khasti@po-box.mcgill.ca.
References
- Hennig R, Lømo T. Firing patterns of motor units in normal rats. . Nature. 1985;314:164–166. doi: 10.1038/314164a0. [DOI] [PubMed] [Google Scholar]
- Pette D, Staron RS. Cellular and molecular diversities of mammalian skeletal muscle fibers. . Rev Physiol Biochem Pharmacol. 1990;116:1–76. doi: 10.1007/3540528806_3. [DOI] [PubMed] [Google Scholar]
- Burke RE. Physiology of motor units. In Myology Basic and Clinical Vol 1 Edited by Engel AG, Franzini-Armstrong C, New York: McGraw-Hill, 1994. pp. 464–483.
- Kelly AM, Rubinstein NA. The diversity of muscle fiber types and its origin during development. . In Myology Basic and Clinical Vol 1 Edited by Engel AG, Franzini-Armstrong C, New York: McGraw-Hill, 1994. pp. 119–133.
- Schiaffino S, Reggiani C. Molecular diversity of myofibrillar proteins: gene regulation and functional significance. Physiol Rev. 1996;76:371–423. doi: 10.1152/physrev.1996.76.2.371. [DOI] [PubMed] [Google Scholar]
- Bottinelli R, Betto R, Schiaffino S, Reggiani C. Maximum shortening velocity and coexistence of myosin heavy chain isoforms in single skinned fast fibres of rat skeletal muscle. J Physiol. 1994;478:341–349. doi: 10.1007/BF00122115. [DOI] [PubMed] [Google Scholar]
- Wu H, Naya FJ, McKinsey TA, Mercer B, Shelton JM, Chin ER, Simard AR, Michel RN, Bassel-Duby R, Olson EN, Williams RS. MEF2 responds to multiple calcium-regulated signals in the control of skeletal muscle fiber type. EMBO J. 2000; 19:1963–1973. doi: 10.1093/emboj/19.9.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Mahoney JV, Guven KL, Lin J, Joya JE, Robinson CS, Wade RP, Hardeman EC. Identification of a novel slow-muscle-fiber enhancer binding protein, MusTRD1. Mol Cell Biol. 1998;18:6641–6652. doi: 10.1128/mcb.18.11.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo S, Venepally P, Cheng J, Buonanno A. Fiber-type-specific transcription of the troponin I slow gene is regulated by multiple elements. . Mol Cell Biol. 1999;19:515–525. doi: 10.1128/mcb.19.1.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boshart M, Weber F, Jahn G, Dorsch-Hasler K, Fleckenstein B, Schaffner W. A very strong enhancer is located upstream of an immediate early gene of human cytomegalovirus. Cell. 1985;41:521–530. doi: 10.1016/S0092-8674(85)80025-8. [DOI] [PubMed] [Google Scholar]
- Foecking MK, Hofstetter H. Powerful and versatile enhancer-promoter unit for mammalian expression vectors. Gene . 1986;45:101–105. doi: 10.1016/0378-1119(86)90137-X. [DOI] [PubMed] [Google Scholar]
- Furth PA, Henninghausen L, Baker C, Beatty B, Woychick R. The variability in activity of the universally expressed human cytomegalovirus immediate early gene 1 enhancer/promoter in transgenic mice. Nucleic Acids Res. 1991;19:6205–6208. doi: 10.1093/nar/19.22.6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koedood M, Fichtel A, Meier P, Mitchell PJ. Human cytomegalovirus (HCMV) immediate-early enhancer/promoter specificity during embryogenesis defines target tissues of congenital HCMV infection. J Virol. 1995;69:2194–2207. doi: 10.1128/jvi.69.4.2194-2207.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskar JF, Smith PP, Nilaver G, Jupp RA, Hoffman S, Peffer NJ, Tenney DJ, Colberg-Poley AM, Ghazal P, Nelson JA. The enhancer domain of the human cytomegalovirus major immediate-early promoter determines cell type-specific expression in transgenic mice. J Virol. 1996;70:3207–3214. doi: 10.1128/jvi.70.5.3207-3214.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TR, Chapman B, Din N, Ingerslev J, Kristensen P, Poulsen K, Hjorth JP. Expression of a cytomegalovirus IE-1-factor VIII cDNA hybrid gene in transgenic mice. Transgenic Res. 1992;1:164–169. doi: 10.1007/BF02522534. [DOI] [PubMed] [Google Scholar]
- Martin DIK, Whitelaw E. The vagaries of variegating transgenes. . BioEssays. 1996;18:919–923. doi: 10.1002/bies.950181111. [DOI] [PubMed] [Google Scholar]
- Garrick D, Fiering S, Martin DIK, Whitelaw E. Repeat-induced gene silencing in mammals. Nature Genetics. 1998;18:56–59. doi: 10.1038/ng0198-56. [DOI] [PubMed] [Google Scholar]
- Hallauer PL, Bradshaw HL, Hastings KEM. Complex fiber-type-specific expression of fast skeletal muscle troponin I gene constructs in transgenic mice. Development. 1993;119:691–701. doi: 10.1242/dev.119.3.691. [DOI] [PubMed] [Google Scholar]
- Donoghue MJ, Alvarez JD, Merlie JP, Sanes JR. Fiber type- and position-dependent expression of a myosin light chain-CAT transgene detected with a novel histochemical stain for CAT. J Cell Biol. 1991;115:423–434. doi: 10.1083/jcb.115.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannerjee-Basu S, Buonanno A. Cis-acting sequences of the rat troponin I slow gene confer tissue- and development-specific transcription in cultured muscle cells as well as fiber type specificity in transgenic mice. Mol Cell Biol. 1993;13:7019–7028. doi: 10.1128/mcb.13.11.7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt LK, O'Mahoney JV, Brennan KJ, Joya JE, Zhu L, Wade RP, Hardeman EC. The human troponin I slow promoter directs slow fiber-specific expression in transgenic mice. DNA & Cell Biol. 1995;14:599–607. doi: 10.1089/dna.1995.14.599. [DOI] [PubMed] [Google Scholar]
- Salminen M, Lopez S, Maire P, Kahn A, Daegelen D. Fast-muscle-specific DNA-protein interactions occurring in vivo at the human aldolase A M promoter are necessary for correct promoter activity in transgenic mice. . Mol Cell Biol. 1996;16:76–85. doi: 10.1128/mcb.16.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox GA, Cole NM, Matsumura K, Phelps SF, Hauschka SD, Campbell KP, Faulkner JA, Chamberlain JS. Overexpression of dystrophin in transgenic mdx mice eliminates dystrophic symptoms without toxicity. Nature. 1993;364:725–729. doi: 10.1038/364725a0. [DOI] [PubMed] [Google Scholar]
- Rafael JA, Sunada Y, Cole NM, Campbell KP, Faulkner JA, Chamberlain JS. Prevention of dystrophic pathology in mdx mice by a truncated dystrophin isoform. Hum Mol Genet. 1994;3:1725–1733. doi: 10.1093/hmg/3.10.1725. [DOI] [PubMed] [Google Scholar]
- Phelps SF, Hauser MA, Cole NM, Rafael JA, Hinkle RT, Faulkner JA, Chamberlain JS. Expression of full-length and truncated dystrophin mini-genes in transgenic mdx mice. Hum Mol Genet. 1995;4:1251–1258. doi: 10.1093/hmg/4.8.1251. [DOI] [PubMed] [Google Scholar]
- Wells DJ, Wells KE, Asante EA, Turner G, Sunada Y, Campbell KP, Walsh FS, Dickson G. Expression of human full-length and minidystrophin in transgenic mdx mice: implications for gene therapy of Duchenne muscular dystrophy. Hum Mol Genet. 1995;4:1245–1250. doi: 10.1093/hmg/4.8.1245. [DOI] [PubMed] [Google Scholar]
- Tinsley JM, Potter AC, Phelps SR, Fisher R, Trickett JI, Davies KE. Amelioration of the dystrophic phenotype of mdx mice using a truncated utrophin transgene. . Nature. 1996;384:349–353. doi: 10.1038/384349a0. [DOI] [PubMed] [Google Scholar]
- Tinsley J, Deconinck N, Fisher R, Kahn D, Phelps S, Gillis J-M, Davies K. Expression of full-length utrophin prevents muscular dystrophy in mdx mice. Nature Med. 1998;4:1441–1444. doi: 10.1038/4033. [DOI] [PubMed] [Google Scholar]
- Fazeli S, Wells DJ, Hobbs C, Walsh FS. Altered secondary myogenesis in transgenic animals expressing the neural cell adhesion molecule under the control of a skeletal muscle alpha-actin promoter. . J Cell Biol. 1996;135:241–251. doi: 10.1083/jcb.135.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manchester J, Skurat AV, Roach P, Hauschka SD, Lawrence JC. Increased glycogen accumulation in transgenic mice overexpressing glycogen synthase in skeletal muscle. Proc Nat'l Acad Sci USA. 1996;93:10707–10711. doi: 10.1073/pnas.93.20.1070710.1073/pnas.93.20.1070710.1073/pnas.93.20.10707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman BS, Thayer RM, Vincent KA, Haigwood NL. Effect of intron A from human cytomegalovirus (Towne) immediate-early gene on heterologous expression in mammalian cells. Nucleic Acids Res. 1991;19:3937–3986. doi: 10.1093/nar/19.14.3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron M, Reynes JP, Stassi D, Tiraby G. A selectable bifunctional beta-galactosidase::phleomycin-resistance fusion protein as a potential marker for eukaryotic cells. Gene. 1992;114:239–243. doi: 10.1016/0378-1119(92)90581-9. [DOI] [PubMed] [Google Scholar]
- Schiaffino S, Gorza L, Sartore S, Saggin L, Ausoni S, Vianello M, Gunderson K, Lømo T. Three myosin heavy chain isoforms in type 2 skeletal muscle fibres. J Muscle Res Cell Motil. 1989;10:197–205. doi: 10.1007/BF01739810. [DOI] [PubMed] [Google Scholar]
- Pin C, Merrifield PAM. Embryonic and fetal rat myoblasts express different phenotypes following differentiation in vitro. Devel Genet. 1993;14:356–368. doi: 10.1002/dvg.1020140505. [DOI] [PubMed] [Google Scholar]
- Brooke MH, Kaiser KK. Three "myosin adenosine triphosphatase" systems: the nature of their pH lability and sulfhydryl dependence. . J Histochem Cytochem. 1970;18:670–672. doi: 10.1177/18.9.670. [DOI] [PubMed] [Google Scholar]
- Hamalainen N, Pette D. The histochemical profiles of fast fiber types IIB, IID, and IIA in skeletal muscles of mouse, rat, and rabbit. J Histochem Cytochem. 1993;41:733–743. doi: 10.1177/41.5.8468455. [DOI] [PubMed] [Google Scholar]