Abstract
Organic fluorophores common to fluorescence-based investigations suffer from unwanted photophysical properties, including blinking and photobleaching, which limit their overall experimental performance. Methods to control such processes are particularly important for single-molecule fluorescence and fluorescence resonance energy transfer imaging where uninterrupted, stable fluorescence is paramount. Fluorescence and FRET-based assays have been carried out on dye-labeled DNA and RNA-based systems to quantify the effect of including small-molecule solution additives on the fluorescence and FRET behaviors of both cyanine and Alexa fluorophores. A detailed dwell time analysis of the fluorescence and FRET trajectories of more than 200,000 individual molecules showed that two compounds identified previously as triplet state quenchers, cyclooctatetraene, and Trolox, as well as 4-nitrobenzyl alcohol, act to favorably attenuate blinking, photobleaching, and influence the rate of photoresurrection in a concentration-dependent and context-dependent manner. In both biochemical systems examined, a unique cocktail of compounds was shown to be optimal for imaging performance. By simultaneously providing the most rapid and direct access to multiple photophysical kinetic parameters, smFRET imaging provides a powerful avenue for future investigations aimed at discovering new compounds, and effective combinations thereof. These efforts may ultimately facilitate tuning organic dye molecule performance according to each specific experimental demand.
Introduction
Fluorescence applications penetrate nearly every field of biological research (1). Bulk fluorimetry, as well as wide-field, scanning confocal, and total internal reflection fluorescence microscopy (TIRF) rely on high-quantum yield, stable fluorescent species. The utility of such fluorophores, including organic dyes, fluorescent proteins, as well as inorganic quantum dots and nanocrystals, is hampered by the undesirable photophysical properties of stochastic “blinking” events and irreversible photobleaching. To understand the origins of dye photophysics, fluorophore properties have been extensively investigated under a variety of conditions (2–23). Blinking describes the tendency of fluorescing molecules to undergo illumination intensity-dependent, reversible losses of fluorescence leading to “dark” states. Photobleaching is an irreversible loss of fluorescence. Photoresurrection describes a photon induced transition from a dark state back to a fluorescing state. A principle determinant of dye photophysics, in particular photobleaching, is molecular oxygen. Oxygen and oxygen-generated molecular species are efficient triplet-state quenchers (TSQs) that reduce the lifetime of dark states but also promote rapid irreversible fluorophore photobleaching (24–26). Solution investigations that depend on long-lived fluorescence, including single molecule fluorescence and fluorescence resonance energy transfer (FRET) imaging, where high illumination intensities are typically used, depend on efficient enzymatic oxygen scavenging systems and/or stringent degassing protocols. Experimental conditions that reduce or remove molecular oxygen, correspondingly exacerbate the prevalence of long-lived temporary dark states (24,27).
The widely used, commercially available cyanine (Cy) and Alexa fluorophores undergo dark state transitions over a broad range of timescales. Fluorophore dark states have been attributed to cis-trans isomerization within the conjugated polyene cores, a build-up of distinct triplet states, charge relay mechanisms, photo-ionization, photo-oxidation, and the absorbance of short wavelength photons from the first excited singlet state (6,8,9,14–16,23,28–35). Although Alexa fluorophores are chemically designed to reduce cis-trans isomerization inherent to the cyanine family of dye molecules, they remain prone to long-lived dark states. Fluorescence resonance energy transfer (FRET) studies, which typically use Cy and Alexa fluorophores, must therefore be interpreted in the context of blinking frequencies where at the single-molecule level, stepwise changes to zero FRET states may be misconstrued as biologically relevant conformational changes (8,36).
The reliable control of such unwanted photophysical processes offers the potential of greatly increasing dye brightness, dye stability and fluorescence lifetime. Such improvements in dye behavior would greatly enhance the robustness of single-molecule FRET (smFRET) observations where high-spatial and -time resolution measurements that demand increased illumination intensities have the potential to show FRET transitions correlated with functionally relevant conformational processes (24,37–51). Photo-induced recovery from dark states, recently leveraged to improve imaging resolution beyond the Rayleigh diffraction limit (52–58), may similarly benefit from control over blinking frequency. To this end, specific compounds such as Trolox, β-mercaptoethanol (BME), mercaptoethylamine (MEA), n-propyl gallate, 1,4-diazabicyclo[2.2.2]octane (DABCO), and cyclooctatetraene (COT) that favorably affect dark state and photobleaching lifetimes have come into increasingly widespread use (3,27,59–63).
Using smFRET imaging, we report a detailed analysis of the kinetic parameters of photophysical processes for cyanine- and Alexa-labeled DNA- and RNA-based systems under oxygen scavenging conditions in the presence and absence of the previously characterized TSQs, Trolox, and COT, as well as the compound, 4-nitrobenzyl alcohol (NBA). These compounds lack complete characterizations despite their increasingly widespread use (38,39,42,61,64–66). Through the quantification of nearly 200,000 single-molecule fluorescence and FRET trajectories using recently developed hidden Markov modeling procedures, specific alterations in blinking, photoresurrection, and photobleaching rates promoted by these compounds have been determined in two environmental contexts. These data, summarized in terms of the total lifetime in nonzero fluorescence/FRET states (Total ton) and zero fluorescence/FRET states (Total toff) before photobleaching, show that all three compounds affect dye photophysics in unique and environment-specific manners without perturbing the observed FRET values.
The data show, as reported previously (27,61), that Trolox improves dye photophysics in the context of a DNA-based system. However, a significant reduction in dye lifetime is observed in a different biochemical context, imaging dye-labeled tRNA molecules within the ribosome. NBA, although tending to reduce Total ton in the DNA system investigated, dramatically increases the Total ton and decreases Total toff in the ribosome system. In both systems, NBA almost completely eliminates long-lived excursions to dark states. More consistent, but attenuated, benefits were observed in the presence of COT. In combination, COT, NBA, and Trolox seem to operate through complementary mechanisms in both systems. By providing a robust means of assessing TSQ function, smFRET experiments provide a potential framework that may be further developed to explore the discovery of new compounds, and/or combinations of compounds, for the specific tuning of the fluorescence lifetime, blinking, and photoresurrection rates of organic fluorophores.
Materials and Methods
Preparation of dye-labeled biomolecules for single-molecule experiments
Two complimentary 12-nucleotide DNAs were chemically synthesized with a 5′-C6-amino linker for dye linkage; one strand possessed an additional 3′-biotin moiety attached via a 22 atom spacer (5′-/5AmM C6/CAT GAC CAT GAC-3′ and 5′-/5AmMC6/GTC ATG GTC ATG/3BioTEG/-3′; IDT, Coralville, IA). Each DNA strand was individually labeled with a commercially available, N-hydroxysuccinimide ester activated dye molecule (Cy3, Cy5; GE Healthcare, Piscataway, NJ; and Alexa 555, Alexa 647N; Molecular Probes, Carlsbad, CA) through the following procedure. The use of long chemical spacers between the dyes and DNA as well as the use of cyanine dyes that possess two negatively charged sulfite groups significantly reduces the contribution of fluorescence anisotropy to the single-molecule measurements (data not shown).
Lyophilized DNAs were resuspended in distilled, deionized water and adjusted to 50 mM potassium borate buffer, pH 8.1, 200 mM KCl. Dye-labeling was achieved by adding a 10-fold molar excess of N-hydroxysuccinimide-activated dye resuspended in dimethyl sulfoxide (DMSO). After a 2-h incubation in the dark at 37°C, unbound dye was removed by two sequential phenol/chloroform extraction steps. Labeled DNA was recovered by ethanol precipitation. Acceptor-labeled, biotinylated strands were hybridized with the complementary donor-labeled strand in the presence of 200 mM KCl by mixing the two in equimolar ratios, briefly heating the solution to 75°C, followed by passive cooling to room temperature. For all experiments, the strand containing the biotin moiety (3BioTEG) was labeled with the acceptor fluorophore. Where acceptor dyes were examined under direct illumination, acceptor-labeled, biotinylated DNA strands were hybridized with a complimentary DNA strand that was unlabeled.
70S ribosomes were isolated from the bacterial strain MRE600 following published procedures (38,39). Briefly, cultures were grown to early log phase and harvested by centrifugation. Cell lysis was achieved using a cell disrupter (Avestin, Ottawa, ON, Canada); insoluble debris was removed by centrifugation at 10,000 × g for 30 min. Ribosomes were isolated from the supernatant fraction by centrifugation through two sequential sucrose cushion steps each containing 20 mM Tris-HCl pH 7.5, 0.5 M NH4Cl, 10 mM MgCl2, 0.5 mM EDTA, 6 mM BME, and 37% sucrose. Ribosomes, resuspended in the same buffer lacking sucrose, were then isolated by centrifugation through a 10%–40% sucrose gradient containing 20 mM Tris-HCl pH 7.5, 100 mM NH4Cl, 10 mM MgCl2, 0.5 mM EDTA, and 6 mM BME. Gradients were fractionated by ultraviolet absorbance and those containing tight coupled 70S (TC70S) particles were aliquoted and stored in liquid nitrogen.
Ribosomal complexes containing site-specifically labeled tRNAs (Cy3-tRNAfMet labeled at 4-thiouridine (s4U) at position 8; Cy5-tRNAPhe labeled at 3-(3-amino-3-carboxypropyl uridine (acp3U) residue at position 47), were prepared using purified translation initiation factors, and synthetic, biotinylated mRNA (Dharmacon, Lafayette, CO) as described previously (38,39,42).
Single-molecule imaging and experimental conditions
All experiments were carried out using a laboratory built, prism-based TIRF apparatus as described previously at specified illumination intensities in Tris-polymix buffer (50 mM Tris-OAc pH 7.5, 100 mM KCl, 5 mM NH4OAc, 0.5 mM Ca(OAc)2, 15 mM Mg(OAc)2, 0.1 mM EDTA, 50 mM β-mercaptoethanol, 5 mM putrescine, and 1 mM spermidine) containing 2 unit/μL glucose oxidase (Sigma-Aldrich, St. Louis, MO), 20 units/μL catalase (Sigma-Aldrich) and 0.1% v/v glucose unless stated otherwise (42). An alternative oxygen scavenging system using protocatechuic acid (PCA) (Sigma-Aldrich)/protocatechuate-3,4-dioxygenase (PCD) (Sigma-Aldrich) was used following published procedures (24). Trolox (97%), COT (98%), and NBA (99%) (Sigma-Aldrich) were resuspended in DMSO and added to the Tris-polymix buffer to a final concentration of 2 mM. Biotinylated DNA molecules and ribosomal complexes were immobilized via a biotin-streptavidin interaction within microfluidic channels constructed on quartz slides (38,39,42,67). Fluorescence from surface-immobilized molecules, illuminated via the evanescent wave generated by total internal reflection of 532 nm (Laser Quantum, Cheshire, UK) and/or 635 nm (Coherent, Auburn, CA) laser sources, was collected using a 1.2 NA 60× water-immersion objective (Nikon, Melville, NY) and imaged onto a Cascade 512B CCD (Roper Scientific, Tucson, AZ). Data were acquired using Metamorph software (Universal Imaging Corporation, Downingtown, PA) collecting at a frame rate of 25/s (40 ms time resolution). For direct 635 nm illumination experiments, data were collected at a frame rate of 10/s (100 ms time resolution).
Kinetic analysis of smFRET time traces
The photophysical properties of dyes were investigated by extracting single-molecule FRET time traces from the acquired CCD images using in-house designed software in MATLAB (The MathWorks, Natick, MA) as described previously (42). FRET efficiencies (EFRET) for each trace were calculated according to the equation: ; where Iacceptor and Idonor correspond to Cy5 or Alexa 647N and Cy3 or Alexa 555 fluorescence intensity, respectively, after correcting for background intensity and cross talk between the donor and acceptor fluorescence signals. In the analysis of smFRET data, only those molecules yielding both donor and acceptor fluorescence were considered for analysis. In fluorescence and FRET imaging experiments molecules yielding a signal/noise (S/N) ratio <7:1 were excluded from analysis. Unless otherwise stated, S/N is defined as the total fluorescence intensity (donor + acceptor)/standard deviation of background fluorescence after photobleaching. The kinetic parameters of blinking and photobleaching were extracted from smFRET trajectories by idealizing fluorescence and FRET data to specific kinetic models using a segmental k-means algorithm implemented in QuB (68) as reported previously (42). Single-molecule data obtained from individual DNA molecules were fit to a two-state kinetic model yielding a series of dwells in the nonzero and zero-fluorescence/FRET states. smFRET data obtained from single ribosome molecules containing donor and acceptor labeled tRNAs was fit to a four-state kinetic model as described previously (42). Because under all smFRET conditions tested the vast majority (>85%) of FRET trajectories were limited by acceptor photobleaching, only dwells before the last acceptor fluorophore dark state were examined. Under all experimental conditions tested, donor fluorophore blinking contributed negligibly to the kinetic parameters of blinking and photobleaching. For direct, dual, and FRET-based illumination experiments, individual dwells in nonzero and zero fluorescent/FRET states were used to estimate ton and toff, the lifetimes of fluorescent and dark states, respectively. From these parameters, the total lifetime that each single-molecule was observed in nonzero fluorescence/FRET (Total ton) and zero- fluorescence/FRET states (Total toff) was estimated by fitting each distribution to exponential decay processes. To simplify the analysis of FRET data obtained from the ribosome system, where multiple nonzero FRET states were observed, all nonzero-FRET states were combined computationally to determine an aggregate nonzero FRET state lifetime. In all cases, ton and Total ton could be well described by a single-exponential decay process: y = y0 + Ae−x/t1. By contrast, toff, and Total toff could only be fit to a multi-exponential decay process consistent with more than one photoresurrection pathway (Supporting Material, Fig. S1 and Fig. S2). Therefore, to estimate Total toff, each distribution was fit to a double exponential decay process: . Exponential fitting was carried out using the Origin software package. The reported values were obtained by calculating a weighted average of fitted lifetimes and therefore represent only an average dark state lifetime, 〈toff〉. To reduce errors introduced by fitting, the percent time each single molecule occupied nonzero FRET states during the duration of fluorescence signal, %Total ton, was calculated from the sum of times occupying FRET and dark state dwells (Total ton/Total ton + Total toff). Maximization of this value and fluorescence lifetime, Total ton, are considered key determinants of optimal experimental conditions. Under each condition examined, an analysis of the average number of dark-state transitions per second is also shown.
Results and Discussion
Single-molecule fluorescence and FRET imaging have been used as a means to extract quantitative information on the blinking, photoresurrection, and photobleaching rates of Cy and Alexa fluorophores under conditions typically used in high-spatial and time resolution single-molecule imaging experiments. To explore the conditions for optimal single-molecule fluorescence imaging, dye photophysical processes have been examined in the presence and absence of solution additives that alter dye performance. Two distinct biochemical systems were tested using specific combinations of oxygen scavenging and solution additives 1), to quantify how Cy and Alexa fluorophores differ in performance in single-molecule imaging; 2), to quantify how experimental conditions effect fluorophore photophysical parameters; 3), to quantify how fluorophore photophysical parameters change in distinct experimental contexts (e.g., DNA versus RNA versus protein); and 4), to establish whether ideal conditions can be obtained that maximize fluorophore performance in single-molecule imaging experiments. Ultimately, such investigations are necessary to establish conditions that maximize the information content of single-molecule fluorescence imaging: minimizing blinking rates by maximizing ton, increasing photoresurrection rates by minimizing toff, and extending the total lifetime of fluorescence, Total ton. Because imaging is predominantly limited by acceptor fluorophore behavior, the focus in this study is given to FRET-based measurements where rapid blinking and photoresurrection rates allow the facile quantitation of acceptor fluorophore behavior.
Our data show that NBA, like COT and Trolox, fit the general criteria of a TSQ, altering the fluorescence lifetime and blinking behavior of the acceptor fluorophore, without otherwise affecting the FRET measurement (e.g., FRET value). However, NBA seems to operate by a unique mechanism, specifically increasing the rates of photoresurrection for both Cy5 and Alexa 647N dye molecules whereas having only modest effects on the lifetime of fluorescence. When used in combination with Trolox and COT, TSQs that specifically enhance fluorescence lifetime, additive effects are observed: an extension of ton and Total ton is afforded by Trolox and COT; a shortened toff and Total toff is afforded by NBA. Thus, under stringent oxygen scavenging conditions, a cocktail of TSQs is shown to effectively circumvent unwanted photophysical dye behavior offering highly optimized conditions for single-molecule imaging. In addition to improving the performance of dyes for single-molecule fluorescence imaging experiments in vitro, barring toxicity effects the means of mitigating fluorophore photophysical processes using such compounds may also prove beneficial to in vivo fluorescence and FRET imaging at both the bulk and single-molecule scale.
Fluorophore blinking is strongly power dependent
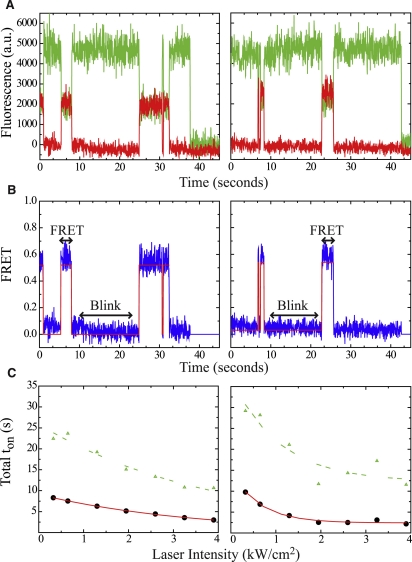
Consistent with previous observations (52), smFRET trajectories obtained for DNA oligonucleotides linked to either Cy3/Cy5 or Alexa-555/Alexa-647N dye pairs showed severe, power-dependent blinking in the absence of triplet state quenchers (Fig. 1). In both dye pairs investigated, blinking events and the total time observed in nonzero FRET states, Total ton, was dominated by the acceptor fluorophore. Although Total ton decreased rapidly with increasing power for both systems, the behavior of the Alexa dye pair was found to be unique in two ways: 1), at very low illumination intensity, Total ton was approximately twice that of the Cy pair; 2), above 1.95 kW/cm2, Total ton plateaued at levels generally below those observed for the Cy pair. Correspondingly, the Alexa pair has generally longer-lived dark states and therefore a longer Total toff than observed for the Cy dye pair. Despite this distinction, the performance of both dye pairs generally parallel each other. Parallel experiments could not be carried out on the ribosome system used in this study because the observed fluorescence lifetimes were too short to be quantified at such illumination intensities in the absence of additives. To quantitatively compare Cy and Alexa performance and their response to solution additives, our studies of fluorescence behavior were carried out at 0.65 kW/cm2 where Cy and Alexa dye pairs show similar Total ton.
Figure 1.
Photophysical processes in Cy and Alexa fluorophores are strongly power-dependent. (A) Blinking observed in the fluorescence trajectories of single Cy (left) and Alexa (right) dye pairs linked to a 12-basepair DNA under oxygen scavenging conditions and without the presence of triplet state quenchers (0.65 kW/cm2). (B) Corresponding smFRET time traces for Cy (left) and Alexa (right) dyes showing dwells in FRET and dark states. FRET is calculated from the relationship:. Idealization of smFRET time traces (red line) using hidden Markov modeling procedures is used to estimate the kinetics of switching between zero and nonzero FRET states. (C) Total ton (circles; red) decreases with increasing laser intensities for Cy (left) and Alexa (right) dye pairs. The photobleaching rate of the donor fluorophore (dotted green line) is shown for reference to indicate that trends in Total ton are largely independent of donor lifetime.
Trolox, COT, and NBA strongly affect the photophysical processes of Cy5 and Alexa-647N fluorophores under direct illumination
Given that triplet dark states span a broad range of lifetimes (microseconds to seconds) and that such processes are intimately linked to photobleaching (15,27,69), it is to be anticipated that compounds acting as TSQs should generally 1), reduce the frequency of blinking events; 2), reduce the observed rate of photobleaching by suppressing both short- and long-lived dark states; and 3), increase the rate exiting dark states by promoting return to the ground and/or singlet states. We examine these predictions under single-wavelength (direct), multiwavelength (dual) and FRET-based (indirect) illumination strategies where particular focus is given to the dye behavior of the acceptor fluorophores, Cy5 and Alexa 647N.
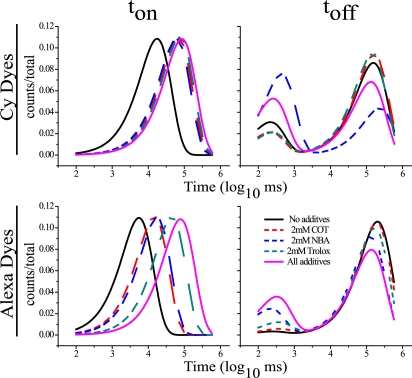
Under direct 635 nm illumination (0.13 kW/cm2), Cy5 and Alexa-647N show severe blinking where dark state dwells, toff, are observed to be both short- (∼200 ms) and long-lived (∼130 sec) (Fig. 2 and Table S1). These data are consistent with dark state lifetimes reported previously (10). By contrast, the fluorescence of Cy5 and Alexa-647N, ton, are well defined by a single exponential distribution of lifetimes (Fig. 2). The addition of COT or NBA in solution caused a ∼4-fold increase in ton for both Cy5 and Alexa-647N; Trolox increased ton ∼4-fold for Cy5 and ∼8-fold for Alexa-647N. For both dye pairs, the addition of all three compounds simultaneously tended to show additive effects beyond that observed for each individual additive.
Figure 2.
Photophysical processes in Cy5 and Alexa-647N are observed to be strongly attenuated under direct 635 nm excitation by the solution additives Trolox, COT, and NBA. The lifetimes of fluorescence, ton, and dark states, toff, are shown as overlaid histograms for both Cy5 and Alexa 647N fluorophores linked DNA oligonucleotides under direct 635 nm illumination in the absence (black), and presence of 2 mM COT (dashed red), 2 mM NBA (dashed blue), 2 mM Trolox (dashed green), and all three additives combined (2 mM each) (magenta). Each histogram fit represents a minimum of 1156 events where R2 values were >0.90 for ton and >0.7 for toff with the exception of 2 mM NBA where the R2 value of toff fit, due to a limited number of observed transitions, was 0.65.
The observed dark states of both dyes are represented by two clearly distinguishable lifetime distributions, short- and long-lived states, which differ in lifetime by more than an order of magnitude. The estimated lifetimes of both short- and long-lived dark states are similar for both types of fluorophores where the long-lived component contributes more substantially to the total dark state lifetime for the Alexa-647N fluorophore. COT, Trolox, or NBA shifted the relative populations of long-lived and short-lived dark states whereas the absolute values of each lifetime remained relatively unchanged. NBA redistributed the lifetime components in favor of short-lived states, resulting in a shorter 〈toff〉 by ∼1.4-fold for Cy5 and ∼1.8-fold for Alexa-647N. Trolox and COT slightly increased the relative population of long-lived dark states for Cy5, increasing 〈toff〉 by ∼1.1-fold. Opposite effects were observed when Trolox or COT was included in direct Alexa-647N illumination experiments. Long-lived dark states decreased and short-lived dark states increased, resulting in a decrease in 〈toff〉 by ∼1.2-fold.
Multiwavelength experiments were next carried out to test the effects of solution additives on dark state lifetimes to provide insights into how 532 nm illumination (0.65 kW/cm2) alters acceptor fluorophore behavior in FRET-based experiments. As reported previously (10), simultaneous 532 nm illumination dramatically increased blinking frequency by reducing toff (Table S1). Both COT and NBA increased ton by ∼2-fold for Cy5 and Alexa-647N. Trolox increased ton by 5.5-fold for Cy5 and nearly sevenfold for Alexa-647N. Under dual illumination, the affects of Trolox, COT, and NBA could not be as easily interpreted as a simple redistribution of short- and long-lived dark states as their presence shifted the absolute values of both lifetime components (Fig. S1). For Cy5, both COT and Trolox reduced 〈toff〉 by ∼1.7-fold, whereas NBA decreased 〈toff〉 by ∼3-fold. For Alexa-647N, COT decreased 〈toff〉 by ∼2.9-fold, NBA decreased 〈toff〉 by ∼4.4-fold, Trolox reduced 〈toff〉 by ∼1.4-fold. Again, for both dye pairs, the addition of all three compounds simultaneously tended to show additive effects beyond that observed for each individual additive.
In general, the changes in ton observed were not as dramatic as those observed without 532 nm illumination; changes in 〈toff〉 were more dramatic than without 532 nm illumination. These data suggest that 532 nm illumination is linked in some manner to the TSQ mechanism.
Trolox, COT, and NBA strongly affect the photophysical processes of Cy5 and Alexa-647N fluorophores under FRET-based illumination
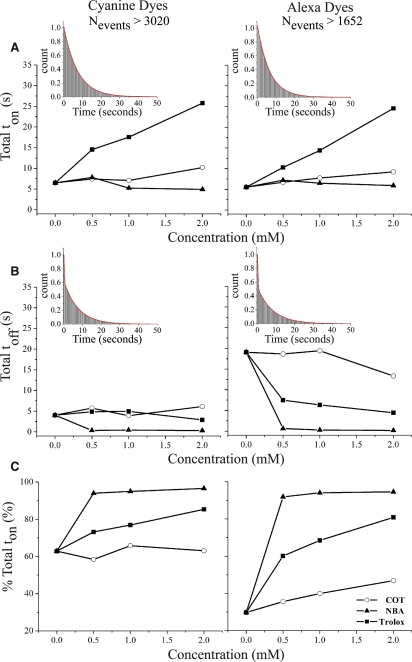
Experiments were carried out on DNA oligonucleotides labeled with either Cyanine (Cy3/5) or Alexa (Alexa555/647N) dye pairs to examine the effects of adding Trolox, COT, and NBA in FRET-based assays. The results, shown in Fig. 3 and summarized in Table 1 and Table S2, show that each compound acts in a concentration-dependent manner to yield dramatic improvements in the quality of smFRET data that stem from dramatic increases in the 〈toff〉 of acceptor dark states as well as increased ton for both donor and acceptor fluorophores. The concentration-dependent improvements in dye photophysical properties are summarized for simplicity as changes in Total ton (Fig. 3 A), Total toff (Fig. 3 B), and the percent time in nonzero FRET state relative to the total fluorescence lifetime (Fig. 3 C). These parameters were calculated from measured ton and toff values for each system (Table S3, A and B) to concisely summarize our findings. Because a large number of transitions were observed in each experiment, a robust and reproducible quantitation of the data were possible as shown by the similarity of results obtained in three separate trials testing the effects of compounds on Cy-labeled DNA (Fig. S4 and Table S4). Maximization of percent time in nonzero FRET states is particularly relevant to smFRET experiments as minimization of contaminating blinking events is highly desirable to preserve the biologically relevant information content of smFRET data. A second related parameter that can be extracted easily from the data is the number of transitions observed per second (ntransitions), which reports directly on the blinking frequency observed.
Figure 3.
Photophysical processes in Cy5 and Alexa-647N are observed to be strongly attenuated in FRET-based experiments by the solution additives Trolox, COT, and NBA. FRET-based experiments (0.65 kW/cm2 532 nm excitation) carried out on donor and acceptor labeled DNA oligonucleotides show that COT (circles), NBA (triangles), and Trolox (squares) effect (A) Total ton, (B) Total toff, and (C) the percent time in nonzero FRET states % Total ton (Total ton/Total ton + Total toff), in a concentration dependent manner. Inset figures show examples of the single exponential decay fittings of Total ton and double exponential decay fittings of Total toff.
Table 1.
Effectiveness of specific compounds is environment sensitive and can be used in combination to minimize photophysical processes in the DNA system
| DNA system |
||||||||
|---|---|---|---|---|---|---|---|---|
| Cy dyes |
Alexa dyes |
|||||||
| Total ton (s) | Total toff (s) | % Total ton (%) | ntransitions (s−1) | Total ton (s) | Total toff (s) | % Total ton (%) | ntransitions (s−1) | |
| No compounds | 6.5 | 4.0 | 62.9 | 0.3 | 5.5 | 19.1 | 29.8 | 0.4 |
| 2 mM COT | 10.2 | 6.1 | 63.1 | 0.2 | 9.2 | 13.3 | 46.9 | 0.2 |
| 2 mM NBA | 4.9 | 0.3 | 96.6 | 0.2 | 5.9 | 0.2 | 94.6 | 0.2 |
| 2 mM Trolox | 25.8 | 2.9 | 85.3 | 0.1 | 24.6 | 4.4 | 80.9 | 0.1 |
| All compounds | 16.6 | 1.0 | 93.6 | 0.1 | 24.1 | 1.7 | 91.0 | 0.1 |
Values of Total ton, Total toff, the percent time each of the systems investigated is in the nonzero FRET state before photobleaching, % Total ton (Total ton/Total ton + Total toff), and the number of transitions per second, ntransitions, are shown in absence and presence of 2 mM COT, 2 mM NBA, or 2 mM Trolox separately and in combination (illumination intensity = 0.65 kW/cm2). As described in the Materials and Methods, the reported values of Total ton and Total toff were calculated by fitting and the percent time in nonzero FRET states, % Total ton, was calculated by dividing the total dwells in the zero FRET state by the total length of the smFRET time trace before photobleaching. In each case a minimum of 1650 number of transitions were observed and fitting resulted in R2 values > 0.995.
As expected from previous reports of Cy3/5-labeled DNA (27), Trolox increases the Total ton in a concentration dependent manner. In line with the predicted qualities of an effective TSQ, Trolox also decreases Total toff. Similar, although attenuated, trends were obtained for Total ton in the case of COT. Under the conditions tested, a ∼4-fold and a ∼1.6-fold increase in Total ton was observed at 2 mM Trolox and COT, respectively (Fig. 3, Table 1). At this concentration, a ∼1.4-fold decrease and ∼1.5-fold increase is also observed in Total toff for Trolox and COT, respectively (Fig. 3, Table 1). Qualitatively similar results were observed for the Alexa dye pair where at 2 mM, a ∼4.5-fold and ∼1.7-fold increase in Total ton, as well as a ∼4.3-fold and ∼1.4-fold decrease in Total toff was observed for Trolox and COT, respectively (Fig. 3, Table 1). NBA also showed clear evidence of TSQ-like behavior for both dye pairs tested. However, distinctions from Trolox and COT are notable. In the Cy-labeled DNA system, 2 mM NBA promoted a modest but reproducible ∼1.3-fold decrease in Total ton. This apparent shortcoming was compensated for by an observed ∼12-fold decrease in Total toff. Given that ntransitions also decreased, we could conclude from these data and the data obtained from the direct illumination experiments described above that NBA has the tendency to strongly suppress long-lived dark states for both Cy5 and Alexa-647N. In the case of the Alexa-labeled DNA system, NBA provided a modest increase in Total ton but a dramatic decrease in Total toff.
By tending to increase Total ton and decrease Total toff, all three compounds tested here have the net effect of increasing the percent time informative data can be obtained from both Cy and Alexa fluorophores (Fig. 3 C). Strikingly, at 2 mM NBA, this percentage increases to >90% for both Cy and Alexa dyes. The observed effectiveness of the compounds used is also observed at greater illumination intensities (1.95 kW/cm2) for both dye pairs (Fig. S3 and Table S2).
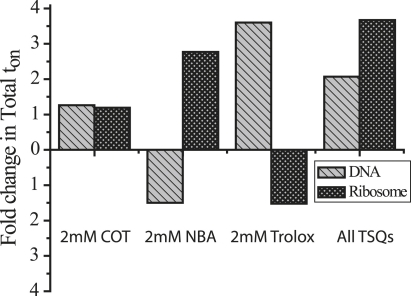
To test whether these results were unique to the DNA system, the response of the Cy-labeled tRNA molecules bound within the ribosome was investigated in the presence of either 2 mM COT, 2 mM NBA, or 2 mM Trolox. The results of these experiments are summarized in Table 2. Unexpectedly, the addition of 2 mM Trolox to the ribosome system decreased the Total ton by ∼1.5-fold. Addition of 2 mM COT increased Total ton by ∼1.2-fold and promoted a modest decrease in Total toff. NBA increased Total ton and decreased Total toff, by ∼1.8-fold. Similar to the DNA system, NBA decreased ntransitions ∼4.5-fold. Thus, as is shown graphically in Fig. 4, the activity of Trolox, COT, and NBA is strongly context dependent where Trolox and NBA have profoundly opposite affects on Total ton in the DNA and ribosome systems. Although TSQs are predicted to improve the mean fluorescence intensities of single molecules, thereby increasing the S/N ratio (where S/N is defined as the total fluorescence intensity (donor + acceptor)/standard deviation of the fluorescence signal), no appreciable changes in these parameters could be detected in our analysis. This observation suggests that the compounds tested either have negligible effects on fast blinking processes that tend to contribute to dye noise and reduce fluorescence intensity, or that the addition of these compounds at high concentrations increases background noise, lowering the S/N ratios to an extent that modest effects on this parameter are masked.
Table 2.
Effectiveness of specific compounds is environment sensitive and can be used in combination to minimize photophysical processes in the Ribosome system
| Ribosome system |
||||
|---|---|---|---|---|
| Cy dyes |
||||
| Total ton(s) | Total toff(s) | % Total ton(%) | ntransitions(s−1) | |
| No compounds | 1.8 | 0.8 | 70.3 | 1.0 |
| 2 mM COT | 2.2 | 0.7 | 76.4 | 0.8 |
| 2 mM NBA | 3.2 | 0.6 | 85.4 | 0.2 |
| 2 mM Trolox | 1.2 | 0.2∗ | 95.5∗ | 0.3 |
| All compounds | 6.3 | 0.4 | 96.7 | 0.1 |
Values of Total ton, Total toff, the percent time each of the systems investigated is in the nonzero FRET state before photobleaching, % Total ton (Total ton/Total ton + Total toff), and the number of transitions per second, ntransitions, are shown in absence and presence of 2 mM COT, 2 mM NBA, or 2 mM Trolox separately and in combination (illumination intensity = 0.65 kW/cm2). As described in the Materials and Methods, the reported values of Total ton and Total toff were calculated by fitting, and the total percent time in nonzero FRET states, % Total ton, was calculated by dividing the total dwells in the zero FRET state by the total length of the smFRET time trace before photobleaching. Exponential fitting was carried out in Origin. In each case a minimum of 532 number of transitions were observed and fitting resulted in R2 values >0.995.
Denotes quantities that could not be accurately determined due to extremely fast photobleaching and/or too few number of transitions.
Figure 4.
Solution additives effect photophysical processes in Cy5 and Alexa-647N in an environment specific manner. Trolox, COT, and NBA differentially impact Total ton in DNA and ribosome systems. Fold changes (increase or decrease) in Total ton for the ribosome and DNA systems are plotted in the presence of 2 mM NBA, 2 mM COT, and 2 mM Trolox added separately or together normalized to those observed in their absence (defined as the x axis).
Trolox, COT, and NBA provide enhanced reductions in dye photophysics when used in combination
To examine whether the compounds provide additive or synergistic reductions in unwanted dye photophysical behaviors, experiments were carried out in the presence of all three additives: 2 mM Trolox, 2 mM COT, and 2 mM NBA. A summary of these results is shown in Table 1 and Table S1 for the DNA system and Table 2 for the ribosome system and shown graphically in Fig. 4. By comparison to dye behaviors in the absence of any compounds, in the presence of all compounds, the Total ton for Cy-labeled DNA increased ∼2.6-fold and Total toff decreased ∼4.2-fold. Thus, the effectiveness of combination quenching is only ∼65% of the increase in Total ton expected from 2 mM Trolox alone and only ∼33% of the expected reduction in Total toff in the presence of 2 mM NBA alone. This result suggests that the individual compounds act competitively. Yet, an improved net outcome of using all the compounds together is evident in the maximized value of percent time in nonzero FRET states, % Total ton. Superior results for combination quenching were also obtained in the case of Alexa-labeled DNA where Total ton increased by ∼4.4-fold and Total toff decreased by ∼10.9-fold compared to experiments lacking the compounds. Total ton remained similar to that observed with 2 mM Trolox alone whereas Total toff was reduced by an additional ∼2.5-fold. Although ntransitions was modestly lower with 2 mM Trolox alone compared to the presence of all triplet state quenchers, the presence of all triplet state quenchers minimized Total toff, the time spent in the dark states. These results are mirrored in experiments carried out on singly labeled Cy5- and Alexa 647N-labeled DNA oligonucleotides where a nonadditive increase in ton and decrease in toff are observed under direct and dual illumination through combination quenching (Fig. 2, Fig. S1, and Table S1).
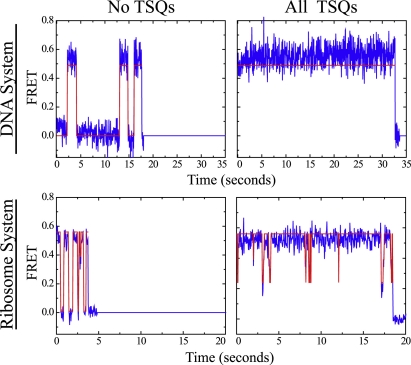
Combination quenching also showed net enhancements in FRET-based measurements for a second biochemical systems. In FRET-based experiments carried out on a previously established system wherein the ribosome contains both Cy3 and Cy5 labeled tRNAs (42), a ∼3.5-fold increase in Total ton was observed with all compounds present −a ∼5-fold increase when compared to data taken in the presence of 2mM Trolox alone. In the presence of all three compounds, Total toff reduced to ∼0.35 s, ∼1.6-fold lower than in the presence of 2mM NBA alone. Compared to the absence of compounds, ntransitions was reduced ∼8 fold. Thus, only partially additive effects were observed for COT and NBA in the ribosome system and the deleterious effects observed for Total ton in the presence of Trolox alone, were absent. The results observed for DNA and ribosome systems under combination quenching conditions are summarized in Fig. 5. At present it is not clear why the presence of NBA and COT seem to counter the negative effects of Trolox in the ribosome system. A protein system containing Cy3 and Cy5 fluorophores, also concurrently under investigation, corroborates the findings that Trolox, COT, and NBA act in an environment-specific manner and support the view that combination quenching can serve to enhance fluorescence lifetimes and reduce dark state lifetimes in FRET-based measurements (S. C. Blanchard, unpublished data).
Figure 5.
Solution additives, when used in combination mitigate unwanted photophysical processes in Cy5 and Alexa-647N. (upper panels) smFRET trajectories observed for Cy-labeled DNA oligonucleotides in the absence (left) and presence (right) of all three solution additives COT, Trolox, and NBA (2 mM each). (lower panels) smFRET trajectories observed for Cy-labeled tRNA molecules on the ribosome in the absence (left) and presence (right) of COT, Trolox, and NBA (2 mM each).
The effect of compounds is also observed with the PCA/PCD oxygen scavenging system
Similar results were obtained in smFRET studies carried out on DNA systems using the PCA/PCD oxygen scavenging system (Table S5) (70). Oxygen scavenging enzymes and most biological molecules containing cysteine residues carry out optimally under reducing conditions. However, reducing conditions are known to diminish the performance of typical single-molecule fluorophores by inducing long lived dark states (27,70). In the presence of all compounds and either 5 mM or 50 mM BME yielded similar results were obtained (Table S6). Experiments were not carried out in the absence of BME. Lower BME concentrations appeared to increase Total ton by a modest 18%. Thus, the compounds tested are effective regardless of the oxygen scavenging system used and their effect is not appreciably affected by high concentrations of reducing agent.
Conclusions
The reliable quenching of dark states offers a powerful and enabling approach to reducing unwanted dye photophysics in Cy- and Alexa-type fluorophores to improve both single-molecule and bulk fluorescence imaging. Our examination of fluorophore stability in multiple biochemical contexts under varied solution conditions, shed light on the mechanisms by which specific compounds affect the lifetimes of fluorescing and dark states under conditions typical of high-spatial and time-resolution single-molecule fluorescence imaging. These data provide a potential framework for the rational optimization of the photophysical properties of fluorescent dye molecules according to experimental demands. Global reductions in dark state transitions and increases in photoresurrection rates helps to preserve the biologically relevant information content present in single-molecule fluorescence and FRET trajectories and increases the effective brightness and photostability of dyes for improved signal/noise ratios. Such improvements may also be particularly important to super-resolution in vivo imaging efforts, where blinking and photoresurrection are leveraged (71).
In this study, we have quantified the effects of three distinct compounds that dramatically alter the photophysical properties of Cy5 and Alexa-647N fluorophores. Under direct, 635 nm illumination, both Cy5 and Alexa-647N fluorophores appear to follow a single pathway to at least two distinct dark states differing in lifetime by more than one order of magnitude. Consistent with previous reports (10), when illuminated simultaneously with 532 nm light the photoresurrection pathways for both Cy5 and Alexa-647N were accelerated, yielding significant reductions in both short- and long-lived dark state lifetimes. The fluorescence lifetimes of both Cy5 and Alexa 647N also decreased. Under direct acceptor fluorophore illumination, the solution additives Trolox, COT, and NBA were each shown to effectively increase the fluorescence lifetime and decrease average lifetime of dark states for both fluorophores. In such experiments, the dominant effect of solution additives was to specifically reduce the relative number of long-lived dark states in favor of short-lived states. Under dual illumination conditions, generally similar results were observed. However, the effects of solution additives were more complex and alterations were observed in both the absolute values and relative number of short- and long-lived dark states consistent with solution additives modifying acceptor fluorophore behavior in an excitation frequency-dependent manner. These observations are consistent with each of the solution additives tested functioning as a triplet state quencher but also show that a complex interplay of factors contributes to their mechanism of action. These factors may include the precise strategy and frequency of illumination, additive concentrations, fluorophore environments and/or subtle differences in the chemical nature of the specific fluorophores used.
As anticipated from our acceptor-only studies, solution additives generally yielded beneficial effects for both dye pairs in FRET-based experiments. A summary of the effects observed on addition of Trolox, COT, and NBA are summarized in Table 1. Dwell time analyses for each system investigated are shown explicitly in Table S1 and Table S2. In FRET-based assays carried out on Cy and Alexa labeled DNA, the lifetime of nonzero FRET states is well defined by a single exponential distribution suggesting the existence of a single, rate-limiting pathway to both temporary (blinking) and permanent (photobleaching) dark states. By contrast, a complex multicomponent lifetime is observed for dark states consistent with observations made in direct and dual illumination acceptor-only experiments showing the existence of at least two mechanistic pathways back to the fluorescing state (6,14–16,35,60).
Through the reduction of photophysical blinking events and the stimulation of photoresurrection, Trolox, COT, and NBA may work individually or in concert to improve the total information content of smFRET data. As shown previously, the action of such compounds is strongly concentration dependent. This observation may be explained if they 1), scavenge oxygen-dependent species that otherwise react with dyes to promote dark states and photobleaching; 2), collide with dyes to mediate electronic events that promote productive excited state relaxation pathways; and/or 3), affect dye relaxation pathways via radiative or non-radiative mechanisms. Further experiments are needed to test these possibilities and to understand how chemically distinct compounds mediate quantitatively distinct effects on Cy- and Alexa-type fluorophores in an environment-dependent manner. The observed distinctions in Trolox, COT, and NBA mechanisms will require more in depth investigations to understand how each compound affects the rates going from lower excited singlet states to higher excited singlet states (kS1 → kSn), the rates of intersystem crossing to triplet states (kS1 → kT1, kSn → kTn), the rates of relaxation (kSn → kS1, kS1 → kS0, kTn → kT1, kT1 → kT0) and rates of back isomerization to the singlet state (kT1 → kS1). It will also be important to understand whether solution additives can affect the blinking behavior of quantum dots and fluorescing protein species as well as their affects on fluorescent molecules in vivo where the extension of dye lifetime and reductions in dark states is paramount to single-molecule fluorescence imaging.
Given the challenges associated with improving dye photophysics through the synthesis of novel organic compounds, the potential to control dye photophysics through the addition of unique TSQs, or combinations of TSQs, may provide the most versatile and generalizable approach to tailoring dye behavior over the broadest range of contexts and experimental systems. Therefore, an extensive search for novel compounds, or combinations of compounds, that improve the photophysical properties of fluorescing molecules is called for. A more diverse repertoire of such reagents will broaden our knowledge of their basic mechanism and will doubtless afford significant further improvements in dye photophysics and the experimental imaging of dye molecules. High-throughput smFRET experiments would aid this endeavor by providing the most direct access to kinetic parameters of photophysical phenomenon with only limited demand for material expense.
Acknowledgments
The authors thank Dr. Peter Geggier for helpful support in data analysis and Roger Altman for providing many valuable reagents required for single-molecule experiments.
This work was supported by the Tri-Institutional Training Program in Chemical Biology (TPCB) and the National Institutes of Health grant 1R01GM079238-01 and the Tri-Institutional Training Program in Computational Biology and Medicine.
Supporting Material
References
- 1.Michalet X., Kapanidis A.N., Laurence T., Pinaud F., Doose S. The power and prospects of fluorescence microscopies and spectroscopies. Annu. Rev. Biophys. Biomol.Struct. 2003;32:161–182. doi: 10.1146/annurev.biophys.32.110601.142525. [DOI] [PubMed] [Google Scholar]
- 2.Dickson R.M., Cubitt A.B., Tsien R.Y., Moerner W.E. On/off blinking and switching behavior of single molecules of green fluorescent protein. Nature. 1997;388:355–358. doi: 10.1038/41048. [DOI] [PubMed] [Google Scholar]
- 3.Dittrich P.S., Schwille P. Photobleaching and stabilization of fluorophores used for single-molecule analysis with one- and two-photon excitation. Appl. Phys. B. 2001;73:829–837. [Google Scholar]
- 4.Eggeling C., Widengren J., Brand L., Schaffer J., Felekyan S. Analysis of photobleaching in single-molecule multicolor excitation and Förster resonance energy transfer measurements. J. Phys. Chem. A. 2006;110:2979–2995. doi: 10.1021/jp054581w. [DOI] [PubMed] [Google Scholar]
- 5.Eggeling C., Widengren J., Rigler R., Seidel C.A. Photobleaching of fluorescent dyes under conditions used for single-molecule detection: evidence of two-step photolysis. Anal. Chem. 1998;70:2651–2659. doi: 10.1021/ac980027p. [DOI] [PubMed] [Google Scholar]
- 6.English D.S., Harbron E.J., Barbara P.F. Probing photoinduced intersystem crossing by two-color, double resonance single molecule spectroscopy. J. Phys. Chem. A. 2000;104:9057–9061. [Google Scholar]
- 7.Ha T., Enderle T., Chemla D.S., Selvin P.R., Weiss S. Quantum jumps of single molecules at room temperature. Chem. Phys. Lett. 1997;271:1–5. [Google Scholar]
- 8.Ha T., Ting A.Y., Liang J., Deniz A.A., Chemla D.S. Temporal fluctuations of fluorescence resonance energy transfer between two dyes conjugated to a single protein. Chem. Phys. 1999;247:107–118. [Google Scholar]
- 9.Kong X., Nir E., Hamadani K., Weiss S. Photobleaching pathways in single-molecule FRET experiments. J. Am. Chem. Soc. 2007;129:4643–4654. doi: 10.1021/ja068002s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sabanayagam C.R., Eid J.S., Meller A. Long time scale blinking kinetics of cyanine fluorophores conjugated to DNA and its effect on Förster resonance energy transfer. J. Chem. Phys. 2005;123:224708. doi: 10.1063/1.2136157. [DOI] [PubMed] [Google Scholar]
- 11.Schenk A., Ivanchenko S., Rocker C., Wiedenmann J., Nienhaus G.U. Photodynamics of red fluorescent proteins studied by fluorescence correlation spectroscopy. Biophys. J. 2004;86:384–394. doi: 10.1016/S0006-3495(04)74114-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song L., Varma C.A., Verhoeven J.W., Tanke H.J. Influence of the triplet excited state on the photobleaching kinetics of fluorescein in microscopy. Biophys. J. 1996;70:2959–2968. doi: 10.1016/S0006-3495(96)79866-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sugisaki M., Ren H.W., Nishi K., Masumoto Y. Fluorescence intermittency in self-assembled InP quantum dots. Phys. Rev. Lett. 2001;86:4883–4886. doi: 10.1103/PhysRevLett.86.4883. [DOI] [PubMed] [Google Scholar]
- 14.Tinnefeld P., Buschmann V., Weston K.D., Sauer M. Direct observation of collective blinking and energy transfer in a bichromophoric system. J. Phys. Chem. A. 2003;107:323–327. [Google Scholar]
- 15.Tinnefeld P., Hofkens J., Herten D.P., Masuo S., Vosch T. Higher-excited-state photophysical pathways in multichromophoric systems revealed by single-molecule fluorescence spectroscopy. ChemPhysChem. 2004;5:1786–1790. doi: 10.1002/cphc.200400325. [DOI] [PubMed] [Google Scholar]
- 16.Zondervan R., Kulzer F., Orlinskii S.B., Orrit M. Photoblinking of Rhodamine 6G in poly(vinyl alcohol): radical dark state formed through the triplet. J. Phys. Chem. 2003;107:6770–6776. [Google Scholar]
- 17.Bates M., Blosser T.R., Zhuang X. Short-range spectroscopic ruler based on a single-molecule optical switch. Phys. Rev. Lett. 2005;94:108101. doi: 10.1103/PhysRevLett.94.108101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fukaminato T., Sasaki T., Kawai T., Tamai N., Irie M. Digital photoswitching of fluorescence based on the photochromism of diarylethene derivatives at a single-molecule level. J. Am. Chem. Soc. 2004;126:14843–14849. doi: 10.1021/ja047169n. [DOI] [PubMed] [Google Scholar]
- 19.Heilemann M., Margeat E., Kasper R., Sauer M., Tinnefeld P. Carbocyanine dyes as efficient reversible single-molecule optical switch. J. Am. Chem. Soc. 2005;127:3801–3806. doi: 10.1021/ja044686x. [DOI] [PubMed] [Google Scholar]
- 20.Irie M., Fukaminato T., Sasaki T., Tamai N., Kawai T. Organic chemistry: a digital fluorescent molecular photoswitch. Nature. 2002;420:759–760. doi: 10.1038/420759a. [DOI] [PubMed] [Google Scholar]
- 21.Kulzer F., Kummer S., Matzke R., Brauchle C., Basche T. Single-molecule optical switching of terrylene in p-terphenyl. Nature. 1997;387:688–691. [Google Scholar]
- 22.Sauer M. Reversible molecular photoswitches: a key technology for nanoscience and fluorescence imaging. Proc. Natl. Acad. Sci. USA. 2005;102:9433–9434. doi: 10.1073/pnas.0504264102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Widengren J., Schwille P. Characterization of photoinduced isomerization and back-isomerization of the cyanine dye Cy5 by fluorescence correlation spectroscopy. J. Phys. Chem. A. 2000;104:6416–6428. [Google Scholar]
- 24.Aitken C.E., Marshall R.A., Puglisi J. An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophys. J. 2008;94:1826–1835. doi: 10.1529/biophysj.107.117689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hübner C.G., Renn A., Renge I., Wild U.P. Direct observation of the triplet lifetime quenching of single dye molecules by molecular oxygen. J. Chem. Phys. 2001;115:9619–9622. [Google Scholar]
- 26.Renn A., Seelig J., Sandoghdar V. Oxygen-dependent photochemistry of fluorescent dyes studied at the single molecule level. Mol. Phys. 2006;104:409–414. [Google Scholar]
- 27.Rasnik I., McKinney S.A., Ha T. Nonblinking and long-lasting single-molecule fluorescence imaging. Nat. Methods. 2006;3:891–893. doi: 10.1038/nmeth934. [DOI] [PubMed] [Google Scholar]
- 28.Davidson R.S. Mechanisms of photo-oxidation reactions. Pestic. Sci. 1979;10:158–170. [Google Scholar]
- 29.Eggeling C., Volkmer A., Seidel C.A. Molecular photobleaching kinetics of Rhodamine 6G by one- and two-photon induced confocal fluorescence microscopy. ChemPhysChem. 2005;6:791–804. doi: 10.1002/cphc.200400509. [DOI] [PubMed] [Google Scholar]
- 30.Jia K., Wan Y., Xia A., Li S., Gong F. Characterization of photoinduced isomerization and intersystem crossing of the cyanine dye Cy3. J. Phys. Chem. A. 2007;111:1593–1597. doi: 10.1021/jp067843i. [DOI] [PubMed] [Google Scholar]
- 31.Tinnefeld P., Herten D.P., Sauer M. Photophysical dynamics of single molecules studied by spectrally-resolved fluorescence lifetime imaging microscopy (SFLIM) J. Phys. Chem. A. 2001;105:7989–8003. [Google Scholar]
- 32.Widengren J., Rigler R. Mechanisms of photobleaching investigated by fluorescence correlation spectroscopy. Bioimaging. 1996;4:149–157. [Google Scholar]
- 33.Wilkinson F., McGarvey D.J., Olea A.F. Excited triplet state interactions with molecular oxygen: influence of charge transfer on the bimolecular quenching rate constants and the yields of singlet oxygen (O;′Ag) for substituted naphthalenes in various solvents. J. Phys. Chem. 1994;98:3762–3769. [Google Scholar]
- 34.Yeh H.C., Puleo C.M., Ho Y.P., Bailey V.J., Lim T.C. Tunable blinking kinetics of cy5 for precise DNA quantification and single-nucleotide difference detection. Biophys. J. 2008;95:729–737. doi: 10.1529/biophysj.107.127530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vogelsang J., Kasper R., Steinhauer C., Person B., Heilemann M. A reducing and oxidizing system minimizes photobleaching and blinking of fluorescent dyes. Angew. Chem. Int. Ed. Engl. 2008;47:5465–5469. doi: 10.1002/anie.200801518. [DOI] [PubMed] [Google Scholar]
- 36.Bagshaw C.R., Cherny D. Blinking fluorophores: what do they tell us about protein dynamics? Biochem. Soc. Trans. 2006;34:979–982. doi: 10.1042/BST0340979. [DOI] [PubMed] [Google Scholar]
- 37.Bai L., Santangelo T.J., Wang M.D. Single-molecule analysis of RNA polymerase transcription. Annu. Rev. Biophys. Biomol. Struct. 2006;35:343–360. doi: 10.1146/annurev.biophys.35.010406.150153. [DOI] [PubMed] [Google Scholar]
- 38.Blanchard S.C., Gonzalez R.L., Kim H.D., Chu S., Puglisi J.D. tRNA selection and kinetic proofreading in translation. Nat. Struct. Mol. Biol. 2004;11:1008–1014. doi: 10.1038/nsmb831. [DOI] [PubMed] [Google Scholar]
- 39.Blanchard S.C., Kim H.D., Gonzalez R.L., Jr., Puglisi J.D., Chu S. tRNA dynamics on the ribosome during translation. Proc. Natl. Acad. Sci. USA. 2004;101:12893–12898. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lipman E.A., Schuler B., Bakajin O., Eaton W.A. Single-molecule measurement of protein folding kinetics. Science. 2003;301:1233–1235. doi: 10.1126/science.1085399. [DOI] [PubMed] [Google Scholar]
- 41.Majumdar D.S., Smirnova I., Kasho V., Nir E., Kong X. Single-molecule FRET reveals sugar-induced conformational dynamics in LacY. Proc. Natl. Acad. Sci. USA. 2007;104:12640–12645. doi: 10.1073/pnas.0700969104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Munro J.B., Altman R.B., O'Connor N., Blanchard S.C. Identification of two distinct hybrid state intermediates on the ribosome. Mol. Cell. 2007;25:505–517. doi: 10.1016/j.molcel.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Munro J.B., Sanbonmatsu K.Y., Blanchard S.C. A new view of protein synthesis: mapping the free energy landscape of the ribosome using single-molecule FRET. Biopolymers. 2008;89:565–577. doi: 10.1002/bip.20961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schuler B. Single-molecule fluorescence spectroscopy of protein folding. ChemPhysChem. 2005;6:1206–1220. doi: 10.1002/cphc.200400609. [DOI] [PubMed] [Google Scholar]
- 45.Schuler B. Application of single molecule Förster resonance energy transfer to protein folding. Methods Mol. Biol. 2007;350:115–138. doi: 10.1385/1-59745-189-4:115. [DOI] [PubMed] [Google Scholar]
- 46.Weiss S. Fluorescence spectroscopy of single biomolecules. Science. 1999;283:1676–1683. doi: 10.1126/science.283.5408.1676. [DOI] [PubMed] [Google Scholar]
- 47.Weiss S. Measuring conformational dynamics of biomolecules by single molecule fluorescence spectroscopy. Nat. Struct. Biol. 2000;7:724–729. doi: 10.1038/78941. [DOI] [PubMed] [Google Scholar]
- 48.Xie X.S., Lu H.P. Single-molecule enzymology. J. Biol. Chem. 1999;274:15967–15970. doi: 10.1074/jbc.274.23.15967. [DOI] [PubMed] [Google Scholar]
- 49.Zhuang X., Bartley L.E., Babcock H.P., Russell R., Ha T. A single-molecule study of RNA catalysis and folding. Science. 2000;288:2048–2051. doi: 10.1126/science.288.5473.2048. [DOI] [PubMed] [Google Scholar]
- 50.Zhuang X., Rief M. Single-molecule folding. Curr. Opin. Struct. Biol. 2003;13:88–97. doi: 10.1016/s0959-440x(03)00011-3. [DOI] [PubMed] [Google Scholar]
- 51.Ditzler M.A., Aleman E.A., Rueda D., Walter N.G. Focus on function: single molecule RNA enzymology. Biopolymers. 2007;87:302–316. doi: 10.1002/bip.20819. [DOI] [PubMed] [Google Scholar]
- 52.Bates M., Huang B., Dempsey G.T., Zhuang X. Multicolor super-resolution imaging with photo-switchable fluorescent probes. Science. 2007;317:1749–1753. doi: 10.1126/science.1146598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Betzig E., Patterson G.H., Sougrat R., Lindwasser O.W., Olenych S. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 54.Gustafsson M.G. Nonlinear structured-illumination microscopy: wide-field fluorescence imaging with theoretically unlimited resolution. Proc. Natl. Acad. Sci. USA. 2005;102:13081–13086. doi: 10.1073/pnas.0406877102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hell S.W. Far-field optical nanoscopy. Science. 2007;316:1153–1158. doi: 10.1126/science.1137395. [DOI] [PubMed] [Google Scholar]
- 56.Hess S.T., Girirajan T.P., Mason M.D. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys. J. 2006;91:4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rust M.J., Bates M., Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat. Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang B., Wang W., Bates M., Zhuang X. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science. 2008;319:810–813. doi: 10.1126/science.1153529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Longin A., Souchier C., Ffrench M., Bryon P.A. Comparison of anti-fading agents used in fluorescence microscopy: image analysis and laser confocal microscopy study. J. Histochem. Cytochem. 1993;41:1833–1840. doi: 10.1177/41.12.8245431. [DOI] [PubMed] [Google Scholar]
- 60.Widengren J., Chmyrov A., Eggeling C., Lofdahl P.A., Seidel C.A. Strategies to improve photostabilities in ultrasensitive fluorescence spectroscopy. J. Phys. Chem. A. 2007;111:429–440. doi: 10.1021/jp0646325. [DOI] [PubMed] [Google Scholar]
- 61.Grunwell J.R., Glass J.L., Lacoste T.D., Deniz A.A., Chemla D.S. Monitoring the conformational fluctuations of DNA hairpins using single-pair fluorescence resonance energy transfer. J. Am. Chem. Soc. 2001;123:4295–4303. doi: 10.1021/ja0027620. [DOI] [PubMed] [Google Scholar]
- 62.Florijn R.J., Slats J., Tanke H.J., Raap A.K. Analysis of antifading reagents for fluorescence microscopy. Cytometry. 1995;19:177–182. doi: 10.1002/cyto.990190213. [DOI] [PubMed] [Google Scholar]
- 63.Pappalardo R., Samelson H., Lempicki A. Long-pulse laser emission from rhodamine 6G. IEEE J Quantum Electron. 1970;6:716–725. [Google Scholar]
- 64.Weninger K., Bowen M.E., Choi U.B., Chu S., Brunger A.T. Accessory proteins stabilize the acceptor complex for synaptobrevin, the 1:1 syntaxin/SNAP-25 complex. Structure. 2008;16:308–320. doi: 10.1016/j.str.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fei J., Kosuri P., MacDougall D.D., Gonzalez R.L., Jr. Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Mol. Cell. 2008;30:348–359. doi: 10.1016/j.molcel.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 66.Cornish P.V., Ermolenko D.N., Noller H.F., Ha T. Spontaneous intersubunit rotation in single ribosomes. Mol. Cell. 2008;30:578–588. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dorywalska M., Blanchard S.C., Gonzalez R.L., Kim H.D., Chu S. Site-specific labeling of the ribosome for single-molecule spectroscopy. Nucleic Acids Res. 2005;33:182–189. doi: 10.1093/nar/gki151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Qin F. Restoration of single-channel currents using the segmental k-means method based on hidden Markov modeling. Biophys. J. 2004;86:1488–1501. doi: 10.1016/S0006-3495(04)74217-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Song L., Hennink E.J., Young I.T., Tanke H.J. Photobleaching kinetics of fluorescein in quantitative fluorescence microscopy. Biophys. J. 1995;68:2588–2600. doi: 10.1016/S0006-3495(95)80442-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Aitken C.E., Marshall R.A., Puglisi J.D. An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophys. J. 2008;94:1826–1835. doi: 10.1529/biophysj.107.117689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Giepmans B.N., Adams S.R., Ellisman M.H., Tsien R.Y. The fluorescent toolbox for assessing protein location and function. Science. 2006;312:217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.