Abstract
VEGF (vascular endothelial growth factor) plays an essential role in angiogenesis during development and in disease largely mediated by signalling events initiated by binding of VEGF to its receptor, VEGFR2 (VEGF receptor 2)/KDR (kinase insert domain receptor). Recent studies indicate that VEGF activates PKD (protein kinase D) in endothelial cells to regulate a variety of cellular functions, including signalling events, proliferation, migration and angiogenesis. To better understand the role of PKD in VEGF-mediated endothelial function, we characterized the effects of a novel pyrazine benzamide PKD inhibitor CRT5 in HUVECs (human umbilical vein endothelial cells). The activity of the isoforms PKD1 and PKD2 were blocked by this inhibitor as indicated by reduced phosphorylation, at Ser916 and Ser876 respectively, after VEGF stimulation. The VEGF-induced phosphorylation of three PKD substrates, histone deacetylase 5, CREB (cAMP-response-element-binding protein) and HSP27 (heat-shock protein 27) at Ser82, was also inhibited by CRT5. In contrast, CRT6, an inactive analogue of CRT5, had no effect on PKD or HSP27 Ser82 phosphorylation. Furthermore, phosphorylation of HSP27 at Ser78, which occurs solely via the p38 MAPK (mitogen-activated protein kinase) pathway, was also unaffected by CRT5. In vitro kinase assays show that CRT5 did not significantly inhibit several PKC isoforms expressed in endothelial cells. CRT5 also decreased VEGF-induced endothelial migration, proliferation and tubulogenesis, similar to effects seen when the cells were transfected with PKD siRNA (small interfering RNA). CRT5, a novel specific PKD inhibitor, will greatly facilitate the study of the role of PKD signalling mechanisms in angiogenesis.
Keywords: angiogenesis, CRT5 inhibitor, endothelium, protein kinase C (PKC), protein kinase D (PKD), vascular endothelial growth factor (VEGF)
Abbreviations: CREB, cAMP-response-element-binding protein; DAG, diacylglycerol; EBM, endothelial basal medium; EGF, epidermal growth factor; ERK, extracellular-signal-regulated kinase; FBS, fetal bovine serum; HDAC, histone deacetylase; HSP27, heat-shock protein 27; HUVEC, human umbilical vein endothelial cell; IMAP, immobilized metal-ion-affinity-based fluorescence polarization; MAPK, mitogen-activated protein kinase; MAKAPK2, MAPK-activated protein kinase 2; MTT, 3-(4,5-dimethylthiaziazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide; PKC, protein kinase C; PKD, protein kinase D; siRNA, small interfering RNA; VEGF, vascular endothelial growth factor
INTRODUCTION
PKDs (Protein kinase Ds) are DAG (diacylglycerol)-stimulated serine/threonine protein kinases, originally classified as a sub-group of the PKC (protein kinase C) family [1,2] but subsequently recognized to be a distinct subgroup of the calcium/calmodulin-dependent protein kinase family [3], only sharing significant homology to PKCs in the DAG-binding domain.
There are currently three known PKD isoforms: PKD1– PKD3. All three isoforms share the same basic structure consisting of a variable N-terminal regulatory domain and a C-terminal kinase domain, the latter with over 90% similarity between the isoforms [4]. PKD2 differs from PKD1 in having a serine-rich linker between the two N-terminal zinc-finger domains, present in all three isoforms, whereas PKD3 lacks the alanine- and proline-rich hydrophobic domain found at the N-terminus of PKD1 and PKD2.
Activation of PKD can occur by multiple pathways, the primary one being non-classical PKC-dependent activation [5,6] resulting from phosphorylation at serine residues within the catalytic domain (Ser744/Ser748 in mouse PKD1, equivalent to Ser738/Ser742 in human PKD1) [7], which occurs at the cell membrane following recruitment of both PKD and PKC by DAG. On PKD1 and PKD2, but not PKD3, there is also an autophosphorylation site near the C-terminus (Ser916/Ser876 in PKD1 and PKD2 respectively), which determines PKD tertiary structure and appears to be important for its interaction with other proteins, including 14-3-3 and PDZ proteins. PKD is predominantly localized in the cytosol, but can be translocated to the nucleus [8] or Golgi [4] after activation at the plasma membrane. The substrate consensus sequence for PKD-mediated phosphorylation is LXR(Q/K/E/M)(M/L/K/E/Q/A)S*XXXX, including a strong preference for a leucine residue at the −5 position, and several target proteins have previously been identified including HSP27 (heat-shock protein 27), HDAC (histone deacetylase) 5 and HDAC7 [9–12].
PKD has been implicated as a mediator in diverse cellular functions, including proliferation, survival, cellular trafficking and regulation of transcription. In endothelial cells PKD is activated by VEGF (vascular endothelial growth factor, also known as VEGF-A), an essential angiogenic factor, during development and in the pathogenesis of human pathologies, including cancer and eye diseases [13,14]. There is growing evidence that endothelial cellular responses to VEGF are mediated at least in part through PKD [15]. We previously showed that PKD is required for VEGF-mediated endothelial migration and tube formation [10], and other studies have demonstrated the role of PKD in VEGF-induced phosphorylation of HDACs, proliferation and migration [11,12]. Given the importance of PKD in VEGF-mediated angiogenesis, a pharmacological inhibitor would be a valuable biological tool for further investigation of PKD-mediated signalling, and could also give rise to novel anti-angiogenic therapeutics. A recent study described the effects of a specific PKD inhibitor in cancer cells [16], although subsequent work indicates that this compound is probably not specific for PKD [17]. In the present study we have characterized a novel specific PKD inhibitor CRT5 and investigate its effects on angiogenic processes in endothelial cells.
MATERIALS AND METHODS
Materials
VEGF-A165 was from R&D Systems. SB203580 and GF109203X were from Calbiochem. Antibodies against total ERK (extracellular-signal-regulated kinase) 1 and 2, total PKD, phospho-PKD (Ser744/Ser748), phospho-PKD1 (Ser916), total CREB (cAMP-response-element-binding protein), phospho-CREB (Ser133), total HSP27 and phospho-HSP27 (Ser82) were from Cell Signaling Technology. The antibody against phospho-HSP27 (Ser78) was from Upstate Biotechnology, and the antibody against phospho-PKD2 (Ser876) was from Universal Biologicals. The antibodies against total and phospho-HDAC5 (Ser498) were obtained from Insight Biotechnology and Autogen Bioclear respectively. CRT5 was synthesized in-house at Cancer Research Technology [18]. CRT6 is a CRT5 analogue with the same core chemical structure indicated in [18]. All other reagents were of the highest grade available.
Cell culture
HUVECs (human umbilical vein endothelial cells) were obtained from TCS CellWorks and were cultured on gelatin-coated plates in EBM (endothelial basal medium; Cambrex BioScience) supplemented with gentamycin–ampicillin, EGF (epidermal growth factor), bovine brain extract (Singlequots; Cambrex BioScience) and 10% (v/v) FBS (fetal bovine serum) (complete EBM). For experimental purposes, fully confluent HUVECs at passages 2–4 were pre-incubated overnight with 0.5% FBS in EBM prior to addition of factors and other treatments.
Compound screening, kinase profiling and IC50 determination
Initially, inhibitors of PKD were identified by screening compounds for their ability to inhibit PKD1 in an in vitro kinase assay using a purified recombinant His6-tagged PKD1 kinase domain expressed in baculovirus (provided by Dr Harold Jefferies, Cancer Research UK London Research Institute, London, U.K.) and an IMAP (immobilized metal-ion-affinity-based fluorescence polarization) detection system (MDS Analytical Technologies). Compound libraries were obtained from the following companies: Maybridge, AsInEx, Bionet Research, Chembridge, Interbioscreen and Specs. Briefly, 12 nM recombinant PKD1 kinase domain [in 20% (v/v) glycerol] was mixed with 200 nM substrate {recombinant MAKAPK2 [MAPK (mitogen-activated protein kinase)]-activated protein kinase 2; MDS Analytical Technologies} and compound for screening [1.2 μM in DMSO; final DMSO concentration of 4% (v/v)] in assay buffer (25 mM Hepes, pH 7.5, and 2 mM MgCl2). ATP was then added (10 μM final concentration) in a total assay volume of 30 μl. The assay mixture was incubated for 1 h at room temperature (21 °C) followed by the addition of 20 μl of IMAP binding solution (MDS Analytical Technologies) and a further 2 h room temperature incubation in the dark. The fluorescence was then read on an Analyst HT plate reader (MDS Analytical Technologies).
Specificity of CRT5 was determined by in vitro kinase assays using a commercial kinase profiling service (Millipore). The IC50 values for CRT5 inhibition with PKD1–PKD3 were determined in vitro using IMAP. Briefly, 1 nM recombinant active PKD [in 20% (v/v) glycerol and assay buffer] was mixed with 200 nM recombinant MAKAPK2 and CRT5 (0.1 nM–1.2 μM) in a total volume of 5 μl in assay buffer. After the addition of 10 μM ATP (in assay buffer), the mixture was incubated for 1 h at room temperature, followed by the addition of 20 μl of IMAP binding solution and a further 2 h of incubation at room temperature in the dark. The fluorescence was then read on an Analyst HT plate reader.
Cell viability assay
HUVECs were seeded in a 96-well plate at a density of 1.5×104 cells/well. Cells were incubated with the PKD inhibitor CRT5 (5 nM–100 μM) in complete EBM. After 24 h 0.5 mg/ml MTT [3-(4,5-dimethylthiaziazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide] was added to each well and incubated for a further 4 h. The medium was then removed and the cells were washed twice with PBS, followed by the addition of 200 μl of DMSO. After 15 min the resulting colour intensity was read at 570 and 650 nm.
Immunoblotting
Cells were pre-treated with inhibitors in serum-free medium for 30 min, followed by a 10 min treatment with VEGF. After treatments, cells were washed with PBS and lysed in buffer containing 50 mM Tris/HCl, pH 7.4, 150 mM NaCl, 1 mM disodium EDTA, 1% (v/v) Igepal CA-630, 0.5% sodium deoxycholate and 0.1% SDS, and supplemented with phosphatase inhibitor cocktails SigmaPhos 1 and 2, and Complete™ protease inhibitor cocktail (Roche Diagnostics). Whole-cell lysates were homogenized by brief sonication, then SDS/PAGE sample buffer [62.5 mM Tris/HCl, pH 6.8, 100 mM DTT (dithiothreitol), 2% (w/v) SDS, 10% (v/v) glycerol and 0.002% Bromophenol Blue] was added and the sample was heated to 95–100 °C for 3 min. Equivalent amounts of protein were separated by SDS/PAGE and transferred on to PVDF membranes (Millipore). Membranes were blocked with 5% (w/v) non-fat dry milk in 0.1% Tween 20 in PBS, for 1–2 h at room temperature, before being probed with the primary antibody by overnight incubation at 4 °C. Detection was via a horseradish peroxidase-linked secondary antibody (Dako) and ECL (enhanced chemiluminescence) plus reagents (GE Healthcare), following the manufacturer's protocol. Autoradiograms of immunoblots were digitized on an Image Scanner (Amersham) and individual protein bands were quantified with ImageJ software (http://rsb.info.nih.gov/nih-image/Default.html). Any differences in protein phosphorylation due to variations in loading were routinely corrected by normalizing to the levels of the appropriate total protein.
Cell migration
Transwell cell culture inserts made of transparent low-pore-density PET (polyethylene terephthalate) with an 8 μm pore size (Falcon; BD Biosciences) were inserted into a 24-well plate. Serum-free medium supplemented with or without 25 ng/ml VEGF-A165 or vehicle was placed in the bottom chamber and HUVECs in suspension (1.5×105 cells/well in serum-free EBM) were added to the top chamber and incubated at 37 °C in for 4 h. HUVECs that had not migrated or had only adhered to the upper side of the membrane were removed before membranes were fixed and stained with a Reastain Quik-Diff kit (IBG Immucor) using the manufacturer's protocol and mounted on to glass slides. HUVECs that had migrated to the lower side of the membrane were counted in four random fields per well at ×20 magnification using an eye-piece-indexed graticule.
Proliferation
Cell proliferation was assessed by direct counting as described by Liu et al. [19]. Briefly, 3.2×104 cells/ml were allowed to attach to gelatin-coated 24-well plates for 4 h. Then the complete medium was then replaced with 0.5% FBS/EBM with 2.5 μM CRT5 or vehicle. After 30 min, cells were treated with 25 ng/ml VEGF or vehicle. After 48 h, cells were trypsinized and counted using a Sysmex CDA-500 cell counter.
Tubulogenesis
Cell culture plates (12-well) were coated (750 μl per well) with PureCol Collagen (Nutacon) solution [80% (v/v) in 0.01 M NaOH and M199 medium], which was allowed to solidify at 37 °C. HUVECs, transiently transfected with siRNA (small interfering RNA) as indicated, were then suspended in EBM containing 0.3% FBS, and overlaid on to the gel (1×105 cells/well) in the presence or absence of 25 ng/ml VEGF and incubated for 16 h at 37 °C. Digital photographs of cells were collected using the OpenLab Improvision system. Representative fields were selected and photos were taken of each well at ×10 magnification. The area of linear tubule formation was calculated using Image J software. Areas of tubule formation were averaged between four fields of view and expressed as percentages of control untreated cells.
Transfection with PKD siRNA
Specific siRNAs targeting PKD1 and PKD2 (Ambion) were resuspended in nuclease-free water to yield a stock concentration of 50 μM. HUVECs in six-well plates were allowed to reach ~70% confluence before transfection. Both PKD1 and PKD2 siRNAs were diluted to 1 μM each and combined with Opti-MEM medium (Invitrogen), containing no serum or antibiotics, and incubated for 25 min with 10 μl of Oligofectamine™ reagent (Invitrogen) in a total volume of 200 μl. Thereafter, HUVEC monolayers were washed with Opti-MEM medium and the complexes were added on to the cells at a final siRNA concentration of 200 nM each. In parallel wells, Silencer™ control siRNA (Ambion), which is a non-targeting scrambled siRNA, was used at 400 nM. After transfection for 4 h, 10% (v/v) FBS was added to the medium and was incubated overnight. The medium was then replaced with complete EBM and the cells incubated for a further 24 h. The knockdown of PKD1 and PKD2 was then confirmed by immunoblotting.
Statistical methods
Statistical analysis of the differences in phosphorylation and migration was performed using one- or two-way ANOVA with post-hoc analysis using Bonferroni's test where appropriate. Results are shown as means±S.E.M. and P<0.05 was considered significant. Where necessary, results were logarithmically transformed prior to analysis, to satisfy the criteria for ANOVA non-linear regression analysis, using GraphPad Prism software.
RESULTS
Toxicity, sensitivity and specificity of the PKD inhibitor
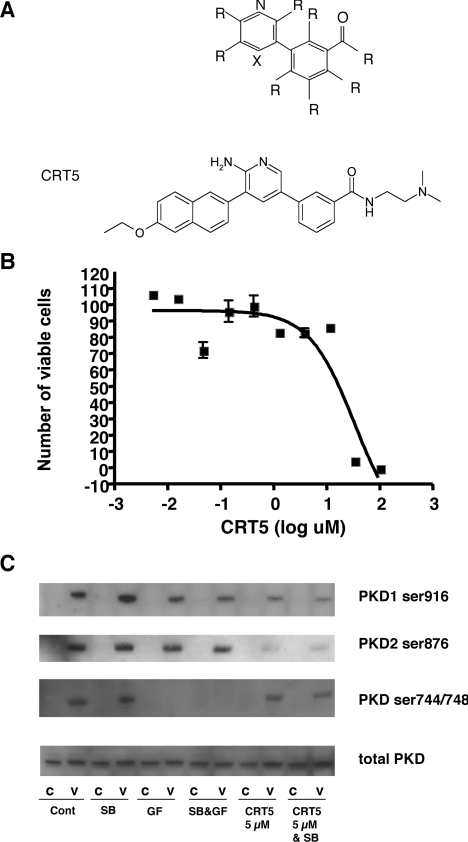
Initial screening of a diverse library containing approx. 55000 compounds in an in vitro assay of PKD1 kinase activity identified several compounds that inhibited the activity of PKD1. Of interest were pyridine benzamides and pyrazine benzamides, all with the core structure depicted in Figure 1(A) [18], including the compound CRT5 (structure shown in Figure 1A). The cytotoxicity of CRT5 was determined in HUVECs using an MTT-based assay. These results showed that CRT5 had an LD50 value of 17 μM as established by non-linear regression analysis (Figure 1B), which was very similar to the cytotoxicity of this compound in cancer cell lines (results not shown). The biochemical IC50 value of CRT5, as determined by inhibition of peptide substrate phosphorylation, was similar for all three PKD isoforms at 1, 2 and 1.5 nM for PKD1, PKD2 and PKD3 respectively. The specificity of CRT5 for PKD was also initially determined in an in vitro kinase assay, which included PKCα, PKCδ and PKCε, the major PKC isoforms expressed in HUVECs [20]. At 1 μM, CRT5 completely inhibited PKD1 and PKD2 as expected, but had little inhibitory effect on any of the PKC isoforms tested (Table 1). In addition, in a multi-kinase screen, CRT5 at 1 μM had little effect (<15% inhibition) on the activity of other serine/threonine and tyrosine protein kinases, including aurora-A, calmodulin-activated kinase 1, Cdk2 (cyclin-dependent kinase 2), c-Raf, c-Src, EGFR (EGF receptor), GSK3β (glycogen synthase kinase 3β), IKKα (IκB kinase α), JAK (Janus kinase)-2, MAPKAPK2, MEK (MAPK/ERK kinase)-1, PAK (p21-activated kinase)-2, PDGFR (platelet-derived growth factor receptor)-β, Akt/PKBα (protein kinase Bα), ROCK (Rho-associated kinase)-2 and RSK (ribosomal S6 kinase)-1. Treatment of HUVECs with 5 μM CRT5 inhibited VEGF-induced phosphorylation of PKD1 at Ser916 and PKD2 at the corresponding site, Ser876 (Figure 1C). In contrast, PKD phosphorylation at Ser744/Ser748 was completely unaffected by CRT5 treatment, whereas the non-selective PKC inhibitor, GF109203X completely inhibited phosphorylation at these sites but caused little decrease in phosphorylation at Ser916/Ser876. The differential effects of CRT5 and GF109203X on PKD phosphorylation are readily explained by the lack of effect of CRT5 on PKC-dependent PKD phosphorylation, and CRT5 inhibition of subsequent PKD activity, demonstrated by the reduction in PKD autophosphorylation at Ser916/Ser876.
Figure 1. Structure, toxicity and specificity of CRT5.
(A) Core structure of PKD pyridine benzamide (upper panel) and pyrazine benzamide inhibitors, where R indicates one of several different functional groups, and X is either a nitrogen (N) or carbon covalently linked to a functional group (C–R). Further details of the structure of these compounds are provided in [18]. The structure of CRT5 is shown in the lower panel. (B) HUVECs were incubated with CRT5 (5 nM–100 μM) for 24 h. Cell viability was estimated by an MTT-based assay and plotted against the log CRT5 concentration. Non-linear regression analysis was used to determine the LD50 value. (C) Confluent HUVECs were pre-incubated for 30 min with vehicle (Cont; 0.1% DMSO), 5 μM SB203580 (SB), 3 μM GF109203X (GF), 5 μM SB203580 plus 3 μM GF109203X (SB&GF) or 5 μM CRT5 with and without 5 μM SB203580 (CRT5&SB and CRT5 respectively) as indicated. This was followed by 10 min stimulation with vehicle (C) or 25 ng/ml VEGF (V). Cell lysates were immunoblotted and probed with antibodies against phospho-PKD1 (Ser916 or Ser744/Ser748), phospho-PKD2 (Ser876) or total PKD as indicated.
Table 1. Kinase specificity of CRT5.
In vitro kinase assays showed that, at 1 μM, CRT5 strongly inhibited PKD but had little effect on a range of PKC isoforms.
| Kinase | Inhibition (% at 1 μM CRT5) |
|---|---|
| PKCα | 5 |
| PKCβI | <1 |
| PKCβII | <1 |
| PKCγ | 23 |
| PKCδ | 1 |
| PKCε | <1 |
| PKCη | <1 |
| PKCι | 10 |
| PKCΦ | 2 |
| PKCξ | <1 |
| PKD1 | 98 |
| PKD2 | 96 |
CRT5 inhibits PKD substrate phosphorylation
To further investigate the effects of CRT5 on PKD activity in the response of endothelial cells to VEGF, we tested its ability to inhibit the phosphorylation of proteins previously identified as PKD substrates: HSP27 (Ser82), HDAC5 and CREB. In endothelial cells two of these substrates, HSP27 and CREB, are also phosphorylated via the p38 MAPK pathway after VEGF activation [10,21]. Therefore to distinguish between the role of PKD and p38 MAPK pathways, we tested CRT5 in the presence or absence of the p38 MAPK inhibitor SB203580.
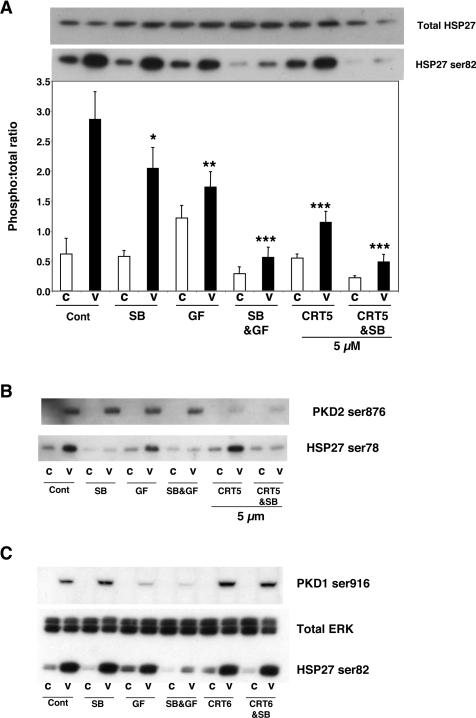
VEGF increased HSP27 phosphorylation at Ser82 approx. 4-fold. CRT5 alone, significantly reduced VEGF-induced HSP27 Ser82 phosphorylation, similar to the effect seen with the PKC inhibitor GF109203X alone (Figure 2A). Importantly, CRT5 in combination with SB203580 completely blocked HSP27 Ser82 phosphorylation, whereas SB203580 and CRT5 alone caused only partial inhibition (Figure 2A). The effect of CRT5 is therefore strikingly similar to the effect of GF109203X, which, in combination with SB203580, also completely abrogated HSP27 Ser82 phosphorylation (Figure 2A), and also similar to the inhibitory effects of PKD knockdown using targeted siRNA, as previously reported by us [10]. Unlike HSP27 Ser82, HSP27 Ser78 is not phosphorylated via PKD but is activated through the p38 MAPK pathway [10]. Consistent with PKD-independent VEGF-induced Ser78 phosphorylation, CRT5 or GF109203X had no effect on Ser78 phosphorylation, whereas SB203580 alone completely inhibited it (Figure 2B). In addition, CRT6, an analogue of CRT5 with the same core structure, that causes no inhibition of PKD at equivalent concentrations in vitro (results not shown), had no effect on either PKD Ser916 or HSP27 Ser82 phosphorylation at 5 μM (Figure 2C).
Figure 2. Effect of CRT5 on HSP27 phosphorylation.
(A) Confluent HUVECs were pre-incubated for 30 min with vehicle (Cont; 0.1% DMSO), 5 μM SB203580 (SB), 3 μM GF109203X (GF), 5 μM SB203580 plus 3 μM GF109203X (SB&GF) or 5 μM CRT5 with and without 5 μM SB203580 (CRT5&SB and CRT5 respectively) as indicated. This was followed by 10 min stimulation with vehicle (C) or 25 ng/ml VEGF (V). Cell lysates were immunoblotted and probed with antibodies against total and phosphorylated HSP27 (Ser82). Results from four independent experiments were quantified by scanning densitometry and are presented as a ratio of HSP27 phosphorylation corrected for total HSP27 (means±S.E.M., arbitrary units). A representative blot corresponding to the quantified results is shown above the histogram. Two-way ANOVA indicated significant treatment, inhibitor and treatment–inhibitor interaction effects (P<0.005). Significant differences of interest between treatments and inhibitors are indicated on the histogram: *P<0.05, **P<0.01 and ***P<0.001 compared with VEGF alone as determined by Bonferroni's post hoc test. (B) HUVECs treated as detailed in (A) were immunoblotted and probed with antibodies against phospho-HSP27 (Ser78) and phospho-PKD2 (Ser876). (C) Confluent HUVECs were pre-incubated for 30 min with vehicle (Cont), 5 μM SB203580 (SB), 3 μM GF109203X (GF), 5 μM SB203580 plus 3 μM GF109203X (SB&GF) or 5 μM CRT6 with and without 5 μM SB203580 (CRT6&SB and CRT6 respectively) followed by 10 min treatment with vehicle (C) or 25 ng/ml VEGF (V). Cell lysates were immunoblotted and probed with antibodies against PKD phosphorylated at Ser916, HSP27 (Ser82) and total ERK as indicated.
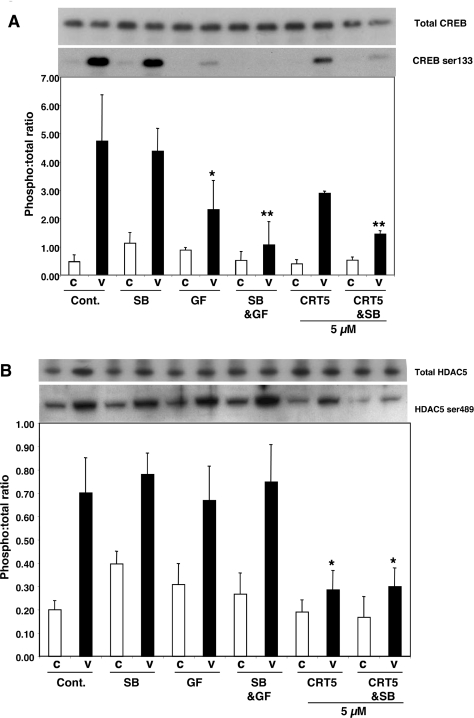
The effect of CRT5 on CREB phosphorylation was similar to that seen with HSP27 Ser82 phosphorylation. CRT5 alone partially reduced the VEGF-induced CREB phosphorylation, in a similar manner to GF109203X, whereas SB203580 caused no significant decrease in CREB phosphorylation. However, CRT5 in combination with SB203580 caused a more marked inhibition of VEGF-induced CREB phosphorylation, and GF109203X with SB203580 also inhibited this response more strongly (Figure 3A).
Figure 3. Effect of CRT5 on CREB and HDAC5 phosphorylation.
(A) Confluent HUVECs were pre-incubated for 30 min with vehicle (Cont.; 0.1% DMSO), 5 μM SB203580 (SB), 3 μM GF109203X (GF), 5 μM SB203580 plus 3 μM GF109203X (SB&GF) or 5 μM CRT5 with and without 5 μM SB203580 (CRT5&SB and CRT5 respectively) as indicated. This was followed by 10 min stimulation with vehicle (C) or 25 ng/ml VEGF (V). Cell lysates were immunoblotted and probed with antibodies against total and phosphorylated CREB (Ser133). Results from three independent experiments were quantified by scanning densitometry and are presented as a ratio of CREB phosphorylation corrected for total CREB (means±S.E.M., arbitrary units). A representative blot corresponding to the quantified results is shown above the histogram. Two-way ANOVA indicated significant treatment and inhibitor effects (P<0.0001 and P<0.05 respectively). Significant differences of interest between treatments and inhibitors are indicated on the histogram: *P<0.05 and **P<0.01 compared with VEGF alone as determined by Bonferroni's post hoc test. (B) Effect of CRT5 on HDAC5 phosphorylation. Cells were treated as detailed for (A). Lysates were immunoblotted and probed with an antibody against phospho-HDAC5 (Ser489). Results from four independent experiments were quantified by scanning densitometry and are presented as means±S.E.M. (arbitrary units). A representative blot corresponding to the quantified results is shown above the histogram. Two-way ANOVA indicated a significant treatment effect (P<0.0001). Significant differences of interest between treatments and inhibitors are indicated on the histogram: *P<0.05 compared with VEGF alone as determined by Bonferroni's post hoc test.
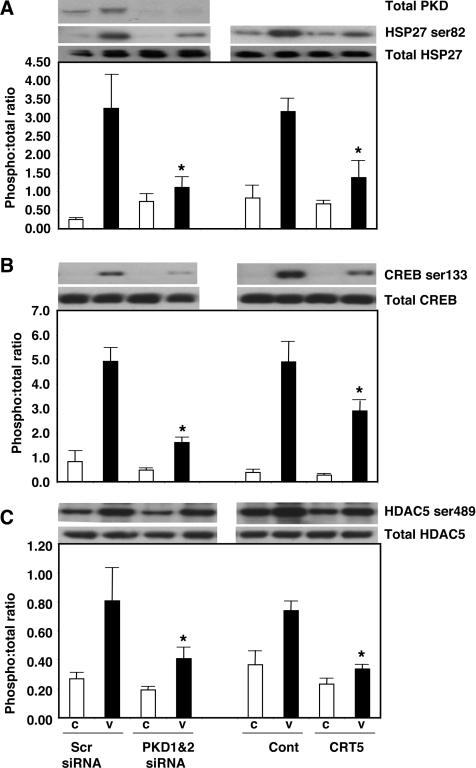
VEGF-induced HDAC5 phosphorylation was strongly inhibited by 5 μM CRT5, but, in contrast with HSP27 and CREB phosphorylation, the combination of CRT5 and SB203580 had no additional inhibitory effect. SB203580 alone had no inhibitory effect, confirming that HDAC5 is not activated via the p38 MAPK pathway. Interestingly, GF109203X, either alone or in combination with SB203580, also had no significant inhibitory effect on VEGF-induced HDAC5 phosphorylation (Figure 3B), in contrast with previous reports [11]. Similar to the effects of CRT5, in parallel cell cultures and treatments, knockdown of PKD1 and PKD2 in HUVECs using siRNA also significantly inhibited VEGF-induced CREB and HDAC5 phosphorylation (Figure 4).
Figure 4. Inhibition of PKD substrate phosphorylation by CRT5 and PKD knockdown.
(A) HUVECs were transfected with 200 nM siRNA targeting PKD1 and PKD2, or 400 nM of a control siRNA (Scr). At 48 h after transfection, cells were then treated for a further 10 min with no addition (C) or with 25 ng/ml VEGF (V). In parallel cell cultures, confluent HUVECs were pre-incubated for 30 min with either vehicle (Cont; 0.1% DMSO) or 5 μM CRT5, followed by 10 min stimulation with vehicle (C) or 25 ng/ml VEGF (V). Cell lysates were then immunoblotted with antibodies against total and phosphorylated HSP27 (Ser82). Knockdown of PKD1 and PKD2 was confirmed using an antibody that recognizes both PKD1 and PKD2 [10]. Results from at least three independent experiments were quantified by scanning densitometry and are presented as the ratios of phospho-HSP27 to total HSP27 (means±S.E.M., arbitrary units). A representative blot corresponding to the quantified results is shown above the histogram. Two-way ANOVA indicated a significant interaction and treatment effects (P<0.05 and P<0.0001 respectively); *P<0.05 compared with VEGF plus Scr siRNA or VEGF plus vehicle as determined by Bonferroni's post hoc test. (B) HUVECs treated as detailed in (A) were immunoblotted with antibodies against total or phosphorylated CREB. Two-way ANOVA indicated a significant interaction, inhibitor and treatment effect (P<0.005); *P<0.01 compared with VEGF control siRNA or VEGF vehicle respectively as determined by Bonferroni's post hoc test. (C) HUVECs treated as detailed in (A) were immunoblotted with antibodies against total or phosphorylated HDAC5. Two-way ANOVA indicated a significant inhibitor and treatment effects (P<0.05 and P<0.001 respectively);*P<0.05 compared with Scr siRNA plus VEGF or vehicle plus VEGF as determined by Bonferroni's post hoc test.
PKD inhibition inhibits VEGF-induced migration, proliferation and in vitro angiogenesis
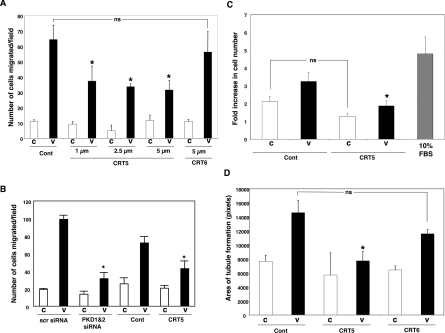
VEGF induced a 5.8-fold increase in directed cell migration compared with vehicle (Figure 5A) and pre-treatment of HUVECs with CRT5 resulted in a significant reduction (by 42–51%) in the migratory response towards VEGF (Figure 5A). In contrast, the inactive CRT5 analogue CRT6 had no significant effect on HUVEC migration. We also verified that, similarly to our previous results [10], knockdown of PKD1 and PKD2 significantly reduced VEGF-stimulated endothelial cell migration with a similar effect to that of CRT5 tested in parallel cell cultures and treatments (Figure 5B). VEGF treatment of HUVECs resulted in a 1.5-fold increase in cell proliferation over 48 h compared with controls (Figure 5C), and this increase was completely inhibited by pre-incubation with 2.5 μM CRT5. CRT5 caused some decrease in the proliferation of control cells not treated with VEGF, but this effect was not statistically significant. VEGF induced a 2-fold increase in HUVEC tubule formation in a collagen-based assay (Figure 5D); CRT5 also markedly inhibited VEGF-induced tubulogenesis, but had little effect on basal levels of tubulogenesis (Figure 5D), whereas CRT6 had no significant effect on tubule formation.
Figure 5. Effect of CRT5 on VEGF-induced migration and tubulogenesis.
(A) HUVECs were trypsinized and incubated for 30 min in the presence of vehicle (Cont; 0.08% DMSO), 1–5 μM CRT5 or 5 μM CRT6 before being added to the upper chamber of a Transwell migration assay chamber. Vehicle (C) or 25 ng/ml VEGF (V) was added to the lower chamber and the cells were allowed to migrate at 37 °C. After 4 h, the inserts were fixed, stained and migrated cells were counted. Quantified results are expressed as the mean number of cells migrating per field for at least three independent experiments. Migration towards vehicle is represented by white bars and towards VEGF by black bars. Results were analysed by one-way ANOVA; *P<0.05 compared with vehicle plus VEGF. (B) HUVECs were transfected with 200 nM PKD1 and PKD2 siRNA, or 400 nM control siRNA (scr), for 48 h. In a parallel set of experiments, non-transfected HUVECs were trypsinized and incubated for 30 min in the presence of vehicle (Cont; 0.08% DMSO) or 5 μM CRT5. Cells were then used to determine migration in response to 25 ng/ml VEGF in a Transwell migration assay as described in the Materials and methods section. Quantified results are expressed as the mean number of cells migrating per field for at least three independent experiments. Migration towards vehicle is represented by white bars and towards VEGF by black bars. Two-way ANOVA indicated a significant inhibitor, interaction and treatment effects (P<0.0001). Significant differences of interest are indicated on the histogram: *P<0.001 compared with scr siRNA plus VEGF or vehicle plus VEGF respectively as determined by Bonferroni's post hoc test. (C) HUVECs were seeded at a density of 3.2×104 cells/ml and allowed to attach to a gelatin-coated 24-well plate for 4 h. The complete medium was then replaced with 0.5% FCS/EBM with 2.5 μM CRT5 or vehicle (Cont) or 10% (v/v) FCS/EBM (grey bar). After 30 min, cells were treated with 25 ng/ml VEGF (black bars) or vehicle (white bars) as indicated. After 48 h, cells were trypsinized and counted using a Sysmex CDA-500 cell counter. The results are expressed as the fold increase in cell number from the original seeding density and are means±S.E.M. for three independent experiments. Results were analysed by one-way ANOVA; *P<0.05 compared with vehicle plus VEGF. (D) HUVECs were trypsinized and incubated for 30 min in the presence of vehicle (Cont; 0.08% DMSO), 5 μM CRT5 or 5 μM CRT6 in EBM containing 0.3% FCS. Cells were then plated on to a collagen base as described in the Materials and Methods section. Cells plated on to collagen were then incubated for 16 h either with no addition (C, white bars) or with 25 ng/ml VEGF (black bars). Tubulogenesis was quantified with ImageJ. ANOVA indicates that CRT5 significantly reduced VEGF-mediated tube formation whereas CRT6 had no significant effect. *P<0.05 compared with VEGF (no inhibitor) as determined by Bonferroni's post hoc test. Values (n≥3) are means±S.E.M., expressed as the area of tubules formed in pixels per field.
DISCUSSION
PKD has recently been implicated in many important cellular processes, such as proliferation, migration and transcription. In particular, recent studies have indicated that PKD plays a major role in VEGF-mediated endothelial cell functions and in angiogenesis. Until recently, however, studies of the role of PKD have been hampered by the lack of specific inhibitors. While the present study was in progress, effects of a PKD inhibitor, CID755673, were reported in a prostate cancer cell line [16]. CRT5 appears to be 100-fold more effective at PKD1 inhibition compared with CID755673 as determined by its IC50 value. Furthermore, a recent study found that CID755673 enhances mitogenic signalling through a PKD-independent pathway and concluded that this compound cannot be regarded as a specific PKD inhibitor [17]. In contrast with CID755673, CRT5, the PKD inhibitor used in the present study, does not prevent PKD phosphorylation by PKC, as shown by the lack of effect on VEGF-induced Ser744/Ser748 phosphorylation. CRT5 does, however, inhibit PKD kinase activity in vitro, and in intact endothelial cells, prevents the autophosphorylation of PKD1 and PKD2 at Ser916 and Ser876 respectively. This is in marked contrast with the effects of the widely used PKC inhibitor GF109203X, which strongly inhibited Ser744/Ser748 phosphorylation but had little effect on PKD autophosphorylation. The concentrations at which CRT5 inhibited PKD in intact endothelial cells are higher than those that inhibited PKD in cell-free assays using recombinant PKD. Such a difference is not unusual for pharmacological agents targeting intracellular enzymes and most likely reflects the cellular metabolism of CRT5 and/or its ability to penetrate the cell membrane. The differential effects of CRT5 and GF109203X, together with the in vitro kinase assay results, support the conclusion that the effect of CRT5 is highly unlikely to be due to inhibition of PKC activity. The specificity of CRT5 was further demonstrated by the observation that, although it partially inhibited HSP27 Ser82 phosphorylation, it did not inhibit VEGF-stimulated HSP27 Ser78 phosphorylation, which is mediated via the p38 MAPK pathway in a PKD-independent manner. We have not investigated whether CRT5 also inhibits PKD3, but results from our previous study indicated that this isoform was not involved in either VEGF-induced HSP27 phosphorylation or migration [10].
It has been proposed recently that Ser916 phosphorylation may not be an unambiguous marker of PKD activity [22]. It was therefore important to test whether CRT5 could block activation of known PKD substrates. HSP27 Ser82 has a strong consensus PKD phosphorylation site and is an established downstream target of PKD in vitro and in cells [9,10]. Consistent with these results, CRT5 pre-incubation reduced HSP27 Ser82 phosphorylation with an effect very similar to that seen with the non-selective PKC inhibitor GF109203X, both in the presence and absence of the specific p38 MAPK inhibitor SB203580, suggesting that both CRT5 and GF109203X act on the same pathway. A notable feature of these findings is that, although CRT5 and SB203580 acting alone caused only partial inhibition of HSP27 Ser82 phosphorylation, the combination of these inhibitors completely blocked Ser82 phosphorylation, consistent with the effects of GF109203X [10]. Furthermore, the effects of PKD inhibition on VEGF-induced HSP27 Ser82 phosphorylation are in agreement with those seen when PKD expression is reduced by siRNA [10]. The VEGF-induced phosphorylation of two other recently identified PKD substrates, CREB and HDAC5, was also inhibited by CRT5. The conclusion that the inhibitory effects of CRT5 on VEGF-stimulated phosphorylation of HSP27 Ser82, CREB and HDAC5 are mediated via inhibition of PKD is supported by the observation that targeted knockdown of PKD1 and PKD2 using siRNAs had a very similar inhibitory effect when compared directly with CRT5 in parallel cell cultures. Although we cannot preclude the possibility that PKD knockdown could have additional effects not caused by kinase inhibition, possibly mediated via kinase-independent protein–protein interactions, the effects of PKD siRNA treatment and CRT5 were in good agreement. The effects of CRT5 on CREB phosphorylation were broadly similar to those seen on HSP27 Ser82, with greater inhibition occurring in the combined presence of CRT5 with SB203580. In contrast, HDAC5 phosphorylation was strongly inhibited by CRT5 alone, with SB203580 having no additional effect. Surprisingly, GF109203X also had no effect on HDAC5 phosphorylation, in contrast with results published previously [11]. The reason for this discrepancy is unclear, but it may be explained by differences in HDAC5 regulation between bovine aortic endothelial cells as used by Ha et al. [11] and HUVECs used in the present study. In addition, it may signify that VEGF can additionally activate PKD via a PKC-independent pathway in these cells. For example, it has been reported that PKD can be activated by phosphorylation at Tyr463, which is mediated by the Src pathway and independent of PKC [23]. This may also explain why GF109203X appears to be less effective than CRT5 at inhibiting PKD phosphorylation at Ser916.
As PKD is thought to play a pivotal role in VEGF-induced angiogenesis [10,11,15,24], we investigated whether CRT5 could disrupt the mechanisms associated with angiogenesis in endothelial cells. CRT5 markedly inhibited the migration and proliferation of HUVECs in response to VEGF stimulation and reduced VEGF-induced in vitro angiogenesis in a collagen-based assay, whereas its inactive analogue CRT6, which does not inhibit PKD at similar concentrations, was unable to significantly disrupt migration or tube formation. The effects of CRT5 on migration and tubulogenesis are similar to those seen when PKD1 or PKD2 are knocked down with siRNA [10].
In conclusion, we have shown that CRT5 acts as a specific inhibitor of PKD in the response of endothelial cells to VEGF, providing further support for the conclusion that PKD is an important mediator of VEGF receptor signalling and biological functions. This compound and/or its derivatives may have great potential, both as a biochemical tool and as a starting point for novel anti-angiogenic therapeutic drug development.
AUTHOR CONTRIBUTION
Ian Evans carried out the research, analysed the data and wrote the paper. Azadeh Bagherzadeh and Alexandra Boakes performed experiments. Mark Charles and Tony Raynham synthesized essential experimental reagents. Lloyd Kelland contributed to the overall design of the study. Chris Ireson helped with writing the paper. Ian Zachary was involved in the study and experimental design and data analysis and wrote the paper.
FUNDING
This work was supported by the British Heart Foundation [grant number RG06/03/003 (to I.Z.)]
References
- 1.Valverde A. M., Sinnett-Smith J., Van Lint J., Rozengurt E. Molecular cloning and characterization of protein kinase D: a target for diacylglycerol and phorbal esters with a distinctive catalytic domain. Proc. Natl. Acad. Sci. U.S.A. 1994;91:8572–8576. doi: 10.1073/pnas.91.18.8572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johannes F. J., Prestle J., Eis S., Oberhagemann P., Pfizenmaier K. PKCμ is a novel, atypical member of the protein kinase C family. J. Biol. Chem. 1994;269:6140–6148. [PubMed] [Google Scholar]
- 3.Manning G., Whyte D. B., Martinez R., Hunter T., Sudar-Sanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 4.Rykx A., De Kimpe L., Mikhalap S., Vantus T., Seufferlein T., Vandenheede J. R., Van Lint J. Protein kinase D: a family affair. FEBS Lett. 2003;546:81–86. doi: 10.1016/s0014-5793(03)00487-3. [DOI] [PubMed] [Google Scholar]
- 5.Zugaza J. L., Sinnett-Smith J., Van Lint J., Rozengurt E. Protein kinase D (PKD) activation in intact cells through a protein kinase C-dependent signal transduction pathway. EMBO J. 1996;15:6220–6230. [PMC free article] [PubMed] [Google Scholar]
- 6.Zugaza J. L., Waldron R. T., Sinnett-Smith J., Rozengurt E. Bombesin, vasopressin, endothelin, bradykinin, and platelet-derived growth factor rapidly activate protein kinase D through a protein kinase C-dependent signal transduction pathway. J. Biol. Chem. 1997;272:23952–23960. doi: 10.1074/jbc.272.38.23952. [DOI] [PubMed] [Google Scholar]
- 7.Iglesias T., Waldron R. T., Rozengurt E. Identification of in vivo phosphorylation sites required for protein kinase D activation. J. Biol. Chem. 1998;273:27662–27667. doi: 10.1074/jbc.273.42.27662. [DOI] [PubMed] [Google Scholar]
- 8.Rey O., Sinnett-Smith J., Zhukova E., Rozengurt E. Rapid protein kinase D translocation in response to G protein-coupled receptor activation: dependence on protein kinase C. J. Biol. Chem. 2001;276:49228–49235. doi: 10.1074/jbc.M109395200. [DOI] [PubMed] [Google Scholar]
- 9.Doppler H., Storz P., Li J., Comb M. J., Toker A. A phosphorylation state-specific antibody recognizes Hsp27, a novel substrate of protein kinase D. J. Biol. Chem. 2005;280:15013–15019. doi: 10.1074/jbc.C400575200. [DOI] [PubMed] [Google Scholar]
- 10.Evans I. M., Britton G., Zachary I. C. Vascular endothelial growth factor induces heat shock protein (HSP) 27 serine 82 phsophorylation and endothelial tubulogenesis via protein kinase D and independent of p38 kinase. Cell. Signalling. 2008;20:1375–1384. doi: 10.1016/j.cellsig.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 11.Ha C. H., Wang W., Jhun B. S., Wong C., Hausser A., Pfizenmaier K., McKinsey T. A., Olsen E. N., Jin Z-G. Protein kinase D-dependent phosphorylation and nuclear export of histone deacetylase 5 mediates vascular endothelial growth factor-induced gene expression and angiogenesis. J. Biol. Chem. 2008;283:14590–14599. doi: 10.1074/jbc.M800264200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang S., Li X., Parra M., Verdin E., Bassel-Duby R., Olsen E. N. Control of endothelial cell proliferation and migration by VEGF signaling to histone deacetylase 7. Proc. Natl. Acad. Sci. U.S.A. 2008;105:7738–7743. doi: 10.1073/pnas.0802857105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferrara N., Gerber H-P., LeCouter J. The biology of VEGF and its receptors. Nat. Med. 2003;9:669–676.. doi: 10.1038/nm0603-669. [DOI] [PubMed] [Google Scholar]
- 14.Holmes D. I. R., Zachary I. The vascular endothelial growth factor family: angiogenic factors in health and disease. Genome Biol. 2005;6:209.1–209.10. doi: 10.1186/gb-2005-6-2-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qin L., Zeng H., Zhao D. Requirement of PKD tyrosine phosphorylation for VEGF-A165-induced angiogenesis through its interaction and regulation of phospholipase Cγ phsophorylation. J. Biol. Chem. 2006;281:32550–32558. doi: 10.1074/jbc.M604853200. [DOI] [PubMed] [Google Scholar]
- 16.Sharlow E. R., Giridhar K. V., LaValle C. R., Chen J., Leimgruber S., Barrett R., Bravo-Altamirano K., Wipf P., Lazo J. S., Wang Q. J. Potent and selective disruption of protein kinase D functionality by a benzoxoloazepinolone. J. Biol. Chem. 2008;283:33516–33526. doi: 10.1074/jbc.M805358200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Torres-Marquez E., Sinnett-Smith J., Guha S., Kui R., Waldron R. T., Rey O., Rozengurt E. CID755673 enhances mitogenic signaling by phorbol esters, bombesin and EGF through a protein kinase D-independent pathway. Biochem. Biophys. Res. Commun. 2010;391:63–68. doi: 10.1016/j.bbrc.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raynham T. M., Hammonds T. R., Charles M. D., Pave G. A., Foxton C. H., Blackaby W. P., Stevens A. P., Ekwuru C. T. Pyridine benzamides and pyrazine benzamides used as PKD inhibitors. Int. Pat. WO/2008/074997. 2008
- 19.Liu D., Evans I., Britton G., Zachary I. The zinc-finger transcription factor, early growth response 3, mediates VEGF-induced angiogenesis. Oncogene. 2008;27:2989–2998. doi: 10.1038/sj.onc.1210959. [DOI] [PubMed] [Google Scholar]
- 20.Gliki G., Abu-Ghazaleh R., Jezequel S., Wheeler-Jones C., Zachary I. Vascular endothelial growth factor-induced prostacyclin production is mediated by a protein kinase C (PKC)-dependent activation of extracellular signal-regulated protein kinases 1 and 2 involving PKC-δ and by mobilization of intracellular Ca2+ Biochem. J. 2001;353:503–512. doi: 10.1042/0264-6021:3530503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mayo L. D., Kesseler K. M., Pincheira R., Warren R. S., Donner D. B. Vascular endothelial cell growth factor activates CRE-binding protein by signaling through the KDR receptor tyrosine kinase. J. Biol. Chem. 2001;276:25184–25189. doi: 10.1074/jbc.M102932200. [DOI] [PubMed] [Google Scholar]
- 22.Rybin V. O., Guo J., Steinberg S. F. Protein kinase D1 autophosphorylation via distinct mechanisms at Ser744/Ser748 and Ser916. J. Biol. Chem. 2009;284:2332–2343. doi: 10.1074/jbc.M806381200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Storz P., Doppler H., Johannes F-J., Toker A. Tyrosine phosphorylation of protein kinase D in the pleckstrin homology domain leads to activation. J. Biol. Chem. 2003;278:17969–17976. doi: 10.1074/jbc.M213224200. [DOI] [PubMed] [Google Scholar]
- 24.Avkiran M., Rowland A. J., Cuello F., Haworth R. S. Protein kinase D in the cardiovascular system: emerging roles in health and disease. Circ. Res. 2008;102:157–163. doi: 10.1161/CIRCRESAHA.107.168211. [DOI] [PubMed] [Google Scholar]