Abstract
In this paper, we report on a general synthetic strategy for the assembly of glycopolymers that capitalizes on the intrinsic reactivity of reducing glycans toward hydrazides to form stable cyclic N-glycosides. We developed a poly(acryloyl hydrazide) (PAH) scaffold to which we conjugated a variety of reducing glycans ranging in structure from simple mono- and disaccharides to considerably more complex human milk and blood oligosaccharides. The conjugation proceeds under mild conditions with excellent ligation efficiencies and in a stereoselective manner, providing glycopolymers with pendant glycans accommodated mostly in their cyclic β-glycosidic form. Utilizing a biotin-terminated PAH scaffold prepared via RAFT polymerization, we quickly assembled a panel of glycopolymers that we microarrayed on streptavidin-coated glass. We then demonstrated that in these microarrays, the glycopolymer ligands bind lectins according to the structures of their pendant glycans. Importantly, glycopolymers containing biologically relevant branched oligosaccharides, such as sialyl Lewisx, as well as sulfated glycosaminoglycan-like epitopes can be readily prepared using our methodology.
Glycopolymers are an important class of synthetic macromolecules that emulate many structural and functional features of cell-surface glycoproteins.(1) These glycoproteins perform key regulatory functions in many essential biological processes, such as innate immunity, cellular communication, and pathogen invasion.(2) With mounting evidence that not only the molecular structure but also the valency and spatial organization of carbohydrate epitopes in glycoproteins influence the specificity and avidity of their interactions with cognate receptors,(3) glycopolymers have increasingly served as a tool for probing the underlying mechanisms of these events.(4) In addition, we have been interested in using glycopolymer mimetics of mucin glycoproteins to control the presentation of carbohydrates in glycan microarrays,(5) which have emerged as a powerful platform for interrogating ligand specificities of carbohydrate-binding proteins.(6)
Although many inventive approaches for the synthesis of glycopolymers have been developed,(1) none is particularly suitable for the rapid, high-throughput assembly of glycopolymer libraries we envisioned for our microarrays. Currently, two general synthetic strategies exist for the generation of glycopolymers that bear glycans in their native form (i.e., as closed cyclic pyranosides appended to the polymer scaffold through a glycosidic linkage). One strategy relies on polymerization of glycan-containing monomers,(7) while the other is based on attachment of appropriately prefunctionalized glycosides to polymer backbones endowed with complementary reactive groups.(8) Both approaches require chemical elaboration of the glycan component, which makes introduction of complex glycans into glycopolymers synthetically challenging and time-consuming.
Free reducing sugars, which are often available from natural sources, react with α-heteroatom nucleophiles (e.g., hydrazide or alkylaminooxy groups) to give cyclic pyranoside adducts in predominantly β-anomeric form.(9) In fact, this intrinsic reactivity of carbohydrates has been skillfully employed in numerous applications, including glycan capture for structural and functional glycomics,(10) assembly of synthetic glycoconjugates,(11) and immobilization of sugars in glycan arrays.(12)
Here we report a general strategy for the synthesis of glycopolymers that is based on the ligation of free reducing sugars to an acryloyl hydrazide polymer scaffold. This approach eliminates the inconvenience and limitations of laborious carbohydrate prefunctionalization presently required for glycopolymer synthesis and offers rapid access to glycopolymers with a broad scope of glycan structures suitable for high-throughput microarray applications.
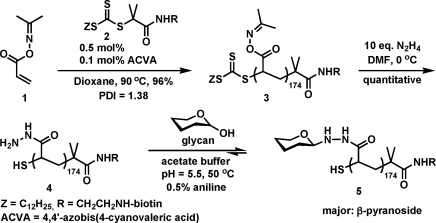
We synthesized biotin-terminated poly(acryloyl hydrazide) scaffold 4 in two steps (Scheme 1). First, we used the reversible addition−fragmentation chain transfer (RAFT) polymerization of acetoxime acrylate (1)(13) in the presence of the biotinylated chain-transfer agent 2 to obtain polymeric intermediate 3. The activated acetoxime esters in 3 were then cleanly converted into the corresponding hydrazides upon treatment with excess hydrazine. Importantly, NMR and elemental analyses of 4 showed no evidence that intramolecular cyclization or cross-linking took place during this step.
Scheme 1. Synthesis of Glycopolymers 5.
Conjugation of a wide range of reducing sugars to 4 proceeded smoothly under acidic conditions in the presence of aniline catalyst(14) to provide glycopolymers 5 (Table 1). The closed β-pyranoside was the predominant glycan form in 5 (Table 1). We obtained glycopolymers containing common mono-, di-, and trisaccharides in good to excellent yields. The ligation reaction is reversible, and its efficiency depends on the sugar size and the stability of the resulting conjugate. We found that 1.1 equiv of sugar at 0.25 M hydrazide group concentration was sufficient to force the equilibrium effectively toward product formation. The ligation efficiency improved with increasing sugar-to-hydrazide ratio and leveled out at ∼2 equiv of sugar (Table 1). The glycopolymers are stable to hydrolysis at pH ≥ 7. Generally, glycans with N-acetylhexosamine reducing ends showed somewhat diminished ligation efficiencies (Table 1, entries 10−14), presumably as a result of the increased steric demands of the C2 acetamido group. Notably, sulfated glycosaminoglycan-like disaccharide building blocks were also compatible with this ligation strategy (Table 1, entry 14).
Table 1. Conjugation of Reducing Sugars to Poly(acryloyl hydrazide) 4.
| isomeric ratio (%)b |
||||||
|---|---|---|---|---|---|---|
| entry | glycan | 5 | l.e. (%)a | β | α | open |
| 1 | glucose | a | 83 (85) | 80 | 18 | 2 |
| 2 | galactose | b | 85 (93) | 74 | 20 | 6 |
| 3 | mannose | c | 78 (90) | 72 | 22 | 6 |
| 4 | fucose | d | 75 (76) | 70 | 25 | 5 |
| 5 | rhamnose | e | 85 (86) | 64 | 28 | 8 |
| 6 | xylose | f | 89 (92) | 88c | 12 | |
| 7 | lactose | g | 75 (95) | 80 | 20 | − |
| 8 | 6α-mannobiose | h | 74 (85) | 73 | 18 | 9 |
| 9 | panose | i | 64 (67) | 88 | 12 | − |
| 10 | N-acetylglucosamine | j | 64 (67) | 74 | 21 | 5 |
| 11 | N-acetylgalactosamine | k | 66 (67) | 89c | 11 | |
| 12 | N-acetyllactosamine | l | 42 (42) | 86 | 14 | − |
| 13 | N,N-diacetylchitobiose | m | 37 (34) | 100c | − | |
| 14 | chondroitin Δdi-6S | n | 44 (44) | 83c | 17 | |
l.e. = ligation efficiency for the reaction of 4 with 1.1 (2.0) equiv of glycan.
Isomeric ratio determined by 1H NMR analysis.
The β/α ratio could not be determined. Glycan structures are shown in the Supporting Information.
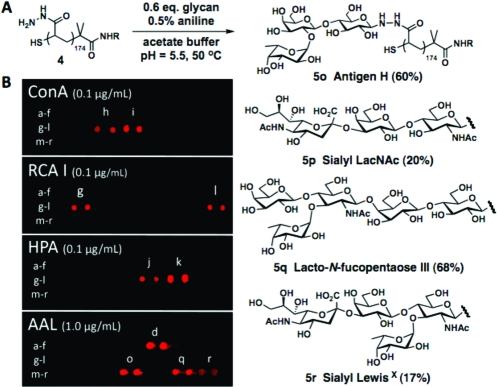
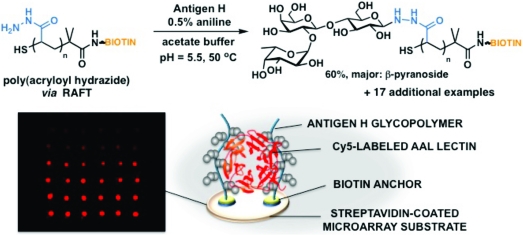
Glycopolymers containing complex glycans are readily accessible using our methodology (Figure 1A). In view of the scant availability of these complex epitopes, we optimized the ligation conditions for substoichiometric amounts of the glycan component (0.6 equiv/hydrazide) (Figure 1A). In agreement with our previous observations, antigen H and lacto-N-fucopentaose III were efficiently conjugated to the polymer backbone through their reducing glucose residues (60 and 68% ligation efficiency, respectively, based on glycan), while sialyl N-acetyllactosamine and sialyl Lewisx glycans bearing N-acetylglucosamine (GlcNAc) at their reducing termini were incorporated to a lesser extent (20 and 17%, respectively).
Figure 1.
Assembly of glycopolymers with complex glycans and microarray display. (A) Conjugation of human milk and blood oligosaccharides (ligation efficiencies based on glycan stoichiometry). (B) Microarrays of glycopolymers 5a−r on streptavidin-coated glass stained with Cy5-conjugated ConA, RCA I, HPA, and AAL lectins.
To evaluate the specificities of their interactions with lectins, we microarrayed polymers 5a−r on streptavidin-coated glass and tested for binding of Cy5-labeled concanavalin A (ConA), Ricinus communis I (RCA I), Helix pomatia agglutinin (HPA), and Aleuria aurantea lectin (AAL) (Figure 1B). As expected, ConA recognized the terminal α-mannose and α-glucose residues in polymers 5h and 5i, respectively.(15) Similarly, RCA I identified polymers 5g and 5l (but not 5q) presenting terminal galactose epitopes.(15) HPA bound to N-acetylgalactosamine-containing polymer 5k and less strongly to polymer 5j displaying GlcNAc, a much weaker HPA ligand.(16) Finally, fucose-specific AAL bound to glycopolymers bearing fucose (5d), antigen H (5o), lacto-N-fucopentaose (5q), and sialyl Lewisx (5r), all of which contain the target residue.(15)
In summary, we have developed a strategy for the expedient synthesis of glycopolymers for microarray applications via ligation of reducing sugars to backbones carrying hydrazide groups and a surface anchor. We used this methodology to assemble a panel of biotinylated glycopolymer ligands, which we microarrayed on streptavidin-coated glass slides. In the microarrays, they were recognized by lectins according to the structures of their pendant glycans, offering a promising new platform for multivalent ligand display in glycan arrays.
Acknowledgments
We thank Prof. Joseph DeRisi and Katherin Hermens for their help with microarray construction. The synthesis of glycopolymers was supported by the Office of Basic Energy Sciences, Division of Materials Sciences, U.S. Department of Energy, under Contract DE-AC03-76SF00098, and their biological evaluation in printed microarrays was supported in part by American Recovery and Reinvestment Act (ARRA) funds through Grant R01GM059907 to C.R.B. from the National Institutes of Health.
Supporting Information Available
Experimental procedures and characterization data. This material is available free of charge via the Internet at http://pubs.acs.org.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- a Spain S. G.; Gibson M. I.; Cameron N. R. J. Polym. Sci., Part A: Polym. Chem. 2007, 45, 2059–2072. [Google Scholar]; b Ladmiral V.; Melia E.; Haddleton D. M. Eur. Polym. J. 2004, 40, 431–449. [Google Scholar]; c Miura Y. J. Polym. Sci., Part A: Polym. Chem. 2007, 45, 5031–5036. [Google Scholar]
- Varki A. Glycobiology 1993, 3, 97–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Dam T. K.; Brewer C. F. Glycobiology 2010, 20, 270–279. [DOI] [PubMed] [Google Scholar]; b Lundquist J. J.; Toone E. J. Chem. Rev. 2002, 102, 555–578. [DOI] [PubMed] [Google Scholar]
- a Kiessling L. L.; Gestwicki J. E.; Strong L. E. Angew. Chem., Int. Ed. 2006, 45, 2348–2368. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Gordon E. J.; Gestwicki J. E.; Strong L. E.; Kiessling L. L. Chem. Biol. 2000, 7, 9–16. [DOI] [PubMed] [Google Scholar]; c Wang Q.; Dordick J. S.; Linhardt R. J. Chem. Mater. 2002, 14, 3232–3244. [Google Scholar]
- Godula K.; Rabuka D.; Nam K.; Bertozzi C. R. Angew. Chem., Int. Ed. 2009, 48, 4973–4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Paulson J. C.; Blixt O.; Collins B. E. Nat. Chem. Biol. 2006, 2, 238–248. [DOI] [PubMed] [Google Scholar]; b Liang P.-H.; Wu C.-Y.; Greenberg W. A. Curr. Opin. Chem. Biol. 2008, 12, 86–92. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Oyelaran O.; Gildersleeve J. C. Curr. Opin. Chem. Biol. 2009, 13, 406–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- For reviews, see ref (1) and:; Okada M. Prog. Polym. Sci. 2001, 26, 67–104. [Google Scholar]
- For representative examples, see:; a Bovin N. V. Glycoconjugate J. 1998, 15, 431–446. [DOI] [PubMed] [Google Scholar]; b Ladmiral V.; Mantovani G.; Clarkson G. J.; Cauet S.; Irwin J. L.; Haddleton D. M. J. Am. Chem. Soc. 2006, 128, 4823–4830. [DOI] [PubMed] [Google Scholar]
- a Takeda Y. Carbohydr. Res. 1979, 77, 9–23. [Google Scholar]; b Bendiak B. Carbohydr. Res. 1997, 304, 85–90. [DOI] [PubMed] [Google Scholar]; c Peri F.; Dumy P.; Mutter M. Tetrahedron 1998, 54, 12269–12278. [Google Scholar]; d Peluso S.; Imperiali B. Tetrahedron Lett. 2001, 42, 2085–2087. [Google Scholar]; e Gudmundsdottir A. V.; Paul C. E.; Nitz M. Carbohydr. Res. 2009, 344, 278–284. [DOI] [PubMed] [Google Scholar]
- a Zhao Y.; Kent S. B. H.; Chait B. T. Proc. Natl. Acad. Sci. U.S.A. 1997, 94, 1629–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Furukawa J. I.; Shinohara Y.; Kuramoto H.; Miura Y.; Shimaoka H.; Kurogochi M.; Nakano M.; Nishimura S. I. Anal. Chem. 2008, 80, 1094–1101. [DOI] [PubMed] [Google Scholar]; c Bohorov O.; Andersson-Sand H.; Hoffmann J.; Blixt O. Glycobiology 2006, 16, 21C–27C. [DOI] [PubMed] [Google Scholar]
- a Cervigni S. E.; Dumy P.; Mutter M. Angew. Chem., Int. Ed. 1996, 35, 1230–1232. [Google Scholar]; b Liu Y.; Feizi T.; Campanero-Rhodes M. A.; Childs R. A.; Zhang Y.; Mulloy B.; Evans P. G.; Osborn H. M. I.; Otto D.; Crocker P. R.; Chai W. Chem. Biol. 2007, 14, 847–859. [DOI] [PubMed] [Google Scholar]
- a Zhi Z.; Powell A. K.; Turnbull J. E. Anal. Chem. 2006, 78, 4786–4793. [DOI] [PubMed] [Google Scholar]; b De Boer A. R.; Hokke C. H.; Deelder A. M.; Wuhrer M. Anal. Chem. 2007, 79, 8107–8113. [DOI] [PubMed] [Google Scholar]; c Park S.; Lee M.-R.; Shin I. Bioconjugate Chem. 2009, 20, 155–162. [DOI] [PubMed] [Google Scholar]; d Zhou X.; Turchi C.; Wang D. J. Proteome Res. 2009, 8, 5031–5040. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Clo E.; Blixt O.; Jensen K. J. Eur. J. Org. Chem. 2010, 540–554. [Google Scholar]
- a Moad G.; Rizzardo E.; Thang S. H. Aust. J. Chem. 2009, 62, 1402–1472. [Google Scholar]; b Metz N.; Theato P. Eur. Polym. J. 2007, 43, 1202–1209. [Google Scholar]
- Dirksen A.; Dirksen S.; Hackeng T. M.; Dawson P. E. J. Am. Chem. Soc. 2006, 128, 15602–15603. [DOI] [PubMed] [Google Scholar]
- Cummings R. D.; Etzler M.. Essentials of Glycobiology, 2nd ed.; CSHL Press: New York, 2009; pp 633−648. [PubMed] [Google Scholar]
- Wu A. M.; Sugii S. Carbohydr. Res. 1991, 213, 127–143. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.