Abstract
HER2 (ErbB2/Neu) is a receptor tyrosine kinase belonging to the epidermal growth factor receptor (EGFR)/ErbB family and is overexpressed in 20–30% of human breast cancers. Although several crystal structures of ErbB kinases have been solved, the precise mechanism of HER2 activation remains unknown, and it has been suggested that HER2 is unique in its requirement for phosphorylation of Y877, a key tyrosine residue located in the activation loop. To elucidate mechanistic details of kinase domain regulation, we performed molecular dynamics simulations of a homology-modeled HER2 kinase structure in active and inactive conformations. Principal component analysis of the atomistic fluctuations reveals a tight coupling between the activation loop and catalytic loop that may contribute to alignment of residues required for catalysis in the active kinase. The free energy perturbation method is also employed to predict a role for phosphorylated Y877 in stabilizing the kinase conformations. Finally, simulation results are presented for a HER2/EGFR heterodimer and reveal that the dimeric interface induces a rearrangement of the αC helix toward the active conformation. Elucidation of the molecular regulatory mechanisms in HER2 will help establish structure-function relationships in the wild-type kinase, as well as predict mutations with a propensity for constitutive activation in HER2-mediated cancers.
Introduction
HER2 (ErbB2/Neu) is a member of the ErbB family of transmembrane receptor tyrosine kinases, which also includes the epidermal growth factor receptor (EGFR/ErbB1), ErbB3 (HER3), and ErbB4 (HER4). ErbB receptors are composed of an extracellular ligand-binding domain, a transmembrane segment, an intracellular protein tyrosine kinase domain, and a C-terminal tail harboring tyrosine phosphorylation sites (1). Under physiological conditions, ligand binding promotes homo- or heterodimerization of the receptors and activation of their cytoplasmic domains (2,3). Dimerization results in auto- or transphosphorylation of tyrosine residues in the C-terminal tail segments, which serve as docking sites for signaling molecules containing SH2 or PTB domains (4). The major signaling cascades activated by the ErbB family include the Ras-Raf-MEK-MAPK and PI3K-Akt pathways, both of which result in transcription of genes involved in cellular proliferation, differentiation, and migration (5). Aberrant activation of the ErbB network is frequently associated with cellular transformation and clinical malignancies such as lung, gastric, and breast cancers (6–12). Overexpression of HER2 results in constitutive, ligand-independent activation of kinase signaling and is found in 20–30% of human breast cancers, where it is correlated with an aggressive tumor phenotype (13,14).
Activation of the ligand-binding domain of the ErbB receptor triggers conformational changes within its cytoplasmic domain. To effect phosphoryl transfer of the γ-phosphate of ATP to tyrosine residues on target substrates, several key loops within the kinase domain must be appropriately positioned (15). Residues 844–850 in HER2 comprise the catalytic loop (C-loop), which is crucial in facilitating the phosphoryl transfer. The αC helix (residues 761–775) and the nucleotide-binding loop, or N-loop (residues 727–732), are responsible for coordination of the ATP and substrate tyrosine. The activation loop (A-loop) comprises residues 863–884 and governs activation of the kinase by regulating accessibility of the target substrate to the C-loop. The A-loop undergoes a significant conformational extension upon activation, uncovering the catalytic machinery and enabling binding of the tyrosine substrate to the C-loop (16).
In most protein kinases, the activation loop assumes its catalytically competent conformation only if it is first phosphorylated on a regulatory tyrosine residue within the A-loop (17). The regulatory tyrosine residue is Y877 in HER2 and Y845 in EGFR. Although phosphorylation of EGFR on Y845 has been observed experimentally, phosphorylation does not seem to be required for catalytic activity, as EGFR possessing a Y845F mutation is fully active (18). In contrast, the role of A-loop phosphorylation in HER2 is controversial, as several studies have highlighted the importance of Y877 phosphorylation for kinase activity (19,20). Xu et al. (21) reported that mutation of Y877 to phenylalanine in COS-7 cells results in decreased autophosphorylation of Y1248, a tyrosine located in the C-terminal tail of HER2. Therefore, it is possible that phosphorylation of Y877 augments HER2 kinase activity.
Several crystal structures of the extracellular and cytoplasmic domains of the ErbB family members have elucidated mechanisms of autoinhibition and activation (16,22–28). Recent structural studies demonstrate that EGFR and ErbB4 are activated through an asymmetric dimerization mechanism analogous to that of a cyclin bound to an activated cyclin-dependent kinase (CDK) (29,30). In this asymmetric arrangement, the C-lobe of the activator kinase interacts with the N-lobe of the CDK-like kinase, activating the CDK-like kinase through allosteric contacts. The dimeric interface is dominated by hydrophobic interactions between the C-lobe of the activator kinase and the αC helix of the kinase undergoing activation. The dimerization model postulates that all members of the ErbB family can serve as the activating kinase, whereas only the catalytically competent members (EGFR, HER2, and ErbB4) can become activated. A sequence alignment of the ErbB kinase domains reveals that the residues involved in the N- and C-lobe faces of the dimer are essentially invariant among the family members, suggesting that HER2 is likely to be activated by a similar asymmetric dimerization scheme.
In this work we performed molecular dynamics (MD) simulations of inactive and active HER2 structures to elucidate details of the mechanism by which the HER2 kinase domain is regulated and activated. We investigated the dimer-mediated allosteric activation of the HER2 kinase through dynamics simulations of a HER2/EGFR heterodimeric system and delineated the role of phosphorylation of the activation loop tyrosine residue, Y877, using the free energy perturbation (FEP) method.
Methods
Homology modeling of the HER2 kinase domain
The HER2 kinase domain was modeled in homology to EGFR, with which it shares 83% sequence identity, using the program MODELLER (31,32). The coordinates of the inactive EGFR kinase domain were downloaded from the Protein Data Bank (PDB code 2GS7) (30) and a sequence alignment between EGFR and HER2 was performed in MODELLER. Missing residues were built using the loop modeling algorithm in MODELLER, and hydrogen atoms were added by employing the hbuild routine in CHARMM27 (33). The structure of the active HER2 kinase domain was based on the model generated by Bagossi et al. (34). The Bagossi model, which was constructed in homology to active EGFR (PDB code 1M14) (16), was refined by applying the loop-modeling routine to segments of missing residues and performing additional energy minimization steps. The final models for the inactive and active HER2 kinase domains were assessed for stereochemical quality using PROCHECK (35). Our homology models are indirectly validated in that the crystal structure of ErbB4, a member of the ErbB family with relatively low percentage sequence identity to EGFR (79%), was recently published (29) and found to be structurally homologous to EGFR, with a root mean-square deviation (RMSD) (backbone atoms) of 1.36 Å for the inactive EGFR and ErbB4 structures, and 1.05 Å for the active structures.
MD simulations
Four different models of the HER2 kinase domain were constructed for analysis. Two models of the active conformation of the kinase were generated (one in which Y877 is phosphorylated and one in which Y877 is unphosphorylated) and analogous models were constructed for the inactive conformation. Each structure was explicitly solvated using the TIP3P model for water (36) and with the buffering distance set to 15 Å. Sodium (Na+) and chloride (Cl−) ions were added to achieve net electroneutrality of the system and an ionic strength of 75 mM. All Na+ and Cl− ions were placed at least 8 Å away from any protein atoms and from each other. The ions were positioned at points of electrostatic extrema using a Debye-Huckel potential calculated within the program Solvate 1.0 (37). The Y877 residue was phosphorylated by applying the phosphotyrosine patch in CHARMM27 (33). The entire system comprised 79,872 atoms for the active structure and 53,751 atoms for the inactive structure, the difference in system size being attributed to the extension of the C-tail in the active system.
All simulations were performed with NAMD (38) using CHARMM27 force-field parameters. Periodic boundary conditions were applied and the particle mesh Ewald algorithm (39) was adopted for the calculation of long-range electrostatic interactions. The integration time step was set to 2.0 fs and the RATTLE algorithm (40) was employed to constrain the lengths of all chemical bonds involving hydrogen atoms at their equilibrium values. Nonbonded van der Waals (VDW) interactions were treated by applying a switching function at 10 Å and truncating the VDW potential energy smoothly at a cutoff distance of 12 Å.
To prepare the system for MD simulation, the solvated structure was energy-minimized using a conjugate gradient algorithm (NAMD) to remove unfavorable contacts. The system was then heated to 300 K using the temperature reassignment method in NAMD. Subsequently, constant pressure and temperature (NPT) simulations were performed at 300 K and 1 atm to equilibrate the volume of the solvation box. Temperature and pressure were maintained using a Langevin piston coupling algorithm (41). After the NPT simulations, constant volume and temperature (NVT) simulations were performed in NAMD. Finally, a 10 ns production run was completed using the same parameters as the NVT simulations. All structural figures were prepared with the program VMD (42).
Construction of the HER2/EGFR heterodimer
The HER2-EGFR heterodimeric system was modeled on the structure of the EGFR homodimer published by Zhang et al. (30). The structure of the inactive HER2 kinase domain was aligned with the activated kinase in the EGFR homodimer, and the activating kinase (EGFR) remained in its crystallographically defined position. The structure was minimized to remove unfavorable contacts, and hydrogen atoms were added using the hbuild routine in CHARMM. The heterodimer was explicitly solvated using the TIP3P water potential with the buffering distance set to 15 Å for a total system size of 124,093 atoms. Sodium (Na+) and chloride (Cl−) ions were added to achieve net electroneutrality of the system and an ionic strength of 75 mM. All Na+ and Cl− ions were placed at least 8 Å away from any protein atoms and from each other. Minimization and MD steps were performed as for the monomeric systems, except that the production simulation was run for a total of 20 ns.
Hydrogen-bonding analysis
CHARMM was used to analyze the hydrogen bonds present in the 10 ns trajectory for each system. Hydrogen bonds were defined by a bond length cutoff of 3.4 Å and an angle cutoff of 150°. Bonds that fulfilled these criteria and were present in at least 60% of the trajectory were tabulated in CHARMM. Salt bridges were defined as hydrogen bonds occurring between an acidic and a basic residue and satisfying a bond length cutoff of 1.6 Å. All hydrogen bonds and salt bridges were also visualized in VMD (42) for the duration of the 10 ns simulation.
Calculation of the RMSD
The RMSD of the backbone atoms of the kinase domain was evaluated for each frame of the 10 ns trajectory using the RMSD Plugin in VMD (42). Each frame was aligned with the relevant reference structure, and the RMS distance between the molecules was then measured. The initial, unminimized conformation was chosen as the reference structure for each system. Calculations were performed for all backbone atoms as well as for specific loops in the kinase domain, including the A-loop and the αC helix.
Principal component analysis
A principal component analysis (PCA) was applied to each of the four monomeric structures to identify the main eigenvectors (3N directions) along which the majority of the complex motion is defined. The calculation is based on the diagonalization of the variance-covariance matrix of the atomic fluctuations along each MD trajectory to yield the set of eigenvectors (PCs) and associated eigenvalues. The eigenvectors represent the independent modes of atomic motion, and the eigenvalues reflect the contribution of the corresponding eigenvectors to the global fluctuation of the protein. PCA computes the covariance matrix as
where (i,j = 1,…,3N), and N is the total number of atoms with positions given by Cartesian coordinates x. The resulting matrix is diagonalized to compute the 3N independent (uncorrelated) eigenvectors, {ξi}, and the eigenvalues, {λi}, sorted in descending order. The angle brackets denote the time average over the entire trajectory. All global translations and rotations about the center of mass are removed before evaluation of σ. PCA of the HER2 structures was performed on an active site region that comprises all domains critical for catalysis, including the A-, C-, and N-loops and the αC helix. Specifically, the Cα atoms of the C-loop/A-loop region (residues 843–888) and the αC helix/N-loop region (residues 725–778) were chosen as the active site, and the analysis was applied to the NVT-equilibrated trajectory for each monomeric structure. The software program CARMA (43) was used to perform PCA on our system and to project out the atomic fluctuations due to the modes along the MD trajectory.
FEP simulations
The FEP method (44) was employed to compute the Helmholtz free energy difference between the Y877-unphosphorylated and phosphorylated structures. Calculations were performed using the alchemical FEP feature in NAMD (38,45). The Helmholtz free energy difference between two thermodynamic states connected by M intermediate, nonphysical substrates in the NVT ensemble is expressed as
where kB is the Boltzmann constant, T is the temperature, and is the potential energy function that depends on the Cartesian coordinates of the system, {x}, and the coupling parameter, λi, that connects the initial and the final states of the transformation. The dual-topology paradigm (46) was utilized, in which the initial and the final state are defined in terms of distinct, noninteracting topologies. Four different simulations were performed, including the transformation of Y877 to pY877 for the solvated unphosphorylated structures (inactive and active) and the transformation of pY877 to Y877 for the solvated phosphorylated systems. Each perturbation was divided into 48 windows with 20 ps of equilibration and 80 ps of data collection per window, producing a total of 4.8 ns of simulation time for each FEP transformation. Harmonic positional restraints were imposed on the sodium and chloride ions, and system electroneutrality was maintained by transforming counterions during the course of the alchemical perturbation. Estimation of error was based on two different sets of coordinates (i.e., the alchemical transformations were performed on configurations obtained after 5 and 10 ns of MD trajectory), and errors are reported in terms of the standard deviation from the mean ΔF. Another source of error involves the end points of the transformation, which correspond to the creation or elimination of a group of atoms and are subject to VDW clashes that result in end-point catastrophes (47,48). To obtain an accurate estimate of the free energy at the diverging end points, we increased the number of windows at the beginning and end of the FEP simulations, collecting data at several points with λ values close to zero or one.
Results
To elucidate the molecular mechanisms of kinase activation, four different models of the HER2 kinase domain were constructed based on homology to EGFR (see the Methods section and Fig. S1 (Supporting Material)). Two models of the active conformation of the kinase were generated, one in which Y877 is phosphorylated and one in which Y877 is unphosphorylated, and analogous models were constructed for the inactive conformation. Each solvated system was subjected to MD simulation for 10 ns and the trajectories were analyzed for conformational shifting as well as for key bonding patterns. All four structures were stable for the duration of the simulation, as indicated by the time-course plots of the RMSD of the backbone atoms in the A-loop and αC helix (Fig. S2 and Fig. S3). The αC helix exhibited greater fluctuations in the active systems but ultimately returned to its initial position at the end of the 10 ns production run.
PCA reveals coupling between the catalytic loop and the activation loop regions of the HER2 kinase
A PCA was applied to each of the four monomeric trajectories to characterize the global motions of the HER2 protein (see the Methods section). The objective of this examination was to delineate the differences in atomistic fluctuations among the four structures, as conformational rearrangement of the kinase domain is expected to correlate with dramatic changes in the dynamical behavior of the protein. The PCA calculation is based on the diagonalization of the variance-covariance matrix of the atomic fluctuations along each MD trajectory to yield the set of eigenvectors (PCs) and associated eigenvalues. The eigenvectors represent the independent modes of atomic motion, and the eigenvalues reflect the contribution of the corresponding eigenvectors to the global fluctuation of the protein. PCA of the HER2 structures was performed on an active site region that comprises all domains critical for catalysis, including the A-, C-, and N-loops and the αC helix. Specifically, the Cα atoms of the C-loop/A-loop region (residues 843–888) and the αC helix/N-loop region (residues 725–778) were chosen as the active site and the analysis was applied to the NVT-equilibrated trajectory for each monomeric structure. Fig. S4 summarizes the contributions of the top 10 PCs to the total motion of each system, and reveals that the first three eigenmodes account for the majority of the atomic fluctuations in each MD trajectory. The top three PCs contribute 65% (both active and inactive Y877-unphosphorylated structures; Fig. S4, A and B), 52% (Y877-phosphorylated active monomer; Fig. S4 C) and 59% (Y877-phosphorylated inactive monomer; Fig. S4 D) to the total motion of the protein. Furthermore, the top 10 eigenmodes account for 75–85% of the total variation in each system, indicating that the strongest correlations are well described by the top few eigenvectors.
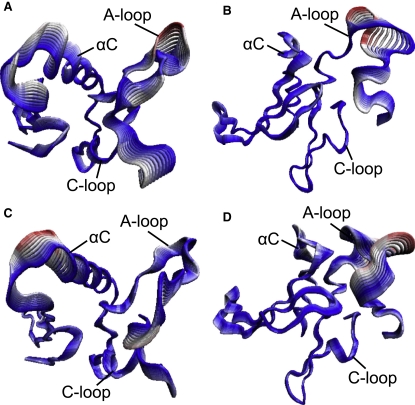
In the Y877-unphosphorylated active structure, the first principal motion involves a shifting of the αC helix in and out of the active site, coupled with a tilting of the N-loop (Fig. 1 A). In concert, the A-loop undergoes a slight bending and the C-loop swings toward the N-loop (an animated image is provided in Movie S1). Motion in the second eigenmode also corresponds to concerted fluctuations of all four domains: the αC helix tilts into the active site while the C-loop and N-loop bend toward each other. In contrast, the first and second eigenmodes for the inactive structure are dominated by A-loop movement and reveal little corresponding motion in the other loops, indicating that the four domains are less coupled in the inactive system (see Fig. 1 B and Movie S2). Analysis of the atomic fluctuations in the Y877-phosphorylated trajectories reveals a similar distinction between the inactive and active structures (compare Fig. 1, C and D; see also Movie S3 and Movie S4). We reason that the interaction among the A-, C-, and N-loops and the αC helix in the active systems is crucial for alignment of the key domains for catalysis. The correlation between the A-loop and C-loop is especially pronounced, ensuring appropriate positioning of the catalytic aspartate, D845, and the coordinating aspartate, D863, for the phosphoryl transfer reaction.
Figure 1.
Global motions of the HER2 structures as determined by the results of PCA. The structures represent displacements along the first eigenmode for (A) the Y877-unphosphorylated active system, (B) the Y877-unphosphorylated inactive system, (C) the Y877-phosphorylated active system, and (D) the Y877-phosphorylated inactive system. The structures are color-coded according to the RMSD (red and white indicate large fluctuations, blue indicates smaller fluctuations) to illustrate the motions in individual residues. The four key domains move in a concerted manner in the active system, whereas the inactive system lacks such correlated fluctuations.
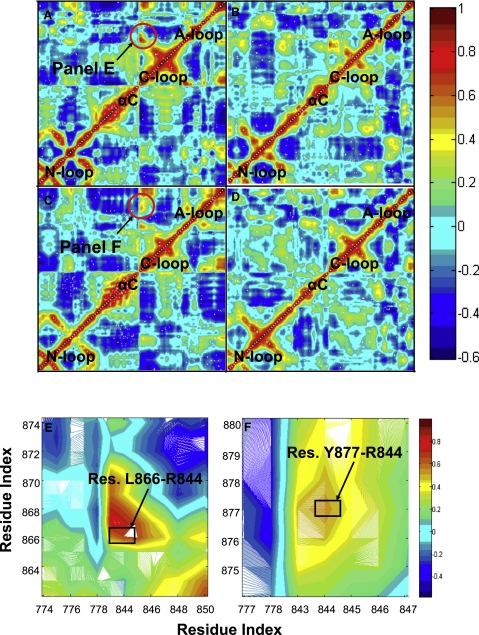
The normalized variance-covariance matrices for vector displacements of atoms further emphasize and quantify the interaction between the A-loop and C-loop in the active system. Fig. 2 displays correlations between vector displacements of atoms (d-〈d〉) for all four systems, where vector d represents the displacement from the origin to the atom of interest and 〈d〉 is the displacement averaged over all frames in the trajectory. For both Y877-unphosphorylated and Y877-phosphorylated structures, it is apparent that the inactive and active systems differ in their respective correlation patterns. The A-loop and C-loop exhibit high levels of correlated atomic fluctuations in the active structures, as indicated by the red/orange regions present at the intersection of the A- and C-loop residues (Fig. 2, A and C). In contrast, the A- and C-loop residues interface at cyan/green regions in the cross-correlation plots for the inactive structures, signifying a lack of concerted motion (Fig. 2, B and D). The expanded figures (Fig. 2, E and F) highlight specific pairs of residues that correspond to the intersection of the A- and C-loops in the cross-correlation plots for the active structures. These residue pairs, which include L866-R844 and Y877-R844, may interact to couple the neighboring residues, D845 and D863, in preparation for catalysis.
Figure 2.
Correlated fluctuations of the Cα atoms in the active site region of (A) the Y877-unphosphorylated active system, (B) the Y877-unphosphorylated inactive system, (C) the Y877-phosphorylated active system, and (D) the Y877-phosphorylated inactive system. The four key domains of the protein (A-loop, C-loop, N-loop, and αC helix) are labeled on the maps, and the regions of high correlation between the C-loop and A-loop are circled and magnified in E and F. The color bars on the right indicate the extent of the correlation as quantified by the normalized correlation coefficient. Residue pairs with a high degree of correlated motion are shown in yellow, orange, and red. Anticorrelated motions are represented by the blue regions. Green and cyan regions indicate little or no correlation.
Rationalizing the differences in global motions in terms of distinctive stabilizing bonding networks in the active and inactive conformations of the kinase
To identify specific interactions that could be contributing to the differences in global motions of the inactive and active systems, and investigate the mechanistic basis for the coupling of the A- and C-loops, individual salt bridges and hydrogen bonds were tabulated for each of the four systems through a hydrogen-bonding analysis of the 10 ns trajectories (see Methods). The hydrogen bonds illustrated in Fig. 3 are labeled according to the numbered residue pairs listed in Table 1. Our analysis reveals that six hydrogen bonds occur in the C-loop of the Y877-unphosphorylated active HER2 structure, the majority of which link the C-loop and A-loop (Fig. 3 B and Table 1). Specifically, R844-L866, N850-T862, V842-R868, and L852-K860 connect the C-loop to the N-terminal end of the A-loop, and the L846-W888 bond couples the C-loop and the C-terminal end of the A-loop. In contrast, only two hydrogen bonds bridge the C-loop and the A-loop in the Y877-unphosphorylated inactive HER2 system (Fig. 3 A and Table 1). Analysis of the bonding patterns in the Y877-phosphorylated trajectories reveals a similar distinction between the inactive and active systems (see Table S4). The extensive hydrogen bonding that preferentially links the A- and C-loops in the active conformations of the kinase provides a rationale for the cooperative fluctuations between these regions, as revealed by the results of the PCA. Indeed, the specific interactions that are identified in the hydrogen-bonding analysis, including R844-L866 and Y877-R844, also appear as correlated residue pairs in the PCA cross-correlation plots (Fig. 2, E and F).
Figure 3.
Hydrogen-bonding analysis of the trajectories for the Y877-unphosphorylated systems in (A) the inactive conformation and (B) the active conformation. Labels correspond to the numbered residue pairs listed in Table 1. Only those bonds that couple the key subdomains of the kinase (A-loop, C-loop, and αC helix) are shown to highlight the preferential bridging of the subdomains in the active conformation. The A-loop is shown in red, C-loop in purple, and αC helix in gray (color online).
Table 1.
Hydrogen-bonding analysis of the MD trajectories for the Y877-unphosphorylated HER2 kinase in the inactive and active conformations
| Label∗ | Subregion | Inactive HER2 | Active HER2 |
|---|---|---|---|
| 1 | αC helix | — | A763 HN, S760 OG |
| 2 | αC helix | N764 HN, S760 O | N764 HN, S760 O |
| 3 | αC helix | — | E766 OE1/2, R756 HE |
| 4 | αC helix/A-loop | — | E766, K883† |
| 5 | αC helix/A-loop | — | D769 OD1/2, R868 HH12 |
| 6 | αC helix | — | E770, K753 |
| 7 | αC helix | Y772 O, G776 HN | — |
| 8 | αC helix | — | V773 O, V777 HN |
| 9 | αC helix | M774 O, L785 HN | — |
| 10 | C-loop/A-loop | V842 O, G865 HN | — |
| 11 | C-loop/A-loop | — | V842 O/HN, R868 HN/0 |
| 12 | C-loop/A-loop | — | R844 HE, L866 O |
| 13 | C-loop/A-loop | — | L846 O, W888 HE 1 |
| 14 | C-loop | A848 O, L807 HN | — |
| 15 | C-loop/A-loop | — | N850 O, T862 HN |
| 16 | C-loop | V851 O, L806 HN | V851 O, L806 HN |
| 17 | C-loop/A-loop | L852 HN, K860 O | L852 HN, K860 O |
| 18 | A-loop | D863, K753 | — |
| 19 | A-loop | — | L870 HN, R840 O |
| 20 | A-loop | — | D871 O, R840 HE/HH12 |
| 21 | A-loop | D873 OD1/2, R897 HE | — |
| 22 | A-loop | — | E876 OE1/2, R898 HE |
| 23 | A-loop | D880, R897 | D880, R897 |
| 24 | A-loop | K883 HZ1/2, E757 OE1 | — |
Labels correspond to the numbered hydrogen bonds illustrated in Fig. 3.
Salt bridges are highlighted in bold.
In cataloging the interactions, we compared the HER2 bonding network to that of EGFR and ErbB4 (49) (Table S1, Table S2, and Table S3). An analysis of the hydrogen bonds and salt bridges in the C-loop region of inactive and active EGFR and ErbB4 reveals several similarities to the HER2 bonding pattern. The active EGFR and ErbB4 structures each contain three bonds connecting to the A-loop, whereas only one such bond occurs in each inactive system (Table S2). Hence the similarity in the pattern of specific interactions that preferentially bridge the A- and C-loops in the active conformations of the EGFR, HER2, and ErbB4 kinases suggests that the tight coupling of the A- and C-loops may be a general feature of the architecture of ErbB receptor kinases that likely aids in the assembly of a catalytically competent active site.
We extended the evaluation of hydrogen bonds to the A-loop and αC helix because these regions undergo the most pronounced structural shifts upon transition from the inactive to the active state of the HER2 kinase. In similarity to the hydrogen-bonding pattern in the C-loop, we observe a considerable difference in the number of interactions in the A-loop and αC helix of the active and inactive systems (see Table 1 and Fig. 3). Specifically, there are four hydrogen bonds and two salt bridges in the A-loop of the Y877-unphosphorylated inactive structure, compared with nine hydrogen bonds and two salt bridges in the active A-loop. The αC helix reflects a more marked distinction between the two systems: two salt bridges occur in the active structure, whereas salt bridges are entirely lacking in the inactive system. The majority of bonds in the active system connect the key domains of the kinase; for instance, the E766-K883 and D769-R868 bonds link the A-loop and the αC helix (Fig. 3 B). Furthermore, the R844-L866, N850-T862, V842-R868, L852-K860, and L846-W888 bonds couple the A-loop and C-loop. Hence, the network of specific hydrogen bonds that bridge the key subregions of the kinase (A-loop, C-loop, and αC helix) provides a rationale for the concerted motion of these subdomains in the active state as depicted in Fig. 1.
Analysis of the stabilizing network of interactions in the active and inactive kinase conformations reveals a novel autoinhibitory mechanism
One of the most crucial interactions for kinase activation is the salt bridge between E770 and K753, which is conserved among ErbB family members (the analogous pair of residues is E738-K721 in EGFR and E743-K726 in ErbB4; see Table S1). This salt bridge is characteristic of the active conformations of kinases, as it facilitates catalysis by interacting with the α- and β-phosphates of ATP (16). Indeed, the bridge is lacking in inactive HER2 because the residues are sequestered by other amino acids. Specifically, K753 is salt-bridged to D863 in the inactive state, preventing the interaction between K753 and E770. It is noteworthy that an analogous inhibitory bond, namely D836-K726, occurs in inactive ErbB4 (see Table S3). We speculate that the autoinhibitory bond, D863-K753, reflects a dual inhibitory mechanism. Not only does it sequester K753, it also engages D863, the coordinating aspartate that facilitates rearrangement of the substrate tyrosine and ATP. The D863-K753 bond is broken in active HER2, allowing the crucial salt bridge to form and freeing the coordinating aspartate for catalysis.
We identify a second autoinhibitory bond that impedes formation of the E770-K753 salt bridge in the inactive state, and note that this mechanism is shared by EGFR. In the inactive EGFR conformation, E738 forms a salt bridge with K836, which corresponds to R868 in HER2. Analogously, R868 sequesters E770 in the inactive HER2 system; however, in the active state, R868 bonds with V842 and D769, allowing E770 to salt-bridge with K753 (see Table S3). Hydrogen bonding of R868 to multiple residues ensures release of E770 and promotion of the active state.
Variation among HER2, EGFR, and ErbB4 in the αC-β4 loop region of the kinase
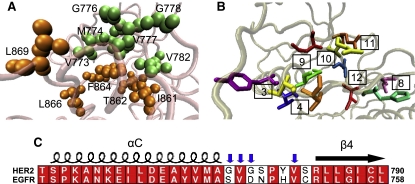
In a recent study, Fan et al. (50) reported that HER2 is strongly autoinhibited relative to EGFR and ErbB4, and that a mechanism for the autoinhibition involves sequence variation in a loop connecting the αC helix and the β4 sheet. The HER2 kinase domain shares 83% sequence identity with EGFR; in the αC-β4 loop, however, five of the eight residues in HER2 differ from those in EGFR. Specifically, the polar residues in the αC-β4 loop of EGFR are replaced by nonpolar residues in HER2, which form a hydrophobic patch that contacts another segment of hydrophobic residues located in the A-loop. Residues comprising the hydrophobic patch in HER2 include V773, M774, G776, V777, G778, and V782 in the αC-β4 loop, and I861, T862, F864, L866, and L869 in the A-loop. This hydrophobic motif has been investigated by several groups in relation to its association with various molecules, such as the molecular chaperone Hsp90 (51,52). Fan et al. (50) postulated that the hydrophobic interactions between the αC-β4 loop and the A-loop stabilize the HER2 kinase in the inactive state, resulting in lower constitutive catalytic activity relative to EGFR and ErbB4.
In light of these experimental findings, we extended our hydrogen-bonding analysis to the αC-β4 loop for HER2, EGFR, and ErbB4. In tabulating the bonds for each system, we observe that both the inactive and active HER2 structures contain only two hydrogen bonds in the αC-β4 region (Table 2). One of the bonds, S783-I861, is shared by both systems and couples the αC-β4 loop to the A-loop. In contrast, EGFR and ErbB4 contain a significantly greater number of hydrogen bonds in the αC-β4 loop. The active EGFR and ErbB4 structures form 10 and eight hydrogen bonds, respectively (Table 2). Several bonds in active EGFR, including H749-I829, C751-I829, and H749-V827, link residues in the αC-β4 loop and the A-loop, whereas other bonds, including S744-Y740, V745-V741, and L753-M742, connect the αC-β4 loop and the αC helix. The active HER2 system lacks many of these hydrogen bonds because hydrophobic interactions, rather than hydrophilic contacts, predominate in the αC-β4 region. In particular, the H749-I829 and H749-V827 bonds are absent in HER2 because the kinase contains a relatively nonpolar tyrosine residue in the position analogous to the positively charged H749 in EGFR. Fig. 4 contrasts the network of hydrogen bonds (in stick representation) in the αC-β4 loop of the active EGFR system with the hydrophobic residues (in VDW representation) in HER2. The individual bonds are labeled according to the numbered residue pairs listed in Table 2.
Table 2.
Comparison of the hydrogen-bonding network in the αC-β4 loop of HER2, EGFR, and ErbB4 kinases in active and inactive conformations
| Label∗ | HER2 active | EGFR active | HER4 active | HER2 inactive | EGFR inactive | HER4 inactive |
|---|---|---|---|---|---|---|
| 1 | — | A743, L679 | A748, Q684 | — | — | — |
| 2 | — | S744, N676 | — | — | — | — |
| 3 | — | S744, Y740 | S749, L745 | G776, Y772 | S744, Y740 | S749, L745 |
| 4 | — | V745, V741 | M750, I746 | — | — | — |
| 5 | — | — | — | — | — | M750, M747 |
| 6 | G778, Y835 | D746, Y803 | — | — | — | — |
| 7 | — | — | H752, Y808 | — | — | H752, Y808 |
| 8 | — | H749, V827 | — | — | — | — |
| 9 | — | H749, I829 | H754, I834 | — | H749, I829 | H754, I834 |
| 10 | S783, I861 | C751, I829 | V756, I834 | S783, I861 | C751, I829 | V756, I834 |
| 11 | — | R752, Q767 | R757, Q772 | — | — | — |
| 12 | — | L753, M742 | L758, M747 | — | L753, M742 | L758, M747 |
Labels correspond to the numbered hydrogen bonds illustrated in Fig. 4B for EGFR kinase in the active conformation.
Figure 4.
Contrast between the hydrophobic interactions in HER2 and the hydrogen bonds in EGFR/ErbB4 in the αC-β4 loop. (A) The patch of hydrophobic residues in the αC-β4 loop of the active HER2 structure. Residues in the αC-β4 loop are colored green and residues in the A-loop are colored orange (color online). (B) The hydrogen-bonding network in the αC-β4 loop of the active EGFR structure. The bonds are labeled according to the numbering scheme in Table 2. (C) Sequence alignment between HER2 and EGFR in the αC-β4 region of the kinase. Blue arrows indicate residues contributing to the hydrophobic patch in HER2.
The bonding analysis of the inactive ErbB systems reveals a trend similar to that observed in the active systems. The inactive EGFR and ErbB4 conformations contain four and six hydrogen bonds, respectively, in contrast to two bonds in the αC-β4 loop of inactive HER2 (Table 2). The lack of hydrogen bonds in the αC-β4 loop of inactive HER2 is consistent with the prominence of hydrophobic interactions in this region as proposed by Fan et al. (50). Although it shares similar bonding patterns with EGFR in the A-loop and αC helix, HER2 differs markedly from EGFR in the αC-β4 region due to the presence of the hydrophobic patch of residues. The dearth of a hydrogen-bonding network and the dominance of hydrophobic interactions surrounding the αC-β4 loop in HER2 is thought to contribute to its interaction with the molecular chaperone Hsp90 (52). As a mature protein, only HER2 among the members of the ErbB family associates with Hsp90, as EGFR and ErbB4 lack the segment of hydrophobic residues in the αC-β4 loop. It has been proposed that binding of Hsp90 to the αC-β4 region in HER2 provides an inhibitory mechanism for regulation of HER2 activity by preventing dimerization and subsequent activation of the HER2 kinase (51).
The hydrophobic association between HER2 and Hsp90 is relevant to the effects of clinical mutations in the HER2 kinase domain. HER2 gene mutations have been identified in a cohort of non-small cell lung cancers that involve in-frame duplications/insertions within exon 20, a region that corresponds to the αC-β4 loop in the kinase domain. The most frequently occurring abnormality is the in-frame YVMA insertion at residue G776 (G776YVMA), which has been shown to undergo markedly higher tyrosine phosphorylation than wild-type HER2, resulting in increased tumorigenicity (14). Another prevalent activating mutation is G776S, which has been found in gastric tumors (53). Such mutations weaken the hydrophobic interactions surrounding the αC-β4 loop and likely promote a hydrogen-bonding network similar to those we have identified in EGFR and ErbB4, which can disrupt the inhibitory stimulus provided by HER2-Hsp90 association.
Heterodimerization with EGFR kinase destabilizes the inactive conformation of HER2 kinase
As postulated by Zhang et al. (30), HER2 is likely to undergo activation by the asymmetric dimer mechanism proposed for EGFR. In the context of a HER2-EGFR heterodimer, the residues comprising the dimeric interface for HER2 (kinase undergoing activation) include P707, Q711, M712, I714, L768, L790, and V794; for EGFR (activating kinase) they include I917, Y920, M921, V924, M928, I929, and V956. To investigate potential mechanisms of activation, we constructed a HER2-EGFR heterodimer in which EGFR is the activator kinase and Y877-unphosphorylated inactive HER2 is the activated kinase (see Fig. S5). The heterodimer was modeled on the structure of the EGFR dimer generated by Zhang et al. (30) (see Methods). A second dimer was constructed in which the Y877-phosphorylated inactive HER2 monomer from the 10 ns equilibrated system was input as the activated kinase. Both heterodimeric systems were solvated and subjected to 20 ns of MD simulation (see Methods). The objective was to determine whether dimerization promotes the active state, and, if so, whether phosphorylation of Y877 facilitates the activation mechanism.
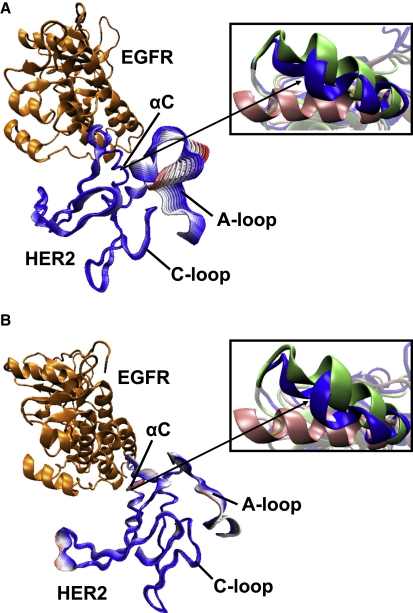
PCA was performed for the 20 ns MD trajectories to characterize the extent of correlation of atomistic fluctuations. As was done for the monomeric systems, the active site region was chosen to include the Cα atoms of the A-, C-, and N-loops and the αC helix of HER2, the kinase that is undergoing activation in the context of the heterodimer. In similarity to the inactive monomeric simulations, the principal eigenmodes are dominated by A-loop movement (Fig. 5, A and B). However, the phosphorylated heterodimer exhibits notable fluctuations in the αC helix of HER2 (Fig. 5 B), in similarity to the active monomeric structures. We also observe a repositioning of the αC helix toward the active conformation during the 20 ns MD simulation, shifting from an RMSD of 2–6 Å relative to the inactive state (Fig. 5 B and Fig. S6 D). The HER2 structure in the unphosphorylated dimer also exhibits a slight conformational rearrangement of its αC helix toward the active state in response to the dimerization interface (Fig. 5 A).
Figure 5.
Global motions of the HER2-EGFR heterodimeric structures as determined by PCA. The structures represent displacements along the first eigenmode for (A) the Y877-unphosphorylated system and (B) the Y877-phosphorylated system. The structures are color-coded according to the RMSD (red and white indicate large fluctuations, blue indicates smaller fluctuations) to illustrate the motions in individual residues (color online). Insets reveal shifting of the αC helix in the dimer (blue) away from the inactive conformation (green) and toward the active conformation (pink).
As in our analysis of the monomeric systems, we aimed to rationalize the pattern of global motions in the heterodimers in terms of specific interactions. To identify stabilizing bonds that are perturbed upon dimerization, we performed a hydrogen-bonding analysis for the 20 ns MD trajectories. An examination of the bonding network in the heterodimeric systems reveals destabilization and severance of several interactions present in the monomeric systems (Table 3). In the dimer featuring Y877-unphosphorylated HER2 as the activated kinase, several bonds responsible for maintaining the inactive state in the monomer are broken, including N764-S760 and Y772-G776 in the αC helix. Indeed, disruption of these bonds is expected as a result of their position within the dimeric interface. Additional bonds that are disturbed include G865-V842 and D873-R897 in the A-loop. In the dimer involving Y877-phosphorylated HER2 as the activated kinase, several bonds expected to break are indeed severed, including T759-E874, L785-M774, and Y772-G776 in the αC helix. Key interactions in the A-loop, such as G865-H843, R868-R840, and V884-K887, have also been perturbed. Our findings indicate that the dimerization interface directly alters the pattern of stabilizing hydrogen bonds in the inactive system, in agreement with the allosteric activation mechanism proposed by Zhang et al. (30). The disruption of specific interactions in the αC helix suggests that the pattern of hydrogen bonds in the dimer is shifting away from the inactive state, and that the kinase domain is more susceptible to perturbations that would enable a conformational rearrangement toward the active state. Moreover, the dimeric interface directly induces the repositioning of the αC helix toward the active conformation even on the relatively short timescale of 20 ns.
Table 3.
Summary of broken and unbroken hydrogen bonds in the MD trajectories for the HER2-EGFR heterodimers
| Y877-unphosphorylated dimer | Y877-phosphorylated dimer | |
|---|---|---|
| Proximal, broken∗ | — | T759-E874 |
| N764-S760 | — | |
| Y772-G776 | Y772-G776 | |
| — | L785-M774 | |
| Proximal, unbroken† | K753-D863 | K753-D863 |
| E757-K883 | E757-K883 | |
| L785-M774 | — | |
| Not proximal, broken‡ | G865-V842 | — |
| — | G865-H843 | |
| D873-R897 | — | |
| — | V884-K887 |
Refers to bonds containing residues that are proximal to the dimer interface (within 3 Å of at least one of the dimeric interface residues for 75% of the production trajectory) and have broken in the dimer trajectory.
Refers to bonds containing residues that are proximal to the dimer interface but have not broken in the dimer trajectory.
Refers to bonds that are not proximal to the dimer interface but have broken in the dimer trajectory.
Several bonds remain unperturbed by the dimerization interface and hence may pose significant free energy barriers to the conformational change accompanying activation. The D863-K753 interaction, which is one of the salt bridges involved in the dual autoinhibitory mechanism in the monomeric simulations, persists throughout the dimeric trajectories. We also note the importance of the K883-E757 bond, which is conserved among all three inactive ErbB kinases, in governing stabilization of the inactive structure, as it must break in order for the K883-E766 salt bridge to assemble in the active state.
FEP analysis of the role of Y877-phosphorylation
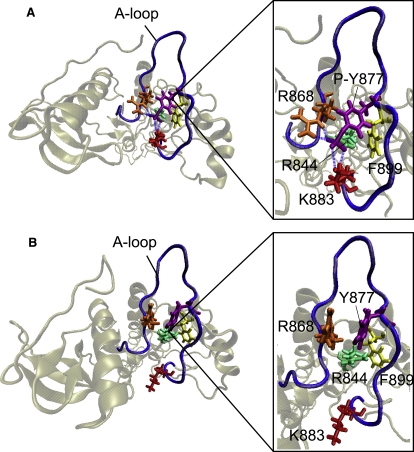
Several experimental studies have highlighted the importance of A-loop phosphorylation in HER2 activation. In the Y877-phosphorylated active system, we identify a network of hydrogen bonds that maintain the A-loop in the open conformation: as the A-loop extends, it uncovers the catalytic loop and promotes access of peptides to the active site. The hydrogen bonds fasten the A-loop to a segment of the αF helix (residues 896–901) and to the region between the αE helix and the C-loop (residues 840–844), ensuring that the A-loop remains in the active state (see Fig. S7). Three hydrogen bonds (L866-R844, V842-R868, and R840-L870) secure the A-loop at its N-terminal end (Fig. S7 B). Likewise, three bonds (Y877-F899, A879-R897, and E876-R898) fasten the A-loop at its C-terminal end (Fig. S7 C). It is noteworthy that several of the bonds, such as L866-R844 and Y877-R844, link residues in the A-loop and C-loop, underscoring the tight coupling of these regions in the active state. However, the key residues required for kinase activity, D863 (the coordinating aspartate) and D845 (the catalytic aspartate), do not participate in the hydrogen-bonding network and hence remain poised for catalysis. Based on these results, we propose an activation model in which residues neighboring D863 and D845 compose hydrogen bonds that stabilize the A-loop in the active state while ensuring the availability of the catalytic aspartate residues for kinase activity.
Intriguingly, we observe that phosphorylated Y877 performs a key function in linking the network of hydrogen bonds that maintain the A-loop in its extended form. The phosphoryl group forms bonds with R844, K883, and R868, thereby bridging the C- and N-terminal ends of the A-loop (Fig. 6 A). Furthermore, the main-chain oxygen of Y877 hydrogen-bonds with F899, contributing to the pattern of fastening interactions in the A-loop. In contrast, fewer hydrogen bonds occur in the A-loop of the Y877-unphosphorylated active system, indicating that the absence of the phosphoryl group results in a decreased number of intra-A-loop interactions (Fig. 6 B and Table S4). We define a similar role for phosphorylated Y845 in the active EGFR system. Y845-phosphorylated EGFR shares eight of nine hydrogen bonds present in the A-loop of Y877-phosphorylated active HER2, including bonds between the phosphoryl group and the A-loop (see Table S5). Our results for HER2 and EGFR suggest that the role of the phosphoryl group in Y877 (or Y845 in EGFR) is to bridge the stabilizing bonds on either side of the A-loop in the active system. In further support of this bridging mechanism of phosphorylated Y877 in HER2 and Y845 in EGFR, we note an analogous function for the phosphorylated tyrosine residue in insulin receptor tyrosine kinase (IRK), for which there exists a crystal structure of the phosphorylated active form of the protein (54). The close alignment of the tyrosine-phosphorylated A-loops of EGFR, HER2, and IRK suggests a structural role for the phosphorylated tyrosine as a bridging residue in the respective active kinase conformations (see Fig. S8).
Figure 6.
(A) Snapshot of the Y877-phosphorylated active system, illustrating the role of phosphorylated Y877 in bridging the network of hydrogen bonds on either side of the A-loop. The phosphoryl group forms bonds with R844, K883, R868, and F899, thereby connecting the N- and C-terminal ends of the A-loop. Hydrogen bonds are depicted as blue dashed lines. (B) Snapshot of the unphosphorylated active system, showing the lack of stabilizing hydrogen bonds due to the absence of the phosphoryl group.
Analysis of the MD trajectories for the inactive systems reveals that phosphorylation of Y877 promotes the formation of additional hydrogen bonds and salt bridges in the A-loop (see Table S4). The phosphoryl group hydrogen-bonds with R844 and K883, spanning the A-loop as it does in the active system. Additional pairs of residues include G865-H843 and R868-R840, which parallel L866-R844 and R868-V842 in the active structure. Although phosphorylation of Y877 in the inactive system alters the hydrogen-bonding pattern so that it more closely resembles the network in the active system, phosphorylation is insufficient for promotion of conformational shifting to the active state within the short timescale of our 10 ns simulations.
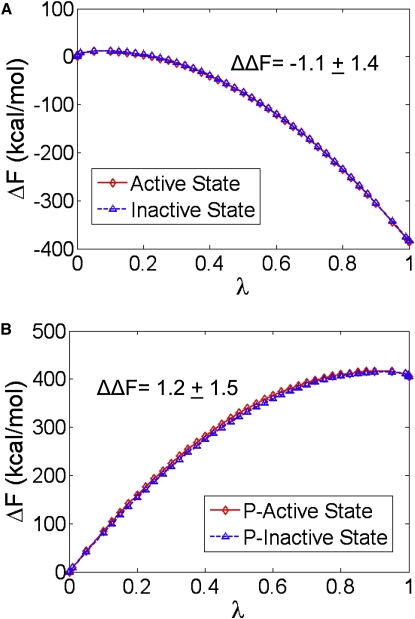
To further investigate the effect of Y877-phosphorylation on kinase activity, we employed the FEP method to calculate the Helmholtz free energy difference between the Y877-unphosphorylated and Y877-phosphorylated states in the NVT ensemble. The alchemical transformations were performed using the dual-topology paradigm (46), in which the initial and the final states are defined in terms of distinct, noninteracting topologies, and the interactions of the transformed atoms with their environment are scaled in terms of a linear parameter, λ (see Methods for details). Four different simulations were performed, including the transformation of Y877 to pY877 in the unphosphorylated structures (inactive and active), and the transformation of pY877 to Y877 in the respective phosphorylated systems. Replacement of Y877 with phosphorylated Y877 resulted in a free energy change of −385.1 ± 1.2 kcal/mol for the active structure and −384.0 ± 0.8 kcal/mol for the inactive structure, yielding a ΔΔF value of −1.1 ± 1.4 kcal/mol (Fig. 7 A). The ΔF values for the reverse transformation, pY877 to Y877, were calculated in a similar manner, and were found to be 405.8 ± 1.1 kcal/mol for the phosphorylated active structure and 404.6 ± 1.1 kcal/mol for the phosphorylated inactive system, resulting in a ΔΔF value of 1.2 ± 1.5 kcal/mol (Fig. 7 B). The individual ΔF values for the forward and reverse transformations differ slightly because the structures contain different numbers of water molecules and ions, yet the error estimates are within the ranges that have been computed for other solvated systems (55–57). The ΔΔF values are significantly close and indicate that phosphorylation of Y877 provides a small increase in stability of the active conformation relative to the inactive state, although it is insufficient to significantly lower the kinase activation barrier. Additional perturbations are required for full catalytic competency of the kinase, suggesting that phosphorylation of Y877 is unlikely to be the primary stimulus for conformational shifting to the active state.
Figure 7.
Evolution of the Helmholtz free energy as a function of the coupling parameter λ. FEP simulations were performed for (A) the transformation of Y877 to pY877 and (B) the transformation of pY877 to Y877 in both the inactive and active systems. Phosphorylation of Y877 stabilizes the inactive and active structures, whereas removal of the phosphoryl group results in an increase in the free energy.
It is noteworthy that differences in specific interactions between phosphorylated and unphosphorylated systems identified in the FEP trajectories also appear in our hydrogen-bonding analysis. In the active structure, replacement of Y877 with pY877 produces several new hydrogen bonds, including R844-Y877, K883-Y877, and R868-Y877, all of which bridge the C- and N-terminal ends of the A-loop. Toward the end of the transformation, the main-chain oxygen of Y877 pairs with F899, which is consistent with the pattern of hydrogen bonds highlighted in the previous section. The perturbation of Y877 into pY877 in the inactive system effects formation of additional hydrogen bonds, such as R844-Y877 and K883-Y877, which link the ends of the A-loop as they do in the active system. Thus, the FEP results are consistent with our hydrogen-bonding analysis, validating the robustness of the identified interactions despite the differences in simulation protocols.
Discussion and Conclusion
Given the involvement of the ErbB receptor tyrosine kinases in a wide range of human diseases, including schizophrenia and various types of cancer (2), it is imperative to understand their mechanism of activation at the molecular level. In this study we investigated the mechanisms that are important in HER2 kinase domain regulation and compared them with two other ErbB family members, EGFR and ErbB4, to determine the molecular basis for HER2's unique mode of activation. Our MD simulations provide us with a framework for studying the atomistic behavior of the HER2 kinase domain in both monomeric and dimeric contexts. We characterized the global motions of the HER2 systems using PCA and rationalized the differences in fluctuations in terms of specific interactions, namely, salt bridges and hydrogen bonds. Specifically, the existence of an extensive bonding network among the four key domains of the kinase in its active state correlates with the concerted motions of the four main loops as revealed by the PCA. The tight coupling of the A-loop and C-loop in the active system is especially pronounced, leading us to postulate the significance of the interaction in alignment of the catalytic residues. We also identify several commonalities in hydrogen-bonding patterns among the three ErbB kinases in the A-loop, C-loop, and αC helix. One such conserved pattern involves the sequestration of D863 (the coordinating aspartate), K753, and E770 in the inactive state, preventing formation of the K753-E770 salt bridge and release of D863 for catalysis. We postulate that this mechanism reflects a means of dual autoinhibition, as both the coordinating aspartate and the crucial salt bridge are sequestered in the inactive state. Despite these shared trends in hydrogen bonds in key loops required for activation, the bonding pattern in HER2 differs markedly in the αC-β4 loop. HER2 lacks many of the hydrogen bonds that occur in the αC-β4 loop in EGFR and ErbB4 due to the presence of the patch of hydrophobic residues in the αC-β4 region. The unique hydrogen-bonding network in the αC-β4 loop in HER2 is relevant to the putative hydrophobic interaction between HER2 and the molecular chaperone Hsp90. The association between HER2 and Hsp90 serves a regulatory role in preventing HER2 dimerization and subsequent activation (51). Unlike the other ErbB family members, the extracellular domain of HER2 is poised for dimerization in the absence of ligand binding (23), rationalizing the requirement for such tight regulatory mechanisms. The decrease in hydrophobic character and concomitant increase in the degree of hydrogen bonding in the αC-β4 region of several clinically identified HER2 mutants alters the mutant HER2 bonding patterns in similarity to those of EGFR and ErbB4. Such mutations are expected to be activating by disrupting the HER2-Hsp90 association.
The analysis of global motions and hydrogen-bonding patterns was extended to a HER2-EGFR heterodimer. MD simulations of the heterodimeric systems result in destabilization of several bonds present in the inactive monomeric structures, including interactions in the αC helix, as the αC helix comprises much of the dimerization interface for the activated kinase. The disruption of the hydrogen-bonding pattern in both Y877-phosphorylated and Y877-unphosphorylated heterodimers provides a rationale for the observed conformational rearrangement of the αC helix toward the active state. Furthermore, we identify several interactions that persist throughout the dimeric trajectories of the inactive states and are hence candidates for further investigation using free energy methods such as umbrella sampling simulations, as the molecular environment surrounding such residues may define the pathway for conformational change and the associated barriers to activation. Since the residues involved in the N- and C-lobe faces of the dimer are essentially invariant among the ErbB family members, our results can in principle be extended to predict the behavior of other ErbB heterodimers.
We also investigated the effect of phosphorylation of the regulatory tyrosine residue, Y877, on kinase activation. The results of our FEP simulations support the hypothesis that phosphorylation of Y877 is unlikely to provide a dominant stimulus for activation. However, our structural and hydrogen-bonding analyses of the MD trajectories strongly suggest that phosphorylated Y877 contributes to the network of fastening bonds in the A-loop by bridging the stabilizing residues at the C- and N-terminal ends of the loop, thereby considerably altering the conformational environment surrounding the A-loop. Consistent with these findings, although phosphorylation of the analogous A-loop tyrosine residue Y845 is unnecessary for catalytic activity of EGFR, it is known to significantly alter downstream signaling events, including activation of STAT5b and EGF-induced DNA synthesis (58,59). Likewise, Ishizawar et al. (60) postulated a role for phosphorylated Y877 in enhancing HER2-ErbB3 heterodimer formation by potentially changing the conformation of the kinase and engaging other molecules. Hence the involvement of phosphorylated Y877 in the network of fastening interactions in the A-loop and the resultant alteration in the conformational environment may influence recruitment of signaling mediators involved in mitogenesis and other downstream processes. We also note that the residues involved in fastening the A-loop, which include R897, R898, and F899 in the αF helix, warrant further investigation because mutation of these residues may destabilize the fastening bonding network, and this effect may further elucidate the role of Y877-phosphorylation in HER2. These predictions can be experimentally tested and validated with the use of HER2 mutation assays.
The protein kinase genes are among the most frequently mutated genes in human cancers, and several HER2 kinase domain mutants have been determined. Our simulations provide insight into the effect of these mutations in the αC-β4 loop region of the HER2 kinase through assessment of structural dynamics and hydrogen-bonding patterns in the kinase domain. Furthermore, we predict the importance of previously overlooked regions, such as the segment of residues directly preceding the αF helix, in regulation of kinase activation. Our results are consistent with a large body of experimental data (29,30,50,52,54) and provide a framework for highlighting the most crucial bonding interactions in the monomeric and dimeric HER2 systems. Elucidation of the molecular regulatory mechanisms will help establish structure-function relationships in the wild-type HER2 kinase, as well as predict mutations with a propensity for constitutive activation. Such molecular variants in HER2 receptors are known to profoundly impact specific therapies targeting HER2-mediated cancers.
Acknowledgments
We thank Mark A. Lemmon, Sung Hee Choi, and Andrew Shih for several helpful discussions, and Péter Bagossi for providing the PDB coordinates for the HER2 model.
We acknowledge funding from the National Science Foundation through grant NSF CBET-0730955. Shannon Telesco received support through a Graduate Research Fellowship from the National Science Foundation and a Graduate Assistantship in Areas of National Need from the Bioengineering Department of the University of Pennsylvania. Computational resources were provided in part by the National Partnership for Advanced Computational Infrastructure through allocation MCB060006.
Supporting Material
References
- 1.Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 2.Linggi B., Carpenter G. ErbB receptors: new insights on mechanisms and biology. Trends Cell Biol. 2006;16:649–656. doi: 10.1016/j.tcb.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 3.Yarden Y., Sliwkowski M.X. Untangling the ErbB signalling network. Nat. Rev. Mol. Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 4.Schulze W.X., Deng L., Mann M. Phosphotyrosine interactome of the ErbB-receptor kinase family. Mol. Syst. Biol. 2005;1 doi: 10.1038/msb4100012. 2005.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Citri A., Yarden Y. EGF-ERBB signalling: towards the systems level. Nat. Rev. Mol. Cell Biol. 2006;7:505–516. doi: 10.1038/nrm1962. [DOI] [PubMed] [Google Scholar]
- 6.Choi S.H., Mendrola J.M., Lemmon M.A. EGF-independent activation of cell-surface EGF receptors harboring mutations found in gefitinib-sensitive lung cancer. Oncogene. 2007;26:1567–1576. doi: 10.1038/sj.onc.1209957. [DOI] [PubMed] [Google Scholar]
- 7.Ji H., Li D., Chen L., Shimamura T., Kobayashi S. The impact of human EGFR kinase domain mutations on lung tumorigenesis and in vivo sensitivity to EGFR-targeted therapies. Cancer Cell. 2006;9:485–495. doi: 10.1016/j.ccr.2006.04.022. [DOI] [PubMed] [Google Scholar]
- 8.Lee J.W., Soung Y.H., Kim S.Y., Nam S.W., Park W.S. ERBB2 kinase domain mutation in the lung squamous cell carcinoma. Cancer Lett. 2006;237:89–94. doi: 10.1016/j.canlet.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 9.Minami Y., Shimamura T., Shah K., LaFramboise T., Glatt K.A. The major lung cancer-derived mutants of ERBB2 are oncogenic and are associated with sensitivity to the irreversible EGFR/ERBB2 inhibitor HKI-272. Oncogene. 2007;26:5023–5027. doi: 10.1038/sj.onc.1210292. [DOI] [PubMed] [Google Scholar]
- 10.Shigematsu H., Takahashi T., Nomura M., Majmudar K., Suzuki M. Somatic mutations of the HER2 kinase domain in lung adenocarcinomas. Cancer Res. 2005;65:1642–1646. doi: 10.1158/0008-5472.CAN-04-4235. [DOI] [PubMed] [Google Scholar]
- 11.Willmore-Payne C., Holden J.A., Layfield L.J. Detection of epidermal growth factor receptor and human epidermal growth factor receptor 2 activating mutations in lung adenocarcinoma by high-resolution melting amplicon analysis: correlation with gene copy number, protein expression, and hormone receptor expression. Hum. Pathol. 2006;37:755–763. doi: 10.1016/j.humpath.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 12.Willmore-Payne C., Holden J.A., Layfield L.J. Detection of EGFR- and HER2-activating mutations in squamous cell carcinoma involving the head and neck. Mod. Pathol. 2006;19:634–640. doi: 10.1038/modpathol.3800552. [DOI] [PubMed] [Google Scholar]
- 13.Slamon D.J., Godolphin W., Jones L.A., Holt J.A., Wong S.G. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244:707–712. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- 14.Wang S.E., Narasanna A., Perez-Torres M., Xiang B., Wu F.Y. HER2 kinase domain mutation results in constitutive phosphorylation and activation of HER2 and EGFR and resistance to EGFR tyrosine kinase inhibitors. Cancer Cell. 2006;10:25–38. doi: 10.1016/j.ccr.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 15.Huse M., Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 16.Stamos J., Sliwkowski M.X., Eigenbrot C. Structure of the epidermal growth factor receptor kinase domain alone and in complex with a 4-anilinoquinazoline inhibitor. J. Biol. Chem. 2002;277:46265–46272. doi: 10.1074/jbc.M207135200. [DOI] [PubMed] [Google Scholar]
- 17.Hubbard S.R., Till J.H. Protein tyrosine kinase structure and function. Annu. Rev. Biochem. 2000;69:373–398. doi: 10.1146/annurev.biochem.69.1.373. [DOI] [PubMed] [Google Scholar]
- 18.Tice D.A., Biscardi J.S., Nickles A.L., Parsons S.J. Mechanism of biological synergy between cellular Src and epidermal growth factor receptor. Proc. Natl. Acad. Sci. USA. 1999;96:1415–1420. doi: 10.1073/pnas.96.4.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bose R., Molina H., Patterson A.S., Bitok J.K., Periaswamy B. Phosphoproteomic analysis of Her2/neu signaling and inhibition. Proc. Natl. Acad. Sci. USA. 2006;103:9773–9778. doi: 10.1073/pnas.0603948103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang H.T., O'Rourke D.M., Zhao H., Murali R., Mikami Y. Absence of autophosphorylation site Y882 in the p185neu oncogene product correlates with a reduction of transforming potential. Oncogene. 1998;16:2835–2842. doi: 10.1038/sj.onc.1201820. [DOI] [PubMed] [Google Scholar]
- 21.Xu W., Yuan X., Beebe K., Xiang Z., Neckers L. Loss of Hsp90 association up-regulates Src-dependent ErbB2 activity. Mol. Cell. Biol. 2007;27:220–228. doi: 10.1128/MCB.00899-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cho H.S., Leahy D.J. Structure of the extracellular region of HER3 reveals an interdomain tether. Science. 2002;297:1330–1333. doi: 10.1126/science.1074611. [DOI] [PubMed] [Google Scholar]
- 23.Cho H.S., Mason K., Ramyar K.X., Stanley A.M., Gabelli S.B. Structure of the extracellular region of HER2 alone and in complex with the Herceptin Fab. Nature. 2003;421:756–760. doi: 10.1038/nature01392. [DOI] [PubMed] [Google Scholar]
- 24.Ferguson K.M., Berger M.B., Mendrola J.M., Cho H.S., Leahy D.J. EGF activates its receptor by removing interactions that autoinhibit ectodomain dimerization. Mol. Cell. 2003;11:507–517. doi: 10.1016/s1097-2765(03)00047-9. [DOI] [PubMed] [Google Scholar]
- 25.Garrett T.P., McKern N.M., Lou M., Elleman T.C., Adams T.E. The crystal structure of a truncated ErbB2 ectodomain reveals an active conformation, poised to interact with other ErbB receptors. Mol. Cell. 2003;11:495–505. doi: 10.1016/s1097-2765(03)00048-0. [DOI] [PubMed] [Google Scholar]
- 26.Garrett T.P., McKern N.M., Lou M., Elleman T.C., Adams T.E. Crystal structure of a truncated epidermal growth factor receptor extracellular domain bound to transforming growth factor α. Cell. 2002;110:763–773. doi: 10.1016/s0092-8674(02)00940-6. [DOI] [PubMed] [Google Scholar]
- 27.Wood E.R., Truesdale A.T., McDonald O.B., Yuan D., Hassell A. A unique structure for epidermal growth factor receptor bound to GW572016 (Lapatinib): relationships among protein conformation, inhibitor off-rate, and receptor activity in tumor cells. Cancer Res. 2004;64:6652–6659. doi: 10.1158/0008-5472.CAN-04-1168. [DOI] [PubMed] [Google Scholar]
- 28.Bouyain S., Longo P.A., Li S., Ferguson K.M., Leahy D.J. The extracellular region of ErbB4 adopts a tethered conformation in the absence of ligand. Proc. Natl. Acad. Sci. USA. 2005;102:15024–15029. doi: 10.1073/pnas.0507591102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiu C., Tarrant M.K., Choi S.H., Sathyamurthy A., Bose R. Mechanism of activation and inhibition of the HER4/ErbB4 kinase. Structure. 2008;16:460–467. doi: 10.1016/j.str.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang X., Gureasko J., Shen K., Cole P.A., Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 31.Fiser A., Do R.K., Sali A. Modeling of loops in protein structures. Protein Sci. 2000;9:1753–1773. doi: 10.1110/ps.9.9.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sali A., Blundell T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 33.MacKerell A.D., Jr., Bashford D., Bellott M., Dunbrack R.L., Evanseck J.D. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 34.Bagossi P., Horvath G., Vereb G., Szollosi J., Tozser J. Molecular modeling of nearly full-length ErbB2 receptor. Biophys. J. 2005;88:1354–1363. doi: 10.1529/biophysj.104.046003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993;26:283–291. [Google Scholar]
- 36.Jorgensen W.L., Chandrasekar J., Madura J., Impey R., Klein M. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 37.Grubmuller H., Heymann B., Tavan P. Ligand binding: molecular mechanics calculation of the streptavidin-biotin rupture force. Science. 1996;271:997–999. doi: 10.1126/science.271.5251.997. [DOI] [PubMed] [Google Scholar]
- 38.Phillips J.C., Braun R., Wang W., Gumbart J., Tajkhorshid E. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Essmann U. A smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 40.Andersen H.C. Rattle: a “velocity” version of the shake algorithm for molecular dynamics calculations. J. Comput. Phys. 1983;52:24–34. [Google Scholar]
- 41.Feller S.E., Zhang Y., Pastor R.W., Brooks B.R. Constant pressure molecular dynamics simulation: The Langevin piston method. J. Chem. Phys. 1995;103:4613–4621. [Google Scholar]
- 42.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–28. [DOI] [PubMed] [Google Scholar]
- 43.Glykos N.M. Software news and updates. CARMA: a molecular dynamics analysis program. J. Comput. Chem. 2006;27:1765–1768. doi: 10.1002/jcc.20482. [DOI] [PubMed] [Google Scholar]
- 44.Zwanzig R.W. High-temperature equation of state by a perturbation method. I. Nonpolar gases. J. Chem. Phys. 1954;22:1420–1426. [Google Scholar]
- 45.Bhandarkar M., Brunner R., Chipot C., Dalke A., Dixit S. Theoretical Biophysics Group, University of Illinois and Beckman Institute; Urbana, IL: 2005. NAMD user's guide, version 2.6b1. [Google Scholar]
- 46.Gao J., Kuczera K., Tidor B., Karplus M. Hidden thermodynamics of mutant proteins: a molecular dynamics analysis. Science. 1989;244:1069–1072. doi: 10.1126/science.2727695. [DOI] [PubMed] [Google Scholar]
- 47.Pitera J.W., van Gunsteren W.F. A comparison of non-bonded scaling approaches for free energy calculations. Mol. Simul. 2002;28:45–65. [Google Scholar]
- 48.Beutler T.C., Mark A.E., van Schaik R.C., Gerber P.R., van Gunsteren W.F. Avoiding singularities and neumerical instabilities in free energy calculations based on molecular simulations. Chem. Phys. Lett. 1994;222:529–539. [Google Scholar]
- 49.Shih A.J., Purvis J., Radhakrishnan R. Molecular systems biology of ErbB1 signaling: bridging the gap through multiscale modeling and high-performance computing. Mol. Biosyst. 2008;4:1151–1159. doi: 10.1039/b803806f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fan Y.X., Wong L., Ding J., Spiridonov N.A., Johnson R.C. Mutational activation of ErbB2 reveals a new protein kinase autoinhibition mechanism. J. Biol. Chem. 2008;283:1588–1596. doi: 10.1074/jbc.M708116200. [DOI] [PubMed] [Google Scholar]
- 51.Citri A., Gan J., Mosesson Y., Vereb G., Szollosi J. Hsp90 restrains ErbB-2/HER2 signalling by limiting heterodimer formation. EMBO Rep. 2004;5:1165–1170. doi: 10.1038/sj.embor.7400300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu W., Yuan X., Xiang Z., Mimnaugh E., Marcu M. Surface charge and hydrophobicity determine ErbB2 binding to the Hsp90 chaperone complex. Nat. Struct. Mol. Biol. 2005;12:120–126. doi: 10.1038/nsmb885. [DOI] [PubMed] [Google Scholar]
- 53.Stephens P., Hunter C., Bignell G., Edkins S., Davies H. Lung cancer: intragenic ERBB2 kinase mutations in tumours. Nature. 2004;431:525–526. doi: 10.1038/431525b. [DOI] [PubMed] [Google Scholar]
- 54.Hubbard S.R. Crystal structure of the activated insulin receptor tyrosine kinase in complex with peptide substrate and ATP analog. EMBO J. 1997;16:5572–5581. doi: 10.1093/emboj/16.18.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Henin J., Maigret B., Tarek M., Escrieut C., Fourmy D. Probing a model of a GPCR/ligand complex in an explicit membrane environment: the human cholecystokinin-1 receptor. Biophys. J. 2006;90:1232–1240. doi: 10.1529/biophysj.105.070599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Donnini S., Mark A.E., Juffer A.H., Villa A. Incorporating the effect of ionic strength in free energy calculations using explicit ions. J. Comput. Chem. 2005;26:115–122. doi: 10.1002/jcc.20156. [DOI] [PubMed] [Google Scholar]
- 57.Zhou R., Das P., Royyuru A.K. Single mutation induced H3N2 hemagglutinin antibody neutralization: a free energy perturbation study. J. Phys. Chem. B. 2008;112:15813–15820. doi: 10.1021/jp805529z. [DOI] [PubMed] [Google Scholar]
- 58.Boerner J.L., Biscardi J.S., Silva C.M., Parsons S.J. Transactivating agonists of the EGF receptor require Tyr 845 phosphorylation for induction of DNA synthesis. Mol. Carcinog. 2005;44:262–273. doi: 10.1002/mc.20138. [DOI] [PubMed] [Google Scholar]
- 59.Kloth M.T., Laughlin K.K., Biscardi J.S., Boerner J.L., Parsons S.J. STAT5b, a mediator of synergism between c-Src and the epidermal growth factor receptor. J. Biol. Chem. 2003;278:1671–1679. doi: 10.1074/jbc.M207289200. [DOI] [PubMed] [Google Scholar]
- 60.Ishizawar R.C., Miyake T., Parsons S.J. c-Src modulates ErbB2 and ErbB3 heterocomplex formation and function. Oncogene. 2007;26:3503–3510. doi: 10.1038/sj.onc.1210138. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.