Abstract
Background
The bacterium Streptococcus pyogenes causes a variety of human diseases that range from relatively mild skin infections to severe invasive diseases, such as acute rheumatic fever, glomerulonephritis, puerperal sepsis, necrotizing fasciitis, meningitis, and streptococcal toxic shock syndrome. Accurate identification and typing of group A hemolytic streptococci (GAS) is essential for epidemiological and pathogenetic studies of streptococcal diseases. For this reason, The genetic diversity of group A streptococcal (GAS) isolates from subjects in the United Arab Emirates with streptococcal disease was studied using emm gene sequence analysis. The emm typing system which is based on sequence analysis of PCR products of the N-terminal hypervariable region of the M protein gene, concurs with M serotyping almost 1:1.
Findings
A total of 38 GAS isolates were analyzed, including 35 isolates from throat and 3 from skin. Among the 38 isolates, a total of 25 different emm/st types were detected: 20 isolates (53%) belonged to 16 validated standard reference emm types and 18 isolates (47%) belonged to 9 recognized sequence types.
Conclusions
This is the first emm typing study in the United Arab Emirates to demonstrate the heterogeneity of the GAS population.
Findings
Introduction
The bacterium Streptococcus pyogenes causes a variety of human diseases that range from relatively mild skin infections to severe invasive diseases, such as acute rheumatic fever, glomerulonephritis, puerperal sepsis, necrotizing fasciitis, meningitis, and streptococcal toxic shock syndrome [1-3].
Accurate identification and typing of group A hemolytic streptococci (GAS) is essential for epidemiological and pathogenetic studies of streptococcal diseases. Rebecca Lancefield developed a serotyping system based on antigenic variation of the cell surface M protein that has been in use since 1928. Although additional serotyping systems, i.e. T and OF typing, were developed as valuable and practical substitutes, the M typing system has been considered the gold standard method. However, a limited supply of antisera and the high nontypeability rate among isolates, in particular among isolates from the tropics, present a challenge to using the M typing system. In recent years, several molecular typing systems have been developed as alternatives to M typing [4-7].
Numerous typing schemes have been used to characterize and measure genetic diversity among S. pyogenes isolates. Perhaps the most common tool used today is emm typing. The emm typing system [4,6,7], which is based on sequence analysis of PCR products of the N-terminal hypervariable region of the M protein gene, concurs with M serotyping almost 1:1. In addition to its simplicity, emm typing has allowed the detection of several previously unknown GAS types from different geographic regions [8-11]. Knowledge of the dynamics of emm types in a given region may shed light on the pathogenesis of GAS infections and is critical for selecting appropriate vaccine candidates.
It has been noted that there are differences in the distribution of GAS M-types in tropical versus temperate regions, since antisera produced against strains from Europe and the United States do not recognize large proportions of isolates from the tropics. This implies that a multivalent vaccine targeting the most common emm types in another part of the world may be ineffective in the tropics. In this study, we surveyed the genetic diversity of GAS in the UAE using emm gene sequence analysis. We wished to better understand the epidemiology of this important pathogen in a country where complications resulting from streptococcal infections are still common. This study is the first to report emm typing results in the UAE. The findings demonstrate the divergent features of GAS in this region.
Materials and methods
Ethical Approval
This study was reviewed and approved by Zayed Military Hospital. Patients were managed under standard approved hospital procedures.
Bacterial isolates
The GAS isolates in the present study were obtained in the UAE from January 2009 to December 2009 from patients with signs of streptococcal disease. Patients are from different ethnic groups (multinational). A total of 38 GAS isolates were included: 35 isolates from throat and 3 from skin. The isolates were associated with cases of acute tonsillitis (n = 35) and impetigo (n = 3). All GAS isolates were subjected to biochemical classification using the VITEK 2 system (bioMérieux) and SLIDEX STREPTO Plus latex agglutination kit (bioMérieux) to further verify that they belonged to Streptococcus pyogenes (group A). This collection of isolates was tested previously for antibiotic resistance to commonly used antibiotics using the disk diffusion method [12].
emm typing
emm typing was performed according to the protocol described by the Centers for Disease Control and Prevention (CDC; http://www.cdc.gov/ncidod/biotech/strep/protocols.html). DNA templates were prepared using the QIAGEN bacterial extraction kit. Briefly, for each isolate, about half a loop of bacterial cells was picked from overnight growth on blood agar plates and suspended in 100 μl of distilled water. This suspension was then used for bacterial extraction according to the instructions for the kit. The eluted DNA was used immediately for PCR or else was stored at -20°C until use. Primers 1 and 2, as specified by the CDC protocol, were used for amplifying the N-terminal region of the emm gene: primer 1, 5-TAT TCG CTT AGA AAA TTA A-3 and primer 2, 5-GCA AGT TCT TCA GCT TGT TT-3. AmpliTaq Gold (Applied Biosystems) PCR reagents were used according to the manufacturer's instructions. Amplification was performed on a PCR thermal cycler (GeneAmp*, PCR System 9700; Applied Biosystems) using cycle parameters specified by the CDC. Amplification products were purified using the JetQuick spin column technique (Germond) according to the manufacturer's instructions. PCR products were analyzed on an agarose gel to ensure the quality of the DNA and to estimate the concentration for sequencing.
Primer emmseq2, 5-TAT TCG CTT AGA AAA TTA AAA ACA GG-3, or primer 1 was used for sequencing the PCR products using the BioDye Terminator Cycle Sequencing Ready Reaction kit (version 2.0; BioSystems). Primer 1 was used when emmseq2 sequencing did not give good results. The sequencing reaction mixtures were prepared according to the kit instructions using the following parameters: 96°C for 1 min; 25 cycles of 96°C for 10 s; 50°C for 5 s; and 60°C for 4 min. The holding temperature was 4°C. The sequencing products were purified using ethanol sodium acetate precipitation and subjected to automated sequence analysis on an ABI Prism 310 Genetic Analyzer. The Geneious genetic software program (version 4.7.5) was used to edit the sequences. The 5' end of the sequences was compared to sequences in the CDC database http://www.cdc.gov/ncidod/biotech/strep/strepblast.htm. Sequences that match sequences in this database were checked for similarity with the published sequences in the GenBank database http://www.ncbi.nlm.nih.gov/BLAST/.
GenBank accession numbers
Sequences from the emm gene that were acquired during this study were deposited in the GenBank under numbers HM015360-HM015397.
Tests for significance
The level of significance was determined by calculating the P value using the chi-square test.
Results
Emm type distribution
Table 1 shows the emm/st type, site of isolation, and disease association of the 38 GAS isolates in this study. "emm" refers to the type-specific sequence of one of the standard reference GAS strains, and "st" stands for sequence types discovered later, including novel sequence types in the present study that await validation by the international committee http://www.cdc.gov/ncidod/biotech/strep/strepblast.htm[13]. A total of 25 different emm/st types were detected among the 38 GAS isolates. Of these 38 isolates, 20 (35%) belonged to 16 validated standard reference emm types [13,14], and 18 isolates (47%) belonged to 9 recognized sequence types http://www.cdc.gov/ncidod/biotech/strep/strepblast.htm.
Table 1.
M protein gene (emm) type distribution in relation to disease and isolation site.
| Site of isolation | Disease status | |||||
|---|---|---|---|---|---|---|
| emm type | Total no. of isolates | % | Skin | Throat | Impetigo | Tonsillitis |
| emm89 | 3 | 7.9 | 3 | 3 | ||
| st4695.0 | 2 | 5.3 | 2 | 2 | ||
| emm83.8 | 1 | 2.6 | 1 | 1 | ||
| emm78.3 | 2 | 5.3 | 2 | 2 | ||
| st3211.0 | 4 | 10.5 | 4 | 4 | ||
| st75.0 | 2 | 5.3 | 2 | 2 | ||
| 112.2 | 1 | 2.6 | 1 | 1 | ||
| stIL103.1 | 1 | 2.6 | 1 | 1 | ||
| emm1 | 1 | 2.6 | 1 | 1 | ||
| stIL62.0 | 2 | 7.9 | 2 | 2 | ||
| emm75.2 | 1 | 2.6 | 1 | 1 | ||
| emm100.5 | 1 | 2.6 | 1 | 1 | ||
| emm4.9 | 1 | 2.6 | 1 | 1 | ||
| emm4.4 | 1 | 2.6 | 1 | 1 | ||
| emm120 | 1 | 2.6 | 1 | 1 | ||
| stC6979.1 | 1 | 2.6 | 1 | 1 | ||
| st0721.0 | 3 | 7.9 | 3 | 3 | ||
| emm75 | 2 | 5.3 | 2 | 2 | ||
| stG97.0 | 1 | 2.6 | 1 | 1 | ||
| emm71.3 | 1 | 2.6 | 1 | 1 | ||
| emm22.1 | 1 | 2.6 | 1 | 1 | ||
| emm22 | 1 | 2.6 | 1 | 1 | ||
| emm83.1 | 1 | 2.6 | 1 | 1 | ||
| st9505.0 | 1 | 2.6 | 1 | 1 | ||
| emm87 | 1 | 2.6 | 1 | 1 | ||
| Total | 38 | 100 | 3 | 35 | 3 | 35 |
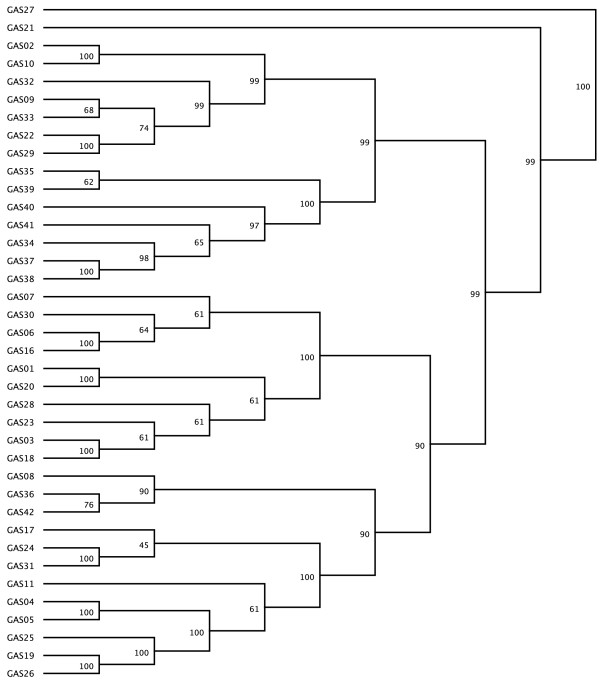
To compare the clonal relationships between the isolates, a dendrogram was generated with the UPGMA algorithm (Fig. 1).
Figure 1.
Dendrogram for the 38 GAS emm gene sequences according the the isolate number. The dendrogram was constructed with Geneious V4.8 sofware, by using the UPGMA algorithm.
In contrast to most studies in which a large proportion of the GAS isolates belong to a few emm types, there were no dominant emm types in the present study. Eight different emm types (listed in descending order: st3211.0, st0721.0, emm89, emm 75, st4695.0, st75.0, stIL62.0, and emm78.3) made up 55.4% of the isolates (Table 1). The most common type, st3211.0, comprised only 10.5% of the total isolates (4 isolates). Interestingly, types emm12, emm18, emm3, emm4, which have remained among the most frequent emm types for decades in the Western world [15-17], were absent in this collection. Type emm89, which is frequent in Canada [17], was also frequent in this study.
emm types of skin and throat isolates
The concept of distinct throat and skin emm types has been widely accepted. M protein serotypes, such as M types 1, 3, 4, 5, 6, 12, 14, 18, 19, and 24 of S. pyogenes, are associated with throat infections [18], while M serotypes such as 2, 49, 57, 59, 60, and 61 are associated with impetigo [19]. The number of isolates of each emm type in the present study is too low make statistical correlations regarding tissue specificity. However, it is worth noting that a number of emm types with ≥3 isolates, namely st3211.0, stIl62.0, st0721.0, and emm89, were isolated only from throat samples. One emm type with two isolates, st4695.0, was isolated only from the skin.
Discussion
The findings in this study demonstrated that the GAS isolates from patients in the UAE are highly heterogeneous, with 25 distinct emm/st types detected among the 38 isolates that were collected over a 1-year period. The number of distinct emm/st types detected in this study, 25, is not much higher than the number reported in emm typing studies conducted in other countries that involved more isolates collected over a longer period of time[2,16,17,19,20]. In Japan, 29 emm/st types were detected among 906 clinical isolates collected from 1990 to 1999 [21]. In Mexico, 31 emm types were detected among 423 isolates collected from symptomatic pharyngeal specimens from 1991 to 2000 [20]. In a population-based study among the Australian aboriginal population, 31 distinct types were reported among 141 isolates collected from the skin and throats of asymptomatic cases over a 25-month period [8]. In a nationwide survey in Canada, 54 M serotypes were reported among 4,760 GAS isolates submitted to the national center for streptococci from 1993 to 1999 [17]. In Spain, only 30 different emm types were detected among 614 pharyngeal isolates collected from eight different hospitals over a 4-year period[22]. The reason for the relatively higher heterogeneity observed among the UAE isolates can be attributed to the heterogeneity of the UAE population due to many international immigrants.
Since the introduction of the GAS sequence typing method based on the heterogeneity of the 5' end of the emm gene [4,6], several previously undocumented emm types have been detected. The number of distinct emm/st types has more than doubled in the last few years, and currently the CDC database contains over 225 distinct emm/st types. In the present study, there were no new emm types among the isolates. This might due to the small number of isolates. The detection rate of undocumented emm types has varied from country to country. In general, more new types have been reported in Malaysia [9], Thailand [10], Brazil [11], the United States [23], and Australia [8] compared than in European countries.
The highly diverse nature of the GAS population in the UAE calls into question the concept of using a multivalent vaccine composed of the N-terminal regions of the most common emm types. A multivalent vaccine composed of 26 M protein N-terminal regions was anticipated to protect against ca. 90% of the invasive GAS infections in the United States [24]. Designing an effective multivalent vaccine based on the N-terminal regions of M protein for countries such as the UAE, where the GAS population is highly heterogeneous and the dynamics of streptococcal infections are poorly understood, presents a challenge. Based on the observations in the present study, a vaccine with at least 25 different emm type-specific epitopes would be required to cover ca. 80% of the GAS infections in the UAE.
The aim of this study was to survey the genetic diversity of GAS isolates from the UAE using emm gene sequence analysis. Our GAS collection contained isolates from the skin and throats of symptomatic individuals, and the findings demonstrated the highly heterogeneous nature of the GAS population in the UAE during the study period. This study is the first of its kind to be conducted in samples from the UAE, and it underscores the unique nature of GAS epidemiology in this region. To better understand the dynamics of GAS epidemiology in the tropics, further surveillance in the region is needed. This may also help elucidate the pathogenesis of this bacterium, as well as help design an appropriate vaccine.
Competing interests
The author declares that he has no competing interests.
Authors' contributions
All the work was conducted by the author himself.
Acknowledgements
This work was financially supported by the Zayed Military Hospital. The author wish to extend special thanks to the medical technologists at the Division of Medical Microbiology and Division of Molecular Microbiology (Department of Pathology & Laboratory Medicine, Zayed Military Hospital, Abudhabi, UAE) for their support in this study.
References
- Efstratiou A. Group A streptococci in the 1990s. J Antimicrob Chemother. 2000;45(Suppl):3–12. doi: 10.1093/jac/45.suppl_1.3. [DOI] [PubMed] [Google Scholar]
- Spencer RC. Invasive streptococci. Eur J Clin Microbiol Infect Dis. 1995;14(Suppl 1):S26–32. [PubMed] [Google Scholar]
- Chiou CS, Liao TL, Wang TH, Chang HL, Liao JC, Li CC. Epidemiology and molecular characterization of Streptococcus pyogenes recovered from scarlet fever patients in central Taiwan from 1996 to 1999. J Clin Microbiol. 2004;42:3998–4006. doi: 10.1128/JCM.42.9.3998-4006.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall B, Facklam R, Thompson T. Sequencing emm-specific PCR products for routine and accurate typing of group A streptococci. J Clin Microbiol. 1996;34:953–958. doi: 10.1128/jcm.34.4.953-958.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner DL, Goodfellow AM, Martin DR, Sriprakash KS. Group A streptococcal Vir types are M-protein gene (emm) sequence type specific. J Clin Microbiol. 1998;36:902–907. doi: 10.1128/jcm.36.4.902-907.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufhold A, Podbielski A, Johnson DR, Kaplan EL, Lutticken R. M protein gene typing of Streptococcus pyogenes by nonradioactively labeled oligonucleotide probes. J Clin Microbiol. 1992;30:2391–2397. doi: 10.1128/jcm.30.9.2391-2397.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewodros W, Kronvall G. M protein gene (emm type) analysis of group A beta-hemolytic streptococci from Ethiopia reveals unique patterns. J Clin Microbiol. 2005;43:4369–4376. doi: 10.1128/JCM.43.9.4369-4376.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessen DE, Carapetis JR, Beall B, Katz R, Hibble M, Currie BJ, Collingridge T, Izzo MW, Scaramuzzino DA, Sriprakash KS. Contrasting molecular epidemiology of group A streptococci causing tropical and nontropical infections of the skin and throat. J Infect Dis. 2000;182:1109–1116. doi: 10.1086/315842. [DOI] [PubMed] [Google Scholar]
- Jamal F, Pit S, Facklam R, Beall B. New emm (M protein gene) sequences of group A streptococci isolated from Malaysian patients. Emerg Infect Dis. 1999;5:182–183. doi: 10.3201/eid0501.990129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruksakorn S, Sittisombut N, Phornphutkul C, Pruksachatkunakorn C, Good MF, Brandt E. Epidemiological analysis of non-M-typeable group A Streptococcus isolates from a Thai population in northern Thailand. J Clin Microbiol. 2000;38:1250–1254. doi: 10.1128/jcm.38.3.1250-1254.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira LM, Barros RR, Castro AC, Peralta JM, Da Gloria SCM, Talkington DF, Vivoni AM, Facklam RR, Beall B. Genetic and phenotypic features of Streptococcus pyogenes strains isolated in Brazil that harbor new emm sequences. J Clin Microbiol. 2001;39:3290–3295. doi: 10.1128/JCM.39.9.3290-3295.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewodros W, Muhe L, Daniel E, Schalen C, Kronvall G. A one-year study of streptococcal infections and their complications among Ethiopian children. Epidemiol Infect. 1992;109:211–225. doi: 10.1017/S0950268800050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facklam RF, Martin DR, Lovgren M, Johnson DR, Efstratiou A, Thompson TA, Gowan S, Kriz P, Tyrrell GJ, Kaplan E, Beall B. Extension of the Lancefield classification for group A streptococci by addition of 22 new M protein gene sequence types from clinical isolates: emm103 to emm124. Clin Infect Dis. 2002;34:28–38. doi: 10.1086/324621. [DOI] [PubMed] [Google Scholar]
- Facklam R, Beall B, Efstratiou A, Fischetti V, Johnson D, Kaplan E, Kriz P, Lovgren M, Martin D, Schwartz B. emm typing and validation of provisional M types for group A streptococci. Emerg Infect Dis. 1999;5:247–253. doi: 10.3201/eid0502.990209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt CM, Spellerberg B, Honscha M, Truong ND, Hoevener B, Lutticken R. Typing of Streptococcus pyogenes strains isolated from throat infections in the region of Aachen, Germany. Infection. 2001;29:163–165. doi: 10.1007/s15010-001-1106-x. [DOI] [PubMed] [Google Scholar]
- Colman G, Tanna A, Efstratiou A, Gaworzewska ET. The serotypes of Streptococcus pyogenes present in Britain during 1980-1990 and their association with disease. J Med Microbiol. 1993;39:165–178. doi: 10.1099/00222615-39-3-165. [DOI] [PubMed] [Google Scholar]
- Tyrrell GJ, Lovgren M, Forwick B, Hoe NP, Musser JM, Talbot JA. M types of group a streptococcal isolates submitted to the National Centre for Streptococcus (Canada) from 1993 to 1999. J Clin Microbiol. 2002;40:4466–4471. doi: 10.1128/JCM.40.12.4466-4471.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stollerman GH. Rheumatic fever in the 21st century. Clin Infect Dis. 2001;33:806–814. doi: 10.1086/322665. [DOI] [PubMed] [Google Scholar]
- Cunningham MW. Pathogenesis of group A streptococcal infections. Clin Microbiol Rev. 2000;13:470–511. doi: 10.1128/CMR.13.3.470-511.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa LE, Li Z, Gomez Barreto D, Calderon Jaimes E, Rodriguez RS, Sakota V, Facklam RR, Beall B. M protein gene type distribution among group A streptococcal clinical isolates recovered in Mexico City, Mexico, from 1991 to and Durango, Mexico, from 1998 to 1999: overlap with type distribution within the United States. J Clin Microbiol. 2000;41:373–378. doi: 10.1128/JCM.41.1.373-378.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka D, Gyobu Y, Kodama H, Isobe J, Hosorogi S, Hiramoto Y, Karasawa T, Nakamura S. emm Typing of group A streptococcus clinical isolates: identification of dominant types for throat and skin isolates. Microbiol Immunol. 2002;46:419–423. doi: 10.1111/j.1348-0421.2002.tb02715.x. [DOI] [PubMed] [Google Scholar]
- Alberti S, Garcia-Rey C, Dominguez MA, Aguilar L, Cercenado E, Gobernado M, Garcia-Perea A. Survey of emm gene sequences from pharyngeal Streptococcus pyogenes isolates collected in Spain and their relationship with erythromycin susceptibility. J Clin Microbiol. 2003;41:2385–2390. doi: 10.1128/JCM.41.6.2385-2390.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall B, Facklam R, Hoenes T, Schwartz B. Survey of emm gene sequences and T-antigen types from systemic Streptococcus pyogenes infection isolates collected in San Francisco, California; Atlanta, Georgia; and Connecticut in 1994 and 1995. J Clin Microbiol. 1997;35:1231–1235. doi: 10.1128/jcm.35.5.1231-1235.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu MC, Walls MA, Stroop SD, Reddish MA, Beall B, Dale JB. Immunogenicity of a 26-valent group A streptococcal vaccine. Infect Immun. 2002;70:2171–2177. doi: 10.1128/IAI.70.4.2171-2177.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]