Abstract
Drawing valid conclusions from an experiment often requires associating statistical confidence measures with the observed data. But these measures can be stated in terms of p-values, false discovery rates or q-values. What are the differences? And how should you decide which one to use?
Imagine that you have just invested a significant amount of time and money in a shotgun proteomics experiment designed to identify proteins involved in a particular biological process. The experiment successfully identifies most of the proteins that you already know to be involved in the process and implicates a few more. Each of these novel candidates will need to be verified with a follow-up assay. How do you decide how many candidates to pursue? The answer lies in the trade-off between the cost associated with a false positive versus the benefit of identifying a novel participant in the biological process that you are studying. A similar cost-benefit tradeoff arises in the context of many genomic or proteomic studies, for example, identifying genes that are differentially expressed on the basis of microarray or RNA-Seq experiments, scanning a genome for occurrences of candidate transcription factor binding sites, searching a protein database for homologs of a query protein, or evaluating the results of a genome-wide association study.
To assess this type of cost-benefit tradeoff, it is helpful to associate with each discovery a statistical confidence measure. However, these measures may variously be stated in terms of p-values, false discovery rates or q-values. The goal of this article is to provide you with an intuitive understanding of these confidence measures, a sense for how they are computed, and some guidelines for how to select an appropriate measure for a given experiment.
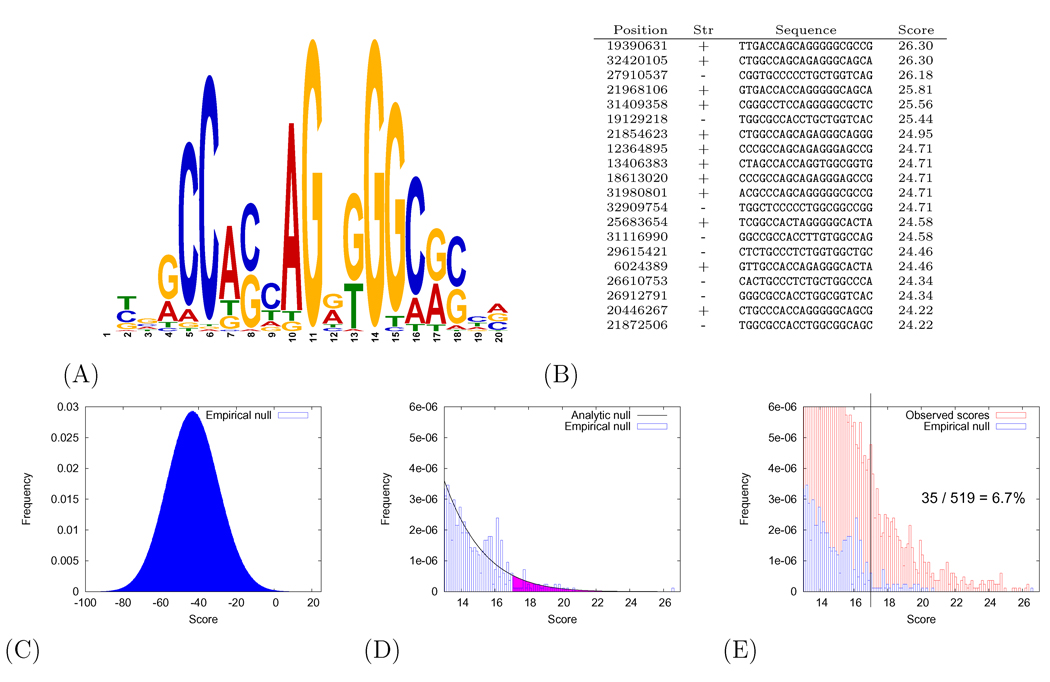
As a motivating example, suppose that you are studying CTCF, a highly conserved zinc finger protein that exhibits diverse regulatory functions and that may play a major role in the global organization of the chromatin architecture of the human genome [1]. To better understand this protein, you want to identify candidate CTCF binding sites in human chromosome 21. Using a previously published model of the CTCF binding motif, shown in Figure 1(A) [2], you compute a score for each length-20 subsequence of chromosome 21. Considering both DNA strands, there are 68 million such subsequences. Figure 1(B) lists the top 20 scores from such a search.
Figure 1. Scanning human chromosome 21 for CTCF binding motifs.
(A) The position-specific scoring matrix is represented as a sequence logo [9], in which the height of each letter is proportional to the information content at that position. The CTCF model is from [2]. (B) The 20 top-scoring occurrences of the CTCF binding site in human chromosome 21. Coordinates of the starting position of each occurrence are given with respect to human genome assembly NCBI 36.1. (C) A histogram of scores produced by scanning a shuffled version of human chromosome 21 with the CTCF motif. (D) This panel zooms in on the right tail of the distribution shown in panel (C). The blue histogram corresponds to the observed score distribution from scanning a shuffled chromosome, and the grey line corresponds to the analytic distribution. The p-value associated with an observed score of 17 is equal to the area under the curve to the right of 17. (E) The FDR is estimated from the empirical null distribution for a score threshold of 17.00. There are 35 null scores > 17 and 519 observed scores > 17, leading to an estimate of 6.7%. This procedure assumes that the number of observed scores equals the number of null scores.
If we consider, for example, the top-scoring sequence with a score of 26.30, then the natural question is whether a score this large is likely to occur by chance. This probability can be estimated by defining a null hypothesis that represents, essentially, the scenario that we are not interested in. In this case, a model of the null hypothesis might be created, for example, by shuffling the bases of chromosome 21. After this shuffling procedure, high-scoring occurrences of the CTCF motif will only appear due to random chance. We can then re-scan this chromosome with the same CTCF matrix. The distribution of the resulting scores is shown in Figure 1(C). Although it is not visible in the figure, out of the 68 million length-20 sequences in this shuffled chromosome, exactly one achieved a score ≥ 26.30. We can say that the probability of observing this score under the null hypothesis is 1/(68 × 106), or 1.5 × 10−8. This probability—the probability that a score at least as large as the observed score would occur in data drawn according to the null hypothesis—is called the p-value.
Of course, we are typically interested not only in the top of the ranked list shown in Figure 1(B). If we consider, for example, a candidate CTCF binding site that receives a score of 17.0, then the corresponding p-value is equal to the percentage of null scores greater than or equal to 17.0. Among the 68 million null scores shown in Figure 1(C), 35 are greater than or equal to 17.0, leading to a p-value of 5.5 × 10−7. The p-value associated with score x corresponds to the area under the null distribution to the right of x, as shown in Figure 1(D).
Shuffling the human genome and re-scanning with the CTCF motif is an example of an empirical null model. In some cases, however, it is possible to analytically calculate the form of the null distribution and calculate corresponding p-values. This approach can be more efficient than explicitly computing a large number of scores. In the case of scanning for CTCF motif occurrences, a dynamic programming algorithm can compute the null distribution assuming that the sequence being scanned is generated randomly with a specified frequency of each of the four nucleotides [3]. The grey line in Figure 1(D) corresponds to this analytic null distribution. This distribution allows us to compute, for example, that the p-value associated with the top score in Figure 1(B) is 2.3 × 10−10. This p-value is more accurate and much cheaper to compute than the p-value estimated from the empirical null model.
In practice, to determine whether an observed score is deemed statistically significant, the corresponding statistical confidence measure (the p-value) must be compared to a confidence threshold α. For historical reasons, many studies use thresholds of α = 0.01 or α = 0.05, though there is nothing magical about these values. In practice, the choice of the significance threshold depends upon the costs associated with false positives and false negatives, and these costs may differ from one experiment to the next.
If, rather than scanning all of chromosome 21, I had only examined a single length-20 sequence, then I could use the p-value directly as a statistical confidence measure. Unfortunately, in the context of an experiment that produces many scores, such as scanning a chromosome for CTCF binding sites, reporting a p-value is inappropriate. This is because the p-value is only computed with respect to a single score. In the example above, a score of 17.0 achieves a p-value of 5.5 × 10−7, which seems very impressive: the chance of obtaining such a p-value from null data is less than one in a million. But this small probability must be adjusted for the fact that we tested 68 million length-20 sequences. The large number of tests explains why, in this particular case, a scan of the shuffled genome produced 35 p-values smaller than 5.5 × 10−7. We need a multiple testing correction procedure to adjust our statistical confidence measures based on the number of tests performed.
Perhaps the simplest and most widely used method of multiple testing correction is the Bonferroni adjustment. If you are using a significance threshold of α, but you perform n separate tests, then the Bonferroni adjustment deems a score significant only if the corresponding p-value is ≤ α/n. In the case of the CTCF scan above, we considered 68 million distinct 20-mers as candidate CTCF sites, so achieving statistical significance at α = 0.01 according to the Bonferroni criterion would require a p-value less than 0.01/(68 × 106) = 1.5 × 10−10. Because the smallest observed p-value in Figure 1(B) is 2.3 × 10−10, no scores are deemed significant in this test.
The Bonferroni adjustment, when applied using a threshold of α to a collection of n scores, controls the family-wise error rate; i.e., the adjustment ensures that the probability that one or more scores were drawn according to the null distribution is α. Practically speaking, this means that, given a set of CTCF sites with a Bonferroni adjusted significance threshold of α = 0.01, we can be 99% sure that none of the scores would be observed by chance when drawn according to the null hypothesis.
In many multiple testing settings, minimizing the family-wise error rate is too strict. Rather than saying that we want to be 99% sure that none of the observed scores is drawn according to the null, it is frequently sufficient to identify a set of scores for which a specified percentage of scores are drawn according to the null. This is the basis of multiple testing correction via false discovery rate (FDR) estimation.
The simplest form of FDR estimation is illustrated in Figure 1(E), again using an empirical null distribution for the CTCF scan. For a specified score threshold t = 17.00, we count the number sb of observed scores ≥ t and the number sn of null scores ≥ t. Assuming that the total number of observed scores and null scores are equal, then the estimated FDR is simply sn/sb. In the case of our CTCF scan, the FDR associated with a score of 17.00 is 35/519 = 6.7%.
Note that, in Figure 1(E), we computed FDR estimates directly from the score. It is also possible to compute FDRs from p-values using the Benjamini-Hochberg procedure, which relies on the p-values being uniformly distributed under the null hypothesis [4]. For example, if the p-values are uniformly distributed, then the p-value 5% of the way down the sorted list should be approximately 0.05. Accordingly, the procedure consists of sorting the p-values in ascending order, and then dividing each observed p-value by its percentile rank to get an estimated FDR. In this way, small p-values that appear far down the sorted list will result in small FDR estimates, and vice versa. In general, when an analytical null model is available, you should use it to compute p-values and then use the Benjamini-Hochberg procedure, because the resulting estimated FDRs will be more accurate. However, if you only have an empirical null model, then there is no need to estimate p-values in an intermediate step.
The simple FDR estimation method shown in Figure 1(E) is sufficient for many studies, and the resulting estimates are provably conservative with respect to a specified null hypothesis; i.e., if the simple method estimates that the FDR associated with a collection of scores is 5%, then on average the true FDR is ≤ 5%. However, a variety of more sophisticated methods have been developed for achieving more accurate FDR estimates (reviewed in [5]). Most of these methods focus on estimating a parameter π0, which represents the percentage of the observed scores that are drawn according to the null distribution. Depending on the data, applying such methods may make a big difference or almost no difference at all. For the CTCF scan, one such method [6] assigns slightly lower estimated FDRs to each observed score, but the number of sites identified at a 5% FDR threshold remains unchanged relative to the simpler method.
Complementary to the FDR, Storey [6] proposed defining the q-value as an analog of the p-value that incorporates FDR-based multiple testing correction. The q-value is motivated, in part, by a somewhat unfortunate mathematical property of the FDR: when considering a ranked list of scores, it is possible for the FDR associated with the first m scores to be higher than the FDR associated with the first m+1 scores. For example, the FDR associated with the first 84 candidate CTCF sites in our ranked list is 0.0119, but the FDR associated with the first 85 sites is 0.0111. Unfortunately, this nonmonotonicity can make the resulting FDR estimates difficult to interpret. Consequently, Storey proposed defining the q-value as the minimum FDR attained at or above a given score. If we use a score threshold of T, then the q-value associated with T is expected proportion of false positives among all of the scores above the threshold. This definition yields a well-behaved measure that is a function of the underlying score. We saw, above, that the Bonferroni adjustment yielded no significant matches at α = 0.05. If we use FDR analysis instead, then we are able to identify a collection of 519 sites at a q-value threshold of 0.05.
In general, for a fixed significance threshold and fixed null hypothesis, performing multiple testing correction via FDR estimation will always yield at least as many significant scores as using the Bonferroni adjustment. In most cases, FDR analysis will yield many more significant scores, as in our CTCF analysis. The question naturally arises, then, whether a Bonferroni adjustment is ever appropriate. Like choosing a significance threshold, choosing which multiple testing correction method to use depends upon the costs associated with false positives and false negatives. In particular, FDR analysis is appropriate if follow-up analyses will depend upon groups of scores. For example, if you plan to perform a collection of follow-up experiments and are willing to tolerate having a fixed percentage of those experiments fail, then FDR analysis may be appropriate. Alternatively, if follow-up will focus on a single example, then the Bonferroni adjustment is more appropriate.
It is worth noting that the statistics literature describes a related probability score, known as the local FDR [7]. Unlike the FDR, which is calculated with respect to a collection of scores, the local FDR is calculated with respect to a single score. The local FDR is the probability that a particular test gives rise to a false positive. In many situations, especially if we are interested in following up on a single gene or protein, this score may be precisely what is desired. However, in general, the local FDR is quite difficult to estimate accurately.
Furthermore, all methods for calculating p-values or for performing multiple testing correction assume a valid statistical model—either analytic or empirical—that captures dependencies in the data. For example, scanning a chromosome with the CTCF motif leads to dependencies among overlapping length-20 sequences. Also, the simple null model produced by shuffling assumes that nucleotides are independent. To the extent that these assumptions are not met, we risk introducing inaccuracies in our statistical confidence measures.
In summary, in any experimental setting in which multiple tests are performed, p-values must be adjusted appropriately. The Bonferroni adjustment controls the probability of making one false positive call. In contrast, false discovery rate estimation, as summarized in a q-value, controls the error rate among a set of tests. In general, multiple testing correction can be much more complex than is implied by the simple methods described here. In particular, it is often possible to design testing strategies that minimize the number of tests performed for a particular hypothesis or set of hypotheses. For more in-depth treatment of multiple testing issues, see [8].
References
- 1.Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim TH, et al. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Staden R. Searching for motifs in nucleic acid sequences. Methods in Molecular Biology. 1994;25:93–102. doi: 10.1385/0-89603-276-0:93. [DOI] [PubMed] [Google Scholar]
- 4.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society B. 1995;57:289–300. [Google Scholar]
- 5.Kerr KF. Comments on the analysis of unbalanced microarray data. Bioinformatics. 2009;25:2035–2041. doi: 10.1093/bioinformatics/btp363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Storey JD. A direct approach to false discovery rates. Journal of the Royal Statistical Society. 2002;64:479–498. [Google Scholar]
- 7.Efron B, Tibshirani R, Storey J, Tusher V. Empirical Bayes analysis of a microarray experiment. Journal of the American Statistical Association. 2001;96:1151–1161. [Google Scholar]
- 8.Dudoit S, van der Laan MJ. Multiple testing procedures with applications to genomics. New York, NY: Springer; 2008. [Google Scholar]
- 9.Schneider TD, Stephens RM. Sequence logos: A new way to display consensus sequences. Nucleic Acids Research. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]