Abstract
myc genes are associated with a wide variety of human cancers including most types of nervous system tumors. While the mechanisms by which myc overexpression causes tumorigenesis are multifaceted and have yet to be clearly elucidated, they are at least in part related to endogenous myc function in normal cells. Knockout (KO) of either c-myc or N-myc genes in neural stem and precursor cells (NSC) driven by nestin-cre impairs mouse brain growth and mutation of N-myc also causes microcephaly in humans in Feingold Syndrome. To further define myc function in NSC and nervous system development, we created a double KO (DKO) for c- and N-myc using nestin-cre. The DKO mice display profoundly impaired overall brain growth associated with decreased cell cycling and migration of NSC, which are strikingly decreased in number. The DKO brain also exhibits specific changes in gene expression including downregulation of genes involved in protein and nucleotide metabolism, mitosis, and chromatin structure as well as upregulation of genes associated with differentiation. Together these data support a model of nervous system tumorigenesis in which excess myc aberrantly locks in a developmentally active chromatin state characterized by overactive cell cycling, and metabolism as well as blocked differentiation.
Keywords: c-myc, N-myc, brain tumors, stem cells, metabolism, mitosis, pluripotency
INTRODUCTION
myc is well-known for its role in tumorigenesis when overexpressed and N-myc (MYCN) is most strongly associated with primitive “blast” nervous system tumors neuroblastoma, medulloblastoma, retinoblastoma, and glioblastoma [1-11]. However, at physiological levels myc genes are important regulators of many aspects of normal cell behavior including metabolism and cycling (reviewed in [12]). myc genes encode members of the basic-helix-loop-helix zipper (bHLHZ) transcription factor superfamily, but Myc proteins are very atypical bHLHZ factors in the diversity of their functions. They can activate and repress the transcription of specific protein coding genes, influence expression of miRNA and rRNA, drive DNA synthesis, and globally influence chromatin structure. More recently myc has been shown to relieve transcriptional pausing in embryonic stem cells (ESC) through a mechanisms involving PTEF-b [13], but in many cases the mechanisms by which myc achieves its diversity of functions are not well understood. One theory is that Myc's wide range of functions is linked to its widespread euchromatic function associated with specific histone modifications such as acetylation of lysine 9 and methylation of lysine 4 of histone H3 [14-19].
There is growing evidence of key roles for myc genes at endogenous levels in both somatic stem cells such as NSC and ESC. Constitutive knockout (KO) of c-myc or N-myc causes embryonic lethality around midgestation [20, 21]. Conditional disruption of N-myc in NSC severely disrupts murine brain growth, particularly that of the cerebellum, while a similar KO of c-myc moderately impairs growth [22, 23]. Disruption of either c-myc or N-myc or both in hematopoietic stem cells (HSC) also alters their normal biological functions, affecting survival and self-renewal [24, 25]. myc genes also are involved in the production of induced pluripotent stem (iPS) cells (reviewed in [26]) [27-31]. While exogenous myc is not formally required for the process [32, 33], it dramatically enhances the efficiency and in its absence its function is likely supplanted by endogenous myc. During iPS cell formation, Myc represses differentiation-associated genes [34] and may not have a key role in directly maintaining expression of pluripotency factors. However, in neuroblastoma some pluripotency genes such as lif, lin28b, Klf2, and Klf4 are N-Myc targets for activation, while a subset of these genes is also regulated in NSC by N-myc [35]. Another role that Myc may play in pluripotency is maintenance of the high levels of cellular metabolism, including protein (reviewed in [37]) and DNA [38] synthesis, observed in highly pluripotent cells.
Perhaps because of the importance of myc genes in normal cellular biology and their ability to cause cancer when in excess, cells have evolved systems to maintain normal total cumulative myc RNA and Myc protein levels. These include cross-regulation, redundancy and compensation between the 3 main myc genes – c-, N-, and L-myc – as well as myc-trigged apoptosis when in excess. Myc protein stability is also tightly controlled (reviewed in [39]). Conditional double knockout (DKO) of c- and N-myc in hematopoietic stem cells yields a far more severe phenotype than disruption of either gene alone, suggesting additive or redundant roles [25]. A large degree of redundancy is also supported by the knockin of N-myc into the c-myc locus largely rescuing the loss of c-myc [40]. The prevailing theory is that what is most critical is the total level of all myc gene expression in each cell. Despite fairly ubiquitous expression in the developing brain and some other regions of the embryo, L-myc constitutive KO was reported to have no phenotype at all [41]. One notion is that this lack of apparent phenotype was due to the continued presence of N-myc and perhaps c-myc, which could fulfill the roles of L-myc in its absence.
During neurogenesis, N-myc plays a particularly important role in NSC to direct brain growth and development [22, 42], consistent with its fairly widespread expression pattern. N-myc is also essential for normal eye and neural retina development [43, 44]. L-myc is also fairly widely expressed, particularly in the early midbrain, which suggests it may function in the brain despite no reported KO phenotype. Although c-myc expression has been less clearly defined, it appears to be more restricted than that of L- and N-myc. Overexpression and c-myc KO studies in rat neurospheres support a crucial role for myc genes overall in NSC function and suggested functional ties to p53/Arf [45], but the relevance for in vivo neurogenesis and tumorigenesis remains unclear. The molecular basis of the c- and N-myc nestin-cre driven brain phenotypes is also not completely clear, but in the N-myc knockout changes in cyclin D2 and cyclin dependent kinase inhibitors such as p18 and p27 were evident suggesting cell cycle regulation plays a role [22, 46], including in the cerebellum. Impeding our understanding is the fact that no unbiased global analysis of gene expression has been conducted for any Myc transcription factors using a loss of function model in the nervous system. Upstream of N-myc are several potential signaling pathways including Shh in the cerebellum and in cerebellar granule neural progenitors (CGNP) [23, 47], while N-myc also appears to be a critical part of a recently defined pathway DLL3 and Notch signaling in which N- Myc protein is targeted by the Huwe1 ubiquitin ligase [11, 48].
To address the endogenous functions of c- and N-myc in brain growth, we created a nervous system specific double c- and N-myc KO (DKO) mouse by crossing doubly homozygously myc floxed mice with nestin-cre, which is expressed specifically in NSC. The myc DKO mice have a nervous system phenotype much more severe than either single myc KO alone, which is ultimately lethal. In terms of mechanisms underlying the phenotype, we found evidence that myc transactivates key gene expression programs in NSC including that of metabolic genes essential for normal stem and precursor fate during neurogenesis, chromatin regulatory genes, and mitosis-related genes. In addition myc appears to repress expression of genes associated with differentiation. Together these findings support the model that excess myc causes nervous system tumors by “locking in” a chromatin-based developmental program in NSC characterized by rapid cell cycling, high cellular metabolism, and blocked differentiation.
RESULTS
Double knockout (DKO) of c- and N-myc in NSC driven by nestin-cre causes a striking impairment of embryonic brain growth
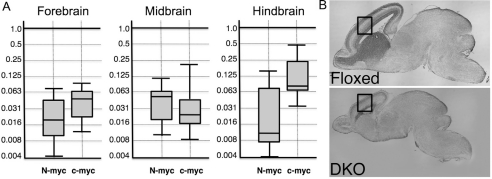
Control (c-myc flox/flox; N-myc flox/flox) and DKO (c-myc flox/flox; n-myc flox/flox nestin-cre+) embryos were produced by timed matings. Embryonic brains were isolated at different stages and analyzed. At E14.5, embryonic brains were microdissected into fore-, mid-, and hindbrains followed by RNA isolation. q RT-PCR was conducted for c- and N-myc. All three main regions of the embryonic brain exhibited pronounced decreases in expression of both myc genes. c- and N-myc levels were reduced by approximately 20 and 50-fold in the forebrain, 40 and 20-fold in the midbrain, and 15 and almost 100-fold in the hindbrain, respectively (Fig. 1A). In terms of phenotype, the E17.5 brain overall was dramatically reduced in size, particularly in the forebrain/neocortex (Fig. 1B). The DKO hindbrain also exhibited a growth phenotype, albeit somewhat less severe at this stage. Surprisingly, the growth of the DKO midbrain was not significantly affected despite the strong reductions in c- and N-myc therein, suggesting midbrain growth prior to midgestation is largely independent of c- and N-myc. In support of this notion, the DKO adult midbrain was also largely normal (not shown).
Figure 1. Nestin-Cre mediates potent reductions in both c- and N-myc expression levels throughout the brain causing dramatically impaired growth of the fore- and hindbrain, but not the midbrain.
(A) q RT-PCR for c- and N-myc expression. RNAs isolated from microdissected E17.5 control (N=4) and DKO (N=3) fore-, mid-, and hindbrains were used for q RT-PCR. Error bars are standard deviations. Mean levels in controls were set as 1.0. Median values are indicated by horizontal bars within vertical rectangles representing the range. p values were < 0.005 in each case. (B) Nissl stained E17.5 sagittal sections of doubly homozygously floxed littermate control and DKO (doubly homozygously floxed nestin-cre+). Boxed regions are shown at higher magnification in Fig. 2A.
myc family expression patterns in the embryonic brain correlate with the general domain specificity of myc knockout phenotypes
We have previously observed high levels of N-Myc protein expression in the developing forebrain and hindbrain/cerebella [22]. These findings correlated with E15.5 data from the brain gene expression website: http://www.stjudebgem.org/web/mainPage/mainPage.php (Supplemental Fig. S1). While c-myc expression at the protein or RNA levels is difficult to detect, there are hints of low-level expression in a number of regions including the developing cortex. N-myc is fairly ubiquitously expressed, but with relatively lower or absent expression in some midbrain regions and very high levels in the neocortex and the developing cerebellum. L-myc, despite no reported KO phenotype, has particularly high expression in the ventricular zone (VZ) of the embryonic midbrain supporting the notion that L-myc may be the central driver of midbrain development, potentially explaining the absence of any clear midbrain phenotype in the c- and N-myc DKO.
Expression of c- and N-myc in NSC is essential for normal architecture of the late embryonic cortical wall
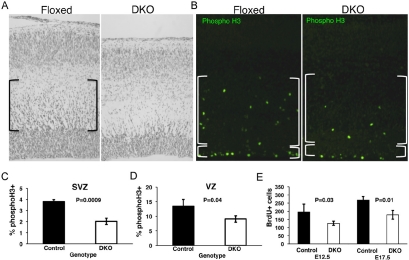
Because the DKO E17.5 cortex exhibited a dramatic overall phenotype visible to the eye, we examined it further by microscopic histological analysis of Nissl stained sagittal sections (Fig. 2A). The DKO cortical wall has a number of phenotypic characteristics including reduced overall thickness. Also evident was a hypocellular VZ and an apparent near complete absence of subventricular zone (SVZ; brackets in the floxed control) cells. These data suggest very few precursors are migrating through and out of the SVZ in the DKO, however the cortical plate at this stage is only modestly reduced in number and not at all in thickness. Together these findings suggest precocious formation of the cortical plate earlier than normal during differentiation.
Figure 2. Loss of c- and N-myc disrupts cortical wall development with a particularly pronounced affect on the SVZ.
(A) Nissl stained E17.5 sagittal sections of cortical wall. Brackets indicate the normal SVZ, largely absent in the DKO. (B) Immunostaining for the mitotic marker phospho-H3 (green) in control and DKO E17.5 sections. Large and small brackets indicate SVZ and VZ respectively. (C) and (D) Quantification of percent mitotic/phosphoH3+ cells in the SVZ and VZ. (E) Quantification of S phase/BrdU+ cells in the entire cortical wall in control and DKO. Error bars are standard deviations. P values are indicated.
Loss of c- and N-myc strongly reduces cell cycling, especially mitosis in the SVZ
To examine the potential effects of the myc DKO on cell cycling we stained for BrdU for S phase cells and phosphoH3 for M phase cells. The DKO cortical wall exhibited a pronounced decrease in the percentage of M phase cells (Fig. 2B), approximately 2-fold in the SVZ and 30% in the VZ (Figs. 2C-D). The more pronounced loss of mitotic cells in the SVZ is consistent with the notion of decreased migrating progenitors suggested by the Nissl staining. The absolute number of mitotic cells was even more dramatically decreased given the hypocellularity of the DKO cortical wall. We also observed a statistically significant loss of BrdU+ cells at all stages examined including as early as E12.5 (Supplemental Fig. S2A, Fig. 2E) and at E17.5.
Loss of c- and N-myc alters cell fate in the SVZ and VZ, consistent with a substantial decrease in stem and progenitors
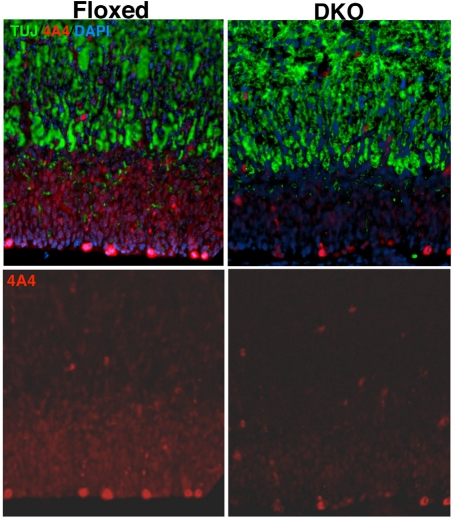
To examine the identity of cells in the cortical wall, we stained for 4A4 (a marker of stem and progenitors) and neural specific B-tubulin III (TUJ) in control and DKO sections (Fig. 3). The absolute size of the TUJ+ subdomain was not substantially altered in the DKO at E14.5 or at E17.5. However, because the overall cortical wall was greatly reduced in thickness in the DKO, the relative proportion of the wall that was TUJ+ was strikingly increased in the DKO (E17.5, not shown; E14.5, Supplemental Fig. S4). Supporting the notion of reduced progenitors, particularly those that are migrating, we saw decreased 4A4+ cells in the DKO (Fig. 3). The decrease in 4A4+ stem and precursor cells was not due to impaired cell survival as TUNEL staining indicated no clear different in apoptosis between control and DKO at E17.5 or at E12.5 (Supplemental Fig. S5; not shown). Interestingly, we found that while anti-4A4 (phospho vimentin) most brightly stains basal stem/precursors immediately adjacent within the VZ, cells in the TUJ- regions of the SVZ also stained 4A4+ (albeit with only moderately intense staining). Such staining is nearly completely absent from the DKO (Fig. 3 bottom). Thus, the SVZ DKO cells are both TUJ and 4A4 negative as well as mostly negative for S and M phase of the cell cycle, suggesting that they are in an abnormal developmental state and location due to the loss of myc.
Figure 3. c- and N-myc are required to maintain normal NSC cell cycling in the forebrain.
E17.5 Sagittal sections of control and myc DKO cortical walls were stained with TUJ (green), 4A4 (red) and DAPI (blue). 40x magnification.
c- and N-myc maintain expression of genes involved in most aspects of cellular metabolism, chromatin regulatory factors, and mitosis
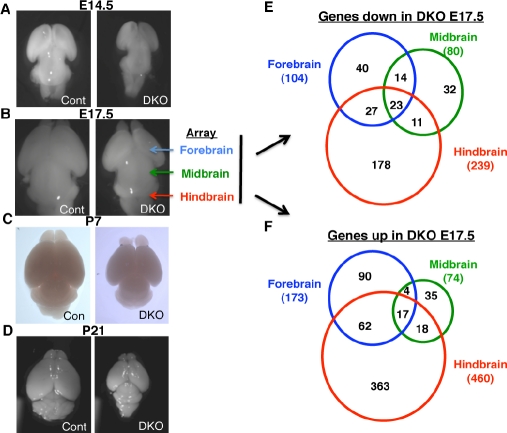
To explore the mechanisms by which Myc proteins acting as transcription factors could regulate brain development and specific NSC functions such as cell cycling and fate, gene expression array studies were conducted on RNAs isolated from E17.5 control and DKO subdomains of the brain including forebrain, midbrain, and hindbrain/cerebellum (Fig. 4). Whole brains were first isolated from E14.5-P21 to compare overall severity of phenotypes (Fig. 4A-D). A phenotype was apparent as early as E12.5 and became progressively more severe. Interestingly, while tangential growth of the brain was only modestly reduced, rostro-caudal growth was severely impaired at all stages. Forebrain, midbrain, and hindbrain were microdissected from control (4 biological replicates) and DKO (3 biological replicates), RNA was produced, and used to probe promoter expression arrays (Fig. 4E-F). There was substantial overlap in the specific genes whose expression was altered in the DKO in the 3 domains, suggesting some conserved transcription functions for Myc in different subdomains of the brain. The magnitude of gene expression changes (total # of genes with increased or decreased expression) not surprisingly was directly linkable to the severity of the phenotype in a given domain. For example, midbrain, which exhibits little if any phenotype had far fewer expression changes. Interestingly no two domains were more similar to each other than the third, suggesting that although the midbrain had no obvious outward phenotype and relatively fewer gene expression changes, a loss of myc gene expression midbrain phenotype was occurring to some extent.
Figure 4. myc DKO whole brain and gene expression phenotypes.
Brains of indicated genotypes (DKO and littermate doubly floxed cre negative controls) were microdissected and photographed from above (A-D). For expression microarrays, RNA was isolated from the indicated domain (fore, mid, or hindbrain; blue, green, and red respectively) in biological triplicate (DKO) or quadruplicate (control, doubly floxed Cre-). (E-F) Venn diagrams indicate the number of genes whose expression was changed 1.3 fold or more in the indicate domain and the overlap.
In addition to examining specific genes, complete lists of up or downregulated gene were subjected to ontological analysis to look for clusters of functionally related genes that are changed by loss of myc using the DAVID algorithm (http://david.abcc.ncifcrf.gov/). Focusing first on the candidate positively regulated Myc targets (genes downregulated in the DKO), the top scoring clusters were functionally related to ribosome biogenesis, protein translation, cellular metabolism, chromosomal binding proteins, and interestingly mitosis (Table 1). Of specific genes whose expression was altered by the DKO, most were verifiable by conventional RT-PCR and qRT-PCR (Supplemental Fig. S5A-B) and several downregulated genes, including Rpl23 and Bcat1 [50], are known myc target genes.
Table 1.
Ontology of genes downregulated in DKO
| Cluster Category | P-Value |
|---|---|
| Forebrain | |
| Ribosome | 5.0 × 10−13 |
| Nucleoside biosynthesis | 8.1 × 10−4 |
| Midbrain | |
| Actin | 4.8 × 10−4 |
| Hindbrain | |
| Ribosome | 1.2 × 10−32 |
| Chromosomal part | 1.2 × 10−8 |
| Nucleolus | 4.8 × 10−8 |
| DNA metabolism | 2.9x 10−7 |
| Mitosis | 1.6 × 10−6 |
Loss of myc leads to gene expression signatures of differentiation including ontological clusters of genes involved in patterning, morphogenesis, migration, and neurogenesis
There were many more genes upregulated by loss of myc than downregulated and correspondingly more functional ontological clusters (Table 2). Of the upregulated genes in the DKO, ontological analysis suggested that Myc most prominently repressed expression of genes associated with differentiation during neurogenesis and that in the DKO differentiation was occurring inappropriately at E17.5 in cells that would otherwise have remained NSC. Interestingly, we also found pronounced upregulation of a number of Wnt family members in the myc DKO hindbrain and N-myc has recently been implicated in cortical basal progenitors as a key effector of Wnt signaling [51]. The upregulation of Wnt signaling with loss of myc could reflect Myc repression of Wnt signaling in a negative feedback loop or alternatively be an indirect consequence of differentiation.
Table 2.
Ontology of genes upregulated in DKO
| Cluster Category | P-Value |
|---|---|
| Forebrain | |
| Signal | 4.9 × 10−11 |
| Branching morphogenesis | 1.4 × 10−6 |
| Cell Migration | 3.2 × 10−6 |
| Pattern Specification | 1.9 × 10−5 |
| Tube Development | 4.4 × 10−5 |
| Extracellular structure | 4.7 × 10−5 |
| Embryonic Organ Development | 8.9 × 10−5 |
| Midbrain | |
| Homeobox | 3.6 × 10−4 |
| Hindbrain | |
| Glycoprotein | 9.3 × 10−38 |
| Extracellular Matrix | 4.2 × 10−34 |
| Vascular Development | 4.2 × 10−16 |
| Tube Development | 1.1 × 10−12 |
| Cell Adhesion | 1.1 × 10−11 |
| Collagen | 2.8 × 10−9 |
| Heart morphogenesis | 8.3 × 10−8 |
| EGF-like region | 8.5 × 10−8 |
| Polysaccharide binding | 1.0 × 10−6 |
| Neurotransmitter transport | 1.5 × 10−6 |
| Fibronectin type III | 2.7 × 10−5 |
| Wnt signaling | 1.2 × 10−5 |
| Pattern Specification | 3.3 × 10−5 |
| Neuron differentiation | 3.5 × 10−5 |
| Hedgehog signaling | 7.5 × 10−5 |
| Bone Development | 8.6 × 10−5 |
| Cell motion | 1.4 × 10−4 |
| Ion Transport | 3.3 × 10−4 |
A subset of genes are consistently altered throughout the brain by loss of myc
When comparing the genes whose expression was changed in the 3 domains of the brain by loss of myc, it is important to note that a significant number had increased or decreased expression in all 3 domains (Supplemental Fig. S6). These genes therefore have the strongest link with myc in overall brain development. Amongst the shared downregulated genes (putative positively Myc regulated genes), although many are non-annotated, ontological analysis indicates that there are 3 kinases (Pkm2, Mapk8, and Adk, p value = 0.07) and 9 genes involved in metabolism (includes the 3 kinases mentioned above plus Arbp, Usp29, Atp6v1d, Ptpre, and Picalm, p value = 0.04), suggesting key Myc targets in precursors are involved in signaling and cell metabolism. Of the shared genes upregulated (putative Myc suppressed genes), ontological analysis found cellular receptors (p value = 0.01), mostly associated with differentiation including Wnt5a. Wnt signaling more generally was also upregulated specifically in the cerebellum, suggesting Myc may not only be a downstream effector of Wnt, but also regulate Wnt expression.
DISCUSSION
Although we and others have found very widespread chromatin and transcriptional functions for Myc proteins [15, 16, 19, 52-55], in our expression array studies here on control and myc DKO brain tissues, we found relatively modest levels of changes in gene expression and in some domains relatively few genes had altered expression with loss of myc. This is somewhat surprising given the striking severity of the DKO phenotype, but it may be reflective of the complexity of cell types in the developing brain (even when divided into fore-, mid-, and hindbrain) compared to more homogeneous cell lines used in previous studies. Nonetheless specific patterns of changes clearly emerged from ontological analyses. We observed that a key function for myc in NSC in the developing brain is to maintain overall cellular metabolism with particularly important roles in ribosome biogenesis and nucleotide metabolism.
We also observed an ontological cluster of downregulated mitotic genes that was highly significant, supporting a potential novel role for myc in regulating mitosis of normal NSC. Remarkably no mitotic genes were upregulated by loss of Myc. Recently Myc-induced genomic instability has been shown to be due to at least in part to mitotic dysfunction [56], consistent with our findings, and together suggesting normal Myc levels, not too high or too low, are essential for mitosis. Since we have also previously observed abnormal, widespread chromatin changes with loss of myc in NSC [16], this altered chromatin state may in addition contribute to problems going through mitosis in myc DKO NSC due to premature chromatin condensation in G2 or to failure to decondense late during mitosis. We also observed a decrease in mitotic cells throughout the developing DKO brain ranging from 50% to 2-fold depending on the region, but it remains unclear if the few mitotic DKO NSC are arrested and unable to undergo normal mitosis, or if they can achieve a normal mitosis. Some degree of mitotic arrest in the DKO NSC is of interest, especially as myc overexpression has been associated with G2 arrest [57] suggesting myc levels may be particularly important for G2/M. Although myc has previously been linked to mitosis, that study demonstrated that high levels of myc disrupted normal mitosis [58]. Since myc has been postulated to dissociate from chromatin during mitosis [59], it may be that during late G2 and early M phases, Myc orchestrates a transcriptional program that then normally carries cells through mitosis. Alternatively some level of Myc still associated with chromatin may directly regulate mitosis during mitosis.
In terms of region-specific brain growth, our model argues that c-, N-, and L-myc have both unique and combinatorial roles in directing regional growth of the murine brain. For example, L-myc is the prime candidate for the myc family member directing midbrain growth, a function that may have remained obscured in the constitutive L-myc KO mice due to redundancy with or compensation by N-myc. Our findings also suggest that myc gene levels and expression patterns may contribute to the differences in relative domain (fore, mid, and hindbrain as well as cerebellar) mass in different species and to brain evolution. For example, echolocators, which are unique in that their cerebellum consists of approximately 20%, instead of the more common 10%, of total brain mass, may have higher total myc expression levels or longer periods of myc expression during cerebellar development [60]. In addition in organisms with a relatively simpler brain structure, one might predict less complexity in myc function, and indeed in Drosophila Melanogaster, for example, there is only one myc gene, dmyc [61].
We did not observe any changes in cell survival and no clear apoptosis-associated gene expression changes were evident in the DKO. Thus, at least c- and N-myc appear largely dispensable for NSC survival in vivo. This sharply contrasts with HSC, where a dominant phenotype of the c- and N-myc DKO is apoptosis manifesting with dramatic changes in apoptosis-related gene expression [25]. Together these findings suggest the intriguing possibility that c and N-myc may have fundamentally distinct roles in NSC and HSC. Recent murine “placental rescue” [62] experiments point to the key role of c-myc in development being hematopoiesis, suggesting N-myc plays the primary role in development of the embryo itself. Consistent with this, mutation of N-myc in humans causes FS with its very large range of birth defects in a host of tissues[63]. Interestingly the range of defects in constitutive N-myc KO mice are remarkably similar to that of FS [64].
It is notable that we observe far more genes upregulated in the DKO brain than downregulated. We have also observed this pattern in gene expression studies in cultured NSC and in ESC (data not shown) [65]. While myc has been shown to have a repressive function through Miz1 [66], it has always been widely assumed that myc nonetheless is predominantly a transcriptional activator. However, growing evidence suggests that repression by myc, direct and/or indirect, is likely to be equally or more important than activation. Consistent with this idea is the notion that a key role for myc in iPS cell formation is suppression of fibroblast specific gene expression [34]. In iPS cells myc appears to potently enhance reprogramming, suggesting an important role in cell fate. In addition to Miz1, another potential mechanism of repression by myc include recruitment of DNMT3A [67].
Enhancing our understanding of myc transcriptional function during normal development and in normal stem cells provides a window into how excess myc may cause tumorigenesis, potentially providing additional targets to treat myc-related tumors. New inhibitors targeting Myc interaction with Max show promise [68], but it may also prove fruitful to target functions downstream of Myc such as transcriptional or chromatin events as well as cellular states maintained by Myc including cellular “stemness”. Consistent with this idea, Myc has recently been linked with maintaining an aberrant pluripotent state in tumor stem cells including those of glioma [69] and medulloblastoma [70, 71]. Forcing changes in chromatin that in turn lead to differentiation of cancer cells that have excess Myc may be a new method of treatment that does not rely upon targeting Myc itself. Future studies on the differences in Myc-regulated chromatin and gene expression between tumor cells and their cognate normal cells of origin should shed additional light on tumorigenesis and potential new chromatin-based treatments.
MATERIALS AND METHODS
Animals
The studies involved knockout mice were approved by UC Davis IACUC.
Breeding pairs of nestin Cre- N-myc(FL/FL) [22]; c-myc(FL/FL) [49] and nestin Cre+ N-myc(FL/WT) c-myc(FL/FL) were set up to generate the four genotypes Nestin Cre- N-myc(FL/FL) c-myc(FL/FL), Nestin Cre- N-myc(FL/WT) c-myc(FL/FL), Nestin Cre+ N-myc(FL/FL) c-myc(FL/FL), and nestin Cre+ N-myc(FL/WT) c-myc(FL/FL). Genotypes of animals were determined via PCR using primer sets for Cre, N-myc and c-myc. Primer sequences were as follow: Cre1: GCC TGC ATT ACC GGT CGA TGC AAC GA; Cre2: GTG GCA GAT GGC GCG GCA ACA ACC ATT; N-myc1: GTC GCG CTA GTA AGA GCT GAG ATC; N-myc2: GGC ACA CAC CTA TAA TCC CAG CTA; N-myc3: CAC AGC TCT GGA AGG TGG GAG AAA GTT GAG CGT CTC C; c-myc1: GCC CCT GAA TTG CTA GGA AGA CTG; c-myc2: CCG ACC GGG TCC GAG TCC CTA TT. Cre1 and 2 primer sets amplified a 700bp band from Cre insertion. The flox specific band for N-myc1, 2 and 3 primers is 260bp, the wild-type band is 217bp, and the deletion band is 350bp. The flox specific band for c-myc1 and 2 primer set is 500bp, and the wild-type band is 400bp. A separate primer set to detect c-myc deletion, c-mycDS: TCG CGC CCC TGA ATT GCT AGG AA, and c-mycDA: TGC CCA GAT AGG GAG CTG TGA TAC TT, were used and amplifies a band at ~750bp.
Immunohisto- and cytochemistry
All E12.5 embryos were immersion-fixed overnight in fresh, buffered 4% paraformaldehyde, paraffin embedded and cut (12μm) sagittally. Pregnant female were anesthetized with 150mg/kg ketamine and 16mg/kg xylazine, then E17.5 embryos were removed individually and perfused with buffered 4% paraformaldehyde. Brains were removed and fixed overnight in fresh, buffered 4% paraformaldehyde, then cryopreserved and cut (12μm) sagittally. For immunostaining, E12.5 embryo sections were deparaffinized then rehydrated and 30mM sodium citrate treated for 10 minutes at 95°C, while E17.5 frozen brain sections were post fixed in −20°C Acetone for 10 minutes followed by three 5 minutes PBS wash. All sections were blocked in 10% normal goat serum in PBS for 1 hour at room temperature then incubated in primary antibody overnight at 4°C. Antibodies used include anti-BrdU (Chemicon MAB3424, 1:150), anti-PhosphoH3 (Upstate 06-570, 1:200), and anti-TubulinßIII (TUJ; Covance PRB-435P, 1:150). The next day sections were washed in PBS three times 10 minutes each, then incubated in secondary antibody for 2 hours at room temperature. Alexa Fluor Goat α Rabbit IgG λ488 (Invitrogen A11008) and Alexa Fluor Goat α Mouse IgG λ546 (Invitrogen A 11003) were used at 1:1000 dilution. Sections were washed again in PBS three times 10 minutes each, then mounted in Vectashield mounting medium with DAPI (VECTOR H-1200). Anti-BrdU staining was conducted on embryos from pregnant females who were IP injected with BrdU (Sigma B5002, 150μg/g) following 2-4 hours BrdU incorporation periods. TUNEL staining was performed using the DeadEnd Fluorometric TUNEL System Kit (Promega G3250) as directed.
Microarray and Quantitative Real-Time PCR
Cortex, midbrain and cerebellum were dissected from brains of freshly sacrificed E17.5 embryos. Total RNA was isolated from these brain areas using RNeasy Mini Kit (Qiagen 74134) with DNaseI digestion (Invitrogen 18068-015) performed post RNA extraction. Quality of RNA was checked and cRNA was produced by UC Davis Gene Expression Analysis facility for hybridization to Sentrix Mouse Ref-8 Expression microarray. qPCR was performed on total RNA from the same batch sent off for array. Samples were processed using Express SYBR Green qPCR Supermix (Invitrogen A10314) according to manufacture protocol. All qPCR data were generated using ß-Actin as reference (house-keeping) gene. This gene was chosen basted on microarray results, which showed no significant expression variation between control and double knockout brains. Primer efficiency was tested by generating standard curves for all primers against each of the 3 brain areas. Cycling was conducted as follows: pre-incubation at 95°C for 5 minutes, 45 cycles of amplification at 95°C for 10 seconds, 60°C for 20 seconds, and 72°C for 30 seconds, followed by one cycle of melting curve at 95°C for 5 seconds, 65°C for 1 minute and 97°C continuous.
Cell Quantification
Ventricular zone of E12.5 and E17.5 embryo cortex were mapped out according to comparable Nissl stained sections and the mouse brain atlas. Ventricular zone size varied depending on size of cortex. Entire ventricular zone was divided into ten sections, lengthwise, but only cells in the middle eight sections were counted. Total number of cells was obtained by counting all the DAPI positive cells. Number of BrdU and Phospho H3 cells was obtained by counting the TRITC and FITC, respectively, positively stained cells. Percentage of BrdU and Phospho H3 cells was calculated by dividing the total number of BrdU or Phospho H3 cells over the total number of cells in the ventricular zone. Furthermore, for Phospho H3 stained sections a separate percentage of positively stained cells were calculated by combing the subventricular and intermediate zone.
SUPPLEMENTAL FIGURES
Acknowledgments
The authors thank Veronica Martinez Cerdeno for advice on histological staining. This work was supported by the following grants to PK: NIH grant 1K01CA114400, a Shriners Hospital for Children grant, and the Basil O'Connor Starter Scholar Award from the March of Dimes.
Footnotes
The authors declare they have no conflict of interest.
REFERENCES
- 1.Kohl NE, Kanda N, Schreck RR, Bruns G, Latt SA, Gilbert F, Alt FW. Transposition and amplification of oncogene-related sequences in human neuroblastomas. Cell. 1983;35:359–367. doi: 10.1016/0092-8674(83)90169-1. [DOI] [PubMed] [Google Scholar]
- 2.Schwab M, Alitalo K, Klempnauer KH, Varmus HE, Bishop JM, Gilbert F, Brodeur G, Goldstein M, Trent J. Amplified DNA with limited homology to myc cellular oncogene is shared by human neuroblastoma cell lines and a neuroblastoma tumour. Nature. 1983;305:245–248. doi: 10.1038/305245a0. [DOI] [PubMed] [Google Scholar]
- 3.Brodeur GM, Seeger RC, Schwab M, Varmus HE, Bishop JM. Amplification of N-myc in untreated human neuroblastoma correlates with advanced disease stage. Science. 1984;224:1121–1124. doi: 10.1126/science.6719137. [DOI] [PubMed] [Google Scholar]
- 4.Garson JA, Pemberton LF, Sheppard PW, Varndell IM, Coakham HB, Kemshead JT. N-myc gene expression and oncoprotein characterisation in medulloblastoma. Br J Cancer. 1989;59:889–894. doi: 10.1038/bjc.1989.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bigner SH, Friedman HS, Vogelstein B, Oakes WJ, Bigner DD. Amplification of the c-myc gene in human medulloblastoma cell lines and xenografts. Cancer Res. 1990;50:2347–2350. [PubMed] [Google Scholar]
- 6.MacGregar DN, Ziff EB. Elevated c-myc expression in childhood medulloblastomas. Pediatr Res. 1990;28:63–68. doi: 10.1203/00006450-199007000-00014. [DOI] [PubMed] [Google Scholar]
- 7.Tomlinson FH, Jenkins RB, Scheithauer BW, Keelan PA, Ritland S, Parisi JE, Cunningham J, Olsen KD. Aggressive medulloblastoma with high-level N-myc amplification. Mayo Clin Proc. 1994;69:359–365. doi: 10.1016/s0025-6196(12)62221-6. [DOI] [PubMed] [Google Scholar]
- 8.Bruggers CS, Tai KF, Murdock T, Sivak L, Le K, Perkins SL, Coffin CM, Carroll WL. Expression of the c-Myc protein in childhood medulloblastoma. J Pediatr Hematol Oncol. 1998;20:18–25. doi: 10.1097/00043426-199801000-00003. [DOI] [PubMed] [Google Scholar]
- 9.Su X, Gopalakrishnan V, Stearns D, Aldape K, Lang FF, Fuller G, Snyder E, Eberhart CG, Majumder S. Abnormal expression of REST/NRSF and Myc in neural stem/progenitor cells causes cerebellar tumors by blocking neuronal differentiation. Mol Cell Biol. 2006;26:1666–1678. doi: 10.1128/MCB.26.5.1666-1678.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee WH, Murphree AL, Benedict WF. Expression and amplification of the N-myc gene in primary retinoblastoma. Nature. 1984;309:458–460. doi: 10.1038/309458a0. [DOI] [PubMed] [Google Scholar]
- 11.Zhao X, D DA, Lim WK, Brahmachary M, Carro MS, Ludwig T, Cardo CC, Guillemot F, Aldape K, Califano A, Iavarone A, Lasorella A. The N-Myc-DLL3 Cascade Is Suppressed by the Ubiquitin Ligase Huwe1 to Inhibit Proliferation and Promote Neurogenesis in the Developing Brain. Dev Cell. 2009;17:210–221. doi: 10.1016/j.devcel.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer. 2008;8:976–990. doi: 10.1038/nrc2231. [DOI] [PubMed] [Google Scholar]
- 13.Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Orian A, Grewal SS, Knoepfler PS, Edgar BA, Parkhurst SM, Eisenman RN. Genomic binding and transcriptional regulation by the Drosophila myc and mnt transcription factors. Cold Spring Harb Symp Quant Biol. 2005;70:1–10. doi: 10.1101/sqb.2005.70.019. [DOI] [PubMed] [Google Scholar]
- 15.Guccione E, Martinato F, Finocchiaro G, Luzi L, Tizzoni L, Dall' Olio V, Zardo G, Nervi C, Bernard L, Amati B. Myc-binding-site recognition in the human genome is determined by chromatin context. Nat Cell Biol. 2006;8:764–770. doi: 10.1038/ncb1434. [DOI] [PubMed] [Google Scholar]
- 16.Knoepfler PS, Zhang XY, Cheng PF, Gafken PR, McMahon SB, Eisenman RN. Myc influences global chromatin structure. Embo J. 2006;25:2723–2734. doi: 10.1038/sj.emboj.7601152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bieda M, Xu X, Singer MA, Green R, Farnham PJ. Unbiased location analysis of E2F1-binding sites suggests a widespread role for E2F1 in the human genome. Genome Res. 2006;16:595–605. doi: 10.1101/gr.4887606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cawley S, Bekiranov S, Ng HH, Kapranov P, Gingeras TR. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- 19.Martinato F, Cesaroni M, Amati B, Guccione E. Analysis of Myc-induced histone modifications on target chromatin. PLoS ONE. 2008;3:e3650. doi: 10.1371/journal.pone.0003650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stanton BR, Perkins AS, Tessarollo L, Sassoon DA, Parada LF. Loss of N-myc function results in embryonic lethality and failure of the epithelial component of the embryo to develop. Genes Dev. 1992;6:2235–2247. doi: 10.1101/gad.6.12a.2235. [DOI] [PubMed] [Google Scholar]
- 21.Davis AC, Wims M, Spotts GD, Hann SR, Bradley A. A null c-myc mutation causes lethality before 10.5 days of gestation in homozygous and reduced fertility in heterozygous female mice. Genes Dev. 1993;7:671–682. doi: 10.1101/gad.7.4.671. [DOI] [PubMed] [Google Scholar]
- 22.Knoepfler PS, Cheng PF, Eisenman RN. N-myc is essential during neurogenesis for the rapid expansion of progenitor cell populations and the inhibition of neuronal differentiation. Genes Dev. 2002;16:2699–2712. doi: 10.1101/gad.1021202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hatton BA, Knoepfler PS, Kenney AM, Rowitch DH, de Alboran IM, Olson JM, Eisenman RN. N-myc is an essential downstream effector of Shh signaling during both normal and neoplastic cerebellar growth. Cancer Res. 2006;66:8655–8661. doi: 10.1158/0008-5472.CAN-06-1621. [DOI] [PubMed] [Google Scholar]
- 24.Wilson A, Murphy MJ, Oskarsson T, Kaloulis K, Bettess MD, Oser GM, Pasche AC, Knabenhans C, Macdonald HR, Trumpp A. c-Myc controls the balance between hematopoietic stem cell self-renewal and differentiation. Genes Dev. 2004;18:2747–2763. doi: 10.1101/gad.313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laurenti E, Varnum-Finney B, Wilson A, Ferrero I, Blanco-Bose WE, Ehninger A, Knoepfler PS, Cheng PF, MacDonald HR, Eisenman RN, Bernstein ID, Trumpp A. Hematopoietic stem cell function and survival depend on c-Myc and N-Myc activity. Cell Stem Cell. 2008;3:611–624. doi: 10.1016/j.stem.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knoepfler PS. Why myc? An unexpected ingredient in the stem cell cocktail. Cell Stem Cell. 2008;2:18–21. doi: 10.1016/j.stem.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 27.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007 doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 28.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 29.Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:260–262. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 30.Yamanaka S, Takahashi K. [Induction of pluripotent stem cells from mouse fibroblast cultures] Tanpakushitsu Kakusan Koso. 2006;51:2346–2351. [PubMed] [Google Scholar]
- 31.Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 32.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced Pluripotent Stem Cell Lines Derived from Human Somatic Cells. Science. 2007 doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 33.Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Mochiduki Y, Takizawa N, Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotech. 2007:1–6. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 34.Sridharan R, Tchieu J, Mason MJ, Yachechko R, Kuoy E, Horvath S, Zhou Q, Plath K. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009;136:364–377. doi: 10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cotterman R, Knoepfler PS. N-Myc regulates expression of pluripotency genes in neuroblastoma including lif, klf2, klf4, and lin28b. PLoS ONE. 2009 doi: 10.1371/journal.pone.0005799. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei D, Kanai M, Huang S, Xie K. Emerging role of KLF4 in human gastrointestinal cancer. Carcinogenesis. 2006;27:23–31. doi: 10.1093/carcin/bgi243. [DOI] [PubMed] [Google Scholar]
- 37.van Riggelen J, Yetil A, Felsher DW. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer. 10:301–309. doi: 10.1038/nrc2819. [DOI] [PubMed] [Google Scholar]
- 38.Dominguez-Sola D, Ying CY, Grandori C, Ruggiero L, Chen B, Li M, Galloway DA, Gu W, Gautier J, Dalla-Favera R. Non-transcriptional control of DNA replication by c-Myc. Nature. 2007 doi: 10.1038/nature05953. [DOI] [PubMed] [Google Scholar]
- 39.Knoepfler PS, Kenney AM. Neural Precursor Cycling at Sonic Speed: N-Myc Pedals, GSK-3 Brakes. Cell Cycle. 2006;5:47–52. doi: 10.4161/cc.5.1.2292. [DOI] [PubMed] [Google Scholar]
- 40.Malynn BA, de Alboran IM, O'Hagen RC, Bronson R, Davidson L, DePinho RA, Alt FW. N-myc can functionally replace c-myc in murine development, cellular growth, and differentiation. Genes Dev. 2000;14:1390–1399. [PMC free article] [PubMed] [Google Scholar]
- 41.Hatton KS, Mahon K, Chin L, Chiu FC, Lee HW, Peng D, Morgenbesser SD, Horner J, DePinho RA. Expression and activity of L-myc in normal mouse development. Molecular & Cellular Biology. 1996;16:1794–1804. doi: 10.1128/mcb.16.4.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hatton B, Knoepfler P, Kenney A, Rowitch D, de Alboran I, Olson J, Eisenman R. N-myc is an essential downstream effector of Shh signaling both during normal and neoplastic cerebellar growth. Cancer Res. 2006;66:1–7. doi: 10.1158/0008-5472.CAN-06-1621. [DOI] [PubMed] [Google Scholar]
- 43.Martins RA, Zindy F, Donovan S, Zhang J, Pounds S, Wey A, Knoepfler PS, Eisenman RN, Roussel MF, Dyer MA. N-myc coordinates retinal growth with eye size during mouse development. Genes Dev. 2008;22:179–193. doi: 10.1101/gad.1608008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen D, Pacal M, Wenzel P, Knoepfler PS, Leone G, Bremner R. Division and apoptosis of E2f-deficient retinal progenitors. Nature. 2009;462:925–929. doi: 10.1038/nature08544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nagao M, Campbell K, Burns K, Kuan CY, Trumpp A, Nakafuku M. Coordinated control of self-renewal and differentiation of neural stem cells by Myc and the p19ARF-p53 pathway. J Cell Biol. 2008;183:1243–1257. doi: 10.1083/jcb.200807130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zindy F, Knoepfler PS, Xie S, Sherr CJ, Eisenman RN, Roussel MF. N-Myc and the cyclin-dependent kinase inhibitors p18Ink4c and p27Kip1 coordinately regulate cerebellar development. Proc Natl Acad Sci U S A. 2006;103:11579–11583. doi: 10.1073/pnas.0604727103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kenney AM, Cole MD, Rowitch DH. Nmyc upregulation by sonic hedgehog signaling promotes proliferation in developing cerebellar granule neuron precursors. Development. 2003;130:15–28. doi: 10.1242/dev.00182. [DOI] [PubMed] [Google Scholar]
- 48.Zhao X, Heng JI, Guardavaccaro D, Jiang R, Pagano M, Guillemot F, Iavarone A, Lasorella A. The HECT-domain ubiquitin ligase Huwe1 controls neural differentiation and proliferation by destabilizing the N-Myc oncoprotein. Nat Cell Biol. 2008;10:643–653. doi: 10.1038/ncb1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Alboran IM, O'Hagan RC, Gartner F, Malynn B, Davidson L, Rickert R, Rajewsky K, DePinho RA, Alt FW. Analysis of C-MYC function in normal cells via conditional gene- targeted mutation. Immunity. 2001;14:45–55. doi: 10.1016/s1074-7613(01)00088-7. [DOI] [PubMed] [Google Scholar]
- 50.Ben-Yosef T, Eden A, Benvenisty N. Characterization of murine BCAT genes: Bcat1, a c-Myc target, and its homolog, Bcat2. Mamm Genome. 1998;9:595–597. doi: 10.1007/s003359900825. [DOI] [PubMed] [Google Scholar]
- 51.Kuwahara A, Hirabayashi Y, Knoepfler PS, Taketo MM, Sakai J, Kodama T, Gotoh Y. Wnt signaling and its downstream target N-myc regulate basal progenitors in the developing neocortex. Development. 2010;137:1035–1044. doi: 10.1242/dev.046417. [DOI] [PubMed] [Google Scholar]
- 52.Zeller KI, Zhao X, Lee CW, Chiu KP, Yao F, Yustein JT, Ooi HS, Orlov YL, Shahab A, Yong HC, Fu Y, Weng Z, Kuznetsov VA, Sung WK, Ruan Y, Dang CV, Wei CL. Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc Natl Acad Sci U S A. 2006;103:17834–17839. doi: 10.1073/pnas.0604129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dang CV, O'Donnell KA, Zeller KI, Nguyen T, Osthus RC, Li F. The c-Myc target gene network. Semin Cancer Biol. 2006;16:253–264. doi: 10.1016/j.semcancer.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 54.Varlakhanova NV, Knoepfler PS. Acting locally and globally: Myc's ever-expanding roles on chromatin. Cancer Res. 2009;69:7487–7490. doi: 10.1158/0008-5472.CAN-08-4832. [DOI] [PubMed] [Google Scholar]
- 55.Cotterman R, Jin VX, Krig SR, Lemen JM, Wey A, Farnham PJ, Knoepfler PS. N-Myc regulates a widespread euchromatic program in the human genome partially independent of its role as a classical transcription factor. Cancer Res. 2008;68:9654–9662. doi: 10.1158/0008-5472.CAN-08-1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li Y, Xu FL, Lu J, Saunders WS, Prochownik EV. Widespread genomic instability mediated by a pathway involving glycoprotein Ib alpha and Aurora B kinase. J Biol Chem. 2010;285:13183–13192. doi: 10.1074/jbc.M109.084913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Felsher DW ZA, Zhu J, Tlsty T, Bishop JM. Overexpression of MYC causes p53-dependent G2 arrest of normal fibroblasts. Proc Natl Acad Sci U S A. 2000;97:10544–10548. doi: 10.1073/pnas.190327097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li Q, Dang CV. C-myc overexpression uncouples DNA replication from mitosis. Mol Cell Biol. 1999;19:5339–5351. doi: 10.1128/mcb.19.8.5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Luscher B, Eisenman RN. Mitosis-specific phosphorylation of the nuclear oncoproteins Myc and Myb. J Cell Biol. 1992;118:775–784. doi: 10.1083/jcb.118.4.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Clark DA, Mitra PP, Wang SS. Scalable architecture in mammalian brains. Nature. 2001;411:189–193. doi: 10.1038/35075564. [DOI] [PubMed] [Google Scholar]
- 61.Gallant P, Shiio Y, Cheng PF, Parkhurst S, Eisenman RN. Myc and Max homologs in Drosophila. Science. 1996;274:1523–1527. doi: 10.1126/science.274.5292.1523. [DOI] [PubMed] [Google Scholar]
- 62.Dubois NC, Adolphe C, Ehninger A, Wang RA, Robertson EJ, Trumpp A. Placental rescue reveals a sole requirement for c-Myc in embryonic erythroblast survival and hematopoietic stem cell function. Development. 2008;135:2455–2465. doi: 10.1242/dev.022707. [DOI] [PubMed] [Google Scholar]
- 63.Teszas A, Meijer R, Scheffer H, Gyuris P, Kosztolanyi G, van Bokhoven H, Kellermayer R. Expanding the clinical spectrum of MYCN-related Feingold syndrome. Am J Med Genet A. 2006;140:2254–2256. doi: 10.1002/ajmg.a.31407. [DOI] [PubMed] [Google Scholar]
- 64.Marcelis CL, Hol FA, Graham GE, Rieu PN, Kellermayer R, Meijer RP, Lugtenberg D, Scheffer H, van Bokhoven H, Brunner HG, de Brouwer AP. Genotype-phenotype correlations in MYCN-related Feingold syndrome. Hum Mutat. 2008;29:1125–1132. doi: 10.1002/humu.20750. [DOI] [PubMed] [Google Scholar]
- 65.Varlakhanova NV, Cotterman RF, de Vries WN, Morgan J, Donahue LR, Murray S, Knowles BB, Knoepfler PS. myc maintains embryonic stem cell pluripotency and self-renewal. Differentiation. 2010 doi: 10.1016/j.diff.2010.05.001. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Staller P, Peukert K, Kiermaier A, Seoane J, Lukas J, Karsunky H, Möröy T, Bartek J, Massague J, Hänel F, Eilers M. Repression of p15INK4b expression by Myc through association with Miz-1. Nature Cell Biology. 2001;3:392–399. doi: 10.1038/35070076. [DOI] [PubMed] [Google Scholar]
- 67.Brenner C, Deplus R, Didelot C, Loriot A, Vire E, De Smet C, Gutierrez A, Danovi D, Bernard D, Boon T, Pelicci PG, Amati B, Kouzarides T, de Launoit Y, Di Croce L, Fuks F. Myc represses transcription through recruitment of DNA methyltransferase corepressor. Embo J. 2005;24:336–346. doi: 10.1038/sj.emboj.7600509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shi J, Stover JS, Whitby LR, Vogt PK, Boger DL. Small molecule inhibitors of Myc/Max dimerization and Myc-induced cell transformation. Bioorg Med Chem Lett. 2009;19:6038–6041. doi: 10.1016/j.bmcl.2009.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang J, Wang H, Li Z, Wu Q, Lathia JD, McLendon RE, Hjelmeland AB, Rich JN. c-Myc is required for maintenance of glioma cancer stem cells. PLoS ONE. 2008;3:e3769. doi: 10.1371/journal.pone.0003769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zheng H, Ying H, Yan H, Kimmelman AC, Hiller DJ, Chen AJ, Perry SR, Tonon G, Chu GC, Ding Z, Stommel JM, Dunn KL, Wiedemeyer R, You MJ, Brennan C, Wang YA, Ligon KL, Wong WH, Chin L, Depinho RA. Pten and p53 Converge on c-Myc to Control Differentiation, Self-renewal, and Transformation of Normal and Neoplastic Stem Cells in Glioblastoma. Cold Spring Harb Symp Quant Biol. 2009 doi: 10.1101/sqb.2008.73.047. [DOI] [PubMed] [Google Scholar]
- 71.Kessler JD, Hasegawa H, Brun SN, Yang ZJ, Dutton JW, Wang F, Wechsler-Reya RJ. N-myc alters the fate of preneoplastic cells in a mouse model of medulloblastoma. Genes Dev. 2009;23:157–170. doi: 10.1101/gad.1759909. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.