Abstract
Problem
The role played by microbial invasion of the amniotic cavity (MIAC) in preterm prelabor rupture of membranes (pPROM) is inadequately characterized, in part because of reliance on cultivation-based methods.
Method of study
Amniotic fluid from 204 subjects with pPROM was analyzed with both cultivation and molecular methods in a retrospective cohort study. Broad-range and group-specific PCR assays targeted small subunit rDNA, or other gene sequences, from bacteria, fungi and archaea. Results were correlated with measurements of host inflammation, and pregnancy and perinatal outcomes.
Results
The prevalence of MIAC was 34% (70/204) by culture, 45% (92/204) by PCR, and 50% (101/204) by both methods combined. The number of bacterial species revealed by PCR (46 species-level phylotypes) was greater than that by culture (14 species) and included as-yet uncultivated taxa. Some taxa detected by PCR have been previously associated with the gastrointestinal tract (e.g., Coprobacillus sp.), the mouth (e.g., Rothia dentocariosa) or the vagina in the setting of bacterial vaginosis (e.g., Atopobium vaginae). The relative risk for histologic chorioamnionitis was 2.1 for a positive PCR (95% confidence interval [CI], 1.4–3.0), and 2.0 for a positive culture (95% CI, 1.4–2.7). Bacterial rDNA abundance exhibited a dose relationship with gestational age at delivery (R2=0.26; P<0.01). A positive PCR was associated with lower mean birthweight, and with higher rates of respiratory distress syndrome and necrotizing enterocolitis (P<0.05 for each outcome).
Conclusion
MIAC in pPROM is more common than previously recognized and is associated in some cases with uncultivated taxa, some of which are typically associated with the gastrointestinal tract. The detection of MIAC by molecular methods has clinical significance.
Keywords: PPROM, Intra-amniotic infection, Intra-amniotic inflammation, preterm delivery, preterm birth, chorioamnionitis, FIRS, 16S, molecular microbiology, IL-6, cytokines
INTRODUCTION
Preterm prelabor rupture of membranes (preterm PROM) causes one-third of preterm births and contributes to significant perinatal morbidity and mortality.1–6 Microbial invasion of the amniotic cavity (MIAC) is found in about 30% of preterm PROM cases,2;7–26 and is associated with earlier gestational age at delivery.2;25 However, the current understanding of MIAC derives largely from cultivation-dependent studies. Because many microbial species are not yet cultivated,27 the role of MIAC in preterm PROM is likely under-recognized and incompletely characterized.
Molecular methods offer a sensitive, cultivation-independent approach for detecting microbes. In particular, broad-range PCR assays that target ribosomal DNA (rDNA) allow for detection and characterization of diverse microbial taxa, including unknown species.28 These methods have been used to assess diversity within the human indigenous microbiota29;30 and to characterize microbes associated with a wide range of clinical syndromes.31;32
Preterm parturition encompasses several distinct clinical phenotypes, including preterm PROM and preterm labor with intact membranes.33 An association between MIAC and preterm parturition has been demonstrated in culture-based34;35 and molecular24;32;36–42 studies. However, the use of molecular techniques — and in particular, broad-range PCR — to investigate preterm parturition syndromes in a rigorous, systematic manner has been more limited.
We used a combination of culture and molecular methods to investigate MIAC in the setting of preterm PROM. Our objective was to determine the frequency, taxonomic diversity and relative abundance of microbes in amniotic fluid of women with preterm PROM, and to examine the relationship between MIAC, host inflammatory response as well as pregnancy/perinatal outcome. Our findings draw attention to the possible role of MIAC in preterm PROM and its association with adverse pregnancy outcome.
METHODS
Study population
A retrospective cohort study was conducted of patients with preterm PROM (defined below) who met the following inclusion criteria: 1) singleton gestation; 2) gestational age between 15 and 36.9 weeks; and 3) amniocentesis with microbiological studies of amniotic fluid. Patients were excluded from the study if: 1) delivery occurred elsewhere and/or clinical metadata were unavailable; or 2) a major fetal chromosomal and/or congenital anomaly was present. All samples were collected in a single institution between December 1997 and March 2007.
All women provided written informed consent prior to the collection of biological samples. The utilization of samples and clinical data for research purposes was approved by the Institutional Review Boards of Sotero del Rio Hospital, Wayne State University, the National Institute of Child Health and Human Development (NICHD/NIH/DHHS), and Stanford University.
Definitions
Membrane rupture was diagnosed by: i) pooling of amniotic fluid in the vagina; ii) a positive nitrazine test; and, iii) a positive ferning test.2 Clinical chorioamnionitis was diagnosed according to criteria previously proposed by Gibbs et al.43 Histologic chorioamnionitis was diagnosed based on the presence of inflammatory cells in the chorionic plate and/or chorioamniotic membranes. 42;44 Acute funisitis was diagnosed by the presence of neutrophils in the wall of the umbilical vessels and/or Wharton’s jelly using criteria previously described.45
Sampling procedures
Patients with preterm PROM were offered amniocentesis to assess the microbial status of the amniotic cavity, and/or fetal lung maturity. Amniocentesis is part of the standard of care of patients with preterm PROM at the participating institution. Amniotic fluid was immediately transported in a capped sterile syringe to the clinical laboratory where it was cultured for aerobic and anaerobic bacteria, and for genital mycoplasmas (Mycotrim® GU Triphasic Culture System, Irvine Scientific, Santa Ana, CA, USA), as described.32 White blood cell (WBC) count46 and Gram stain47 of amniotic fluid were also performed shortly after collection using methods previously described. Shortly after the amniocentesis, amniotic fluid not required for clinical assessment was centrifuged at 1300 × g for 10 minutes at 4°C, and the supernatant was aliquoted into gamma-irradiated nonpyrogenic DNase/RNase-free cryovials (Corning, Acton, MA, USA), and immediately frozen at −70°C. Amniotic fluid interleukin-6 (IL-6) concentrations were determined using a specific and sensitive immunoassay which had been validated for amniotic fluid. IL-6 determinations were performed after all patients were delivered and were not used in clinical management.
Genomic DNA extraction
After a storage interval of 1 to 9 years, amniotic fluid that was not required for clinical purposes (≈120 μl from each sample) was shipped on dry ice to Stanford, CA, where genomic DNA was extracted as described,32 with the exception that the lysozyme preparation was replaced with a recombinant form (Epicentre Biotechnologies, Madison, WI, USA). Extracted DNA was eluted into a final volume of 100 μl of QIAamp® AE buffer and stored at −20°C or colder until thawing for molecular analyses. Strategies to prevent, detect and neutralize potential contamination were implemented at critical steps,48 according to a previously described protocol that included mock extraction blanks to monitor potential contamination (at least one mock per 17 processed samples).32
Qualitative analysis by end-point PCR
DNA extracts from each amniotic fluid sample were analyzed by broad-range end-point PCR using broad-range bacterial 16S rDNA primers, and by group-specific end-point PCR using primers specific for six taxonomic groups (Table 1). PCRs were performed as described,32 with the exception that each reaction contained 2 μl of prepared DNA template and was carried out in a Veriti thermal cycler (Applied Biosystems). Ten microliters from each PCR well was electophoresed through a 1.5% (wt/vol) Tris-acetate-EDTA-agarose gel containing GelGreen nucleic acid stain (Biotium, Inc, Hayward, CA, USA). Amplicons producing visible bands upon scanning with a Typhoon 9410 variable mode imager (Amersham Biosciences, Piscataway, NJ, USA) were purified and, if from broad-range PCR, cloned as described.32 Sequencing of amplicons from group-specific PCRs, and of positive recombinants from broad-range PCRs (up to 10 clones per reaction) was performed as described.32
Table 1.
PCR assays used in this study.
| Approximate taxonomic level | End-point PCR taxonomic specificity | Lower detection limit (gene copies per μL) | Oligonucleotide name | Use | Sequence (5′ – > 3′) | Gene target | Reference |
|---|---|---|---|---|---|---|---|
| domain | Bacteria | 100 | Bact-8FM | FP | AGAGTTTGATCMTGGCTCAG | 16S rDNA | 53 |
| Bact-806R | RP | GGACTACCAGGGTATCTAAT | 54 | ||||
| genus | Ureaplasma | 10 | Urease185F | FP | GCTGCTGACGTTGCAAGAAG | urease gene | present study |
| Urease756R | RP | CTCCTGGTTCAAAACGAATAGC | present study | ||||
| genus | Fusobacterium | 100 | Fuso-422F | FP | CGGAATGTAAAGTGCTTTC | 16S rDNA | present study |
| Fuso-710R | RP | CCCATCGGCATTCCTAC | present study | ||||
| genus | Sneathia/Leptotrichia | 10 | SsLa-140F | FP | TAGACTGGGATAACAGAGG | 16S rDNA | present study |
| SsLa-406R | RP | AGTCCTAAAACCTTCTTCACAC | present study | ||||
| species | Streptococcus agalactiae | 10 | Sag059F | FP | TTTCACCAGCTGTATTAGAAGTA | cfb | 55 |
| Sag190R | RP | GTTCCCTGAACATTATCTTTGAT | 55 | ||||
| species | Mycoplasma hominis | 10 | Mh-148F | FP | CAATGGCTAATGCCGGATACG | 16S rDNA | mod. from 56 |
| Mh-463R | RP | GGTACCGTCAGTCTGCAATC | mod. from 56 | ||||
| genus | Candida | 10 | Cand-ITS2-42F | FP | GGGTTTGCTTGAAAGACGGTA | ITS2 | present study |
| Cand-ITS2-125R | RP | TTGAAGATATACGTGGTRGACGTTA | present study | ||||
| Real-time PCR taxonomic specificity | Dynamic range (gene copies per μL) | ||||||
| domain | Bacteria | 15 to 1e8 | Bact-8FM | FP | AGAGTTTGATCMTGGCTCAG | 16S rDNA | 53 |
| Bact-338K* | Probe | CCAKACTCCTACGGGAGGCAGCAG | 53 | ||||
| Bact-515R | RP | TTACCGCGGCKGCTGGCAC | 57 | ||||
| domain | Archaea | 100 to 1e8 | Arch333F | FP | TCCAGGCCCTACGGG | 16S rDNA | 58 |
| Univ-515F* | Probe | GTGCCAGCMGCCGCGGTAA | 57 | ||||
| Arch958R | RP | YCCGGCGTTGAMTCCAATT | 59 |
FP = forward primer, RP = reverse primer, Probe = TaqMan probe
Conjugated on the 5′ end to 6-carboxyfluorescein, and on the 3′ end to 6-carboxy-tetramethylrhodamine
Sequence alignment and phylogenetic analysis
Forward and reverse sequence reads were assembled into contigs as described.32 Assembled sequences from group-specific PCR were queried against NCBI’s GenBank database using a basic local alignment search tool (BLAST) algorithm49 to confirm specificity. Assembled sequences from broad-range end-point PCR were aligned using the Greengenes NAST aligner50 and imported into the Greengenes version of the Arb software package,51;52 where they were compared to a database of over 200,000 small subunit rRNA sequences. Alignments were manually inspected and edited based on the original chromatograms and inserted into the Greengenes phylogeny according to a maximum parsimony algorithm. Sequences with no close relative in the Greengenes database were queried against NCBI’s GenBank database using a BLAST algorithm49 to determine approximate phylogenetic affiliation, and their closest neighbors were added to the alignment. Sequences without close neighbors were screened using RDP’s ‘Chimera Check’ program (http://35.8.164.52/cgis/chimera.cgi?su=SSU). After removal of chimeric, vector, human, and poor-quality sequences from the alignment, a neighbor-joining tree was generated based on Felsenstein correction and 682 unambiguous filter positions. Phylotypes were defined using a 99% sequence similarity threshold, which approximates species-level classification.
Quantitative analysis by real-time PCR
Extracted DNA from each sample was analyzed by means of two real-time PCR assays, each of which was designed to specifically amplify and quantify 16S rDNA of domain Bacteria or domain Archaea (Table 153–59). Reactions were carried out as described32 with the exception that amplifications were carried out in a StepOnePlus real-time PCR system (Applied Biosystems), and absolute rDNA abundance was estimated from the standard curves using StepOne software version 2.0 (Applied Biosystems).
Outcome Measures
In order to assess the clinical significance of MIAC detected with our approach, outcome variables from four broad categories were measured: 1) intra-amniotic inflammation at presentation (including amniotic fluid WBC count46 and IL-6 concentration60); 2) histopathologic inflammation of the placenta and chorioamniotic membranes after delivery; 3) pregnancy outcomes (including gestational age at delivery, and amniocentesis-to-delivery interval); and, 4) perinatal outcomes (including respiratory distress syndrome, pneumonia, necrotizing enterocolitis, intraventricular hemorrhage ≥ grade III, sepsis and bronchopulmonary dysplasia), which were diagnosed according to previously described criteria,21 as well as birthweight, admission to the neonatal intensive care unit and perinatal death.
Statistical analysis
Statistical analyses were performed using ‘R’ (open source, www.r-project.org) version 2.4.1, including the ‘Epi’ and ‘Survival’ packages. Differences in the mean between two groups were computed using Student’s t-test and assuming unequal variances. Differences in the median between two groups were computed using the Mann Whitney U test, and the Kruskal-Wallis analysis of variance test for more than two groups. Differences in proportions were computed using Fisher’s exact test when samples were independent. Correlation coefficients for continuous outcomes were estimated by least squares linear regression modeling. Time-to-event outcomes were modeled by Kaplan-Meier survival methods and differences between survival curves were evaluated by means of the Mantel-Haenszel log-rank test. Prior to survival analysis, influential outliers, as defined by a dfbetas residual >3 standard deviations, were excluded61 (n=3). In addition, patients who delivered preterm for maternal or fetal indications (except clinical chorioamnionitis) were included in the analysis with a censored time that was equal to the amniocentesis-to-delivery interval. For all analyses, a two-tailed P value <0.05 was considered significant.
RESULTS
Table 2 presents baseline characteristics of the 230 enrolled subjects. For 26 subjects, results were unavailable from at least one of the three culture assays used in the study (i.e., for aerobes, anaerobes or genital mycoplasmas); therefore, the remaining 204 subjects were used for comparisons of cultivation and molecular methods.
Table 2.
Baseline characteristics of analyzable subjects (n=204), according to results of PCR and culture of amniotic fluid.
| Characteristic | Negative culture and negative PCR (n=103) | P* | Negative culture but positive PCR (n=31) | P† | Positive culture (n=70) | Missing Culture Data (n=26) |
|---|---|---|---|---|---|---|
| Maternal age (y, mean ± SD) | 29.4 ± 7.3 | 0.2 | 27.5 ± 8.0 | 0.6 | 28.4 ± 7.6 | 28.9 ± 7.1 |
| Nulliparity (N=55) | 19/103 (19%) | <0.05 | 12/31 (39%) | 0.2 | 18/70 (26%) | 6/26 (23%) |
| Cervical insufficiency (N=6) | 1/103 (1%) | 0.1 | 2/31 (7%) | 0.6 | 3/70 (4%) | 0/26 (0%) |
| Smoking (N=44) | 18/103 (18%) | 0.6 | 7/31 (23%) | 0.2 | 8/70 (11%) | 11/26 (42%) |
| Alcohol use (N=14) | 7/103 (7%) | 0.7 | 1/31 (3%) | 1 | 2/70 (3%) | 4/26 (15%) |
| Drug use (N=2) | 2/103 (2%) | 1 | 0/31 (0%) | NA | 0/70 (0%) | 0/26 (0%) |
Comparison of a negative culture and negative PCR with a negative culture but positive PCR.
Comparison of a negative culture but positive PCR with a positive culture.
P values for comparison of mean maternal age were calculated using Student’s t-test, and assumed unequal variances.
P values for proportions were calculated using Fisher’s exact test.
Culture methods underestimate microbial prevalence and diversity
The rate of MIAC in preterm PROM was 34% (70/204) based on cultivation, 45% (92/204) based on PCR, and 50% (101/204) based on the combined results of both methods (Figure 1). PCR was positive in 87% (61/70) of culture-positive subjects, and culture was positive in 66% (61/92) of PCR-positive subjects. Thus, findings from culture alone — which is the conventional diagnostic approach — underestimated MIAC prevalence by at least 30% (31/101).
Figure 1. Distribution of subjects (n=204) according to results of PCR and culture of amniotic fluid.
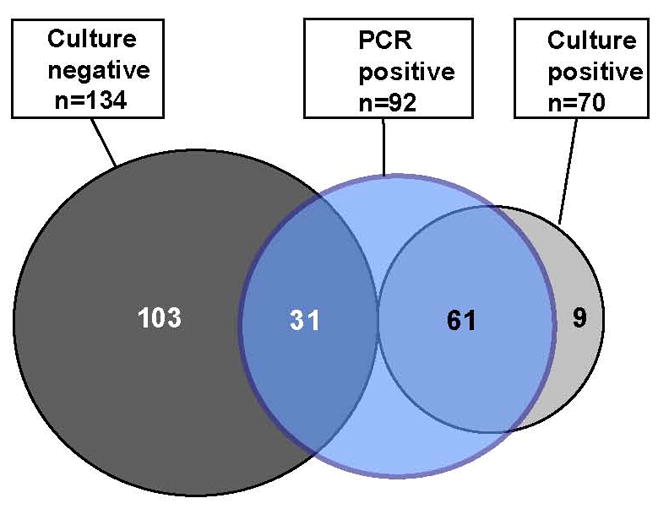
Data are from amniocentesis at presentation of 204 subjects for whom results from all culture assays were available. Culture refers to the aggregate results from routine cultivation methods for bacteria (aerobes, anaerobes and genital mycoplasmas) and for fungi. PCR refers to the aggregate results from end-point or real-time PCR targeting domain Bacteria, domain Archaea, genus Candida, and five specific bacterial groups (see Methods). Circle areas are not to scale.
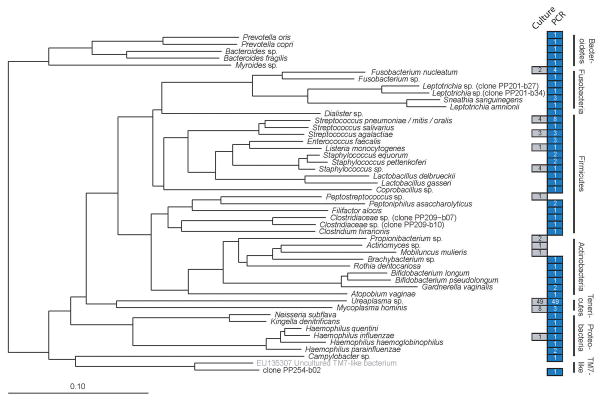
Figure 2 presents the bacterial taxa encountered in this study. The species richness revealed by PCR (n=44) was three times that found by culture (n=14). In addition, the types of bacterial taxa revealed by molecular methods were notable for three reasons. First, some taxa, including a Coprobacillus species, an uncultured Bacteroides species (clone PP209-b04), an uncultured Clostridiaceae bacterium (clone PP209-b07), and another uncultured Clostridiaceae bacterium (clone PP209-b10) appear to be commensals of the gastrointestinal tract (see Discussion). Second, one phylotype is previously-uncharacterized: clone PP254-b02 (<94% sequence similarity to its nearest database relative) clustered near the phylum TM7, a deeply-branching taxonomic group from which no members have been cultivated to purity. Third, to our knowledge, a number of additional bacterial taxa have never been detected in amniotic fluid, including species associated recently with bacterial vaginosis (e.g., Atopobium vaginae), with the oral cavity (e.g., Rothia dentocariosa), or with rare infections of the urogenital tract (e.g., Myroides sp.). A total of 34 cases (34%) were found to be polymicrobial; 12 of these were found to be polymicrobial by culture alone, and 22 were found to be polymicrobial by PCR alone.
Figure 2. Bacterial taxa detected by PCR or culture.
Phylogeny of the bacterial taxa identified in this study, based on a neighbor-joining algorithm with Felsenstein correction and a 682-column filter. The scale bar represents evolutionary distance (10 substitutions per 100 nucleotides). The taxon in brackets and gray type (uncultured TM7-like bacterium) is a public database sequence included for reference and was not detected in this study. Colored boxes indicate the number of subjects who were positive for a given taxon by culture (gray) or PCR (blue) (some samples were polymicrobial). Because culture isolates were not sequenced, each is represented by a GenBank sequence that corresponds to the taxonomic resolution to which culture isolates were phenotypically identified. Two culture isolates are not represented because they were not characterized to a sufficiently narrow taxonomic resolution to allow tree placement (viridans group streptococcus, and gram positive bacillus, each detected in a separate subject). Candida was the lone fungal genus detected in the study population (not shown in this figure).
Five bacterial species were detected in the study population by culture but not by PCR. One was recovered from more than one subject (Propionibacterium sp. (n=2)), and the remaining four species were found in one subject each (Actinomyces sp., Mobiluncus mulieris, Peptostreptococcus sp., and a Gram-positive bacillus that was not further identified). However, in all but one instance (one case with Propionibacterium sp.), the infection was polymicrobial and the PCR was positive, but for a different bacterial species. It is therefore unclear whether the failure of PCR to detect these five taxa reflected bias in DNA amplification, the limited number of clones sequenced (n=10), or other factors (see Discussion).
For 26 additional subjects, results from one or more culture assays were unavailable. Although excluded from outcomes analyses (below), these subjects provided an opportunity to investigate further the molecular microbial diversity of MIAC. PCR was positive in 54% (14/26) of subjects and revealed 16 phylotypes. The most common taxa were Ureaplasma spp. (5 subjects; 23 clones), Sneathia sanguinegens (3; 17) and Fusobacterium nucleatum (2; 7). One sample contained several taxa, each found once, that are typical commensals of the gastrointestinal tract: Bacteroides xylanisolvens (>99% similarity), Eubacterium halii (100%), Faecalibacterium sp. (98.8%), and Finegoldia magna (>99%) (Figure 3).
Figure 3. Bacterial taxa detected by PCR in those samples (n=26) for which culture data were incomplete.
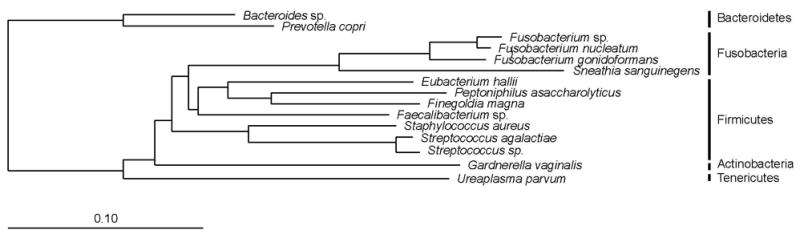
Phylogeny is based on a neighbor-joining algorithm with Felsenstein correction and a 682-column filter. The scale bar represents evolutionary distance (10 substitutions per 100 nucleotides).
Candida species were under-recognized by culture, and were associated with pregnancies with an intrauterine device (IUD)
Results from fungal culture methods were available for 221 subjects. The overall rate of MIAC due to Candida species — as detected by PCR or culture — was 57% higher than the rate detected by culture methods alone (5% [11/221] vs. 3.2% [7/221], respectively). PCR for Candida was positive in 82% (9/11) of all detected cases, and culture was positive in 64% (7/11). MIAC due to Candida occurred significantly more frequently in subjects with an IUD than in subjects without an IUD (28% [5/18] vs. 3% [6/203], respectively; P<0.01). By contrast, the rate of MIAC due to bacteria was equivalent in women with and without an IUD (56% [10/18] vs. 45% [92/203], respectively; P=0.5).
Association of MIAC with host inflammation
The median amniotic fluid WBC count at presentation was higher in women who had PCR-positive results only, than in those who had PCR-negative and culture-negative results (median [cells/ml]: 15 vs. 5; P<0.01) (Figure 4A). Similarly, the median amniotic fluid IL6 concentration was higher in patients who had PCR-positive results of amniotic fluid, than in those who were negative by both PCR and culture; however, this difference did not reach statistical significance (median [ng/ml]: 1.38 vs. 1.0; P=0.16) (Figure 4B). For both AF WBCs and IL6, there was no difference between the groups that were positive by each method alone; also, levels of each marker of inflammation were higher in the group of patients that was positive by both methods (PCR and culture) combined, than in the other clinical groups (see Figure 4).
Figure 4. Inflammatory markers in amniotic fluid at presentation, according to PCR and culture results.
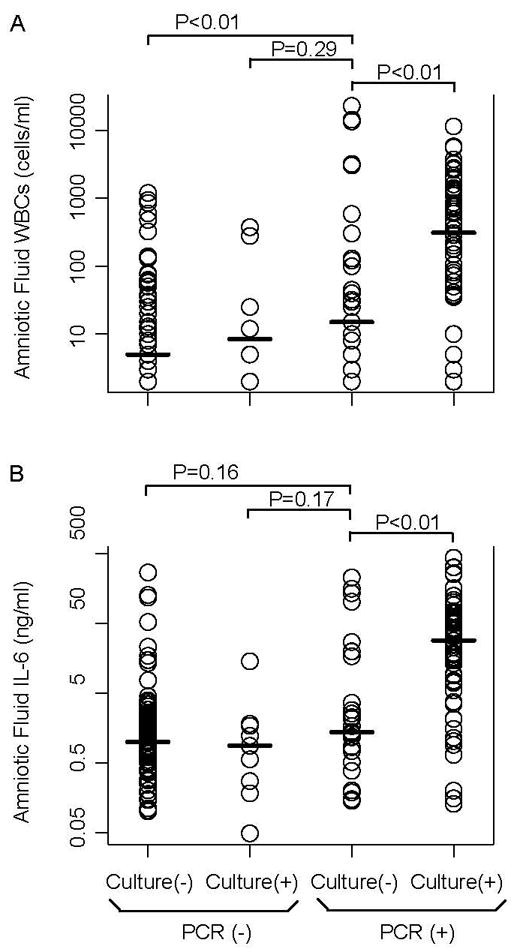
Panel A presents white blood cell counts. Panel B presents interleukin-6 concentrations. For both panels, P values were calculated using the Mann-Whitney U test. Data are from amniocentesis at presentation of 204 subjects for whom results from all culture assays were available.
Table 3 presents the relative risk for histologic inflammation of maternal and fetal membranes at delivery. The analysis included 146 subjects for whom histologic data were available. The relative risk for histologic chorioamnionitis was 2.1 for a positive PCR (95% confidence interval [CI], 1.4 – 3.0), and 2.0 for a positive culture (95% CI, 1.4 – 2.7). The relative risk for funisitis was 2.2 for a positive PCR (95% CI, 1.3 – 3.9), and 2.8 for a positive culture (95% CI, 1.7 – 4.7).
Table 3. Relative risk of a positive PCR or culture for histologic chorioamnionitis or funisitis.
Analysis was based on 146 subjects for whom data on histologic inflammation were available.
| Histologic Chorioamnionitis | Funisitis | Histologic Chorioamnionitis and/or Funisitis | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Present (n=61) | Absent (n=85) | Relative risk (95% CI) | Present (n=39) | Absent (n=107) | Relative risk (95% CI) | Present (n=62) | Absent (n=84) | Relative risk (95% CI) | |
| PCR positive (n=65) | 38 | 27 | 2.1 (1.4 – 3.0) | 25 | 40 | 2.2 (1.3 – 3.9) | 39 | 26 | 2.1 (1.4 – 3.1) |
| Culture (n=46) | 29 | 17 | 2.0 (1.4 – 2.7) | 22 | 24 | 2.8 (1.7 – 4.7) | 30 | 16 | 2.0 (1.4 – 2.8) |
Associations of microbial DNA in amniotic fluid with adverse pregnancy outcomes, and with perinatal morbidity and mortality
Bacterial 16S rRNA gene copy number exhibited a statistically significant correlation with early gestational age at presentation (R2=0.17; P<0.01) (Figure 5, Panel A), and with early gestational age at delivery (R2=0.26; P<0.01) (Figure 5, Panel B). Subjects with a negative amniotic fluid culture but positive PCR delivered at an earlier median gestational age than did subjects with a negative amniotic fluid culture and negative PCR (31.9 vs. 33.4 weeks, respectively, P=0.02) (Table 4). There was no significant difference in the median gestational age at delivery between subjects with a negative amniotic fluid culture but positive PCR and subjects with a positive culture (31.9 vs. 29.6 weeks, respectively, P=0.08).
Figure 5. Correlation of bacterial 16S rDNA concentration with pregnancy outcomes.
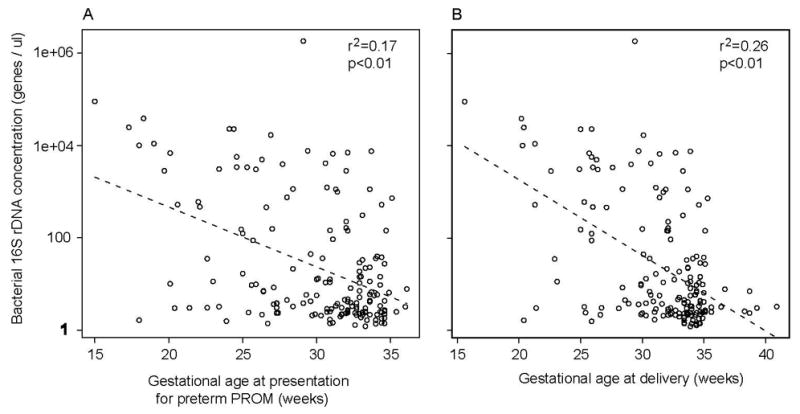
Panel A presents results for gestational age at which subjects presented with preterm PROM. Panel B presents results for gestational age at delivery.
Table 4.
Pregnancy outcomes according to results of PCR and culture of amniotic fluid.
| Negative culture and negative PCR (n=103) | P* | Negative culture but positive PCR (n=31) | P† | Positive culture (n=70) | |
|---|---|---|---|---|---|
| Gestational age at presentation for pPROM | 31.7 (18.0 – 36.1) | 0.08 | 30.6 (18.3 – 35.4) | 0.3 | 28.2 (15.0 – 35.1) |
| Gestational age at delivery | 33.4 (20.4 – 40.9) | <0.05 | 31.9 (20.2 – 35.6) | 0.08 | 29.6 (15.6 – 35.3) |
| Clinical Chorioamnionitis | 4/103 (4%) | 0.6 | 2/31 (7%) | 0.3 | 10/70 (14%) |
Gestational age reported as median (range)
Comparison of a negative culture and negative PCR with a negative culture but positive PCR.
Comparison of a negative culture but positive PCR with a positive culture.
P values for comparison of median gestational age at delivery were calculated using the Mann Whitney U test.
P values for proportions were calculated using Fisher’s exact test.
Furthermore, subjects with a negative amniotic fluid culture but positive PCR had a significantly shorter amniocentesis-to-delivery interval than did subjects with a negative amniotic fluid culture and negative PCR (P=0.01) (Figure 6). In contrast, there was no difference in the amniocentesis-to-delivery intervals of subjects with a negative amniotic fluid culture but positive PCR and those with a positive culture (regardless of PCR result) (P=0.8).
Figure 6. Kaplan-Meier survival plot of amniocentesis-to-delivery interval according to results of PCR and culture of amniotic fluid.
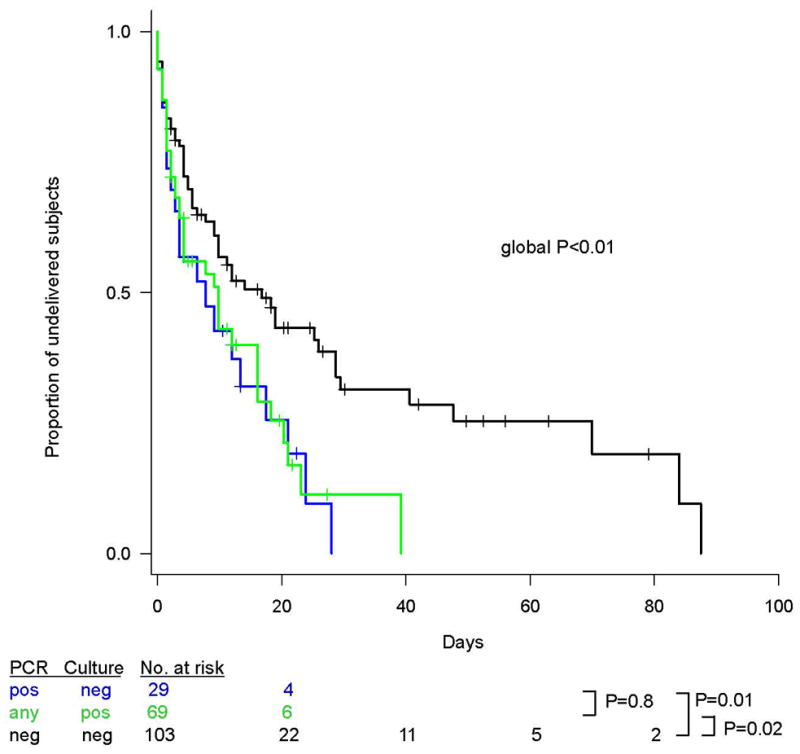
Observations were right-censored for subjects who underwent cesarean section prior to labor onset, or who underwent labor induction. Prior to analysis, influential outliers, as defined by a dfbetas residual >3 standard deviations, were excluded (n=3). P values for differences in survival curves were calculated by means of the Mantel-Haenszel log-rank test.
A negative amniotic fluid culture but positive PCR was associated with a lower mean birthweight, and with higher rates of respiratory distress syndrome and necrotizing enterocolitis, as compared with the finding of a negative amniotic fluid culture and negative PCR (P<0.05 for each outcome) (Table 5).
Table 5.
Perinatal outcomes according to results of PCR and culture of amniotic fluid.
| Outcome | Negative culture and negative PCR n=103 | P* | Negative culture but positive PCR n=31 | P† | Positive culture n=70 |
|---|---|---|---|---|---|
| Birth weight (g, mean ± SD) | 2021 ± 596 | <0.05 | 1682 ± 641 | 0.2 | 1488 ± 703 |
| Admission to neonatal intensive care unit | 81/101 (81%) | 0.1 | 28/30 (93%) | 1 | 54/58 (93%) |
| Bronchopulmonary dysplasia | 5/101 (5%) | <0.05 | 5/30 (17%) | 0.5 | 6/58 (10%) |
| Respiratory distress syndrome | 16/101 (16%) | <0.05 | 11/30 (37%) | 0.6 | 18/58 (31%) |
| Pneumonia | 6/101 (6%) | 1 | 1/30 (3%) | 1 | 2/58 (3%) |
| Sepsis | 37/101 (37%) | 0.5 | 13/30 (43%) | 0.2 | 34/58 (59%) |
| Necrotizing Enterocolitis | 1/101 (1%) | <0.05 | 3/30 (10%) | 0.3 | 2/58 (3%) |
| Intraventricular hemorrhage (≥ grade III) | 10/101 (10%) | 0.7 | 2/30 (7%) | 0.3 | 9/58 (16%) |
| Perinatal death | 4/101 (4%) | 0.08 | 4/30 (13%) | 0.4 | 15/68 (22%) |
Comparison of a negative culture and negative PCR with a negative culture but positive PCR.
Comparison of a negative culture but positive PCR with a positive culture.
P values for comparison of mean birth weight were calculated using Student’s t-test, and assumed unequal variances.
P values for proportions were calculated using Fisher’s exact test.
DISCUSSION
Principal findings of the study
By including molecular methods in our approach we found that MIAC in preterm PROM: 1) affected half of all subjects, yet was frequently undetected by culture-based methods; 2) was caused by diverse microbes, including taxa that are as-yet uncultivated, uncharacterized, or not previously found in amniotic fluid; 3) was associated in some women with microbial species that may have originated from the gastrointestinal tract; 4) was more frequently associated with Candida species than previous believed, especially in the presence of an IUD; 5) exhibited temporal and dose-response associations (based on microbial DNA presence or abundance) with early delivery, suggesting a causal relationship; 6) was associated with a fetal inflammatory response as reflected by the amniotic fluid white blood cell count; and 7) was associated with adverse perinatal outcomes.
Microbial invasion of the amniotic cavity detected by cultivation methods vs. molecular methods
Culture is the standard approach for the identification of MIAC and was positive in one-third of subjects in this study. This frequency is similar to the average frequency reported by others. 2 However, when molecular methods were included, MIAC was found in one-half of subjects. Interestingly, the rate at which culture underestimated MIAC in the present study (31% [31/101]) was comparable to that found by our group in a recent study of preterm labor with intact membranes, using a similar molecular approach (36% [9/25]).32 In addition to facilitating a more accurate estimation of microbial prevalence, the molecular findings from this study advance our understanding of microbial diversity associated with MIAC.
Microbial diversity in preterm PROM
The conventional view is that microorganisms from the lower genital tract ascend into the amniotic cavity. This can occur in patients with either intact, or ruptured membranes. However, our data are the first, to our knowledge, to implicate the gastrointestinal tract as a potential source of the microbes that invade the amniotic cavity. This identification of source is based on reviewing: i) the Human Oral Microbiome Database62 (to help ascertain the possibility of a given sequence having an oral source); ii) systematic studies of human indigenous microbiota; iii) reports identified by relevant PubMed queries; and, iv) GenBank metadata of close sequence neighbors (to review the anatomic source reported by the submitting investigator).
Although the concept of microbial hematogenous dissemination from the gastrointestinal tract to the amniotic cavity is relatively unexplored, it is consistent with accepted or emerging paradigms of pathogenesis. For example, it has long been established that the gastrointestinal tract can serve as a portal for pathogens to enter the bloodstream and cause distant infection (e.g., endocarditis caused by Streptococcus bovis in the setting of colonic neoplasia). And more recent data suggest that some microbes invade the amniotic cavity from the bloodstream after dissemination from remote sites (e.g., the mouth63–65). It therefore stands to reason that residents of the gastrointestinal microbiota may be capable of entering and transiting the bloodstream to cause MIAC. Among women undergoing cesarean delivery, bacteremia was found to be common in women who were in labor or had membrane rupture, especially if these occurred preterm.66 Our data suggest the need for studies that define more clearly the time-course and sources of bacteremia in this setting.
One taxon detected by PCR, but not culture, represented a novel species based on sequence divergence from database relatives. This sequence type (clone PP254-b02) clustered near TM7, a candidate phylum from which no representatives have yet been propagated to purity in culture.67 TM7 members have been detected in the oral cavity,68 including in association with periodontitis,57 and in the vagina in the setting of bacterial vaginosis.31 Our data expand the human habitats and clinical syndromes with which TM7-like taxa are associated.
Other taxa — in addition to those discussed above — were also detected in amniotic fluid for the first time. These taxa comprised three categories. The first has been associated with bacterial vaginosis in recent molecular studies,31;69 and includes Atopobium sp., Dialister sp., and Peptoniphilus sp. Bacterial vaginosis is associated with a perturbed microbial ecosystem and an increased risk of preterm birth.70 Our data support the hypothesis that members of the vaginal microbiota associated with bacterial vaginosis invade the amniotic cavity. The second category includes taxa known to be associated with the oral cavity. Filifactor alocis has been linked to primary endodontic infections,71 and Rothia dentocariosa has been associated not only with odontogenic abscesses but also with intrauterine fetal death72 and neonatal septicemia.73 Our identification of these species expands the census of oral-associated taxa previously found in amniotic fluid32;63–65 and strengthens the link between the oral microbiota, MIAC and preterm birth. The third category includes rare pathogens whose role in human disease appears to be incompletely characterized, such as Myroides (formerly Flavobacterium) spp.,74 which have been implicated in cases of endocarditis,74 urogenital disease,75 and ventriculitis and bacteremia in a six-week old premature infant.76
We did not detect members of the domain Archaea, although some members have been found in the vagina of some women with bacterial vaginosis in a small study77 Our data regarding the absence of Archaea in amniotic fluid mirror findings of a study of women in preterm labor.32 This suggests that Archaea invade the amniotic cavity either very rarely or never, or that their abundance or sequence diversity is not detected by our assays.
Our data on Candida species and MIAC in preterm PROM corroborate findings from a recent large study that demonstrated a significantly higher rate of intra-amniotic Candida infection in pregnancies associated with an IUD.78 Although a link between Candida infection and IUDs was described decades ago,79 Candida biofilm formation on IUDs was reported only recently.80 Our findings suggest a need to re-think the clinical management of pregnancies with preterm PROM associated with an IUD.
The host response to MIAC
To determine the clinical relevance of a positive molecular assay result, we compared the findings from PCR with those from culture in relation to several clinical outcomes. With respect to host inflammation (e.g., amniotic fluid WBC count and IL-6 concentration at presentation, histologic chorioamnionitis and funisitis at delivery), earlier gestational age at delivery, and perinatal morbidity (e.g., respiratory distress syndrome, necrotizing enterocolitis), the strength of association was equivalent for both PCR and culture. In addition, PCR results exhibited both temporal (e.g., shortened amniocentesis-to-delivery interval) and dose-response relationships (e.g., gestational age at delivery as a function of bacterial rDNA abundance), in support of a possible causal link with clinical outcomes.32;81–83
Microbial footprints detected by molecular techniques were associated with a fetal inflammatory response21;84–86 as gauged by the number of WBCs in the amniotic cavity (which are of fetal origin87). This observation is consistent with previous observations that fetal inflammation, as assessed by fetal plasma IL-6 concentration or by histologic inflammation of fetal membranes, is associated with a shorter interval-to-delivery, a higher neonatal morbidity, and evidence of multi-systemic involvement (adrenal,88 central nervous system89, thymus,90–93 lung,94–96 etc.). Studies that used a primate model of intra-uterine infection and included measurements of amniotic fluid cytokines have reported similar findings.97–99 However, regulation of cytokines in amniotic fluid appears to be complex. Not only are some cytokines constitutively produced by non-inflammatory amnion cells (e.g., fibroblasts and epithelial cells),100;101 but cytokine release by fetal membranes in response to bacteria appears to be heterogeneous.102 In addition, little is known regarding the time-course of cytokine release in the human amniotic cavity, or the effect of factors such as duration of infection, inoculum size, and the number or virulence of microbial species present. Further studies are warranted.
Strengths and limitations of the study
The current study is one of the largest studies reported to date of patients with preterm PROM, presenting to a single institution, with an interest in the role of infection in preterm birth. We used state-of-the-art molecular techniques and we assessed the inflammatory response, as well as pregnancy and neonatal outcome. The limitations include the high rate of oligohydramnios in patients with preterm PROM which limited the sample volumes available for research assays (average of ≈120 μl); specifically, the equivalent of only ≈2 μl of amniotic fluid was used in each PCR assay (by contrast, 150 μl was used for aerobic and anaerobic cultures, and 250 μl for mycoplasma cultures). Second, although culture was performed immediately after sample collection, DNA extraction and molecular analyses were performed 1 to 9 years later, increasing the likelihood of DNA degradation.103 In fact, the median time interval to DNA extraction for the group that was positive by culture but negative by PCR was 88 months [range 65–113], as compared to 74 months [18–115] for the group that was positive by PCR regardless of culture (P=0.03; Figure 7). Third, our molecular approach may have failed to detect phylotypes present in polymicrobial samples at low relative abundance because we sequenced only a limited number of clones per sample. Fourth, molecular findings may have been affected by PCR inhibition,104 or by biases in DNA extraction,104 PCR amplification efficiency,104 or PCR primer specificity.105 Last, we did not target viruses or non-fungal eukaryotic microbes. It is possible that some of these limitations (e.g., sample volumes) may have diminished the yield from PCR disproportionately; thus, our data are likely to underestimate the true microbial prevalence and diversity in preterm PROM. Further studies should include exploration of the role of viruses.
Figure 7.
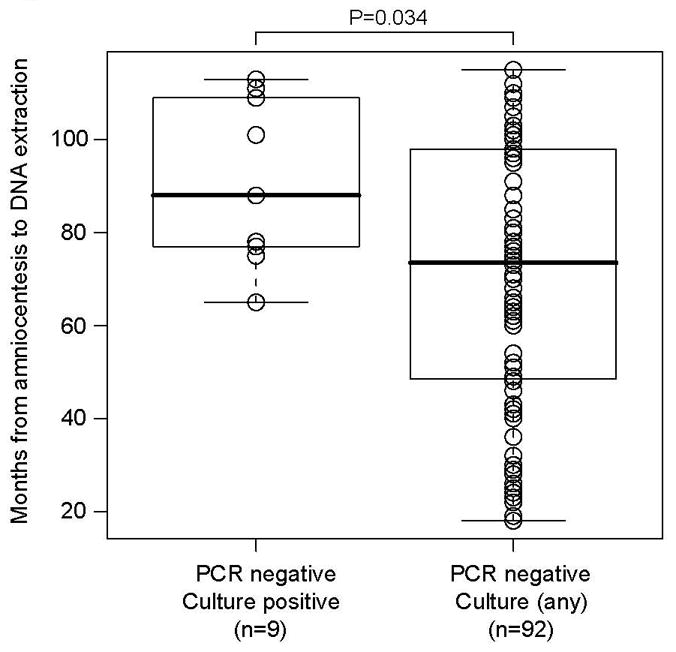
Time interval from amniotic fluid collection to DNA extraction.
Implications of the findings
Randomized clinical trials of antibiotic administration to women with preterm PROM106–114 indicate that antimicrobial agents can prolong pregnancy, and reduce the rate of proven neonatal sepsis and of clinical chorioamnionitis. However, recent observations indicate that the standard treatment recommended by professional societies does not eradicate or prevent MIAC in patients with preterm PROM.115
Knowledge of the microbial species involved in MIAC may be important to inform prognosis and therapy. First, inflammatory cytokines have been implicated in the pathogenesis of preterm birth;17;116–140 however, fetal membrane cytokine response profiles are highly variable and may reflect stereotypic responses to divergent bacterial species.102;141;142 Second, biofilm formation — a process known to diminish antibiotic efficacy — requires the expression of diverse genes143 that may vary between microbial species. Indeed, recent observations suggest that biofilm formation occurs in MIAC144 but the prevalence and nature of this association with respect to various microbial taxa remains largely unexplored. Third, polymicrobial infection may promote pathogen synergy as a result of the particular microbes involved,145;146 or may enable horizontal transfer of antibiotic resistance genes between co-located species.147;148 Our data indicate that most polymicrobial MIAC cases are misclassified by cultivation methods as either monomicrobial or free of microbial invasion altogether. Fourth, other poorly characterized factors, including the role of microbial products,46;149–152 amniotic fluid “sludge”153–155 and the poor transplacental passage of certain antibiotics such as erythromycin,156;157 may have implications for pathogenesis or therapy of MIAC that vary with respect to the microbial species present. A deeper understanding of the microbial diversity of MIAC in preterm PROM and improved risk stratification may be necessary to develop more effective clinical strategies.
Our findings argue for additional detailed molecular studies of MIAC in the preterm parturition syndromes, and may inform the rational design of prophylactic and therapeutic interventions in patients with preterm PROM or who are otherwise at risk for preterm delivery (e.g. an elevated IL-6 or MMP-8 in amniotic fluid at the time of midtrimester amniocentesis135).
Conclusions
1) Molecular methods enable the detection of MIAC cases that are undiagnosed by cultivation techniques. 2) Greater microbial diversity is found in MIAC by sequence-based methods than by cultivation techniques. 3) Culture-negative cases of MIAC are associated with fetal inflammation, as assessed by amniotic fluid white blood cell count. 4) MIAC detected by molecular methods carries clinical significance, even in the setting of a negative culture; pregnancy and neonatal outcomes were similar whether MIAC was detected by molecular methods alone, or by cultivation methods.
Acknowledgments
This work was supported, in part, by the Intramural Research Program of the National Institute of Child Health and Human Development, NIH, DHHS, and by a grant from the March of Dimes Foundation to DAR. DAR is supported by an NIH Director’s Pioneer Award (NIH DP1OD000964).
We are grateful to study subjects and their families. We thank Harold Amogan for work in optimizing some of the PCR assays used in this study.
Reference List
- 1.Mercer BM. Preterm premature rupture of the membranes: current approaches to evaluation and management. Obstet Gynecol Clin North Am. 2005;32:411–428. doi: 10.1016/j.ogc.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 2.Santolaya-Forgas J, Romero R, Espinoza J, Erez O, Friel AL, Kusanovic JP, Bahado-Singh R, Nien JK. Prelabor rupture of membranes. In: Reece EA, Hobbins JC, editors. Clinical Obstetrics: The Fetus and the Mother. Blackwell Publishing; 2007. pp. 1130–1188. [Google Scholar]
- 3.Goldenberg RL, Culhane JF, Iams JD, Romero R. Epidemiology and causes of preterm birth. Lancet. 2008;371:75–84. doi: 10.1016/S0140-6736(08)60074-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iams JD, Romero R, Culhane JF, Goldenberg RL. Primary, secondary, and tertiary interventions to reduce the morbidity and mortality of preterm birth. Lancet. 2008;371:164–175. doi: 10.1016/S0140-6736(08)60108-7. [DOI] [PubMed] [Google Scholar]
- 5.Saigal S, Doyle LW. An overview of mortality and sequelae of preterm birth from infancy to adulthood. Lancet. 2008;371:261–269. doi: 10.1016/S0140-6736(08)60136-1. [DOI] [PubMed] [Google Scholar]
- 6.March of Dimes white paper on preterm birth: The global and regional toll. March of Dimes. 2009 Ref Type: Report. [Google Scholar]
- 7.Garite TJ, Freeman RK, Linzey EM, Braly P. The use of amniocentesis in patients with premature rupture of membranes. Obstet Gynecol. 1979;54:226–230. [PubMed] [Google Scholar]
- 8.Garite TJ, Freeman RK. Chorioamnionitis in the preterm gestation. Obstet Gynecol. 1982;59:539–545. [PubMed] [Google Scholar]
- 9.Cotton DB, Hill LM, Strassner HT, Platt LD, Ledger WJ. Use of amniocentesis in preterm gestation with ruptured membranes. Obstet Gynecol. 1984;63:38–43. [PubMed] [Google Scholar]
- 10.Broekhuizen FF, Gilman M, Hamilton PR. Amniocentesis for gram stain and culture in preterm premature rupture of the membranes. Obstet Gynecol. 1985;66:316–321. [PubMed] [Google Scholar]
- 11.Vintzileos AM, Campbell WA, Nochimson DJ, Weinbaum PJ, Escoto DT, Mirochnick MH. Qualitative amniotic fluid volume versus amniocentesis in predicting infection in preterm premature rupture of the membranes. Obstet Gynecol. 1986;67:579–583. [PubMed] [Google Scholar]
- 12.Feinstein SJ, Vintzileos AM, Lodeiro JG, Campbell WA, Weinbaum PJ, Nochimson DJ. Amniocentesis with premature rupture of membranes. Obstet Gynecol. 1986;68:147–152. [PubMed] [Google Scholar]
- 13.Romero R, Quintero R, Oyarzun E, Wu YK, Sabo V, Mazor M, Hobbins JC. Intraamniotic infection and the onset of labor in preterm premature rupture of the membranes. Am J Obstet Gynecol. 1988;159:661–666. doi: 10.1016/s0002-9378(88)80030-9. [DOI] [PubMed] [Google Scholar]
- 14.Gauthier DW, Meyer WJ, Bieniarz A. Correlation of amniotic fluid glucose concentration and intraamniotic infection in patients with preterm labor or premature rupture of membranes. Am J Obstet Gynecol. 1991;165:1105–1110. doi: 10.1016/0002-9378(91)90480-f. [DOI] [PubMed] [Google Scholar]
- 15.Coultrip LL, Grossman JH. Evaluation of rapid diagnostic tests in the detection of microbial invasion of the amniotic cavity. Am J Obstet Gynecol. 1992;167:1231–1242. doi: 10.1016/s0002-9378(11)91694-9. [DOI] [PubMed] [Google Scholar]
- 16.Gauthier DW, Meyer WJ. Comparison of gram stain, leukocyte esterase activity, and amniotic fluid glucose concentration in predicting amniotic fluid culture results in preterm premature rupture of membranes. Am J Obstet Gynecol. 1992;167:1092–1095. doi: 10.1016/s0002-9378(12)80044-5. [DOI] [PubMed] [Google Scholar]
- 17.Romero R, Yoon BH, Mazor M, Gomez R, Gonzalez R, Diamond MP, Baumann P, Araneda H, Kenney JS, Cotton DB. A comparative study of the diagnostic performance of amniotic fluid glucose, white blood cell count, interleukin-6, and gram stain in the detection of microbial invasion in patients with preterm premature rupture of membranes. Am J Obstet Gynecol. 1993;169:839–851. doi: 10.1016/0002-9378(93)90014-a. [DOI] [PubMed] [Google Scholar]
- 18.Averbuch B, Mazor M, Shoham-Vardi I, Chaim W, Vardi H, Horowitz S, Shuster M. Intra-uterine infection in women with preterm premature rupture of membranes: maternal and neonatal characteristics. Eur J Obstet Gynecol Reprod Biol. 1995;62:25–29. doi: 10.1016/0301-2115(95)02176-8. [DOI] [PubMed] [Google Scholar]
- 19.Font GE, Gauthier DW, Meyer WJ, Myles TD, Janda W, Bieniarz A. Catalase activity as a predictor of amniotic fluid culture results in preterm labor or premature rupture of membranes. Obstet Gynecol. 1995;85:656–658. doi: 10.1016/0029-7844(95)00026-n. [DOI] [PubMed] [Google Scholar]
- 20.Carroll SG, Papaioannou S, Ntumazah IL, Philpott-Howard J, Nicolaides KH. Lower genital tract swabs in the prediction of intrauterine infection in preterm prelabour rupture of the membranes. Br J Obstet Gynaecol. 1996;103:54–59. doi: 10.1111/j.1471-0528.1996.tb09515.x. [DOI] [PubMed] [Google Scholar]
- 21.Gomez R, Romero R, Ghezzi F, Yoon BH, Mazor M, Berry SM. The fetal inflammatory response syndrome. Am J Obstet Gynecol. 1998;179:194–202. doi: 10.1016/s0002-9378(98)70272-8. [DOI] [PubMed] [Google Scholar]
- 22.Hussey MJ, Levy ES, Pombar X, Meyer P, Strassner HT. Evaluating rapid diagnostic tests of intra-amniotic infection: Gram stain, amniotic fluid glucose level, and amniotic fluid to serum glucose level ratio. Am J Obstet Gynecol. 1998;179:650–656. doi: 10.1016/s0002-9378(98)70059-6. [DOI] [PubMed] [Google Scholar]
- 23.Rizzo G, Capponi A, Vlachopoulou A, Angelini E, Grassi C, Romanini C. Ultrasonographic assessment of the uterine cervix and interleukin-8 concentrations in cervical secretions predict intrauterine infection in patients with preterm labor and intact membranes. Ultrasound Obstet Gynecol. 1998;12:86–92. doi: 10.1046/j.1469-0705.1998.12020086.x. [DOI] [PubMed] [Google Scholar]
- 24.Yoon BH, Romero R, Kim M, Kim EC, Kim T, Park JS, Jun JK. Clinical implications of detection of Ureaplasma urealyticum in the amniotic cavity with the polymerase chain reaction. Am J Obstet Gynecol. 2000;183:1130–1137. doi: 10.1067/mob.2000.109036. [DOI] [PubMed] [Google Scholar]
- 25.Goncalves LF, Chaiworapongsa T, Romero R. Intrauterine infection and prematurity. Ment Retard Dev Disabil Res Rev. 2002;8:3–13. doi: 10.1002/mrdd.10008. [DOI] [PubMed] [Google Scholar]
- 26.Shim SS, Romero R, Hong JS, Park CW, Jun JK, Kim BI, Yoon BH. Clinical significance of intra-amniotic inflammation in patients with preterm premature rupture of membranes. Am J Obstet Gynecol. 2004;191:1339–1345. doi: 10.1016/j.ajog.2004.06.085. [DOI] [PubMed] [Google Scholar]
- 27.Hugenholtz P. Exploring prokaryotic diversity in the genomic era. Genome Biol. 2002;3:REVIEWS0003. doi: 10.1186/gb-2002-3-2-reviews0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Relman D, Loutit J, Schmidt T, Falkow S, Tompkins L. The agent of bacillary angiomatosis: An approach to the identification of uncultured pathogens. N Engl J Med. 1990;323:1573–1580. doi: 10.1056/NEJM199012063232301. [DOI] [PubMed] [Google Scholar]
- 29.Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43:5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fredricks DN, Fiedler TL, Marrazzo JM. Molecular identification of bacteria associated with bacterial vaginosis. N Engl J Med. 2005;353:1899–1911. doi: 10.1056/NEJMoa043802. [DOI] [PubMed] [Google Scholar]
- 32.DiGiulio DB, Romero R, Amogan HP, Kusanovic JP, Bik EM, Gotsch F, Kim CJ, Erez O, Edwin S, Relman DA. Microbial prevalence, diversity and abundance in amniotic fluid during preterm labor: a molecular and culture-based investigation. PLoS One. 2008;3:e3056. doi: 10.1371/journal.pone.0003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Romero R, Espinoza J, Kusanovic JP, Gotsch F, Hassan S, Erez O, Chaiworapongsa T, Mazor M. The preterm parturition syndrome. BJOG. 2006;113(Suppl 3):17–42. 17–42. doi: 10.1111/j.1471-0528.2006.01120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bobitt JR, Ledger WJ. Unrecognized amnionitis and prematurity: a preliminary report. J Reprod Med. 1977;19:8–12. [PubMed] [Google Scholar]
- 35.Martinez MA, Ovalle A, Ulloa MT, Vidal RM. Role of Haemophilus influenzae in intra-amniotic infection in patients with preterm rupture of membranes. Eur J Clin Microbiol Infect Dis. 1999;18:890–892. doi: 10.1007/s100960050425. [DOI] [PubMed] [Google Scholar]
- 36.Jalava J, Mantymaa ML, Ekblad U, Toivanen P, Skurnik M, Lassila O, Alanen A. Bacterial 16S rDNA polymerase chain reaction in the detection of intra-amniotic infection. Br J Obstet Gynaecol. 1996;103:664–669. doi: 10.1111/j.1471-0528.1996.tb09835.x. [DOI] [PubMed] [Google Scholar]
- 37.Oyarzun E, Yamamoto M, Kato S, Gomez R, Lizama L, Moenne A. Specific detection of 16 micro-organisms in amniotic fluid by polymerase chain reaction and its correlation with preterm delivery occurrence. Am J Obstet Gynecol. 1998;179:1115–1119. doi: 10.1016/s0002-9378(98)70115-2. [DOI] [PubMed] [Google Scholar]
- 38.Gerber S, Vial Y, Hohlfeld P, Witkin SS. Detection of Ureaplasma urealyticum in second-trimester amniotic fluid by polymerase chain reaction correlates with subsequent preterm labor and delivery. J Infect Dis. 2003;187:518–521. doi: 10.1086/368205. [DOI] [PubMed] [Google Scholar]
- 39.Yoon BH, Romero R, Lim JH, Shim SS, Hong JS, Shim JY, Jun JK. The clinical significance of detecting Ureaplasma urealyticum by the polymerase chain reaction in the amniotic fluid of patients with preterm labor. Am J Obstet Gynecol. 2003;189:919–924. doi: 10.1067/s0002-9378(03)00839-1. [DOI] [PubMed] [Google Scholar]
- 40.Gardella C, Riley DE, Hitti J, Agnew K, Krieger JN, Eschenbach D. Identification and sequencing of bacterial rDNAs in culture-negative amniotic fluid from women in premature labor. Am J Perinatol. 2004;21:319–323. doi: 10.1055/s-2004-831884. [DOI] [PubMed] [Google Scholar]
- 41.Miralles R, Hodge R, McParland PC, Field DJ, Bell SC, Taylor DJ, Grant WD, Kotecha S. Relationship between antenatal inflammation and antenatal infection identified by detection of microbial genes by polymerase chain reaction. Pediatr Res. 2005;57:570–577. doi: 10.1203/01.PDR.0000155944.48195.97. [DOI] [PubMed] [Google Scholar]
- 42.Kim MJ, Romero R, Gervasi MT, Kim JS, Yoo W, Lee DC, Mittal P, Erez O, Kusanovic JP, Hassan SS, Kim CJ. Widespread microbial invasion of the chorioamniotic membranes is a consequence and not a cause of intra-amniotic infection. Lab Invest. 2009;89:924–936. doi: 10.1038/labinvest.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gibbs RS, Blanco JD, St Clair PJ, Castaneda YS. Quantitative bacteriology of amniotic fluid from women with clinical intraamniotic infection at term. J Infect Dis. 1982;145:1–8. doi: 10.1093/infdis/145.1.1. [DOI] [PubMed] [Google Scholar]
- 44.Redline RW, Faye-Petersen O, Heller D, Qureshi F, Savell V, Vogler C. Amniotic infection syndrome: nosology and reproducibility of placental reaction patterns. Pediatr Dev Pathol. 2003;6:435–448. doi: 10.1007/s10024-003-7070-y. [DOI] [PubMed] [Google Scholar]
- 45.Pacora P, Chaiworapongsa T, Maymon E, Kim YM, Gomez R, Yoon BH, et al. Funisitis and chorionic vasculitis: The histological counterpart of the fetal inflammatory response syndrome. J Matern Fetal Neonatal Med. 2002;11:18–25. doi: 10.1080/jmf.11.1.18.25. [DOI] [PubMed] [Google Scholar]
- 46.Romero R, Quintero R, Nores J, Avila C, Mazor M, Hanaoka S, Hagay Z, Merchant L, Hobbins JC. Amniotic fluid white blood cell count: a rapid and simple test to diagnose microbial invasion of the amniotic cavity and predict preterm delivery. Am J Obstet Gynecol. 1991;165:821–830. doi: 10.1016/0002-9378(91)90423-o. [DOI] [PubMed] [Google Scholar]
- 47.Romero R, Emamian M, Quintero R, Wan M, Hobbins JC, Mazor M, Edberg S. The value and limitations of the Gram stain examination in the diagnosis of intraamniotic infection. Am J Obstet Gynecol. 1988;159:114–119. doi: 10.1016/0002-9378(88)90503-0. [DOI] [PubMed] [Google Scholar]
- 48.Borst A, Box AT, Fluit AC. False-positive results and contamination in nucleic acid amplification assays: suggestions for a prevent and destroy strategy. Eur J Clin Microbiol Infect Dis. 2004;23:289–299. doi: 10.1007/s10096-004-1100-1. [DOI] [PubMed] [Google Scholar]
- 49.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 50.DeSantis TZ, Jr, Hugenholtz P, Keller K, Brodie EL, Larsen N, Piceno YM, Phan R, Andersen GL. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 2006;34:W394–W399. doi: 10.1093/nar/gkl244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, Buchner A, Lai T, Steppi S, Jobb G, Forster W, Brettske I, Gerber S, Ginhart AW, Gross O, Grumann S, Hermann S, Jost R, Konig A, Liss T, Lussmann R, May M, Nonhoff B, Reichel B, Strehlow R, Stamatakis A, Stuckmann N, Vilbig A, Lenke M, Ludwig T, Bode A, Schleifer KH. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilson KH, Blitchington R, Frothingham R, Wilson JA. Phylogeny of the Whipple’s-disease-associated bacterium. Lancet. 1991;338:474–475. doi: 10.1016/0140-6736(91)90545-z. [DOI] [PubMed] [Google Scholar]
- 54.DeLong EF. Archaea in coastal marine environments. Proc Natl Acad Sci U S A. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ke D, Menard C, Picard FJ, Boissinot M, Ouellette M, Roy PH, Bergeron MG. Development of conventional and real-time PCR assays for the rapid detection of group B streptococci. Clin Chem. 2000;46:324–331. [PubMed] [Google Scholar]
- 56.Zariffard MR, Saifuddin M, Sha BE, Spear GT. Detection of bacterial vaginosis-related organisms by real-time PCR for Lactobacilli, Gardnerella vaginalis and Mycoplasma hominis. FEMS Immunol Med Microbiol. 2002;34:277–281. doi: 10.1111/j.1574-695X.2002.tb00634.x. [DOI] [PubMed] [Google Scholar]
- 57.Brinig MM, Lepp PW, Ouverney CC, Armitage GC, Relman DA. Prevalence of bacteria of division TM7 in human subgingival plaque and their association with disease. Appl Environ Microbiol. 2003;69:1687–1694. doi: 10.1128/AEM.69.3.1687-1694.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lepp PW, Brinig MM, Ouverney CC, Palm K, Armitage GC, Relman DA. Methanogenic Archaea and human periodontal disease. Proc Natl Acad Sci U S A. 2004;101:6176–6181. doi: 10.1073/pnas.0308766101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yoon BH, Romero R, Kim CJ, Jun JK, Gomez R, Choi JH, Syn HC. Amniotic fluid interleukin-6: a sensitive test for antenatal diagnosis of acute inflammatory lesions of preterm placenta and prediction of perinatal morbidity. Am J Obstet Gynecol. 1995;172:960–970. doi: 10.1016/0002-9378(95)90028-4. [DOI] [PubMed] [Google Scholar]
- 61.Fox J. Applied Regression Analysis, Linear Models, and Related Methods. Thousand Oaks, CA: Sage Publications; 1997. [Google Scholar]
- 62.Dewhirst FE, Izard J, Paster BJ, et al. The Human Oral Microbiome Database. 2008 Ref Type. Data File. [Google Scholar]
- 63.Hill GB. Preterm birth: associations with genital and possibly oral microflora. Ann Periodontol. 1998;3:222–232. doi: 10.1902/annals.1998.3.1.222. [DOI] [PubMed] [Google Scholar]
- 64.Bearfield C, Davenport ES, Sivapathasundaram V, Allaker RP. Possible association between amniotic fluid micro-organism infection and microflora in the mouth. BJOG. 2002;109:527–533. doi: 10.1111/j.1471-0528.2002.01349.x. [DOI] [PubMed] [Google Scholar]
- 65.Han YW, Ikegami A, Bissada NF, Herbst M, Redline RW, Ashmead GG. Transmission of an uncultivated Bergeyella strain from the oral cavity to amniotic fluid in a case of preterm birth. J Clin Microbiol. 2006;44:1475–1483. doi: 10.1128/JCM.44.4.1475-1483.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Boggess KA, Watts DH, Hillier SL, Krohn MA, Benedetti TJ, Eschenbach DA. Bacteremia shortly after placental separation during cesarean delivery. Obstet Gynecol. 1996;87:779–784. doi: 10.1016/0029-7844(96)00037-3. [DOI] [PubMed] [Google Scholar]
- 67.Hugenholtz P, Tyson GW, Webb RI, Wagner AM, Blackall LL. Investigation of candidate division TM7, a recently recognized major lineage of the domain Bacteria with no known pure-culture representatives. Appl Environ Microbiol. 2001;67:411–419. doi: 10.1128/AEM.67.1.411-419.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, Levanos VA, Sahasrabudhe A, Dewhirst FE. Bacterial diversity in human subgingival plaque. J Bacteriol. 2001;183:3770–3783. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ferris MJ, Masztal A, Aldridge KE, Fortenberry JD, Fidel PL, Jr, Martin DH. Association of Atopobium vaginae, a recently described metronidazole resistant anaerobe, with bacterial vaginosis. BMC Infect Dis. 2004;4(5):5. doi: 10.1186/1471-2334-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hillier SL, Nugent RP, Eschenbach DA, Krohn MA, Gibbs RS, Martin DH, Cotch MF, Edelman R, Pastorek JG, Rao AV. Association between bacterial vaginosis and preterm delivery of a low-birth-weight infant. The Vaginal Infections and Prematurity Study Group. N Engl J Med. 1995;333:1737–1742. doi: 10.1056/NEJM199512283332604. [DOI] [PubMed] [Google Scholar]
- 71.Siqueira JF, Jr, Rocas IN. Detection of Filifactor alocis in endodontic infections associated with different forms of periradicular diseases. Oral Microbiol Immunol. 2003;18:263–265. doi: 10.1034/j.1399-302x.2003.00073.x. [DOI] [PubMed] [Google Scholar]
- 72.Karlsson MD, Jacobsson B. Intrauterine fetal death associated with Rothia dentocariosa: a case report. Am J Obstet Gynecol. 2007;197:e6–e7. doi: 10.1016/j.ajog.2007.08.024. [DOI] [PubMed] [Google Scholar]
- 73.Shin JH, Shim JD, Kim HR, Sinn JB, Kook JK, Lee JN. Rothia dentocariosa septicemia without endocarditis in a neonatal infant with meconium aspiration syndrome. J Clin Microbiol. 2004;42:4891–4892. doi: 10.1128/JCM.42.10.4891-4892.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Green BT, Green K, Nolan PE. Myroides odoratus cellulitis and bacteremia: case report and review. Scand J Infect Dis. 2001;33:932–934. doi: 10.1080/00365540110077065. [DOI] [PubMed] [Google Scholar]
- 75.Yagci A, Cerikcioglu N, Kaufmann ME, Malnick H, Soyletir G, Babacan F, Pitt TL. Molecular typing of Myroides odoratimimus (Flavobacterium odoratum) urinary tract infections in a Turkish hospital. Eur J Clin Microbiol Infect Dis. 2000;19:731–732. doi: 10.1007/s100960070001. [DOI] [PubMed] [Google Scholar]
- 76.Macfarlane DE, Baum-Thureen P, Crandon I. Flavobacterium odoratum ventriculitis treated with intraventricular cefotaxime. J Infect. 1985;11:233–238. doi: 10.1016/s0163-4453(85)93228-1. [DOI] [PubMed] [Google Scholar]
- 77.Belay N, Mukhopadhyay B, Conway de ME, Galask R, Daniels L. Methanogenic bacteria in human vaginal samples. J Clin Microbiol. 1990;28:1666–1668. doi: 10.1128/jcm.28.7.1666-1668.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim SK, Romero R, Kusanovic JP, Erez O, Vaisbuch E, Mazaki-Tovi S, Gotsch F, Mittal P, Chaiworapongsa T, Pacora P, Ogge G, Gomez R, Yoon BH, Yeo L, Lamont RF, Hassan SS. The prognosis of pregnancy conceived despite the presence of an intrauterine device (IUD) J Perinat Med. 2010;38:45–53. doi: 10.1515/JPM.2009.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schweid AI, Hopkins GB. Monilial chorionitis associated witn an intrauterine contraceptive device. Obstet Gynecol. 1968;31:719–721. doi: 10.1097/00006250-196805000-00021. [DOI] [PubMed] [Google Scholar]
- 80.Auler ME, Morreira D, Rodrigues FF, Abrao MS, Margarido PF, Matsumoto FE, Silva EG, Silva BC, Schneider RP, Paula CR. Biofilm formation on intrauterine devices in patients with recurrent vulvovaginal candidiasis. Med Mycol. 2009:1–6. doi: 10.3109/13693780902856626. [DOI] [PubMed] [Google Scholar]
- 81.HILL AB. THE ENVIRONMENT AND DISEASE: ASSOCIATION OR CAUSATION? Proc R Soc Med. 1965;58:295–300. 295–300. doi: 10.1177/003591576505800503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fredericks DN, Relman DA. Sequence-based identification of microbial pathogens: a reconsideration of Koch’s postulates. Clin Microbiol Rev. 1996;9:18–33. doi: 10.1128/cmr.9.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Romero R, Gomez R, Chaiworapongsa T, Conoscenti G, Kim JC, Kim YM. The role of infection in preterm labour and delivery. Pediatr Perinat Epidemiol. 2001;15 (Suppl 2):41–56. doi: 10.1046/j.1365-3016.2001.00007.x. [DOI] [PubMed] [Google Scholar]
- 84.Romero R, Gomez R, Ghezzi F, Yoon BH, Mazor M, Edwin SS, Berry SM. A fetal systemic inflammatory response is followed by the spontaneous onset of preterm parturition. Am J Obstet Gynecol. 1998;179:186–193. doi: 10.1016/s0002-9378(98)70271-6. [DOI] [PubMed] [Google Scholar]
- 85.Gotsch F, Romero R, Kusanovic JP, Mazaki-Tovi S, Pineles BL, Erez O, Espinoza J, Hassan SS. The fetal inflammatory response syndrome. Clin Obstet Gynecol. 2007;50:652–683. doi: 10.1097/GRF.0b013e31811ebef6. [DOI] [PubMed] [Google Scholar]
- 86.Madsen-Bouterse SA, Romero R, Tarca AL, Kusanovic JP, Espinoza J, Kim CJ, Kim JS, Edwin SS, Gomez R, Draghici S. The transcriptome of the fetal inflammatory response syndrome. Am J Reprod Immunol. 2010;63:73–92. doi: 10.1111/j.1600-0897.2009.00791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sampson JE, Theve RP, Blatman RN, Shipp TD, Bianchi DW, Ward BE, Jack RM. Fetal origin of amniotic fluid polymorphonuclear leukocytes. Am J Obstet Gynecol. 1997;176:77–81. doi: 10.1016/s0002-9378(97)80015-4. [DOI] [PubMed] [Google Scholar]
- 88.Yoon BH, Romero R, Jun JK, Maymon E, Gomez R, Mazor M, Park JS. An increase in fetal plasma cortisol but not dehydroepiandrosterone sulfate is followed by the onset of preterm labor in patients with preterm premature rupture of the membranes. Am J Obstet Gynecol. 1998;179:1107–1114. doi: 10.1016/s0002-9378(98)70114-0. [DOI] [PubMed] [Google Scholar]
- 89.Yoon BH, Romero R, Park JS, Kim CJ, Kim SH, Choi JH, Han TR. Fetal exposure to an intra-amniotic inflammation and the development of cerebral palsy at the age of three years. Am J Obstet Gynecol. 2000;182:675–681. doi: 10.1067/mob.2000.104207. [DOI] [PubMed] [Google Scholar]
- 90.De Felice C, Toti P, Santopietro R, Stumpo M, Pecciarini L, Bagnoli F. Small thymus in very low birth weight infants born to mothers with subclinical chorioamnionitis. J Pediatr. 1999;135:384–386. doi: 10.1016/s0022-3476(99)70140-x. [DOI] [PubMed] [Google Scholar]
- 91.Toti P, De FC, Stumpo M, Schurfeld K, Di LL, Vatti R, Bianciardi G, Buonocore G, Seemayer TA, Luzi P. Acute thymic involution in fetuses and neonates with chorioamnionitis. Hum Pathol. 2000;31:1121–1128. doi: 10.1053/hupa.2000.16676. [DOI] [PubMed] [Google Scholar]
- 92.Di Naro E, Cromi A, Ghezzi F, Raio L, Uccella S, D’Addario V, Loverro G. Fetal thymic involution: a sonographic marker of the fetal inflammatory response syndrome. Am J Obstet Gynecol. 2006;194:153–159. doi: 10.1016/j.ajog.2005.05.036. [DOI] [PubMed] [Google Scholar]
- 93.Yinon Y, Zalel Y, Weisz B, Mazaki-Tovi S, Sivan E, Schiff E, Achiron R. Fetal thymus size as a predictor of chorioamnionitis in women with preterm premature rupture of membranes. Ultrasound Obstet Gynecol. 2007;29:639–643. doi: 10.1002/uog.4022. [DOI] [PubMed] [Google Scholar]
- 94.Watterberg KL, Demers LM, Scott SM, Murphy S. Chorioamnionitis and early lung inflammation in infants in whom bronchopulmonary dysplasia develops. Pediatrics. 1996;97:210–215. [PubMed] [Google Scholar]
- 95.Yoon BH, Romero R, Kim KS, Park JS, Ki SH, Kim BI, Jun JK. A systemic fetal inflammatory response and the development of bronchopulmonary dysplasia. Am J Obstet Gynecol. 1999;181:773–779. doi: 10.1016/s0002-9378(99)70299-1. [DOI] [PubMed] [Google Scholar]
- 96.Yoon BH, Romero R, Shim JY, Lim JH, Choe G, Kadar N, Park M. “Atypical” chronic lung disease of the newborn is linked to fetal systemic inflammation. Am J Obstet Gynecol. 2002;187:S129. [Google Scholar]
- 97.Gravett MG, Witkin SS, Haluska GJ, Edwards JL, Cook MJ, Novy MJ. An experimental model for intraamniotic infection and preterm labor in rhesus monkeys. Am J Obstet Gynecol. 1994;171:1660–1667. doi: 10.1016/0002-9378(94)90418-9. [DOI] [PubMed] [Google Scholar]
- 98.Gravett MG, Haluska GJ, Cook MJ, Novy MJ. Fetal and maternal endocrine responses to experimental intrauterine infection in rhesus monkeys. Am J Obstet Gynecol. 1996;174:1725–1731. doi: 10.1016/s0002-9378(96)70203-x. [DOI] [PubMed] [Google Scholar]
- 99.Novy MJ, Duffy L, Axthelm MK, Sadowsky DW, Witkin SS, Gravett MG, Cassell GH, Waites KB. Ureaplasma parvum or Mycoplasma hominis as sole pathogens cause chorioamnionitis, preterm delivery, and fetal pneumonia in rhesus macaques. Reprod Sci. 2009;16:56–70. doi: 10.1177/1933719108325508. [DOI] [PubMed] [Google Scholar]
- 100.Keelan JA, Sato T, Mitchell MD. Regulation of interleukin (IL)-6 and IL-8 production in an amnion-derived cell line by cytokines, growth factors, glucocorticoids, and phorbol esters. Am J Reprod Immunol. 1997;38:272–278. doi: 10.1111/j.1600-0897.1997.tb00514.x. [DOI] [PubMed] [Google Scholar]
- 101.Keelan JA, Sato T, Mitchell MD. Interleukin (IL)-6 and IL-8 production by human amnion: regulation by cytokines, growth factors, glucocorticoids, phorbol esters, and bacterial lipopolysaccharide. Biol Reprod. 1997;57:1438–1444. doi: 10.1095/biolreprod57.6.1438. [DOI] [PubMed] [Google Scholar]
- 102.Menon R, Peltier MR, Eckardt J, Fortunato SJ. Diversity in cytokine response to bacteria associated with preterm birth by fetal membranes. Am J Obstet Gynecol. 2009;201:306. doi: 10.1016/j.ajog.2009.06.027. [DOI] [PubMed] [Google Scholar]
- 103.Moncla BJ, Braham P, Dix K, Watanabe S, Schwartz D. Use of synthetic oligonucleotide DNA probes for the identification of Bacteroides gingivalis. J Clin Microbiol. 1990;28:324–327. doi: 10.1128/jcm.28.2.324-327.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.von Wintzingerode F, Gobel UB, Stackebrandt E. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev. 1997;21:213–229. doi: 10.1111/j.1574-6976.1997.tb00351.x. [DOI] [PubMed] [Google Scholar]
- 105.Baker GC, Smith JJ, Cowan DA. Review and re-analysis of domain-specific 16S primers. J Microbiol Methods. 2003;55:541–555. doi: 10.1016/j.mimet.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 106.Mercer BM, Moretti ML, Prevost RR, Sibai BM. Erythromycin therapy in preterm premature rupture of the membranes: a prospective, randomized trial of 220 patients. Am J Obstet Gynecol. 1992;166:794–802. doi: 10.1016/0002-9378(92)91336-9. [DOI] [PubMed] [Google Scholar]
- 107.Lewis DF, Fontenot MT, Brooks GG, Wise R, Perkins MB, Heymann AR. Latency period after preterm premature rupture of membranes: a comparison of ampicillin with and without sulbactam. Obstet Gynecol. 1995;86:392–395. doi: 10.1016/0029-7844(95)00181-P. [DOI] [PubMed] [Google Scholar]
- 108.Grable IA, Garcia PM, Perry D, Socol ML. Group B Streptococcus and preterm premature rupture of membranes: a randomized, double-blind clinical trial of antepartum ampicillin. Am J Obstet Gynecol. 1996;175:1036–1042. doi: 10.1016/s0002-9378(96)80049-4. [DOI] [PubMed] [Google Scholar]
- 109.Mercer BM, Miodovnik M, Thurnau GR, Goldenberg RL, Das AF, Ramsey RD, Rabello YA, Meis PJ, Moawad AH, Iams JD, Van Dorsten JP, Paul RH, Bottoms SF, Merenstein G, Thom EA, Roberts JM, McNellis D. Antibiotic therapy for reduction of infant morbidity after preterm premature rupture of the membranes. A randomized controlled trial. National Institute of Child Health and Human Development Maternal-Fetal Medicine Units Network. JAMA. 1997;278:989–995. [PubMed] [Google Scholar]
- 110.Lovett SM, Weiss JD, Diogo MJ, Williams PT, Garite TJ. A prospective, double-blind, randomized, controlled clinical trial of ampicillin-sulbactam for preterm premature rupture of membranes in women receiving antenatal corticosteroid therapy. Am J Obstet Gynecol. 1997;176:1030–1038. doi: 10.1016/s0002-9378(97)70398-3. [DOI] [PubMed] [Google Scholar]
- 111.Kenyon SL, Taylor DJ, Tarnow-Mordi W. Broad-spectrum antibiotics for preterm, prelabour rupture of fetal membranes: the ORACLE I randomised trial. ORACLE Collaborative Group. Lancet. 2001;357:979–988. doi: 10.1016/s0140-6736(00)04233-1. [DOI] [PubMed] [Google Scholar]
- 112.Lewis DF, Adair CD, Robichaux AG, Jaekle RK, Moore JA, Evans AT, Fontenot MT. Antibiotic therapy in preterm premature rupture of membranes: Are seven days necessary? A preliminary, randomized clinical trial. Am J Obstet Gynecol. 2003;188:1413–1416. doi: 10.1067/mob.2003.382. [DOI] [PubMed] [Google Scholar]
- 113.Kenyon S, Pike K, Jones DR, Brocklehurst P, Marlow N, Salt A, Taylor DJ. Childhood outcomes after prescription of antibiotics to pregnant women with preterm rupture of the membranes: 7-year follow-up of the ORACLE I trial. Lancet. 2008;372:1310–1318. doi: 10.1016/S0140-6736(08)61202-7. [DOI] [PubMed] [Google Scholar]
- 114.Hutzal CE, Boyle EM, Kenyon SL, Nash JV, Winsor S, Taylor DJ, Kirpalani H. Use of antibiotics for the treatment of preterm parturition and prevention of neonatal morbidity: a metaanalysis. Am J Obstet Gynecol. 2008;199:620–628. doi: 10.1016/j.ajog.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 115.Gomez R, Romero R, Nien JK, Medina L, Carstens M, Kim YM, Espinoza J, Chaiworapongsa T, Gonzalez R, Iams JD, Rojas I. Antibiotic administration to patients with preterm premature rupture of membranes does not eradicate intra-amniotic infection. J Matern Fetal Neonatal Med. 2007;20:167–173. doi: 10.1080/14767050601135485. [DOI] [PubMed] [Google Scholar]
- 116.Romero R, Manogue KR, Mitchell MD, Wu YK, Oyarzun E, Hobbins JC, Cerami A. Infection and labor. IV. Cachectin-tumor necrosis factor in the amniotic fluid of women with intraamniotic infection and preterm labor. Am J Obstet Gynecol. 1989;161:336–341. doi: 10.1016/0002-9378(89)90515-2. [DOI] [PubMed] [Google Scholar]
- 117.Romero R, Brody DT, Oyarzun E, Mazor M, Wu YK, Hobbins JC, Durum SK. Infection and labor. III. Interleukin-1: a signal for the onset of parturition. Am J Obstet Gynecol. 1989;160:1117–1123. doi: 10.1016/0002-9378(89)90172-5. [DOI] [PubMed] [Google Scholar]
- 118.Romero R, Avila C, Santhanam U, Sehgal PB. Amniotic fluid interleukin 6 in preterm labor. Association with infection. J Clin Invest. 1990;85:1392–1400. doi: 10.1172/JCI114583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Santhanam U, Avila C, Romero R, Viguet H, Ida N, Sakurai S, Sehgal PB. Cytokines in normal and abnormal parturition: elevated amniotic fluid interleukin-6 levels in women with premature rupture of membranes associated with intrauterine infection. Cytokine. 1991;3:155–163. doi: 10.1016/1043-4666(91)90037-e. [DOI] [PubMed] [Google Scholar]
- 120.Romero R, Mazor M, Tartakovsky B. Systemic administration of interleukin-1 induces preterm parturition in mice. Am J Obstet Gynecol. 1991;165:969–971. doi: 10.1016/0002-9378(91)90450-6. [DOI] [PubMed] [Google Scholar]
- 121.Romero R, Ceska M, Avila C, Mazor M, Behnke E, Lindley I. Neutrophil attractant/activating peptide-1/interleukin-8 in term and preterm parturition. Am J Obstet Gynecol. 1991;165:813–820. doi: 10.1016/0002-9378(91)90422-n. [DOI] [PubMed] [Google Scholar]
- 122.Romero R, Tartakovsky B. The natural interleukin-1 receptor antagonist prevents interleukin-1-induced preterm delivery in mice. Am J Obstet Gynecol. 1992;167:1041–1045. doi: 10.1016/s0002-9378(12)80035-4. [DOI] [PubMed] [Google Scholar]
- 123.Romero R, Mazor M, Sepulveda W, Avila C, Copeland D, Williams J. Tumor necrosis factor in preterm and term labor. Am J Obstet Gynecol. 1992;166:1576–1587. doi: 10.1016/0002-9378(92)91636-o. [DOI] [PubMed] [Google Scholar]
- 124.Romero R, Mazor M, Brandt F, Sepulveda W, Avila C, Cotton DB, Dinarello CA. Interleukin-1 alpha and interleukin-1 beta in preterm and term human parturition. Am J Reprod Immunol. 1992;27:117–123. doi: 10.1111/j.1600-0897.1992.tb00737.x. [DOI] [PubMed] [Google Scholar]
- 125.Romero R, Sepulveda W, Kenney JS, Archer LE, Allison AC, Sehgal PB. Interleukin 6 determination in the detection of microbial invasion of the amniotic cavity. Ciba Found Symp. 1992;167:205–20. doi: 10.1002/9780470514269.ch13. discussion 220–3.:205–220. [DOI] [PubMed] [Google Scholar]
- 126.Romero R, Yoon BH, Kenney JS, Gomez R, Allison AC, Sehgal PB. Amniotic fluid interleukin-6 determinations are of diagnostic and prognostic value in preterm labor. Am J Reprod Immunol. 1993;30:167–183. doi: 10.1111/j.1600-0897.1993.tb00618.x. [DOI] [PubMed] [Google Scholar]
- 127.Cherouny PH, Pankuch GA, Romero R, Botti JJ, Kuhn DC, Demers LM, Appelbaum PC. Neutrophil attractant/activating peptide-1/interleukin-8: association with histologic chorioamnionitis, preterm delivery, and bioactive amniotic fluid leukoattractants. Am J Obstet Gynecol. 1993;169:1299–1303. doi: 10.1016/0002-9378(93)90297-v. [DOI] [PubMed] [Google Scholar]
- 128.Hillier SL, Witkin SS, Krohn MA, Watts DH, Kiviat NB, Eschenbach DA. The relationship of amniotic fluid cytokines and preterm delivery, amniotic fluid infection, histologic chorioamnionitis, and chorioamnion infection. Obstet Gynecol. 1993;81:941–948. [PubMed] [Google Scholar]
- 129.Gomez R, Romero R, Galasso M, Behnke E, Insunza A, Cotton DB. The value of amniotic fluid interleukin-6, white blood cell count, and gram stain in the diagnosis of microbial invasion of the amniotic cavity in patients at term. Am J Reprod Immunol. 1994;32:200–210. doi: 10.1111/j.1600-0897.1994.tb01115.x. [DOI] [PubMed] [Google Scholar]
- 130.Fidel PL, Jr, Romero R, Wolf N, Cutright J, Ramirez M, Araneda H, Cotton DB. Systemic and local cytokine profiles in endotoxin-induced preterm parturition in mice. Am J Obstet Gynecol. 1994;170:1467–1475. doi: 10.1016/s0002-9378(94)70180-6. [DOI] [PubMed] [Google Scholar]
- 131.Gomez R, Ghezzi F, Romero R, Munoz H, Tolosa JE, Rojas I. Premature labor and intra-amniotic infection. Clinical aspects and role of the cytokines in diagnosis and pathophysiology. Clin Perinatol. 1995;22:281–342. [PubMed] [Google Scholar]
- 132.Andrews WW, Hauth JC, Goldenberg RL, Gomez R, Romero R, Cassell GH. Amniotic fluid interleukin-6: correlation with upper genital tract microbial colonization and gestational age in women delivered after spontaneous labor versus indicated delivery. Am J Obstet Gynecol. 1995;173:606–612. doi: 10.1016/0002-9378(95)90290-2. [DOI] [PubMed] [Google Scholar]
- 133.Yoon BH, Romero R, Park JS, Chang JW, Kim YA, Kim JC, Kim KS. Microbial invasion of the amniotic cavity with Ureaplasma urealyticum is associated with a robust host response in fetal, amniotic, and maternal compartments. Am J Obstet Gynecol. 1998;179:1254–1260. doi: 10.1016/s0002-9378(98)70142-5. [DOI] [PubMed] [Google Scholar]
- 134.Pacora P, Romero R, Maymon E, Gervasi MT, Gomez R, Edwin SS, Yoon BH. Participation of the novel cytokine interleukin 18 in the host response to intra-amniotic infection. Am J Obstet Gynecol. 2000;183:1138–1143. doi: 10.1067/mob.2000.108881. [DOI] [PubMed] [Google Scholar]
- 135.Yoon BH, Oh SY, Romero R, Shim SS, Han SY, Park JS, Jun JK. An elevated amniotic fluid matrix metalloproteinase-8 level at the time of mid-trimester genetic amniocentesis is a risk factor for spontaneous preterm delivery. Am J Obstet Gynecol. 2001;185:1162–1167. doi: 10.1067/mob.2001.117678. [DOI] [PubMed] [Google Scholar]
- 136.Yoon BH, Romero R, Moon JB, Shim SS, Kim M, Kim G, Jun JK. Clinical significance of intra-amniotic inflammation in patients with preterm labor and intact membranes. Am J Obstet Gynecol. 2001;185:1130–1136. doi: 10.1067/mob.2001.117680. [DOI] [PubMed] [Google Scholar]
- 137.Chaiworapongsa T, Romero R, Espinoza J, Kim YM, Edwin S, Bujold E, Gomez R, Kuivaniemi H. Macrophage migration inhibitory factor in patients with preterm parturition and microbial invasion of the amniotic cavity. J Matern Fetal Neonatal Med. 2005;18:405–416. doi: 10.1080/14767050500361703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hirsch E, Filipovich Y, Mahendroo M. Signaling via the type I IL-1 and TNF receptors is necessary for bacterially induced preterm labor in a murine model. Am J Obstet Gynecol. 2006;194:1334–1340. doi: 10.1016/j.ajog.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 139.Hanna N, Bonifacio L, Weinberger B, Reddy P, Murphy S, Romero R, Sharma S. Evidence for interleukin-10-mediated inhibition of cyclo-oxygenase-2 expression and prostaglandin production in preterm human placenta. Am J Reprod Immunol. 2006;55:19–27. doi: 10.1111/j.1600-0897.2005.00342.x. [DOI] [PubMed] [Google Scholar]
- 140.Gotsch F, Romero R, Kusanovic JP, Erez O, Espinoza J, Kim CJ, Vaisbuch E, Than NG, Mazaki-Tovi S, Chaiworapongsa T, Mazor M, Yoon BH, Edwin S, Gomez R, Mittal P, Hassan SS, Sharma S. The anti-inflammatory limb of the immune response in preterm labor, intra-amniotic infection/inflammation, and spontaneous parturition at term: a role for interleukin-10. J Matern Fetal Neonatal Med. 2008;21:529–547. doi: 10.1080/14767050802127349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lamont RF, Rose M, Elder MG. Effect of bacterial products on prostaglandin E production by amnion cells. Lancet. 1985;2:1331–1333. doi: 10.1016/s0140-6736(85)92628-5. [DOI] [PubMed] [Google Scholar]
- 142.Lamont RF, Anthony F, Myatt L, Booth L, Furr PM, Taylor-Robinson D. Production of prostaglandin E2 by human amnion in vitro in response to addition of media conditioned by microorganisms associated with chorioamnionitis and preterm labor. Am J Obstet Gynecol. 1990;162:819–825. doi: 10.1016/0002-9378(90)91017-7. [DOI] [PubMed] [Google Scholar]
- 143.Prince AS. Biofilms, antimicrobial resistance, and airway infection. N Engl J Med. 2002;347:1110–1111. doi: 10.1056/NEJMcibr021776. [DOI] [PubMed] [Google Scholar]
- 144.Romero R, Schaudinn C, Kusanovic JP, Gorur A, Gotsch F, Webster P, Nhan-Chang CL, Erez O, Kim CJ, Espinoza J, Goncalves LF, Vaisbuch E, Mazaki-Tovi S, Hassan SS, Costerton JW. Detection of a microbial biofilm in intraamniotic infection. Am J Obstet Gynecol. 2008;198:135. doi: 10.1016/j.ajog.2007.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Rotstein OD, Pruett TL, Simmons RL. Mechanisms of microbial synergy in polymicrobial surgical infections. Rev Infect Dis. 1985;7:151–170. doi: 10.1093/clinids/7.2.151. [DOI] [PubMed] [Google Scholar]
- 146.Sibley CD, Duan K, Fischer C, Parkins MD, Storey DG, Rabin HR, Surette MG. Discerning the complexity of community interactions using a Drosophila model of polymicrobial infections. PLoS Pathog. 2008;4:e1000184. doi: 10.1371/journal.ppat.1000184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Nijssen S, Florijn A, Top J, Willems R, Fluit A, Bonten M. Unnoticed spread of integron-carrying Enterobacteriaceae in intensive care units. Clin Infect Dis. 2005;41:1–9. doi: 10.1086/430711. [DOI] [PubMed] [Google Scholar]
- 148.Wu HM, Harcourt BH, Hatcher CP, Wei SC, Novak RT, Wang X, Juni BA, Glennen A, Boxrud DJ, Rainbow J, Schmink S, Mair RD, Theodore MJ, Sander MA, Miller TK, Kruger K, Cohn AC, Clark TA, Messonnier NE, Mayer LW, Lynfield R. Emergence of ciprofloxacin-resistant Neisseria meningitidis in North America. N Engl J Med. 2009;360:886–892. doi: 10.1056/NEJMoa0806414. [DOI] [PubMed] [Google Scholar]
- 149.Bennett PR, Rose MP, Myatt L, Elder MG. Preterm labor: stimulation of arachidonic acid metabolism in human amnion cells by bacterial products. Am J Obstet Gynecol. 1987;156:649–655. doi: 10.1016/0002-9378(87)90070-6. [DOI] [PubMed] [Google Scholar]
- 150.Romero R, Kadar N, Hobbins JC, Duff GW. Infection and labor: the detection of endotoxin in amniotic fluid. Am J Obstet Gynecol. 1987;157:815–819. doi: 10.1016/s0002-9378(87)80061-3. [DOI] [PubMed] [Google Scholar]
- 151.Romero R, Hobbins JC, Mitchell MD. Endotoxin stimulates prostaglandin E2 production by human amnion. Obstet Gynecol. 1988;71:227–228. [PubMed] [Google Scholar]
- 152.Bennett PR, Elder MG, Myatt L. Secretion of phospholipases by bacterial pathogens may initiate preterm labor. Am J Obstet Gynecol. 1990;163:241–242. doi: 10.1016/s0002-9378(11)90709-1. [DOI] [PubMed] [Google Scholar]
- 153.Espinoza J, Goncalves LF, Romero R, Nien JK, Stites S, Kim YM, Hassan S, Gomez R, Yoon BH, Chaiworapongsa T, Lee W, Mazor M. The prevalence and clinical significance of amniotic fluid ‘sludge’ in patients with preterm labor and intact membranes. Ultrasound Obstet Gynecol. 2005;25:346–352. doi: 10.1002/uog.1871. [DOI] [PubMed] [Google Scholar]
- 154.Romero R, Kusanovic JP, Espinoza J, Gotsch F, Nhan-Chang CL, Erez O, Kim CJ, Khalek N, Mittal P, Goncalves LF, Schaudinn C, Hassan SS, Costerton JW. What is amniotic fluid ‘sludge’? Ultrasound Obstet Gynecol. 2007;30:793–798. doi: 10.1002/uog.5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kusanovic JP, Espinoza J, Romero R, Goncalves LF, Nien JK, Soto E, Khalek N, Camacho N, Hendler I, Mittal P, Friel LA, Gotsch F, Erez O, Than NG, Mazaki-Tovi S, Schoen ML, Hassan SS. Clinical significance of the presence of amniotic fluid ‘sludge’ in asymptomatic patients at high risk for spontaneous preterm delivery. Ultrasound Obstet Gynecol. 2007;30:706–714. doi: 10.1002/uog.4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Philipson A, Sabath LD, Charles D. Transplacental passage of erythromycin and clindamycin. N Engl J Med. 1973;288:1219–1221. doi: 10.1056/NEJM197306072882307. [DOI] [PubMed] [Google Scholar]
- 157.Heikkinen T, Laine K, Neuvonen PJ, Ekblad U. The transplacental transfer of the macrolide antibiotics erythromycin, roxithromycin and azithromycin. BJOG. 2000;107:770–775. doi: 10.1111/j.1471-0528.2000.tb13339.x. [DOI] [PubMed] [Google Scholar]