Abstract
Chemical crosslinking combined with mass spectrometry can be a powerful approach for the identification of protein-protein interactions and for providing constraints on protein structures. However, enrichment of crosslinked peptides is crucial to reduce sample complexity before mass spectrometric analysis. In addition compact crosslinkers are often preferred to provide short spacer lengths, surface accessibility to the protein complexes, and must have reasonable solubility under condition where the native complex structure is stable. In this study, we present a novel compact crosslinker that contains two distinct features: 1) an alkyne tag and 2) a small molecule detection tag (NO2-) to maintain reasonable solubility in water. The alkyne tag enables enrichment of the crosslinked peptide after proteolytic cleavage after coupling of an affinity tag using alkyne-azido click chemistry. Neutral loss of the small NO2- moiety provides a secondary means of detecting crosslinked peptides in MS/MS analyses, providing additional confidence in peptide identifications. We show the labeling efficiency of this crosslinker, which we termed CLIP (Click-enabled Linker for Interacting Proteins) using ubiquitin. The enrichment capability of CLIP is demonstrated for crosslinked ubiquitin in highly complex E. coli cell lysates. Sequential CID-MS/MS and ETD-MS/MS of inter-crosslinked peptides (two peptides connected with a crosslinker) are also demonstrated for improved automated identification of crosslinked peptides.
Keywords: Mass spectrometry, proteomics, protein crosslinking, click chemistry
Introduction
Chemical crosslinking combined with mass spectrometric analysis is emerging as a powerful technique for protein-protein interaction and protein structure elucidation studies.1 Crosslinkers covalently link two interacting proteins, often with chemistries specific to certain amino acid side chains. After enzymatic digestion of the proteins, the resulting crosslinked peptides can be analyzed by LC-MS(/MS) to identify crosslinked species.2 There are two limitations which need to be addressed in order to convert the targeted approaches into global discovery-based methods for identifying protein interactions. The first limitation could be addressed by the development of crosslinkers that are multifunctional but still compact, while the second limitation could be addressed by methods or tools that enable better interpretation of large numbers of complex LC-MS/MS spectra. Due to the low relative abundance of crosslinked products compared to unmodified species, enrichment of crosslinked species to improve sensitivity and data quality, and thus the likelihood of unambiguous identification of crosslinked peptides is another highly desirable feature. A number of research groups have demonstrated different MS-based approaches and applications towards crosslinking in complex systems. Approaches to reduce the complexity include adding affinity tags, using high-resolution mass spectrometry, adding reporter groups which are unique to the crosslinked species, using isotopic labeling techniques and developing new bioinformatics analysis methods. Herein we present a new small, amine-reactive crosslinker, which includes a reporter group and a handle which permits the coupling of a reagent for affinity purification, and demonstrate that its presence results in readily-interpretable MS/MS-spectra.
This crosslinker builds on previous approaches coupling chemical protein crosslinking with mass spectrometry. Sinz and co-authors first utilized high-resolution Fourier transfer ion cyclotron resonance (FTICR)-MS to understand the structure of calmodulin and its binding partners using amine-reactive crosslinking approaches.3,4 In an effort to extend these methods to more complex samples, amine-reactive homo-bifunctional crosslinkers with an affinity group to enrich crosslinked peptides have been developed. For example, Trester-Zedlitz et al. have introduced novel crosslinking reagents, which contain affinity tags and cleavable isotopic tags to reduce the complexity of crosslinked samples.5 However, most of the currently available “enrichable” crosslinkers are bulky, reducing the number of crosslinked species identifiable by LC-MS/MS.
New Crosslinkers, which result in unique signatures in mass spectra after being cleaved either chemically or in the mass spectrometer, have been used to pinpoint crosslinked species in high complexity biological samples. Petrotchenko et al. reported an isotope-coded crosslinker that can be cleaved under base treatment. The hydrolytic cleavage helped to identify crosslinked species in complex mixtures by producing characteristic mass shift patterns.6 Another interesting approach uses crosslinkers that selectively fragment cleavable bonds during MS, which generates reporter ions for highlighting spectra that contain crosslinked peptides in complex biological samples.7-9 These MS-identifiable crosslinkers, referred to as PIRs (Protein Interaction Reporters), generally consist of two low energy MS-cleavable bonds and NHS ester reactive groups that produce distinct signature ions.8,9 Several other researchers are also utilizing this strategy to construct more effective MS-identifiable crosslinkers.10,11 Goshe and co-workers recently reported a crosslinker that consists of a single MS-cleavable bond that can be selectively fragmented in the mass spectrometer, which enables identification of crosslinked peptides by commercial proteomics software tools.11 Recent efforts include a sulfonium ion containing crosslinking reagent, which was developed by Reid and co-workers.10 The crosslinked peptides produced from this strategy have shown facile cleavage of the C-S bonds in collision-induced dissociation (CID)-MS in model peptide labeling studies. Additionally, an infra-red chromogenic crosslinker was developed by Broadbelt and co-workers, which utilized a phosphate functional group in the crosslinker for selectively dissociating crosslinked peptides by infrared multiphoton dissociation from unmodified species.12
In spite of the variety of the approaches described above, few are broadly used. A critical challenge that remains for most of these crosslinking approaches is unambiguous assignment of the location of crosslinking, especially identifying the inter-crosslinked peptides (two peptides connected with the crosslinker). Crosslinked peptides in most datasets are typically present in low abundance and/or do not fragment well. While several crosslinking strategies and software tools are available, the large data volume complicates analysis.13-16 Rinner et al recently developed a software tool (XQuest) that utilizes isotopically labeled crosslinkers to reduce the data complexity of a crosslinked dataset, but point out that enrichment functionality on the crosslinker would further aid identification.17 In order to effectively and efficiently discover protein-protein interactions, chemical crosslinkers are needed that are small in size, can effectively crosslink protein or protein complexes, are amenable to mass spectrometry-based bottom-up proteomics analysis, and contain functionality for enrichment.
Herein, we present “CLIP” (Click-enabled Linker for Interacting Proteins), a compact amine-reactive crosslinker augmented with an alkyne tag incorporated into the crosslinker to enable subsequent enrichment of crosslinked peptides and/or proteins through alkyne-azido click chemistry.18 This crosslinker generates crosslinked species that were confidently identified using liquid chromatography-tandem mass spectrometry (LC-MS/MS), both before and after click coupling to a biotin affinity reagent. Additional confidence in the data analysis was achieved by utilizing a neutral loss tag in the crosslinker and sequential LC-CID-MS/MS and LC- electron transfer dissociation (ETD)-MS/MS of inter-crosslinked peptides.
Materials and Methods
Reagents
All chemicals were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise specified. TCEP was purchased from Thermo Fisher Scientific (Waltham, MA) and Peptide Substance P, from AnaSpec (San Hose, CA).
Crosslinking of Substance P
1 μL of Substance P (0.5 μM final concentration) was placed in a centrifuge tube, to which 200 μL of PBS buffer was added. Crosslinker was then added in a 1:5 peptide–to-crosslinker molar ratio and the reaction was allowed to proceed with constant shaking for 30 min at room temperature. After desalting by ZipTip (Millipore, St. Charles, MO), the solution was dried by Speed-vac and then resuspended in ESI buffer (49:49:2, methanol: water: acetic acid) for direct infusion into the FTICR mass spectrometer.
Crosslinking of Ubiquitin
Crosslinking and enrichment reagent stock solutions were prepared in DMSO (100 mM). The crosslinking reaction was performed utilizing a 1:25 protein-to-crosslinker molar ratio in the presence of 200 μL of 20 mM PBS buffer (pH 7.5, Thermo Fisher Scientific). The final concentrations were 10 μM for ubiquitin (bovine) and 250 μM for the crosslinking reagent, respectively. The reaction was allowed to proceed for 30 min after which the reaction was quenched with 50 μL of 50 mM Tris-HCl (pH 8.0). Excess crosslinker was removed using an excellulose dextran column (5 kDa MWCO cut off, Thermo Fisher Scientific), and buffer exchange was accomplished using 50 mM NH4HCO3 (pH 8.0). Protein concentrations were determined by BCA protein assay (Thermo Fisher Scientific). Initial characterization of ubiquitin crosslinking was performed by one dimensional SDS - PAGE, using a precast gradient gel (bio-rad, 4-20%, precision plus blue standard, 10-250 kDa).
Digestion
In-solution digestion was performed after removing the excess crosslinker. Trypsin (Promega, Madison, WI) was added to the solution at a 1: 50 protease-to-protein ratio. Proteolysis was conducted at 37 °C for 6 h, and the trypsin digestion was stopped by using 1% trifluoroacetic acid (TFA) in water. The final solution was SPE purified (C18, reverse phase) and a BCA assay was performed to determine peptide concentrations. In-gel digestion involved excising a band that corresponded to the higher molecular weight species in the crosslinked samples, destaining, and digesting according to an existing protocol.19
Cell Culture and Complex Biological Sample Preparation
We prepared a protein mixture containing three proteins to evaluate enrichment from a moderately complex mixture. Bovine serum albumin (BSA), ribonuclease S (RNase S) and ubiquitin (1 μL, 1 mM) were placed in a centrifuge tube and mixed with crosslinked Substance P in a peptide molar ratio of 10:1.
To prepare a more complex biological sample, E. coli TOP10 containing an empty PBAD vector conferring kanamycin resistance were grown at 37 °C in 200 mL Luria Bertani media with 50 μg/mL kanamycin to a cell density of 1.5 at 600 nm. The cells were harvested by centrifugation and the pellet resuspended in 2 mL ammonium bicarbonate buffer. Cells were lysed by sonication and bead-beating with zirconia silica beads and then centrifuged to clear the lysate. After centrifugation, the supernatant was collected and a BCA protein assay was performed to determine the protein concentration of the E. coli cell lysate.
A total of six samples, including controls, were prepared to demonstrate enrichment from a complex sample. Control samples consisted of 1) E. coli cell lysate (135 μL, 3.70 μg/μL, 500 μg); 2) crosslinked ubiquitin (56 μL, 0.9 μg/μL, 50 μg); and 3) a mixture of E. coli cell lysate and un-crosslinked ubiquitin (10:1 wt:wt). Three additional samples were prepared by mixing E. coli cell lysate and crosslinked ubiquitin at different ratios (wt:wt basis). These samples consisted of E. coli and crosslinked ubiquitin in ratios of 1:1, 10:1, and 100:1 ratio. The samples were reduced and alkylated with dithiothreitol and iodoacetamide, using commonly utilized protocols. No alkylation was performed for the sample containing E. coli cell lysate only, but the reduction step was performed utilizing 40 μL of 10 mM dithiothreitol.
All the samples were diluted to the same volume (~1.4 mL in a 2 mL centrifuge tube) with 50 mM solution of NH3HCO3. Trypsin digestions were performed overnight, utilizing a protein to protease ratio of 100:1.
After trypsinization the volume was reduced using a speed vac to 500 μL. A portion of the tryptic digest (100 μL) was placed in a centrifuge tube for LC-MS/MS analysis. Desalting of this sample was performed utilizing 100 μL solid phase micro-extraction tips (C18 reverse phase, Varian, Palo Alto, CA). Concentrations were determined by BCA protein assay. Click labeling and affinity purification were performed on the remainder of the samples.
Click Labeling and Affinity Enrichment
Procedures by Speers et al. and Galan et al.20,21 were modified for click labeling of the enrichment reagent. Briefly, the peptide mixture produced from the in-solution digestion was reduced to dryness and reconstituted in PBS buffer (100 mM sodium phosphate, 0.15 M NaCl, pH 7.5) that was supplemented with 1 μL 100 mM Tris-[1-benzyl-1H-1,2,3-triazol-4-yl)methylamine (TTA) (0.25 mM final concentration), 20 μL of 50 mM CuSO4 (2.5 mM final concentration), and 40 μL of 50 mM Tris (2-carboxyethyl) phosphine (TCEP) (5 mM final concentration) to a final volume of 400 μL. Biotin-azide enrichment reagent was added in a 1:10 molar ratio of peptide-to-enrichment reagent and allowed to react for 1-2 h at room temperature and with constant shaking.
After click labeling the solution was incubated with 100 μL ultra-link monomeric avidin bead suspension (Thermo Fisher Scientific) after which, the beads were washed with PBS and biotin elution and regeneration buffers, as described in the manufacturer’s protocol. At 0.5 h incubation, the beads were washed three times with PBS buffer. Next, the bead solution was taken up on a 50 kDa cut-off spin column (Thermo Fisher Scientific) and washed five times with 200 μL of PBS and 100 mM NH4HCO3 solution. The spin column allowed for automated washing, its membrane filter that held the beads in place minimized bead loss during washing. Peptides were eluted from the beads with a mixture of 50% acetonitrile: 50% H2O: 0.4 % TFA.22 Final peptide concentrations were determined by BCA peptide assay.
Instrumental Analysis and Methods
LC-MS/MS analysis was performed using a custom LC platform coupled to a LTQ or LTQ-Orbitrap mass spectrometer (Thermo Scientific, San Jose, CA). 23 LTQ-Orbitrap mass spectrometer is outfitted with a custom ion funnel based API source and electrospray ionization (ESI) interface.24 Electrospray emitters were custom made using 150 um o.d. × 20 um i.d. chemically etched fused silica.25 The heated capillary temperature and spray voltage were 200°C and 2.2 kV, respectively. Data was acquired for 100 min, beginning 65 min after sample injection (15 min into gradient). Orbitrap spectra (AGC 1×106) were collected from 400-2000 m/z at a resolution of 100k followed by data dependant ion trap MS/MS spectra (AGC 1×104) of the six most abundant ions using a collision energy of 35%. A dynamic exclusion time of 60 sec was used to discriminate against previously analyzed ions. ETD-MS/MS was performed using an LTQ mass spectrometer (Thermo Fisher Scientific). For parallel CID-ETD MS/MS analysis of crosslinked samples a manual capillary RPLC system was used for peptide separations. For ETD experiments, a precursor cation target was set to 30,000, and the reagent anion target was set to 100,000. The ETD reaction time was set to 125 ms. Data-dependent data sets were collected for the four most abundant species after each MS scan using sequential CID and ETD collision modes. Direct infusion studies were performed using an 11T FTICR mass spectrometer (Bruker, Billerica, MA, modified in house).
Data Analysis
MS/MS spectra were automatically analyzed for crosslinked peptides using Xlink-Identifier software developed in-house (to be published in detail elsewhere). Briefly, Xlink-Identifier consists of two sequential steps. First, a database of theoretical crosslinked peptides is generated from theoretical tryptic cleavages of all proteins under consideration. Note that crosslinker-labeled lysines are not considered for tryptic cleavage. Crosslinked peptides are then identified by calculating a score indicating the similarity between observed and theoretical MS/MS spectra. An additional level of confidence is established by utilizing the neutral fragment of the nitro group (45.99 Da) from the precursor m/z of the crosslinked peptides. Because the nitro group was not very labile, only the loss from the precursor m/z was considered. Initially, only ‘b’ and ‘y’ ions (general peptide fragmentation patterns, cleavage in the amide carbonyl bonds) were searched for CID fragmentations. To confirm identification of inter-crosslinked peptides, a minimum of two crosslinked ‘b’ or ‘y’ fragments were necessary in addition to the free ‘b’ and ‘y’ ions identified from two crosslinked peptides. For ETD fragmentations, ‘c’ and ‘z’ ions were considered, i.e., general ETD fragmentation rules, as well as charge neutralized peaks from the precursor m/z due to electron transfers.
Inter-crosslinked peptides were manually validated utilizing another software called Xlink-Explorer that also was developed in house. This software compares sequence masses, peptide fragment ions, water and ammonia losses, and internal peptide fragments generated from simultaneous fragmentation of two peptides. The tool is written as an MSExcel (Microsoft) VBA function and makes use of ICR2LS (developed in house) and Xcalibur (Thermo Fisher Scientific) libraries. Both ICR2LS and Xlink-Explorer are available with instructions for general use on http://omics.pnl.gov.
Results and Discussion
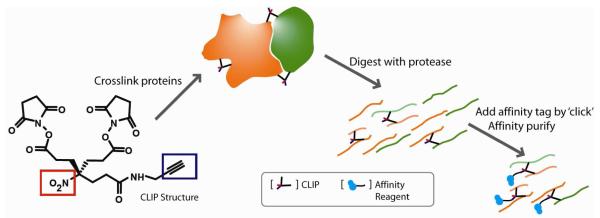
In this study, we designed, synthesized, and evaluated a novel compact crosslinker (spacer chain length, 9-10 Å) that incorporates two distinct features: 1) an alkyne tag to enable enrichment of crosslinked peptides after coupling of an affinity group by way of alkyne-azido click chemistry and 2) a small molecule detection tag (NO2 group) to maintain solubility in water (Figure 1). Because alkyne functional groups are typically not found in proteins or other biomolecules, the alkyne tag represents a bio-orthogonal chemical reporter26 and allows addition of a biotin enrichment moiety through click chemistry. Neutral loss of the small molecule NO2 group provides a secondary means of detecting crosslinked peptides in MS/MS spectra, thereby providing additional confidence in peptide identifications.
Figure 1.
CLIP structure and enrichment strategy of crosslinked peptides after addition of an affinity tag through alkyne-azido click chemistry (alkyne tag for enrichment, blue rectangle, and NO2 tag for neutral loss validation, red rectangle).
In our CLIP crosslinking strategy depicted in Figure 1, proteins or protein complexes in a sample are crosslinked prior to proteolytic digestion of the sample. The Huisgen reaction, also known as alkyne–azido click chemistry is employed to label crosslinked peptides with biotin, followed by enrichment using biotin-avidin affinity chromatography prior to analysis using LC-MS/MS, and identification using in-house informatics tools. We demonstrated the applicability of this strategy in a series of increasingly complex experiments.
Demonstration of CLIP Crosslinking and CID-MS/MS without Enrichment
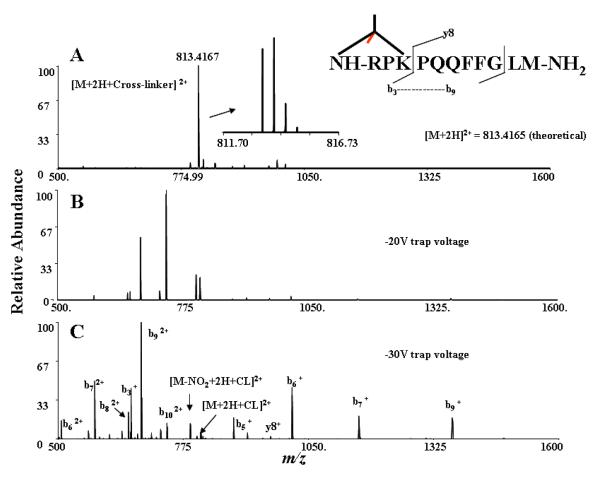
CLIP was synthesized and purified as described in the supporting information. The labeling efficiency of CLIP was initially tested using Substance P (RPKPQQFFGLM-NH2). This model peptide was crosslinked with CLIP between two primary amine residues in the peptide; one situated in the N-terminus and the other in the lysine side chain (an intra-peptide crosslink). Modified m/z was observed with 0.3 ppm mass accuracy (please see the calculation procedure in the supplementary information). As shown in Figure 2, substance P was completely labeled with the crosslinker, as evidenced by the fact that no unmodified m/z corresponding to Substance P was observed in the parent spectrum. MS/MS spectra of this modified peptide also showed efficient fragmentation. Variation of the collisional trap voltage revealed no evidence of fragmentation for the crosslinker backbone. Loss of the NO2 (nitro) group from the precursor was observed in MS/MS spectra and is characteristic in most spectra containing the CLIP reagent; however, loss of the nitro group from fragment ions was not observed.
Figure 2.
A simple example of CID fragmentation patterns being maintained after addition of CLIP. Shown is an ESI-FTICR-MS/MS analysis of Substance P labeled with crosslinker. There is no peak corresponding to substance peak alone (MW: 1346.7281 Da) A) Substance P, labeled with CLIP. B) MS/MS of intra-crosslinked Substance P with −20V trap voltage. C) MS/MS of the modified peptide with −30V collisional trap voltage.
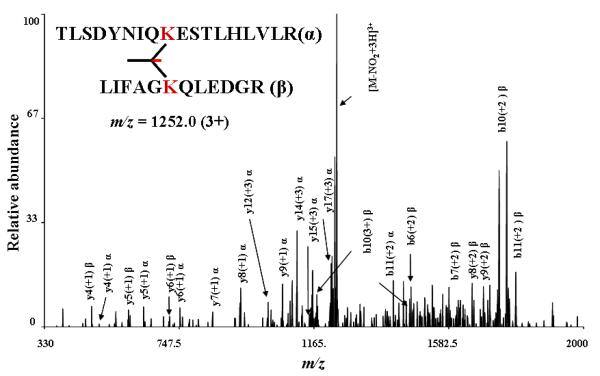
Next, we used CLIP with ubiquitin, a small protein (~8.5 kDa) with 7 lysines that has been extensively characterized by MS. After crosslinking for 30 min, the reaction was quenched and excess CLIP was removed. A portion of the sample was digested with trypsin and analyzed by LC-MS/MS. The other portion was analyzed using SDS-PAGE. The crosslinked sample in the gel exhibited a second higher molecular weight band compared to the uncrosslinked control (Supplementary figure S2). The higher molecular band was excised from the gel, digested with trypsin, and analyzed by LC-MS/MS. Several crosslinked peptides were identified by assigning ions of the MS/MS fragmentation ladder (shown in Figure 3> for the intra protein species containing inter-crosslinks between lysine 48 (LIFAGK48QLEDGR) and lysine 63 (TLSDYNIQK63ESTLHLVLR)) with the help of in-house developed software. Confident identifications from ubiquitin (Supporting information figures S3, Table T1) included three peptides with attached crosslinker, where one end of the crosslinker was hydrolyzed (dead-end), as well as three species that were crosslinked between two peptides (inter-peptide crosslinking) and peptide self crosslinked (intra-peptide crosslinking).
Figure 3.
An annotated example of an intra-protein inter-crosslinked peptide identified after in-solution digestion of crosslinked ubiquitin before enrichment to demonstrate the ability to interpret complex spectra for inter-crosslinked species.
Table T1.
Table of confident identifications of inter-, intra--, and dead-end peptides from ubiquitin before or after click labeling utilizing our automated data analysis platform for data run on CID-MS/MS alone or CID and ETD-MS/MS together, where CS is the charge state and labeled lysines are designated K^. NA – not applicable, i.e., experiments not performed.
| Type of Crosslinking |
Sequence 1 | Sequence 2 | Work up condition |
Before click |
CS | NO2 loss Before click |
After click |
CS | NO2 loss After click |
|---|---|---|---|---|---|---|---|---|---|
| Inter | LIFAGK^QLEDG R |
IQDK^EGIPPDQQ R |
In gel | Yes | 3 | No | No | --- | No |
| LIFAGK^QLEDG R |
LIFAGK^QLEDGR | In solution |
No | 3 | Yes | Yes | 4 | Yes | |
| LIFAGK^QLEDG R |
LIFAGK^QLEDGR | In gel | Yes | 3 | Yes | NA | 4 | Yes | |
| LIFAGK^QLEDG R |
TLSDYNIQK^EST LHLVLR |
In solution |
Yes | 3 | Yes | Yes | 5 | Yes | |
| Intra- | AK^IQDK^EGIPP DQQR |
-------------- | In solution |
Yes | 2 | Yes | No search performed |
NA | NA |
| Dead-end | LIFAGK^QLEDG R |
------------ | In solution |
Yes | 2 | Yes | Yes | 2 | Yes |
| LIFAGK^QLEDG R |
------------ | In gel | Yes | 2 | Yes | Yes | 2 | Yes | |
| TLSDYNIQK^ES TLHLVLR |
------------ | In solution |
Yes | 2,3 | Yes | Yes | 2 | Yes | |
| MQIFVK^TLTGK | ------------ | In solution |
No | --- | Yes | Yes | 3 | Yes |
Demonstration of CLIP Crosslinking and CID-MS/MS after Coupling of Enrichment Agent
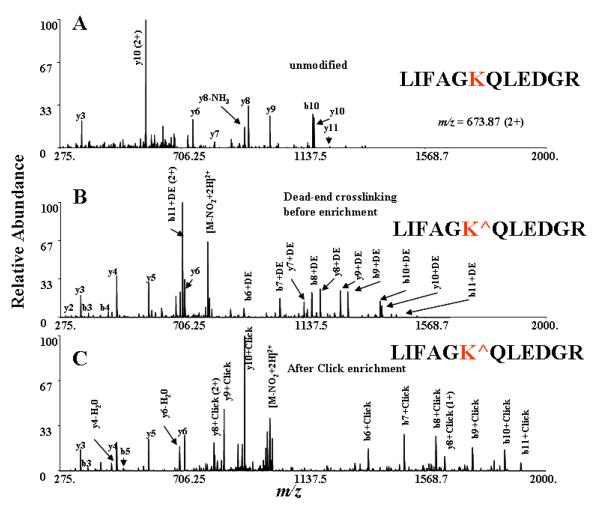
To add enrichment functionality to the small CLIP crosslinker through alkyne-azido click chemistry, an affinity agent containing an azide moiety and biotin was synthesized (see supporting information). To ensure that the crosslinker containing the enrichment reagent did not disrupt MS/MS fragmentation patterns, we performed MS/MS after coupling the enrichment agent to in-solution digested crosslinked ubiquitin using copper-catalyzed alkyne-azido click chemistry.21 The complete strategy is illustrated in Figure 4 using the spectrum of a peptide incorporating hydrolyzed CLIP (dead-end crosslinking). After CLIP labeling, the added mass of the crosslinker is apparent in the MS/MS spectrum (Figure 4B). After coupling of the enrichment reagent, the newly added mass is readily observed in the dead-end crosslinking form of this peptide (Figure 4C). As summarized in table T1, we observed the same inter-crosslinked and dead-end crosslinked peptides before and after click labeling, with the exception of one inter-crosslinked peptide. One crosslinked species stemming from an ubiquitin-ubiquitin interaction could only be confidently identified after enrichment, either by analyzing the higher apparent molecular weight band of the SDS-PAGE gel or with biotin/avidin affinity chromatography (Supplementary figure S4A). This demonstrates that enrichment of crosslinked peptides is critical to sensitively and specifically identifying crosslinked species.
Figure 4.
Demonstration of crosslinker stability with and without the enrichment moiety during MS/MS of a dead-end modified peptide from ubiquitin. A) MS/MS of unmodified peptide is shown for comparison. B) MS/MS of modified peptide after CLIP labeling. C) MS/MS modified peptide after click labeling and enrichment. Mass spectra clearly show the mass shift due to modification before and after addition of the enrichment reagent. K^ - dead-end labeled lysine residue.
Enrichment and Identification of Crosslinked Species from Complex Backgrounds
Next, we demonstrated the enrichment and identification of crosslinked peptides from both moderately and highly complex biological samples. First, a moderately complex protein sample was prepared by spiking crosslinked Substance P (10:1 molar ratio) into a mixture of three uncrosslinked proteins (BSA, RNase S and Ubiquitin) of different molecular weights. The sample was trypsinized and reacted using click chemistry with a biotin-azide enrichment agent, and then enriched using monomeric avidin. CID-MS/MS revealed that Substance P contained the additional masses of the combined crosslinking and enrichment reagents (see Figure 2 for MS/MS spectrum before enrichment), and clearly identified this click-labeled and affinity purified peptide (Supplementary Figure S5).
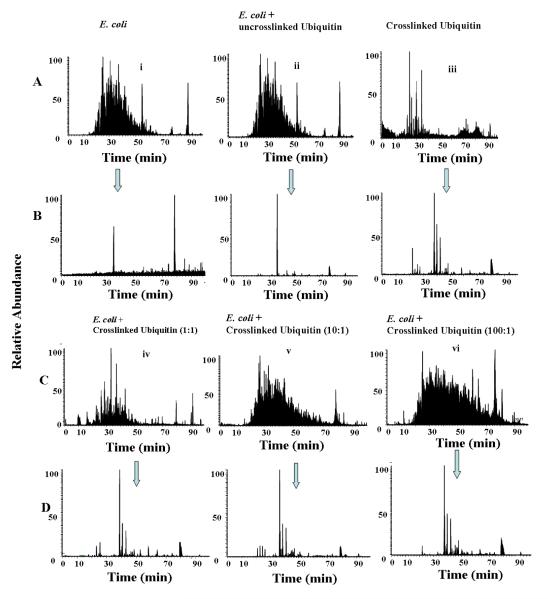
Demonstrating the ability to enrich from a highly complex biological sample involved three samples that contained a mixture of E. coli proteins and crosslinked ubiquitin prepared in different known ratios (1:1, 10:1, and 100:1 wt:wt), as well as three control samples to characterize the isolated components of the mixtures. The three control samples were 1) E. coli cell lysate, 2) mixture of E. coli and uncrosslinked ubiquitin, and 3) crosslinked ubiquitin without enrichment. After proteolytic digestion, click labeling and biotin-avidin enrichment, LC-MS/MS were performed in replicate. The LC-MS chromatograms in Figure 5 illustrate a clear reduction in data complexity after click labeling and enrichment. A comparison of the chromatograms reveals that crosslinker-containing peptides are greatly enriched, even from a sample in which the crosslinked protein represents only 1% of the total protein (Figure 5D). The chromatograms containing crosslinked peptides are dominated by peaks originating from the enrichment agent and the ligand catalyst used for click chemistry, but after enrichment the LC traces of the four samples containing crosslinked ubiquitin peptides show remarkably constant features, implying the high quality of enrichment even of the most dilute crosslinked species. As a consequence, data analysis of MS/MS spectra using XLink-Identifier identified the same peptides commonly observed after crosslinking purified ubiquitin, including the homodimeric crosslinked species (Supporting Figures S6 and S4). The repeated identification of the same crosslinked peptides in biological samples with varying complexities clearly illustrates the utility of the enrichment ability of CLIP.
Figure 5.
Demonstration of the “CLIP” enrichment strategy from complex biological samples. Arrows indicate corresponding LC-MS/MS datasets before and after enrichment. A) LC-MS/MS chromatogram of three controls- i) E. coli ii) E. coli and uncrosslinked ubiquitin spiked in the sample and iii) Crosslinked ubiquitin. B) LC-MS/MS chromatogram after click labeling and biotin-avidin enrichment of control samples. C) Three complex biological samples with different complexity ratios iv) cell lysate of E.coli spiked with crosslinked ubiquitin, 1:1 weight ratio. v) E. coli cell lysate spiked with crosslinked ubiquitin, 10:1 weight ratio. vi) E. coli cell lysate spiked with crosslinked ubiquitin, 100:1 weight ratio. D) LC-MS/MS chromatogram of each datasets after click labeling and biotin-avidin enrichment. Raw data associated with this paper are available at http://omics.pnl.gov.
Application of Sequential CID and ETD to Aid MS/MS Interpretation
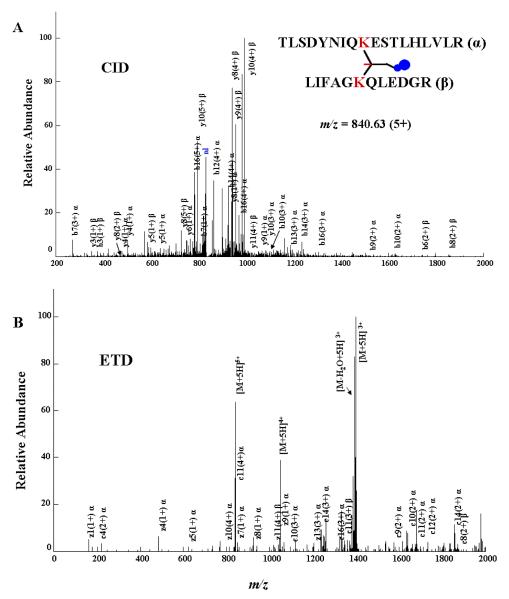
While enrichment of the crosslinked species increases the confidence of the identification, it seemed useful to test the amenability of CLIP to an orthogonal mass spectrometric method designed for ions of higher charge states. With common ion charge states of ≤3+, intra- and dead-end crosslinked peptides are amenable to analysis using CID-MS/MS and general software such as SEQUEST.27 However, inter-crosslinked peptides have ion charge states ≥3+, which alone complicates data analysis. Because ETD fragmentation results in more interpretable spectra than CID fragmentation for ≥3+ ion charge states,28 we utilized ETD-MS/MS as a complementary technique for obtaining additional confidence in inter-crosslinked peptide identifications. ETD is a novel fragmentation technique which is reported to keep labile modifications intact in a fashion similar to ECD (electron capture dissociation).29 ECD on an intra crosslinked peptide has been reported previously,8 but ETD of an inter crosslinked peptide has not previously been shown. Figure 6 (A, B) exemplifies sequential CID-MS/MS and ETD-MS/MS spectra for an enriched inter-crosslinked species. A second example is provided in Supplemental Figure S4. The CID spectrum is dominated by fragment ions from both chains, whereas the highest-intensity species in the ETD spectrum are different ionization states of the parent ion. As with CID, CLIP-containing peptides showed no unexpected fragmentations and were amenable to fragmentation. Although fragmentation by CID and ETD resulted in different spectral characteristics, they both unambiguously identified the same crosslinked species.
Figure 6.
An example of sequential CID and ETD-MS/MS of the same inter-crosslinked species with annotation of the associated spectra to demonstrate the interpretability of the fragmentation patterns following the enrichment procedure. nl: neutral loss peak (diagnostic loss of NO2 group).
Validation of Crosslinked Sites with Published Crystal Structure of Ubiquitin
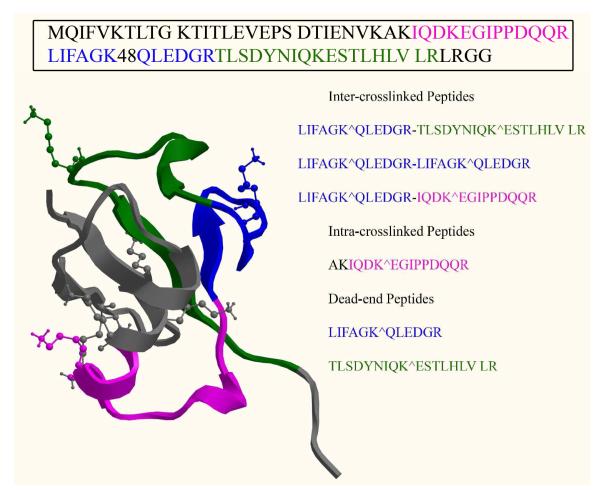
As shown in table T1, we have identified multiple crosslinked peptide species from ubiquitin both before and after enrichment. Measuring distance constraints between of the confidently identified crosslinked peptides is a ubiquitin crystal structure shows that the distances between the crosslinked lysine α-carbons of ubiquitin are within 20Å (Figure 7). This finding is consistent with a calculated fully extended backbone length for similar crosslinkers of about 10Å with lysine side chain of ~6 Å. Commercial crosslinkers DSS and BS3 that have similar backbone lengths have been reported to crosslink primary amines at a distance of <24 Å.16,30 A species from dimeric ubiquitin crosslinked through lysine 48 was confidently identified only after enrichment and confirmed by sequential CID and ETD-MS/MS. This crosslinked species is consistent with transient dimerization that permits crosslinking at lysine 48. Note that lysine 48 is highly reactive and located in a hydrophobic patch that promotes interactions among ubiquitin accessory and substrate proteins.31 The identified crosslinks are thus consistent with the known structure and reactivity of ubiquitin.
Figure 7.
Ubiquitin sequence, crystal structure (pdb:1V80) and color coded crosslinking sites identified by using CLIP.
Conclusion
We have presented here a characterization of a new crosslinking strategy. Most crosslinker designs have specific shortcomings that we believe have been overcome in our design, specifically: 1) the crosslinking reagent is compact to allow optimal crosslinking with folded proteins, 2) with or without addition of the affinity moiety the CID fragmentation patterns do not show crosslinker fragmentation except for loss of a small nitro spectrum-detection tag from the parent ion, 3) enrichment has been shown to be specific to permit mass-spectrometric identification of crosslinked peptides, and 4) application of ETD in conjunction with CID yields complementary results for improved identification and confidence levels for crosslinked species. We believe that this novel crosslinker will aid the discovery of crosslinked peptides in an automated high-throughput manner to facilitate characterization of protein interactions, as well as aid the elucidation of protein structures through distance constraints. This CLIP crosslinker and associated analysis strategy are one step towards developing a mass spectrometry oriented-global approach to studying protein-protein interactions.
Supplementary Material
Acknowledgment
We thank Thomas C. Squier for advice and suggestions. This work was supported by the Laboratory Directed Research and Development program at Pacific Northwest National Laboratory (PNNL) and the NIH National Center for Research Resources (RR18522). Significant portions of this work were performed in the Environmental Molecular Science Laboratory, a U. S. Department of Energy (DOE) national scientific user facility located at PNNL (Richland, WA). Battelle Memorial Institute operates PNNL for the DOE under contract DE-AC05-76RLO01830.
Footnotes
Supporting Information Available Materials and methods, MS/MS spectra of dead-end, intra- and other inter-crosslinked peptides are available. This material are available at free of charge at http://pubs.acs.org.
References
- (1).Sinz A. Mass Spectrom Rev. 2006;25:663–82. doi: 10.1002/mas.20082. [DOI] [PubMed] [Google Scholar]
- (2).Collins CJ, Schilling B, Young M, Dollinger G, Guy RK. Bioorg Med Chem Lett. 2003;13:4023–6. doi: 10.1016/j.bmcl.2003.08.053. [DOI] [PubMed] [Google Scholar]
- (3).Ihling C, Schmidt A, Kalkhof S, Schulz DM, Stingl C, Mechtler K, Haack M, Beck-Sickinger AG, Cooper DM, Sinz A. J Am Soc Mass Spectrom. 2006;17:1100–13. doi: 10.1016/j.jasms.2006.04.020. [DOI] [PubMed] [Google Scholar]
- (4).Kalkhof S, Ihling C, Mechtler K, Sinz A. Anal Chem. 2005;77:495–503. doi: 10.1021/ac0487294. [DOI] [PubMed] [Google Scholar]
- (5).Trester-Zedlitz M, Kamada K, Burley SK, Fenyo D, Chait BT, Muir TW. J Am Chem Soc. 2003;125:2416–25. doi: 10.1021/ja026917a. [DOI] [PubMed] [Google Scholar]
- (6).Petrotchenko EV, Olkhovik VK, Borchers CH. Mol Cell Proteomics. 2005;4:1167–79. doi: 10.1074/mcp.T400016-MCP200. [DOI] [PubMed] [Google Scholar]
- (7).Back JW, Hartog AF, Dekker HL, Muijsers AO, de Koning LJ, de Jong L. J Am Soc Mass Spectrom. 2001;12:222–7. doi: 10.1016/S1044-0305(00)00212-9. [DOI] [PubMed] [Google Scholar]
- (8).Chowdhury SM, Munske GR, Tang X, Bruce JE. Anal Chem. 2006;78:8183–93. doi: 10.1021/ac060789h. [DOI] [PubMed] [Google Scholar]
- (9).Tang X, Munske GR, Siems WF, Bruce JE. Anal Chem. 2005;77:311–8. doi: 10.1021/ac0488762. [DOI] [PubMed] [Google Scholar]
- (10).Lu Y, Tanasova M, Borhan B, Reid GE. Anal Chem. 2008 doi: 10.1021/ac801625e. [DOI] [PubMed] [Google Scholar]
- (11).Soderblom EJ, Goshe MB. Anal Chem. 2006;78:8059–68. doi: 10.1021/ac0613840. [DOI] [PubMed] [Google Scholar]
- (12).Gardner MW, Vasicek LA, Shabbir S, Anslyn EV, Brodbelt JS. Anal Chem. 2008;80:4807–19. doi: 10.1021/ac800625x. [DOI] [PubMed] [Google Scholar]
- (13).Anderson GA, Tolic N, Tang X, Zheng C, Bruce JE. J Proteome Res. 2007;6:3412–21. doi: 10.1021/pr070035z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Schilling B, Row RH, Gibson BW, Guo X, Young MM. J Am Soc Mass Spectrom. 2003;14:834–50. doi: 10.1016/S1044-0305(03)00327-1. [DOI] [PubMed] [Google Scholar]
- (15).Young MM, Tang N, Hempel JC, Oshiro CM, Taylor EW, Kuntz ID, Gibson BW, Dollinger G. Proc Natl Acad Sci U S A. 2000;97:5802–6. doi: 10.1073/pnas.090099097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Lee YJ, Lackner LL, Nunnari JM, Phinney BS. J Proteome Res. 2007;6:3908–17. doi: 10.1021/pr070234i. [DOI] [PubMed] [Google Scholar]
- (17).Rinner O, Seebacher J, Walzthoeni T, Mueller L, Beck M, Schmidt A, Mueller M, Aebersold R. Nat Methods. 2008;5:315–8. doi: 10.1038/nmeth.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Diaz DD, Rajagopal K, Strable E, Schneider J, Finn MG. J Am Chem Soc. 2006;128:6056–7. doi: 10.1021/ja061251w. [DOI] [PubMed] [Google Scholar]
- (19).Shevchenko A, Wilm M, Vorm O, Mann M. Anal Chem. 1996;68:850–8. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- (20).Galan JA, Guo M, Sanchez EE, Cantu E, Rodriguez-Acosta A, Perez JC, Tao WA. Mol Cell Proteomics. 2008;7:785–99. doi: 10.1074/mcp.M700321-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Speers AE, Cravatt BF. Chem Biol. 2004;11:535–46. doi: 10.1016/j.chembiol.2004.03.012. [DOI] [PubMed] [Google Scholar]
- (22).Goshe MB, Conrads TP, Panisko EA, Angell NH, Veenstra TD, Smith RD. Anal Chem. 2001;73:2578–86. doi: 10.1021/ac010081x. [DOI] [PubMed] [Google Scholar]
- (23).Livesay EA, Tang K, Taylor BK, Buschbach MA, Hopkins DF, LaMarche BL, Zhao R, Shen Y, Orton DJ, Moore RJ, Kelly RT, Udseth HR, Smith RD. Anal Chem. 2008;80:294–302. doi: 10.1021/ac701727r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Tang K, Tolmachev AV, Nikolaev E, Zhang R, Belov ME, Udseth HR, Smith RD. Anal Chem. 2002;74:5431–7. doi: 10.1021/ac0202583. [DOI] [PubMed] [Google Scholar]
- (25).Kelly RT, Page JS, Luo Q, Moore RJ, Orton DJ, Tang K, Smith RD. Anal Chem. 2006;78:7796–801. doi: 10.1021/ac061133r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Prescher JA, Bertozzi CR. Nat Chem Biol. 2005;1:13–21. doi: 10.1038/nchembio0605-13. [DOI] [PubMed] [Google Scholar]
- (27).Yates JR, 3rd, Eng JK, McCormack AL, Schieltz D. Anal Chem. 1995;67:1426–36. doi: 10.1021/ac00104a020. [DOI] [PubMed] [Google Scholar]
- (28).Good DM, Wirtala M, McAlister GC, Coon JJ. Mol Cell Proteomics. 2007;6:1942–51. doi: 10.1074/mcp.M700073-MCP200. [DOI] [PubMed] [Google Scholar]
- (29).Chi A, Huttenhower C, Geer LY, Coon JJ, Syka JE, Bai DL, Shabanowitz J, Burke DJ, Troyanskaya OG, Hunt DF. Proc Natl Acad Sci U S A. 2007;104:2193–8. doi: 10.1073/pnas.0607084104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Kruppa GH, Schoeniger J, Young MM. Rapid Commun Mass Spectrom. 2003;17:155–62. doi: 10.1002/rcm.885. [DOI] [PubMed] [Google Scholar]
- (31).Haririnia A, D’Onofrio M, Fushman D. J Mol Biol. 2007;368:753–66. doi: 10.1016/j.jmb.2007.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.