SUMMARY
FoxO transcription factors control development and longevity in diverse species. Although FoxO regulation via changes in its subcellular localization is well established, little is known about how FoxO activity is regulated in the nucleus. Here we show that the conserved C. elegans protein EAK-7 acts in parallel to the serine/threonine kinase AKT-1 to inhibit the FoxO transcription factor DAF-16. Loss of EAK-7 activity promotes diapause and longevity in a DAF-16/FoxO-dependent manner. Whereas akt-1 mutation activates DAF-16/FoxO by promoting its translocation from the cytoplasm to the nucleus, eak-7 mutation increases nuclear DAF-16/FoxO activity without influencing DAF-16/FoxO subcellular localization. Thus, EAK-7 and AKT-1 inhibit DAF-16/FoxO activity via distinct mechanisms. Our results implicate EAK-7 as a FoxO regulator and highlight the biological impact of a new regulatory pathway that governs the activity of nuclear FoxO without altering its subcellular location.
INTRODUCTION
FoxO transcription factors (TFs) promote lifespan extension, stress resistance and metabolic homeostasis in diverse species (Accili and Arden, 2004; Arden, 2008; Calnan and Brunet, 2008; Gross et al., 2008; Partridge and Bruning, 2008). FoxO knockout mice develop tumors and exhibit abnormalities in glucose metabolism and bone mineral density (Ambrogini et al., 2010; Dong et al., 2008; Matsumoto et al., 2007; Paik et al., 2007; Rached et al., 2010), suggesting that dysregulation of FoxO TFs may contribute to the pathophysiology of common human diseases associated with aging such as cancer, type 2 diabetes, and osteoporosis. Intriguingly, FoxO3 polymorphisms are associated with extreme longevity in humans (Flachsbart et al., 2009; Li et al., 2009; Willcox et al., 2008). Thus, understanding how FoxO TFs are regulated has the potential to yield fundamental insights into both the pathophysiology of human disease as well as the physiology of normal aging.
Activation of the phosphoinositide 3-kinase (PI3K)/Akt pathway by insulin and IGF-1 signaling (IIS) results in direct phosphorylation of FoxO by Akt and its subsequent association with 14-3-3 proteins and nuclear export (Van Der Heide et al., 2004), whereupon it is targeted for ubiquitin-mediated proteasomal degradation (Huang et al., 2005; Matsuzaki et al., 2003). The paradigm of FoxO inhibition by IIS first emerged from genetic analysis in C. elegans, where a conserved IIS pathway controls lifespan, stress resistance, and entry into a developmentally arrested larval stage known as dauer. The C. elegans insulin-like receptor (InsR), DAF-2 (Kimura et al., 1997), activates the PI3K AGE-1 (Morris et al., 1996), PDK-1 (Paradis et al., 1999), and the AGC family kinases AKT-1, AKT-2, and SGK-1 (Hertweck et al., 2004; Paradis and Ruvkun, 1998). AKT phosphorylation of the C. elegans FoxO transcription factor DAF-16 promotes its binding to the 14-3-3 protein FTT-2 and subsequent nuclear exclusion (Berdichevsky et al., 2006; Li et al., 2007a). The PI3K/Akt pathway is antagonized by the C. elegans phosphatase and tensin (PTEN) ortholog DAF-18 (Gil et al., 1999; Mihaylova et al., 1999; Ogg and Ruvkun, 1998; Rouault et al., 1999). Reduction of IIS results in lifespan extension, increased stress resistance, and constitutive dauer arrest, and these phenotypes are suppressed by DAF-16/FoxO loss-of-function mutations (Finch and Ruvkun, 2001; Kenyon, 2005). Thus, DAF-16/FoxO promotes longevity, stress resistance, and dauer arrest and is inhibited by IIS.
DAF-16/FoxO localizes to the nucleus in daf-2/InsR mutants (Henderson and Johnson, 2001; Lee et al., 2001; Lin et al., 2001); however, nuclear localization per se is insufficient for full DAF-16/FoxO activation, as a DAF-16/FoxO mutant lacking all AKT phosphorylation sites exhibits constitutive nuclear localization but does not promote dauer arrest or longevity (Hertweck et al., 2004; Lin et al., 2001). Furthermore, although loss-of-function alleles of daf-18/PTEN and gain-of-function alleles of akt-1 or pdk-1 fully suppress the dauer-constitutive phenotype of age-1/PI3K null mutants, they weakly suppress dauer arrest caused by a partial loss-of-function mutation in daf-2/InsR (Gil et al., 1999; Ogg and Ruvkun, 1998; Paradis et al., 1999; Paradis and Ruvkun, 1998). These data suggest that a second pathway acts in parallel to the PI3K/Akt pathway to regulate the activity of nuclear DAF-16/FoxO (Figure 1A).
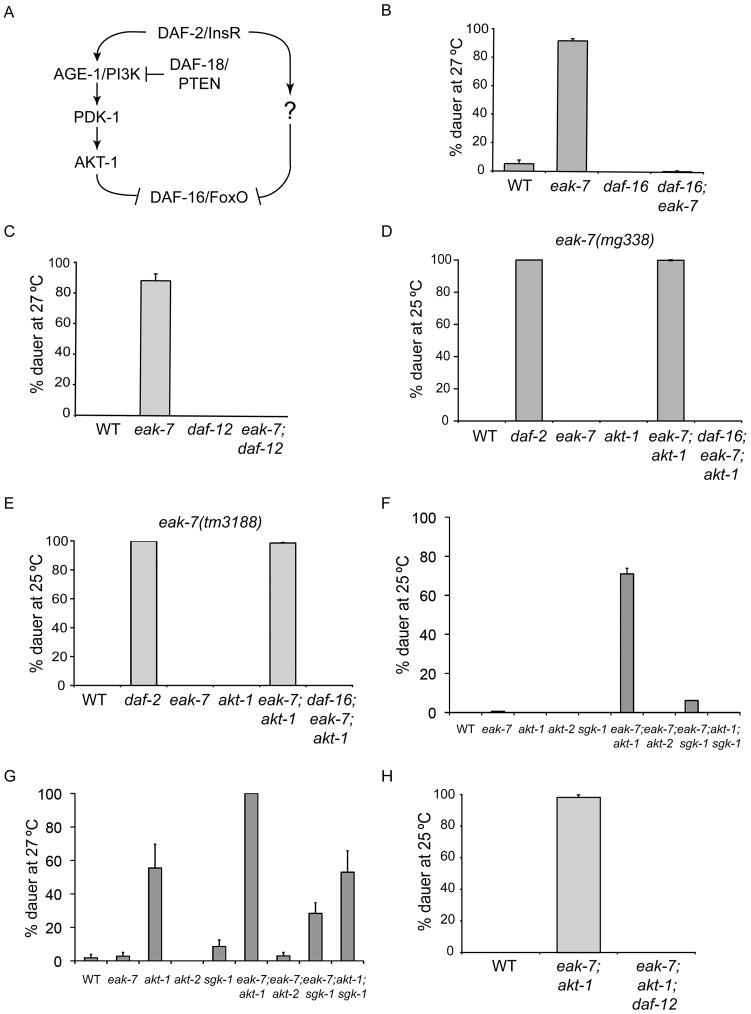
Figure 1. EAK-7 acts in parallel to AKT-1 to inhibit dauer arrest via DAF-16/FoxO and DAF-12.
A. Schematic of the DAF-2/InsR pathway. B. daf-16/FoxO null and C. daf-12 null mutations suppress the dauer arrest phenotype of an eak-7 null mutant at 27°C on NGM plates lacking supplemental cholesterol. D. Dauer arrest phenotypes of eak-7(mg338) at 25°C. The null allele daf-16(mgDf47) was used in this experiment. E. Dauer arrest phenotypes of the null allele eak-7(tm3188) at 25°C. F. and G. Genetic interactions of eak-7 null mutation with akt-1, akt-2, and sgk-1 null mutations at F. 25°C and G. 27°C on standard NGM plates containing supplemental cholesterol. H. A daf-12 null mutation suppresses the dauer arrest phenotype of eak-7;akt-1 double mutants at 25°C. All measurements in this figure represent the mean + S.D. See Table S1 for raw data and numbers of animals scored in each dauer assay.
To identify genes encoding regulators of nuclear DAF-16/FoxO activity, we performed a screen for mutants that enhance the weak dauer arrest phenotype of an akt-1 null mutant (i.e., eak mutants; Hu et al., 2006). Here we describe the identification and characterization of eak-7, which encodes a conserved protein that regulates development, lifespan, and stress resistance by controlling nuclear DAF-16/FoxO activity.
RESULTS
At 25°C, akt-1 null mutants exhibit nuclear enrichment of DAF-16/FoxO but do not arrest as dauers (Hu et al., 2006; Zhang et al., 2008). We mutagenized akt-1 null mutant animals and identified rare F2 progeny that arrested as dauers at 25°C. eak mutants were defined as those mutants whose constitutive dauer arrest phenotype required the presence of the akt-1 mutation (Hu et al., 2006; Zhang et al., 2008). The eak-7 gene is defined by the missense allele mg338 and the independently isolated deletion allele tm3188, which is a null allele (Figures S1A–B).
EAK-7 acts in parallel to AKT-1 to regulate dauer arrest
daf-2/InsR mutants arrest as dauers at 25°C (Gems et al., 1998; Kimura et al., 1997). In contrast, whereas eak-7 mutants undergo dauer arrest at 27°C on plates lacking supplemental cholesterol (Figures 1B–C), they develop into adults on standard NGM plates at 25°C (Figures 1D–F). However, eak-7 mutation strongly enhances dauer arrest in an akt-1 null mutant (Figures 1D–F) as well as in age-1/PI3K and pdk-1 partial loss-of-function mutants (Figure S1C) and daf-2(e1370) mutants (data not shown). The enhancement of the dauer arrest phenotype of an akt-1 null mutant by an eak-7 null mutation indicates that EAK-7 acts in parallel to AKT-1 to regulate dauer arrest.
EAK-7 acts in the EAK pathway to regulate dauer arrest
Four other EAK proteins act in a single pathway in parallel to AKT-1 to regulate dauer arrest (Hu et al., 2006; Zhang et al., 2008). To determine whether EAK-7 also acts in this pathway, we examined the effect of eak-7 mutation on the dauer arrest phenotype of other eak mutants. eak-7 mutation did not enhance the dauer arrest phenotype of eak-3, sdf-9/eak-5, or eak-6 mutants (Figure S1D).
Dauer arrest is also regulated by dafachronic acids (DAs), which are steroid hormone ligands for the nuclear receptor DAF-12 (Motola et al., 2006). Mutations in the DA biosynthetic proteins DAF-9 and DAF-36 induce dauer arrest (Gerisch et al., 2001; Jia et al., 2002; Rottiers et al., 2006), and eak-3 mutations enhance the dauer arrest phenotype of daf-9 and daf-36 mutants (Zhang et al., 2008). Similarly, eak-7 mutation strongly enhanced the dauer arrest phenotype of daf-9 and daf-36 mutants (Figure S1E). Taken together, these data suggest that EAK-7 functions in the same pathway as other EAK proteins in dauer regulation.
EAK-7 regulates dauer arrest via DAF-16/FoxO
IIS and EAK proteins regulate dauer arrest by inhibiting DAF-16/FoxO activity (Gottlieb and Ruvkun, 1994; Hu et al., 2006; Ohkura et al., 2003; Vowels and Thomas, 1992; Zhang et al., 2008). To determine whether EAK-7 inhibits DAF-16/FoxO, we tested whether dauer arrest phenotypes in eak-7 mutants were daf-16/FoxO-dependent. Dauer arrest phenotypes of eak-7;akt-1 double mutants and eak-7 single mutants were fully suppressed by a daf-16/FoxO null mutation (Figures 1B, 1D, and 1E). Dauer arrest in eak-7 single mutants was also suppressed by a gain-of-function mutation in akt-1 (Paradis and Ruvkun, 1998) and a loss-of-function mutation in daf-18/PTEN (Ogg and Ruvkun, 1998) (Figure S1F), both of which are predicted to inhibit DAF-16/FoxO via increased AKT activity. Thus, EAK-7 regulates dauer arrest by inhibiting DAF-16/FoxO activity.
eak-7 mutation does not enhance dauer arrest in akt-2 mutants
AKT-1, AKT-2, and SGK-1 physically associate with each other (Hertweck et al., 2004) and are thought to inhibit DAF-16/FoxO activity by phosphorylating DAF-16/FoxO at canonical Akt/PKB phosphorylation sites (Hertweck et al., 2004; Lee et al., 2001; Lin et al., 2001). Since EAK-7 acts in parallel to AKT-1 to regulate dauer arrest (Figures 1D–F), we sought to determine whether EAK-7 also acts in parallel to AKT-2 and/or SGK-1. To this end, we constructed eak-7;akt-2, akt-1;akt-2, eak-7;sgk-1, and akt-1;sgk-1 double mutants using the candidate null alleles akt-2(ok393) (Hertweck et al., 2004) and sgk-1(mg455) (Soukas et al., 2009). If EAK-7 acts in parallel to AKT-2 or SGK-1, then eak-7 mutation should enhance the dauer arrest phenotype of akt-2 or sgk-1 mutants.
sgk-1 mutants did not undergo significant dauer arrest at 25°C or 27°C on standard NGM plates containing cholesterol and did not enhance the dauer arrest phenotype of akt-1 mutants at either temperature (Figures 1F–G). Thus, in agreement with previous results (Hertweck et al., 2004), SGK-1 plays a minor role in dauer regulation. sgk-1 mutation weakly enhanced the dauer arrest phenotype of eak-7 mutants at both temperatures (Figures 1F–G), suggesting that SGK-1 acts in parallel to EAK-7.
In contrast, although akt-2 mutants did not arrest as dauers at 25°C or 27°C (Figures 1F–G), akt-2 mutation strongly enhanced dauer arrest in an akt-1 mutant (Figure S1G), as previously reported (Oh et al., 2005). In fact, akt-1;akt-2 double mutants undergo nonconditional dauer arrest and do not recover (Oh et al., 2005) (Figure S1G and data not shown). Thus, like eak-7 mutations, akt-2 mutations are strong enhancers of the dauer arrest phenotype of akt-1 mutants. The severity of the akt-1;akt-2 double mutant dauer arrest phenotype, i.e., non-conditional constitutive dauer arrest, is similar to that observed in age-1/PI3K and daf-2/InsR null mutants (Morris et al., 1996; Patel et al., 2008) and suggests that AKT-1 and AKT-2 constitute the major dauer regulatory output of IIS.
In contrast to eak-7;akt-1 double mutants, which have a strong dauer arrest phenotype at 25°C (Figures 1D–F), eak-7;akt-2 double mutants did not arrest as dauers at 25°C or 27°C (Figures 1F–G). Thus, eak-7 mutation does not enhance the dauer arrest phenotype of akt-2 mutants. This result is consistent with at least two models of EAK-7 action. EAK-7 could act in the same pathway as AKT-2 to inhibit DAF-16/FoxO activity. Alternatively, since akt-2 mutation has a relatively modest effect on DAF-16::GFP nuclear localization in an akt-1 wild-type background (Hertweck et al., 2004), the amount of DAF-16/FoxO present in nuclei of eak-7;akt-2 double mutant animals may be below the threshold necessary for eak-7 mutation to have a phenotypic effect.
EAK-7 regulates dauer arrest via DAF-12
DAF-12 is required for dauer arrest in all dauer-constitutive mutants studied to date (Fielenbach and Antebi, 2008; Hu, 2007). Dauer-constitutive phenotypes of eak-7 single mutants and eak-7;akt-1 double mutants were fully suppressed by a daf-12 null mutation (Figures 1C and 1H), indicating that EAK-7 promotes reproductive development by inhibiting DAF-12 activity.
EAK-7 controls lifespan via DAF-16/FoxO
IIS mutants also exhibit DAF-16/FoxO-dependent lifespan extension and stress resistance during adulthood (Gems et al., 1998; Honda and Honda, 1999; Kenyon et al., 1993; Morris et al., 1996; Murakami and Johnson, 1996; Paradis et al., 1999). eak-7 mutants also live longer than wild-type animals (Figures 2A–F, S2A–E, and Table S2), and this phenotype requires daf-16/FoxO (Figures 2B, S2A, and Table S2) as well as the protein phosphatase 4 regulatory subunit SMK-1 (Kim et al., 2007; Wolff et al., 2006) and the heat-shock transcription factor HSF-1 (Hsu et al., 2003) (Figures S2B–C and Table S2), both of which are required for increased longevity in daf-2/InsR mutants. eak-7 mutants also exhibited DAF-16/FoxO-dependent resistance to ultraviolet, heat, and oxidative stress (Figures S2F–H and Table S2). Thus, EAK-7 controls lifespan and stress resistance by inhibiting DAF-16/FoxO activity.
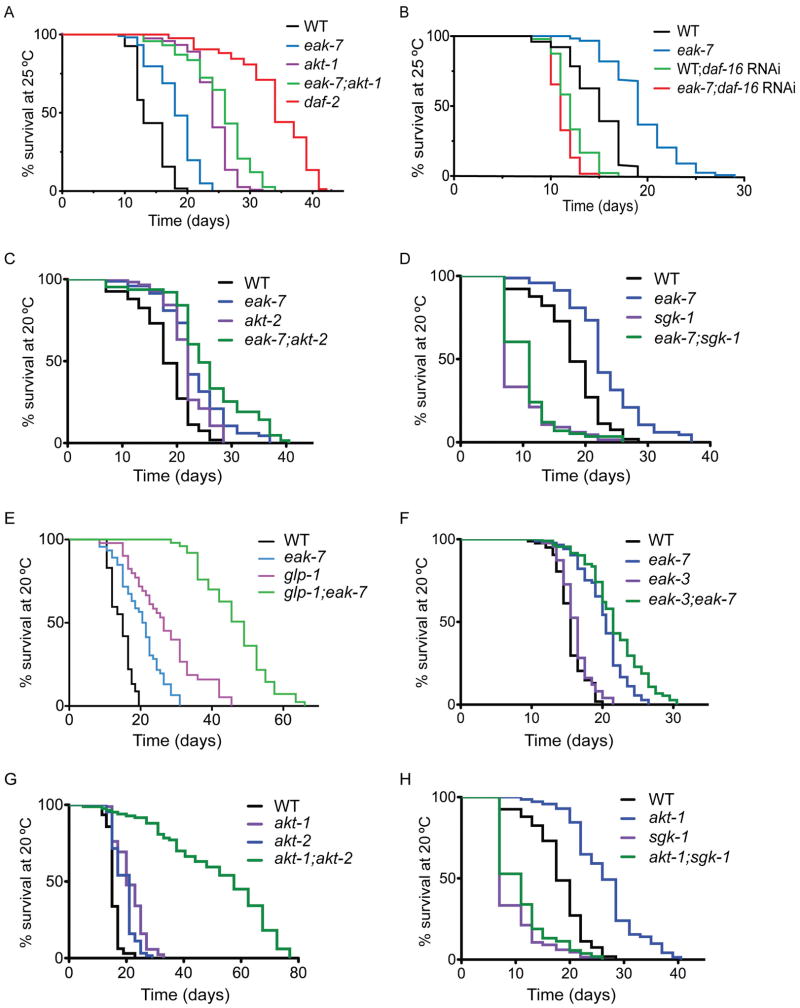
Figure 2. Genetic interactions of eak-7 mutants in lifespan control.
A. Lifespan phenotype of eak-7 null mutants at 25°C. B. daf-16/FoxO RNAi suppresses lifespan extension in eak-7 null mutants. C. eak-7 null mutation enhances the lifespan extension observed in akt-2 null mutants. D. sgk-1 activity is required for lifespan extension in eak-7 null mutants. E. eak-7 null mutation enhances lifespan extension of glp-1(e2141) mutants. F. Lifespan extension in eak-7 null mutants does not require eak-3 activity. G. akt-1;akt-2 double mutants are extremely long-lived compared to single mutants. H. sgk-1 activity is required for lifespan extension in akt-1 null mutants. See Table S2 for numbers of animals assayed and p-values for all lifespan experiments.
EAK-7 acts in parallel to AKT-1 and AKT-2 to control lifespan
In contrast to the strong enhancement of dauer arrest in akt-1 mutants caused by eak-7 mutation (Figures 1D–F), eak-7 mutation enhanced the extended lifespan phenotype of akt-1 mutants modestly (Figure 2A and Table S2; mean lifespans in days +/− s.d. of 24.0+/−3.3 for akt-1 vs. 25.5+/−4.9 for eak-7;akt-1, p < 0.0001 by the log-rank test). eak-7 mutation enhanced the lifespan extension phenotype of akt-2 mutants to a comparable extent (Figure 2C and Table S2; mean lifespans +/− s.d. of 21.8+/−3.8 for akt-2 vs. 25.7+/−7.4 for eak-7;akt-2, p < 0.0001 by the log-rank test). akt-1;akt-2 double mutants exhibited a profound extension of lifespan compared to akt-1 and akt-2 single mutants (Figure 2G). This is reminiscent of the extreme longevity of adult age-1/PI3K null mutants (Ayyadevara et al., 2008) and suggests that, as is the case for dauer regulation, AKT-1 and AKT-2 are the major outputs of IIS in lifespan control.
Taken together, these results suggest that, whereas EAK-7 acts in parallel to AKT-1 to regulate dauer arrest, EAK-7 acts in parallel to both AKT-1 and AKT-2 to control adult lifespan. Notably, the magnitude of the enhancement of akt-1 and akt-2 mutant lifespan extension phenotypes by eak-7 mutation is substantially smaller than the effect of eak-7 mutation on the akt-1 mutant dauer arrest phenotype.
EAK-7 acts in parallel to the germline to control lifespan
Germline ablation extends lifespan by inducing the nuclear translocation of DAF-16/FoxO in intestinal cells (Berman and Kenyon, 2006; Hsin and Kenyon, 1999; Libina et al., 2003). Thus, one possible explanation for the relatively modest effect of eak-7 mutation on lifespan extension in akt-1 and akt-2 mutants (Figures 2A and 2C) compared to its effect on dauer arrest in akt-1 mutants (Figures 1D–F) is that in eak-7;akt-1 and eak-7;akt-2 double mutant adult animals, signals from the germline that are not present in early larvae inhibit DAF-16/FoxO nuclear translocation. Therefore, we wished to determine whether eak-7 mutation enhances lifespan extension in animals lacking a germline, in which relative concentrations of nuclear DAF-16/FoxO are increased. To this end, we constructed double mutants with glp-1(e2141) animals, which lack a germline when raised at 25°C (Priess et al., 1987). Lifespan extension caused by eak-7 mutation was comparable at 20°C and 25°C (compare Figures 2C–F to Figures 2A–B; Table S2). The magnitude of lifespan extension caused by the glp-1(e2141) mutation was consistent with previous reports (Berman and Kenyon, 2006) and greater than that caused by eak-7 mutation (Figure 2E and Table S2). Strikingly, glp-1;eak-7 double mutants lived nearly twice as long as glp-1 single mutants (Figure 2E and Table S2). Thus, EAK-7 acts in parallel to germline signals to control lifespan.
SGK-1 is required for lifespan extension in eak-7 mutants
Culturing worms on E. coli HT115 that express sgk-1 double-stranded RNA promotes long life due to a food avoidance behavior induced by reduction of sgk-1 activity that results in dietary restriction (Hertweck et al., 2004; Soukas et al., 2009). In contrast, sgk-1 null mutants are short-lived when cultured on the standard E. coli OP50 strain (Soukas et al., 2009). To determine whether SGK-1 is required for lifespan extension in eak-7 mutants, we assayed the lifespans of eak-7;sgk-1 double mutants. The double mutant lived slightly longer than sgk-1 single mutants and substantially shorter than eak-7 single mutants (Figure 2D and Table S2), indicating that SGK-1 is necessary for lifespan extension in eak-7 mutants. Surprisingly, SGK-1 was also required for lifespan extension in akt-1 mutants (Figure 2H and Table S2). Thus, the requirement for SGK-1 in lifespan extension is not specific to eak-7 mutants. SGK-1 may either be required for full DAF-16/FoxO activation in lifespan control, or SGK-1 may promote lifespan extension independently of DAF-16/FoxO.
Lifespan extension in eak-7 mutants does not require other EAK proteins
Although EAK-7 acts in the same pathway as other EAK proteins to regulate dauer arrest (Figure S1D), eak-7 mutants are distinct from other eak mutants in that they are long-lived (Figure 2F and S2D–E; Hu et al., 2006; Zhang et al., 2008). To determine whether the activity of other eak genes is required for lifespan extension in eak-7 mutants, we performed lifespan assays on various eak-7;eak double mutants. Mutations in eak-3, sdf-9/eak-5, and eak-6 did not suppress lifespan extension in eak-7 mutants (Figures 2F and S2D–E; Table S2). Thus, other EAK proteins are not required for lifespan extension caused by eak-7 mutation.
EAK-7 inhibits DAF-16/FoxO target gene expression
To determine whether eak-7 mutation influences DAF-16/FoxO activity, we assayed endogenous transcript levels of three DAF-16/FoxO target genes: sod-3, mtl-1, and dod-3 (Murphy et al., 2003; Oh et al., 2006) (Figures 3A–D and S3). In early larval stages, both eak-7 and akt-1 single mutants had increased DAF-16/FoxO target gene mRNA levels relative to wild-type animals. akt-1 mutation caused a consistently larger increase than eak-7 mutation did. Strikingly, eak-7;akt-1 double mutants exhibited a synergistic increase in mRNA levels of all three DAF-16/FoxO target genes that either was comparable to or exceeded that observed in daf-2/InsR mutants (Figures 3A–C). DAF-16/FoxO target gene expression in eak-7;akt-1 double mutants was approximately 50–100-fold greater than in wild-type animals and approximately 6–8-fold greater than in akt-1 single mutants (Figures 3A–C). As expected, DAF-16/FoxO target gene expression in this context was completely dependent upon daf-16/FoxO.
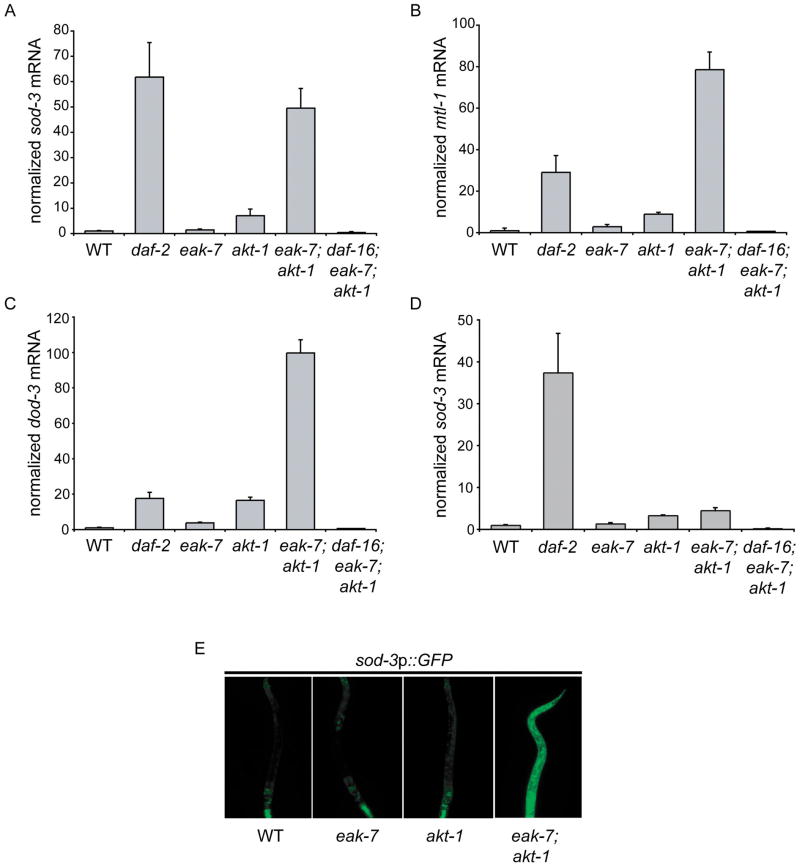
Figure 3. DAF-16/FoxO target gene expression in eak-7 null mutants.
A. sod-3, B. mtl-1, and C. dod-3 mRNA quantification using qRT-PCR on total RNA from L2 larvae. Measurements represent the mean + S.D. D. sod-3 mRNA quantification in adult animals. E. sod-3p::GFP expression in various strains. The anterior of the animal is facing down in all images.
Whereas adult eak-7 and akt-1 mutant animals exhibited increases in sod-3 mRNA comparable to increases observed in larvae, adult eak-7;akt-1 double mutant animals did not exhibit a synergistic increase in sod-3 transcript levels (Figure 3D). Furthermore, sod-3 transcript levels in adult eak-7;akt-1 double mutants were substantially lower than those observed in adult daf-2/InsR mutants (Figure 3D). This difference in DAF-16/FoxO target gene expression in distinct life stages of eak-7;akt-1 double mutants is commensurate with the magnitude of enhancement of akt-1 mutant dauer arrest and lifespan extension phenotypes by eak-7 mutation (Figures 1D–F and 2A) and may be a consequence of the inhibitory effect of the adult germline on DAF-16/FoxO activity (Lin et al., 2001).
In order to determine whether EAK-7 regulates DAF-16/FoxO target gene expression by influencing promoter activity, we examined the effect of eak-7 mutation on the expression of a GFP reporter under the control of the sod-3 promoter (Libina et al., 2003) in early stage larvae. Whereas mutations in eak-7 and akt-1 alone did not result in substantial changes in sod-3::GFP expression, eak-7;akt-1 double mutants exhibited a dramatic increase in GFP expression throughout the animal relative to wild-type animals (Figure 3E). These findings are consistent with the effects of eak-7 and akt-1 mutations on endogenous DAF-16/FoxO target gene expression (Figures 3A–C). Thus, EAK-7 inhibits DAF-16/FoxO target gene transcription, and in larvae the effect of eak-7 mutation on DAF-16/FoxO target gene transcription is magnified in an akt-1 mutant background.
EAK-7 inhibits nuclear DAF-16/FoxO activity
Because eak-7 mutation enhances akt-1 mutant phenotypes and DAF-16/FoxO target gene expression more strongly during larval development than in adulthood (compare Figures 1D–F to Figure 2A and Figure 3A to 3D), we explored the mechanism by which EAK-7 inhibits DAF-16/FoxO activity in larvae. Since IIS inhibits DAF-16/FoxO by promoting its cytoplasmic sequestration (Henderson and Johnson, 2001; Lee et al., 2001; Lin et al., 2001), we assayed the subcellular localization of a functional DAF-16::GFP fusion protein (Henderson and Johnson, 2001) in various mutant backgrounds (Figure 4A). As expected, we observed diffuse fluorescence corresponding to cytoplasmic localization in wild-type animals and increased punctate fluorescence corresponding to nuclear localization in akt-1 mutants (Figure 4A; Hertweck et al., 2004; Zhang et al., 2008). In contrast to akt-1 mutants, eak-7 mutant animals exhibited diffuse fluorescence indistinguishable from that observed in wild-type animals (Figure 4A). These results suggest that, unlike AKT-1, which inhibits DAF-16/FoxO by promoting its translocation from the nucleus to the cytoplasm, EAK-7 inhibits nuclear DAF-16/FoxO activity without inducing DAF-16/FoxO nuclear exclusion.
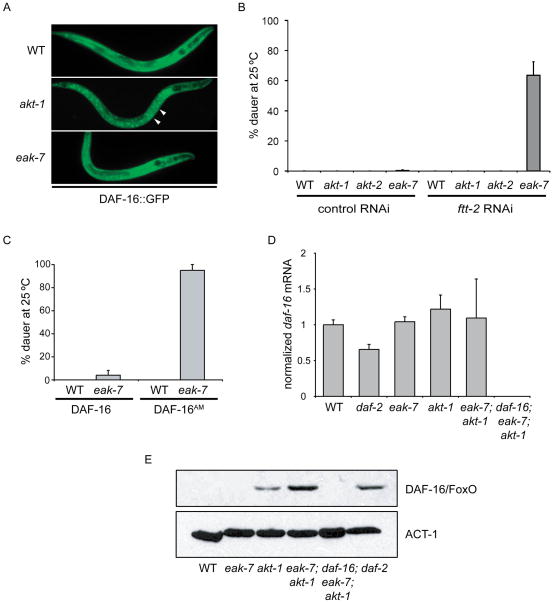
Figure 4. EAK-7 regulation of DAF-16/FoxO.
A. An eak-7 null mutation does not affect the subcellular localization of DAF-16::GFP. White arrows denote nuclear fluorescence in akt-1 mutants. The anterior of the animal is facing right in all images. B. ftt-2 RNAi enhances dauer arrest at 25°C in an eak-7 null mutant but not in akt-1 or akt-2 null mutants. C. An eak-7 null mutation enhances dauer arrest at 25°C in animals harboring DAF-16AM, a constitutively nuclear DAF-16/FoxO mutant GFP fusion protein lacking all canonical Akt/PKB phosphorylation sites. D. eak-7 null mutation does not affect endogenous daf-16/FoxO mRNA levels in L2 larvae. E. An eak-7 null mutation increases endogenous DAF-16/FoxO protein levels in an akt-1 null mutant. DAF-16/FoxO protein was quantified by anti-DAF-16/FoxO immunoblot of whole worm lysates. Anti-ACT-1 immunoblotting was used to confirm equal protein loading. Measurements in dauer assays represent the mean + S.D.
To examine the possibility that EAK-7 inhibits nuclear DAF-16/FoxO activity in more detail, we determined the effect of 14-3-3 protein loss-of-function on the dauer arrest phenotype of eak-7 mutants. The 14-3-3 protein FTT-2 binds to AKT-phosphorylated DAF-16/FoxO and sequesters it in the cytoplasm, and reduction of FTT-2 function promotes DAF-16/FoxO nuclear localization (Berdichevsky et al., 2006; Li et al., 2007a). Since ftt-2 deletion is lethal (Berdichevsky et al., 2006; Li et al., 2007a), we inactivated ftt-2 using RNAi. To clarify whether EAK-7 acts in the same pathway as AKT-2 in DAF-16/FoxO regulation (see Figures 1F–G), we also tested the effect of ftt-2 RNAi on the dauer arrest phenotype of akt-2 mutants. If EAK-7 and AKT-2 act in the same pathway to regulate DAF-16/FoxO activity, then ftt-2 RNAi should have the same effect on the dauer arrest phenotype of both eak-7 and akt-2 mutants.
As expected, ftt-2 RNAi did not induce dauer arrest in wild-type or akt-1 mutant animals (Figure 4B). Whereas ftt-2 RNAi strongly promoted dauer arrest in eak-7 mutant animals, it did not induce dauer arrest in an akt-2 null mutant. Thus, EAK-7 and AKT-2 act in distinct pathways to regulate DAF-16/FoxO activity. The lack of phenotypic enhancement observed in eak-7;akt-2 double mutants (Figures 1F–G) is likely a consequence of the relatively modest effect of akt-2 mutation on DAF-16/FoxO nuclear translocation in a wild-type akt-1 background (Hertweck et al., 2004). Although this result supports a model whereby EAK-7 inhibits nuclear DAF-16/FoxO activity, it does not exclude the possibility that EAK-7 may contribute to DAF-16/FoxO cytoplasmic retention in an FTT-2-independent manner.
To further investigate this issue, we determined the effect of eak-7 mutation on dauer arrest in animals expressing a constitutively nuclear DAF-16/FoxO mutant fused to GFP that lacks all consensus Akt/PKB phosphorylation sites (DAF-16AM; Lin et al., 2001). As previously reported (Lin et al., 2001), wild-type animals expressing DAF-16AM did not arrest as dauers (Figure 4C). In contrast, eak-7 mutation promoted highly penetrant dauer arrest in animals expressing DAF-16AM but not in animals expressing a wild-type DAF-16::GFP transgene (Figure 4C). This result provides further support for the hypothesis that EAK-7 inhibits nuclear DAF-16/FoxO activity and indicates that the ability of eak-7 mutation to enhance dauer arrest in akt-1 mutants is a function of nuclear enrichment of DAF-16/FoxO caused by akt-1 mutation as opposed to the dysregulation of other AKT-1 targets such as SKN-1 and CEP-1/p53 (Quevedo et al., 2007; Tullet et al., 2008). Furthermore, it demonstrates that EAK-7 inhibition of DAF-16/FoxO activity does not require the canonical Akt/PKB phosphorylation sites present in wild-type DAF-16/FoxO.
Thus, EAK-7 inhibits the activity of DAF-16/FoxO that localizes to the nucleus by virtue of akt-1 mutation (Figures 1D–F, 3A–C, and 3E), depletion of FTT-2 activity (Figure 4B), or mutation of its Akt/PKB phosphorylation sites (Figure 4C).
EAK-7 reduces steady-state DAF-16/FoxO protein levels
To further elucidate the mechanism by which EAK-7 inhibits DAF-16/FoxO activity, we determined the effect of eak-7 mutation on endogenous DAF-16/FoxO transcript and protein levels. Mutation of eak-7 and akt-1, either alone or in combination, did not significantly affect daf-16/FoxO mRNA levels (Figure 4D). Endogenous DAF-16/FoxO protein was undetectable in lysates from wild-type and eak-7 single mutants (Figure 4E). In contrast, DAF-16/FoxO was detectable in lysates from akt-1 single mutants, and eak-7;akt-1 double mutants exhibited a synergistic increase in DAF-16/FoxO protein levels compared to eak-7 and akt-1 single mutants (Figure 4E). Increased DAF-16/FoxO protein levels were also observed in daf-2/InsR mutants (Figure 4E). Thus, EAK-7 reduces steady-state DAF-16/FoxO protein levels in akt-1 mutants without influencing daf-16/FoxO mRNA levels. Relative DAF-16/FoxO protein levels in lysates from various mutants correlated with both the magnitude of dauer arrest (Figures 1D–F) as well as relative levels of DAF-16/FoxO target gene expression in the same mutants (Figures 3A–C and 3E).
eak-7 encodes a novel conserved protein
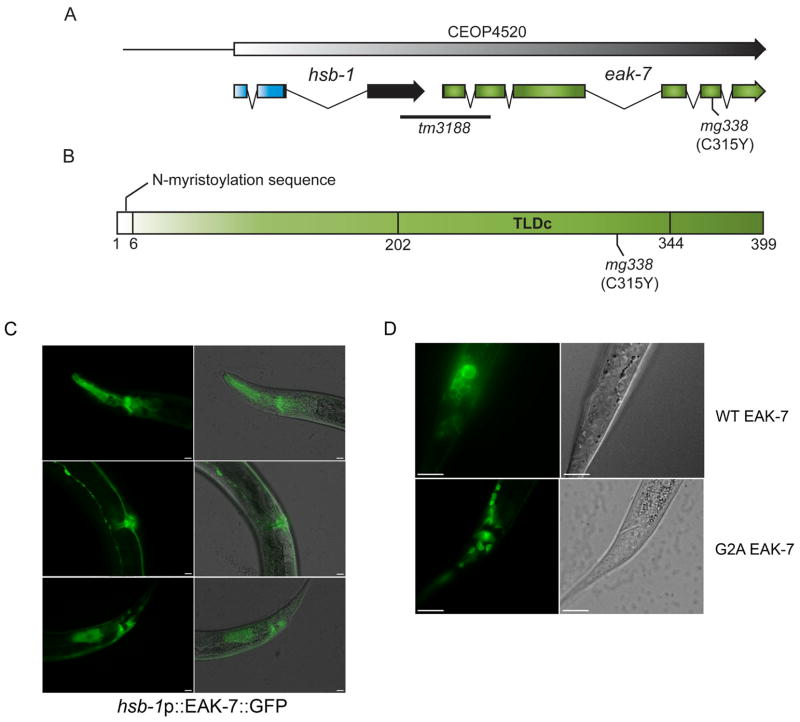
eak-7 encodes a conserved protein with a consensus N-myristoylation motif (Farazi et al., 2001) and a TLDc (TBC and LysM domain-containing) domain (Doerks et al., 2002), the function of which is obscure (Figures 5A–B and S4A–B). The mg338 allele is a missense mutation in the TLDc domain (Figures 5A–B and S4A). The eak-7(tm3188) null mutation harbors a deletion that spans the entire first exon and part of the second exon of K08E7.1 (Figure 5A). The eak-7 gene lies in an operon downstream of hsb-1 (Figure 5A), which encodes a conserved protein that binds to and inhibits HSF-1 (Satyal et al., 1998).
Figure 5. eak-7 gene structure, protein domain organization, and expression pattern.
A. eak-7 gene structure. eak-7 is the downstream gene in an operon with hsb-1. The deletion in the eak-7(tm3188) allele begins 249 bp upstream of the eak-7 initiator methionine codon and deletes portions of exons 1 and 2 of eak-7. The mg338 allele is a C/T transition that results in a cysteine-to-tyrosine mutation at amino acid 315 (C315Y). B. EAK-7 domain organization. The N-myristoylation motif and TLDc domain are shown. C. hsb-1p::EAK-7::GFP expression pattern in L4 larvae. hsb-1p::EAK-7::GFP is strongly expressed in the pharynx and nerve ring (top panels), the vulva and ventral nerve cord (middle panels), and in intestinal cells and cells surrounding the anus (bottom panels). Images were taken at 400× magnification. The scale bar is 10 μm. D. Subcellular localization of wild-type EAK-7::GFP (top panels) and EAK-7::GFP harboring a mutation of glycine 2 to alanine (G2A; bottom panels) at 1000× magnification. The scale bar is 10 μm.
EAK-7 is expressed in multiple tissues
To determine the expression pattern and subcellular localization of EAK-7, we analyzed transgenic animals that expressed an EAK-7::GFP fusion protein under the control of the hsb-1 operon promoter (hsb-1p::EAK-7::GFP). This transgene rescued the lifespan extension phenotype of an eak-7 null mutant (Figure S5). GFP was first expressed during embryogenesis (Figure S6A). At all stages of post-embryonic development, transgenic animals exhibited fluorescence in the pharynx, nervous system, intestine, body wall muscle, hypodermis, vulva, and a group of cells near the anus (Figures 5C, S6B–G, and data not shown). This expression pattern is consistent with that reported for a K08E7.1 promoter fusion constructed as part of a high-throughput analysis of C. elegans gene expression (Hunt-Newbury et al., 2007). EAK-7::GFP is also expressed in the XXX cells (Figure S6H), where eak-3, eak-4, sdf-9/eak-5, and eak-6 are specifically expressed (Hu et al., 2006; Ohkura et al., 2003; Zhang et al., 2008).
Consistent with the presence of an N-myristoylation motif, EAK-7::GFP localized to the plasma membrane (Figures 5D and S6H). Mutation of the glycine residue (G2A) that is required for N-myristoylation (Farazi et al., 2001) resulted in diffuse cytoplasmic fluorescence (Figure 5D), suggesting that the motif is functional. The G2A EAK-7::GFP mutant partially rescued dauer arrest in eak-7;akt-1 double mutants (data not shown), indicating that N-myristoylation is not an absolute requirement for EAK-7 function.
EAK-7 controls dauer arrest and lifespan nonautonomously
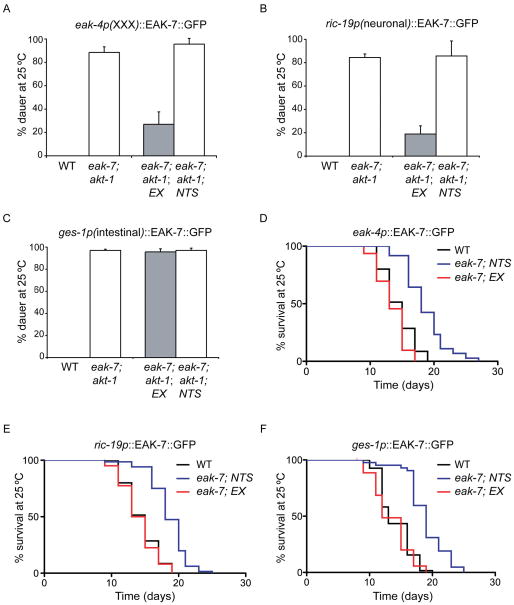
To determine the site of EAK-7 action in the control of dauer arrest and lifespan, we generated tissue-specific EAK-7::GFP expression constructs and tested their ability to rescue eak-7 mutant phenotypes. Since both EAK-7 and DAF-16/FoxO are expressed in neurons and intestine (Figure 5C; Henderson and Johnson, 2001; Lee et al., 2001; Lin et al., 2001) and DAF-16/FoxO activity in neurons and intestine promotes dauer arrest and lifespan extension (Apfeld and Kenyon, 1998; Libina et al., 2003; Wolkow et al., 2000), we expressed EAK-7::GFP using neuron (ric-19p)- and intestine (ges-1p)-specific promoters (Aamodt et al., 1991; Pilon et al., 2000). Since EAK-7 is also expressed in the XXX cells (Figure S6H) and acts in the same pathway as other EAK proteins that are expressed specifically in the XXX cells (Figure S1D; Hu et al., 2006; Ohkura et al., 2003; Zhang et al., 2008), we also tested an EAK-7::GFP transgene expressed under the control of the XXX-specific eak-4 promoter (Hu et al., 2006). Transgenic lines harboring these constructs exhibited fluorescence patterns consistent with the reported promoter specificity (data not shown).
XXX- and neuron-specific EAK-7::GFP expression rescued the dauer arrest phenotype of eak-7;akt-1 mutants, whereas intestinal EAK-7::GFP expression failed to rescue dauer arrest (Figures 6A–C and Table S3). In contrast, expression of EAK-7::GFP in the XXX cells, neurons, or the intestine was sufficient to rescue the lifespan extension phenotype of eak-7 mutants (Figures 6D–F and Table S4). Thus, similar to DAF-2/InsR (Apfeld and Kenyon, 1998; Wolkow et al., 2000), EAK-7 acts nonautonomously to control both dauer arrest and lifespan.
Figure 6. Rescue of eak-7 mutant phenotypes by tissue-specific EAK-7::GFP expression.
Rescue of dauer arrest at 25°C in eak-7;akt-1 double mutants (A.-C.) and rescue of lifespan extension in an eak-7 null mutant (D.–F.) by EAK-7::GFP transgenes under the control of eak-4 (XXX-specific; A. and D.), ric-19 (neuronal; B. and E.), and ges-1 (intestinal; C. and F.) promoters. “EX” and “NTS” denote transgenic animals carrying the extrachromosomal array and their non-transgenic siblings, respectively. Measurements represent the mean + S.D. Data are from a single representative transgenic line. Data from additional lines are in Tables S3 and S4.
DISCUSSION
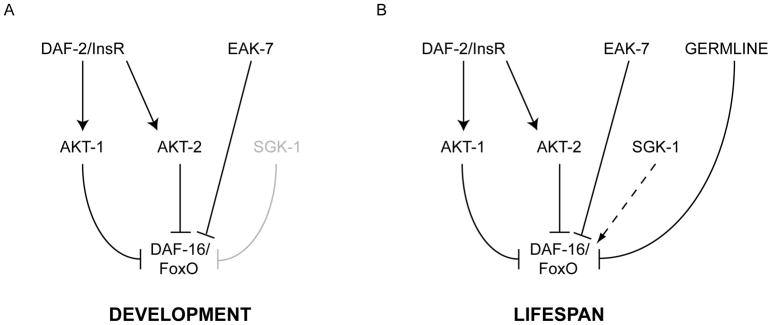
Our results indicate that EAK-7 defines a conserved pathway that acts in parallel to signals that inhibit DAF-16/FoxO nuclear translocation (primarily AKT-1 in early larvae and AKT-1, AKT-2, and the germline in adults) to control DAF-16/FoxO-dependent dauer arrest and lifespan extension (Figure 7). Since an eak-7 null mutation promotes dauer arrest in three distinct contexts characterized by increases in relative nuclear concentrations of DAF-16/FoxO (Figures 1 and 4B–C) and enhances lifespan extension in germline-deficient animals (Figure 2E), which exhibit increased DAF-16/FoxO nuclear localization (Lin et al., 2001), we favor a model whereby EAK-7 inhibits the activity of nuclear DAF-16/FoxO.
Figure 7. Genetic model of EAK-7 function during development and adulthood.
A. During development, EAK-7 acts in parallel to AKT-1 to inhibit DAF-16/FoxO activity. SGK-1 plays a minor role in dauer regulation. B. In adults, EAK-7 acts in parallel to AKT-1, AKT-2, and the germline to inhibit DAF-16/FoxO activity. SGK-1 may be required for maximal DAF-16/FoxO activity.
The increase in endogenous DAF-16/FoxO protein levels caused by eak-7 mutation in an akt-1 mutant background (Figure 4E) suggests that EAK-7 inhibits nuclear DAF-16/FoxO activity by reducing steady-state DAF-16/FoxO protein levels. This could occur via regulation of DAF-16/FoxO synthesis or turnover. Our data suggest that the DAF-16/FoxO E3 ubiquitin ligase RLE-1 (Li et al., 2007b) and the cullins CUL-5 and CUL-6 (Ghazi et al., 2007) do not mediate potential effects of EAK-7 on DAF-16/FoxO turnover (Figure S7 and data not shown). At this time we cannot exclude the possibility that EAK-7 also inhibits DAF-16/FoxO activity through mechanisms that are independent of DAF-16/FoxO protein levels.
EAK-7 controls lifespan and dauer arrest nonautonomously (Figure 6), as do DAF-2/InsR and DAF-16/FoxO (Apfeld and Kenyon, 1998; Libina et al., 2003; Wolkow et al., 2000). Intestinal EAK-7::GFP expression rescued lifespan extension but not dauer arrest in eak-7 mutants (Figures 6C and 6F), consistent with the role of intestinal DAF-16/FoxO in lifespan control (Libina et al., 2003). Interestingly, expression of EAK-7::GFP in the XXX cells, where a DAF-16::GFP fusion protein is not expressed (Hu et al., 2006), suffices to rescues both dauer arrest and lifespan extension in eak-7 mutants (Figures 6A and 6D). Thus, EAK-7 may regulate DAF-16/FoxO activity via both cell-autonomous and cell-nonautonomous mechanisms. Since a functional EAK-7::GFP fusion protein localizes to the plasma membrane (Figures 5D and S6H), cell-autonomous regulation of nuclear DAF-16/FoxO by EAK-7 is likely to be indirect.
The conservation of EAK-7 and IIS throughout animal phylogeny (Figure S4B) suggests that mechanisms by which EAK-7 regulates DAF-16/FoxO activity may also be conserved in mammals. In light of recent reports demonstrating that FoxO transcription factors are tumor suppressors and critical regulators of bone mass and glucose homeostasis in mice (Ambrogini et al., 2010; Dong et al., 2008; Matsumoto et al., 2006; Paik et al., 2007; Rached et al., 2010), dysregulation of human EAK-7 may play a role in the pathogenesis of type 2 diabetes, osteoporosis, and cancer. EAK-7 may also play a role in human longevity control, as FoxO3 polymorphisms are associated with extreme longevity in three independent cohorts of long-lived individuals (Flachsbart et al., 2009; Li et al., 2009; Willcox et al., 2008). Thus, functional human EAK-7 polymorphisms could influence metabolism, tumor survival, bone mineral density, and lifespan based on their effect on nuclear FoxO activity.
EXPERIMENTAL PROCEDURES
Strains and reagents
The following strains were used: N2 Bristol (wild-type), daf-2(e1370) (Kimura et al., 1997), eak-7(mg338), eak-7(tm3188), akt-1(mg306) (Hu et al., 2006), daf-16(mgDf47) (Ogg et al., 1997), daf-16(mu86) (Lin et al., 2001), akt-2(ok393) (Hertweck et al., 2004), sgk-1(mg455) (Soukas et al., 2009), daf-12(rh61rh411) (Antebi et al., 2000), glp-1(e2141) (Priess et al., 1987), eak-3(mg344) (Zhang et al., 2008), rle-1(tm2447) (Li et al., 2007b), TJ356 (Henderson and Johnson, 2001), CF1553 (Libina et al., 2003), CF1371, and CF1330 (Lin et al., 2001). Unless otherwise indicated, the akt-1 mutant allele used was mg306, the eak-7 mutant allele used was tm3188, and the daf-16/FoxO mutant allele used was mu86. Double and triple mutants were constructed using standard genetic techniques. Genotypes were confirmed using either restriction fragment length or PCR polymorphisms.
Dauer arrest assays
Dauer arrest assays were performed as described (Zhang et al., 2008). 27 °C dauer assays were performed using NGM plates with or without supplemental cholesterol as indicated.
Lifespan assays
Lifespan assays were performed at 20°C or 25°C as described (Zhang et al., 2008). Briefly, L4 larvae were placed onto NGM plates containing 25 μg/ml fluorodeoxyuridine (FUDR) and 10 μg/ml nystatin that had been seeded with 20× concentrated E. coli OP50. Animals were assayed for viability visually or with mild prodding. GraphPad Prism® (GraphPad Software, La Jolla, CA) was used for graphing and statistical analysis.
Quantitative RT-PCR
For L2 animals, wild-type and mutant animals grown at 25 °C were harvested 24h after a two-hour egg lay, and total RNA was prepared from 300–400 animals per strain using Trizol (Invitrogen, Carlsbad, CA). For adults, 50 L4 animals were placed on FUDR/nystatin plates at 25 °C and harvested four days later. qPCR primer sequences are available upon request.
sod-3p::GFP and DAF-16::GFP localization assays
sod-3::GFP (muIs84) (Libina et al., 2003) and DAF-16::GFP (zIs356) (Henderson and Johnson, 2001) were introduced into various mutant backgrounds and visualized at 200× magnification immediately after mounting.
RNAi
Feeding RNAi was performed using standard procedures. For dauer assays, 6-cm plates containing NGM + 5 mM IPTG + 25 μg/ml carbenicillin were spotted with 500 μl of overnight culture of E. coli HT115 harboring either control L4440 vector or ftt-2 RNAi plasmid. Plates were allowed to dry overnight at room temperature. Gravid animals cultured on standard NGM plates containing E. coli OP50 were picked to RNAi plates for egglays. Dauers were scored after progeny had been incubated at 25°C for 48–60 hours.
For lifespan assays, 6-cm plates containing NGM + 5 mM IPTG + 25 μg/ml carbenicillin + 25 μg/ml FUDR + 10 μg/ml nystatin were spotted with 400 μl of 5× concentrated overnight culture of E. coli HT115 harboring either control L4440 vector or smk-1 RNAi plasmid. Plates were allowed to dry overnight at room temperature. Young adult animals were picked to plates and scored for viability as described.
DAF-16AM dauer assay
CF1371 and CF1330 (Lin et al., 2001) carry extrachromosomal arrays containing either a GFP::DAF-16A transgene with all AKT phosphorylation sites mutated (CF1371) or a wild-type GFP::DAF-16A transgene (CF1330) with the rol-6 co-injection marker. (Lin et al., 2001). Transgenic animals and their non-transgenic siblings were identified using a fluorescence dissecting microscope and/or the Rol phenotype. Measurements represent the percentage of Rol animals that arrested as dauers.
Immunoblotting
L2 larvae grown at 25 °C were harvested 24h after a two-hour egg lay and washed three times in M9 buffer. Protein lysates were prepared by boiling 600–800 animals per strain in sample buffer (Bio-Rad, Hercules, CA). Proteins were separated by SDS-PAGE, and immunoblots were performed using standard procedures. Anti-DAF-16/FoxO antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA), anti-actin antibodies from Millipore (Billerica, MA), anti-mouse-HRP from GE (Piscataway, NJ), and anti-goat-HRP from Jackson Labs (Bar Harbor, MA). The EAK-7 antibody was raised in rabbits against a peptide corresponding to the last 19 residues of C. elegans of EAK-7 (Proteintech Group Inc., Chicago, IL).
Cloning of eak-7
Isolation, mapping, and sequencing of eak-7(mg338) were performed as described for eak-3 (Zhang et al., 2008).
Transgenes and transgenic rescue experiments
The hsb-1p::EAK-7::GFP translational fusion was generated using overlap extension PCR (Hobert, 2002). The following fragments were amplified and fused: a 3391 bp genomic fragment corresponding to nucleotides 6205–9595 of cosmid K08E7 encoding the hsb-1 promoter, a 1884 bp genomic fragment corresponding to nucleotides 3090–4973 of cosmid K08E7 that includes the open reading frame of K08E7.1 up to but not including the translation termination codon, and a DNA fragment containing GFP and the unc-54 3′ UTR that was amplified from pPD95.75 (a gift from Dr. Andrew Fire). The EAK-7::GFP G2A N-myristoylation mutant was also constructed using overlap extension PCR by incorporating nucleotide changes into the primers resulting in mutation of glycine at residue 2 of EAK-7 to alanine.
Tissue-specific EAK-7::GFP transgenes contained the same K08E7.1 genomic sequences as the hsb-1p::EAK-7::GFP construct and were also generated by overlap extension PCR. The following regions were used in place of the hsb-1 promoter: 1054 nucleotides upstream of the eak-4 start site amplified from cosmid F53B2 for the XXX-specific construct, 1547 nucleotides upstream of the ric-19 start site amplified from pJK325 for the neuron-specific construct, and 2256 nucleotides upstream of the ges-1 start site amplified from pJK332 for the intestine-specific construct. All fusion products were verified by restriction digest or sequencing. Fusion PCR products were purified using the Qiaquick PCR purification kit (Qiagen, Venlo, The Netherlands).
Transgenic animals were generated and localization studies performed as described previously (Zhang et al., 2008). Animals were injected with a 100 ng/μl mixture containing the transgene construct (25 ng/μl), the rol-6(pRF4) coinjection marker (12.5ng/μl), and pBluescript. Transgenic animals were visualized using an Olympus BX61 upright microscope and analyzed using SlideBook 4.1 digital microscopy software (Intelligent Imaging Innovations, Inc., Denver, CO). To determine if EAK-7 was expressed in the XXX cells, hsb-1p::EAK-7::GFP was injected into worms carrying an integrated sdf-9p::RFP promoter fusion (Hu et al., 2006). Animals were visualized using a Leica DM6000 confocal microscope and analyzed using Leica LAS AF Version 1.8.2. Animals were mounted on 2% agarose pads in the presence of 10 mM levamisole or 10 mM sodium azide, visualized, and photographed immediately after mounting.
Transgenic animals and their non-transgenic siblings were distinguished using a fluorescence dissecting microscope and/or the Rol phenotype.
Highlights for Alam et al., D-10-00082.
eak-7 emerged from a C. elegans screen for enhancers of the akt-1 mutant phenotype
EAK-7 controls larval development and adult lifespan by inhibiting DAF-16/FoxO
EAK-7 and AKT-1 inhibit DAF-16/FoxO via distinct mechanisms
EAK-7 inhibits nuclear DAF-16/FoxO without promoting its cytoplasmic translocation
Supplementary Material
Acknowledgments
We thank John Kim, Shawn Xu, and Lois Weisman for discussions and comments on the manuscript, Sylvia Lee for useful suggestions, Cheng-Yu Lee for assistance with confocal microscopy, Yuji Kohara for eak-7 cDNAs, Andrew Fire for pPD95.75, John Kim for pJK325 and pJK332, Alex Soukas for sgk-1(mg455), and Muazzum Shah for outcrossing eak-7(tm3188). Some strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR). These studies were supported by the Life Sciences Institute at the University of Michigan as well as by grants from the Nathan Shock Center for the Biology of Aging and the Michigan Diabetes Research and Training Center at the University of Michigan Medical School, the Sidney Kimmel Foundation for Cancer Research, NIH grants DK62884 and DK78183 (P.J.H.), and an American Cancer Society postdoctoral fellowship (#113680) to H.A.
Footnotes
The authors disclose no conflicts of interest with this study.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aamodt EJ, Chung MA, McGhee JD. Spatial control of gut-specific gene expression during Caenorhabditis elegans development. Science. 1991;252:579–582. doi: 10.1126/science.2020855. [DOI] [PubMed] [Google Scholar]
- Accili D, Arden KC. FoxOs at the crossroads of cellular metabolism, differentiation, and transformation. Cell. 2004;117:421–426. doi: 10.1016/s0092-8674(04)00452-0. [DOI] [PubMed] [Google Scholar]
- Ambrogini E, Almeida M, Martin-Millan M, Paik JH, Depinho RA, Han L, Goellner J, Weinstein RS, Jilka RL, O’Brien CA, et al. FoxO-mediated defense against oxidative stress in osteoblasts is indispensable for skeletal homeostasis in mice. Cell Metab. 2010;11:136–146. doi: 10.1016/j.cmet.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antebi A, Yeh WH, Tait D, Hedgecock EM, Riddle DL. daf-12 encodes a nuclear receptor that regulates the dauer diapause and developmental age in C. elegans. Genes Dev. 2000;14:1512–1527. [PMC free article] [PubMed] [Google Scholar]
- Apfeld J, Kenyon C. Cell nonautonomy of C. elegans daf-2 function in the regulation of diapause and life span. Cell. 1998;95:199–210. doi: 10.1016/s0092-8674(00)81751-1. [DOI] [PubMed] [Google Scholar]
- Arden KC. FOXO animal models reveal a variety of diverse roles for FOXO transcription factors. Oncogene. 2008;27:2345–2350. doi: 10.1038/onc.2008.27. [DOI] [PubMed] [Google Scholar]
- Ayyadevara S, Alla R, Thaden JJ, Shmookler Reis RJ. Remarkable longevity and stress resistance of nematode PI3K-null mutants. Aging Cell. 2008;7:13–22. doi: 10.1111/j.1474-9726.2007.00348.x. [DOI] [PubMed] [Google Scholar]
- Berdichevsky A, Viswanathan M, Horvitz HR, Guarente L. C. elegans SIR-2.1 interacts with 14-3-3 proteins to activate DAF-16 and extend life span. Cell. 2006;125:1165–1177. doi: 10.1016/j.cell.2006.04.036. [DOI] [PubMed] [Google Scholar]
- Berman JR, Kenyon C. Germ-cell loss extends C. elegans life span through regulation of DAF-16 by kri-1 and lipophilic-hormone signaling. Cell. 2006;124:1055–1068. doi: 10.1016/j.cell.2006.01.039. [DOI] [PubMed] [Google Scholar]
- Calnan DR, Brunet A. The FoxO code. Oncogene. 2008;27:2276–2288. doi: 10.1038/onc.2008.21. [DOI] [PubMed] [Google Scholar]
- Doerks T, Copley RR, Schultz J, Ponting CP, Bork P. Systematic identification of novel protein domain families associated with nuclear functions. Genome Res. 2002;12:47–56. doi: 10.1101/gr.203201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong XC, Copps KD, Guo S, Li Y, Kollipara R, DePinho RA, White MF. Inactivation of hepatic Foxo1 by insulin signaling is required for adaptive nutrient homeostasis and endocrine growth regulation. Cell Metab. 2008;8:65–76. doi: 10.1016/j.cmet.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farazi TA, Waksman G, Gordon JI. The biology and enzymology of protein N-myristoylation. J Biol Chem. 2001;276:39501–39504. doi: 10.1074/jbc.R100042200. [DOI] [PubMed] [Google Scholar]
- Fielenbach N, Antebi A. C. elegans dauer formation and the molecular basis of plasticity. Genes Dev. 2008;22:2149–2165. doi: 10.1101/gad.1701508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch CE, Ruvkun G. The genetics of aging. Annu Rev Genomics Hum Genet. 2001;2:435–462. doi: 10.1146/annurev.genom.2.1.435. [DOI] [PubMed] [Google Scholar]
- Flachsbart F, Caliebe A, Kleindorp R, Blanche H, von Eller-Eberstein H, Nikolaus S, Schreiber S, Nebel A. Association of FOXO3A variation with human longevity confirmed in German centenarians. Proc Natl Acad Sci U S A. 2009;106:2700–2705. doi: 10.1073/pnas.0809594106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gems D, Sutton AJ, Sundermeyer ML, Albert PS, King KV, Edgley ML, Larsen PL, Riddle DL. Two pleiotropic classes of daf-2 mutation affect larval arrest, adult behavior, reproduction and longevity in Caenorhabditis elegans. Genetics. 1998;150:129–155. doi: 10.1093/genetics/150.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerisch B, Weitzel C, Kober-Eisermann C, Rottiers V, Antebi A. A hormonal signaling pathway influencing C. elegans metabolism, reproductive development, and life span. Dev Cell. 2001;1:841–851. doi: 10.1016/s1534-5807(01)00085-5. [DOI] [PubMed] [Google Scholar]
- Ghazi A, Henis-Korenblit S, Kenyon C. Regulation of Caenorhabditis elegans lifespan by a proteasomal E3 ligase complex. Proc Natl Acad Sci U S A. 2007;104:5947–5952. doi: 10.1073/pnas.0700638104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil EB, Malone Link E, Liu LX, Johnson CD, Lees JA. Regulation of the insulin-like developmental pathway of Caenorhabditis elegans by a homolog of the PTEN tumor suppressor gene. Proc Natl Acad Sci U S A. 1999;96:2925–2930. doi: 10.1073/pnas.96.6.2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb S, Ruvkun G. daf-2, daf-16 and daf-23: genetically interacting genes controlling Dauer formation in Caenorhabditis elegans. Genetics. 1994;137:107–120. doi: 10.1093/genetics/137.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross DN, van den Heuvel AP, Birnbaum MJ. The role of FoxO in the regulation of metabolism. Oncogene. 2008;27:2320–2336. doi: 10.1038/onc.2008.25. [DOI] [PubMed] [Google Scholar]
- Henderson ST, Johnson TE. daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Curr Biol. 2001;11:1975–1980. doi: 10.1016/s0960-9822(01)00594-2. [DOI] [PubMed] [Google Scholar]
- Hertweck M, Gobel C, Baumeister R. C. elegans SGK-1 is the critical component in the Akt/PKB kinase complex to control stress response and life span. Dev Cell. 2004;6:577–588. doi: 10.1016/s1534-5807(04)00095-4. [DOI] [PubMed] [Google Scholar]
- Hobert O. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques. 2002;32:728–730. doi: 10.2144/02324bm01. [DOI] [PubMed] [Google Scholar]
- Honda Y, Honda S. The daf-2 gene network for longevity regulates oxidative stress resistance and Mn-superoxide dismutase gene expression in Caenorhabditis elegans. Faseb J. 1999;13:1385–1393. [PubMed] [Google Scholar]
- Hsin H, Kenyon C. Signals from the reproductive system regulate the lifespan of C. elegans. Nature. 1999;399:362–366. doi: 10.1038/20694. [DOI] [PubMed] [Google Scholar]
- Hsu A, Murphy CT, Kenyon C. Regulation of Aging and Age-Related Disease by DAF-16 and Heat-Shock Factor. Science. 2003;300:1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- Hu PJ. WormBook. 2007. Dauer; pp. 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu PJ, Xu J, Ruvkun G. Two membrane-associated tyrosine phosphatase homologs potentiate C. elegans AKT-1/PKB signaling. PLoS Genet. 2006;2:e99. doi: 10.1371/journal.pgen.0020099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Regan KM, Wang F, Wang D, Smith DI, van Deursen JM, Tindall DJ. Skp2 inhibits FOXO1 in tumor suppression through ubiquitin-mediated degradation. Proc Natl Acad Sci U S A. 2005;102:1649–1654. doi: 10.1073/pnas.0406789102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt-Newbury R, Viveiros R, Johnsen R, Mah A, Anastas D, Fang L, Halfnight E, Lee D, Lin J, Lorch A, et al. High-throughput in vivo analysis of gene expression in Caenorhabditis elegans. PLoS Biol. 2007;5:e237. doi: 10.1371/journal.pbio.0050237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia K, Albert PS, Riddle DL. DAF-9, a cytochrome P450 regulating C. elegans larval development and adult longevity. Development. 2002;129:221–231. doi: 10.1242/dev.129.1.221. [DOI] [PubMed] [Google Scholar]
- Kenyon C. The plasticity of aging: insights from long-lived mutants. Cell. 2005;120:449–460. doi: 10.1016/j.cell.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- Kim SH, Holway AH, Wolff S, Dillin A, Michael WM. SMK-1/PPH-4.1-mediated silencing of the CHK-1 response to DNA damage in early C. elegans embryos. J Cell Biol. 2007;179:41–52. doi: 10.1083/jcb.200705182. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- Lee RY, Hench J, Ruvkun G. Regulation of C. elegans DAF-16 and its human ortholog FKHRL1 by the daf-2 insulin-like signaling pathway. Curr Biol. 2001;11:1950–1957. doi: 10.1016/s0960-9822(01)00595-4. [DOI] [PubMed] [Google Scholar]
- Li J, Tewari M, Vidal M, Lee SS. The 14-3-3 protein FTT-2 regulates DAF-16 in Caenorhabditis elegans. Dev Biol. 2007a;301:82–91. doi: 10.1016/j.ydbio.2006.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Gao B, Lee SM, Bennett K, Fang D. RLE-1, an E3 ubiquitin ligase, regulates C. elegans aging by catalyzing DAF-16 polyubiquitination. Dev Cell. 2007b;12:235–246. doi: 10.1016/j.devcel.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Li Y, Wang WJ, Cao H, Lu J, Wu C, Hu FY, Guo J, Zhao L, Yang F, Zhang YX, et al. Genetic association of FOXO1A and FOXO3A with longevity trait in Han Chinese populations. Hum Mol Genet. 2009;18:4897–4904. doi: 10.1093/hmg/ddp459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libina N, Berman JR, Kenyon C. Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell. 2003;115:489–502. doi: 10.1016/s0092-8674(03)00889-4. [DOI] [PubMed] [Google Scholar]
- Lin K, Hsin H, Libina N, Kenyon C. Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat Genet. 2001;28:139–145. doi: 10.1038/88850. [DOI] [PubMed] [Google Scholar]
- Matsumoto M, Han S, Kitamura T, Accili D. Dual role of transcription factor FoxO1 in controlling hepatic insulin sensitivity and lipid metabolism. J Clin Invest. 2006;116:2464–2472. doi: 10.1172/JCI27047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto M, Pocai A, Rossetti L, Depinho RA, Accili D. Impaired regulation of hepatic glucose production in mice lacking the forkhead transcription factor Foxo1 in liver. Cell Metab. 2007;6:208–216. doi: 10.1016/j.cmet.2007.08.006. [DOI] [PubMed] [Google Scholar]
- Matsuzaki H, Daitoku H, Hatta M, Tanaka K, Fukamizu A. Insulin-induced phosphorylation of FKHR (Foxo1) targets to proteasomal degradation. Proc Natl Acad Sci U S A. 2003;100:11285–11290. doi: 10.1073/pnas.1934283100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaylova VT, Borland CZ, Manjarrez L, Stern MJ, Sun H. The PTEN tumor suppressor homolog in Caenorhabditis elegans regulates longevity and dauer formation in an insulin receptor-like signaling pathway. Proc Natl Acad Sci U S A. 1999;96:7427–7432. doi: 10.1073/pnas.96.13.7427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris JZ, Tissenbaum HA, Ruvkun G. A phosphatidylinositol-3-OH kinase family member regulating longevity and diapause in Caenorhabditis elegans. Nature. 1996;382:536–539. doi: 10.1038/382536a0. [DOI] [PubMed] [Google Scholar]
- Motola DL, Cummins CL, Rottiers V, Sharma KK, Li T, Li Y, Suino-Powell K, Xu HE, Auchus RJ, Antebi A, et al. Identification of ligands for DAF-12 that govern dauer formation and reproduction in C. elegans. Cell. 2006;124:1209–1223. doi: 10.1016/j.cell.2006.01.037. [DOI] [PubMed] [Google Scholar]
- Murakami S, Johnson TE. A genetic pathway conferring life extension and resistance to UV stress in Caenorhabditis elegans. Genetics. 1996;143:1207–1218. doi: 10.1093/genetics/143.3.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, Ruvkun G. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- Ogg S, Ruvkun G. The C. elegans PTEN homolog, DAF-18, acts in the insulin receptor-like metabolic signaling pathway. Mol Cell. 1998;2:887–893. doi: 10.1016/s1097-2765(00)80303-2. [DOI] [PubMed] [Google Scholar]
- Oh SW, Mukhopadhyay A, Dixit BL, Raha T, Green MR, Tissenbaum HA. Identification of direct DAF-16 targets controlling longevity, metabolism and diapause by chromatin immunoprecipitation. Nat Genet. 2006;38:251–257. doi: 10.1038/ng1723. [DOI] [PubMed] [Google Scholar]
- Oh SW, Mukhopadhyay A, Svrzikapa N, Jiang F, Davis RJ, Tissenbaum HA. JNK regulates lifespan in Caenorhabditis elegans by modulating nuclear translocation of forkhead transcription factor/DAF-16. Proc Natl Acad Sci U S A. 2005;102:4494–4499. doi: 10.1073/pnas.0500749102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkura K, Suzuki N, Ishihara T, Katsura I. SDF-9, a protein tyrosine phosphatase-like molecule, regulates the L3/dauer developmental decision through hormonal signaling in C. elegans. Development. 2003;130:3237–3248. doi: 10.1242/dev.00540. [DOI] [PubMed] [Google Scholar]
- Paik JH, Kollipara R, Chu G, Ji H, Xiao Y, Ding Z, Miao L, Tothova Z, Horner JW, Carrasco DR, et al. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128:309–323. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis S, Ailion M, Toker A, Thomas JH, Ruvkun G. A PDK1 homolog is necessary and sufficient to transduce AGE-1 PI3 kinase signals that regulate diapause in Caenorhabditis elegans. Genes Dev. 1999;13:1438–1452. doi: 10.1101/gad.13.11.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis S, Ruvkun G. Caenorhabditis elegans Akt/PKB transduces insulin receptor-like signals from AGE-1 PI3 kinase to the DAF-16 transcription factor. Genes Dev. 1998;12:2488–2498. doi: 10.1101/gad.12.16.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge L, Bruning JC. Forkhead transcription factors and ageing. Oncogene. 2008;27:2351–2363. doi: 10.1038/onc.2008.28. [DOI] [PubMed] [Google Scholar]
- Patel DS, Garza-Garcia A, Nanji M, McElwee JJ, Ackerman D, Driscoll PC, Gems D. Clustering of genetically defined allele classes in the Caenorhabditis elegans DAF-2 insulin/IGF-1 receptor. Genetics. 2008;178:931–946. doi: 10.1534/genetics.107.070813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilon M, Peng XR, Spence AM, Plasterk RH, Dosch HM. The diabetes autoantigen ICA69 and its Caenorhabditis elegans homologue, ric-19, are conserved regulators of neuroendocrine secretion. Mol Biol Cell. 2000;11:3277–3288. doi: 10.1091/mbc.11.10.3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priess JR, Schnabel H, Schnabel R. The glp-1 locus and cellular interactions in early C. elegans embryos. Cell. 1987;51:601–611. doi: 10.1016/0092-8674(87)90129-2. [DOI] [PubMed] [Google Scholar]
- Quevedo C, Kaplan DR, Derry WB. AKT-1 regulates DNA-damage-induced germline apoptosis in C. elegans. Curr Biol. 2007;17:286–292. doi: 10.1016/j.cub.2006.12.038. [DOI] [PubMed] [Google Scholar]
- Rached MT, Kode A, Silva BC, Jung DY, Gray S, Ong H, Paik JH, DePinho RA, Kim JK, Karsenty G, et al. FoxO1 expression in osteoblasts regulates glucose homeostasis through regulation of osteocalcin in mice. J Clin Invest. 2010;120:357–368. doi: 10.1172/JCI39901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottiers V, Motola DL, Gerisch B, Cummins CL, Nishiwaki K, Mangelsdorf DJ, Antebi A. Hormonal control of C. elegans dauer formation and life span by a Rieske-like oxygenase. Dev Cell. 2006;10:473–482. doi: 10.1016/j.devcel.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Rouault JP, Kuwabara PE, Sinilnikova OM, Duret L, Thierry-Mieg D, Billaud M. Regulation of dauer larva development in Caenorhabditis elegans by daf-18, a homologue of the tumour suppressor PTEN. Curr Biol. 1999;9:329–332. doi: 10.1016/s0960-9822(99)80143-2. [DOI] [PubMed] [Google Scholar]
- Satyal SH, Chen D, Fox SG, Kramer JM, Morimoto RI. Negative regulation of the heat shock transcriptional response by HSBP1. Genes Dev. 1998;12:1962–1974. doi: 10.1101/gad.12.13.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukas AA, Kane EA, Carr CE, Melo JA, Ruvkun G. Rictor/TORC2 regulates fat metabolism, feeding, growth, and life span in Caenorhabditis elegans. Genes Dev. 2009;23:496–511. doi: 10.1101/gad.1775409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullet JM, Hertweck M, An JH, Baker J, Hwang JY, Liu S, Oliveira RP, Baumeister R, Blackwell TK. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell. 2008;132:1025–1038. doi: 10.1016/j.cell.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Heide LP, Hoekman MF, Smidt MP. The ins and outs of FoxO shuttling: mechanisms of FoxO translocation and transcriptional regulation. Biochem J. 2004;380:297–309. doi: 10.1042/BJ20040167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vowels JJ, Thomas JH. Genetic analysis of chemosensory control of dauer formation in Caenorhabditis elegans. Genetics. 1992;130:105–123. doi: 10.1093/genetics/130.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willcox BJ, Donlon TA, He Q, Chen R, Grove JS, Yano K, Masaki KH, Willcox DC, Rodriguez B, Curb JD. FOXO3A genotype is strongly associated with human longevity. Proc Natl Acad Sci U S A. 2008;105:13987–13992. doi: 10.1073/pnas.0801030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff S, Ma H, Burch D, Maciel GA, Hunter T, Dillin A. SMK-1, an essential regulator of DAF-16-mediated longevity. Cell. 2006;124:1039–1053. doi: 10.1016/j.cell.2005.12.042. [DOI] [PubMed] [Google Scholar]
- Wolkow CA, Kimura KD, Lee MS, Ruvkun G. Regulation of C. elegans life-span by insulinlike signaling in the nervous system. Science. 2000;290:147–150. doi: 10.1126/science.290.5489.147. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Xu J, Puscau C, Kim Y, Wang X, Alam H, Hu PJ. Caenorhabditis elegans EAK-3 inhibits dauer arrest via nonautonomous regulation of nuclear DAF-16/FoxO activity. Dev Biol. 2008;315:290–302. doi: 10.1016/j.ydbio.2007.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.