Abstract
Intrinsic transcription termination sites generally contain a tract of adenines in the DNA template that yields a tract of uracils at the 3’- end of the nascent RNA. To understand how this base sequence contributes to termination of transcription, we have investigated two nucleic acid structures. The first is the RNA-DNA hybrid that contains the uracil tract 5’-rUUUUUAU-3’ from the tR2 intrinsic terminator of bacteriophage λ. The second is the homologous DNA-DNA duplex that contains the adenine tract 5’-dATAAAAA-3’. This duplex is present at the tR2 site when the DNA is not transcribed. The opening and the stability of each rU-dA/dT-dA base pair in the two structures are characterized by imino proton exchange and nuclear magnetic resonance spectroscopy. The results reveal concerted opening of the central rU-dA base pairs in the RNA-DNA hybrid. Furthermore, the stability profile of the adenine tract in the RNA-DNA hybrid is very different from that of the tract in the template DNA-DNA duplex. In the RNA-DNA hybrid, the stabilities of rU-dA base pairs range from 4.3 to 6.5 kcal/mol (at 10°C). The sites of lowest stability are identified at the central positions of the tract. In the template DNA-DNA duplex, the dT-dA base pairs are more stable than the corresponding rU-dA base pairs in the hybrid by 0.9 to 4.6 kcal/mol and, in contrast to the RNA-DNA hybrid, the central base pairs have the highest stability. These results suggest that the central rU-dA/dT-dA base pairs in the adenine tract make the largest energetic contributions to transcription termination by promoting both, the dissociation of the RNA transcript and the closing of the transcription bubble. The results also suggest that the high stability of dT-dA base pairs in the DNA provides a signal for the pausing of RNA polymerase at the termination site.
Keywords: Intrinsic transcription termination, base-pair opening, RNA-DNA hybrids, adenine tracts, proton exchange
Introduction
Transcription is the first step in gene expression. The transfer of base sequence information from the DNA to the nascent messenger RNA occurs through a transient RNA-DNA hybrid structure in which RNA bases form complementary pairs with bases in the DNA template strand. The hybrid generally contains 7-9 base pairs.1, 2 Its formation requires separation of the two DNA strands, through at least 12 base pairs, into a transcription bubble. The transcription process is initiated by binding of RNA polymerase to the promoter of the corresponding gene(s). During the subsequent RNA synthesis, the polymerase remains bound to the DNA and to the nascent RNA. This elongation complex maintains its high stability as it advances through thousands of base pairs at speeds up to 100 base pairs/s. The stability of the complex is, however, lost at the DNA sites where transcription must end. At these sites, the elongation complex disassembles by the release of the RNA transcript and the dissociation of the polymerase from DNA. Transcription termination sites are mostly found at the end of operons.3 They are also present near promoters and between genes in an operon. In the latter case, the termination sites function as regulators of gene expression: they dictate whether or not transcription of the downstream genes will occur.
In prokaryotes, transcription termination sites are generally classified according to their requirements. Factor-dependent (or Rho-dependent) sites require auxiliary factors, such as Rho protein, to terminate transcription in vitro. In contrast, at intrinsic (or Rho-independent) sites, termination is triggered solely by signals encoded in the base sequence and in the structure of the RNA and DNA. In E. Coli, about half of the transcription termination sites belongs to this class.3
Intrinsic transcription termination sites share two base sequence motifs: a dGC-rich sequence with hyphenated inverted symmetry and a dA-rich tract in the DNA template strand (Figure 1a).4 The two motifs work in concert with each other to induce termination.5-10 The GC-rich sequence acts in the RNA transcript where it forms a hairpin. The hairpin disrupts the hybrid downstream from it and alters key contacts of the RNA polymerase with the nucleic acid framework. The second motif exerts its action through the RNA-DNA hybrid. When the polymerase reaches the termination point, the hybrid generated by the dA-rich tract consists of consecutive rU-dA base pairs. Such hybrids are very unstable, as demonstrated previously by optical melting experiments.11 The instability of the hybrid weakens the conformational constraints for the upstream formation of the hairpin and allows for ready release of the RNA transcript.
Figure 1.
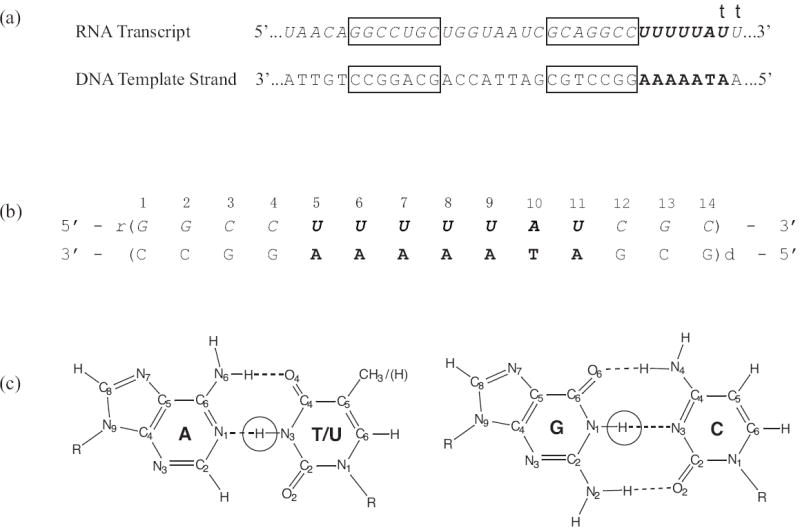
(a) Base sequences of the DNA template strand and of the RNA transcript in tR2 transcription termination site from bacteriophage λ. The GC-rich sequence with inverted symmetry is boxed. The sequences corresponding to the A-rich tract are shown in bold. Transcription terminates at one of the two positions indicated by a “t”. The most frequent termination point is at the first of these positions.
(b) Base sequence and numbering of base pairs in the RNA-DNA hybrid investigated.
(c) The structures of GC and AU/T base pairs with the imino protons highlighted.
Our laboratory has been working toward defining the energetic contributions of individual base pairs from the RNA transcript and the DNA template to the destabilization of the elongation complex at transcription termination sites. The system studied is the tR2 terminator of bacteriophage λ shown in Figure 1. This terminator has been a model system for investigations of the molecular mechanisms of intrinsic transcription termination.5, 8, 10, 12 The methods of choice in our work are nuclear magnetic resonance (NMR) spectroscopy and proton exchange. The combined use of these two techniques allows us to define the stability of each base pair in several structural states of the nucleic acid framework involved in termination of transcription at the λ tR2 site. In a previous paper, we have reported the structural energetic maps for the two homologous RNA-DNA hybrids formed by the GC-rich sequence.13 In the present work we report our results on the RNA-DNA hybrid formed when the dA-rich tract is transcribed, and on the homologous DNA-DNA duplex present in the template when the DNA is not transcribed. The RNA-DNA hybrid investigated, henceforth-abbreviated rU-dA RNA-DNA hybrid, is shown in Figure 1b. The main interest of the work is on the rU-dA base pairs of the molecule, which reproduce the hybrid formed at the tR2 site just before the termination point (Figure 1a). The rG-dC/rC-dG base pairs are added at the ends of the duplex to increase the stability of the structure and to prevent the effects of fraying upon proton exchange in the rU-dA base pairs of interest, while maintaining the base sequence context upstream from the hybrid.
Results
Assignment of the Imino Proton Resonances in the rU-dA RNA-DNA Hybrid
Characterization of the stabilities of individual base pairs in nucleic acid structures by NMR spectroscopy relies upon observation of the imino proton in each base pair (Methods section and Figure 1c). The NMR resonances of the imino protons in the rU-dA RNA-DNA hybrid investigated are shown in Figure 2. The spectrum exhibits severe overlap especially for the resonances of rU-dA base pairs, which are of interest in the present work. Due to this overlap, the methods generally used for resonance assignments, such as 1H-1H NOESY, are of limited value in this case. To circumvent this problem, and assign the uracil imino proton resonances, we have constructed RNA-DNA hybrid molecules in which the N3 atom of single uracil bases was 15N-labeled. The 15N-labeled uracil was placed at each of the following positions: 5, 6, 7, 8, 9, and 11. The 15N-edited spectra of these six singly labeled hybrids are shown in Figure 3. As one can see, the resonance of (rU-dA)5 is the only uracil resonance that is resolved (14.14 ppm). The imino proton resonances of (rU-dA)6 and (rU-dA)11 both occur at 13.95 ppm, while those of (rU-dA)7 and (rU-dA)8 occur at 13.88 ppm. The resonance of (rU-dA)9 is at 13.12 ppm and, in the unedited spectrum, overlaps with other imino proton resonances (Figure 2).
Figure 2.
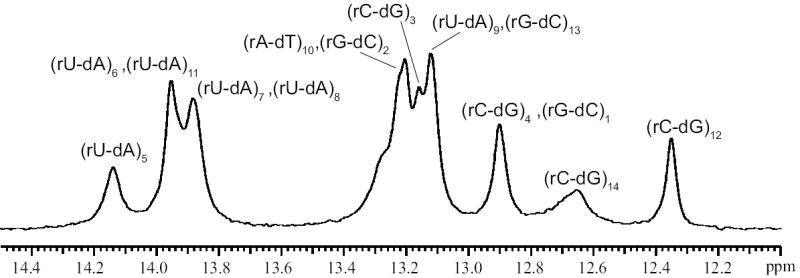
Imino proton resonances of the rU-dA RNA-DNA hybrid in 10 mM phosphate buffer at pH 7.8 and at 10°C.
Figure 3.
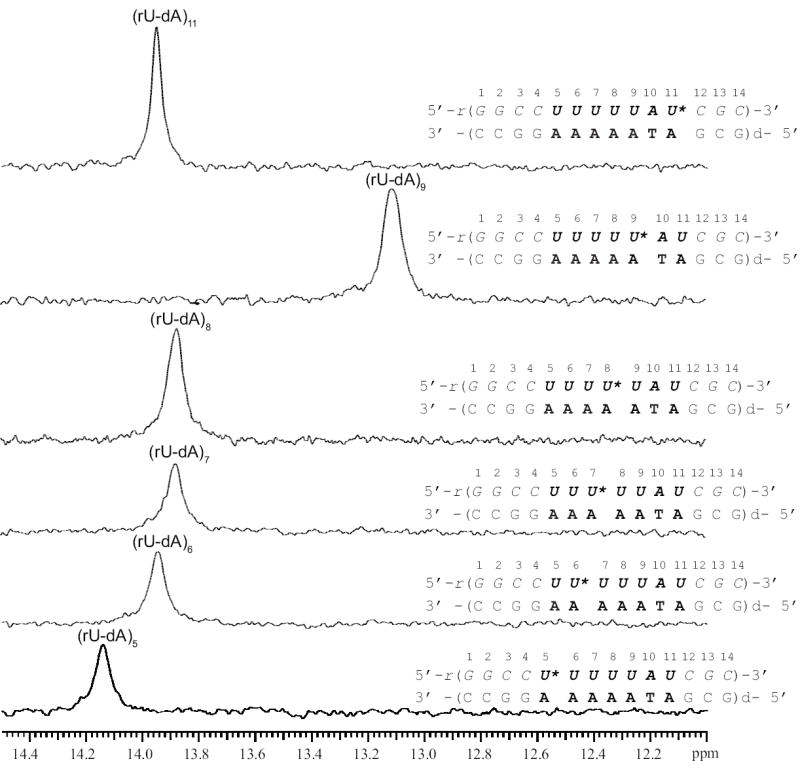
15N-edited NMR resonances of imino protons for rU-dA RNA-DNA hybrid molecules in which a single uracil was labeled with 15N. The 15N-labeled uracil in each molecule is indicated by an asterisk.
The imino proton resonances of (rA-dT)10 and of guanines were assigned from 1H-1H NOESY and from uniform 15N-labeling, as detailed in the Supplementary Data.
Imino Proton Exchange in the rU-dA RNA-DNA Hybrid
The exchange rates of the imino protons in the rU-dA hybrid were measured on the singly labeled samples illustrated in Figure 3 and on a triply labeled sample. In the latter sample, the uracils at positions 6, 8, and 9 were labeled with 15N. The resonances of the labeled uracils were observed in the 15N-edited spectrum (Figure 4a), while those of the remaining uracils (i.e., positions 5, 7 and 11) were observed in the 14N-edited spectrum (Figure 4b). This triply labeling strategy allowed us to measure the exchange rates of all uracil imino protons on the same sample.
Figure 4.
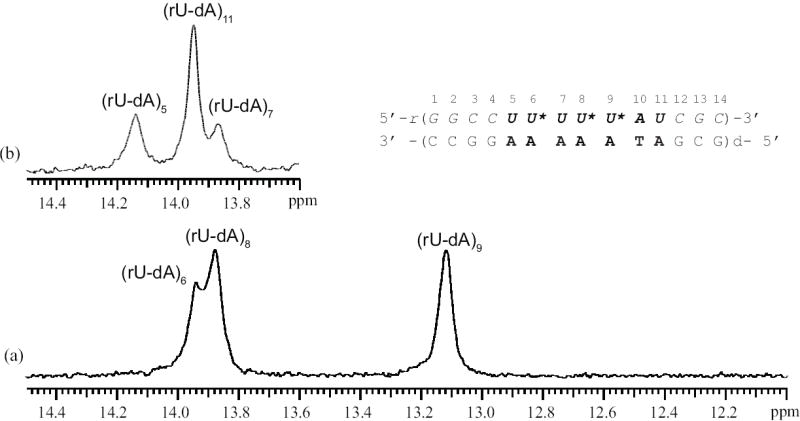
NMR resonances of the uracil imino protons for the rU-dA RNA-DNA hybrid containing 15N-labeled uracil in positions 6, 8 and 9. (a) 15N-edited uracil imino proton resonances. (b) 14N-edited uracil imino proton resonances.
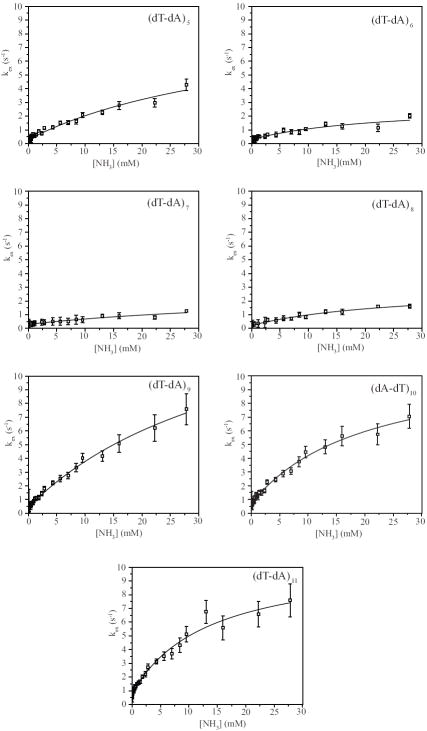
The exchange rates were measured as a function of the concentration of ammonia (Methods section). The dependence of the exchange rates on the concentration of ammonia base, shown in Figure 5, was analyzed according to the exchange model presented in the Methods section. For all rU-dA base pairs, the exchange is fast even at low ammonia concentrations. As the ammonia concentration is increased, the exchange rates become higher than ~80-90 s-1, and cannot be measured by the NMR transfer of magnetization technique. The equilibrium constants of the base-pair opening reactions were obtained by fitting the exchange rates as a function of ammonia base concentration to Eq. 7 (Methods section). The obtained values are reported in Table 1.
Figure 5.
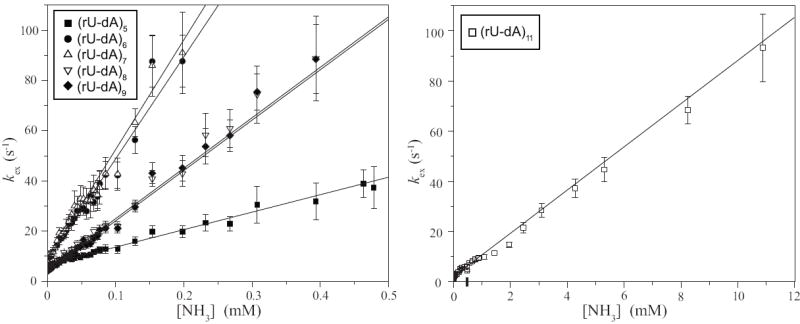
Dependence of the exchange rates of individual uracil imino protons in the rU-dA RNA-DNA hybrid on the concentration of ammonia base. The exchange rates for (rU-dA)11 are shown separately due to the different range of ammonia base concentrations in which the exchange can be measured. The lines represent least-squares fits to Eq. 7.
Table 1.
Equilibrium Constants and Free Energy Changes for Base-Pair Opening in the rU-dA RNA-DNA Hybrid and dT-dA DNA Duplexes Investigated
| rU-dA RNA-DNA Hybrid | dT-dA DNA Duplex | |||||
|---|---|---|---|---|---|---|
| Base pair | Kop (×106) | ΔGopa | Base pair | Kop (×106) | ΔGopa | ΔΔGopa |
| (rU-dA)5 | 79 ± 2 | 5.30±0.01 | (dT-dA)5 | 0.5±0.2 | 8.1±0.2 | -2.8±0.2 |
| (rU-dA)6 | 462 ± 14 | 4.31±0.02 | (dT-dA)6 | 0.3±0.1 | 8.4±0.2 | -4.1±0.2 |
| (rU-dA)7 | 497 ± 19 | 4.27±0.02 | (dT-dA)7 | 0.1±0.1 | 9.0±0.6 | -4.7±0.6 |
| (rU-dA)8 | 228 ± 7 | 4.70±0.02 | (dT-dA)8 | 0.3±0.1 | 8.4±0.2 | -3.7±0.2 |
| (rU-dA)9 | 228 ± 5 | 4.70±0.01 | (dT-dA)9 | 1.0±0.2 | 7.7±0.1 | -3.0±0.1 |
| (rA-dT)10 | b | b | (dA-dT)10 | 1.4±0.2 | 7.6±0.1 | |
| (rU-dA)11 | 9.8 ± 0.4 | 6.47±0.02 | (dT-dA)11 | 1.9±0.3 | 7.4±0.1 | -0.9±0.1 |
in kcal/mol at 10°C
Imino proton resonance is not resolved in the NMR spectrum
Imino Proton Exchange in the Homologous DNA-DNA Duplex
We have also measured the imino proton exchange in the DNA-DNA duplex with the same base sequence as that of the rU-dA RNA-DNA hybrid (except for the replacement of rU with dT, Figure 6). This duplex, henceforth-abbreviated dT-dA DNA, is of interest because it represents the state of the template when the DNA is not transcribed.
Figure 6.
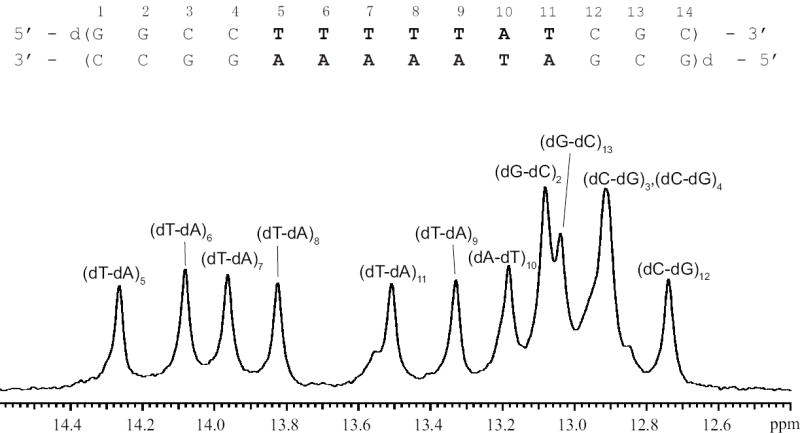
The base sequence and the imino proton resonances of the dT-dA DNA duplex in 10 mM phosphate buffer at pH 7.8 and at 10°C.
The imino proton resonances of the dT-dA DNA duplex (Figure 6) are well resolved and can be assigned to specific imino protons using standard 1H-1H NOESY experiments. The NOESY spectrum and the resonance assignments for this duplex are discussed in Supplementary Data.
The exchange rates of the imino protons in the dT-dA DNA were measured as a function of the concentration of ammonia under the same experimental conditions as those used for the rU-dA RNA-DNA hybrid. Representative examples of the dependence of the exchange rates on ammonia base concentration are shown in Figure 7. In contrast to the hybrid, the exchange of thymine imino protons in the DNA duplex is slow, i.e., exchange rates less than 10 s-1. The dependence of the exchange rates on ammonia base concentration was analyzed according to Eq. 4 (Methods section). The equilibrium constants for the opening reactions of the dT-dA base pairs in the DNA duplex, obtained from this analysis and Eq. 5, are compared to those of rU-dA base pairs in the RNA-DNA hybrid in Table 1.
Figure 7.
Dependence of the exchange rates of thymine imino protons in the dT-dA DNA duplex on the concentration of ammonia base. The curves represent nonlinear least-squares fits to Eq. 4.
Discussion
The results obtained in the present work allow us to define the stability of each base pair in the RNA-DNA and DNA-DNA duplexes investigated. The exchange of an imino proton with solvent protons monitors the conformational transition of the corresponding base pair from its native state inside the double helix to an open, solvent accessible state (Methods section). The equilibrium constant of this opening reaction (Kop) provides a measure of the stability of the base pair in its native, intra-helical state. The equilibrium constant Kop is related to the free energy change in the opening reaction by
| (1) |
where T is the absolute temperature and R is the universal gas constant. Base pairs with high structural stability are characterized by small values of the equilibrium constant Kop and large values of the free energy change ΔGop. Alternatively, high Kop and small ΔGop values reflect low structural stability of the corresponding base pairs.
The equilibrium constants for the opening of the rU-dA base pairs in the RNA-DNA hybrid range from 9.8·10-6 to 497·10-6 (Table 1). The base pairs in positions 5 through 9 are contiguous in the sequence, while that in position 11 represents a single, isolated rU-dA base pair. It is readily seen that the opening equilibrium constants of the contiguous base pairs are 8-to 50-fold greater than that of the isolated (rU-dA)11 base pair. Accordingly, for these base pairs, the opening free energy changes are smaller than that of (rU-dA)11 by 1.17 to 2.20 kcal/mol. Hence, contiguous rU-dA base pairs are structurally less stable than a rU-dA base pair placed in a mixed sequence context. This finding is in agreement with optical melting studies, which showed that the stability of RNA-DNA hybrid structures containing contiguous rU-dA base pairs is very low.11 Our results describe this low overall stability at the level of individual rU-dA base pairs. The most stable is the first base pair in the tract, (rU-dA)5, while the next two base pairs, (rU-dA)6 and (rU-dA)7, have the lowest stabilities. For the last base pairs, (rU-dA)8 and (rU-dA)9, the stability is increased by ~0.4 kcal/mol relative to that of the preceding two base pairs. This energetic profile of the RNA-DNA hybrid provides new insight into features of transcription termination observed in previous mutational and biochemical studies of the λ tR2 terminator.
The efficiency of transcription termination at the tR2 site is strongly dependent on the rU-dA base pairs in the 2nd to the 5th positions of the tract.8, 10, 14, 15 Large changes in the efficiency of termination are observed when one or two of these rU-dA base pairs are replaced with rG-dC base pairs. These substitutions also inhibit formation of the hairpin by the upstream inverted sequence in nascent RNA. Our present results suggest that this part of the tract plays such an important functional role due, at least in part, to the structural stability of the base pairs therein. Among all positions in the tract, the rU-dA base pairs in positions 2 to 5 of the tract [i.e., (rU-dA)6, (rU-dA)7, (rU-dA)8 and (rU-dA)9 in our construct] have the lowest stabilities (Table 1). Therefore, these base pairs represent the sites of the hybrid where separation of the RNA and DNA strands is energetically least costing. This energetic cost (i.e., 4.3-4.7 kcal/mol) is much less than the costs of opening rU-dA base pairs in mixed sequence contexts (e.g., 6.47 kcal/mol, Table 1) or rG-dC/rC-dG base pairs (i.e., 7.8-9.75 kcal/mol13). Hence, substitutions of the rU-dA base pairs in the tract’s positions 2 to 5 by stronger base pairs, like in the previous studies, should affect the most the stability of the rU-dA hybrid at the site of transcription termination.
Our proton exchange results reveal that, in addition to low structural stability, the tract of rU-dA base pairs also has unique dynamic properties. These properties are inferred from the observation that the exchange rate of the imino proton in (rU-dA)6 is the same as the exchange rate of the imino proton in (rU-dA)7; and the exchange rate of the imino proton in (rU-dA)8 is the same as the exchange rate of the imino proton in (rU-dA)9. This observation is presented in Figure 8 by plots of the exchange rates for one base pair against the exchange rates for the other base pair at all concentrations of proton acceptor investigated (similar plots for the other base pairs are given in the Supplementary Data). Both plots in Figure 8 have a slope equal or close to unity indicating that the exchange rates are the same over the entire range of proton acceptor concentrations. This fact explains why the opening equilibrium constant for (rU-dA)6 is, within experimental errors, the same as that for (rU-dA)7; and the opening equilibrium constant for (rU-dA)8 is the same as that for (rU-dA)9 (Table 1).
Figure 8.
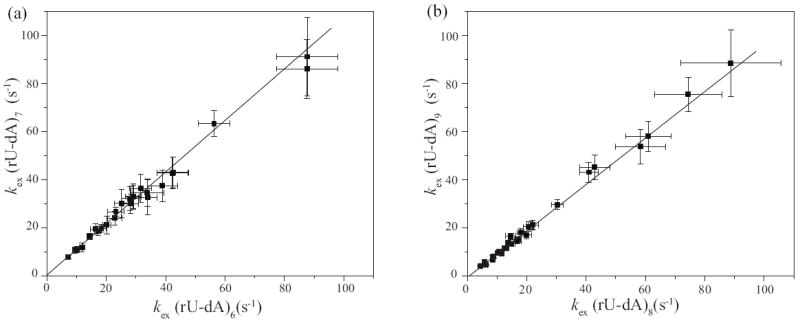
(a) Comparison of the exchange rates of the imino proton in (rU-dA)7 to the exchange rates of the imino proton in (rU-dA)6; the slope of the least-squares fitted line is 1.07±0.02.
(b) Comparison of the exchange rates of the imino proton in (rU-dA)9 to the exchange rates of the imino proton in (rU-dA)8; the slope of the least-squares fitted line is 0.97±0.03.
In general, in nucleic acid duplexes, imino protons in adjacent base pairs exchange at different rates.16, 17 This general observation has led to the postulate that base pairs open one at a time, independently of the opening of their neighbors.16 Our results suggest that, in the rU-dA tract of the RNA-DNA hybrid investigated, some base pairs may not follow this general rule. Instead, it is likely that the exchange of imino protons in (rU-dA)6 and (rU-dA)7 occurs in a single opening reaction involving both base pairs. Similarly, for (rU-dA)8 and (rU-dA)9, the identity of imino proton exchange rates also suggests concerted opening of the two base pairs. These concerted opening reactions must clearly originate from specific structural features of the tract that couple one base pair to its neighbor. The nature of these structural features is unknown at present. Nevertheless, it is likely that the same structural features aid neighboring bases in the RNA transcript to separate from the DNA template strand in a concerted manner.
Further insight into the energetics of intrinsic transcription termination is provided by our results on the dT-dA DNA duplex (Table 1). The equilibrium constants for opening of the dT-dA base pairs in this duplex are consistently smaller than those of the corresponding rU-dA base pairs in the RNA-DNA hybrid. When the base pair is in a mixed sequence context, like in position 11 of the two duplexes studied here, the Kop value for dT-dA is ~ 5-fold lower than that for rU-dA, corresponding to an increase in the opening free energy of 0.9 kcal/mol. Much larger differences are, however, observed for contiguous rU-dA/dT-dA base pairs, namely, the Kop values for dT-dA base pairs are 160- to 5,000-fold lower than those for rU-dA base pairs, and the corresponding opening free energies are higher by up to 4.7 kcal/mol. From these energetic profiles (Figure 9a) we conclude that, within the same tract, dT-dA base pairs are much more stable than rU-dA base pairs.
Figure 9.
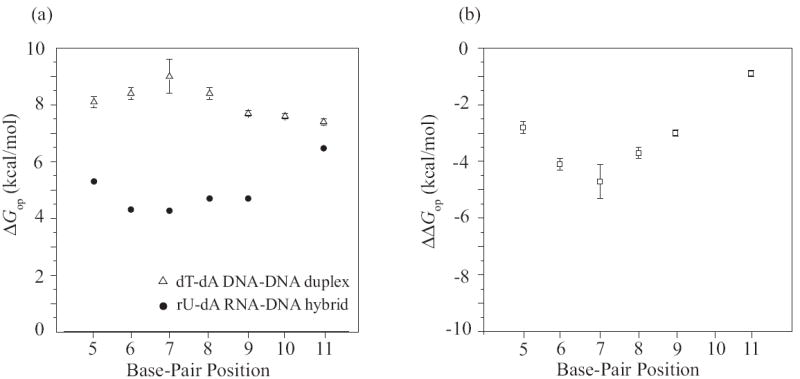
(a) Opening free energy changes (ΔGop) for rU-dA/dT-dA base pairs in the RNA-DNA and DNA-DNA duplexes investigated. The experimental errors for the values in the RNA-DNA hybrid are smaller than the symbols.
(b) The free energy contributions (ΔΔGop) of individual rU-dA/dT-dA base pairs to the destabilization of the elongation complex at the transcription termination site, calculated based on the thermodynamic model (Eq. 3).
The relevance of our findings for the energetics of transcription termination is best understood in the framework of the thermodynamic model for elongation, termination and editing in transcription, proposed by von Hippel and coworkers.6, 15, 18 According to this model, formation of the transcription complex from free RNA polymerase and double-stranded DNA is driven by the following free energy change:
| (2) |
where is the unfavorable free energy change for opening of base pairs in double-stranded DNA to form the transcription bubble; is the favorable free energy change for formation of the base pairs in the RNA-DNA hybrid; and the term includes the free energies of all interactions of RNA polymerase with the nucleic acid (NA) components of the transcription complex. During elongation, the favorable and terms outweigh the cost of opening the DNA duplex ( ), thus ensuring the processivity of transcription. The model postulates that, at the site of an intrinsic terminator, the favorable interactions between the RNA transcript and the DNA template ( ) are weakened, and those between the polymerase and the nucleic acid components ( ) are perturbed. As a result, the transcription complex is destabilized so that the RNA-DNA hybrid separates into strands and liberates the newly made RNA, the polymerase is released, and the transcription bubble closes up.
In the framework of this thermodynamic model, our data provide the contribution of each base pair of the double-stranded DNA and of the RNA-DNA hybrid to the destabilization of the elongation complex at the termination site. The contributions are calculated as the total free energy change that occurs when a base pair in the RNA-DNA hybrid opens (or “melts”) and the corresponding base pair re-forms in double-stranded DNA:
| (3) |
This equation assumes that the presence of the polymerase in the elongation complex does not affect significantly the stabilities of the base pairs in the hybrid structure. The results of the calculations are shown in the last column of Table 1 and in Figure 9b. Clearly, transition of a base pair from the RNA-DNA hybrid to double-stranded DNA is energetically favorable. The free energy change in the transition varies from -0.9 to -4.7 kcal/mol. The smallest free energy contribution is made by the isolated base pair, (rU-dA)11. The most favorable free energy contributions (-3.7 to -4.7 kcal/mol) are made by the base pairs in the 2nd, 3rd and 4th positions in the tract, i.e., (rU-dA)6, (rU-dA)7 and (rU-dA)8. A large contribution from these base pairs is expected since, as discussed above, these are among the weakest base pairs in the hybrid (Figure 9a). One notes also that the contributions of these base pairs are enhanced by the fact that, in the DNA duplex, the corresponding dT-dA base pairs are the most stable base pairs. Hence, these base pairs should experience the largest change in stabilization free energy upon dissociation of the RNA-DNA hybrid and re-formation of double-stranded DNA.
Current molecular models for intrinsic transcription termination attribute the selection of the terminator site by RNA polymerase to the hairpin formed by the inverted GC-rich sequence in nascent RNA, and to the rU-dA hybrid formed when the downstream part of the terminator is transcribed.8-10, 19 Our present results suggest that the base sequence of the DNA itself could also play a role in the recognition of the termination site by the enzyme. This suggestion is explained in Figure 10. The figure compares the opening free energies (ΔGop) for dT-dA base pairs in two DNA duplexes: (i) the dT-dA DNA duplex studied in the present work, and (ii) a DNA duplex recently studied by our laboratory.20 The latter duplex (henceforth-abbreviated GC-rich DNA duplex) incorporates in positions 4 through 12 the downstream half of the inverted GC-rich sequence and the first two dT-dA base pairs from the dA-rich sequence of the tR2 site. As one can see, in the GC-rich DNA duplex, the ΔGop values for dT-dA base pairs range from 5.28 to 6.2 kcal/mol. In contrast, in the dT-dA DNA duplex, the ΔGop values are much higher, i. e., 7.4 to 9 kcal/mol. This increase is clearly due to the tract of consecutive dT-dA base pairs in the dT-dA duplex. For example, in the GC-rich duplex, (dT-dA)11 and (dT-dA)12 have opening free energies of 6.2 and 5.28 kcal/mol, respectively. When the same base pairs are placed in the tract of the dT-dA duplex [i.e., (dT-dA)5 and (dT-dA)6], their opening free energies become 8.1 and 8.4 kcal/mol, respectively (Table 1). Therefore, when several dT-dA base pairs are consecutive, their stabilities greatly increase relative to that of dT-dA base pairs in a mixed sequence context. Similar energetic features have been previously observed in other DNA tracts of dT-dA base pairs21-23. The high stabilities of consecutive dT-dA base pairs could contribute to the recognition of the terminator by the polymerase by increasing the energetic cost for formation of the transcription bubble at this site beyond that required at a site of random base sequence. The high cost of unwinding the DNA helix could, in turn, lower the rate of translocation into the terminator, thus allowing the time necessary for early termination-specific conformational changes in the enzyme and in the RNA to take place.8, 24
Figure 10.
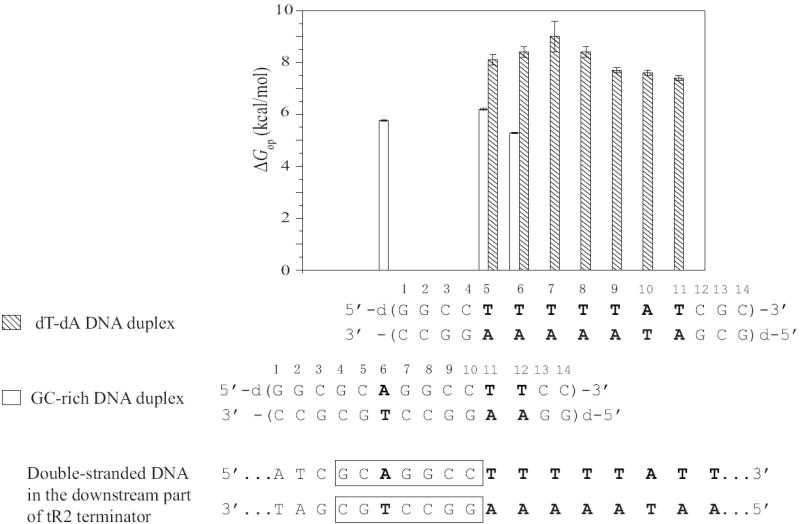
Opening free energy changes (ΔGop in kcal/mol at 10°C) for dT-dA base pairs in the GC-rich and dT-dA DNA duplexes. Empty bars represent the values in the GC-rich duplex, and filled bars represent the values in the dT-dA duplex. The downstream part of the double-stranded DNA in the tR2 terminator is shown at the bottom.
In summary, the results presented in this paper suggest that an intrinsic transcription terminator uses the same base sequence, i.e., a tract of dA bases in the DNA template strand, to specify termination in two concurrent ways. When the dA bases pair with dT, like in the double-stranded DNA template, the stability of the resulting base pairs is higher than that in a random base sequence. When the dA bases pair with rU, like in the transcription RNA-DNA hybrid, the resulting base pairs are much less stable than those in a hybrid of random sequence. The energetic balance between these alternative paired states of dA bases thus favors the dissociation of the RNA-DNA hybrid as well as the closing of the transcription bubble into a double-stranded DNA helix. Resolution of the structural stability at the level of individual bases also suggests that rU-dA/dT-dA base pairs in the central positions of the dA-rich tract are the main contributors to this energetic balance.
Materials and Methods
Materials
(3-15N)-labeled 2’-O-tom-protected uridine phosphoramidite was prepared via a 7-step synthesis as previously described.25-27 The scheme for this synthesis is shown in Supplementary Data. The labeled phosphoramidite was used to synthesize the RNA strands for the six samples of singly 15N-labeled RNA-DNA hybrid, and for the sample of the triply 15N-labeled RNA-DNA hybrid. The DNA and RNA strands were synthesized on an automated DNA synthesizer using the solid-support phosphoramidite method. They were purified by HPLC on a semipreparative column in 50 mM triethylamine acetate buffer with a gradient of 5%-32% acetonitrile over 46 minutes at 60 °C. The RNA strand containing 15N-guanine (Cambridge Isotope Laboratory, MA) was synthesized by in vitro transcription using T7 RNA polymerase.28 For both DNA and RNA strands the counterions were replaced with Na+ ions by repeated centrifugation in Amicon Ultra-4 centrifugal filter units using 0.5 M NaCl, followed by centrifugation against water. The final NMR samples contained 0.3 to 0.8 mM RNA-DNA or DNA-DNA duplex in 10 mM Na2HPO4/NaH2PO4 and 0.5 mM EDTA in 90% H2O/10% D2O at pH 7.8 ± 0.1 (at 10 °C). All samples also contained 0.5 mM triethanolamine, which was used to determine the pH of the samples directly in the NMR tube as we have described.29
Methods
NMR Experiments
The NMR experiments were performed at 10 °C on a Varian INOVA 500 spectrometer operating at 11.75 T. The exchange rates of imino protons were measured by transfer of magnetization from water. The water resonance was selectively inverted using a Gaussian 180 pulse (5.8ms) followed by a variable delay for the exchange of magnetization between water and imino protons. A gradient of 0.21 G/cm was applied during the exchange delay to prevent the effects of radiation damping upon the recovery of water magnetization to equilibrium. A second selective pulse on water was applied to bring the water magnetization back onto the z-axis before observation. Twenty-five values of the exchange delay in the range from 1 to 800 ms were used in each experiment. The exchange rates were calculated from the dependence of the intensity of the imino proton resonance on the exchange delay as described previously.17 For the unlabeled samples the observation was with the Jump-and-Return pulse sequence.30 For the 15N-labeled samples the observation was with the one-dimensional version of the fast HSQC (fHSQC) pulse sequence to edit the resonances of protons attached to 15N and eliminate all other proton resonances.31 A modified, 14N-editing, fHSQC pulse sequence was used to filter out the resonances of protons attached to 15N and retain all other proton resonances.32 The highest exchange rates that can be accurately measured by these methods is ~80 - 90 s-1. The 1H-1H NOESY spectra were obtained using the Watergate pulse sequence33 with a mixing time of 250 ms.
Imino Proton Exchange in Nucleic Acid Duplexes
The exchange of imino protons in nucleic acids is a two-step process. In the first step, the hydrogen bonds in the base pair break and the base flips out of the structure into an open state. In this state, the imino proton is accessible to proton acceptors present in the solvent. The second step is the actual transfer of the proton to an acceptor, for example, NH3. The exchange rate observed experimentally is given as:16, 34
| (4) |
where kop and kcl are the rates of opening and closing, respectively, of the base containing the imino proton, [B] is the concentration of proton acceptor B, and kB is the rate constant for the transfer of the imino proton to the proton acceptor in isolated nucleotides. The rates of opening and closing define the equilibrium constant of the opening reaction:
| (5) |
Two kinetic regimes for imino proton exchange can be distinguished depending on how the rate of exchange from the open state compares with the rate of closing. When the concentration of proton acceptor is sufficiently high to make the exchange from the open state very fast (kex,open ≫ kcl, EX1 regime), the exchange is limited by the rate of base-pair opening. In this case, Eq. 4 becomes:
| (6) |
At low concentrations of proton acceptor, the rate of exchange from the open state is much smaller than the rate of base-pair closing (kex,open ≪ kcl, EX2 regime). In this regime, the observed exchange rate is proportional to the concentration of proton acceptor:
| (7) |
In this work, we have used ammonia base (NH3) as the imino proton acceptor in the exchange. Previous work from this and other laboratories has shown that, due to its small size, good solubility, high pK, and lack of charge, ammonia base is the acceptor of choice for proton exchange studies of nucleic acids.16, 17 The rate constants for transfer of imino protons to NH3 (kB) were previously calculated as: 8.8 × 108 M-1s-1 for the uracil proton and 4.1 × 108 M-1s-1 for the thymine proton, at 10 °C.13 The total ammonia concentration was varied from 0 to 150 mM for the experiments on the rU-dA RNA-DNA hybrid, and from 0 to 500 mM for the experiments on the dT-dA DNA duplex. The concentration of ammonia base NH3 was calculated from the total ammonia concentration C0 and the pH as:
| (8) |
The pH was measured at each ammonia concentration, directly in the NMR tube, using the proton resonances of triethanolamine.29 The pK value of ammonia at 10 °C is 9.73.35
Supplementary Material
Acknowledgments
This work was supported by a grant from the National Institutes of Health (GM077188). We would like to thank Dr. Smita Patel (University of Medicine and Dentistry of New Jersey-Robert Wood Johnson Medical School) for a gift of T7 RNA polymerase, and Dr. Michael Calter (Wesleyan University) for his help with the synthesis of (3-15N)-labeled 2’-O-tom protected uridine phosphoramidite.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nudler E, Mustaev A, Lukhtanov E, Goldfarb A. The RNA-DNA Hybrid Maintains the Register of Transcription by Preventing Backtracking of RNA Polymerase. Cell. 1997;89:33–41. doi: 10.1016/s0092-8674(00)80180-4. [DOI] [PubMed] [Google Scholar]
- 2.Korzheva N, Mustaev A, Kozlov M, Malhotra A, Nikiforov V, Goldfarb A, Darst SA. A Structural Model of Transcription Elongation. Science. 2000;289:619–625. doi: 10.1126/science.289.5479.619. [DOI] [PubMed] [Google Scholar]
- 3.Lesnik EA, Sampath R, Levene HB, Henderson TJ, McNeil JA, Ecker DJ. Prediction of rho-independent transcriptional terminators in Escherichia Coli. Nucleic Acids Res. 2001;29:3583–3594. doi: 10.1093/nar/29.17.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.d’Aubenton Carafa Y, Brody E, Thermes C. Prediction of Rho-independent Escherichia coli Transcription Terminators. A Statistical Analysis of their RNA Stem-Loop Structures. J Mol Biol. 1990;216:835–858. doi: 10.1016/s0022-2836(99)80005-9. [DOI] [PubMed] [Google Scholar]
- 5.Yarnell WS, Roberts JW. Mechanism of Intrinsic Transcription Termination and Antitermination. Science. 1999;284:611–615. doi: 10.1126/science.284.5414.611. [DOI] [PubMed] [Google Scholar]
- 6.von Hippel PH. An Integrated Model of the Transcription Complex in Elongation, Termination, and Editing. Science. 1998;281:660–665. doi: 10.1126/science.281.5377.660. [DOI] [PubMed] [Google Scholar]
- 7.Greive SJ, von Hippel PH. Thinking quantitatively about transcription regulation. Nature Rev Mol Cell Biol. 2005;6:221–232. doi: 10.1038/nrm1588. [DOI] [PubMed] [Google Scholar]
- 8.Gusarov I, Nudler E. The Mechanism of Intrinsic Transcription Termination. Mol Cell. 1999;3:495–504. doi: 10.1016/s1097-2765(00)80477-3. [DOI] [PubMed] [Google Scholar]
- 9.Nudler E, Gottesman ME. Transcription termination and anti-termination in E. Coli. Genes Cells. 2002;7:755–768. doi: 10.1046/j.1365-2443.2002.00563.x. [DOI] [PubMed] [Google Scholar]
- 10.Komissarova N, Becker J, Solter S, Kireeva M, Kashlev M. Shortening of RNA:DNA Hybrid in the Elongation Complex of RNA Polymerase Is a Prerequisite for Transcription Termination. Mol Cell. 2002;10:1151–1162. doi: 10.1016/s1097-2765(02)00738-4. [DOI] [PubMed] [Google Scholar]
- 11.Martin FH, Tinoco IJ. RNA-DNA hybrid duplexes containing oligo (dA:rU) sequences are exceptionally unstable and may facilitate termination of transcription. Nucleic Acids Res. 1980;8:2295–2299. doi: 10.1093/nar/8.10.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson KS, von Hippel PH. Transcription termination at intrinsic terminators: The role of the RNA hairpin. Proc Natl Acad Sci US. 1995;92:8793–8797. doi: 10.1073/pnas.92.19.8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang Y, Chen C, Russu IM. Dynamics and Stability of Individual Base Pairs in Two Homologous RNA-DNA Hybrids. Biochemistry. 2009;48:3988–3997. doi: 10.1021/bi900070f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Epshtein V, Cardinale CJ, Ruckenstein AE, Borukhov S, Nudler E. An Alosteric Path to Transcription Termination. Mol Cell. 2007;28:991–1001. doi: 10.1016/j.molcel.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 15.Datta K, von Hippel PH. Direct Spectroscopic Study of Reconstituted Transcription Complexes Reveals That Intrinsic Termination is Driven Primarily by Thermodynamic Destabilization of the Nucleic Acid Framework. J Biol Chem. 2008;283:3537–3549. doi: 10.1074/jbc.M707998200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gueron M, Leroy J-L. Studies of Base Pair Kinetics by NMR Measurement of Proton Exchange. Methods Enzymol. 1995;261:383–413. doi: 10.1016/s0076-6879(95)61018-9. [DOI] [PubMed] [Google Scholar]
- 17.Russu IM. Probing Site-Specific Energetics in Proteins and Nucleic Acids by Hydrogen Exchange and NMR Spectroscopy. Methods Enzymol. 2004;379:152–175. doi: 10.1016/S0076-6879(04)79009-6. [DOI] [PubMed] [Google Scholar]
- 18.Yager TD, von Hippel PH. A Thermodynamic Analysis of RNA Transcript Elongation and Termination in Escherichia coli. Biochemistry. 1991;30:1097–1118. doi: 10.1021/bi00218a032. [DOI] [PubMed] [Google Scholar]
- 19.Toulokhonov I, Artsimovitch I, Landick R. Allosteric control of RNA polymerase by a site that contacts nascent RNA hairpins. Science. 2001;292:730–733. doi: 10.1126/science.1057738. [DOI] [PubMed] [Google Scholar]
- 20.Huang Y, Weng X, Russu IM. In preparation 2010 [Google Scholar]
- 21.Leroy JL, Charretier E, Kochoyan M, Gueron M. Evidence from Base-Pair Kinetics for Two Types of Adenine Tract Structures in Solution: Their Relation to DNA Curvature. Biochemistry. 1988;27:8894–8898. doi: 10.1021/bi00425a004. [DOI] [PubMed] [Google Scholar]
- 22.Moe JG, Russu IM. Proton Exchange and Base Pair Opening Kinetics in 5′-d (CGCGAATTCGCG)-3′ and Related Dodecamers. Nucleic Acids Res. 1990;18:821–827. doi: 10.1093/nar/18.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moe JG, Folta-Stogniew E, Russu IM. Energetics of Base-Pair Opening in a DNA Dodecamer Containing an A3T3 Tract. Nucleic Acids Res. 1995;23:1984–1989. doi: 10.1093/nar/23.11.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tadigotla VR, Maoileidigh DO, Sengupta AM, Epshtein V, Ebright RH, Nudler E, Ruckenstein AE. Thermodynamic and kinetic modeling of transcriptional pausing. Proc Natl Acad Sci U S A. 2006;103:4439–4444. doi: 10.1073/pnas.0600508103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ariza X, Bou V, Vilarrasa J. A new Route to 15N-labeled, N-alkyl, and N-Amino Nucleosides via N-Nitration of Uridines and Inosines. J Am Chem Soc. 1995;117:3665–3673. [Google Scholar]
- 26.Pitsch S, Weiss PA, Jenny L, Stutz A, Wu XL. Reliable chemical synthesis of oligoribonucleotides (RNA) with 2′-O-[(triisopropylsilyl)oxy]methyl(2′-O-tom)-protected phosphoramidites. Helvetica Chimica Acta. 2001;84:3773–3795. [Google Scholar]
- 27.Wenter P, Pitsch S. Synthesis of selectively 15N-labeled 2′-O-{[(triisopropylsilyl)oxy]methyl}(=tom)-protected ribonucleoside phosphoramidites and their incorporation into a bistable 32mer RNA sequence. Helvetica Chimica Acta. 2003;86:3955–3974. [Google Scholar]
- 28.Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen C, Russu IM. Sequence-Dependence of the Energetics of Opening of AT Base Pairs in DNA. Biophys J. 2004;87:1–7. doi: 10.1529/biophysj.104.045179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Plateau P, Gueron M. Exchangeable proton NMR without base-line distortion, using new strong-pulse sequences. J Am Chem Soc. 1982;104:7310–7311. [Google Scholar]
- 31.Mori S, Abeygunawardana C, Johnson MO, van Zijl PCM. Improved Sensitivity of HSQC Spectra of Exchanging Protons at Short Interscan Delays Using a new Fast HSQC(FHSQC) Detection Scheme That Avoids Water Saturation. J Magn Reson B. 1995;108:94–98. doi: 10.1006/jmrb.1995.1109. [DOI] [PubMed] [Google Scholar]
- 32.Chen C, Jiang L, Michalczyk R, Russu IM. Structural Energetics and Base-Pair Opening Dynamics in Sarcin-Ricin Domain RNA. Biochemistry. 2006;45:13606–13613. doi: 10.1021/bi060908n. [DOI] [PubMed] [Google Scholar]
- 33.Lippens G, Dhalluin C, Wieruszeski J-M. Use of a water flip-back pulse in the homonuclear NOESY experiment. J Biomol NMR. 1995;5:327–331. doi: 10.1007/BF00211762. [DOI] [PubMed] [Google Scholar]
- 34.Englander SW, Kallenbach NR. Hydrogen exchange and structural dynamics of proteins and nucleic acids. Q Rev Biophys. 1984;16:521–655. doi: 10.1017/s0033583500005217. [DOI] [PubMed] [Google Scholar]
- 35.Weast RC. CRC Handbook of Chemistry and Physics. 67. CRC Press; Boca Raton, FL: 1987. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.