Abstract
Arg96 is a highly conservative residue known to catalyze spontaneous green fluorescent protein (GFP) chromophore biosynthesis. To understand a role of Arg96 in conformational stability and structural behavior of EGFP, the properties of a series of the EGFP mutants bearing substitutions at this position were studied using circular dichroism, steady state fluorescence spectroscopy, fluorescence lifetime, kinetics and equilibrium unfolding analysis, and acrylamide-induced fluorescence quenching. During the protein production and purification, high yield was achieved for EGFP/Arg96Cys variant, whereas EGFP/Arg96Ser and EGFP/Arg96Ala were characterized by essentially lower yields and no protein was produced when Arg96 was substituted by Gly. We have also shown that only EGFP/Arg96Cys possessed relatively fast chromophore maturation, whereas it took EGFP/Arg96Ser and EGFP/Arg96Ala about a year to develop a noticeable green fluorescence. The intensity of the characteristic green fluorescence measured for the EGFP/Arg96Cys and EGFP/Arg96Ser (or EGFP/Arg96Ala) was 5- and 50-times lower than that of the nonmodified EGFP. Intriguingly, EGFP/Arg96Cys was shown to be more stable than EGFP toward the GdmCl-induced unfolding both in kinetics and in the quasi-equilibrium experiments. In comparison with EGFP, tryptophan residues of EGFP/Arg96Cys were more accessible to the solvent. These data taken together suggest that besides established earlier crucial catalytic role, Arg96 is important for the overall folding and conformational stability of GFP.
Keywords: green fluorescent protein, enhanced green fluorescent protein, fluorescent protein, point mutation, chromophore structure, conformational stability, circular dichroism
INTRODUCTION
Green fluorescent protein, GFP, from jellyfish Aequorea victoria, is a member of a family of colored globular proteins known for their unique ability to spontaneously synthesize their own chromophore from three buried residues.1–3 Crystallographic structures resolved for the wild-type GFP, a protein consisting of 238 amino acid residues, and its enhanced colored mutants (ECFP, EGFP, and EYFP)1,2,4 revealed that this protein resembles an 11 stranded β-can wrapped around a single central helix in the middle of which is the chromophore [see Fig. 1(A)]. There are also short helical segments on the end of the β-can. The cylinder has a diameter of about 30 Å and a length of about 40 Å.2 Several physical–chemical studies have been performed on the GFP chromophore formation and fluorescence emission (e.g., see Refs. 7–12). Furthermore, it has been shown that GFP fluorescence is stable under a variety of conditions, including a treatment with detergents,13,14 proteases,15 GdmCl,16,17 and temperature.17,18
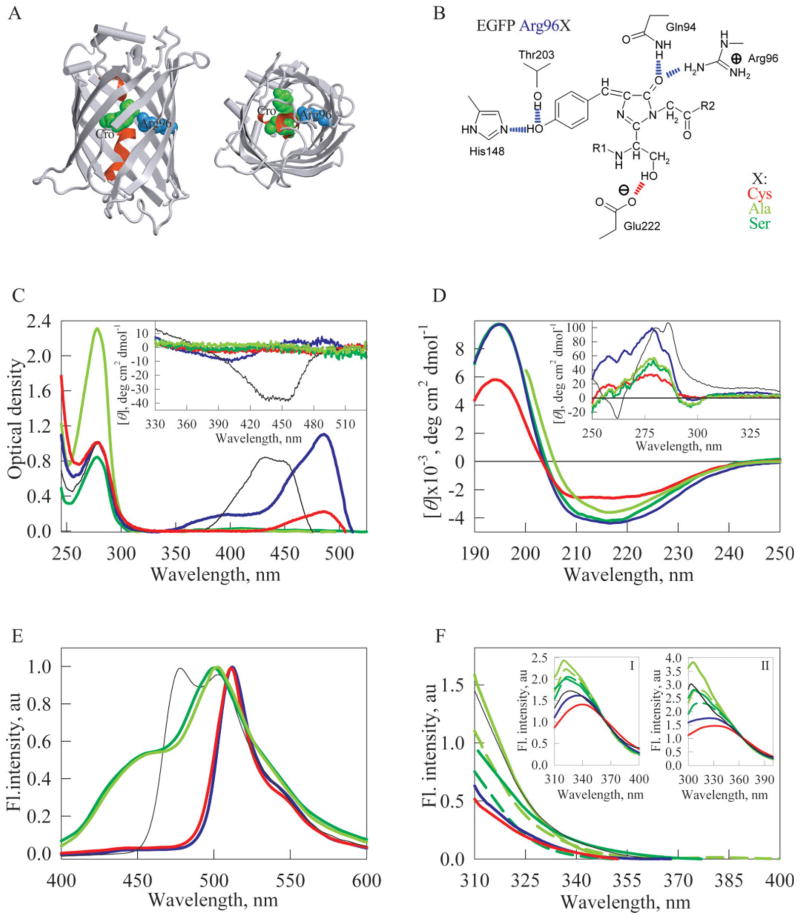
Figure 1.
Spectral properties of the original EGFP and its mutant variants. (A) X-ray crystal structure of Aequorea GFP with S65T and Q80R substitutions [PDB code 1EMA] in two projections. Chromophore is shown as a green space-filling union. A central α-helix which includes chromophore is shown in red. The residue Arg96 is shown in blue. The drawing was generated by the graphic programs VMD5 and Raster3D.6 (B) Some of the interactions of fluorescent protein’s chromophore with the surrounding side chains. Hydrogen bonds are shown in blue, except for the one presumably stabilizing the respective form of the chromophore, which is shown in red. Modified from [Silke Jonda’s Principles of Protein Structure Using Internet project entitled “Structure and Function of GFP” updated on 28.11.96 (http://www.cryst.bbk.ac.uk/PPS2/projects/jonda/)]. (C) UV/VIS absorbance spectra of EGFP (blue line), EGFP/Arg96Cys (red line), EGFP/Arg96Ala (light green line), EGFP/Arg96Ser (green line). Inset represents near UV/VIS spectra of these proteins. (D) Far-UV CD spectra of EGFP (blue line), EGFP/Arg96Cys (red line), EGFP/Arg96Ala (light green line), EGFP/Arg96Ser (green line). Inset: near-UV CD spectra of these proteins. (E) Fluorescence emission spectra of EGFP (blue line), EGFP/Arg96Cys (red line), EGFP/Arg96Ala (light green line), EGFP/Arg96Ser (green line). Spectra have been normalized to have similar maximal intensity. (F) Tryptophan fluorescence spectra of EGFP (blue line), EGFP/Arg96Cys (red line), EGFP/Arg96Ala (light green line), EGFP/Arg96Ser (green line). For EGFP/Arg96Ala and EGFP/Arg96Ser spectra have been measured just after purification (dashed line) and after the incubation for 1 year (solid line). λex = 297 nm.
Many mutants of GFP with useful characteristics such as enhanced fluorescence intensity,19,20 shifted emission wavelength,1,21 and pH-sensitive fluorescence changes22 have been described. The unrivalled capability of FPs to emit a visible light without the use of any substrate determines their wide use as specific reporters in studies on gene expression, protein dynamics, and localization within the living cells.3,15 The discovery of a red GFP-like protein DsRed23,24 from corallomorph Discosoma sp., and development of its improved mutants, DsRed-Timer,25 DsRed2,26 and fast-maturating DsRed-Express,27 have significantly increased the range of FP applications including multicolor protein tagging,28 intracellular reporting,29 and resonance energy transfer.30 The GFP-based Ca2+ indicators have been elaborated, which comprise the camgaroo and pericam probes based on a circularly permutated GFP.31 Another type of Ca2+ indicators, called chameleons, are genetically-encoded fluorescent indicators for Ca2+ based on green fluorescent protein variants and calmodulin (CaM).32,33 These chameleons are chimeric proteins consisting of a blue or cyan mutant of green fluorescent protein (GFP), calmodulin (CaM), a glycylglycine linker, the CaM-binding domain of myosin light chain kinase (M13), and a green or yellow version of GFP.32,33 Binding of Ca2+ makes calmodulin wrap around the M13 domain, increasing the fluorescence resonance energy transfer (FRET) between the flanking GFPs. These chameleon probes have been found to be useful in for intracellular calcium measurements.31–33
The remarkable feature of GFP is a unique chromophore, a p-hydroxybenzylidene-imidazolidone, which is located almost at the center of the cylinder [Fig. 1(A)]. It is completely protected from the bulk solvent, which makes it difficult for an enzyme to access the chromophore and catalyze its formation, underlining the hypothesis of the autocatalytic cyclization of the polypeptide backbone.1,2,4 Upon protein folding, chromophore formation is initiated by a backbone crosslinking reaction that involves the residues Ser65, Tyr66, and Gly67 in wild-type GFP, leading to the formation of the imidazolidone ring produced by the cyclized backbone of these residues.19,21 Part of the chromophore’s π-system is derived from the phenol of Tyr66, which plays an important function in determining the color of the mature chromophore, and replacement of which with other aromatic residues was shown to lead to visible fluorescence with altered emission wavelengths.3 Although the amino acid sequence Ser-Tyr-Gly could be found in different non-FPs, it is neither cyclized in any of these, nor is the tyrosine oxidized, implying that the tendency to form such a chromophore does not represent the intrinsic property of this tripeptide, and is dependent on its specific local environment.
Intriguingly, a recent study of α-synuclein, a protein involved in Parkinson’s disease and a number of other neurodegenerative diseases known as synucleinopathies,34,35 uncovered that aggregation of this protein is accompanied by the development of a progressive photo-activity in the visible range of the electromagnetic spectrum.36 Some parameters of this photo-activity resembled those typical of the family of green fluorescent proteins. On the basis of these observations it has been hypothesized that the fibrillation-induced photo-activity is governed by the same mechanism as seen for the intrinsic chromophore of 4-(p-hydroxybenzylidene)-5-imidazolinone-type in GFPs and involves several steps of chain cyclization, amino acid dehydration, and aerial oxidation.36
Four residues in the interior of FPs are highly conserved and apparently participate in generating the fluorescent entity: Tyr66, Gly67, Arg96, and Glu222.24 Ty66 and Gly67 are the crucial parts of the chromophore five-membered heterocycle, whereas Arg96 and Glu222 appear to play catalytic roles. In fact, Arg96 and Glu222 are found to be in close proximity to the chromophore in the tertiary structure of the protein [Fig. 1(B)]. The Arg96 guanidinium group is hydrogen bonded to the Tyr66-derived carbonyl oxygen of the chromophore, and the Glu222 carboxylic acid is positioned near the opposite face of the chromophore.3 Arg96 is believed to facilitate the cyclization reaction by increasing the nucleophilic reactivity of the amide nitrogen of Gly67,37–39 whereas the carboxylate of Glu222 was shown to play the role of a general base, facilitating proton abstraction from the Gly67 amide nitrogen or the Tyr66 α-carbon.37 On the other hand, neither Arg96 nor Glu222 is absolutely necessary for chromophore biosynthesis37–40 and proposed functions for these residues range from electrostatic, steric, and catalytic roles to contributions to protein folding and stability.3,41 In this study, we analyzed the effect of the amino acid substitutions at position 96 on structural properties and conformational stability of EGFP. In addition to the previously established catalytic role, Arg96 is shown here to be crucial for the proper protein folding and stability.
METHODS
Plasmid construction and protein expression
The plasmids encoding original EGFP, as well as its Arg96Cys, Arg96Ser, Arg96Ala, and Arg96Gly mutants with polyhistidine tags, were constructed as previously described.29 Briefly, the 0.75-kb NcoI-BamHI fragment of EGFP was amplified from the pEGFP-N1 plasmid (BD Clontech) using polymerase chain reaction (PCR), and cloned into the pET-11d vector (Invitrogen). The polyhistidine tag was added to the C-termini of the protein in the course of PCR-amplification. The site-directed mutagenesis of Arg96Cys, Arg96Ser, Arg96Ala, and Arg96Gly of the EGFP gene was performed using Quik-Change Site-Directed Mutagenesis kit (Stratagene), and verified by direct sequencing.
The resulting plasmids, pET-11d-EGFP, pET-11d-EGFP/96Cys, pET-11d-EGFP/96Ser, pET-11d-EGFP/96Ala, and pET-11d-EGFP/96Gly were transformed into an E. coli BL21(DE3) host (Invitrogen). Protein expression was induced by an incubation of the cells with 0.5 mM IPTG (isopropyl-β-D-thiogalactopyranozide, Sigma) at 37°C for 6 h, and the proteins were purified with Ni-NTA agarose (Qiagen). The protein solutions were concentrated to 12 mg/mL in 100 mM citrate-phosphate buffer, pH 7.6, using Ultrafree-4 centrifugal filters (Millipore). The purity of the recombinant proteins was not less than 95%, as indicated by SDS-PAGE. Concentrations of FPs have been determined by the Bio-Rad protein assay kit.
Fluorescence spectroscopy
To analyze the characteristic green fluorescence, original EGFP, EGFP/Arg96Cys, EGFP/Arg96Ser, EGFP/Arg96Ala, and EGFP/Arg96Gly were excited at 365 nm, and emission was detected at 510 nm. Intrinsic tryptophan fluorescence was excited at 280 or 297 nm (to exclude the tyrosine contribution). The position and form of the fluorescence spectra were characterized by the parameter: A = (I320/I365)297, where I320 and I365 are fluorescence intensities at λem = 320 and 365 nm, respectively, and λex = 297 nm.42 The values of parameter A and of fluorescence spectrum were corrected by the instrument sensitivity. The contribution of tyrosine residues to the bulk fluorescence of protein usually is characterized by the value42:
| (1) |
where I365 is fluorescence intensity at 365 nm and subscripts 280 and 297 indicate excitation wavelength. The fluorescence spectrophotometers described in Ref. 42 or FluoroMax-2 (Jobin Yvon) were used for steadystate spectroscopic analysis. Measurements were performed with temperature of samples adjusted to 25°C.
Analysis of the fluorescence decay
The fluorescence decay curves were recorded with homemade spectrofluorimeter with time-resolved excitation.42 To analyze the decay curves a special program was developed. The fitting routine was based on the nonlinear least-squares method. Minimization was accomplished according to Marquardt.43 P-terphenyl in ethanol and N-acetyl-tryptophanamide in water were used as reference compounds.44 Experimental data were analyzed using the multiexponential approach:
| (2) |
where αi and τi are amplitude and lifetime of component i, Σαi = 1. The root-mean square value of fluorescent lifetimes, 〈τ〉, for multiexponential decay is determined as , where fi = αiτi/Σi αiτi.
Stern-Volmer quenching and estimation of the bimolecular quenching rates
The conformational state of proteins was further characterized by acrylamide-induced fluorescence quenching. Samples were prepared in 50 mM TrisHCl buffer, pH = 8.0, 150 mM NaCl, pH 7.5 and the protein concentrations were adjusted to provide an optical density at the excitation wavelength less than 0.1. Aliquots of a 3.0M acrylamide stock solution were consecutively added to 1 mL protein solution in order to increase acrylamide concentration. Experiments were performed using excitation at 297 nm with fluorescence emission at 340 nm (for the Trp accessibility determination) and excitation at 365 nm with fluorescence emission at 510 nm (for the Cro accessibility evaluation), and the fluorescence intensities were recorded for 30 s. Experiments were performed in triplicate, and the data were corrected for the dilution effects. Quenching data were plotted as the ratio of fluorescence in the absence of quencher (I0) to the intensity in the presence of quencher (I) against quencher concentration. The resulting data were fit to dynamic parameters according to the Stern-Volmer equation I0/I = 1 + KSV[Q], where KSV is the Stern-Volmer quenching constant and [Q] the quencher concentration.45 The bimolecular quenching rates, kq, have been calculated from KSV and fluorescence lifetimes, <τ>, as kq = KSV/<τ>(M−1 s−1).45
Circular dichroism measurements
CD spectra were obtained with J-810 spectropolarimeter (Jasco, Tokyo, Japan) equipped with the Neslab RTE-110 temperature-controlled liquid system (Neslab Instruments, Portsmouth, NH). The instrument was calibrated with a standard solution of (+)-10-camphorsulfonic acid. Sealed cuvettes with a path length of 0.1 cm (Helma, Germany) were used. The photomultiplier voltage never exceeded 600 V in the spectral regions that were measured. Each spectrum was averaged five times and smoothed with spectropolarimeter system software, version 1.00 (Jasco). All measurements were performed under a nitrogen flow. Before undergoing CD analyses, all samples were kept at the temperature being studied for 10 min. Protein concentration was ~0.25 and 0.5 mg/mL for far and near UV CD measurements, respectively. Far UV CD spectra were recorded in a 0.1 mm pathlength cell from 250 to 190 nm with a step size of 0.5 nm, a bandwidth of 1.5 nm, and an averaging time of 10 s. Near UV CD spectra were recorded in a 1.0 cm pathlength cell from 600 to 250 nm with a step size of 1.0 nm, a bandwidth of 1.5 nm, and an averaging time of 10 s. CD spectra of the appropriate buffers were recorded and subtracted from the protein spectra.
Kinetics measurements of protein unfolding
All kinetic experiments were performed in microcells 101.016-QS 5 mm × 5 mm (Hellma, Germany). Unfolding of the protein was initiated by manual mixing of protein solution with buffer containing desired GdmCl concentrations. About 350 μL of the GdmCl solution of appropriate concentration was injected in the cell with 50 μL of the solution of native protein. The dead time was determined from the control experiments and was determined to be about 4 s. The spectrofluorimeter was equipped with thermostat that held a constant temperature of 25°C in the cell and in the special box where the solutions are held before mixing.
Kinetic curve was detected in the continuous regime for first 10 min after the mixing the protein and GdmCl solutions. Then, data were taken periodically during the first 120 h after the beginning of the unfolding reaction. All these data were used for the unfolding-refolding rate constant determination. Analysis of the kinetic curves was performed assuming the one step transition:
| (3) |
Here, N is the native state, U is the unfolded state, k1 and k2 are the rate constants of the forward and back reactions, respectively. The relative fluorescence intensity I(t) is determined as follows:
| (4) |
where I∞ is fluorescence intensity at the infinity time and ΔI and kobs are the amplitude and the observed rate constant, respectively. When model (3) failed to fit measured kinetic dependencies, those dependencies were analyzed assuming the existence of intermediate state(s)46:
| (5) |
The values of the rate constants ki, or kobs were determined by the nonlinear least-squares method, as the values that fit the minimum value:
| (6) |
Here, Iexp(t) and I(t) are experimental and calculated values of relative intensity, respectively.
GdmCl-induced equilibrium unfolding
Protein samples were incubated for the desired amounts of time at 25°C in the presence of various concentrations of GdmCl. Unfolding curves were determined by monitoring the fluorescence spectra at 25°C. The pH was checked to ensure a constant value throughout the whole transition, and the denaturant concentration was determined from refractive index measurements,47 using an Abbe-3L refractometer from Spectronic Instruments.
Protein stability in the absence of denaturant ΔG0([0]) was determined using the following relationship48:
| (7) |
where [D] is the denaturant concentration. In the middle of transition [D] = [D]50%, ΔG0([D]) = 0 and therefore,
| (8) |
The ΔG0([D]) value is connected with the equilibrium constant K as:
| (9) |
where T is the absolute temperature and, R is the universal gas constant.
Protein stabilities were evaluated from the dependencies of the green fluorescence on denaturant concentration, which can be described as:
| (10) |
Equations (7)–(10) were used to derive relationship that was used to fit the quasi-equilibrium dependencies of fluorescence intensity:
| (11) |
Approximation of experimental data was performed via the nonlinear regression method using Sigma Plot program. Characteristic feature of the FP unfolding is a very low rate of their N → U transitions. Therefore, the shape of the denaturation curves depended significantly on the incubation time. Denaturation curves were characterized by the C1/2(t) values, which are the denaturant concentrations, corresponding to the middle of transition curves measured after the incubation of a protein in the presence of various denaturant concentrations for the time t. These data were used to plot C1/2 versus incubation time dependencies. Thermodynamic parameters were evaluated from the denaturation curves retrieved after the incubation of the corresponding solutions for time ti, which corresponds to the incubation time at which the C1/2(t) curve reaches the plateau. In practice, the thermodynamic constants were calculated using the quasi-equilibrium dependencies obtained after the incubation of a given protein in the presence of various GdmCl concentrations for 5 days.
RESULTS AND DISCUSSION
Expression and purification of original EGFP and EGFP/Arg96Cys, EGFP/Arg96Ser, EGFP/Arg96Ala, and EGFP/Arg96Gly mutants from bacteria
In our studies, besides original EGFP, a high yield of green protein was achieved only for the EGFP/Arg96Cys mutant. The yields of EGFP/Arg96Ser and EGFP/Arg96Ala mutants were essentially lower and these proteins were not colored. No protein was produced when the pET-11d-EGFP/Arg96Gly plasmid was transformed into an E. coli and protein expression was induced.
Kinetics of the chromophore maturation in original EGFP, EGFP/Arg96Cys, EGFP/Arg96Ser, and EGFP/Arg96Ala mutants
When original EGFP and its mutants were purified, we attempted to analyze the rate of the chromophore maturation. To do so, such spectral properties as absorption spectra, tryptophan and green fluorescence spectra were tested twice a week for 3 weeks. Both, the original EGFP and its EGFP/Arg96Cys mutant possessed the characteristic absorption spectra and the characteristic green fluorescence immediately after the purification and none of their spectral properties changed during the repetitive measurements, suggesting that the chromophore maturation is a relatively fast process for these two proteins. As it has been mentioned earlier, even being incubated for a about year EGFP/Arg96Ser and EGFP/Arg96Ala mutants did not develop noticeable absorption in the visible region neither just after purification, nor after a year of storage. However, after this prolonged incubation, these mutants were shown to possess some characteristic green fluorescence. This indicates that the rate of chromophore formation is significantly altered in these proteins, which is in accord with earlier studies.37,39 It is important to remember that fluorescence can be reliably detected even in a case when no detectable absorption spectrum is observed. This is due to the fact that optical density measurements rely on the detection of a small difference between two large signals which are proportional to the intensities of the light beams passing through the sample and reference cells. Registration of weak fluorescence implies registration of a low signal in the “dark field”. In a spectrofluorometer with an optimized alignment of the excitation and emission channels, no exciting light reaches photo-receiver. This creates the mentioned “dark field” which allows registration of a very weak fluorescent signal, eventually even observation of single fluorescence photons.
Effect of Arg96 substitutions on spectral properties of EGFP
Figure 1(C) represents UV/VIS absorbance spectra of original EGFP and its Arg96Cys, Arg96Ser, and Arg96Ala mutants. Figure 1(C) shows that all the proteins were characterized by comparable UV absorption spectra. Among the EGFP mutants studied, only Arg96Cys had a noticeable absorption band in the visible region (λmax = 485 nm), whereas EGFP/Arg96Ser and EGFP/Arg96Ala did not absorb in this region even after the 1 year incubation. The absorption of the EGFP/Arg96Cys in the visible region was noticeably lower (~5 times) than that of the original EGFP.
Insets to Figure 1(C,D) show a considerable difference in the near-UV/visible CD spectra measured for the proteins studied, with the most dramatic discrepancy being observed in the visible region. Dichroic bands of FPs in the visible region are known to have small negative Cotton effects, and are visible at high optical density of the chromophore.49 In agreement with earlier studies, a comparison of the absorption and visible CD spectra for the original EGFP indicates that although the absorption band at 400 nm has much smaller oscillator strength than the one arising at 490 nm, the rotational strength of bands show the opposite behavior [Fig. 1(C), inset]. These data indicate that the chromophore microenvironment in EGFP is asymmetric. Visible CD spectrum of the EGFP/Arg96Cys mutant differs significantly from that of EGFP, for example, its CD band at 490 nm is more pronounced than the CD band at 400 nm. This suggests that the introduction of the Arg96 to Cys substitutions introduces noticeable changes in the chromophore microenvironment asymmetry. Visible CD spectra of the EGFP/Arg96Ser and EGFP/Arg96Ala mutants were not recorded because of the very low optical density of their solutions in this spectral region (see above).
Inset to Figure 1(D) shows that the aromatic residues in the original EGFP is characterized by the rigid and unique environment. Near-UV CD spectra of EGFP/Arg96Cys, EGFP/Arg96Ser, and EGFP/Arg96Ala, being significantly less intensive than the spectrum of the original protein, have a set of peaks similar to that of EGFP. This indicates that the amino acid substitutions do induce noticeable alterations in the rigid tertiary structure of EGFP, with the Arg96Cys mutation inducing the largest structural distortion. The fact that the near-UV CD spectra of the original EGFP and its Arg96Cys, Arg96Ser, and Arg96Ala mutants are characterized by a similar fine structure (all of them have a similar set of peaks) indicates that these mutations likely affect the rigidity of the environment of aromatic residues rather than induce structural changes.
Figure 1(D) shows that Arg96Ser and Arg96Ala mutations do not affect secondary structure of the protein to a significant degree. This is manifested by the almost indistinguishable far-UV CD spectra measured for original EGFP and EGFP/Arg96Ser and EGFP/Arg96Ala. However, the far-UV CD spectrum of the EGFP/Arg96Cys indicates that this amino acid substitution does induce noticeable alterations in the EGFP secondary structure, as reflected in the profound changes in the spectral shape.
As it follows from the almost identical shapes of the EGFP and EGFP/Arg96Cys green fluorescence spectra, Arg96Cys mutation does not affect the chromophore structure [see Fig. 1(E)]. However, the fluorescence intensity of EGFP/Arg96Cys is approximately 5 times lower than that of the original EGFP. This likely is due to the lower absorption in the visible spectrum. It has been noted that the maturation of the chromophore in the EGFP/Arg96Ser and EGFP/Arg96Ala mutants was an extremely slow process, as they possessed noticeable green fluorescence only after the incubation for a year. The fluorescence of these mutants was ~50 lower than the green fluorescence of the original EGFP. Interestingly, the fluorescence maximum of these mutants was shifted from 512 to 500–504 nm and coincided with the position of the long-wavelength maximum of the ECFP fluorescence (502 nm, not shown).
Figure 1(F) shows fluorescence spectra of single Trp residue of EGFP and its mutants. The Trp fluorescence spectrum of EGFP/Arg96Cys mutant is red-shifted, whereas fluorescence spectra of the EGFP/Arg96Ser and EGFP/Arg96Ala mutants are blue-shifted in comparison with that of EGFP. The Trp fluorescence spectra of EGFP/Arg96Ala mutant measured after the incubation for 1 year was blue-shifted in comparison with the spectra measured just after this protein was purified (see also Table I). This reflects the existence of the time-dependent changes in the environment of tryptophan residue; that is, very slow folding process.
Table I.
Fluorescence Characteristics of the Original EGFP and Its Mutant Variants
| Timea | Tryptophanb |
Green chromophorec |
|||||
|---|---|---|---|---|---|---|---|
| λmax (nm) | Parameter Ad | 〈τ〉e (ns) | λmax (nm) | Imax | 〈τ〉f (ns) | ||
| EGFP | 1,2 | 336 | 1.49 | 4.0 ± 0.2 | 512 | 1 | 2.6 ± 0.3 |
| EGFP/Arg96Cys | 1,2 | 340 | 1.18 | 4.2 ± 0.2 | 512 | 0.2 | 2.5 ± 0.3 |
| EGFP/Arg96Ser | 1 | 324 | 2.07 | –– | –– | –– | |
| 2 | 323 | 2.08 | 2.7 ± 0.3 | 500 | 0.02 | 1.8 ± 0.4 | |
| EGFP/Arg96Ala | 1 | 323 | 2.34 | –– | –– | –– | |
| 2 | 320 | 2.52 | 2.2 ± 0.3 | 502 | 0.02 | 2.8 ± 0.4 | |
In column “time” 1 indicates that experiment was performed just after purification, 2 indicates that experiment was performed after samples incubation for one year.
Tryptophan fluorescence was excited at 297 nm.
Green chromophore fluorescence was excited at 365 nm.
A = (I320/I365)297, where I320 and I365 are fluorescence intensities at λem 320 and 365 nm, respectively, and λex 297 nm.
The lifetime of tryptophan fluorescence decay was determined upon fluorescence recording at 340 nm.
The lifetime of green chromophore fluorescence decay was determined upon recording fluorescence at wavelength corresponding to maximum of emission spectrum.
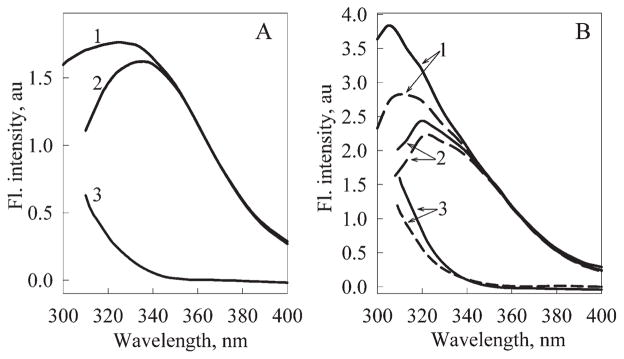
Figure 2 shows that intrinsic fluorescence spectra measured at the excitation at 280 and 297 nm are different at their short-wavelength edges. This is due to the fact tyrosine residues contribute to the total fluorescence excited at 280 nm. In addition to a single Trp, EGFP contains 9 Tyr residues. The major contribution to the total fluorescence is provided by Tyr 39, Tyr 74, Tyr 151, Tyr 182, and Tyr 200 residues as, according to our calculations based on the analysis of the crystallographic data, whereas other Tyr residues transfer their excitation energy to Trp 57 rather efficiently. The efficiency of energy transfer from Tyr 143, Tyr 145, Tyr 92, and Tyr 106 to Trp 57 is of 96, 91, 49 and 47% respectively. There is practically no energy transfer between the Tyr residues.
Figure 2.
The fluorescence spectra excited at 280 nm (curves 1) and 297 nm (curves 2) and the contribution of the Tyr residues (curves 3) to the bulk fluorescence of EGFP (panel A) and EGFP/Arg96Ala (panel B). For EGFP/Arg96Ala spectra have been measured just after purification (dashed line) and after the incubation for 1 year (solid line).
Interestingly, Tyr fluorescence was utilized recently for the characterization of the GFP denaturation processes.50 However, it is important to remember that there is a significant potential contribution of the Trp residue to the total fluorescence signal under conditions used in that study (excitation at 276 nm and registration at 307 nm). Usually, the contribution of the tyrosine residues to the total protein fluorescence is described by the Δ(λ) parameter values [see Eq. (1)]. However, as I(λ) = I(λ)Tyr + I(λ)Trp, then Δ(λ) = (I(λ)Tyr/I365)280. This means that the Δ(λ) value is determined not only by the intensity of Tyr fluorescence but it also depends on the intensity of the Trp fluorescence (I365). In other words, the value of the Δ(λ) parameter might reflect the changes in the contribution of the tyrosine residues to the total fluorescence only in a case when Trp fluorescence remains unchanged. Thus, this parameter cannot be utilized to characterize the Tyr contribution unambiguously if the Trp fluorescence in the analyzed protein states is different. Therefore, the increase in the Δ(λ) values for EGFP/Arg96Ala [Fig. 2(B)] and EGFP/Arg96Ser (data not shown) can be due not only to the increase in the Tyr fluorescence contribution (usually, the contribution of Tyr residues increases upon unfolding as the efficiency of the energy transfer from Tyr to Trp decreases), but also can be determined by the decrease in the Trp contribution, for example, due to the increase in the efficiency of Trp to Cro energy transfer.
Accessibility of a green chromophore and a Trp residue to solvent
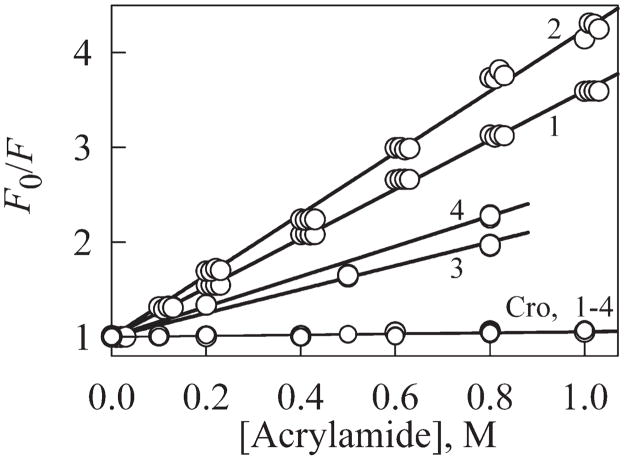
To get an idea about changes in the chromophore and tryptophan accessibility to solvent induced by the Arg96 substitution, the efficiency of acrylamide quenching of green and tryptophan fluorescence has been analyzed for the original EGFP and EGFP/Arg96Cys, EGFP/Arg96Ser, and EGFP/Arg96Ala mutants. Figure 3 and Table II show that chromophore is practically inaccessible to quencher in all proteins, that is, this point mutations did not affect the accessibility of the green chromophore to solvent.
Figure 3.
Acrylamide quenching of tryptophan fluorescence (curves 1–4) and chromophore fluorescence (curves Cro, 1–4) for EGFP, EGFP/Arg96Cys, EGFP/Arg96Ala, EGFP/Arg96Ser, curves 1, 2, 3, and 4, respectively. Quenching experiments were carried out as described under “Materials and Methods.” The slopes of the regression lines, corresponding to the Stern-Volmer constants (KSV), are given in Table II.
Table II.
Quenching Constants of the Original EGFP and Its Mutant Proteins
| Tryptophan accessibility |
Chromophore accessibility |
|||
|---|---|---|---|---|
| KSV (M−1) | kq (108M−1 s−1) | KSV (M−1) | kq (108M−1 s−1) | |
| EGFP | 2.59 ± 0.03 | 6.5 ± 0.4 | 0.06 ± 0.01 | 0.23 ± 0.07 |
| EGFP/Arg96Cys | 3.24 ± 0.05 | 7.7 ± 0.5 | 0.05 ± 0.01 | 0.19 ± 0.06 |
| EGFP/Arg96Ser | 1.59 ± 0.01 | 5.9 ± 0.6 | 0.05 ± 0.02 | 0.30 ± 0.07 |
| EGFP/Arg96Ala | 1.25 ± 0.04 | 5.6 ± 0.8 | 0.06 ± 0.01 | 0.22 ± 0.07 |
Tryptophan residues in EGFP/Arg96Cys were more solvent accessible as it follows from the quenching data (Fig. 3, Table II), positions of the tryptophan fluorescence excited at 297 nm [Fig. 1(F), Table I] and the parameter A values (Table I). These data show that the EGFP tryptophan fluorescence spectrum is characterized by the shorter wavelength maximum position in comparison with EGFP/Arg96Cys (A297EGFP = 1.49; A297EGFP/Arg96Cys = 1.18).
Table II shows that the mutations (except to Arg96Ser) generally do not affect the lifetime of green fluorescence. In fact, all measured lifetimes are in a good agreement with a value of 3 ns reported earlier for the fluorescence lifetime of wild-type GFP and GFP/Ser65Thr.51 The lifetime of tryptophan fluorescence was not affected only in a case of the Arg96Cys mutation, whereas all other substitutions induced measurable decrease in this parameter. These data as well as the rather blue-shifted intrinsic fluorescence spectra suggest that the microenvironments of Trp in these mutants are different from the Trp microenvironment in EGFP. The measured values of the fluorescence lifetime were used to estimate the bimolecular quenching rates for EGFP and its mutant (see above).
Effect of Arg96 substitution on conformational stability of EGFP
A unique characteristic feature of the green florescent proteins unfolding is that this process is very slow.15,50,52,53 Therefore, the kinetic measurements were performed for 5 days. Kinetic curves describing the FP unfolding at low GdmCl concentrations cannot be approximated by a mono-exponential fluorescence decay law. Likely, this reflects the fact that unfolding is accompanied by the formation of several intermediate states.46,50,52 In the presence of high denaturant concentrations (5.6M GdmCl), the processes of the intermediate formation and unfolding are fast and significantly overlap. As a result, corresponding kinetic curves are approximated well by a single exponent. Corresponding unfolding rate constants are shown in Table III.
Table III.
Kinetic Rates of EGFP and Its Mutant Proteins Denaturation at 5.6M GdmCl
| kapp (10−3 s−1) | |
|---|---|
| EGFP | 3.9 ± 0.1 |
| EGFP/Arg96Cys | 2.0 ± 0.1 |
| EGFP/Arg96Ser | 5.3 ± 0.1 |
| EGFP/Arg96Ala | 4.8 ± 0.1 |
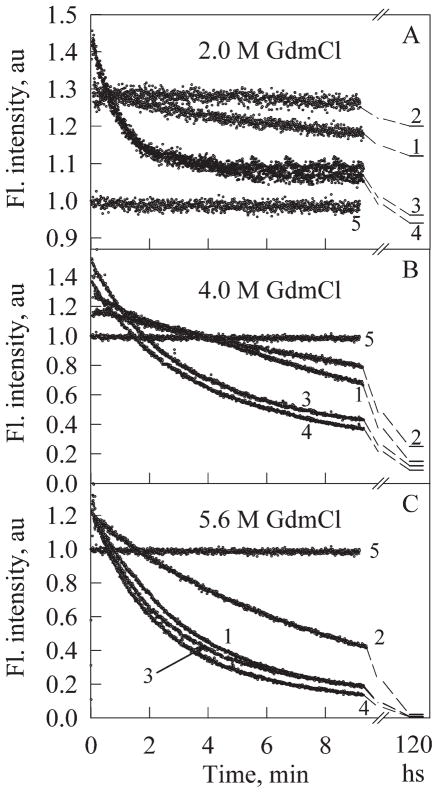
Kinetic analysis revealed that the EGFP/Arg96Cys possesses higher resistance toward the GdmCl unfolding than the original EGFP (Fig. 4 and Table III). It is seen that at 5.6M GdmCl it is required correspondingly 2.7 min to reach the 50%-reduction of the EGFP green fluorescence intensity. In the case of EGFP/Arg96Cys, this value is almost tripled (7.8 min). Interestingly, the green fluorescence intensity observed immediately after the mixing of a fluorescent protein with a denaturant solution exceeds significantly the fluorescence level of a given protein in the absence of denaturant. This effect was observed for all proteins studied and was essentially more pronounced when protein was transferred to a solution containing low GdmCl concentrations. This might be determined by the fact that when protein is transferred to the solution with high denaturant concentration both fast initial increase in the fluorescence intensity and the subsequent fast decrease in green fluorescence caused by protein unfolding might rapidly occur during the experiment dead-time. The initial increase in the fluorescence intensity is then accompanied by the monotonous decrease likely determined by the unfolding-induced increase in the chromophore accessibility to solvent.16
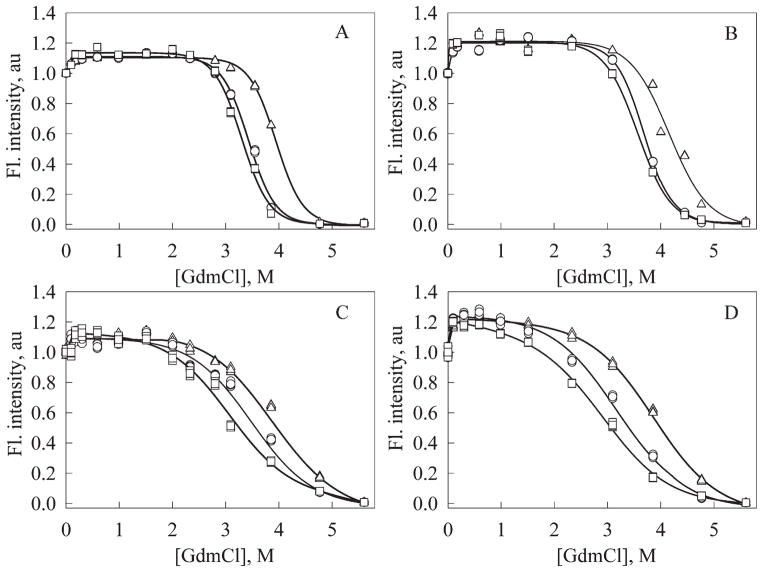
Figure 4.
Kinetics of green fluorescence changes accompanying protein unfolding in 2.0M (A), 4.0M (B), and 5.6M GdmCl (C) for EGFP (1), EGFP/Arg96Cys (2), EGFP/Arg96Ala (3), EGFP/Arg96Ser (4). Kinetic curves for each protein have been normalized to its fluorescence intensity in native state. Curves 5 in Figures 4(A–C) are kinetic dependencies for EGFP in 0M GdmCl.
Additional proof of the enhanced conformational stability of EGFP/Arg95Cys followed from the analysis of quasi-equilibrium curves of their GdmCl-induced unfolding (see Fig. 5). As the establishing of the unfolding equilibrium in FPs is very slow process [see below and Refs. 17, 50, 52, and 53], data for this plot have been accumulated after the incubation of both proteins in the presence of desired GdmCl concentration for 5 days. The unfolding curves in Figure 5 reflect the GdmCl-induced changes in fluorescence intensity, measured for a given protein after incubation for the desired amount of time in the presence of the desired GdmCl concentration.
Figure 5.
Conformational stability of original EGFP (A), EGFP/Arg96Cys (B), EGFP/Arg96Ala (C), EGFP/Arg96Ser (D) as manifested by the resistance of proteins toward the GdmCl-induced unfolding monitored by changes in green fluorescence. Proteins have been incubated in the presence of the desired GdmCl concentration for 1 day (triangles), 4 days (circles), and 5 days (squares).
Figure 5 shows that the addition of small concentrations of denaturant (0.1–0.2M GdmCl) induces considerable increase in the green fluorescence intensity. This phenomenon likely has the same nature as discussed earlier, rapid increase in the fluorescence intensity taking place just after the exposure of a protein to the denaturant solution. Similar effects were reported for other proteins earlier. In some cases, this phenomenon was due to the low denaturant concentration-induced dissociation of dimers (e.g. creatine kinase54), in other cases it was due to the so-called GdmCl stabilizing effects.55 This phenomenon can be explained assuming that although low GdmCl concentrations do not induce protein denaturation the denaturant molecules are able to form hydrogen bonds with the carbonyl groups at the protein surface. This new net of hydrogen bonds releases tensions that present in the protein backbone due to the charge–charge interactions. In several cases we detected not only changes in the fluorescence intensity but also observed the blue-shift of the fluorescence maximum likely due to the some compaction of a protein molecule (unpublished data).
Shown in Figure 5 dependencies of the green fluorescence intensity on GdmCl concentrations suggest that under the different denaturant concentrations the fluorescent proteins can be in at least three states: the native state (N), the state N1, which is induced by low GdmCl concentrations and the completely unfolded state (U). Earlier we described the formation of the N1 state for other EGFP mutants and for other fluorescent proteins.17,53 The observed increase in the green fluorescence in the presence of low GdmCl concentrations could be due to denaturant-induced removal of the protein scaffold tension. Under such conditions, the chromophore remains to be nonaccessible to the solvent, but becomes less “twisted” and, thus, more planar. This results in the increased conjugation of the chromophore π-bonds finally leading to the increase in the fluorescence quantum yield.
In a number of recent studies,46,50,52 it is indicated that the denaturation of green fluorescent proteins is accompanied by the formation of several intermediate states. The appearance of such intermediates was attributed to the fact that the structure of GFP has two characteristics that may lead to a rough landscape.52 First, the chromophore is slightly twisted in the native form, whereas it is planar in the unfolded form. Second, the central helix shows many deviations from optimal geometry. These structural GFP peculiarities may be responsible for the noticeable hysteresis between the unfolding and refolding curves. Interestingly, such hysteresis was absent for proteins lacking chromophore.52 As an example, the authors indicated the lack of hysteresis for the unfolding–refolding of the super-folder GFP (sfGFP), a protein that contained an Arg96Ala point mutation.52 However, according to our data, this mutation does not preclude the chromophore formation, but results in the dramatic reduction of its maturation rate.
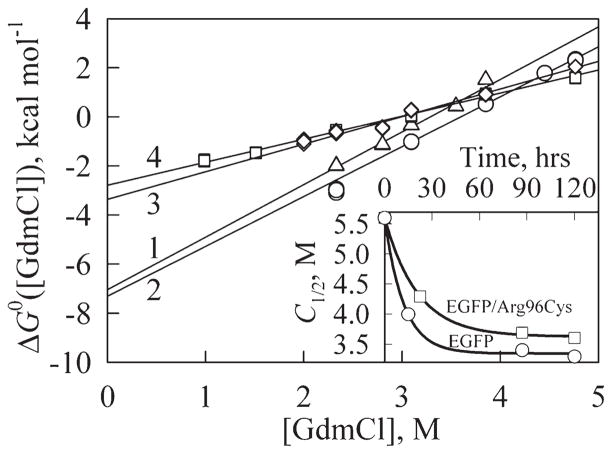
The analysis of quasi-stationary dependencies shown in Figure 5 was performed assuming the all-or-none mechanism for the N1 → U transitions. The value of the C1/2 parameter corresponding to the half-transition point for the N1 → U transition was determined for all unfolding curves. These data were used to plot the dependencies of C1/2 on the incubation time (see inset in Fig. 6). These data suggest that the quasi-equilibrium is reached for the EGFP and EGFP/Arg96Cys within 5 days. We cannot exclude that approaching of equilibrium for EGFP/Arg96Ala and EGFP/Arg96Ser mutants requires longer incubation times. Therefore, values of D50%, m, and parameters shown in Table IV can be overestimated.
Figure 6.
The dependences of ΔG0 on the GdmCl concentration for EGFP (1, triangles), EGFP/Arg96Cys (2, circles), EGFP/Arg96Ala (3, diamonds), EGFP/Arg96Ser (4, squares). The corresponding thermodynamics constants are given in Table IV. Insert represents kinetics of the approaching of unfolding equilibrium determined as C1/2 versus incubation time dependences for EGFP (circles) and EGFP/Arg96Cys (squares).
Table IV.
Thermodynamic Parameters of GdmCl-Induced Unfolding of EGFP and Its Mutant Proteins, as Obtained from the Analysis of the Equilibrium Transitions
| m (kcal mol−1M−1) | D50% (M) | (kcal mol−1) | |
|---|---|---|---|
| EGFP | 2.1 ± 0.2 | 3.3 ± 0.1 | −7.0 ± 0.5 |
| EGFP/Arg96Cys | 2.0 ± 0.2 | 3.6 ± 0.1 | −7.3 ± 0.8 |
| EGFP/Arg96Ser | 1.0 ± 0.2 | 3.1 ± 0.1 | −2.8 ± 0.6 |
| EGFP/Arg96Ala | 1.1 ± 0.2 | 2.8 ± 0.1 | −3.4 ± 0.7 |
Overall, data presented in Figures 4–6 confirm our conclusion that EGFP/Arg96Cys is noticeably more stable toward the GdmCl-induced unfolding than the original EGFP, whereas both EGFP/Arg96Ala and EGFP/Arg96Ser are significantly less stable. For EGFP/Arg96Cys, this is manifested by the increased equilibrium C1/2 value ([D]50% = 3.6 vs. 3.3M GdmCl, see Table IV), and by the increase in the time required to reach the unfolding equilibrium.
CONCLUSIONS
The unique spectral properties of GFP are due to the presence of a specific chromophore, which is assembled by the self-catalyzed covalent modification of amino acids Ser-Tyr-Gly at positions 65–67 to form a p-hydroxybenzylidene-imidazolidinone species backbone.1,2,4 The chromophore, being located inside the protein [see Fig. 1(A)], is not exposed to the solvent. This is in a good agreement with our quenching experiments, which show very low accessibility of the chromophore to acrylamide. However, chromophore interacts with the side chains of many surrounding residues, Gln94, Arg96, His148, Thr203, and Glu222, via hydrogen bonds [Fig. 1(B)]. These interactions determine the spectroscopic behavior of GFP. For example, the basic residues of His148, Gln94, and Arg96 may stabilize and further delocalize the charge on the chromophore.2 On the other hand, the hydroxyl of Thr203 may donate a hydrogen-bond to Tyr66, stabilizing the phenolate form,1 whereas the anionic form of Glu222 inhibits ionization of Tyr66, thus favoring the hydroxyl form for electrostatic reasons.1 The crucial role of the chromophore interactions with these surrounding residues has been confirmed by the extensive mutational analysis. For example, the Glu222Gly variant, which lacks the negative charge of Glu222, favors the phenolate form of Tyr66, and gives rise to a single excitation maximum at 481 nm.10 The Thr203Ile variant was shown to have a single absorption maximum at 400 nm and the same fluorescence emission spectrum as the wild type. This indicates that Thr203 indeed stabilizes the phenolate form of Tyr66, which is responsible for the appearance of the 470 nm absorption band. Since Ile cannot form the hydrogen-bond, Tyr66 is predominantly in its hydroxyl form, absorbing light at 400 nm but not at 470 nm.10
Arg96 and Glu222, being located in the immediate proximity to the GFP chromophore, are highly conserved24 and were proposed to have either electrostatic, steric, or catalytic roles in the chromophore biosynthesis and were suggested to contribute to protein folding and stability.3,41 However, neither Arg96 nor Glu222 are absolutely necessary for chromophore biosynthesis.40 In fact, GFP mutants with the substitutions at these positions were shown to exhibit altered spectral properties.3,10 The role of Arg96 in the chromophore formation was intensively analyzed.4,37–39,56,57 On the basis of this analysis it has been concluded that the major function of this residue is to induce structural rearrangements important in aligning the molecular orbitals for ring cyclization where Arg96 was proposed to facilitate formation of an sp2-hybridized N3 atom through electrostatic destabilization and deprotonation.56 Accordingly, the mutations of Arg96 were shown to greatly retard the GFP chromophore maturation through loss of steric and electrostatic interactions.56,57 The crystal structure of GFP variant Ser65Ala/Tyr66Ser (GFPhal) bearing an additional Arg96Ala mutation was solved at 1.90 Å resolution. The analysis of this structure revealed a noncyclized chromophore conformation and supported a role for Arg96 in organizing the GFP central helix for backbone cyclization.56 Kinetic analysis revealed that the rate of chromophore formation for the EGFP/Arg96Ala variant was greatly attenuated compared to wild-type GFP, occurring on the time frame of weeks to months.56
Subsequent analysis of the Arg96Met mutant revealed that the rate of chromophore formation was greatly reduced possessing the time constant of 7.5 × 103 h at pH 7.0 and exhibited pH dependence.37 This was in contrast to the original EGFP which maturated with a time constant of 1 h in the pH-independent manner (pH 7.0–10.0). In agreement with these earlier observations, we are showing here that it took EGFP/Arg96Ser and EGFP/Arg96Ala mutants about a year to develop a noticeable green fluorescence. Surprisingly, the rate of the chromophore maturation in the EGFP/Arg96Cys was comparable with that of the original EGFP, although the mutant protein possessed reduced fluorescence intensity. Importantly, Arg96Cys mutation did not affect the chromophore structure as it followed from the almost identical shapes of the green fluorescence spectra of the original EGFP and EGFP/Arg96Cys.
Near-UV CD spectra of EGFP/Arg96Cys, EGFP/Arg96Ser, and EGFP/Arg96Ala, being significantly less intensive than the spectrum of the original EGFP, still preserved a set of the characteristic peaks. Therefore, substitutions at the Arg96 position were shown to alter the EGFP rigid tertiary structure. Secondary structure of the protein was not affected by the Arg96Ser and Arg96Ala mutations, whereas Arg96Cys substitution induced noticeable alterations in the EGFP secondary structure.
Finally, both kinetic and quasi-equilibrium experiments revealed that EGFP/Arg96Cys was more stable than the original EGFP toward the GdmCl-induced unfolding. Furthermore, in comparison with EGFP, tryptophan residue of EGFP/Arg96Cys was more accessible to the solvent. Taken together our data suggest that besides established earlier crucial catalytic role, Arg96 is important for the overall folding and conformational stability of GFP.
Acknowledgments
Russian Academy of Science Program MCB, Program of President of Russian Federation “Leading Scientific Schools of Russia” 1961.2008.4, Russian Foundation of Basic Research 07-04-01038 (KKT), Russian Science Support Foundation, Administration of St. Petersburg, (OVS), National Institutes of Health GM070358 and GM073913 (VVV).
Measurements were performed using equipment of the Joint Research Center “Material Science and Characterization in High Technology.”
Abbreviations
- CD
circular dichroism
- Cro
green chromophore
- DsRed
red fluorescent protein
- EGFP/Arg96Cys, EGFP/Arg96Ala, EGFP/Arg96Ser, and EGFP/Arg96Gly
mutant forms of EGFP with point amino acid substitutions of Arg96 to Cys, Ala, Ser, and Gly, respectively
- EGFP
enhanced green fluorescent protein
- EYFP
enhanced yellow fluorescent protein
- ECFP
enhanced cyan fluorescent protein
- FP
fluorescent protein
- GdmCl
guanidine hydrochloride
- GFP
green fluorescent protein
- UV
ultra violet
- VIS
visible
References
- 1.Ormo M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 2.Yang F, Moss LG, Phillips GN., Jr The molecular structure of green fluorescent protein. Nat Biotechnol. 1996;14:1246–1251. doi: 10.1038/nbt1096-1246. [DOI] [PubMed] [Google Scholar]
- 3.Tsien RY. The green fluorescent protein. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 4.Wachter RM, Elsliger MA, Kallio K, Hanson GT, Remington SJ. Structural basis of spectral shifts in the yellow-emission variants of green fluorescent protein. Structure. 1998;6:1267–1277. doi: 10.1016/s0969-2126(98)00127-0. [DOI] [PubMed] [Google Scholar]
- 5.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. 27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 6.Merritt EA, Murphy ME. Raster3D Version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr D Biol Crystallogr. 1994;50(Pt 6):869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- 7.Prendergast FG, Mann KG. Chemical and physical properties of aequorin and the green fluorescent protein isolated from Aequorea forskalea. Biochemistry. 1978;17:3448–3453. doi: 10.1021/bi00610a004. [DOI] [PubMed] [Google Scholar]
- 8.Prasher DC, Eckenrode VK, Ward WW, Prendergast FG, Cormier MJ. Primary structure of the Aequorea victoria green-fluorescent protein. Gene. 1992;111:229–233. doi: 10.1016/0378-1119(92)90691-h. [DOI] [PubMed] [Google Scholar]
- 9.Cody CW, Prasher DC, Westler WM, Prendergast FG, Ward WW. Chemical structure of the hexapeptide chromophore of the Aequorea green-fluorescent protein. Biochemistry. 1993;32:1212–1218. doi: 10.1021/bi00056a003. [DOI] [PubMed] [Google Scholar]
- 10.Ehrig T, O’Kane DJ, Prendergast FG. Green-fluorescent protein mutants with altered fluorescence excitation spectra. FEBS Lett. 1995;367:163–166. doi: 10.1016/0014-5793(95)00557-p. [DOI] [PubMed] [Google Scholar]
- 11.Reid BG, Flynn GC. Chromophore formation in green fluorescent protein. Biochemistry. 1997;36:6786–6791. doi: 10.1021/bi970281w. [DOI] [PubMed] [Google Scholar]
- 12.Prendergast FG. Biophysics of the green fluorescent protein. Methods Cell Biol. 1999;58:1–18. doi: 10.1016/s0091-679x(08)61945-7. [DOI] [PubMed] [Google Scholar]
- 13.Bokman SH, Ward WW. Renaturation of Aequorea green-fluorescent protein. Biochem Biophys Res Commun. 1981;101:1372–1380. doi: 10.1016/0006-291x(81)91599-0. [DOI] [PubMed] [Google Scholar]
- 14.Ward WW, Bokman SH. Reversible denaturation of Aequorea green-fluorescent protein: physical separation and characterization of the renatured protein. Biochemistry. 1982;21:4535–4540. doi: 10.1021/bi00262a003. [DOI] [PubMed] [Google Scholar]
- 15.Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 16.Fukuda H, Arai M, Kuwajima K. Folding of green fluorescent protein and the cycle3 mutant. Biochemistry. 2000;39:12025–12032. doi: 10.1021/bi000543l. [DOI] [PubMed] [Google Scholar]
- 17.Verkhusha VV, Kuznetsova IM, Stepanenko OV, Zaraisky AG, Shavlovsky MM, Turoverov KK, Uversky VN. High stability of Discosoma DsRed as compared to Aequorea EGFP. Biochemistry. 2003;42:7879–7884. doi: 10.1021/bi034555t. [DOI] [PubMed] [Google Scholar]
- 18.Ward WW, Prentice HJ, Roth AF, Cody CW, Reeves SC. Spectral perturbations of the Aequorea green-fluorescent protein. Photochem Photobiol. 1982;35:803–808. [Google Scholar]
- 19.Cubitt AB, Heim R, Adams SR, Boyd AE, Gross LA, Tsien RY. Understanding, improving and using green fluorescent proteins. Trends Biochem Sci. 1995;20:448–455. doi: 10.1016/s0968-0004(00)89099-4. [DOI] [PubMed] [Google Scholar]
- 20.Heim R, Cubitt AB, Tsien RY. Improved green fluorescence. Nature. 1995;373:663–664. doi: 10.1038/373663b0. [DOI] [PubMed] [Google Scholar]
- 21.Heim R, Prasher DC, Tsien RY. Wavelength mutations and post-translational autoxidation of green fluorescent protein. Proc Natl Acad Sci USA. 1994;91:12501–12504. doi: 10.1073/pnas.91.26.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miesenbock G, De Angelis DA, Rothman JE. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature. 1998;394:192–195. doi: 10.1038/28190. [DOI] [PubMed] [Google Scholar]
- 23.Fradkov AF, Chen Y, Ding L, Barsova EV, Matz MV, Lukyanov SA. Novel fluorescent protein from Discosoma coral and its mutants possesses a unique far-red fluorescence. FEBS Lett. 2000;479:127–130. doi: 10.1016/s0014-5793(00)01895-0. [DOI] [PubMed] [Google Scholar]
- 24.Matz MV, Fradkov AF, Labas YA, Savitsky AP, Zaraisky AG, Markelov ML, Lukyanov SA. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat Biotechnol. 1999;17:969–973. doi: 10.1038/13657. [DOI] [PubMed] [Google Scholar]
- 25.Terskikh A, Fradkov A, Ermakova G, Zaraisky A, Tan P, Kajava AV, Zhao X, Lukyanov S, Matz M, Kim S, Weissman I, Siebert P. “Fluorescent timer”: protein that changes color with time. Science. 2000;290:1585–1588. doi: 10.1126/science.290.5496.1585. [DOI] [PubMed] [Google Scholar]
- 26.Yanushevich YG, Staroverov DB, Savitsky AP, Fradkov AF, Gurskaya NG, Bulina ME, Lukyanov KA, Lukyanov SA. A strategy for the generation of non-aggregating mutants of Anthozoa fluorescent proteins. FEBS Lett. 2002;511:11–14. doi: 10.1016/s0014-5793(01)03263-x. [DOI] [PubMed] [Google Scholar]
- 27.Bevis BJ, Glick BS. Rapidly maturing variants of the Discosoma red fluorescent protein (DsRed) Nat Biotechnol. 2002;20:83–87. doi: 10.1038/nbt0102-83. [DOI] [PubMed] [Google Scholar]
- 28.Lauf U, Lopez P, Falk MM. Expression of fluorescently tagged connexins: a novel approach to rescue function of oligomeric DsRed-tagged proteins. FEBS Lett. 2001;498:11–15. doi: 10.1016/s0014-5793(01)02462-0. [DOI] [PubMed] [Google Scholar]
- 29.Verkhusha VV, Otsuna H, Awasaki T, Oda H, Tsukita S, Ito K. An enhanced mutant of red fluorescent protein DsRed for double labeling and developmental timer of neural fiber bundle formation. J Biol Chem. 2001;276:29621–29624. doi: 10.1074/jbc.C100200200. [DOI] [PubMed] [Google Scholar]
- 30.Mizuno H, Sawano A, Eli P, Hama H, Miyawaki A. Red fluorescent protein from Discosoma as a fusion tag and a partner for fluorescence resonance energy transfer. Biochemistry. 2001;40:2502–2510. doi: 10.1021/bi002263b. [DOI] [PubMed] [Google Scholar]
- 31.Demaurex N, Frieden M. Measurements of the free luminal ER Ca(2+) concentration with targeted “cameleon” fluorescent proteins. Cell Calcium. 2003;34:109–119. doi: 10.1016/s0143-4160(03)00081-2. [DOI] [PubMed] [Google Scholar]
- 32.Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, Ikura M, Tsien RY. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature. 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- 33.Miyawaki A, Griesbeck O, Heim R, Tsien RY. Dynamic and quantitative Ca2+ measurements using improved cameleons. Proc Natl Acad Sci USA. 1999;96:2135–2140. doi: 10.1073/pnas.96.5.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uversky VN. A protein-chameleon: conformational plasticity of alpha-synuclein, a disordered protein involved in neurodegenerative disorders. J Biomol Struct Dyn. 2003;21:211–234. doi: 10.1080/07391102.2003.10506918. [DOI] [PubMed] [Google Scholar]
- 35.Uversky VN. Neuropathology, biochemistry, and biophysics of α-synuclein aggregation. J Neurochem. 2007;103:17–37. doi: 10.1111/j.1471-4159.2007.04764.x. [DOI] [PubMed] [Google Scholar]
- 36.Tcherkasskaya O. Photo-activity induced by amyloidogenesis. Protein Sci. 2007;16:561–571. doi: 10.1110/ps.062578307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sniegowski JA, Lappe JW, Patel HN, Huffman HA, Wachter RM. Base catalysis of chromophore formation in Arg96 and Glu222 variants of green fluorescent protein. J Biol Chem. 2005;280:26248–26255. doi: 10.1074/jbc.M412327200. [DOI] [PubMed] [Google Scholar]
- 38.Wood TI, Barondeau DP, Hitomi C, Kassmann CJ, Tainer JA, Getz-off ED. Defining the role of arginine 96 in green fluorescent protein fluorophore biosynthesis. Biochemistry. 2005;44:16211–16220. doi: 10.1021/bi051388j. [DOI] [PubMed] [Google Scholar]
- 39.Sniegowski JA, Phail ME, Wachter RM. Maturation efficiency, trypsin sensitivity, and optical properties of Arg96, Glu222, and Gly67 variants of green fluorescent protein. Biochem Biophys Res Commun. 2005;332:657–663. doi: 10.1016/j.bbrc.2005.04.166. [DOI] [PubMed] [Google Scholar]
- 40.Jung G, Wiehler J, Zumbusch A. The photophysics of green fluorescent protein: influence of the key amino acids at positions 65, 203, and 222. Biophys J. 2005;88:1932–1947. doi: 10.1529/biophysj.104.044412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Branchini BR, Nemser AR, Zimmer M. A computational analysis of the unique protein-induced tight turn that results in posttranslational chromophore formation in green fluorescent protein. J Am Chem Soc. 1998;120:1–6. [Google Scholar]
- 42.Turoverov KK, Biktashev AG, Dorofeiuk AV, Kuznetsova IM. A complex of apparatus and programs for the measurement of spectral, polarization and kinetic characteristics of fluorescence in solution. Tsitologiia. 1998;40:806–817. [PubMed] [Google Scholar]
- 43.Marquardt DW. An algorithm for least-squares estimation of nonlinear parameters. J Soc Indust Appl Math. 1963;11:431–441. [Google Scholar]
- 44.Zuker M, Szabo AG, Bramall L, Krajcarski DT, Selinger B. Delta-function convolution method (DFCM) for fluorescence decay experiments. Rev Sci Instrum. 1985;56:14–22. [Google Scholar]
- 45.Lakowicz JR. Principles of fluorescence spectroscopy. New York: Plenum Press; 1983. [Google Scholar]
- 46.Enoki S, Maki K, Inobe T, Takahashi K, Kamagata K, Oroguchi T, Nakatani H, Tomoyori K, Kuwajima K. The equilibrium unfolding intermediate observed at pH 4 and its relationship with the kinetic folding intermediates in green fluorescent protein. J Mol Biol. 2006;361:969–982. doi: 10.1016/j.jmb.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 47.Pace CN. Determination and analysis of urea and guanidine hydrochloride denaturation curves. Methods Enzymol. 1986;131:266–280. doi: 10.1016/0076-6879(86)31045-0. [DOI] [PubMed] [Google Scholar]
- 48.Nolting B. Protein folding kinetics: biophysical methods. Berlin: Springer Verlag; 1999. p. 203. [Google Scholar]
- 49.Visser NV, Hink MA, Borst JW, van der Krogt GN, Visser AJ. Circular dichroism spectroscopy of fluorescent proteins. FEBS Lett. 2002;521:31–35. doi: 10.1016/s0014-5793(02)02808-9. [DOI] [PubMed] [Google Scholar]
- 50.Huang JR, Craggs TD, Christodoulou J, Jackson SE. Stable intermediate states and high energy barriers in the unfolding of GFP. J Mol Biol. 2007;370:356–371. doi: 10.1016/j.jmb.2007.04.039. [DOI] [PubMed] [Google Scholar]
- 51.Volkmer A, Subramaniam V, Birch DJ, Jovin TM. One- and two-photon excited fluorescence lifetimes and anisotropy decays of green fluorescent proteins. Biophys J. 2000;78:1589–1598. doi: 10.1016/S0006-3495(00)76711-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Andrews BT, Schoenfish AR, Roy M, Waldo G, Jennings PA. The rough energy landscape of superfolder GFP is linked to the chromophore. J Mol Biol. 2007;373:476–490. doi: 10.1016/j.jmb.2007.07.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stepanenko OV, Verkhusha VV, Kazakov VI, Shavlovsky MM, Kuznetsova IM, Uversky VN, Turoverov KK. Comparative studies on the structure and stability of fluorescent proteins EGFP, zFP506, mRFP1, “dimer2”, and DsRed1. Biochemistry. 2004;43:14913–14923. doi: 10.1021/bi048725t. [DOI] [PubMed] [Google Scholar]
- 54.Kuznetsova IM, Stepanenko OV, Turoverov KK, Zhu L, Zhou JM, Fink AL, Uversky VN. Unraveling multistate unfolding of rabbit muscle creatine kinase. Biochim Biophys Acta. 2002;1596:138–155. doi: 10.1016/s0167-4838(02)00212-1. [DOI] [PubMed] [Google Scholar]
- 55.Bhuyan AK. Protein stabilization by urea and guanidine hydrochloride. Biochemistry. 2002;41:13386–13394. doi: 10.1021/bi020371n. [DOI] [PubMed] [Google Scholar]
- 56.Barondeau DP, Putnam CD, Kassmann CJ, Tainer JA, Getzoff ED. Mechanism and energetics of green fluorescent protein chromophore synthesis revealed by trapped intermediate structures. Proc Natl Acad Sci USA. 2003;100:12111–12116. doi: 10.1073/pnas.2133463100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barondeau DP, Kassmann CJ, Tainer JA, Getzoff ED. Understanding GFP chromophore biosynthesis: controlling backbone cyclization and modifying post-translational chemistry. Biochemistry. 2005;44:1960–1970. doi: 10.1021/bi0479205. [DOI] [PubMed] [Google Scholar]