Abstract
The maternal-to-zygotic transition (MZT) is a universal step in animal development characterized by two major events: activation of zygotic transcription and degradation of maternally provided mRNAs. How zygotic gene products instruct the degradation of maternal messages remains a long-standing question in biology. MicroRNAs (miRNAs) have recently emerged as widespread regulators of gene expression. miRNAs control temporal and spatial gene expression by both accelerating the decay of mRNAs from previous developmental stages, as well as modulating the levels of actively transcribed genes. In this review, I discuss recent studies of the roles of miRNAs during the maternal-to-zygotic transition and cellular reprogramming, where they reshape transcriptional landscapes to facilitate the establishment of novel cellular states.
Introduction
The earliest stages of embryonic development depend on maternal instructions loaded into the oocyte in the form of mRNA and proteins [1-3]. This maternal program is ultimately responsible for the activation of the zygotic genome. Within hours of fertilization, a large fraction of maternally deposited mRNAs is eliminated via two cooperative, yet distinct, programs [3,4]. First, maternally encoded products initiate the destruction of maternal mRNAs. In Drosophila, the maternally provided RNA-binding protein Smaug is responsible for the deadenylation and clearance of the majority of unstable transcripts following egg activation [5,6]. In Xenopus, maternal mRNAs possessing an embryonic deadenylation element (EDEN) within their 3’UTRS are targeted for deadenylation and translational silencing by the maternally provided EDEN-binding protein [7].
In addition to these maternal factors, which have been thoroughly reviewed elsewhere [2,3,7], a second degradation program is initiated by, and is dependent on, zygotic transcription. In particular, zygotically expressed miRNAs have been shown to dramatically enhance the efficiency of maternal mRNA clearance in zebrafish [8], Xenopus [9] and Drosophila [10]. miRNAs are small ~22nt RNAs that regulate gene expression post-transcriptionally [11-13]. Mature miRNAs are generated from longer primary transcripts through sequential cleavage by the RNAseIII enzymes Drosha and Dicer. The mature miRNA, once incorporated into a silencing complex (miRISC), guides the miRISC to target mRNAs, resulting in their deadenylation, repression, and decay [8,14-17]reviewed in [13,18,19]. Functional analyses have shown that miRNAs shape gene expression within multiple developmental contexts (reviewed in [20]. miRNAs have been shown to control temporal gene expression by downregulating mRNAs transcribed during previous developmental stages [8,21-23]. On the other hand, miRNAs can shape spatial expression of a given gene, by modulating the levels of actively transcribed genes in a specific domain [24-27]. In the case of the maternal-zygotic transition, the removal of pre-existing mRNAs prevents their interference with zygotic development [8,10]. For example, by “wiping the slate clean”, the zygotic counterpart of a maternally-provided ubiquitous mRNA can be expressed in a restricted pattern [28-30]. In addition, the post-transcriptional nature of miRNA-mediated regulation provides an ideal mechanism to precisely modulate mRNA dosage of zygotic [31] and pre-existing maternal mRNAs deposited in the oocyte.
In this review, we highlight recent contributions to the molecular regulation of the maternal-to-zygotic transition by miRNAs, and how they function in development to clear maternal mRNAs, facilitate tissue specific expression of maternal-ubiquitously provided mRNAs and improve cellular reprogramming by erasing the cell’s transcriptional history.
miRNAs clear maternal mRNAs during the maternal-to-zygotic transition in vertebrates
Activation of zygotic transcription is intimately linked to the degradation of maternal messages [4,32-35] Reviewed in [3]. Indeed, inhibition of zygotic transcription, results in the stabilization of a large fraction of maternal mRNAs [32]. Yet, the factors responsible for this selective and active degradation have remained mostly elusive. The orthologous miRNAs, miR-430 and miR-427, are abundantly expressed during the maternal-to-zygotic transition in zebrafish [36,37] and Xenopus [38,39], respectively. In the fish, transcriptional profiling of maternal-zygotic dicer mutants (MZdicer) and in vivo validation experiments have identified more than 200 miR-430 target mRNAs in vivo [8]. These targets are strongly enriched for maternally provided mRNAs (~4 fold). In addition, analysis of the maternal mRNA population (in which ~70% of all zebrafish genes are represented) reveals a ~4-fold enrichment for the presence of miR-430 complementary sites when compared to mRNAs that are strictly zygotic. Further, loss of miR-430 slows the decay of several hundred maternal mRNAs [8] (Figure 1). miR-430-mediated mRNA degradation is achieved through the accelerated deadenylation of target transcripts, and has provided an entry point for understanding the molecular mechanisms behind miRNA-mediated target mRNA turnover.
Figure 1. MicroRNAs clear the cell’s transcriptional landscape during developmental transitions.
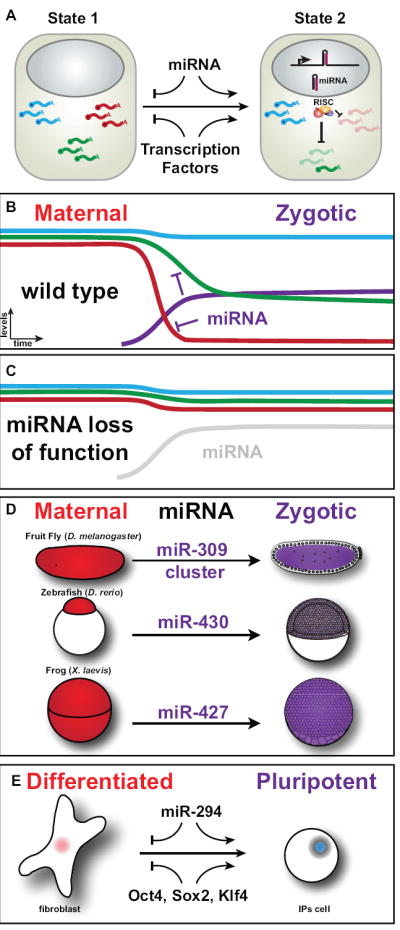
(A) Diagram of a cell in two different states. The expression of a miRNA (hairpin) in the second state leads to the clearance of some transcripts (red) and the partial downregulation of other targets (green). (B,C) Schematic representation of levels for different mRNAs and a miRNA in the maternal-to-zygotic transition in the presence (B) and the absence (C) of a miRNA. The different curves represent the degradation profiles of maternal (red, blue) and maternal-zygotic (green) transcripts that are regulated by the miRNA during the maternal-to-zygotic transition or during reprogramming., resulting in a rapid decay of the mRNA levels. Targeting of the green transcript by the miRNA allows the cell to regulate the steady state levels of this mRNA. (D) Diagram representing known examples of miRNAs that regulate the clearance of maternal transcripts during the maternal-to-zygotic transition in different organisms. (E) Reprogramming of somatic cells to a pluripotent state. Introducing the mouse ortholog of miR-430 (miR-294) into differentiated cells together with Oct4 Sox2, and Klf4 enhances the reprogramming efficiency.
These above findings support the hypothesis that miR-430 accelerates the deadenylation and decay of several hundred maternally loaded mRNAs. A similar scenario has been observed for the Xenopus miR-430 orthologue, miR-427[9]. miR-427 is highly transcribed by RNA Pol II prior to general zygotic genome activation [38,39]. As a result, cells accumulate high levels of miR-427, with ~109 copies of mature miRNA/embryo [9]. Lund et al. provide evidence that miR-427 directly accelerates the deadenylation of maternally deposited cyclin B2 and A1 mRNAs. Although additional targets need to be validated, 3’-UTR sequence analysis in Xenopus has identified conserved miR-427 target sites in eight additional genes. The miR-427 target list is likely to increase once the genome-wide functions of miR-427 are examined. It is important to note that multiple mammalian orthologues of miR-430 (miR-295 in mice, and miR-302, miR-372, miR-516-520 in humans and primates) are expressed during early embryogenesis and could potentially regulate the clearance of maternal transcripts in mammals. Despite the sequence homology among the miR-430 orthologues and their important roles during maternal clearance (Zebrafish and Xenopus) and stem cell maintenance (mouse and human), little is known about the upstream factors that activate their expression during development. A recent study has shown that the stem cell factors Oct4 and Sox2 bind the promoter of miR-302 and activate its expression in EScells [40]. Since these factors are also expressed in the early embryo, and at least Oct4 is maternally provided, it is tantalizing to speculate that the same factors might drive zygotic activation of miR-430/427 to initiate maternal degradation.
Based on the occurrence of complementary target sites, miR-430 has the potential to regulate up to 40% of all maternal messages present in the early fish embryo [8]. However, most of these putative targets await experimental validation [8]. The rules governing miRNA-target regulation are not yet fully understood [13]. The mere presence of a putative miRNA target site does not guarantee miRNA-mediated regulation. On the other hand, many bona fide targets that are primarily regulated at the level of translation will be missed in expression profiling studies. For many targets, it is likely that much finer-grained analyses will be required, especially in cases where the miRNA functions together with other RNA-binding proteins such as Smaug or Pumillio to confer an additional layer of repression [3].
The role of miRNAs in the maternal-to-zygotic transition in Drosophila
How do other animals that lack miR-430 orthologues remove maternally-deposited messages? The clearance of maternally provided mRNAs has been studied using chromosomal deletions to eliminate the confounding effect of transcription of the zygotic contribution of that gene. In contrast to that observed for miR-430 in the fish, De Renzis and colleagues did not find any predominant miRNA target site sequence that was enriched within the unstable maternal mRNA pool. This might be explained by the potential diluting effects of analyzing each miRNAs separately [28]. Interestingly, the generation of a miRNA cluster knock out in Drosophila revealed that zygotic expression of the miR-309 cluster, which encodes miR-3, -4, -5, -6 and -309, directs the degradation of a subset of maternal mRNAs at the MZT [10]. Loss of the miR-309 clusters results in the stabilization of a large set of maternal mRNAs that are rapidly degraded during the MZT. Interestingly, a comparison of the occurrence of all putative miRNA target sites versus those specific to the miR-309 cluster, suggests that maternal genes are enriched for, and zygotic genes are depleted of, miR-309 cluster target sites in their 3’UTRs. This finding suggests that the expression of the miR-309 cluster (zygotically-provided) tends to be temporally anticorrelated with the expression of its targets (maternally-provided)[10].
The above findings clearly implicate a family of miRNAs in the clearance of maternal mRNAs in the fly [10]. But how is this miRNA cluster activated in the first place to ensure the timely degradation of the maternal mRNAs? A recent study has identified a maternally deposited transcription factor, Zelda, is required for the activation of the miR-309 cluster in addition to other zygotic genes [41]. Interestingly, there is a link between miR-309 activation and Smaug-mediated degradation of maternally deposited mRNAs. Smaug is a conserved RNA-binding protein required for the destruction of maternal mRNAs during the MZT in Drosophila. Intriguingly, roughly 85% of the 410 maternal mRNAs upregulated in the absence of the miR-309 cluster are also stabilized in smaug mutants. This result is likely due to reduced miR-309 expression in smaug mutant embryos, suggesting that maternal mRNA clearance (Smaug-mediated) is required for high-level zygotic activation, including the miR-309 cluster, which in turn leads to further destabilization of a subset of maternal mRNAs [6].
Loading of the miRNA processing machinery in the egg allows timely regulation of maternal mRNAs
miRNAs must be processed into their mature forms to mediate repression. Interestingly, the miRNA processing machinery, including Dicer and Drosha, appears to be maternally supplied in frog, fish, and fly embryos. As a result, embryos are “primed” for rapid processing once zygotic miRNAs are transcribed at the MZT. This triggers the deadenylation and clearance of a large fraction of the maternal mRNAs with the appropriate timing and decay rate. Consistent with this hypothesis, the lethality observed from maternal loss of zebrafish dicer cannot be rescued by zygotically provided dicer (inherited from the male). The time required to transcribe and translate zygotic Dicer results in an insurmountable delay in the processing of miR-430 and subsequent maternal clearance. However, direct injection of processed miR-430 into these embryos rescues the lethal phenotype (Mdicer-/-, Zdicer-/+) to make viable adults, suggesting that miR-430 is the only miRNA that requires the maternal miRNA processing machinery for survival (Giraldez unpublished results). Furthermore, in Xenopus, providing miR-427 before endogenous miR-427 is transcribed, causes the premature deadenylation of its targets [9]. These results indicate that the miRNA processing and effector machinery are maternally provided, thereby allowing the timely repression, deadenylation and decay of miRNA targets at the onset of zygotic transcription.
MicroRNAs regulate steady state mRNA levels
miRNAs not only function to clear the target mRNAs expressed in a previous developmental state. A fraction of the target mRNAs is also zygotically transcribed. In this case, miR-430-mediated regulation (i) tunes down the activation of the zygotic mRNA, and (ii) modulates the steady state mRNA levels for these transcripts (Figure 1). Indeed, more than 70% of the vertebrate mRNAs are thought to be under miRNA-mediated regulation. This might reflect a role for miRNAs to control the rate of target mRNA decay and maintain mRNA homeostasis [8,27,42]
A common theme for miRNA function, forget your past
The finding that unrelated miRNA clusters (miR-309 in drosophila and miR-430/427 in Zebrafish/Xenopus) have taken on similar functions during MZT illustrates a striking example of convergent evolution. In both cases, the expression of the miRNA serves to clear the cell of previously expressed transcripts, thereby setting the stage for subsequent developmental stages (Figure 1). In many ways, the functions of miR-309 and miR-430 during MZT are reminiscent of those observed for the founding miRNAs lin-4 and let-7 in C. elegans. Both lin-4 and let-7 clear and repress previously expressed transcripts and facilitate progression to the following developmental stage [21] [22,43,44]. In hindsight, the use of miRNAs in shaping the temporal dynamics underlying developmental transitions provides a versatile system with multiple advantages for the embryo. Maternaly-deposited mRNAs can only be regulated post-transcriptionaly and as such, miRNAs can easily regulate mRNAs that have been previously generated. In addition, the small “seed” size of miRNA target sites (6-8nt long), allows for a miRNA to simultaneously control a large number of unrelated transcripts. Simply by acquiring few mutations in their 3’UTRs during evolution, genes can gain and lose target sites, with no impact on their coding sequences. The level of regulation can be modified by changing the extent of complementarity between the target and the miRNA as well as the number of complementary sites. Additional specificity can be conferred by RNA-binding proteins, discussed below, which modulate miRNA-mediated regulation in cell-type specific manner. Taken together, this inherent versatility allows the same miRNA to both remove previously transcribed mRNAs that are no longer needed, as well as precisely regulate steady-state levels of those that are still being transcribed.
Antagonizing miRNA function during maternal clearance shapes gene expression
Recent studies have provided interesting insights into how RNA-binding proteins can modulate miRNA-mediated regulation depending on cell type or cellular state (Figure 2). In the germ line, the RNA-binding proteins, Deadend [45] and Dazl [30], protect some maternal mRNAs from the clearing effects of miR-430 [29]. These germ cell-specific factors counteract the effects of miR-430 on nanos1 and Tudor-domain-containing-protein 7 (Tdrd-7) mRNA through two different mechanisms. The Deadend-binding site in the nanos1 3’UTR overlaps with the miR-430 site, such that interaction with Deadend provides steric protection from miR-430 [45]. In contrast, Dazl antagonizes the activity of miR-430 by inducing mRNA polyadenylation when bound to the Tdrd-7 3’UTR [30]. In both cases, the modulation of miRNA activity facilitates tissue-specific expression of ubiquitously provided mRNAs.
Figure 2. RNA binding proteins modulate miRNA-mediated repression of maternal mRNAs in germ cells.

Model for the post-transcriptional regulation of mRNA targets by miRNAs. (A) Target mRNA translation: interaction between poly(A) binding protein (PABP) on poly(A) tail with translation initiation factors eIF4G/eIF4E on Cap stimulates translation. (B) miRNA-mediated target mRNA deadenylation: The miRNA induced silencing complex (miRISC) is recruited to the 3’ UTR of target mRNA and accelerates deadenylation. (C) Binding of the DAZL to the 3’UTR of the target (Tdrd7) antagonizes miRNA-mediated repression by promoting polyadenylation. (D) Binding of Dead end (Dnd) to the 3’UTR of nanos1 blocks the binding of the miRISC to the target, and antagonizes miRNA-mediated repression of nanos1 in germ cells.
MicroRNAs clear the cell’s history during cellular reprogramming
The maternal-to-zygotic transition is in some ways analogous to the process of cellular reprogramming. The nucleus of a differentiated cell, when exposed to the cytoplasm of the fertilized egg can be reprogrammed to a totipotent state [46,47]. In both cases, during the maternal-to-zygotic transition and during cellular reprogramming, the cell’s history is erased to facilitate the establishment of novel cellular states by specific transcription factors (zygotic state or pluripotency). Intriguingly, the mammalian orthologs of miR-430 (miR-294 in mice and miR-302/-372 in humans) are abundantly expressed in embryonic stem cells and embryonic tissues [48,49]. While it is tempting to speculate that these miRNAs might play important roles in the clearance of the maternal transcripts in mammals, several lines of evidence suggest that these miRNAs contribute to pluripotency and have important roles in cellular reprogramming. Recent studies have shown that defined transcription factors can reprogram differentiated cells to adopt pluripotency (induced pluripotent stem cells: iPS cells). These factors include Oct4, Klf4, Sox2 and c-myc [50]. While only Oct4 appears to be critical [51], all of these factors increase the reprogramming efficiency. Interestingly, co-introduction of a miRNA (miR-294) with Oct4, Sox2 and Klf4 in differentiated fibroblasts dramatically enhances the reprogramming efficiency 10-fold compared to the three factors alone [52] (Figure 1). What could make the miR-294 family such an efficient reprogramming factor? ES cells defective in miRNA processing show defects in proliferation, differentiation and self-renewal [53]. Interestingly, Blelloch and colleagues have shown that replenishing dicer mutant ES cells with miR-430/302/294 family members rescues proliferation defects [53]. However, promoting cell proliferation in differentiated cells only modestly increases the reprogramming efficiency, suggesting that there are additional functions of the miR-294/302 family other than accelerating proliferation. Because miRNAs shape gene expression in both spatial and temporal dimensions, they make ideal candidates to clear the cells’ transcriptional memory. Indeed, it has been shown that many miRNAs tend to be expressed in an anticorrelative pattern with their targets [24,26,27,42,54]. Genes required for a specific cellular state tend to avoid strong repression by co-expressed miRNAs. For example, miRNAs expressed in embryonic stem cells will typically not target essential stem cell factors. In this scenario, introducing these ES cell miRNAs (miR-294) into differentiated cells is likely helping to erase the transcriptional landscape of the differentiated cell. This creates a clear slate where the specific reprogramming transcription factors can return the cell to a pluripotent state
Many miRNAs have a tissue specific expression in the embryo [55] and shape the gene expression during differentiation [24,26,27,42]. As a consequence, miRNAs not only help cells to forget their past transcriptional history during reprogramming, but can also redirect their path during differentiation. For example, providing miR-145 into multipotent neural crest stem cells can influence their downstream differentiation path to vascular smooth muscle fate [56], suggesting that expression of specific miRNA can favor differentiation into specific fates. miRNAs might stabilize not only a cell fate, but also the differentiated state per se. In contrast to stem cell specific miRNAs, other miRNAs such as let-7 inhibit pluripotency once the cell becomes committed to a specific fate therefore stabilizing this decision [57]. Conversely, loss-of-let7 has been associated with cancer [58], a state where the cell might return to an embryonic state by stimulating proliferation and de-repressing the pluripotent state.
Future outlook
Despite the wide variety of biological contexts where miRNAs function, a common theme emerges, whereby miRNAs shape spatial and temporal gene expression: (i) modulate the levels of actively transcribed genes and (ii) accelerate the clearance of previously transcribed messages. However, miRNAs only correspond to a small fraction of the non-coding genome and, as such, represent just the tip of the non-coding iceberg. Future studies will be needed to shed light on the functions of additional small and long non-coding RNAs during the maternal-to-zygotic transition, in DNA integrity surveillance, and epigenetic regulation. Additional proteins are likely to modulate miRNA function in cells and the transcription factors responsible for their expression are largely unknown. RNA binding proteins can antagonize or potentiate miRNA activity. The accessibility of the miRNA to the target mRNA could be regulated by modifying the secondary structure of the RNA, blocking the target site, or altering the polyadenylation site to change the length of the 3’UTR to include or exclude specific miRNA target sites. These modifiers can modulate the temporal and spatial activity of miRNAs providing a highly versatile system to regulate gene expression during embryogenesis. While some miRNAs are ubiquitously expressed, the majority shows restricted spatial expression within particular tissues and organs. Future studies will provide important insights in the use of miRNAs during cellular reprogramming to tailor the differentiation path of cells to specific fates in vivo.
Acknowledgments
Thanks to Carter Takacs for critical reading of this review. AJG is a Lois and Franklin Top Yale Scholar. The Giraldez Lab is funded by the Yale Scholar program, the Pew Scholars Program, Muscular Dystrophy Association and NIH R01GM081602-03 /03S1.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Abrams EW, Mullins MC. Early zebrafish development: it’s in the maternal genes. Curr Opin Genet Dev. 2009;19:396–403. doi: 10.1016/j.gde.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farley BM, Ryder SP. Regulation of maternal mRNAs in early development. Crit Rev Biochem Mol Biol. 2008;43:135–162. doi: 10.1080/10409230801921338. [DOI] [PubMed] [Google Scholar]
- xx3.Tadros W, Lipshitz HD. The maternal-to-zygotic transition: a play in two acts. Development. 2009;136:3033–3042. doi: 10.1242/dev.033183.. This review provides an extensive and thorough overview of the maternal-to-zygotic transition beyond the role of miRNAs, including our current understanding of the zygotic genome activation and other mechanisms of maternal clearance.
- 4.Bashirullah A, Halsell SR, Cooperstock RL, Kloc M, Karaiskakis A, Fisher WW, Fu W, Hamilton JK, Etkin LD, Lipshitz HD. Joint action of two RNA degradation pathways controls the timing of maternal transcript elimination at the midblastula transition in Drosophila melanogaster. Embo J. 1999;18:2610–2620. doi: 10.1093/emboj/18.9.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tadros W, Houston SA, Bashirullah A, Cooperstock RL, Semotok JL, Reed BH, Lipshitz HD. Regulation of maternal transcript destabilization during egg activation in Drosophila. Genetics. 2003;164:989–1001. doi: 10.1093/genetics/164.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- x6.Benoit B, He CH, Zhang F, Votruba SM, Tadros W, Westwood JT, Smibert CA, Lipshitz HD, Theurkauf WE. An essential role for the RNA-binding protein Smaug during the Drosophila maternal-to-zygotic transition. Development. 2009;136:923–932. doi: 10.1242/dev.031815.. This paper provides an interesting link between he degradation of the zygotic mRNAs and the high level activation of the zygotic genome
- 7.Paillard L, Osborne HB. East of EDEN was a poly(A) tail. Biol Cell. 2003;95:211–219. doi: 10.1016/s0248-4900(03)00038-8. [DOI] [PubMed] [Google Scholar]
- xx8.Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689.. This paper describes for the first time the role of miRNAs in the clearance of maternal mRNAs, and provides a molecular mechanism for the action of miRNAs that implicates the deadenylation of the target mRNAs.
- 9.Lund E, Liu M, Hartley RS, Sheets MD, Dahlberg JE. Deadenylation of maternal mRNAs mediated by miR-427 in Xenopus laevis embryos. Rna. 2009;15:2351–2363. doi: 10.1261/rna.1882009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- x10.Bushati N, Stark A, Brennecke J, Cohen SM. Temporal reciprocity of miRNAs and their targets during the maternal-to-zygotic transition in Drosophila. Curr Biol. 2008;18:501–506. doi: 10.1016/j.cub.2008.02.081.. This paper provides an interesting example of convergent evolution where a miRNA cluster (miR-309) accelerates the clearance of maternal mRNAs like miR-430 does in zebrafish
- 11.Bushati N, Cohen SM. microRNA Functions. Annu Rev Cell Dev Biol. 2007 doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 12.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 13.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci U S A. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eulalio A, Huntzinger E, Izaurralde E. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat Struct Mol Biol. 2008;15:346–353. doi: 10.1038/nsmb.1405. [DOI] [PubMed] [Google Scholar]
- 16.Wakiyama M, Takimoto K, Ohara O, Yokoyama S. Let-7 microRNA-mediated mRNA deadenylation and translational repression in a mammalian cell-free system. Genes Dev. 2007;21:1857–1862. doi: 10.1101/gad.1566707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Standart N, Jackson RJ. MicroRNAs repress translation of m7Gppp-capped target mRNAs in vitro by inhibiting initiation and promoting deadenylation. Genes Dev. 2007;21:1975–1982. doi: 10.1101/gad.1591507. [DOI] [PubMed] [Google Scholar]
- 18.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 19.Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol. 2007;17:118–126. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 20.Takacs CM, Giraldez AJ. MicroRNAs as genetic sculptors: Fishing for clues. Semin Cell Dev Biol. doi: 10.1016/j.semcdb.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 22.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 23.Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 24.Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3’UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 25.Woltering JM, Durston AJ. MiR-10 represses HoxB1a and HoxB3a in zebrafish. PLoS One. 2008;3:e1396. doi: 10.1371/journal.pone.0001396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 27.Mishima Y, Abreu-Goodger C, Staton AA, Stahlhut C, Shou C, Cheng C, Gerstein M, Enright AJ, Giraldez AJ. Zebrafish miR-1 and miR-133 shape muscle gene expression and regulate sarcomeric actin organization. Genes Dev. 2009;23:619–632. doi: 10.1101/gad.1760209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Renzis S, Elemento O, Tavazoie S, Wieschaus EF. Unmasking activation of the zygotic genome using chromosomal deletions in the Drosophila embryo. PLoS Biol. 2007;5:e117. doi: 10.1371/journal.pbio.0050117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mishima Y, Giraldez AJ, Takeda Y, Fujiwara T, Sakamoto H, Schier AF, Inoue K. Differential regulation of germline mRNAs in soma and germ cells by zebrafish miR-430. Curr Biol. 2006;16:2135–2142. doi: 10.1016/j.cub.2006.08.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takeda Y, Mishima Y, Fujiwara T, Sakamoto H, Inoue K. DAZL relieves miRNA-mediated repression of germline mRNAs by controlling poly(A) tail length in zebrafish. PLoS One. 2009;4:e7513. doi: 10.1371/journal.pone.0007513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choi WY, Giraldez AJ, Schier AF. Target Protectors Reveal Dampening and Balancing of Nodal Agonist and Antagonist by miR-430. Science. 2007 doi: 10.1126/science.1147535. [DOI] [PubMed] [Google Scholar]
- 32.Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: I. characterization and timing of cellular changes at the midblastula stage. Cell. 1982;30:675–686. doi: 10.1016/0092-8674(82)90272-0. [DOI] [PubMed] [Google Scholar]
- 33.Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: II. Control of the onset of transcription. Cell. 1982;30:687–696. doi: 10.1016/0092-8674(82)90273-2. [DOI] [PubMed] [Google Scholar]
- 34.Bouvet P, Paris J, Phillippe M, Osborne HB. Degradation of a developmentally regulated mRNA in Xenopus embryos is controlled by the 3’ region and requires the translation of another maternal mRNA. Mol Cell Biol. 1991;11:3115–3124. doi: 10.1128/mcb.11.6.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edgar BA, Datar SA. Zygotic degradation of two maternal Cdc25 mRNAs terminates Drosophila’s early cell cycle program. Genes Dev. 1996;10:1966–1977. doi: 10.1101/gad.10.15.1966. [DOI] [PubMed] [Google Scholar]
- 36.Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, Hammond SM, Bartel DP, Schier AF. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- 37.Chen PY, Manninga H, Slanchev K, Chien M, Russo JJ, Ju J, Sheridan R, John B, Marks DS, Gaidatzis D, et al. The developmental miRNA profiles of zebrafish as determined by small RNA cloning. Genes Dev. 2005;19:1288–1293. doi: 10.1101/gad.1310605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Watanabe T, Imai H, Minami N. Identification and expression analysis of small RNAs during development. Methods Mol Biol. 2008;442:173–185. doi: 10.1007/978-1-59745-191-8_13. [DOI] [PubMed] [Google Scholar]
- 39.Watanabe T, Takeda A, Mise K, Okuno T, Suzuki T, Minami N, Imai H. Stage-specific expression of microRNAs during Xenopus development. FEBS Lett. 2005;579:318–324. doi: 10.1016/j.febslet.2004.11.067. [DOI] [PubMed] [Google Scholar]
- 40.Card DA, Hebbar PB, Li L, Trotter KW, Komatsu Y, Mishina Y, Archer TK. Oct4/Sox2-regulated miR-302 targets cyclin D1 in human embryonic stem cells. Mol Cell Biol. 2008;28:6426–6438. doi: 10.1128/MCB.00359-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- xx41.Liang HL, Nien CY, Liu HY, Metzstein MM, Kirov N, Rushlow C. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature. 2008;456:400–403. doi: 10.1038/nature07388.. This paper provides one of the first examples of a maternally deposited transcription factor that is responsible for the activation of the zygotic genome, one of the long sought holy grials of developmental biology.
- 42.Shkumatava A, Stark A, Sive H, Bartel DP. Coherent but overlapping expression of microRNAs and their targets during vertebrate development. Genes Dev. 2009;23:466–481. doi: 10.1101/gad.1745709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 44.Abbott AL, Alvarez-Saavedra E, Miska EA, Lau NC, Bartel DP, Horvitz HR, Ambros V. The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev Cell. 2005;9:403–414. doi: 10.1016/j.devcel.2005.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- xx45.Kedde M, Strasser MJ, Boldajipour B, Oude Vrielink JA, Slanchev K, le Sage C, Nagel R, Voorhoeve PM, van Duijse J, Orom UA, et al. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell. 2007;131:1273–1286. doi: 10.1016/j.cell.2007.11.034.. This paper identifies the RNA binding protein able to protect specific transcripts from miRNA-mediated repression.
- 46.Gurdon JB. The developmental capacity of nuclei taken from intestinal epithelium cells of feeding tadpoles. J Embryol Exp Morphol. 1962;10:622–640. [PubMed] [Google Scholar]
- 47.Gurdon J. Nuclear reprogramming in eggs. Nat Med. 2009;15:1141–1144. doi: 10.1038/nm1009-1141. [DOI] [PubMed] [Google Scholar]
- 48.Suh MR, Lee Y, Kim JY, Kim SK, Moon SH, Lee JY, Cha KY, Chung HM, Yoon HS, Moon SY, et al. Human embryonic stem cells express a unique set of microRNAs. Dev Biol. 2004;270:488–498. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 49.Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific MicroRNAs. Dev Cell. 2003;5:351–358. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- 50.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 51.Kim JB, Sebastiano V, Wu G, Arauzo-Bravo MJ, Sasse P, Gentile L, Ko K, Ruau D, Ehrich M, van den Boom D, et al. Oct4-induced pluripotency in adult neural stem cells. Cell. 2009;136:411–419. doi: 10.1016/j.cell.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 52.Judson RL, Babiarz JE, Venere M, Blelloch R. Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat Biotechnol. 2009;27:459–461. doi: 10.1038/nbt.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, Blelloch R. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat Genet. 2008;40:1478–1483. doi: 10.1038/ng.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 55.Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 56.Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, Lee TH, Miano JM, Ivey KN, Srivastava D. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705–710. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- xx57.Melton C, Judson RL, Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 463:621–626. doi: 10.1038/nature08725.. This paper demonstrates an antagonistic interaction between stem cell specific miRNAs (miR-294) and miRNAs present in differentiated cells (let-7)
- 58.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]


