Abstract
Rhomboids are an ancient and conserved family of intramembrane-cleaving proteases, a small group of proteolytic enzymes capable of hydrolyzing a peptide bond within a transmembrane helix that anchors a substrate protein to the membrane. Mitochondrial rhomboids evolved in eukaryotes to coordinate a critical aspect of cell biology, the regulation of mitochondrial membranes dynamics. This function appears to have required the emergence of a structural feature that is unique among all other rhomboids: an additional transmembrane helix (TMH) positioned at the N-terminus of six TMHs that form the core proteolytic domain of all prokaryotic and eukaryotic rhomboids. This “1+6” structure, which is shared only among mitochondrial rhomboids, defines a subfamily of rhomboids with the prototypical family member being mammalian Parl. Here, we present the findings that in eleven years have elevated mitochondrial rhomboids as the gatekeepers of mitochondrial dynamics and apoptosis; further, we discuss the aspects of their biology that are bound to introduce new paradigm shifts in our understanding of how the organelle uses this unique type of protease to govern stress, signaling to the nucleus, and other key mitochondrial activities in health and disease.
Keywords: Mitochondria, rhomboids, serine protease, regulated intramembrane proteolysis, catalytic dyad, membrane dynamics, apoptosis, Parl, Opa1, Hax1, Omi, Htr2A, Mgm1, mitochondrial stress, retrograde signaling, neurodegenerative disease, Parkinson's disease, cancer
1. Introduction
Parl was discovered in 1998 in the laboratory of Luciano D'Adamio at the NIH following a yeast two-hybrid (Y2H) screening done with the objective of finding what at that time was the “Holy Grail” in Alzheimer's research, the protease implicated in the intramembraneous cleavage of AβPP: the γ-secretase. Since the Alzheimer's Presenilin protein interacts with AβPP and is necessary for its cleavage, this protein was chosen as bait to identify the elusive protease (it has subsequently been shown that presenilin itself provides, as part of a multiprotein complex, the proteolytic component for the cleavage of AβPP [1]). That Y2H screening yielded several clones encoding a near-full length, uncharacterized polytopic membrane protein. Bioinformatic analysis conducted by Eugene Koonin at the NIH revealed that this protein was a member of the Rhomboid family of proteins, a conserved family of membrane proteins that includes a developmental regulator from Drosophila. It was, therefore, named Parl, for Presenilin-associated Rhomboid-like protein. Using approaches to predict protein function from conserved sequence motifs [2,3], Koonin further predicted that Parl and, by implication, other rhomboids, are proteases [4]. This prediction, which was promptly shared with leading scientists of the Rhomboids and Alzheimer's field, set in motion a series of events that, years later, led to a new understanding of the rhomboids as a novel class of intramembrane-cleaving proteases important in signaling, with Parl as a gatekeeper of apoptosis [5].
Thus, a decade after its discovery, Parl has proved to have a name that is only partly justified: indeed, it is a Rhomboid but not a functional interacting partner for Presenilin. However, its predicted proteolytic activity and critical role for the biology of the cell has stood the test of time. This review will address many facets of Parl biology, with the aim to support the future elucidation on its puzzling structure, complex mechanism of regulation and possible role in mitochondrial stress, retrograde signaling, and more.
2. Mitochondrial Rhomboids Evolution
Parl is a member of the Rhomboid family of regulated intramembrane proteases (RIP) [4,6-8], which are integral membrane enzymes that catalyze peptide bond hydrolysis of membrane-anchored substrates through cleavage of either the transmembrane spanning helix or adjacent region [1,9-11].
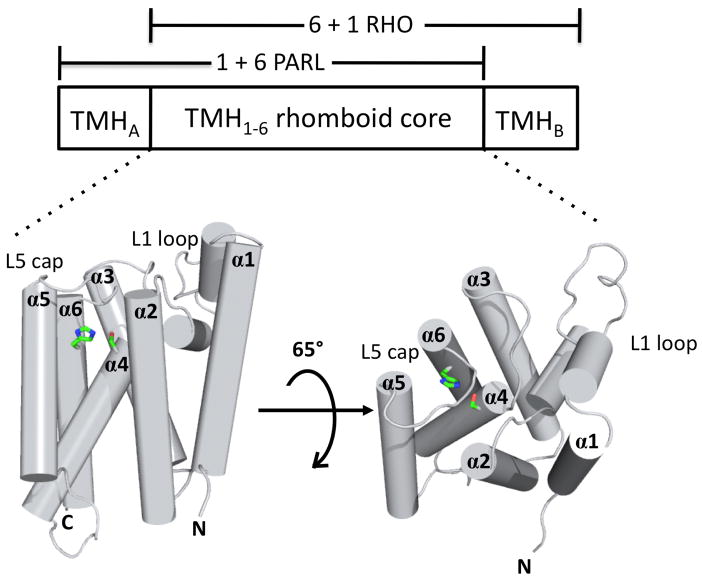
Rhomboids are among the most conserved family of polytopic membrane proteins known to date, as they are represented in most of the sequenced bacterial, archaeal and eukaryotic genomes [7]. The exact sequence similarity between eukaryotic and prokaryotic rhomboid family proteins is relatively low, around 10-15% identity in the conserved catalytic rhomboid domain. This is not surprising since polytopic transmembrane proteins are, in general, not particularly strongly conserved during evolution. The notable conserved feature of all rhomboid proteins is within the core rhomboid domain, which is composed of 6 TMH, with the strictly conserved catalytic serine and histidine residues located in TMH-4 and TMH-6, respectively (Figure 1) [7].
Figure 1.
Domain architecture of rhomboid proteases. Rhomboids are composed of a 6 TMH core domain found in prokaryotes with two subfamilies that differ in the placement of an additional TMH at either the N-terminus (“1+6” mitochondrial PARL subfamily) or the C-terminus (“6+1” RHO subfamily). High resolution structures of the 6 TMH core rhomboid domain from bacterial rhomboids have been determined [12-15,35] and show the architecture of the active site with the absolutely conserved Ser and His arranged in a manner akin to classical serine proteases. Based on this and other evidence, Ser is thought to act as a nucleophile and initiate catalysis on the polypeptide substrate. Histidine plays essential acid-base roles in catalysis. The figure was made using the software PyMOL [110] and 3B45.pdb [35].
Phylogenetic analysis revealed that eukaryotic rhomboids are split between two major subfamilies, which are positioned in the midst of different prokaryotic branches. The first subfamily was designated the RHO subfamily after its prototype the Drosophila developmental regulator Rho-1.The RHO subfamily typically have an extra TMH added at the carboxy-terminus of the 6-TMH rhomboid core (“6+1” structure, Figure 1). The second eukaryotic subfamily was designated the PARL subfamily after its prototype, the mammalian Parl protein. The PARL subfamily consists of rhomboid proteins with a domain architecture characterized by an extra TMH added to the amino terminus of the 6-TMH rhomboid domain (“1+6” structure) [7].
Most of the bacterial and archaeal genomes have a single gene coding for a rhomboid-family protein. In contrast, eukaryotes show expansion of the rhomboid family, with 2 members in yeast, 7 in Drosophila, 5 in humans and as many as 13 in Arabidopsis. However, animal genomes contain only one gene encoding for a member of the PARL protein subfamily, which studies in yeast, Drosophila and the mammalian system have shown to encode for a mitochondrial protein of the inner membrane(discussed below) [7]. Thus, the “6+1” (RHO) and “1+6” (PARL) rhomboids differ in domain architecture and sub-cellular localization, with the PARL subfamily being composed by members that are exclusively localized in the mitochondria.
Phylogenetic analysis also reveals a selective expansion of the RHO but not of the PARL subfamily, which is typically represented by only one member per genome [7]. What evolutionary constraints have led, in animals, to the selective expansion of the RHO subfamily and, possibly, to the selected elimination of additional members of the PARL subfamily are unknown. Understanding the evolutionary constraints that, in animals, led the PARL subfamily to be represented by a single member may help to establish the role of mitochondrial rhomboids in the functional and structural organization of the organelle. Future studies on Parl and its eukaryotic orthologs, Rho-7 in Drosophila and Pcp1 in S. cerevisiae, might clarify this intriguing aspect of the peculiar evolution of mitochondrial rhomboids in animals.
3. Insights into Mitochondrial Rhomboid Structure
Before discussing the mitochondrial rhomboids in detail, we highlight recent advances in the structural biology of the rhomboid core domain. Membrane protein structure determination is notoriously challenging, however, the phenomenal progress that has occurred in the rhomboid biology since their identification has been matched by the determination to atomic resolution of four bacterial rhomboid structures from two species [12-15]. These structures are quite similar and confirm earlier predictions that the rhomboid domain consists of six TMHs [7]. These structures revealed the previously proposed catalytic serine and histidine spatially arranged in a manner similar to that seen in classical serine proteases. In the mechanism of classical serine proteases, the substrate undergoes a nucleophilic attack from the catalytic Ser with the His plays essential acid-base roles. In many, but not all, serine proteases a third catalytic residue, often Asp or Asn, is found and plays an important but not essential role. In some rhomboids, biochemical evidence supported a highly conserved Asn as a third catalytic residue, however, the high-resolution structures clearly show that this third residue, Asn154 on TMH 2 of GlpG, is spatially not in position to play the classical role of the third catalytic residue in stabilizing His conformation necessary for efficient catalysis. This Asn residue is likely conserved for some other yet unknown purpose.
These rhomboid structures have answered many questions raised from genetic and biochemical studies, but have also generated a debate on the mechanism of substrate entry (entry from helices 1 and 3 or from helices 2 and 5) and on the exact location of catalysis (adjacent to or inside the acyl region of the transbilayer). We refer the interested reader to two excellent reviews on this controversy [10,16] and focus here on common features important for catalysis and how they might inform on the PARL subfamily of rhomboids.
The bacterial rhomboid structures address two key questions on how intramembrane proteolysis occurs. The first question is how H2O enters a lipid bilayer, which is essential for performing substrate hydrolysis. The structures show the catalytic dyad lying buried in the detergent micelles up to 10 Å from the surface, but despite this burial, H2O is able to penetrate into the dyad and its entry may be regulated by the conserved L1 loop and/or less conserved L5 cap loop (Figure 1). The exact mechanism of H2O entry is unknown and this question is difficult to address especially given that membrane protein structure can be highly dependent on the membrane environment. Structural analysis is also dependent on choice of membrane mimetic used [17], and the structures available to date have been determined using detergent micelles as the membrane mimetic. Indeed, the sensitivity of GlpG to chemical modification differs between detergent micelle and membrane environments [18], suggesting a sensitivity of the rhomboid structure to the surrounding lipidic milieu.
The second key question concerns how the enzyme gains access to the peptide backbone, which is necessary for proteolysis. The geometry of the α-helix naturally shields the peptide backbone because the amino acid side chains “sprout away” from the helical axis preventing a close approach to the backbone. Therefore, an α-helix likely unfolds first in order for a protease to gain access to its peptide backbone substrate. The requirement for the unfolding of the TMH substrate sets up a dilemma as TMHs in the membrane are unlikely to unfold as the thermodynamics strongly favors the folded helical conformation (exposure of the polar peptide bond to the acyl bilayer is unfavorable) [19-21]. The presence of water in the structures and the substrate preference for Gly/Val/Ile residues (see below) offers a reasonable explanation for how local unfolding might occur: the helical propensities of these residues are dramatically different in a membrane vs. aqueous environment [22,23]. In the membrane, these residues have a high helical propensity, whereas in aqueous solution it is quite low. Therefore, upon substrate binding these Gly/Val/Ile residues likely become exposed to water in some manner under debate [10,16], and by doing so likely become unfolded allowing the enzyme access to the peptide backbone.
Addressing the structure of the rhomboid domain of eukaryotic rhomboids is imperative to understand their biology and potential as therapeutic targets. Although one would expect that eukaryotic rhomboids share a similar core structure to that of prokaryotic members. Eukaryotic rhomboids have 7 and not 6 TMH. Parl has the additional TMH appended N-terminally to the 6-TMH rhomboid domain (TMH-A). TMH-A is presumably docked against the rhomboid 6-TMH core, and from a cursory analysis of the GlpG structure is likely constrained topologically to lie at the helical interface between helices 1-2 or 2-5, which could poise it to play an important role in substrate recognition if substrate entry is through the proposed helix 2-5 gate (Hill and Pellegrini, unpublished observations). Such an orientation might allow Parl further modes of discriminating substrates and regulation. It is intriguing to speculate that the paucity of substrates of the mitochondrial rhomboids might be explained by the presence of TMH-A, although the repertoire of substrates recognized by the mitochondrial rhomboids is not firmly established.
4. Insights into Mitochondrial Rhomboid Mechanism and Substrate Specificity
The structural studies above have helped to interpret a body of genetic and biochemical studies on rhomboid substrate specificity. Studies in bacteria and drosophila have shown that RHO subfamily members Aar from Providencia stuarti and Rho-1 from Drosophila melanogaster can functionally complement each other. Expression of Providencia AarA triggers EGFR signalling when expressed in flies, whereas Drosophila Rhomboid-1 rescuea the Providencia phenotype caused by AarA deficiency, thereby indicating that the fly and bacterial RHO enzymes act by similar mechanisms including shared substrate specificity [24,25]. The same appears to be true for PARL subfamily members, as yeast strains lacking the mitochondrial rhomboid Pcp1Δ can be rescued by expression of mammalian Parl [8]. However, this finding is not entirely general as human thrombomodulin is cleaved by the human, mouse and zebrafish RHBDL2, but not by the drosophila Rhomboid-1 and the bacterial Aara rhomboid proteases [26]. These studies suggest that some discrimination of the substrate must occur.
The substrate specificity of the RHO subfamily is better defined than that of the PARL subfamily, and appears to have a rather weak consensus motif. For the RHO subfamily, studies using Drosophila Rho-1 and mutagenesis of two natural substrates, the TMH of Spitz and Keren, have shown that Rho-1 recognizes a common region of the TMH substrate that contains small residues (Gly,Ser,Ala), some of which are known to destabilize water-soluble helices (Gly,Ser,Ile,Val,Thr) [18,25]. The position of this region in the TMH is noteworthy as in some rhomboids it appears to have a strict requirement for being in the first 25% of residues that define the TMH, which places the cleavage site at the “top” of the TMH lying adjacent to the extracellular region of the substrate [27]. However, for human thrombomodulin, it appears that the TMH plays less of a role than its attached soluble domain in determining cleavage by RHBDL2 [26], which is quite interesting in comparison with others that support a model in which rhomboids cleave any transmembrane helix that contains a Gly-Gly type motif clustered at the “top” of the TMH [27,28]. Grafting a Gly-Gly motif onto a TMH from TGFα resulted in cleavage of this protein that is not normally a substrate for Rho-1 [25], and grafting the thrombomodulin soluble domain onto this TM also conferred cleavage [26]. These differences likely highlight the differences between Rho-1 and RHBDL2 and suggest that a substrate specificitiy for rhomboids may be promiscuous but not identical, even though studies with rhomboids from different species can functionally complement each other in some cases [24,25].
These general substrate requirements have also been put to the test by their use in predicting novel rhomboid targets in other species. Indeed, by using this substrate consensus motif and searching genomes for potential rhomboid targets, Urban and colleagues discovered rhomboid substrates in Toxoplasma gondii that appear important in host cell invasion of this pathogenic species [27]. Subsequent elegant work from the Urban laboratory has similarly identified rhomboid substrates in the malarial parasite Plasmodium falciparum [29,30]. These studies nicely demonstrate the utility of defining rhomboid substrate specificity and the widespread use of rhomboids in signaling. However, the importance of further defining the substrate specificity is underscored by the fact that the natural substrate for the E. coli rhomboid, GlpG, is still not known.
To address this issue and the general question of substrate specificity, Akiyama and co-workers used a combinatorial approach in E. coli that allowed randomization of a reporter substrate LY2 for the GlpG rhomboid [32,109]. They found that a negatively charged residue was the primary determinant of cleavage [32,109]. In this work, the amino acid preceding peptide bond hydrolysis (the P1 position) had a preference for the small and polar Ser residue; surprisingly, they found the amino acid succeeding peptide bond hydrolysis (the P1′ position) had a preference for negatively charged Asp, which has not been observed in other rhomboid studies. This work is notable because it suggests that peptide bond hydrolysis may also occur outside of the plane of the bilayer, a possibility supported by the demonstration of peptide bond hydrolysis in non-TMH regions of casein, a soluble protein [31,32,109], a finding that, extended to mitochondrial rhomboids, could explain why the cleavage of Mgm1p occurs outside of the TMH of the GTPase([33]; see chapter 5). More recently, the bacterial rhomboid GlpG was shown to cleave outside of the TMH [34]. Thus, it appears that rhomboids can cleave within a TMH and the adjacent region, which has mechanistic implications in terms of how this exactly occurs and is important for the identification of rhomboid substrates and the development of therapeutics directed against rhomboid activity. It is anticipated, therefore, that computational modeling and structural studies on co-complexes between enzyme and non-hydrolyzable substrate analogs will be required to address this crucial issue.
The mechanistic details of rhomboids are currently being debated in the field, with two models proposed for both substrate entry into the active site (helices 1-3 vs. helices 2-5), and for where hydrolysis takes place (discussed above). Additionally, it appears that the irregular shape and small hydrophobic thickness of the bacterial rhomboid protein may cause significant bilayer deformations, which could play an important role in substrate entry into the active site [35,36]. Certainly the activities of rhomboids exhibit some dependence on the composition of the lipid bilayer in which they are embedded [18,28], suggesting that lipids (or membrane microdomains) could play a role in the regulation of the activity of these proteases in vivo.
An open and fundamental question for the field is the regulation of hydrolytic activity. The above studies have elegantly demonstrated activity independent of other accessory protein factors [18], yet proteolysis is generally an activity that is highly regulated within the cell to prevent unregulated proteolysis that would likley be detrimental. Given the role of rhomboids in cellular signaling, one would also expect these enzymes to be highly regulated to prevent improper signaling. Unlike most proteases, the rhomboids are not synthesized as inactive precursors (zymogens). Therefore, it seems likely that rhomboid activity is mediated through accessory proteins or postranslational modifications, especially given the broad substrate specificity that would suggest a large number of potential substrates. We expect the regulation of these enzymes will be a fruitful area of investigation in the future.
For the PARL subfamily members, little is known about substrate specificity, mainly because very few substrates are known and cleavage sites of these have not been identified with a notable exception being the inner membrane dynamin-related protein Mgm1p [33]. In Mgm1p, substituting GlyGlyMet in the predicted TMH with bulkier ValValLeu blocks Pcp1/Rbd1-mediated cleavage, suggesting that the GlyGly substrate motif of RHO rhomboids is also important for the PARL rhomboids [8]. Interestingly, however, in the Mgm1p homologue of the fission yeast Schizosaccharomyces pombe, Msp1p (NP_596452), the putative GlyGly substrate motif is placed differently and lies one helical turn away from the predicted TMH. This suggests that the GlyAla/GlyGly motif may not be the primary determinant for the substrates of mitochondrial rhomboids. Notably, the TMH of the other Pcp1/Rbd1 substrate, cytochrome c peroxidase Ccp1, does not share structural similarity to that of Mgm1p, which raises the possibility that mitochondrial rhomboids either do not require their TMH substrates to have the same structural determinants as RHO family members.
The possibility that the TMH of Mgm1p from S. cerevisiae and S. pombe may be cleaved at different places along the TMH points to unusual protein dynamics being important for catalysis. The importance of protein dynamics is further highlighted by the ability of many rhomboids from both families to hydrolyze substrates in two positions that only differ by two residues, placing them on opposite faces of a canonical α-helix [28,37]. Two possible mechanisms come to mind: one in which dual cleavages arise from a screw-type motion of the substrate helix, and the other from local unfolding of the substrate. The local unfolding mechanism is perhaps more plausible based on mechanistic concerns discussed above. Either mechanism requires intrinsic dynamics of the enzyme, and it is important that biochemical studies on substrate specificity in detergent micelles also be performed in native membranes given selective differences that these environments present for protein structure and dynamics [38]. Importantly, recent molecular dynamics simulations of the E. coli GlpG indicates that the enzyme undergoes significant coupled motions between regions that are remote in tertiary structure [15,36]. Future studies using molecular dynamics simulations of enzyme-inhibitor complexes should be revealing and lead to experiments that may discriminate between these different models for substrate recognition and cleavage.
Given the importance of processing Mgm1p and its human homologue, Opa1, for the regulation of mitochondria dynamics in yeast and apoptosis in mammalian cells, cleavage site identification of Opa1 (and of its orthologs) and the substrate specificity of mitochondrial rhomboids are worthwhile goals. The elucidation of this aspect of mitochondrial rhomboids biology is of outstanding importance in finding inhibitors of mitochondrial rhomboid proteases.
5. The Yeast Mitochondrial Rhomboid, Pcp1/Rbd1
With the above structural and mechanistic information in mind, we know focus on recent advances in mitochondrial rhomboid biology. In Saccharomyces cerevisiae, two rhomboid genes exist, Rbd1 and Rbd2. In 2002, Esser and Pratje were the first to report that one of them, Rbd1, encodes a mitochondrial rhomboid protease, which they showed to be required for the processing of cytochrome c peroxidase (Ccp1p; NP_012992) and named, therefore, Pcp1 (NP_011615). Pcp1/Rbd1 codes for a polypeptide of 346 amino acid residues (38.8 kDa) that contains a MitoProt-predicted mitochondrial targeting sequence (score: 0.99), a 6-TMH rhomboid domain (spanning a.a.143-331), and a predicted TMH (106PLGSMTILGLSLMAGIYFG124) appended near its N-terminus. The protease is inserted in the inner mitochondrial membrane, where it serves to cleave Ccp1p [39] and, as later jointly reported by the Freeman and Reichert labs, the inner membrane dynamin-related protein Mgm1p [8,33] (NP_014854).
Ablation of Pcp1/Rbd1 activity has a profound effect on mitochondrial shape. Morphological analysis of the organelle in a yeast strain lacking the rhomboid protease showed that 72% of Pcp1Δ cells contained partially fragmented mitochondria, although each cell carried a few tubular-shaped organelles. In 17% of the cells, mitochondria were completely fragmented, a three-fold increase versus the amount of disorganized mitochondria observed in wild-type cells (∼5%). The mitochondrial morphology in Pcp1Δ cells is similar to yeast strains lacking other regulators of mitochondrial fusion, namely Fzo1, Ugo1, and Mgm1p, although the fragmentation of mitochondria in Fzo1Δ, Ugo1Δ, and Mgm1Δ cells is somewhat more severe, and tubular structures are rarely observed [40-43]. The fragmentation of mitochondria in Fzo1-, Ugo1-, and Mgm1-deficient cells is rescued by disrupting Dnm1, a gene encoding a dynamin-related GTPase essential for mitochondrial fission. In double null Pcp1Δ Dnm1Δ cells, mitochondria appeared as elongated tubules, similar to those seen in Mgm1Δ Dnm1Δ mutants, thereby indicating that fragmentation of mitochondria by loss of the mitochondrial rhomboid protease in yeast cells depends on loss of mitochondrial fusion [44]. Surprisingly, in contrast to Mgm1Δ cells, which are completely defective in mitochondrial fusion, Pcp1Δ cells can fuse their mitochondria after yeast cell mating. Interestingly, loss of Rbd1/Pcp1p is more detrimental to yeast cells than loss of Ccp1: Pcp1Δ cells lose mtDNA and cannot grow on nonfermentable carbon sources, whereas Ccp1Δ mutants maintain mtDNA and grow on nonfermentable medium [44,45]. These findings may indicate that the yeast mitochondrial rhomboid protease has multiple substrates.
In wild-type Saccharomyces cerevisiae mitochondria, the dynamin-related protein Mgm1p exists in two forms of 100 kDa (l-Mgm1) and a 90kDa (s-Mgm1), respectively; under steady state conditions these two forms are present in roughly equal amounts [46,47]. Dynamins are a superfamily of large GTPase mechanoenzymes involved in membrane remodeling. Mgm1 and its animal homologs contain an N-terminal mitochondrial import sequence (MIS), followed by a predicted TMH (70IIRLPIYVGGGMAAAGSYIAY90), a coiled-coiled domain, a GTPase domain, a middle domain, and a C-terminal coiled-coil domain known as the GTPase Effector Domain (GED). The MIS targets full length Mgm1p to the mitochondria, where it is cleaved upon import by the mitochondrial processing peptidase (MPP), to generate l-Mgm1 [33,48]. While the sub-mitochondrial localization of this dynamin has been for sometime the subject of controversy, it is now widely accepted that l-Mgm1 is inserted in the inner mitochondrial membrane, with the C-terminal GTPase domain exposed in the inter-membrane space. N-terminal sequencing of the two mitochondria-imported forms of Mgm1p identified an N-terminus of 60ISHFPKII for l-Mgm1 and of 142LIAATS for s-Mgm1[29]. Interestingly, two species of the s-Mgm1 that differ by 2 amino acids (140ATLIAATS and 142LIAATS) have been found in similar amounts [33]. It has been suggested that the initial cleavage takes place after Thr139 and that the further removal of two residues is caused by other peptidases in the intermembrane space or during the preparation of cell extracts; however, it is also plausible that the Pcp1/Rbd1 cleaves Mgm1p after Thr139 as well as after Thr141 [33]; the 79GGM81→79VVL81 substitution in the predicted TMH of Mgm1p blocks the generation of sMgm1 [8].
Cells lacking Pcp1p are defective in the 90-kDa s-Mgm1 form of the dynamin protein which is, therefore, present in the mitochondria of yeast Pcp1Δ cells only as the 100 kDa l-Mgm1 form. Mutations in any of the two catalytic residues in Pcp1/Rbd1 rhomboid domain (S256A and H313A) lead to complete loss of Pcp1/Rbd1 function and s-Mgm1 generation, indicating that its proteolytic activity is required for the processing of l-Mgm1, which in turn controls normal mitochondrial shape, and mtDNA maintenance via the generation of the 90 kDa form (s-Mgm1) [8,33,44]. However, the processing of Mgm1 is not strictly required for mitochondrial fusion, indicating that the 100 kDa form is sufficient to promote yeast mitochondrial fusion [44]. Yeast Pcp1Δ cells carrying the plasmid coding for s-Mgm1 grow on YPG in contrast to the Pcp1Δ strain lacking it, indicating that s-Mgm1 can partially complement the respiration-deficient phenotype of the Pcp1Δ strain [33]. Also, maintenance of mtDNA and restoration of the mitochondrial morphology defect in Pcp1Δ cells depends on the expression of s-Mgm1, further supporting the conclusion that the activity of the mitochondrial rhomboid protease is to generate s-Mgm1 [33], a notion supported by the finding that the mammalian ortholog of Pcp1, Parl [7], can rescue the defects observed in Pcp1Δ yeast strains [8]. Importantly, only the co-expression of the l-Mgm1 and s-Mgm1 forms of this dynamin protein rescued the mitochondrial morphology defects shown by Mgm1Δ yeast cells, although differences in the expression levels of s-Mgm1 and l-Mgm1, or the ratio of both isoforms, leads to different degrees of complementation and differentially rescues the loss of mtDNA and mitochondrial morphology [33]. Recent studies have led to propose that the primary function of l-Mgm1 is to act, in a cardiolipin-dependent manner, as an anchor in the inner mitochondrial membrane where, in concert with the GTPase activity of s-Mgm1, co-assembles with s-Mgm1 to mediate the fusion of inner membranes [49,50].
Another substrate of Pcp1 is the mitochondrial protein Ccp1, which also exists in two forms [39]: a mature, membrane-bound form and a IMS soluble form similar to Mgm1. During maturation, Ccp1 is initially cleaved by Yta10/Yta12 [39], a mitochondrial AAA-protease anchored in the inner membrane with its active site in the matrix [51]. As mentioned above, yeast Pcp1Δ strains lack a low molecular weight form of Ccp1 (similar to Mgm1p). Cleavage of Ccp1 by Pcp1/Rbd1 occurs directly after or within its hydrophobic sorting sequence, which has been suggested to serve as a translocation-arrest signal. Thereby, Ccp1 becomes localized to the intermembrane space [52,53]. Unlike Ccp1, the processing of Mgm1 is not dependent on Yta10/Yta12. Therefore, processing by Yta10/Yta12 appears not to be a general prerequisite for proteolytic processing by the mitochondrial rhomboid protease Pcp1/Rbd1.
Whether the mitochondrial rhomboid protease Pcp1/Rbd1 is involved in the processing of additional proteins important for yeast respiration competence, maintenance of mtDNA, and mitochondrial morphology, remains to be elucidated. However, as discussed in the elegant work from the Reichert's group, this possibility appears unlikely in light of their finding that the Pcp1Δ phenotype is rescued by expression of s-Mgm1 [33]. Therefore, the phenotypic effects observed can currently be explained without claiming a requirement of Pcp1/Rbd1 for processing of other substrates relevant for the maintenance of mtDNA and mitochondrial morphology. In agreement with this, the deletion of Ccp1, the only other putative substrate for Pcp1/Rbd1 described to date [39], did not show any defects in mitochondrial morphology or respiration competence [45]. Thus, the phenotype observed in Pcp1Δ yeast cells appears to be due to the specific loss of s-Mgm1 biogenesis, a possibility supported by the observation that the TMH of Ccp1 is structurally different from that of Mgm1.
6. The Fly Mitochondrial Rhomboid, Rho-7
The member of the PARL subfamily in Drosophila is Rho-7 [7] (AAF58598). The protein is 351 amino acids long and, like all other mitochondrial rhomboids, has a predicted N-terminal mitochondrial localization signal and the signature “1+6” structure, with the proximal TMH spanning amino acid 77-95 (77AVAFTGAFTVGCFAGATIL95). Functional studies in vivo have shown that approximately 90% of Rho-7 mutant flies that lack the transcriptional start site and the first 18 codons of the protease die before pupariation [54]. Death occurs during both embryonic and larval stages, but those flies that survive to pupariation develop to adults (10% of these die during the process of eclosion, as they get stuck while crawling from the pupal case). Surviving flies appear morphologically normal but live for only 3 days whereas control flies live for an average of 60 days. Males are sterile but the females are fertile. Surviving adult Rho-7 mutants are morphologically normal, with the exception of a wing-position defect, in which the wings of about 60% of individuals hang down on either side of the abdomen, as opposed to control flies that have their wings tucked and positioned on top of the abdomen. However, these flies are unable to fly, have extreme difficulty walking, and display erratic twitching in their legs and head, indicating severe synaptic defects, which appear to be the primary consequences of Rho-7 loss. Apoptosis is not believed to have a role in the observed massive embryonic/larval lethality, as also substantiated by the observation that Drosophila S2 cells in which Rho-7 was reduced by RNAi showed no extra sensitivity to cycloheximide-induced apoptosis [54]. Importantly, this phenotype is similar to that observed in flies null for PINK1 and Parkin, two genes linked to human Parkisonism. Recent studies in flies have shown a genetic link between Rho-7, PINK1, and Parkin, suggesting a role of the mitochondrial rhomboid in this disease [55]. However, since in the mammalian system the pathogenic activity of Parkin and PINK1 mutants in Parkinson's diseases is localized in the cytosol [56-58], not inside the mitochondrion, this genetic interaction remains orphan of a molecular mechanism that could explain the putative role of the mammalian mitochondrial rhomboid protease, Parl, in this disease.
Electron microscopy analysis on Rho-7 mutant flies showed many small mitochondria in the space between the myofibrils, as compared to larger mitochondria that completely filled the intramyofibril space in control flies. Significant loss of inner membrane cristae was also observed as compared to control flies. Further, nebenkerns undergo normal mitochondrial aggregation but fail to undergo membrane fusion, thereby supporting a functional participation of Rho-7 in this process [54].
Taken together these data demonstrate a functional conservation of the yeast and fly mitochondrial rhomboid in the regulation of mitochondria dynamics. However, they also expose striking phenotypic and functional differences with respect to what observed in the mammalian system. Mice null for Parl do not show embryonic lethality and synaptic defects are not observed [5]. Moreover, the function of the mammalian mitochondrial rhomboid protease in homeostatic control of apoptosis is unequivocal [5,59]. In this respect, the structural differences existing between Rho-7 and mammalian Parl are noteworthy. Unlike mammals, in insects the N-terminal domain and the loop that connects the first two TMHs (TMH-A and TMH-1) are small and not conserved, thereby confirming that in-depth protein domain analysis is necessary to understand the complex and diverse functionality of rhomboids across evolution [60,61].
7. The Mammalian Mitochondrial Rhomboid, Parl
Parl was discovered following a yeast two-hybrid screening done using as bait the C-terminal region of Presenilin-2 (PSEN2 or PS2), a proapoptotic familial Alzheimer's disease protein that is highly homologous to Presenilin-1 (PSEN1 or PS1) [62]. The bait, spanning amino acid 330-448 and termed PS2-C-cas (produced by caspase cleavage during apoptosis [63]), includes part of the large hydrophilic loop and the last 3 transmembrane domains of PS2; it shares a high degree of homology to PS1, it is highly conserved in mammals, and has antiapoptotic activity [63]. A human brain cDNA library screened with PS2-C-cas yielded multiple clones of a near-full length unknown protein which, in the Y2H system, showed very strong interaction also with a C-terminal fragment spanning amino acids 264-467 of PS1. Bioinformatic analysis revealed that this uncharacterized protein was a member of the Rhomboid family of proteins, a conserved family of developmental regulators in Drosophila. It was, therefore, named Parl, for Presenilin-associated Rhomboid-like protein [4]. Subsequent studies indicated, however, that the reported interaction between Parl and the Presenilin proteins must be artifactual, possibly due to the highly hydrophobic nature of these two polytopic membrane proteins.
Parl is a 379 amino acids-long protein containing seven transmembrane helices, organized in a “1+6” structure, where the first TMH is not part of the 6 TMHs that form the core rhomboid domain (spanning amino acid 178-353). The first, proximal TMH (101PLFFTVGFTGCAFGSAAIWQ120) is here proposed to be termed TMH-A. Instead, the distal 6 TMHs forming the rhomboid fold are here proposed to be termed TMH-1 to -6, to allow an easy comparison with the six TMHs that constitute the catalytic serine protease domain found in all prokaryotic and eukaryotic rhomboids.
Although preliminary investigations located Parl in both mitochondrial membranes [60], further studies refined this finding and conclusively localized the protease only in the inner mitochondrial membrane, with the N-terminus exposed to the matrix and the C-terminus to the IMS [61]. Unlike the yeast and fly PARL orthologues, the N-terminal domain of Parl, termed Pβ domain, undergoes two consecutive processing events, termed α- and β-cleavage. The proximal α-cleavage (Gly52↓Phe53) is constitutive and linked to the import of the full length Parl protein in the mitochondria; therefore, it is probably executed by one of the two proteases that typically are responsible for the import of most IMM proteins, the mitochondrial processing peptidase (MPP) and the inner membrane peptidase (IMP). This cleavage produces MAMP, for mature mitochondrial Parl (spanning amino acid 53-379 of full length Parl). The distal β-cleavage (Ser77↓Ala78) is not constitutive and appears to be subjected to a very complex mechanism of regulation (discussed below, chapter 8); it produces PACT, for Parl C-terminal fragment (spanning amino acid 78-379 of Parl; [60]).
The human and murine Parl gene are located on chromosome 3 (location: 3q27.1) and 16, respectively. Variation in Parl sequence and/or expression have been suggested to represent a risk factor for type 2 diabetes and other components of the metabolic syndrome, a hypothesis based on the observation that in 1,031 human subjects a conserved amino acid substitution (L262V) in Parl was associated with increased plasma insulin concentration, a key risk factor for diabetes [64]. However, subsequent studies on a significantly larger cohort disputed the association between this polymorphism and two measures of insulin resistance (fasting plasma insulin and blood glucose levels), suggesting that this variant is highly unlikely to be an important contributor to insulin resistance [65,66]. To date, no other genetic association between Parl and any human disease or metabolic condition have surfaced; their identification will require a better understanding of Parl structure, function, and mechanisms of regulation, to allow geneticists to predict hypomorphic, hypermorphic, gain of function, and loss of function mutations.
Studies from the De Strooper group have shown that ablation of the Parl gene in the mouse does not affect development, yet has a major impact on postnatal growth and lifespan. Mice lacking Parl die between weeks 8-12 from cachexia sustained by multisystemic atrophy; however, Parl +/- mice do not show any obvious phenotype [5], suggesting the existence of compensatory mechanisms to gene dosage effects. Surprisingly, the expected role of Parl in mitochondrial morphology and oxidative phosphorylation was not observed. In every tissue analyzed, Parl ablation did not alter the morphology of the mitochondrial reticulum or mitochondrial respiration, irrespective of the substrate used by the organelle; further, it failed to be required for mitochondria fusion [5]. Although a role of Parl in mitochondria morphology regulation exists [61] (see Chapter 7.3), these data call into question, in the mammalian system, the importance of mitochondrial rhomboids in inner mitochondrial membrane fusion, a function which is instead well documented in yeast [33,49,67]. Since we still don't known whether loss of Rho-7 in drosophila blocks Opa1 cleavage and mitochondria fusion [54], it is unclear at which point in metazoan evolution rhomboid regulation of mitochondria fusion might have been lost.
7.1 Parl and Apoptosis
Elegant studies from the Scorrano lab showed that cells lacking Parl were more susceptible to apoptosis, suggesting a mechanism for the massive atrophy observed in the Parl knockout mouse [5]. Further investigations from this group revealed that Parl is required to regulate the kinetics of cytochrome c release from mitochondria. More specifically, Parl does not influence the actual efflux of cytochrome c from the organelle, but influences the pathway of cristae remodelling controlled by Opa1 [5,59]. This function requires the integrity of the catalytic dyad of Parl, as substantiated by the inability of a catalytically dead Parl mutant to complement Parl-/- cells. Genetic analysis show that Opa1 and Parl are part of the same pathway, with Parl positioned upstream of Opa1 in the control of apoptosis; physical association between Parl and Opa1 were also reported in Y2H experiments and biochemical assays [5]. In Parl-/- mitochondria, Opa1 oligomer-containing complexes are reduced and their disassembly accelerated during apoptosis. Biochemical analysis showed that levels of IMS Opa1 are reduced by approximately 50% in Parl-/- mitochondria [5]; this reduction could explain why in the absence of Parl the cristae are not significantly disrupted under basal conditions, yet undergo increased remodelling during apoptosis.
The retrieval of a substantial fraction of IMS-soluble Opa1 form in Parl-/- mitochondria implies that Parl is probably not the only protease involved in Opa1 processing. Consistent with this possibility, Paraplegin, an IMM mAAA metalloprotease, Yme1 [68,69], and possibly other metalloproteases [70] were also shown to participate in the cleavage of Opa1, to produce a short isoform, s-Opa1, that is mitochondria fusion incompetent [71,72]. Further, processing of OPA1 to s-OPA1 is sensitive to metalloprotease inhibitors and is not affected by the KO or the silencing of Parl [68,70,71] (for reviews on the biology of Opa1 see [73-79]). Thus, in steady state conditions the function of Parl is to execute the intramembraneous cleavage of Opa1, either directly or indirectly, to liberate an IMS-soluble form of the protein (IMS-Opa1) that assembles in macromolecular complexes with Parl and with the uncleaved, IMM-bound form of Opa1. This structure “staples” cristae junctions together [5,59], thereby precluding cytochrome c access to the IMS. Consistent with this model, expression of IMS-Opa1 in the mitochondrion was shown to protect MEF cells from several intrinsic pro-apoptotic stimuli [5]. Whether in some cancer cells Parl activity is upregulated is not known, although this may be a mechanism tumour cells develop to acquire insensitivity to intrinsic proapoptotic stimuli or resistance to anti-cancer drugs. The analysis of the Parl +/- mouse could prove to be a precious genetic model to address this possibility.
A recent study has claimed that Parl has an additional role in apoptosis [80]. According to this report, the congenital neutropenia protein Hax1 [81] (NP_035956) presents the serine protease Omi/HtrA2 to Parl, to facilitate the generation of an active form of Omi/HtrA2 that prevents the accumulation of active Bax on the mitochondrial outer-membrane [80], which is required for cytochrome c release to the cytosol. However, a model in which Omi/HtrA2 regulates Bax is not supported by available data. It is also not clear that Hax1-mediates substrate presentation of Omi/HtrA2 to Parl because it is difficult to reconcile with the following observations: Hax1 is not part of the mitochondrial proteome [82,83], it appears peripherally associated to the ER where it is functionally implicated in SERCA activity [84,85], it does not appear to be a bona fide Parl-interacting protein [84], and it is not required for any other Parl-related cleavage (Opa1 [80] and Parl N-terminus [84] - discussed in chapter 7.3). Therefore, to date the established role of Parl in apoptosis is the one linked to the Opa1-mediated mechanism of cristae remodelling [5,59]. However, the possibility that Parl could cleave Omi/Htr2A [80], independent of Hax1, is plausible and deserving of further investigation.
7.2 Parl and Mitochondrial Stress
Omi/HtrA2 is a mitochondrial serine protease [86]. In rodents, loss of Omi/HtrA2 activity causes mitochondrial dysfunctions that lead to a progressive neuromuscular disorder and death by 40 days of age [87,88]. Further, it induces the transcriptional upregulation of nuclear genes characteristic of the integrated stress response and results in the accumulation of unfolded proteins in the mitochondria, defective mitochondrial respiration and enhanced production of reactive oxygen species that contribute to neuronal cell death [89]. A mutation in the gene encoding Omi/HtrA2 has been linked to Parkisonian neurodegeneration in humans [90], but its actual role in the etiology of the disease is disputed [91,92]. The functional participation of Omi/HtrA2 in the Parkinson's PINK/Parkin pathway is also contested [58,93,94] and highlights the difficulties in dissecting and comparing complex cellular stress response pathways in different organisms.
In humans, the Omi/Htr2A transcript is expressed as two splice variants, one encoding a protein of 458 amino acids (NP_037379), and the other a lower molecular weight polypeptide of 361 residues which is missing amino acid 238-302 and 372-403 (NP_659540). Mouse (NP_062726) and drosophila (NP_650366) appear instead to express only one isoform of the protein. The structural organization of Omi/HtrA2 is highly conserved in fly and mammals, and exhibit a strongly predicted TMH located around amino acid 75 and 115, respectively. The protein is localized in the mitochondrial intermembrane space, where it exists in two forms, one that is integrated to the inner mitochondrial membrane and one of lower molecular weight that is soluble in the IMS (IMS-Omi/HtrA2). Since loss of Parl expression halves the amount of IMS-Omi/HtrA2, it has been proposed that the membrane-bound form is cleaved by the rhomboid protease [80]. Similar data were also observed in Drosophila where, loss of Rho-7 activity seems to have a more profound effect on the elimination of the processed form of Omi/HtrA2 [55]; the reason why ablation of Parl does not suppress IMS-Omi/HtrA2 remains unexplained [80]. Thus, the possibility that PARL members could participate in the mitochondrial stress response via Omi/HtrA2 cleavage awaits validation from additional detailed biochemical and functional analysis. In this respect, a noteworthy feature of the poorly conserved TMH from the fly, fish (CAX13510) and mouse orthologues of Omi/HtrA2 is the presence of a conserved Gly residue, which is reminiscent of the Gly present within the consensus cleavage site detected in substrates of the RHO subfamily [27]. This observation encourages further investigation on the potential link between Parl and Omi/HtrA2 cleavage.
7.3 Parl and Mitochondria Morphology
The N-terminal domain of Parl is exposed to the mitochondrial matrix [61] and undergoes to two consecutive cleavage events, termed α- and β-cleavage [60]. The first, α-cleavage, is constitutive and removes the mitochondria targeting sequence, 1MAWRGWAQRGWGCGQAWGASVGGRSCEELTAVLTPPQLLGRRFNFFIQQKCG52, as demonstrated by the observation that, fused to GFP, this peptide targets the chimeric fluorescent protein to the organelle [60]. By constrast, β-cleavage is not constitutive: in transfected HeLa and HEK 293 cells it occurs when the cells become confluent [60,61,84].
Four lines of evidence suggest that β-cleavage of Parl (70SGEAYKRS77↓78ALIPPVEETVFYP90), being regulated and occurring in a domain of Parl that is strictly conserved only in mammals [60], has a critical function in mitochondria biology.
First, β-cleavage requires a mechanism of proteolysis that depends on Parl rhomboid activity supplied in trans: in transfected cells, a catalytically dead Parl S277G mutant is not subjected to β-cleavage unless wild-type Parl is co-expressed [60]. The identity of the protease that cleaves Parl at the β-cleavage site remains elusive, although one interpretation of available data is consistent with a model in which Parl cleaves itself in trans ([60], Jeyaraju and Pellegrini, unpublished observations).
Second, Parl β-cleavage is sensitive to amino acid substitutions: mutant Parl R76E, S77E, A78E and L79E have rhomboid activity but are not subjected to β-cleavage ([60,61,84]; Jeyaraju and Pellegrini, unpublished observations). Such substrate specificity is typical of proteases whose activity must be tightly controlled, like those of caspases; it is, therefore, evidence that a key biological process depends on it.
Third, β-cleavage regulation is contingent on the the structure of an intact N-terminal Pβ-domain: deletion of amino acids 84EETV87 produces a form of Parl that when expressed in human HeLa and HEK293 cells is subjected to constitutive β-cleavage [61]. This finding is consistent with the observation that in the Pβ domain of marsupial and placental mammals orthologues of Parl there are no insertions or deletions; this indicates that during the ∼100 million years of mammalian evolution the Pβ domain must have been subjected to strong purifying selection, which can only be explained by functional constraints linked to the role that the mitochondrial rhomboid protease has acquired in higher vertebrates [61].
Fourth, β-cleavage of Parl is blocked by phosphorylation of residues located in close proximity to the cleavage site: phosphomimetic substitutions of these amino acids (S65D, T69D, and S70D) impair the processing without, however, affecting Parl rhomboid activity [61]. Since both endogenous and transfected wild type Parl are constitutively hyperphosphorylated [61], it is conceivable to speculate that a phosphorylation switch mediated by yet uncharacterized matrix kinase and phosphatase are an integral component of the mechanism of regulation of Parl β-cleavage, thereby increasing the degree of complexity of the mechanism proposed to its regulation in vivo (which we have observed in multiple tissues [61,84]).
The function of Parl β-cleavage has just started to emerge. In transfection studies, this processing cause mitochondria to fragment, even in the presence of dominant negative Drp1, indicating that the observed remodelling of the mitochondrial reticulum occurs through a block of mitochondrial fusion([61]; Pellegrini and McBride, unpublished observations). Mutant Parl, where β-cleavage has been abolished by removing (Δ75KRSAL79) or mutating the β-cleavage site (L79E) do not induce fragmentation, indicating that the processing is a gain of function, a finding consistent with the observation that Parl-/- MEF cells show only minor defects in mitochondria morphology [5]. What triggers β-cleavage remains to be determined, although in HeLa cells a number of stimuli that are known to trigger mitochondria fragmentation do not induce β-cleavage of endogenous or transfected Parl (Jeyaraju and Pellegrini, unpublished observations). Ongoing studies on a mouse mutant deficient in Parl β-cleavage will contribute to elucidating the function of such an intricate mitochondrial processing event that impacts on mitochondrial morphology in a unique way.
8. Mitochondrial Rhomboids and Cell Signaling
The signaling activity identified in rhomboids of the RHO subfamily [9] raises the question of whether rhomboids of the PARL subfamily share a similar signaling function. A careful analysis of the structural innovations acquired during vertebrate evolution by mammalian members of the PARL subfamily, and the study of its complex mechanism of regulation, strongly support the concept that, at least in mammals, mitochondrial rhomboids have likely been recruited to execute an important signaling function.
Mitochondria fusion and fission are two independent but intertwined processes directed by highly conserved proteins [74,77,95-97]. During vertebrate evolution the mechanisms governing fusion and fission were recruited to coordinate new mitochondrial functions, like apoptosis [5,59,73,76,98-102], a process that cannot have happened without the parallel emergence of novel mechanisms of regulation of the “old” core machinery that fuse and divide the mitochondrion. Despite their conservation, the mitochondrial rhomboid proteins in yeast (Pcp1/Rbd1 and Msp1) and mammals (Parl) have unrelated N-termini. In fact, Parl N-terminus shows no detectable similarity to any other available protein sequence. This region of Parl, designated Pβ domain (spanning amino acid 40–100 of full-length Parl), is vertebrate-specific, as indicated by the notable conservation among mammals and, to a lesser extent, other vertebrates, but not between vertebrates and insects [60]. Although the function of the Pβ domain has now emerged [61], its functional importance is evident from its sequence conservation. Indeed, in the four available mammalian Parl sequences, 58 of the 62 residues of the Pβ domain are invariant, and there are no insertions or deletions, which suggests that at least during mammalian evolution, the N-terminal region of Parl was subject to strong purifying selection, which can only be explained by functional constraints. In unconstrained sequences evolving neutrally, very few, if any, invariant residues would be expected to survive the 100 million years of evolution separating mammalian orders.
As discussed above, the N-terminal Pβ domain of Parl is processed upon the protein's import and insertion in the IMM, followed by a second cleavage whose sophisticated regulation through phosphorylation/dephosphorylation of its N-terminus underscores its importance for mitochondria biology. The latter cleavage releases within the mitochondrial matrix a 25 amino acid-long peptide termed Pβ-peptide [60,61]. Recent pioneering studies have shown that, in yeast, 6- to 27-amino acid-long peptides are constantly exported from the mitochondria matrix to the cytosol in an ATP- and temperature-dependent manner [103]. The biogenesis of these peptides is an intrinsic necessity for the organelle because linked to the catabolism of matrix and IMM proteins [103]. Therefore, the machinery transporting them across the two mitochondrial membranes must be conserved in all eukaryotes, indicating that the Pβ-peptide has a way to get expelled to the cytosol. Once in the cytosol, it has been shown that this peptide is then directed to the nucleus [60]. Two lines of evidence support this finding. First, a potent monopartite nuclear localization signal (NLS) is embedded within its sequence (54RKxxRKxxxRR64 [60]). In transfected HEK293 and HeLa cells, the Pβ-peptide fused to GFP efficiently transports the fusion protein inside the nucleus, a targeting that is disrupted when the basic residues of its NLS are mutated (e.g. RK/AA or RR/AA substitutions). Second, in primary cultures of differentiating mouse neurons, immunohistochemical analysis done with an antibody raised against the Pβ-peptide paints the nucleus of these cells [60]. Taken together these data suggest that Parl β-clevage is monitored by the nucleus through the Pβ-peptide, which therefore appears to execute mitochondrial retrograde signaling (MRS).
MRS is a recently emerged concept in cell biology, and defines a pathway of communication from the organelle to the nucleus under normal and pathophysiological conditions [104,105]. Functionally, it implies the participation of mitochondria in the regulation of nuclear activities, and it connotes the ability of the cell to feedback on the modulation of mitochondrial function [105]. MRS senses mitochondrial activities/dysfunctions and relay this information to the nucleus in order to initiate appropriate physiological readjustments. Accordingly, in yeast MRS coordinates a wide assortment of cellular activities, including nutrient sensing, growth control, aging, metabolism, and organelle homeostasis [105]. In mammals, MRS coordinates the mitochondrial stress response [106]; whether additional functions of the organelle are orchestrated through retrograde regulation remains unclear. Furthermore, the identity of the signals released from the mitochondria, the molecular mechanisms that sense them in the nucleus, and the response triggered there remain largely unexplored [107,108]. In this respect, although further studies are required to identify the role and mechanism of activity of the Pβ-peptide in MRS, its existence brings mammalian Parl within the mainstream concept of cell signaling via regulated intramembrane proteolysis. The case of Parl Pβ-peptide signaling is, however, unique because unlike other intramembrane-cleaving proteases, including RHO family members, Parl is the only intramembrane-cleaving protease where the putative signaling moiety is also part of the protease itself. It can be expected, therefore, that the study of the Pβ-peptide-mediated MRS will yield novel paradigms on how the nucleus monitors and coordinates structural and functional transitions in mitochondria, a process which is central to the life and death of the cell.
Acknowledgments
We thank Heidi McBride (University of Ottawa) for critical reading of the manuscript. This work was supported by a grant from the Canadian Institutes of Health Research to LP (MOP-82718). LP is a FRSQ Chercheur-Boursier Senior scholar.
Abbreviations
- MOMP
mitochondrial outer membrane permeabilization
- IMS
mitochondrial intermembrane space
- OMM
outer mitochondrial membrane
- IMM
inner mitochondrial membrane
- a.a.
amino acid
- TMH
transmembrane helix
- RIP
regulated intramembrane proteolysis
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tolia A, De Strooper B. Structure and function of gamma-secretase. Semin Cell Dev Biol. 2009;20:211–218. doi: 10.1016/j.semcdb.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 2.Bork P, Koonin EV. Predicting functions from protein sequences--where are the bottlenecks? Nat Genet. 1998;18:313–318. doi: 10.1038/ng0498-313. [DOI] [PubMed] [Google Scholar]
- 3.Koonin EV, Galperin MY. Sequence-evolution-function: Computational approaches in comparative genomics. Kluwer Academic Publishers Group; Dordrecht, The Netherlands: 2003. [PubMed] [Google Scholar]
- 4.Pellegrini L, Passer BJ, Canelles M, Lefterov I, Ganjei JK, Fowlkes BJ, Koonin EV, D'Adamio L. PAMP and PARL, two novel putative metalloproteases interacting with the COOH-terminus of Presenilin-1 and -2. J Alzheimers Dis. 2001;3:181–190. doi: 10.3233/jad-2001-3203. [DOI] [PubMed] [Google Scholar]
- 5.Cipolat S, Rudka T, Hartmann D, Costa V, Serneels L, Craessaerts K, Metzger K, Frezza C, Annaert W, D'Adamio L, Derks C, Dejaegere T, Pellegrini L, D'Hooge R, Scorrano L, De Strooper B. Mitochondrial rhomboid PARL regulates cytochrome c release during apoptosis via OPA1-dependent cristae remodeling. Cell. 2006;126:163–175. doi: 10.1016/j.cell.2006.06.021. [DOI] [PubMed] [Google Scholar]
- 6.Urban S, Lee JR, Freeman M. Drosophila rhomboid-1 defines a family of putative intramembrane serine proteases. Cell. 2001;107:173–182. doi: 10.1016/s0092-8674(01)00525-6. [DOI] [PubMed] [Google Scholar]
- 7.Koonin EV, Makarova KS, Rogozin IB, Davidovic L, Letellier MC, Pellegrini L. The rhomboids: a nearly ubiquitous family of intramembrane serine proteases that probably evolved by multiple ancient horizontal gene transfers. Genome Biol. 2003;4:R19. doi: 10.1186/gb-2003-4-3-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McQuibban GA, Saurya S, Freeman M. Mitochondrial membrane remodelling regulated by a conserved rhomboid protease. Nature. 2003;423:537–541. doi: 10.1038/nature01633. [DOI] [PubMed] [Google Scholar]
- 9.Urban S. Rhomboid proteins: conserved membrane proteases with divergent biological functions. Genes Dev. 2006;20:3054–3068. doi: 10.1101/gad.1488606. [DOI] [PubMed] [Google Scholar]
- 10.Ha Y. Structure and mechanism of intramembrane protease. Semin Cell Dev Biol. 2009;20:240–250. doi: 10.1016/j.semcdb.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Golde TE, Wolfe MS, Greenbaum DC. Signal peptide peptidases: a family of intramembrane-cleaving proteases that cleave type 2 transmembrane proteins. Semin Cell Dev Biol. 2009;20:225–230. doi: 10.1016/j.semcdb.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y, Zhang Y, Ha Y. Crystal structure of a rhomboid family intramembrane protease. Nature. 2006;444:179–180. doi: 10.1038/nature05255. [DOI] [PubMed] [Google Scholar]
- 13.Wu Z, Yan N, Feng L, Oberstein A, Yan H, Baker RP, Gu L, Jeffrey PD, Urban S, Shi Y. Structural analysis of a rhomboid family intramembrane protease reveals a gating mechanism for substrate entry. Nat Struct Mol Biol. 2006;13:1084–1091. doi: 10.1038/nsmb1179. [DOI] [PubMed] [Google Scholar]
- 14.Ben-Shem A, Fass D, Bibi E. Structural basis for intramembrane proteolysis by rhomboid serine proteases. Proc Natl Acad Sci U S A. 2007;104:462–466. doi: 10.1073/pnas.0609773104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lemieux MJ, Fischer SJ, Cherney MM, Bateman KS, James MN. The crystal structure of the rhomboid peptidase from Haemophilus influenzae provides insight into intramembrane proteolysis. Proc Natl Acad Sci U S A. 2007;104:750–754. doi: 10.1073/pnas.0609981104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Urban S, Shi Y. Core principles of intramembrane proteolysis: comparison of rhomboid and site-2 family proteases. Curr Opin Struct Biol. 2008;18:432–441. doi: 10.1016/j.sbi.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sulistijo ES, Mackenzie KR. Structural basis for dimerization of the BNIP3 transmembrane domain. Biochemistry. 2009;48:5106–5120. doi: 10.1021/bi802245u. [DOI] [PubMed] [Google Scholar]
- 18.Urban S, Wolfe MS. Reconstitution of intramembrane proteolysis in vitro reveals that pure rhomboid is sufficient for catalysis and specificity. Proc Natl Acad Sci U S A. 2005;102:1883–1888. doi: 10.1073/pnas.0408306102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sachs JN, Engelman DM. Introduction to the membrane protein reviews: the interplay of structure, dynamics, and environment in membrane protein function. Annu Rev Biochem. 2006;75:707–712. doi: 10.1146/annurev.biochem.75.110105.142336. [DOI] [PubMed] [Google Scholar]
- 20.Curran AR, Engelman DM. Sequence motifs, polar interactions and conformational changes in helical membrane proteins. Curr Opin Struct Biol. 2003;13:412–417. doi: 10.1016/s0959-440x(03)00102-7. [DOI] [PubMed] [Google Scholar]
- 21.Popot JL, Engelman DM. Helical membrane protein folding, stability, and evolution. Annu Rev Biochem. 2000;69:881–922. doi: 10.1146/annurev.biochem.69.1.881. [DOI] [PubMed] [Google Scholar]
- 22.O'Neil KT, DeGrado WF. A thermodynamic scale for the helix-forming tendencies of the commonly occurring amino acids. Science. 1990;250:646–651. doi: 10.1126/science.2237415. [DOI] [PubMed] [Google Scholar]
- 23.Senes A, Gerstein M, Engelman DM. Statistical analysis of amino acid patterns in transmembrane helices: the GxxxG motif occurs frequently and in association with beta-branched residues at neighboring positions. J Mol Biol. 2000;296:921–936. doi: 10.1006/jmbi.1999.3488. [DOI] [PubMed] [Google Scholar]
- 24.Gallio M, Sturgill G, Rather P, Kylsten P. A conserved mechanism for extracellular signaling in eukaryotes and prokaryotes. Proc Natl Acad Sci U S A. 2002;99:12208–12213. doi: 10.1073/pnas.192138799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Urban S, Schlieper D, Freeman M. Conservation of intramembrane proteolytic activity and substrate specificity in prokaryotic and eukaryotic rhomboids. Curr Biol. 2002;12:1507–1512. doi: 10.1016/s0960-9822(02)01092-8. [DOI] [PubMed] [Google Scholar]
- 26.Lohi O, Urban S, Freeman M. Diverse substrate recognition mechanisms for rhomboids; thrombomodulin is cleaved by Mammalian rhomboids. Curr Biol. 2004;14:236–241. doi: 10.1016/j.cub.2004.01.025. [DOI] [PubMed] [Google Scholar]
- 27.Urban S, Freeman M. Substrate specificity of rhomboid intramembrane proteases is governed by helix-breaking residues in the substrate transmembrane domain. Mol Cell. 2003;11:1425–1434. doi: 10.1016/s1097-2765(03)00181-3. [DOI] [PubMed] [Google Scholar]
- 28.Lei X, Ahn K, Zhu L, Ubarretxena-Belandia I, Li YM. Soluble oligomers of the intramembrane serine protease YqgP are catalytically active in the absence of detergents. Biochemistry. 2008;47:11920–11929. doi: 10.1021/bi800385r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brossier F, Jewett TJ, Sibley LD, Urban S. A spatially localized rhomboid protease cleaves cell surface adhesins essential for invasion by Toxoplasma. Proc Natl Acad Sci U S A. 2005;102:4146–4151. doi: 10.1073/pnas.0407918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baker RP, Wijetilaka R, Urban S. Two Plasmodium rhomboid proteases preferentially cleave different adhesins implicated in all invasive stages of malaria. PLoS Pathog. 2006;2:e113. doi: 10.1371/journal.ppat.0020113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maegawa S, Ito K, Akiyama Y. Proteolytic action of GlpG, a rhomboid protease in the Escherichia coli cytoplasmic membrane. Biochemistry. 2005;44:13543–13552. doi: 10.1021/bi051363k. [DOI] [PubMed] [Google Scholar]
- 32.Maegawa S, Koide K, Ito K, Akiyama Y. The intramembrane active site of GlpG, an E. coli rhomboid protease, is accessible to water and hydrolyses an extramembrane peptide bond of substrates. Mol Microbiol. 2007;64:435–447. doi: 10.1111/j.1365-2958.2007.05679.x. [DOI] [PubMed] [Google Scholar]
- 33.Herlan M, Vogel F, Bornhovd C, Neupert W, Reichert AS. Processing of Mgm1 by the rhomboid-type protease Pcp1 is required for maintenance of mitochondrial morphology and of mitochondrial DNA. J Biol Chem. 2003;278:27781–27788. doi: 10.1074/jbc.M211311200. [DOI] [PubMed] [Google Scholar]
- 34.Erez E, Bibi E. Cleavage of a Multispanning Membrane Protein by an Intramembrane Serine Protease. Biochemistry. 2009 doi: 10.1021/bi901648g. [DOI] [PubMed] [Google Scholar]
- 35.Wang Y, Maegawa S, Akiyama Y, Ha Y. The role of L1 loop in the mechanism of rhomboid intramembrane protease GlpG. J Mol Biol. 2007;374:1104–1113. doi: 10.1016/j.jmb.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bondar AN, del Val C, White SH. Rhomboid protease dynamics and lipid interactions. Structure. 2009;17:395–405. doi: 10.1016/j.str.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baker RP, Young K, Feng L, Shi Y, Urban S. Enzymatic analysis of a rhomboid intramembrane protease implicates transmembrane helix 5 as the lateral substrate gate. Proc Natl Acad Sci U S A. 2007;104:8257–8262. doi: 10.1073/pnas.0700814104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duong MT, Jaszewski TM, Fleming KG, MacKenzie KR. Changes in apparent free energy of helix-helix dimerization in a biological membrane due to point mutations. J Mol Biol. 2007;371:422–434. doi: 10.1016/j.jmb.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Esser K, Tursun B, Ingenhoven M, Michaelis G, Pratje E. A novel two-step mechanism for removal of a mitochondrial signal sequence involves the mAAA complex and the putative rhomboid protease Pcp1. J Mol Biol. 2002;323:835–843. doi: 10.1016/s0022-2836(02)01000-8. [DOI] [PubMed] [Google Scholar]
- 40.Guan K, Farh L, Marshall TK, Deschenes RJ. Normal mitochondrial structure and genome maintenance in yeast requires the dynamin-like product of the MGM1 gene. Curr Genet. 1993;24:141–148. doi: 10.1007/BF00324678. [DOI] [PubMed] [Google Scholar]
- 41.Hermann GJ, Thatcher JW, Mills JP, Hales KG, Fuller MT, Nunnari J, Shaw JM. Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J Cell Biol. 1998;143:359–373. doi: 10.1083/jcb.143.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rapaport D, Brunner M, Neupert W, Westermann B. Fzo1p is a mitochondrial outer membrane protein essential for the biogenesis of functional mitochondria in Saccharomyces cerevisiae. J Biol Chem. 1998;273:20150–20155. doi: 10.1074/jbc.273.32.20150. [DOI] [PubMed] [Google Scholar]
- 43.Sesaki H, Jensen RE. UGO1 encodes an outer membrane protein required for mitochondrial fusion. J Cell Biol. 2001;152:1123–1134. doi: 10.1083/jcb.152.6.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sesaki H, Southard SM, Hobbs AE, Jensen RE. Cells lacking Pcp1p/Ugo2p, a rhomboid-like protease required for Mgm1p processing, lose mtDNA and mitochondrial structure in a Dnm1p-dependent manner, but remain competent for mitochondrial fusion. Biochem Biophys Res Commun. 2003;308:276–283. doi: 10.1016/s0006-291x(03)01348-2. [DOI] [PubMed] [Google Scholar]
- 45.Dimmer KS, Fritz S, Fuchs F, Messerschmitt M, Weinbach N, Neupert W, Westermann B. Genetic basis of mitochondrial function and morphology in Saccharomyces cerevisiae. Mol Biol Cell. 2002;13:847–853. doi: 10.1091/mbc.01-12-0588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shepard KA, Yaffe MP. The yeast dynamin-like protein, Mgm1p, functions on the mitochondrial outer membrane to mediate mitochondrial inheritance. J Cell Biol. 1999;144:711–720. doi: 10.1083/jcb.144.4.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wong ED, Wagner JA, Gorsich SW, McCaffery JM, Shaw JM, Nunnari J. The dynamin-related GTPase, Mgm1p, is an intermembrane space protein required for maintenance of fusion competent mitochondria. J Cell Biol. 2000;151:341–352. doi: 10.1083/jcb.151.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pelloquin L, Belenguer P, Menon Y, Gas N, Ducommun B. Fission yeast Msp1 is a mitochondrial dynamin-related protein. J Cell Sci. 1999;112(Pt 22):4151–4161. doi: 10.1242/jcs.112.22.4151. [DOI] [PubMed] [Google Scholar]
- 49.DeVay RM, Dominguez-Ramirez L, Lackner LL, Hoppins S, Stahlberg H, Nunnari J. Coassembly of Mgm1 isoforms requires cardiolipin and mediates mitochondrial inner membrane fusion. J Cell Biol. 2009;186:793–803. doi: 10.1083/jcb.200906098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zick M, Duvezin-Caubet S, Schäfer A, Vogel F, Neupert W, Reichert AS. Distinct roles of the two isoforms of the dynamin-like GTPase Mgm1 in mitochondrial fusion. FEBS Lett. 2009;583:2237–2243. doi: 10.1016/j.febslet.2009.05.053. [DOI] [PubMed] [Google Scholar]
- 51.Arnold I, Langer T. Membrane protein degradation by AAA proteases in mitochondria. Biochim Biophys Acta. 2002;1592:89–96. doi: 10.1016/s0167-4889(02)00267-7. [DOI] [PubMed] [Google Scholar]
- 52.Daum G, Böhni PC, Schatz G. Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J Biol Chem. 1982;257:13028–13033. [PubMed] [Google Scholar]
- 53.Reid GA, Yonetani T, Schatz G. Import of proteins into mitochondria. Import and maturation of the mitochondrial intermembrane space enzymes cytochrome b2 and cytochrome c peroxidase in intact yeast cells. J Biol Chem. 1982;257:13068–13074. [PubMed] [Google Scholar]
- 54.McQuibban GA, Lee JR, Zheng L, Juusola M, Freeman M. Normal mitochondrial dynamics requires rhomboid-7 and affects Drosophila lifespan and neuronal function. Curr Biol. 2006;16:982–989. doi: 10.1016/j.cub.2006.03.062. [DOI] [PubMed] [Google Scholar]
- 55.Whitworth AJ, Lee JR, Ho VM, Flick R, Chowdhury R, McQuibban GA. Rhomboid-7 and HtrA2/Omi act in a common pathway with the Parkinson's disease factors Pink1 and Parkin. Dis Model Mech. 2008;1:168–74. 173. doi: 10.1242/dmm.000109. discussion. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou C, Huang Y, Shao Y, May J, Prou D, Perier C, Dauer W, Schon EA, Przedborski S. The kinase domain of mitochondrial PINK1 faces the cytoplasm. Proc Natl Acad Sci U S A. 2008;105:12022–12027. doi: 10.1073/pnas.0802814105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sha D, Chin LS, Li L. Phosphorylation of parkin by Parkinson disease-linked kinase PINK1 activates parkin E3 ligase function and NF-{kappa}B signaling. Hum Mol Genet. 2009 doi: 10.1093/hmg/ddp501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Frezza C, Cipolat S, Martins de Brito O, Micaroni M, Beznoussenko GV, Rudka T, Bartoli D, Polishuck RS, Danial NN, De Strooper B, Scorrano L. OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell. 2006;126:177–189. doi: 10.1016/j.cell.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 60.Sík A, Passer BJ, Koonin EV, Pellegrini L. Self-regulated cleavage of the mitochondrial intramembrane-cleaving protease PARL yields Pbeta, a nuclear-targeted peptide. J Biol Chem. 2004;279:15323–15329. doi: 10.1074/jbc.M313756200. [DOI] [PubMed] [Google Scholar]
- 61.Jeyaraju DV, Xu L, Letellier MC, Bandaru S, Zunino R, Berg EA, McBride HM, Pellegrini L. Phosphorylation and cleavage of presenilin-associated rhomboid-like protein (PARL) promotes changes in mitochondrial morphology. Proc Natl Acad Sci U S A. 2006;103:18562–18567. doi: 10.1073/pnas.0604983103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wolozin B, Iwasaki K, Vito P, Ganjei JK, Lacanà E, Sunderland T, Zhao B, Kusiak JW, Wasco W, D'Adamio L. Participation of presenilin 2 in apoptosis: enhanced basal activity conferred by an Alzheimer mutation. Science. 1996;274:1710–1713. doi: 10.1126/science.274.5293.1710. [DOI] [PubMed] [Google Scholar]
- 63.Vito P, Ghayur T, D'Adamio L. Generation of anti-apoptotic presenilin-2 polypeptides by alternative transcription, proteolysis, and caspase-3 cleavage. J Biol Chem. 1997;272:28315–28320. doi: 10.1074/jbc.272.45.28315. [DOI] [PubMed] [Google Scholar]
- 64.Walder K, Kerr-Bayles L, Civitarese A, Jowett J, Curran J, Elliott K, Trevaskis J, Bishara N, Zimmet P, Mandarino L, Ravussin E, Blangero J, Kissebah A, Collier GR. The mitochondrial rhomboid protease PSARL is a new candidate gene for type 2 diabetes. Diabetologia. 2005;48:459–468. doi: 10.1007/s00125-005-1675-9. [DOI] [PubMed] [Google Scholar]
- 65.Fawcett KA, Wareham NJ, Luan J, Syddall H, Cooper C, O'Rahilly S, Day IN, Sandhu MS, Barroso I. PARL Leu262Val is not associated with fasting insulin levels in UK populations. Diabetologia. 2006;49:2649–2652. doi: 10.1007/s00125-006-0443-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Powell BL, Wiltshire S, Arscott G, McCaskie PA, Hung J, McQuillan BM, Thompson PL, Carter KW, Palmer LJ, Beilby JP. Association of PARL rs3732581 genetic variant with insulin levels, metabolic syndrome and coronary artery disease. Hum Genet. 2008;124:263–270. doi: 10.1007/s00439-008-0552-2. [DOI] [PubMed] [Google Scholar]
- 67.Meeusen S, DeVay R, Block J, Cassidy-Stone A, Wayson S, McCaffery JM, Nunnari J. Mitochondrial inner-membrane fusion and crista maintenance requires the dynamin-related GTPase Mgm1. Cell. 2006;127:383–395. doi: 10.1016/j.cell.2006.09.021. [DOI] [PubMed] [Google Scholar]
- 68.Griparic L, Kanazawa T, van der Bliek AM. Regulation of the mitochondrial dynamin-like protein Opa1 by proteolytic cleavage. J Cell Biol. 2007;178:757–764. doi: 10.1083/jcb.200704112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Song Z, Chen H, Fiket M, Alexander C, Chan DC. OPA1 processing controls mitochondrial fusion and is regulated by mRNA splicing, membrane potential, and Yme1L. J Cell Biol. 2007;178:749–755. doi: 10.1083/jcb.200704110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guillery O, Malka F, Landes T, Guillou E, Blackstone C, Lombès A, Belenguer P, Arnoult D, Rojo M. Metalloprotease-mediated OPA1 processing is modulated by the mitochondrial membrane potential. Biol Cell. 2008;100:315–325. doi: 10.1042/BC20070110. [DOI] [PubMed] [Google Scholar]
- 71.Ishihara N, Fujita Y, Oka T, Mihara K. Regulation of mitochondrial morphology through proteolytic cleavage of OPA1. EMBO J. 2006;25:2966–2977. doi: 10.1038/sj.emboj.7601184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Duvezin-Caubet S, Koppen M, Wagener J, Zick M, Israel L, Bernacchia A, Jagasia R, Rugarli EI, Imhof A, Neupert W, Langer T, Reichert AS. OPA1 processing reconstituted in yeast depends on the subunit composition of the m-AAA protease in mitochondria. Mol Biol Cell. 2007;18:3582–3590. doi: 10.1091/mbc.E07-02-0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pellegrini L, Scorrano L. A cut short to death: Parl and Opa1 in the regulation of mitochondrial morphology and apoptosis. Cell Death Differ. 2007;14:1275–1284. doi: 10.1038/sj.cdd.4402145. [DOI] [PubMed] [Google Scholar]
- 74.Berman SB, Pineda FJ, Hardwick JM. Mitochondrial fission and fusion dynamics: the long and short of it. Cell Death Differ. 2008;15:1147–1152. doi: 10.1038/cdd.2008.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tatsuta T, Langer T. Quality control of mitochondria: protection against neurodegeneration and ageing. EMBO J. 2008;27:306–314. doi: 10.1038/sj.emboj.7601972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Suen DF, Norris KL, Youle RJ. Mitochondrial dynamics and apoptosis. Genes Dev. 2008;22:1577–1590. doi: 10.1101/gad.1658508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Soubannier V, McBride HM. Positioning mitochondrial plasticity within cellular signaling cascades. Biochim Biophys Acta. 2008 doi: 10.1016/j.bbamcr.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 78.Liesa M, Palacín M, Zorzano A. Mitochondrial dynamics in mammalian health and disease. Physiol Rev. 2009;89:799–845. doi: 10.1152/physrev.00030.2008. [DOI] [PubMed] [Google Scholar]
- 79.Lenaers G, Reynier P, Elachouri G, Soukkarieh C, Olichon A, Belenguer P, Baricault L, Ducommun B, Hamel C, Delettre C. OPA1 functions in mitochondria and dysfunctions in optic nerve. Int J Biochem Cell Biol. 2009;41:1866–1874. doi: 10.1016/j.biocel.2009.04.013. [DOI] [PubMed] [Google Scholar]
- 80.Chao JR, Parganas E, Boyd K, Hong CY, Opferman JT, Ihle JN. Hax1-mediated processing of HtrA2 by Parl allows survival of lymphocytes and neurons. Nature. 2008;452:98–102. doi: 10.1038/nature06604. [DOI] [PubMed] [Google Scholar]
- 81.Klein C, Grudzien M, Appaswamy G, Germeshausen M, Sandrock I, Schäffer AA, Rathinam C, Boztug K, Schwinzer B, Rezaei N, Bohn G, Melin M, Carlsson G, Fadeel B, Dahl N, Palmblad J, Henter JI, Zeidler C, Grimbacher B, Welte K. HAX1 deficiency causes autosomal recessive severe congenital neutropenia (Kostmann disease) Nat Genet. 2007;39:86–92. doi: 10.1038/ng1940. [DOI] [PubMed] [Google Scholar]
- 82.Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, Hill DE, Vidal M, Evans JG, Thorburn DR, Carr SA, Mootha VK. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yamada A, Yamamoto T, Yamazaki N, Yamashita K, Kataoka M, Nagata T, Terada H, Shinohara Y. Differential permeabilization effects of Ca2+ and valinomycin on the inner and outer mitochondrial membranes as revealed by proteomics analysis of proteins released from mitochondria. Mol Cell Proteomics. 2009;8:1265–1277. doi: 10.1074/mcp.M800377-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jeyaraju DV, Cisbani G, De Brito OM, Koonin EV, Pellegrini L. Hax1 lacks BH modules and is peripherally associated to heavy membranes: implications for Omi/HtrA2 and PARL activity in the regulation of mitochondrial stress and apoptosis. Cell Death Differ. 2009 doi: 10.1038/cdd.2009.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhao W, Waggoner JR, Zhang ZG, Lam CK, Han P, Qian J, Schroder PM, Mitton B, Kontrogianni-Konstantopoulos A, Robia SL, Kranias EG. The anti-apoptotic protein HAX-1 is a regulator of cardiac function. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0906998106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vande Walle L, Lamkanfi M, Vandenabeele P. The mitochondrial serine protease HtrA2/Omi: an overview. Cell Death Differ. 2008;15:453–460. doi: 10.1038/sj.cdd.4402291. [DOI] [PubMed] [Google Scholar]
- 87.Jones JM, Datta P, Srinivasula SM, Ji W, Gupta S, Zhang Z, Davies E, Hajnóczky G, Saunders TL, Van Keuren ML, Fernandes-Alnemri T, Meisler MH, Alnemri ES. Loss of Omi mitochondrial protease activity causes the neuromuscular disorder of mnd2 mutant mice. Nature. 2003;425:721–727. doi: 10.1038/nature02052. [DOI] [PubMed] [Google Scholar]
- 88.Martins LM, Morrison A, Klupsch K, Fedele V, Moisoi N, Teismann P, Abuin A, Grau E, Geppert M, Livi GP, Creasy CL, Martin A, Hargreaves I, Heales SJ, Okada H, Brandner S, Schulz JB, Mak T, Downward J. Neuroprotective role of the Reaper-related serine protease HtrA2/Omi revealed by targeted deletion in mice. Mol Cell Biol. 2004;24:9848–9862. doi: 10.1128/MCB.24.22.9848-9862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Moisoi N, Klupsch K, Fedele V, East P, Sharma S, Renton A, Plun-Favreau H, Edwards RE, Teismann P, Esposti MD, Morrison AD, Wood NW, Downward J, Martins LM. Mitochondrial dysfunction triggered by loss of HtrA2 results in the activation of a brain-specific transcriptional stress response. Cell Death Differ. 2009;16:449–464. doi: 10.1038/cdd.2008.166. [DOI] [PubMed] [Google Scholar]
- 90.Strauss KM, Martins LM, Plun-Favreau H, Marx FP, Kautzmann S, Berg D, Gasser T, Wszolek Z, Müller T, Bornemann A, Wolburg H, Downward J, Riess O, Schulz JB, Krüger R. Loss of function mutations in the gene encoding Omi/HtrA2 in Parkinson's disease. Hum Mol Genet. 2005;14:2099–2111. doi: 10.1093/hmg/ddi215. [DOI] [PubMed] [Google Scholar]
- 91.Simón-Sánchez J, Singleton AB. Sequencing analysis of OMI/HTRA2 shows previously reported pathogenic mutations in neurologically normal controls. Hum Mol Genet. 2008;17:1988–1993. doi: 10.1093/hmg/ddn096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bras JM, Singleton A. Genetic susceptibility in Parkinson's disease. Biochim Biophys Acta. 2009;1792:597–603. doi: 10.1016/j.bbadis.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yun J, Cao JH, Dodson MW, Clark IE, Kapahi P, Chowdhury RB, Guo M. Loss-of-function analysis suggests that Omi/HtrA2 is not an essential component of the PINK1/PARKIN pathway in vivo. J Neurosci. 2008;28:14500–14510. doi: 10.1523/JNEUROSCI.5141-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tain LS, Chowdhury RB, Tao RN, Plun-Favreau H, Moisoi N, Martins LM, Downward J, Whitworth AJ, Tapon N. Drosophila HtrA2 is dispensable for apoptosis but acts downstream of PINK1 independently from Parkin. Cell Death Differ. 2009;16:1118–1125. doi: 10.1038/cdd.2009.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hoppins S, Lackner L, Nunnari J. The machines that divide and fuse mitochondria. Annu Rev Biochem. 2007;76:751–780. doi: 10.1146/annurev.biochem.76.071905.090048. [DOI] [PubMed] [Google Scholar]
- 96.Westermann B. Molecular machinery of mitochondrial fusion and fission. J Biol Chem. 2008;283:13501–13505. doi: 10.1074/jbc.R800011200. [DOI] [PubMed] [Google Scholar]
- 97.Hoppins S, Nunnari J. The molecular mechanism of mitochondrial fusion. Biochim Biophys Acta. 2009;1793:20–26. doi: 10.1016/j.bbamcr.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 98.Frank S, Gaume B, Bergmann-Leitner ES, Leitner WW, Robert EG, Catez F, Smith CL, Youle RJ. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev Cell. 2001;1:515–525. doi: 10.1016/s1534-5807(01)00055-7. [DOI] [PubMed] [Google Scholar]
- 99.Cereghetti GM, Scorrano L. The many shapes of mitochondrial death. Oncogene. 2006;25:4717–4724. doi: 10.1038/sj.onc.1209605. [DOI] [PubMed] [Google Scholar]
- 100.Cheung EC, McBride HM, Slack RS. Mitochondrial dynamics in the regulation of neuronal cell death. Apoptosis. 2007;12:979–992. doi: 10.1007/s10495-007-0745-5. [DOI] [PubMed] [Google Scholar]
- 101.James DI, Martinou JC. Mitochondrial dynamics and apoptosis: a painful separation. Dev Cell. 2008;15:341–343. doi: 10.1016/j.devcel.2008.08.011. [DOI] [PubMed] [Google Scholar]
- 102.Benard G, Karbowski M. Mitochondrial fusion and division: Regulation and role in cell viability. Semin Cell Dev Biol. 2009;20:365–374. doi: 10.1016/j.semcdb.2008.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Augustin S, Nolden M, Müller S, Hardt O, Arnold I, Langer T. Characterization of peptides released from mitochondria: evidence for constant proteolysis and peptide efflux. J Biol Chem. 2005;280:2691–2699. doi: 10.1074/jbc.M410609200. [DOI] [PubMed] [Google Scholar]
- 104.Butow RA, Avadhani NG. Mitochondrial signaling: the retrograde response. Mol Cell. 2004;14:1–15. doi: 10.1016/s1097-2765(04)00179-0. [DOI] [PubMed] [Google Scholar]
- 105.Liu Z, Butow RA. Mitochondrial retrograde signaling. Annu Rev Genet. 2006;40:159–185. doi: 10.1146/annurev.genet.40.110405.090613. [DOI] [PubMed] [Google Scholar]
- 106.Biswas G, Adebanjo OA, Freedman BD, Anandatheerthavarada HK, Vijayasarathy C, Zaidi M, Kotlikoff M, Avadhani NG. Retrograde Ca2+ signaling in C2C12 skeletal myocytes in response to mitochondrial genetic and metabolic stress: a novel mode of inter-organelle crosstalk. EMBO J. 1999;18:522–533. doi: 10.1093/emboj/18.3.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Curtis C, Landis GN, Folk D, Wehr NB, Hoe N, Waskar M, Abdueva D, Skvortsov D, Ford D, Luu A, Badrinath A, Levine RL, Bradley TJ, Tavaré S, Tower J. Transcriptional profiling of MnSOD-mediated lifespan extension in Drosophila reveals a species-general network of aging and metabolic genes. Genome Biol. 2007;8:R262. doi: 10.1186/gb-2007-8-12-r262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Owusu-Ansah E, Yavari A, Mandal S, Banerjee U. Distinct mitochondrial retrograde signals control the G1-S cell cycle checkpoint. Nat Genet. 2008;40:356–361. doi: 10.1038/ng.2007.50. [DOI] [PubMed] [Google Scholar]
- 109.Akiyama Y, Maegawa S. Sequence features of substrates required for cleavage by GlpG, an Escherichia coli rhomboid protease. Mol Microbiol. 2007;64:1028–37. doi: 10.1111/j.1365-2958.2007.05715.x. [DOI] [PubMed] [Google Scholar]
- 110.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]