Abstract
Missense alterations of the mismatch repair gene MLH1 have been identified in a significant proportion of individuals suspected of having Lynch syndrome, a hereditary syndrome which predisposes for cancer of colon and endometrium. The pathogenicity of many of these alterations, however, is unclear. A number of MLH1 alterations are located in the C-terminal domain (CTD) of MLH1, which is responsible for constitutive dimerization with PMS2. We analyzed which alterations may result in pathogenic effects due to interference with dimerization. We used a structural model of CTD of MLH1-PMS2 heterodimer to select 19 MLH1 alterations located inside and outside two candidate dimerization interfaces in the MLH1-CTD. Three alterations (p.Gln542Leu, p.Leu749Pro, p.Tyr750X) caused decreased co-expression of PMS2, which is unstable in the absence of interaction with MLH1, suggesting that these alterations interfere with dimerization. All three alterations are located within the dimerization interface suggested by our model. They also compromised mismatch repair, suggesting that defects in dimerization abrogate repair and confirming that all three alterations are pathogenic. Additionally, we provided biochemical evidence that four alterations with uncertain pathogenicity (p.Ala586Pro, p.Leu636Pro, p.Thr662Pro, and p.Arg755Trp) are deleterious because of poor expression or poor repair efficiency, and confirm the deleterious effect of eight further alterations.
Keywords: Lynch syndrome, HNPCC, MLH1, PMS2, MutL, missense mutation, dimerization
Introduction
Lynch syndrome (also known as HNPCC: hereditary non-polyposis colorectal cancer) is a hereditary predisposition for developing cancer of colon and endometrium, and to lesser extent of other organs (Meyer, et al., 2009; OMIM; Schmeler and Lu, 2008). It is the most important heritable colorectal cancer syndrome characterized as yet and accounts for 3% of all colon cancer cases (Burt and Neklason, 2005). The primary cause of Lynch syndrome is dysfunction of the DNA mismatch repair system (MMR), which is responsible for correction of replication errors (mismatches and small insertions and deletions) that escape the proofreading activity of a DNA polymerase.
Mutations in one of the main MMR genes, MLH1 (MIM# 120436), a member of the MutL family, account for half of all Lynch syndrome cases, and one third of the mutations identified in this gene result in amino acid replacements (Peltomaki and Vasen, 2004). The classification of these missense variants as either polymorphism or disease-causing mutation often is very difficult, since they occur infrequently and data on mutation co-segregation with disease is scarce. Immense efforts therefore have been made during the last decade to solve this problem by biochemical analyses of corresponding protein variants. Several databases have been created to assemble the information gained in these studies together with clinical references of the individual mutations: the International Society for Gastrointestinal Hereditary Tumors (InSIGHT) maintains a general database of all mutations reported in Lynch syndrome (including unpublished ones) (Peltomaki and Vasen, 2004), while the Mismatch Repair Genes Variant Database assembles literature references (Woods, et al., 2007) and the MMR Gene Unclassified Variants Database has specialized in missense mutations (Ou, et al., 2008).
While these databases greatly facilitate access to information, they cannot give simple and reliable pathogenicity information in many cases. Recently, MLH1 missense mutations included in these databases have been carefully re-classified as deleterious, neutral, or as variants of uncertain significance (VUS) in respect to causative effect on familial colorectal cancer (Chao, et al., 2008). The classification was based on clinical information from the literature and biochemical data. The criteria used for classification were very strict, resulting in that many mutations previously described as deleterious/pathogenic got classified as VUS, which further underlined that more studies are needed to determine their pathogenicity.
In the absence of sufficient clinical data for unequivocal pathogenicity assessment of MLH1 alterations, their classification to a large extent relies on biochemical studies. These studies typically rely on the determination of protein expression, mismatch repair function, subcellular distribution, and heterodimerization with another MutL paralog, PMS2. This heterodimerization is of special interest, since MLH1 needs to bind PMS2 to form a catalytically functional and correctly localized heterodimer called MutLα (Li and Modrich, 1995; Wu, et al., 2003).
Constitutive dimerization of MLH1 with PMS2 occurs via their C-terminal domains (CTD) (Guerrette, et al., 1999; Nystrom-Lahti, et al., 2002; Plotz, et al., 2003). The three-dimensional structure of MutLα-CTD heterodimer is not known, but recently we have constructed its structural model based on the crystal structure of the E. coli MutL-CTD (Kosinski, et al., 2008). In our model, the dimeric interface is formed by the external (Ex) subdomains of MLH1 and PMS2. However, investigations on the dimer interface of the closely related yeast MutLα (Cutalo, et al., 2006) suggested that in yMutLα the dimeric interface is different than the interface proposed by us for MutLα, and corresponds to the interface located in the internal (In) subdomains, also proposed originally for E. coli MutL (Guarne, et al., 2004). Therefore, the question about the location of the dimeric interface in MutLα is not yet finally resolved. This knowledge, however, is required for interpreting the potential effect of MLH1 alterations on dimerization.
In this work, we asked which Lynch syndrome alterations result in pathogenic effects due to direct interference with dimerization and thus mismatch repair function. First, we have evaluated the two potential dimerization sites with a bioinformatic analysis and selected a series of MLH1 alterations identified in Lynch syndrome patients that are located either inside or outside the two alternative dimerization interfaces. Then we analyzed the selected alterations with respect to their effect on protein expression, dimerization, and MMR activity. Finally, we discuss our findings in the light of previously published biochemical data.
Materials and Methods
Bioinformatic analysis
Structural modeling of MutLα-CTD dimer has been described previously (Kosinski, et al., 2008). Calculation and mapping of evolutionary rates onto the structural model were performed using Consurf (Landau, et al., 2005) and multiple sequence alignment of MutL family created previously (Kosinski, et al., 2008). Protein structures were visualized using PyMol (Warren DeLano, http://www.pymol.org/). The dimeric interface residues were defined by the PROTORP server (Reynolds, et al., 2009) based on the alternative dimer models.
Cell lines, expression vectors and reagents
For production of recombinant MutLα we used MutLα-deficient HEK293T cells which had been kindly provided by Prof. Josef Jiricny, Zürich, Switzerland, and maintained in DMEM nut mix F-12 (HAM) with 10% FCS. Oligonucleotides were from Eurofins (Ebersberg, Germany). The pcDNA3 expression vector (Invitrogen, Carlsbad, CA) containing the entire open reading frame of MLH1 was a gift of Dr. Hong Zhang (Huntsman Cancer Institute, University of Utah, Salt Lake City, UT). The pSG5 expression vector (Stratagene, La Jolla, CA) containing full-length PMS2 cDNA was provided by Prof. Bert Vogelstein (Johns Hopkins Oncology Center, Baltimore, MD). Nucleotide and amino-acid positions refer to the 2484 bp MLH1 mRNA (GenBank accession: U07343.1) and the 756 amino-acid MLH1 sequence (GenBank accession: AAC50285), respectively. Correct description of all alterations investigated in this study was confirmed by the Mutalyzer Sequence Variations Nomenclature Checker (Wildeman, et al., 2008). The plasmids for the missense and deletion mutations used in this study were generated by site-directed mutagenesis using the QuikChange kit (Stratagene, La Jolla, CA) according to the manufacturer’s instructions, and verified by direct sequencing. Immunodetections and immunoprecipitations were performed with the following antibodies: anti-MLH1 (G168–728) and anti-PMS2 (A16-4) from BD Biosciences, Heidelberg, Germany, and anti-MLH1 (N-20) from Santa Cruz Biotechnologies, Santa Cruz, CA, U.S.A.
Protein expression and co-immunoprecipitation
HEK293T cells were transfected either using calcium phosphate precipitation as described before (Plotz, et al., 2006) or with polyethyleneimine (PEI). For this purpose, PEI (linear, 25 kDa, from Polysciences, Warrington, PA, USA) was dissolved in water (1 mg/ml) at 85°C and sterile filtered. 5 µg plasmid DNA were incubated with 20 µg PEI solution in serum-free medium for 10 min. Subsequently, transfection mixes were applied to 9 cm dishes of HEK293T cells with 10 ml of standard growth medium. After 48 h, extracts were prepared as described before (Plotz, et al., 2006). Expression was analyzed by separation of 50 µg of protein extract on SDS-PAGE and immunoblotting. Expression levels of protein variants were quantified in comparison to wild-type protein as detailed below. β-Actin levels were assessed in parallel to ensure identical loading of all lanes.
Immunoprecipitations were carried out using 150 µg of extract in a total volume of 500 µl precipitation buffer (50 mM HEPES-KOH (pH 7.6), 100 mM NaCl, 0.5 mM EDTA, 0.2 mM PMSF, 0.5 mM DTT, 1% Triton X-100) with 1 µg of anti-MLH1 N-20. After one hour of agitated incubation at 4°C, protein G sepharose slurry (20 µl) were added and incubation continued for 3 h. Precipitates were extensively washed in cold precipitation buffer. Success of washing was always confirmed by running samples without antibody in parallel. The sepharose was boiled in SDS-PAGE sample buffer and the samples were separated by 10% SDS-PAGE and blotted. Proteins were detected by antibody overlay using a horseraddish-peroxidase-conjugated secondary antibody. Chemiluminescence was detected by exposition to x-ray films or with the LAS-4000 mini chemiluminesence detection camera (Fujifilm). It was taken care that the signals of the resulting films were in the grayscale area to enable accurate quantification, which for the films was performed with GelScan 5.0 software (BioSciTec, Frankfurt, Germany). Camera images were quantified using MultiGauge v3.2, Fujifilm. Co-precipitation was calculated by dividing the quotient of the PMS2/MLH1 signal of the variant by the respective wild-type quotient.
Statistical evaluation of expression data
Expression data of the MLH1 constructs carrying alterations and of the co-transfected PMS2 constructs was compared with wildtype transfections using a two-sample t-test. Tests were two-sided and p-values were corrected for multiple testing. P-values below 0.05 were considered significant.
MMR assay
Mismatch repair reactions were performed in vitro as described before (Plotz, et al., 2006) using a plasmid substrate with a G–T mismatch within an AseI restriction site which is restored when repair occurs directed by a 3’ single strand nick in 83 bp distance to the mismatch. Digestion with AseI was used to assess repair efficiency. Restriction digests were separated on 2% agarose gels, stained with ethidium bromide and bands were quantified using Quantity One Software v4.6.1 (Bio-Rad, Hercules, CA). While the amount of mismatched plasmid DNA present in parallel incubations is always identical (aliquoted from one master mix), phenol-extraction/ethanol precipitation of the processed plasmid can show differences in recovery and therefore the overall DNA amounts can differ from lane to lane. However, repair efficiency is measured as quotient of the intensities of bands indicating repair and the sum of all band intensities, therefore gives an accurate repair value independent of the amount of DNA actually recovered during plasmid extraction. Relative repair efficiency was calculated by dividing the value of the variant through the value of a wild-type protein preparation that had been expressed, processed and tested in parallel.
Results
Dimeric interface in MutLα-CTD
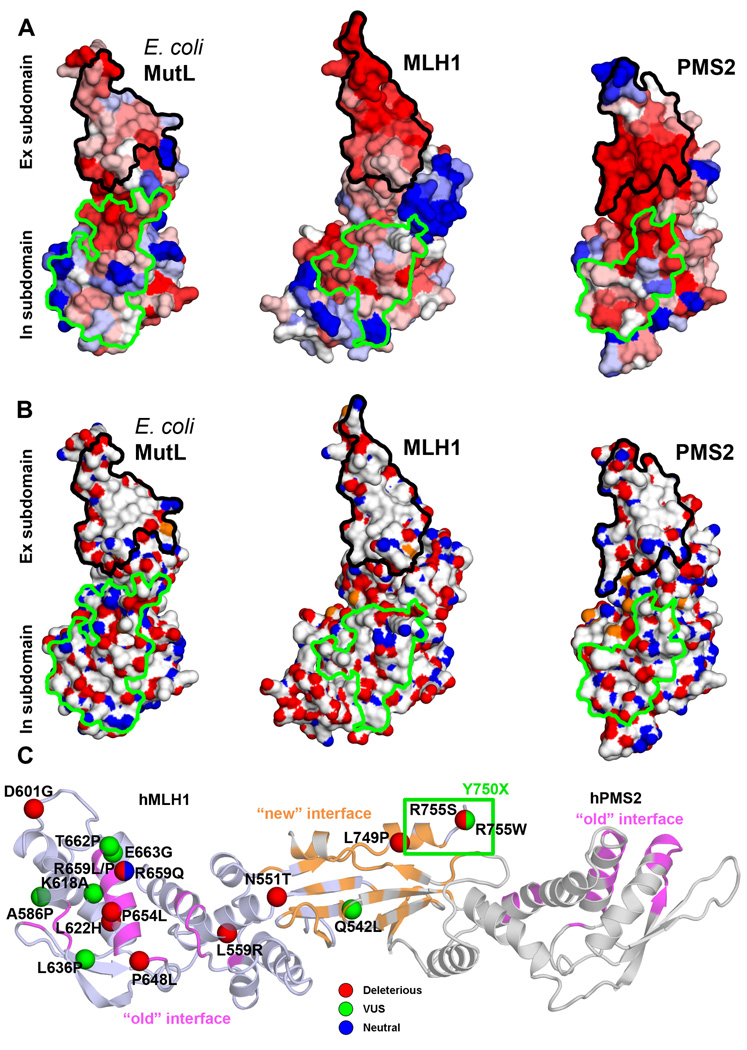
We first performed a bioinformatic analysis of the candidate dimerization interfaces of the MutL-CTD. Mapping of sequence conservation onto structures of the CTDs of monomeric E. coli MutL, and models of PMS2 and MLH1 (Figure 1A), reveals that residues in the presumed dimeric interface in the Ex subdomain are conserved between close homologs of E. coli MutL, and MLH1 and PMS2 families, which is typical for protein-protein interaction sites (Valdar and Thornton, 2001). In contrast, residues corresponding to the previously suggested dimerization interface (Guarne, et al., 2004), which are located in the In subdomain, are not conserved in MLH1 and PMS2 families (Figure 1A). Moreover, the dimerization interface suggested by us includes a highly hydrophobic surface patch often found in interaction sites (Nooren and Thornton, 2003; Tsai, et al., 1997; Young, et al., 1994) (Figure 1B). On the contrary, the alternative interface does not contain a hydrophobic patch. Overall, this theoretical analysis strongly suggests that dimerization occurs by the interface proposed by us (Kosinski, et al., 2008; Kosinski, et al., 2005), and therefore is formed by residues 531–549 and 740–756 in MLH1, and residues 679–699 and 847–862 in PMS2.
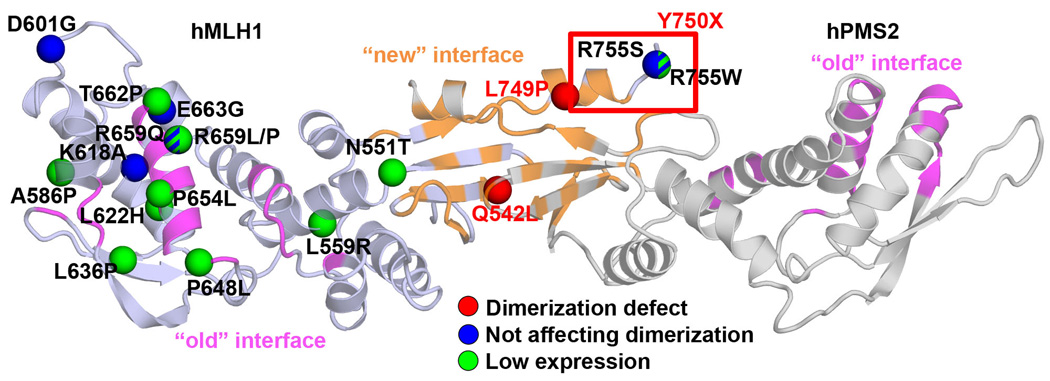
Figure 1. Analysis of dimerization interface in the CTD of E. coli MutL, MLH1, and PMS2.
(A) Molecular surface of MutL/MLH1/PMS2 CTD colored according to sequence conservation with a color gradient from blue (variable) to red (conserved). In (A) and (B) the "new" interface corresponding to the dimerization interface proposed in this and previous work (Kosinski, et al., 2008; Kosinski, et al., 2005) is encircled in black, the "old" interface proposed by others for E. coli MutL (Guarne, et al., 2004) and yMutLα (Cutalo, et al., 2006) encircled in green. (B) Molecular surface of MutL/MLH1/PMS2 CTDs colored according to atom type: carbon atoms colored gray, oxygen – red, nitrogen – blue, and sulfur atoms – orange. (C) Amino acid substitutions related to Lynch syndrome studied in this work mapped on the structural model of MutLα-CTD. Mutations are indicated as spheres corresponding to Cα atoms of corresponding residues, and colored according to their classification (Chao, et al., 2008): deleterious – red, VUS – green, and neutral - blue. Residues deleted in the p.Tyr750X variant (VUS) are indicated with a green rectangle. Structures are shown in cartoon representation; MLH1 is colored dark blue, PMS2 is colored gray. The "new" interface corresponding to the dimerization interface proposed in this and previous work (Kosinski, et al., 2008; Kosinski, et al., 2005) is colored orange, the "old" interface proposed by others for E. coli MutL (Guarne, et al., 2004) and yMutLα (Cutalo, et al., 2006) is colored magenta. The interface residues were defined by PROTORP server (Reynolds, et al., 2009) based on the alternative dimer models.
Selection of substitutions caused by Lynch syndrome mutations in the MLH1-CTD
To analyze the effect of alterations observed in Lynch syndrome on dimerization, we selected alterations that are located inside and outside the two candidate dimeric interfaces (Figure 1C). Selected alterations included those located in the area of the dimerization interface proposed by us (p.Gln542Leu, p.Leu749Pro, p.Tyr750X, p.Arg755Ser, p.Arg755Trp) or its close neighborhood (p.Asn551Thr). Other mutations locate in the area of the alternative dimer interface reported before (p.Pro648Leu, p.Pro654Leu, and p.Thr662Pro) or its direct neighborhood (p.Arg659Leu, p.Arg659Pro, p.Arg659Gln, and p.Glu663Gly). The remaining mutations are located outside any of the interfaces (p.Leu559Arg, p.Ala586Pro, p.Asp601Gly, p.Lys618Ala, p.Leu622His, p.Leu636Pro). Ten of these alterations have been classified as deleterious, seven as uncertain (VUS) and one as neutral (Chao, et al., 2008); see Table 1 and Figure 1C. The p.Tyr750X variant was not included in this analysis by Chao et al., but is present in the InSiGHT database as VUS (Peltomaki and Vasen, 2004).
Table 1.
Comparison of biochemical data obtained in this and previous analyses for Lynch-syndrome-related hMLH1 variants analyzed in this work
| Variant * | DNA change ** | Class | Activity | Expr. | Dimerization | This work | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DME | MMR activ. |
Y2H | Co-IP | Affin- ity |
PMS2 stabil. |
Expr. | PMS2 stabil. |
MMR activ. |
Class (new) | ||||
| Gln542Leu | c.1625A>T | VUS | ***1 |
*1 (*5) |
***1 |
**2 * 9 |
*9 |
***2 ***3 |
*** | - | ** | Del | |
| Asn551Thr | c.1652A>C | Del | ***1 | **1 | ** | n.a. | Del | ||||||
| Leu559Arg | c.1676T>G | Del | - 6 | - 6 | ** | n.a. | Del | ||||||
| Ala586Pro | c.1756G>C | VUS | - 1 | *1 | **1 | ** | n.a. | * | Del | ||||
| Asp601Gly | c.1802A>G | Del | *** | + | *** | VUS | |||||||
| Lys618Ala | c.1852AA>GC | VUS | *1 |
***1 ***4 |
**1 ***4 **6 **7 * 8 |
**2 | ***4 |
*2 * 3 **6 -7 |
***7 | *** | + | *** | VUS |
| Leu622His | c.1865T>A | Del | - 1 | ***1 | **1 | ** | n.a. | Del | |||||
| Leu636Pro | c.1907T>C | VUS | ** | n.a. | *** | Del | |||||||
| Pro648Leu | c.1943C>T | Del | - 1 | **1 |
**1 * 4 |
***4 | ** | n.a. | Del | ||||
| Pro654Leu | c.1961C>T | Del | - 1 |
**1 ***4 |
**1 * 4 |
***4 | ** | n.a. | Del | ||||
| Arg659Leu | c.1976G>T | Del | ** | n.a. | Del | ||||||||
| Arg659Pro | c.1976G>C | Del | - 1 |
*1 **4 *10 |
*1 ***4 |
*2 * 9 |
*4 * 9 * 10 |
*2 * 3 |
* | n.a. | Del | ||
| Arg659Gln | c.1976G>A | Neutral | **1 |
***1 ***4 |
**1 ***4 |
***4 | ** | + | *** | VUS | |||
| Thr662Pro | c.1984A>C | VUS | - 1 | ***1 | **1 | * | n.a. | *** | Del | ||||
| Glu663Gly | c.1988A>G | VUS | ***1 | ***1 | ***1 | *** | + | *** | VUS | ||||
| Leu749Pro | c.2246T>C | Del | ***8 | *** | - | ** | Del | ||||||
| Tyr750X | c.2250C>A | - | ***7 | *7 | *** | - | * | Del | |||||
| Arg755Ser | c.2265G>T | Del | ***1 | - 1 | ***1 | *** | + | Del | |||||
| Arg755Trp | c.2263A>T | VUS | ***7 | ***7 | ** | + | * | Del | |||||
Codon numbering corresponds to the protein sequence of MLH1 (GenBank accession: AAC50285)
Nucleotide numbering reflects cDNA numbering with +1 corresponding to the A of the ATG translation initiation codon in the MLH1 nucleotide sequence (GenBank accession U07343.1)
"Class" – classification of missense mutations according to Chao et al. (Chao, et al., 2008). "Del" – Deleterious, "VUS" – Variant of Uncertain Significance
"DME" – dominant mutator effect of hMLH1 variant interference on yeast MMR (Shimodaira, et al., 1998; Takahashi, et al., 2007). "***" - DME in all three different assays performed (i.e. variant fully functional), "**" - in two, "*" – one, "-" – in none (variant non-functional in MMR)
"MMR activity" – 1,4,10 in vitro MMR activity: *** corresponds to MMR activity > 60% comparing to wild-type, ** - 30–60%, * - 10–30% - - < 10%, 5in vivo activity of equivalent yeast variant
"Expr." – relative expression *** corresponds to >75%, ** - 25–75%, * - <25%
"Dimerization": "Y2H" - yeast-two-hybrid for human homologs; "coIP" - co-immunoprecipitation from insect or human cell extracts; "Affinity" - affinity chromatography using fusion proteins expressed using in vitro transcription-translation systems or in heterologous systems such as E. coli"PMS2 stabil." – MLH1-dependent PMS2 stability in mouse cells or in human cells (this work);
corresponds to > 70% comparing to wild-type, ** - 30–70%, * - 0–30%. n.a. – not applicable, since insufficient levels of MLH1 were present to assess PMS2 stabilization.
References:1 Takahashi, 2007; 2 Kondo, 2003; 3 Guerette, 1999; 4 Raevaara, 2005; 5 Ellison, 2001; 6 Belvedersi, 2006; 7Mohd, 2006; 8 Perera, 2008; 9 Fan, 2007; 10 Nystrom-Lahti, 2002.
Effects of selected Lynch syndrome mutations on the expression and dimerization of MLH1 and PMS2
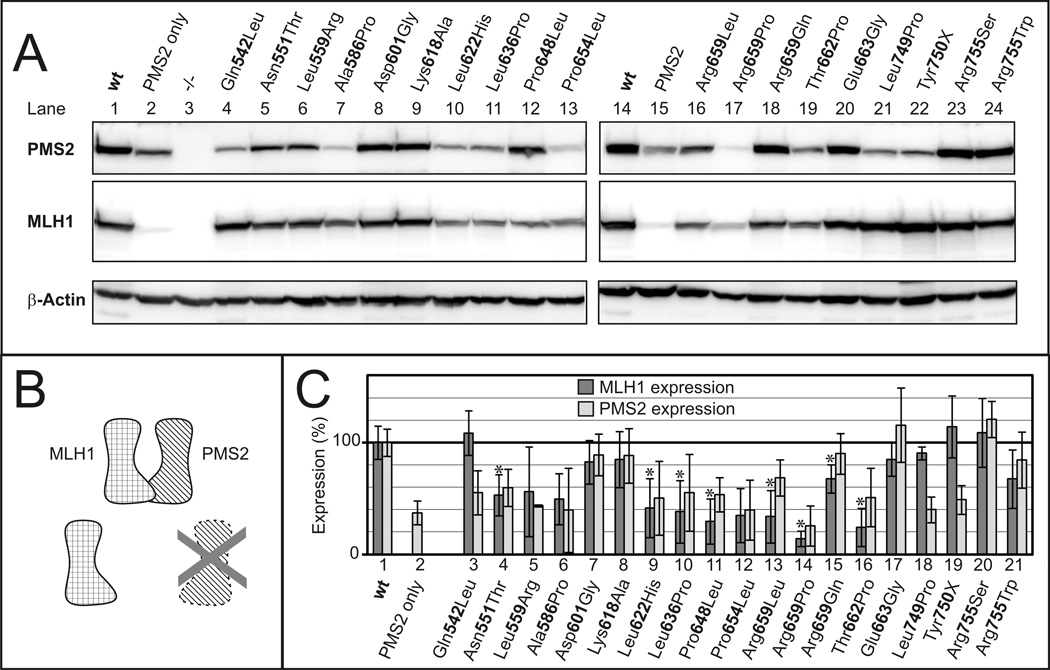
To assess the effects of the selected alterations on protein stability and dimerization, we expressed all protein variants together with PMS2 in HEK293T cells, which do not express endogenous MutLα (Trojan, et al., 2002). Many of the investigated MLH1 missense alterations affected expression (Figure 2A, middle panel). Importantly, the amount of PMS2 also varied significantly in the individual transfections although identical amounts of PMS2 plasmid were transfected (Figure 2A, top panel). It has been observed before that PMS2 is efficiently expressed only in the presence of its dimeric partner MLH1 (Brieger, et al., 2005; Chang, et al., 2000; Mohd, et al., 2006). Cell lines lacking MLH1 expression are practically devoid of PMS2 protein, although normal PMS2 mRNA levels are produced (Chang, et al., 2000). Therefore, PMS2 expression is reduced because of its low protein stability in the absence of dimerization with MLH1 (Figure 2B), and this is also evident in our transient transfection experiments (compare lanes 1/2 and 14/15 of Figure 2A).
Figure 2. Expression of MLH1 variants.
A. MLH1 wild-type (wt) or MLH1 variants were co-transfected with PMS2 into HEK293T cells. After 24h, extracts were prepared and 50 µg of extract was analyzed by SDS-PAGE and western blotting. β-Actin signals were used as loading controls. B. PMS2 and MLH1 form a stable heterodimer, and MLH1 also forms a stable protein when expressed alone. In contrast, PMS2 alone is quickly degraded. C. Several independent transfections of plasmids encoding wildtype MLH1 or its alterations into HEK293T cells using different transfection techniques were performed. Extracts were prepared and expression of MLH1 and PMS2 was analyzed by SDS-PAGE and western blotting. Expression was quantified as detailed in Materials and Methods, and average expression levels as well as standard deviations (n=4 to 10) were determined. MLH1 and PMS2 expression levels are shown with dark and light gray bars respectively, standard deviations are shown by black lines. Corrected p-values were determined for the expression data of all alterations of MLH1. Statistically significant reductions of expression (p<0.05 after correction for multiple testing) are marked by asterisks.
To precisely evaluate the effect of the alterations on expression of MLH1 and PMS2, we performed multiple independent transfection experiments (Figure 2C). Seven variants reproducibly showed MLH1 expression levels similar to the wild-type (>75% of the wild-type level): p.Glu542Leu, p.Asp601Gly, p.Lys618Ala, p.Glu663Gly, p.Leu749Pro, p.Tyr750X and p.Arg755Ser. In contrast, for the following eight alterations a statistically significant reduction of MLH1 expression was observed (p<0.05 after correction for multiple testing): p.Asn551Thr, p.Leu622His, p.Leu636Pro, p.Pro648Leu, p.Arg659Leu, p.Arg659Pro, p.Arg659Gln and p.Thr662Pro.
The expression levels of PMS2 always corresponded to those of MLH1 except for three cases: expression of the MLH1 proteins carrying the alterations p.Glu542Leu, p.Leu749Pro and p.Tyr750X was indistinguishable from wild-type MLH1, but the corresponding PMS2 levels were similar to those achieved when PMS2 was expressed in the absence of MLH1 (compare bars 1 and 2 with 3, 18 and 19 in Figure 2C). This reduction was statistically significant (p<0.05 after correction for multiple testing). This suggests that PMS2 was not efficiently stabilized by MLH1 p.Glu542Leu, p.Leu749Pro and p.Tyr750X.
To assess directly the impact of the mutations on MLH1-PMS2 heterodimerization, we measured the amount of PMS2 that is co-precipitated when MLH1 is immunoprecipitated from the extract (Figure 3). However, none of the MLH1 variants displayed a strong defect in PMS2 binding, since the levels of co-precipitated PMS2 corresponded to their expression levels (compare Figure 3 with Figure 2C, PMS2 expression). Therefore, while the defect of dimerization is readily detectable by the loss of PMS2 stabilization in expression experiments, a significant reduction of affinity was not detectable under the applied conditions. This is consistent with previous findings showing that dimerization-defective MLH1 mutations may retain affinity in vitro (Mohd, et al., 2006), suggesting that PMS2 stabilization may be the best parameter for investigating the effect of MLH1 alterations on dimerization with PMS2.
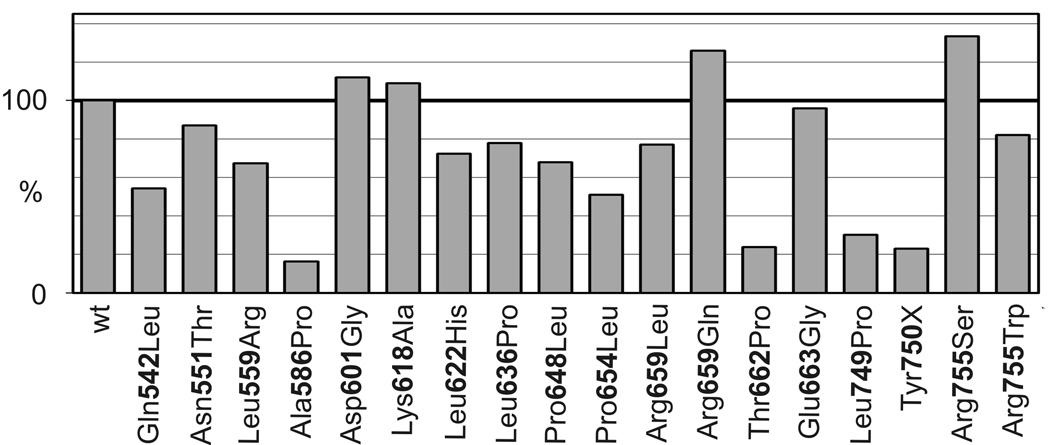
Figure 3. Co-immunoprecipitation of MutLα.
MLH1 was precipitated from extracts with an antibody binding the N-terminus of MLH1 as detailed in Materials and Methods. The amount of co-precipitated PMS2 was determined in relation to wild-type (100%) (average; standard deviation when more than one experiment was performed): Gln542Leu (54; 26); Asn551Thr (87; 7); Leu559Arg (68; 25); Ala586Pro (17; 16); Asp601Gly (112; 7); Lys618Ala (109; 3); Leu622His (73; 8); Leu636Pro (78; 1); Pro648Leu (68); Pro654Leu (51); Arg659Leu (77); Arg659Gln (126); Thr662Pro (24); Glu663Gly (96); Leu749Pro (30; 24); Tyr750X (23;33); Arg755Ser (134;9); Arg755Trp (82; 37). One variant (p.Arg659Pro) was omitted from the analysis due to low expression.
Mismatch repair activity of dimerization-deficient MLH1 variants and of variants with uncertain significance
We next tested whether the defect in dimerization of the MLH1 variants p.Glu542Leu, p.Leu749Pro and p.Tyr750X also affected their mismatch repair activity. Additionally, we included in this analysis all variants of uncertain significance (VUS; see Table 1). We also included p.Asp601Gly, which is a poorly characterized variant with normal expression and Arg659Gln, which is a well-expressed variant within the alternative dimerization interface.
The repair efficiency of the three variants with faulty PMS2 stabilization (MLH1 p.Glu542Leu, p.Leu749Pro and p.Tyr750X) was severely compromised (Figure 4). This was also true for the VUS p.Ala586Pro and p.Arg755Trp. In contrast, the VUS p.Lys618Ala, p.Thr662Pro, and p.Glu663Gly, and neutral p.Arg659Gln showed repair activities similar to wild-type MLH1. The variant p.Asp601Gly, which has been classified as deleterious, also showed normal repair activity.
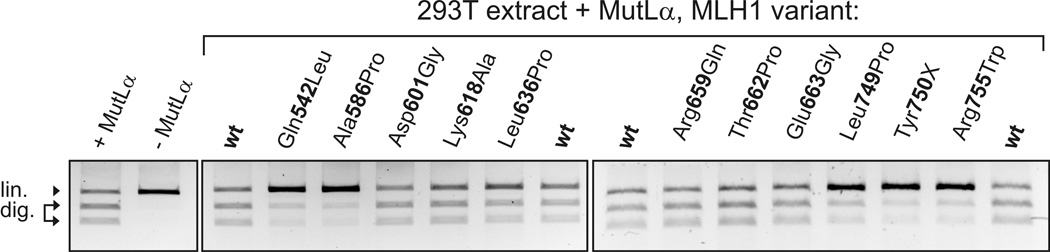
Figure 4. MMR activity of MLH1 variants.
MMR activity of MutLα heterodimer variants was assessed in vitro in parallel to wild-type MutLα as detailed in Materials and Methods. The mismatch is formed by the third thymine of an AseI restriction sequence (ATTAAT) within a 2 kbp plasmid. The unrecognizable mismatched AseI restriction site will be restored when subjected to a MMR reaction. The plasmid contains a second AseI restriction site, therefore unrepaired plasmids will be linearized by AseI ("lin."), while repaired plasmids will be cut into two fragments (1200 bp and 800 bp, "dig."). Numerical values of 4 independent measurements of the individual alterations were (mean and standard deviation): Gln542Leu, 44(18); Ala586Pro 24(27); Asp601Gly 96(5); Lys618Ala 92(8); Leu636Pro 77(9); Arg659Gln 97(4); Thr662Pro 89(11); Glu663Gly 92(7); Leu749Pro 33(34); Tyr750X 16(22); Arg755Trp 7(8).
Discussion
In this work, we have analyzed alterations observed in (suspected) Lynch syndrome patients in the region of the MLH1 gene corresponding to the CTD in the MLH1 protein, in order to test if defects in MLH1-PMS2 dimerization may underlie their (confirmed or questionable) pathogenicity. We have selected 19 alterations (Table 1) that fall inside and outside the two predicted dimerization interfaces (Figure 1C).
From the six variants located within or in proximity of the predicted dimerization interface, three (p.Gln542Leu, p.Leu749Pro, p.Tyr750X) showed a defect in PMS2 stabilization, suggesting that they confer a pathogenic effect due to direct interference with dimerization. These also severely compromised mismatch repair activity. Therefore, our data confirm that p.Leu749Pro is deleterious (Table 1). The unclassified p.Tyr750X variant, which lacks seven residues at the MLH1 C-terminus, has been identified in one Lynch syndrome patient in the United Kingdom (Syngal, et al., 1999) and in further patients without clinical confirmation of Lynch syndrome in the United Kingdom and China (Stone, et al., 2001; Wang, et al., 2006). Co-segregation data, which can provide the most reliable clinical information on pathogenicity, is unavailable for this mutation. However, the current findings confirm that p.Tyr750X is pathogenic. The p.Gln542Leu variant has been identified in Korean kindreds with confirmed Lynch syndrome but without co-segregation information (Han, et al., 1995; Shin, et al., 2004). It has been classified as VUS due to incongruent biochemical data: it has either been found to have no effect on MLH1 function (Guerrette, et al., 1999; Kondo, et al., 2003; Shimodaira, et al., 1998) or be deleterious (Ellison, et al., 2001; Takahashi, et al., 2007). Even excess amounts of MLH1 Gln542Leu repaired mismatches less than half as efficiently as wildtype MLH1 in our experiments, and the significant reduction of MLH1-PMS2 heterodimer formation in cells will decrease the repair efficiency even more. The repair activity of this mutant (44%) is clearly below the limit that previous comprehensive analyses have established as non-pathogenic even for variants which have no defect in expression of MLH1 or stabilization of PMS2: this minimum repair efficiency has been 70% (Raevaara, et al., 2005) or 75% (Takahashi, et al., 2007). Therefore, p.Gln542Leu must be considered deleterious.
Altogether, the current study included seven alterations with uncertain effect (VUS, Table 1, third column). Pathogenicity of a variant can be caused by compromised repair activity as well as by low stability (Raevaara, et al., 2005). According to our analysis, five of the VUS must be considered deleterious (Table 1, last column): p.Ala586Pro, p.Leu636Pro and p.Thr662Pro were severely compromised in expression, while p.Gln542Leu and p.Arg755Ser were defective in repair activity (Table 1, Figure 2 and Figure 4). Both p.Lys618Ala and p.Arg659Gln, classified as VUS and neutral, respectively, displayed mild reductions of repair or expression; although these alterations therefore seem largely neutral, a final judgement on their pathogenicity is not possible from the current data. Therefore, we classified both as VUS. Interestingly, one variant classified as deleterious was indistinguishable from the wild-type in our analyses: p.Asp601Gly has been identified in an Arab kindred with microsatellite-unstable carcinoma but without co-segregation information (Chen-Shtoyerman, et al., 2003). As yet, this alteration has not been tested experimentally. Since the current study has not found any evidence of a repair defect, application of the criteria suggested by Chao et al. would result in re-classification of this alteration to a VUS.
The data of the current study also confirmed that eight further MLH1 alterations have a deleterious effect, mostly due to defects in expression (Table 1).
Our bioinformatic analyses showed that the interaction interface in the Ex subdomain is conserved in MutL and in MLH1 and PMS2 families (Figure 1A), and that it contains a hydrophobic patch (Figure 1B). Both these features are typical hallmarks of strong protein-protein interaction sites (Tsai, et al., 1997; Valdar and Thornton, 2001; Young, et al., 1994). In contrast, the originally suggested dimerization interface in the In subdomain is not conserved in all MutL subfamilies and is not hydrophobic (Figure 1). The three variants that affected PMS2 stabilization and mismatch repair (p.Gln542Leu, p.Leu749Pro, and p.Tyr750X) are located in the dimerization interface of the Ex subdomain (Figure 5). Conversely, two alterations located within or in proximity of the alternative dimer interface (p.Arg659Gln and p.Glu663Gly) affected neither PMS2 stabilization nor repair activity. These observations confirm that the dimerization interface is located in the Ex subdomain (Kosinski, et al., 2008; Kosinski, et al., 2005) and not in the In subdomain (Cutalo, et al., 2006; Guarne, et al., 2004).
Figure 5. Mapping of amino acid substitutions onto the structural model of MutLα-CTD with their effect on protein expression and dimerization.
Substitutions are indicated as spheres corresponding to Cα atoms of corresponding residues, and colored according to their effect on protein expression and dimerization (red: interfering with dimerization, blue: not affecting dimerization and with good expression, green: significantly decreasing expression). Substitutions resulting in only moderately compromised expression and having no effect on PMS2 dimerization (p.Arg659Gln and p.Arg755Trp) are indicated as dashed green-blue spheres. Residues corresponding to p.Tyr750X variant are indicated as red rectangle. Structures are shown in cartoon representation; MLH1 is colored dark blue, PMS2 is colored gray. The "new" interface corresponding to the dimerization interface proposed in this and previous work (Kosinski, et al., 2008; Kosinski, et al., 2005) is colored orange, the "old" interface proposed by others for E. coli MutL (Guarne, et al., 2004) and yMutLα (Cutalo, et al., 2006) is colored magenta.
The major biochemical evidence that dimerization occurs by the In subdomain has come from investigations of yMutLα using chemical surface modification experiments (Cutalo, et al., 2006). In that study, three lysine residues that become buried only in the yMutLα dimer (as opposed to monomer) were identified. However, these results can be also explained by additional interactions of CTD with NTD or the linker in the dimeric state. Importantly, these lysine residues are near the hinge region between CTD subdomains (Kosinski, et al., 2005), so they could become buried after dimerization solely due to conformational changes.
Previous analyses have also investigated the effect of small MLH1 alterations (missense type and deletions of few residues) on its dimerization with PMS2 by affinity methods, yeast two-hybrid analyses and coimmunoprecipitation (Table 1). Only one study has as yet used the stabilization of PMS2 as a measure of interaction (Mohd, et al., 2006). While all methods seemed to work acceptably well in deletion studies, many results concerning missense alterations are conflicting. For example, p.Arg659Pro was frequently found to disturb dimerization (Table 1), seemingly supporting that dimerization occurs via the In domain. However, our data and other investigations showed that this is a very destabilizing alteration (introducing the helix-breaking Pro residue into an α-helix, Figure 5), therefore a (partial) unfolding of MLH1 probably causes the loss of dimerization without p.Arg659 being actually within the interaction interface. This is corroborated by two other substitutions of this residue (p.Arg659Leu and p.Arg659Gln) which showed no effect on dimerization in our analysis.
Data gained with affinity methods frequently are conflicting with other methods (Table 1). Both MLH1 and PMS2 have been routinely expressed in bacteria for this investigation, therefore PMS2 will lack its extensive post-translational modifications (Raschle, et al., 2002), which may affect its interacting properties. Additionally, it has been observed that suitable, stringent washing conditions may be required to detect a decrease in affinity in vitro (Mohd, et al., 2006). For these reasons, detecting the stabilization of PMS2 after MLH1-PMS2 expression in mammalian cells seems to be the most reliable method for identifying a defect of dimerization, and it is probably the test giving best biological (and diagnostic) information.
In conclusion, the current work demonstrates that three MLH1 variants (p.Gln542Leu, p.Leu749Pro, and p.Tyr750X) observed in Lynch syndrome patients disturb MLH1-PMS2 dimerization. They are all located within a conserved hydrophobic surface area suggested as dimerization interface based on our bioinformatic analysis using the recently constructed model of MutLα-CTD. These alterations also severely affected mismatch repair, confirming that they are pathogenic and suggesting that defective dimerization underlies their deleterious effect. Moreover, the current work provides strong evidence that five MLH1 variants with uncertain significance (VUS) are deleterious and confirms the deleterious effect of eight further alterations, suggesting that all 13 variants can be causative for Lynch syndrome.
Acknowledgments
We are grateful to Prof. Eva Herrmann, Institute of Biostatistics and Mathematical Modeling, Faculty of Medicine, Johann Wolfgang Goethe University, Frankfurt am Main, Germany, for help in the statistical evaluation. We want to thank curators of the InSiGHT (International Society for Gastrointestinal Hereditary Tumors) and MMR gene unclassified variants (MMRUV) databases, which were very useful for conducting this work. J.K. was a recipient of a scholarship from the Postgraduate School of Molecular Medicine at the Medical University of Warsaw and had a young investigator award ("START" programme) from Foundation for Polish Science. G.P. and I.H. have been supported by grant 2007.030.1 from the Wilhelm Sander-Stiftung. J.M.B. has been supported by grants from the Polish Ministry of Science and Higher Education (188/N-DFG/2008/0 and PBZ-MNiI-2/1/2005) and by the NIH (1R01GM081680-01). J.K. and J.M.B. were also supported by the DNA-ENZYMES (MRTN-CT-2005-019566) and HEALTH-PROT (contract number 229676) grants from the 6th and 7th Framework Programmes of the E.U. P.F. was supported by DNA-ENZYMES (6FP EU grant number MOBILITY-1 19566) and by the German Science foundation (GRK1384).
References
- Brieger A, Plotz G, Raedle J, Weber N, Baum W, Caspary WF, Zeuzem S, Trojan J. Characterization of the nuclear import of human MutLalpha. Mol.Carcinog. 2005;43(1):51–58. doi: 10.1002/mc.20081. [DOI] [PubMed] [Google Scholar]
- Burt R, Neklason DW. Genetic testing for inherited colon cancer. Gastroenterology. 2005;128(6):1696–1716. doi: 10.1053/j.gastro.2005.03.036. [DOI] [PubMed] [Google Scholar]
- Chang DK, Ricciardiello L, Goel A, Chang CL, Boland CR. Steady-state regulation of the human DNA mismatch repair system. J.Biol.Chem. 2000;275(24):18424–18431. doi: 10.1074/jbc.M001140200. [DOI] [PubMed] [Google Scholar]
- Chao EC, Velasquez JL, Witherspoon MS, Rozek LS, Peel D, Ng P, Gruber SB, Watson P, Rennert G, Anton-Culver H, et al. Accurate classification of MLH1/MSH2 missense variants with multivariate analysis of protein polymorphisms-mismatch repair (MAPP-MMR) Hum Mutat. 2008;29(6):852–860. doi: 10.1002/humu.20735. [DOI] [PubMed] [Google Scholar]
- Chen-Shtoyerman R, Theodor L, Harmati E, Friedman E, Dacka S, Kopelman Y, Sternberg A, Zarivach R, Bar-Meir S, Fireman Z. Genetic analysis of familial colorectal cancer in Israeli Arabs. Hum Mutat. 2003;21(4):446–447. doi: 10.1002/humu.9123. [DOI] [PubMed] [Google Scholar]
- Cutalo JM, Darden TA, Kunkel TA, Tomer KB. Mapping the dimer interface in the C-terminal domains of the yeast MLH1-PMS1 heterodimer. Biochemistry. 2006;45(51):15458–15467. doi: 10.1021/bi061392a. [DOI] [PubMed] [Google Scholar]
- Ellison AR, Lofing J, Bitter GA. Functional analysis of human MLH1 and MSH2 missense variants and hybrid human-yeast MLH1 proteins in Saccharomyces cerevisiae. Hum Mol Genet. 2001;10(18):1889–1900. doi: 10.1093/hmg/10.18.1889. [DOI] [PubMed] [Google Scholar]
- Guarne A, Ramon-Maiques S, Wolff EM, Ghirlando R, Hu X, Miller JH, Yang W. Structure of the MutL C-terminal domain: a model of intact MutL and its roles in mismatch repair. EMBO J. 2004 doi: 10.1038/sj.emboj.7600412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrette S, Acharya S, Fishel R. The interaction of the human MutL homologues in hereditary nonpolyposis colon cancer. J.Biol.Chem. 1999;274(10):6336–6341. doi: 10.1074/jbc.274.10.6336. [DOI] [PubMed] [Google Scholar]
- Han HJ, Maruyama M, Baba S, Park JG, Nakamura Y. Genomic structure of human mismatch repair gene, hMLH1, and its mutation analysis in patients with hereditary non-polyposis colorectal cancer (HNPCC) Hum Mol Genet. 1995;4(2):237–242. doi: 10.1093/hmg/4.2.237. [DOI] [PubMed] [Google Scholar]
- Kondo E, Suzuki H, Horii A, Fukushige S. A yeast two-hybrid assay provides a simple way to evaluate the vast majority of hMLH1 germ-line mutations. Cancer Res. 2003;63(12):3302–3308. [PubMed] [Google Scholar]
- Kosinski J, Plotz G, Guarne A, Bujnicki JM, Friedhoff P. The PMS2 subunit of human MutLalpha contains a metal ion binding domain of the iron-dependent repressor protein family. J Mol Biol. 2008;382(3):610–627. doi: 10.1016/j.jmb.2008.06.056. [DOI] [PubMed] [Google Scholar]
- Kosinski J, Steindorf I, Bujnicki JM, Giron-Monzon L, Friedhoff P. Analysis of the quaternary structure of the MutL C-terminal domain. J.Mol.Biol. 2005;351(4):895–909. doi: 10.1016/j.jmb.2005.06.044. [DOI] [PubMed] [Google Scholar]
- Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, Ben-Tal N. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–W302. doi: 10.1093/nar/gki370. (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li GM, Modrich P. Restoration of mismatch repair to nuclear extracts of H6 colorectal tumor cells by a heterodimer of human MutL homologs. Proc.Natl.Acad.Sci.U.S.A. 1995;92(6):1950–1954. doi: 10.1073/pnas.92.6.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer LA, Broaddus RR, Lu KH. Endometrial cancer and Lynch syndrome: clinical and pathologic considerations. Cancer Control. 2009;16(1):14–22. doi: 10.1177/107327480901600103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohd AB, Palama B, Nelson SE, Tomer G, Nguyen M, Huo X, Buermeyer AB. Truncation of the C-terminus of human MLH1 blocks intracellular stabilization of PMS2 and disrupts DNA mismatch repair. DNA Repair (Amst) 2006;5(3):347–361. doi: 10.1016/j.dnarep.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Nooren IM, Thornton JM. Structural characterisation and functional significance of transient protein-protein interactions. J Mol Biol. 2003;325(5):991–1018. doi: 10.1016/s0022-2836(02)01281-0. [DOI] [PubMed] [Google Scholar]
- Nystrom-Lahti M, Perrera C, Raschle M, Panyushkina-Seiler E, Marra G, Curci A, Quaresima B, Costanzo F, D'Urso M, Venuta S, et al. Functional analysis of MLH1 mutations linked to hereditary nonpolyposis colon cancer. Genes Chromosomes.Cancer. 2002;33(2):160–167. [PubMed] [Google Scholar]
- OMIM. National Center for Biotechnology Information (NCBI); Online Mendelian Inheritance of Man: Lynch Syndrome, #120435. http://www.ncbi.nlm.nih.gov/entrez/dispomim.cgi?id=120435.
- Ou J, Niessen RC, Vonk J, Westers H, Hofstra RM, Sijmons RH. A database to support the interpretation of human mismatch repair gene variants. Hum Mutat. 2008;29(11):1337–1341. doi: 10.1002/humu.20907. [DOI] [PubMed] [Google Scholar]
- Peltomaki P, Vasen H. Mutations associated with HNPCC predisposition -- Update of ICG-HNPCC/INSiGHT mutation database. Dis Markers. 2004;20(4–5):269–276. doi: 10.1155/2004/305058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotz G, Raedle J, Brieger A, Trojan J, Zeuzem S. N-terminus of hMLH1 confers interaction of hMutLalpha and hMutLbeta with hMutSalpha. Nucleic Acids Res. 2003;31(12):3217–3226. doi: 10.1093/nar/gkg420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotz G, Welsch C, Giron-Monzon L, Friedhoff P, Albrecht M, Piiper A, Biondi RM, Lengauer T, Zeuzem S, Raedle J. Mutations in the MutSalpha interaction interface of MLH1 can abolish DNA mismatch repair. Nucleic Acids Res. 2006;34(22):6574–6586. doi: 10.1093/nar/gkl944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raevaara TE, Korhonen MK, Lohi H, Hampel H, Lynch E, Lonnqvist KE, Holinski-Feder E, Sutter C, McKinnon W, Duraisamy S, et al. Functional significance and clinical phenotype of nontruncating mismatch repair variants of MLH1. Gastroenterology. 2005;129(2):537–549. doi: 10.1016/j.gastro.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Raschle M, Dufner P, Marra G, Jiricny J. Mutations within the hMLH1 and hPMS2 Subunits of the Human MutLalpha Mismatch Repair Factor Affect Its ATPase Activity, but Not Its Ability to Interact with hMutSalpha. J.Biol.Chem. 2002;277(24):21810–21820. doi: 10.1074/jbc.M108787200. [DOI] [PubMed] [Google Scholar]
- Reynolds C, Damerell D, Jones S. ProtorP: a protein-protein interaction analysis server. Bioinformatics. 2009;25(3):413–414. doi: 10.1093/bioinformatics/btn584. [DOI] [PubMed] [Google Scholar]
- Schmeler KM, Lu KH. Gynecologic cancers associated with Lynch syndrome/HNPCC. Clin Transl Oncol. 2008;10(6):313–317. doi: 10.1007/s12094-008-0206-9. [DOI] [PubMed] [Google Scholar]
- Shimodaira H, Filosi N, Shibata H, Suzuki T, Radice P, Kanamaru R, Friend SH, Kolodner RD, Ishioka C. Functional analysis of human MLH1 mutations in Saccharomyces cerevisiae. Nat Genet. 1998;19(4):384–389. doi: 10.1038/1277. [DOI] [PubMed] [Google Scholar]
- Shin YK, Heo SC, Shin JH, Hong SH, Ku JL, Yoo BC, Kim IJ, Park JG. Germline mutations in MLH1, MSH2 and MSH6 in Korean hereditary non-polyposis colorectal cancer families. Hum Mutat. 2004;24(4):351. doi: 10.1002/humu.9277. [DOI] [PubMed] [Google Scholar]
- Stone JG, Coleman G, Gusterson B, Marossy A, Lakhani SR, Ward A, Nash A, McKinna A, A'Hern R, Stratton MR, et al. Contribution of germline MLH1 and MSH2 mutations to lobular carcinoma in situ of the breast. Cancer Lett. 2001;167(2):171–174. doi: 10.1016/s0304-3835(01)00448-7. [DOI] [PubMed] [Google Scholar]
- Syngal S, Fox EA, Li C, Dovidio M, Eng C, Kolodner RD, Garber JE. Interpretation of genetic test results for hereditary nonpolyposis colorectal cancer: implications for clinical predisposition testing. Jama. 1999;282(3):247–253. doi: 10.1001/jama.282.3.247. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Shimodaira H, Andreutti-Zaugg C, Iggo R, Kolodner RD, Ishioka C. Functional Analysis of Human MLH1 Variants Using Yeast and In vitro Mismatch Repair Assays. Cancer Res. 2007;67(10):4595–4604. doi: 10.1158/0008-5472.CAN-06-3509. [DOI] [PubMed] [Google Scholar]
- Trojan J, Zeuzem S, Randolph A, Hemmerle C, Brieger A, Raedle J, Plotz G, Jiricny J, Marra G. Functional analysis of hMLH1 variants and HNPCC-related mutations using a human expression system. Gastroenterology. 2002;122(1):211–219. doi: 10.1053/gast.2002.30296. [DOI] [PubMed] [Google Scholar]
- Tsai CJ, Lin SL, Wolfson HJ, Nussinov R. Studies of protein-protein interfaces: a statistical analysis of the hydrophobic effect. Protein Sci. 1997;6(1):53–64. doi: 10.1002/pro.5560060106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdar WS, Thornton JM. Protein-protein interfaces: analysis of amino acid conservation in homodimers. Proteins. 2001;42(1):108–124. [PubMed] [Google Scholar]
- Wang XL, Yuan Y, Zhang SZ, Cai SR, Huang YQ, Jiang Q, Zheng S. Clinical and genetic characteristics of Chinese hereditary nonpolyposis colorectal cancer families. World J Gastroenterol. 2006;12(25):4074–4077. doi: 10.3748/wjg.v12.i25.4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildeman M, van Ophuizen E, den Dunnen JT, Taschner PE. Improving sequence variant descriptions in mutation databases and literature using the Mutalyzer sequence variation nomenclature checker. Hum Mutat. 2008;29(1):6–13. doi: 10.1002/humu.20654. [DOI] [PubMed] [Google Scholar]
- Woods MO, Williams P, Careen A, Edwards L, Bartlett S, McLaughlin JR, Younghusband HB. A new variant database for mismatch repair genes associated with Lynch syndrome. Hum Mutat. 2007 doi: 10.1002/humu.20502. [DOI] [PubMed] [Google Scholar]
- Wu X, Platt JL, Cascalho M. Dimerization of MLH1 and PMS2 limits nuclear localization of MutLalpha. Mol.Cell Biol. 2003;23(9):3320–3328. doi: 10.1128/MCB.23.9.3320-3328.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young L, Jernigan RL, Covell DG. A role for surface hydrophobicity in protein-protein recognition. Protein Sci. 1994;3(5):717–729. doi: 10.1002/pro.5560030501. [DOI] [PMC free article] [PubMed] [Google Scholar]