Abstract
An important component of the pathologic process underlying Alzheimer’s disease (AD) is oxidative stress. Met35 in amyloid β-protein (Aβ) is prone to participating in redox reactions promoting oxidative stress, and therefore is believed to contribute significantly Aβ-induced toxicity. Thus, substitution of Met35 by residues that do not participate in redox chemistry would be expected to decrease Aβ toxicity. Indeed, substitution of Met35 by norleucine (Nle) was reported to reduce Aβ toxicity. Surprisingly, however, substitution of Met35 by Val was reported to increase toxicity. Aβ toxicity is known to be strongly related to its self-assembly. However, neither substitution is predicted to affect Aβ assembly substantially. Thus, the effect of these substitutions on toxicity is difficult to explain. We revisited this issue and compared Aβ40 and Aβ42 with analogues containing Met35→Nle or Met35→Val substitutions using multiple biophysical and toxicity assays. We found that substitution of Met35 by Nle or Val had moderate effects on Aβ assembly. Surprisingly, despite these effects, neither substitution changed Aβ neurotoxicity significantly in three different assays. These results suggest that the presence of Met35 in Aβ is not important for Aβ toxicity, challenging to the prevailing paradigm, which suggests that redox reactions involving Met35 contribute substantially to Aβ-induced toxicity.
Keywords: Alzheimer’s disease, amyloid β-protein, oxidative stress, neurotoxicity, structure–activity relationship
INTRODUCTION
Alzheimer’s disease (AD) is a progressive, age-related neurodegenerative disorder, which gradually impairs cognitive abilities, causes difficulties in execution of routine tasks, and finally leads to dementia and death (Selkoe 2001, Cummings 2004). Amyloid plaques, neurofibrillary tangles, neurite dystrophy, synapse loss, and neurodegeneration in the cerebral cortex and hippocampus are pathologic hallmarks of AD (Selkoe 2001). Genetic, physiologic, and biochemical data indicate that self-assembly of amyloid β-protein (Aβ) initiates the disruption of interneuronal communication in AD (Roher et al. 1993, Hardy & Selkoe 2002, Roychaudhuri et al. 2009). Though initially Aβ fibrils, the predominant component of amyloid plaques, were thought to be the culprit, now soluble Aβ oligomers are believed to be the major neurotoxic species in AD (Dahlgren et al. 2002, White et al. 2005, Haass & Selkoe 2007, Tomic et al. 2009). Plaque formation is thought be an attempt of the brain to sequester the toxic oligomers in a less harmful fibrillar structure (Bravo et al. 2008, Josephs et al. 2008, Reiman et al. 2009), though plaques also may serve as reservoirs of oligomeric Aβ (Koffie et al. 2009).
The C-terminal region of Aβ plays a key role in controlling Aβ oligomerization and aggregation (Bitan et al. 2003a, Jarrett et al. 1993, Bitan et al. 2003c). Within this region, oxidation of Met35 to sulfoxide has been reported to alter substantially Aβ oligomerization (Palmblad et al. 2002, Hou et al. 2002, Hou et al. 2004, Bitan et al. 2003b) and neurotoxicity (Varadarajan et al. 1999, Varadarajan et al. 2001, Varadarajan et al. 2000, Barnham et al. 2003, Ciccotosto et al. 2004). The sulfoxide form of Aβ has been found in cerebral tissue extracts derived from patients with AD and transgenic mice (Kuo et al. 2001, Näslund et al. 1994). Whether these findings point to a causative effect of oxidation of Aβ to the sulfoxide in AD or are merely a result of the oxidative environment in the AD brain, or a protein-extraction-induced artifact, is an open question.
Redox reactions in the AD brain generate highly reactive free radicals (Butterfield 2002, Halliwell 2006). To regain stability, these radicals abstract electrons or hydrogen atoms from neighboring molecules or groups, leading to perturbation of the chemical structure, and destruction of biological molecules (Butterfield et al. 2007). Free radicals normally form in a controlled manner in the mitochondrial respiratory chain during oxidative phosphorylation when molecular oxygen is reduced to water (Halliwell 2006). During this process, two reactive oxygen species (ROS) form: superoxide anion (O2−) and hydrogen peroxide (H2O2). In addition, O2− or H2O2 may be generated by multiple other enzymatic and non-enzymatic cellular mechanisms (Halliwell 1989). H2O2 may further form hydroxyl radicals (OH•) via Fenton chemistry (Halliwell 2006, Markesbery & Lovell 1998). When the control mechanisms that maintain these and other ROS are compromised, as happens in AD, the result is oxidative damage, including lipid peroxidation, protein carbonylation, and other modifications of essential biomolecules (Crouch et al. 2008, Markesbery & Lovell 1998, Halliwell 2006, Butterfield et al. 2007).
Under the highly oxidative environment in the AD brain, oxidation of Met35 in Aβ may lead to formation of Met-sulfuranyl radicals (MetS•), which may initiate, or participate in, the destructive chemistry described above. Thus, oxidation of Met35 in Aβ has been postulated to be directly involved in Aβ toxicity. Alternatively, neurotoxic mechanisms caused by Aβ oligomers, including disruption of Ca2+ homeostasis and excitotoxicity (Piacentini et al. 2008), may cause oxidative stress that would, among other things, oxidize Met35 in Aβ, as a result, not cause, of Aβ toxicity.
If Met35 oxidation causes Aβ toxicity, substitution of Met35 by aliphatic residues that lack sulfur, do not form radicals easily, and are not substrate for ROS, would be expected to suppress Aβ toxicity. In line with this hypothesis, substitution of Met35 by Nle has been reported to decrease Aβ toxicity (Varadarajan et al. 1999, Butterfield & Kanski 2002, Clementi et al. 2006, Yatin et al. 1999). Surprisingly, however, substitution of Met35 by Val had the opposite effect (Ciccotosto et al. 2004). Complicating matters further, Ciccotosto et al reported that [Val35]Aβ42 produces similar amounts of H2O2 as Aβ42, whereas Murray et al. found that [Val35]Aβ42 showed reduced lipid peroxidation relative to Aβ42 (Murray et al. 2005).
Because Met35 resides in the middle of the hydrophobic C-terminus of Aβ, and because Met, Nle, and Val all are hydrophobic residues, the Met35→Nle or Met35→Val substitutions are not expected to have a strong effect on Aβ assembly. Indeed, substitution of Met35 by Nle had little effect on the oligomer size distribution of Aβ (Bitan et al. 2003b).
Because substantial efforts have been dedicated to understanding the role of Met35 in Aβ in AD, we felt that the discrepancy between previous studies using similar strategies to answer the same question merited a re-examination. Here, we addressed this discrepancy by investigating the effect of substituting Met35 in Aβ40 and Aβ42 by either Nle or Val. We describe a systematic, side-by-side comparison of each substituted Aβ analogue using assays for oligomer size distribution, conformational change, assembly size, fibril morphology, and toxicity in primary neurons.
MATERIALS AND METHODS
Chemicals and supplies
Silver-staining kit, 10–20% gradient Tris-tricine gels, penicillin/streptomycin, and APO-BrdU Apoptosis Detection kit were purchased from Invitrogen (Carlsbad, CA). Tris(2,2’-bipyridyl)ruthenium dichloride (Ru(Bpy)), ammonium persulfate, glutaraldehyde, uranyl acetate, cytosine arabinofuranoside, and poly D-lysine were from Sigma (St. Louis, MO). 1,1,1,3,3,3-Hexafluoroisopropanol (HFIP) was from TCI America (Portland, OR). Trypsin-EDTA solution, Dulbecco’s Modified Eagle’s Medium (DMEM), Leibovitz L15 medium, and fetal bovine serum were from ATCC (Manassas, VA). Electron microscopy (EM) grids were purchased from Electron Microscopy Science (Hatfield, PA). Cover slips and 96-well black and white plates were from Fisher scientific (Tustin, CA). CytoTox-ONE™ Homogeneous Membrane Integrity Assay kits and 2,5-diphenyltetrazolium bromide were from Promega (Madison, WI).
Animals
All experiments were performed in accordance with the National Research Council Guide for the Care and Use of Laboratory Animals and were approved by the UCLA Institutional Animal Care Use Committee. Pregnant, E18, Sprague-Dawley rats were purchased from Charles River laboratory (Wilmington, MA).
Peptides synthesis
Aβ40, [Nle35]Aβ40, [Val35]Aβ40, Aβ42, [Nle35]Aβ42, and [Val35]Aβ42 were synthesized, purified and characterized by the UCLA Biopolymers Laboratory as described previously (Lomakin et al. 1996). Briefly, peptides were synthesized on an automated peptide synthesizer (Model 433A, Applied Biosystems, Foster City, CA) using 9-fluorenylmethoxycarbonyl chemistry and purified by reverse-phase high-performance liquid chromatography (HPLC). Quantitative amino acid analysis and mass spectrometry were used to characterize the expected compositions and molecular masses, respectively, for each peptide.
Preparation of peptide solutions
Purified peptides were stored as lyophilized powders at −20°C. Before use, peptides were treated with HFIP and stored as dry films at −20°C as described previously (Rahimi et al. 2009). For biophysical measurements, immediately before use, films were dissolved in 60 mM NaOH at 10% of the desired volume. The solution then was diluted to 50% of the desired volume with deionized water (18.2 MΩ produced by a Milli-Q system, Millipore, Bedford, MA) and sonicated for 1 min. Then, the solution was diluted with 20 mM sodium phosphate, pH 7.4, to the final peptide concentration, which was 10 µM unless stated otherwise. For toxicity experiments, peptides were diluted with cell-culture media after initial dissolution in 10% NaOH, and then sonicated for 1 min.
Photo-cross-linking and SDS-PAGE analysis
The experimental protocol was described previously (Rahimi et al. 2009, Vollers et al. 2005, Bitan 2006). Briefly, peptide solutions were centrifuged at 14,000 g for 10 min. The supernates were subjected to Photo-Induced Cross-linking of Unmodified Proteins (PICUP) (Fancy & Kodadek 1999). For each cross-linking reaction, 2 µl of 1 mM Ru(Bpy) and 2 µl of 20 mM ammonium persulfate (APS) were added to 18 µl of peptide solution. The mixtures were irradiated with visible light for 1 s and the reaction was quenched immediately with 10 µl of Tricine sample buffer (Invitrogen) containing 5% β-mercaptoethanol. The cross-linked peptides were boiled for 5 min and analyzed by SDS-PAGE, silver stained, and subjected to densitometric analysis using Image J (NIH software, http://rsb.info.nih.gov). The data are an average of six independent experiments.
Circular dichroism spectroscopy (CD)
Samples were incubated at 25°C with continuous agitation using an orbital shaker at 200 rpm. Spectra were recorded every 2 h during the first 12 h, and then at 24, 48, and 72 h, using a J-810 spectropolarimeter (Jasco, Easton, MD) equipped with a thermostable sample cell at 25°C using 1-mm path-length cuvettes. Spectra were collected from 190–260 nm with 1-s response time, 50-nm/min scan speed, 0.2-nm resolution and 2-nm bandwidth, and averaged after background subtraction. The data are representative of three independent experiments.
Dynamic light scattering (DLS)
Samples prepared as described above, filtered immediately before the first measurement through 20-nm cutoff, Anotop filters (Whatman, Florham Park, NJ), and incubated at room temperature without agitation. Measurements were performed using an in-house-built system with a He-Ne laser model 127 (wavelength 633 nm, power 60 mW, Spectra Physics lasers, Mountain View, CA). Light scattered at 90° was collected using image transfer optics and detected by an avalanche photodiode built into a PD4047 multitau correlator (Precision Detectors, Bellingham, MA). The size distribution of scattering particles was reconstructed from the correlation function of the scattered light using PrecisionDeconvolve (Precision Detectors) based on the regularization method by Tikhonov and Arsenin (Tikhonov, 1977). The data are representative of two independent experiments.
Electron microscopy (EM)
Samples were incubated at 25°C with continuous agitation using an orbital shaker at 200 rpm. Eight-µl aliquots were applied to glow-discharged, carbon-coated Formvar grids for 20 min. The solution was wicked gently with filter paper. Then the samples were fixed with 5 µl of 2.5% glutaraldehyde for 4 min and stained with 5 µl of 1% uranyl acetate for 3 min. The solution was wicked off and the grids were air-dried. The morphology was visualized using a CM120 (FEI, Philips) transmission electron microscope.
Cell culture
Primary neurons were prepared from E18 rat embryos as described previously (Segal & Manor 1992). Briefly, E18 pregnant rats were euthanized with CO2 and the pups were collected immediately. The brains were dissected in chilled Leibovitz’s L15 medium in the presence of penicillin/streptomycin (1 µg/ml). The tissue was incubated with 0.25% trypsin-EDTA solution for 30 min and then mechanically dissociated in a small volume of Leibovitz’s L15 media using a fire-polished Pasteur pipette. The cells were suspended in DMEM containing 10% heat-inactivated fetal bovine serum and penicillin/streptomycin (1 µg/ml), and plated in poly D-lysine (0.1 mg %)-coated 96-well plates (COSTAR, Corning, USA) at a density of 3×105 cells/ml. The cultures were maintained at 37°C in a humidified atmosphere of 5% CO2 for 6 d before treatment with peptides. Twenty-four hours after plating, the medium was replaced with fresh medium supplemented with 5 µM cytosine β-D-arabinofuranoside to inhibit the proliferation of glial cells.
MTT assay
Cells were treated with freshly prepared Aβ analogues for 48 h. Cell viability was measured using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) cell-metabolism assay, as described previously (Fradinger et al. 2008). Briefly, following treatment, 15 µl of MTT were added to each well and incubated for 4 h at 37°C. Then, stop solution was added and kept overnight at room temperature. The OD was measured using a Synergy plate reader (Bio-TEK instruments, Winooski, VT). The cell viability results of three independent experiments (6 wells per condition) were normalized to the medium control group and expressed as mean ± SEM.
LDH assay
Neurons were incubated with freshly prepared Aβ analogues for 48 h and cell death was assayed by measuring the release of lactate dehydrogenase (LDH) using CytoTox-ONE™ Homogeneous Membrane Integrity Assay kit (Promega) according to the manufacturer’s instructions. Data from three independent experiments (6 wells per condition) were normalized to medium control and expressed as mean ± SEM.
TUNEL assay
Neurons were grown on poly D-lysine-coated cover slips and treated with freshly prepared Aβ analogues for 48 h. The cells were then washed with phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde. Cover slips then were treated with ethanol for 30 min at −20°C, followed by 3×5 min washes with the “wash buffer” supplied in the kit, and incubated with terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) DNA labeling solution for 1 h at 37°C. The cells were washed and treated with an anti-BrdU antibody for 30 min at room temperature in the dark. Cover slips then were washed with the “rinse buffer” supplied in the kit and counter-stained with 1% propidium iodide for 15 min in the dark, washed with deionized water, dehydrated, cleared, and mounted on glass slides using glycerol. Fluorescent signals were visualized using a Nikon Eclipse E400 microscope (Nikon Instruments Inc., Melville, NY) at λex = 480 nm and λem = 530 nm. Images were merged using the bundled software “Picture Frame” (Optronics, Goleta, CA). Images were taken from multiple fields in at least three independent experiments and the number of TUNEL-positive cells divided by the total number of counted cells was expressed as % apoptotic death (mean ± SEM).
Data analysis
Data were analyzed using a one-way analysis of variance (ANOVA) with Tukey’s pair-wise comparison test as a post hoc test using Prism 5.0b (GraphPad, La Jolla, CA).
RESULTS
Effect of substitution of Met35 by Nle or Val on Aβ oligomerization
A possible explanation of the different effects of substitution of Met35 by Nle or Val on Aβ toxicity is that the substitutions have a profound effect on Aβ assembly. Though this would be unexpected given the similar hydrophobic nature of all three-side chains, we felt that it was an important hypothesis to examine.
To test the effect of substituting Met35 by Nle or Val on the oligomer size distribution of Aβ, the WT and substituted Aβ40 and Aβ42 analogues each were cross-linked using PICUP, fractionated by SDS-PAGE and silver-stained (Fig. 1A). Uncross-linked WT and substituted analogues of Aβ40 migrated as monomers, whereas Aβ42 analogues migrated as a combination of monomer and a broad and smeary trimer/tetramer band (data not shown) as described previously (Bitan et al. 2003a, Bitan et al. 2003b, Bitan et al. 2003c). Substitution of Met35 by Nle or Val in Aβ40 had little effect on the oligomer size distributions obtained using PICUP (Fig. 1A, B). All three peptides had comparable abundance of monomer through trimer, followed by a lower abundance of tetramer band (Fig. 1A), similar to data reported previously (Bitan et al. 2001, Bitan et al. 2003a).
Fig. 1. Oligomer size distribution of Aβ analogues.
Peptides were cross-linked using PICUP immediately after preparation, fractionated by SDS-PAGE, and the gels silver-stained. A) A representative gel showing the oligomer size distribution of each analogue. The migration of molecular weight markers is shown on the left. B) Relative band abundance of cross-linked Aβ40 analogues. C) Relative band abundance of cross-linked Aβ42 analogues. The relative abundance of each band was calculated as a percentage of the entire lane. The data in panels B and C are an average of 6 independent experiments. Statistical significance: * p<0.05, ** p<0.01, and ***p<0.001 applicable to all figures.
Aβ42 analogues showed high abundance of pentameric and hexameric paranuclei as described previously (Bitan et al. 2003a) (Fig. 1A). Substitution of Met35 by Nle or Val yielded lower abundance of dimer and trimer and higher abundance of tetramer, pentamer, and hexamer (Fig. 1A, C). These differences, particularly the decrease in dimer and increase in tetramer abundance, were subtle for [Nle35]Aβ42 and more pronounced for [Val35]Aβ42. Statistical analysis of densitometric data showed that the differences between WT Aβ42 and [Val35]Aβ42 were statistically significant for monomer and all the oligomers except heptamer (Fig. 1C). The difference between WT Aβ42 and [Nle35]Aβ42 was significant only for dimer, trimer, and hexamer.
Effect of substitution of Met35 by Nle or Val on β-sheet formation
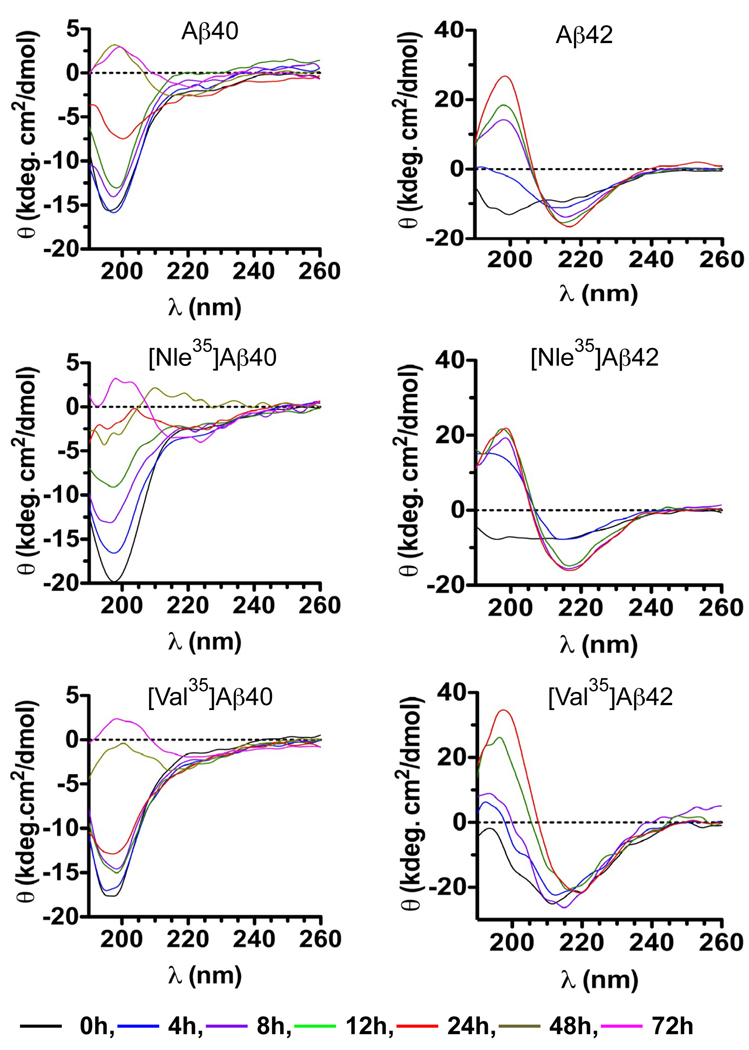
We used CD spectroscopy to study the effect of substituting Met35 by Nle or Val on conformational changes during Aβ assembly (Fig. 2). In all cases except [Val35]Aβ42, the initial spectrum was characterized by a minimum at 196–200 nm, suggesting that the peptide conformation was predominantly a statistical coil (SC). With incubation, in all cases the minimum at 196–200 nm was replaced by a maximum at 198–199 nm and, in the spectra of Aβ42 analogues, was accompanied by formation of a minimum at 215–217 nm, which is indicative of formation of β-sheet. A similar minimum typically is observed for Aβ40 analogues at higher peptide concentrations (Kirkitadze et al. 2001). We chose to keep the concentration consistent across all assays and therefore did not observe the typical minimum at 215 nm for Aβ40 analogues here.
Fig. 2. Conformational change kinetics of Aβ analogues.
Aβ40, Aβ42, and their Nle35- or Val35-substituted analogues were incubated at 25°C with agitation and the solutions monitored periodically by CD spectroscopy. Aβ40 analogues were measured up to 72 h and Aβ42 analogues up to 24 h. The spectra are representative of three independent experiments.
Substitution of Met35 Nle slightly increased, whereas substitution by Val slightly decreased the kinetics of conformational change in Aβ40. The kinetics of conformational transition in [Nle35]Aβ42 was slightly increased relative to WT Aβ42, whereas β-sheet formation by [Val35]Aβ42 was apparent already at the first time point (~5 min after dissolution) and was faster than the kinetics of conformational change in [Nle35]Aβ42 or WT Aβ42 (Fig. 2). These observations correlated with the trends observed in the PICUP experiments, i.e., higher abundance of larger oligomers correlates with faster kinetics of β-sheet formation.
Effect of substitution of Met35 by Nle or Val on assembly size
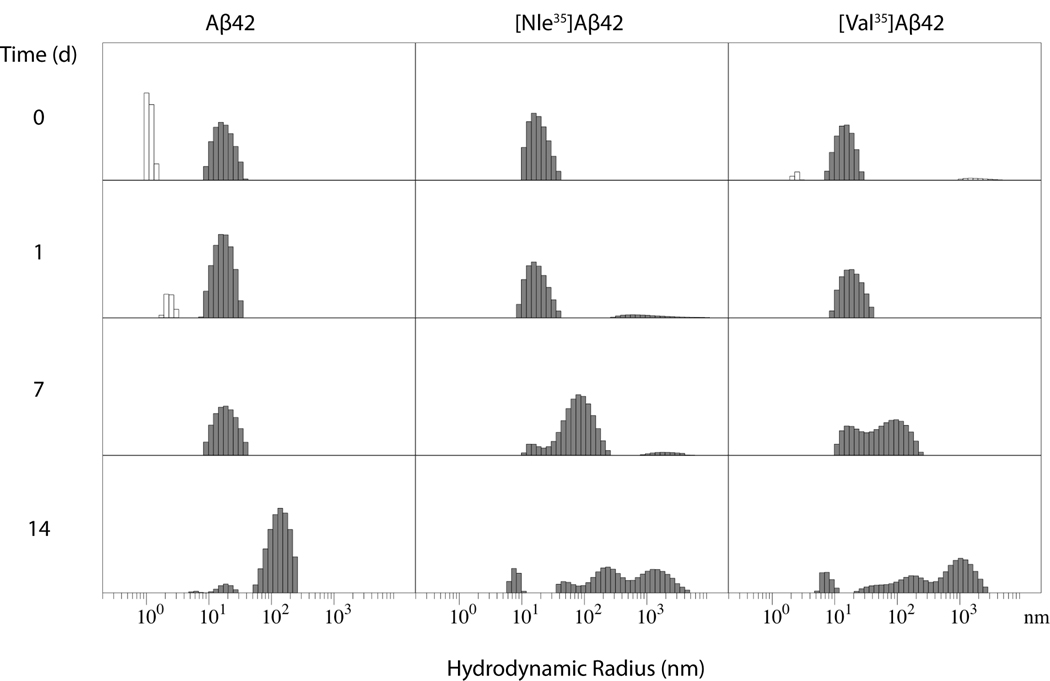
We used DLS to monitor the initial assembly size distribution of the Aβ analogues and the progressive growth of aggregation size. In our experience, unlike Aβ42, which shows formation of intermediate particle sizes during aggregation, in DLS experiments, Aβ40 initially shows only particles of RH ~1–2 nm and following incubation (typically ≥1 week), very large particles appear, without accumulation of intermediate size particles (Bitan et al. 2003a). Based on this experience and because the effect of substitution by Nle or Val on Aβ40 was found using PICUP or CD was relatively small, we did no expect DLS experiments with Aβ40 to add useful information and studied only Aβ42 analogues by DLS. We measured both the change in particle size and the frequency of intensity spikes that occur when very large particles cross the laser beam. For aggregating peptides, the frequency of intensity spikes can be used to estimate the rate of fibril formation (Fradinger et al. 2008).
Initially the WT and substituted Aβ42 analogues formed predominantly particles of hydrodynamic radius (RH) =10–30 nm (Fig. 3) and no intensity spikes. This situation was unchanged after 24 h of incubation and remained stable for several days. After 7 days, the same distribution and no intensity spikes still were observed for WT Aβ42, whereas particles of RH =100–200 nm appeared for both substituted analogues, which also displayed occasional intensity spikes, indicating the beginning of fibril formation. By 14 days of incubation, particles of RH =100–200 nm were observed also for WT Aβ42. At the same time, particles of RH =1000–2000 nm were detected for the two substituted analogues. The faster kinetics of [Nle35]Aβ42 and [Val35]Aβ42 relative to WT Aβ42 correlated with the PICUP and CD data.
Fig. 3. Particle size growth Aβ analogues.
Aβ42 and its Nle35- and Val35-substituted analogues were filtered through a 20-nm pore size filter, incubated at 25°C under quiescent conditions and monitored by DLS for up to 14 days.
It is important to note that because in DLS measurements the intensity is proportional to the square of the mass, the populations of larger particles are highly overrepresented in Fig. 3. Particles of RH =10–15 nm still existed at all time points in all cases but they were overshadowed by the intensity of larger particles.
The overall slower kinetics of aggregation in the DLS experiments compared to CD and EM (see below) reflects the different preparation and incubation conditions. Samples measured by DLS must be filtered to remove dust particles and are incubated without agitation. Without these measures, large particles render the measurements useless already at the very early time points.
Effect of substitution of Met35 by Nle or Val on morphology
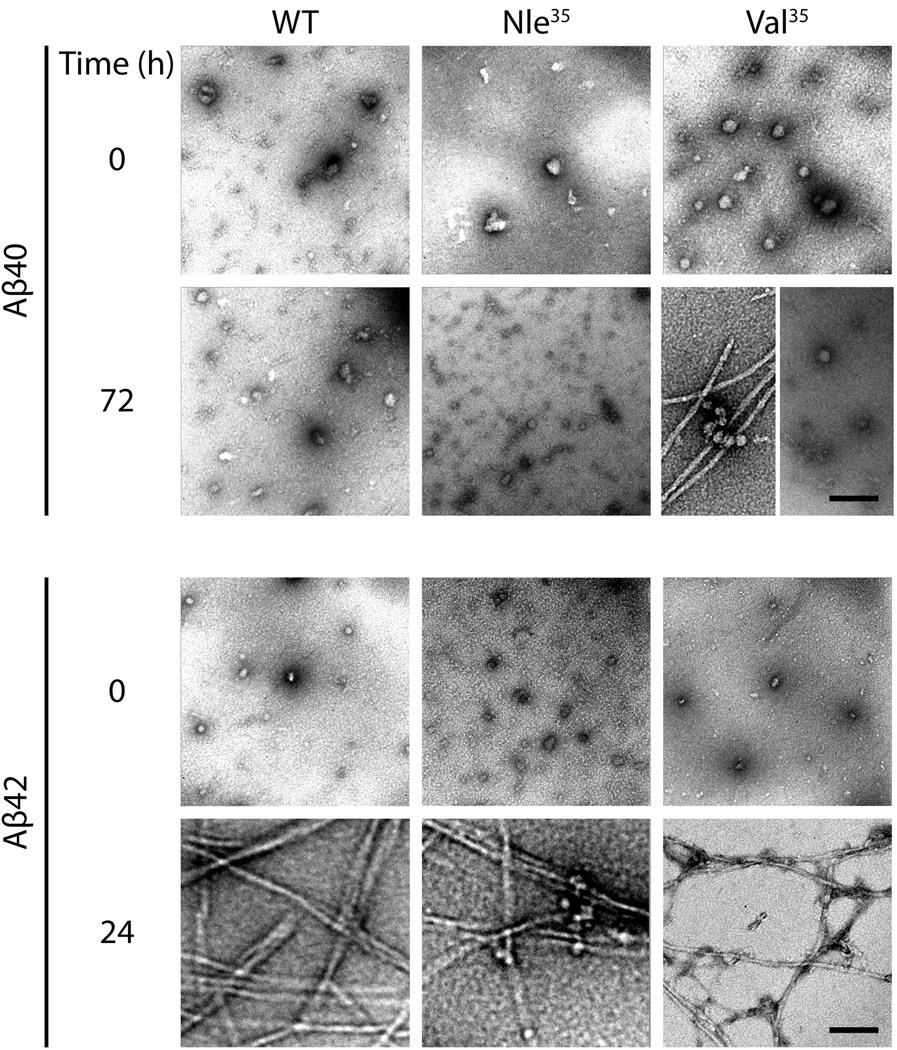
The morphology of the WT and substituted peptides was examined by EM immediately after preparation and after 72 h (Aβ40) or 24 h (Aβ42) of incubation (Fig 4). All the peptides initially had non-fibrillar morphology and showed structures consistent with oligomers. Following 72 h of incubation, the morphology of Aβ40 and [Nle35]Aβ40 did not change. The behavior of [Val35]Aβ40 was less consistent. In a some experiments, this peptide showed mixture of oligomers and fibrils whereas in others its morphology was similar to Aβ40 and [Nle35]Aβ40 (Fig 4). Aβ42 and both its substituted analogues showed abundant fibrils after 24 h of incubation. Quasi-spherical oligomers still could be observed in all cases (Fig 4). Taken together, the results of most of the biophysical experiments show that substitution of Met by Nle or Val increases the tendency of Aβ to self-assemble. This tendency correlates with the higher hydrophobicity of Nle and Val relative to Met.
Fig. 4. Time-dependent morphological change in Aβ analogues.
Aβ analogues were incubated at 25°C for 72 h (Aβ40) or 24 h (Aβ42) with agitation. Aliquots were spotted at the beginning and end of the measurement on glow-discharged, carbon-coated grids, stained with uranyl acetate, and examined by EM. The images are representative of three independent experiments. The scale bars indicate 100 nm and are applicable to all images.
Effect of substitution of Met35 by Nle or Val on Aβ neurotoxicity
To study the structure–activity relationships of substituted Aβ analogues, we treated rat primary cortical or hippocampal neurons with WT, Nle-, or Val-substituted Aβ40 or Aβ42 and measured neurotoxicity using the MTT reduction, LDH release, and TUNEL staining assays. We used these three different assays because each measures a different aspect of cell viability: The MTT assay measures mitochondrial activity of viable cells (Wang et al. 2006), the LDH assay signifies membrane integrity and is a direct measurement of cell death (Decker & Lohmann-Matthes 1988), and the TUNEL assay indicates DNA fragmentation and denotes apoptosis.
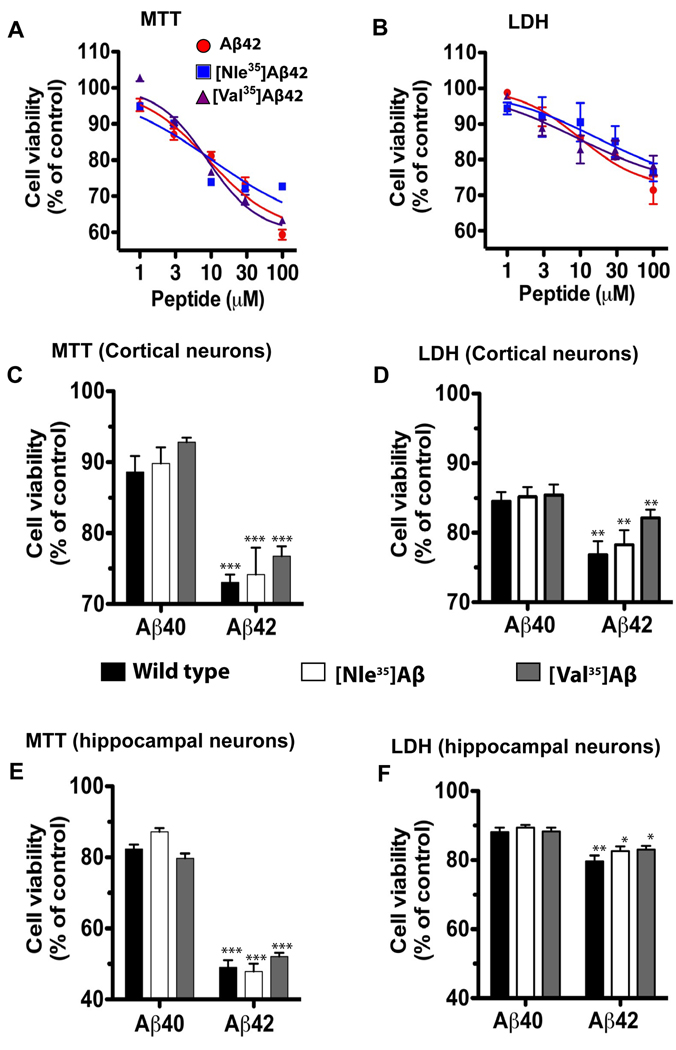
Because Aβ42 is substantially more toxic than Aβ40, we performed full dose-response analysis (at 1–100 µM) for Aβ42 analogues using both the MTT and LDH assays, whereas the toxicity of Aβ40 analogues was assessed at a single concentration—10 µM, for comparison with Aβ42 analogues and to facilitate correlating the data with the results of the biophysical measurements described above.
Using the MTT assay, the concentration at which 50% of the maximal toxicity (EC50) was observed were 10±2 µM, 11±3, and 9±1 fo r Aβ42, [Nle35]Aβ42, and [Val35]Aβ42, respectively (Fig. 5A). The EC50 values measured using the LDH assay were 11±5, 11±5, and 12±8 for Aβ42, [Nle35]Aβ42, and [Val35]Aβ42, respectively (Fig 5B). Comparison of the neurotoxicity at 10 µM showed that the toxicity of the substituted analogues was similar to that of the WT peptides for both Aβ40 and Aβ42 alloforms using both the MTT (Fig. 5C) and LDH (Fig 5D) assays. In all cases, the only statistically significant differences were found between Aβ42 and Aβ40 analogues, whereas the differences between the substituted analogues and the corresponding WT Aβ alloforms were small and statistically insignificant. Cortical and hippocampal neurons showed similar sensitivity to Aβ analogues in most experiments.
Fig. 5. Aβ-induced neurotoxicity measured by the MTT and LDH assays.
Rat primary cortical or hippocampal neurons were treated with Aβ40, Aβ42, or their Nle35- or Val35-substituted analogues for 48 h. Cell viability was assayed by measuring MTT reduction by active cells or by LDH release. A) Dose–response analysis of Aβ42 analogues by the MTT assay in cortical neuron. B) Dose–response analysis of Aβ42 analogues by the LDH assay in cortical neuron. C) Cell viability following treatment with 10 µM of each peptide measured by the MTT assay in cortical neurons. D) Cell viability following treatment with 10 µM of each peptide measured by the LDH assay in cortical neurons. E) Cell viability following treatment with 10 µM of each peptide measured by the MTT assay in hippocampal neurons. F) Cell viability following treatment with 10 µM of each peptide measured by the LDH assay in hippocampal neurons. The data are average of three independent experiments with 6 data points per condition (n=18). *p<0.05, **p<0.01, and ***p<0.001 vs the corresponding Aβ40 analogue.
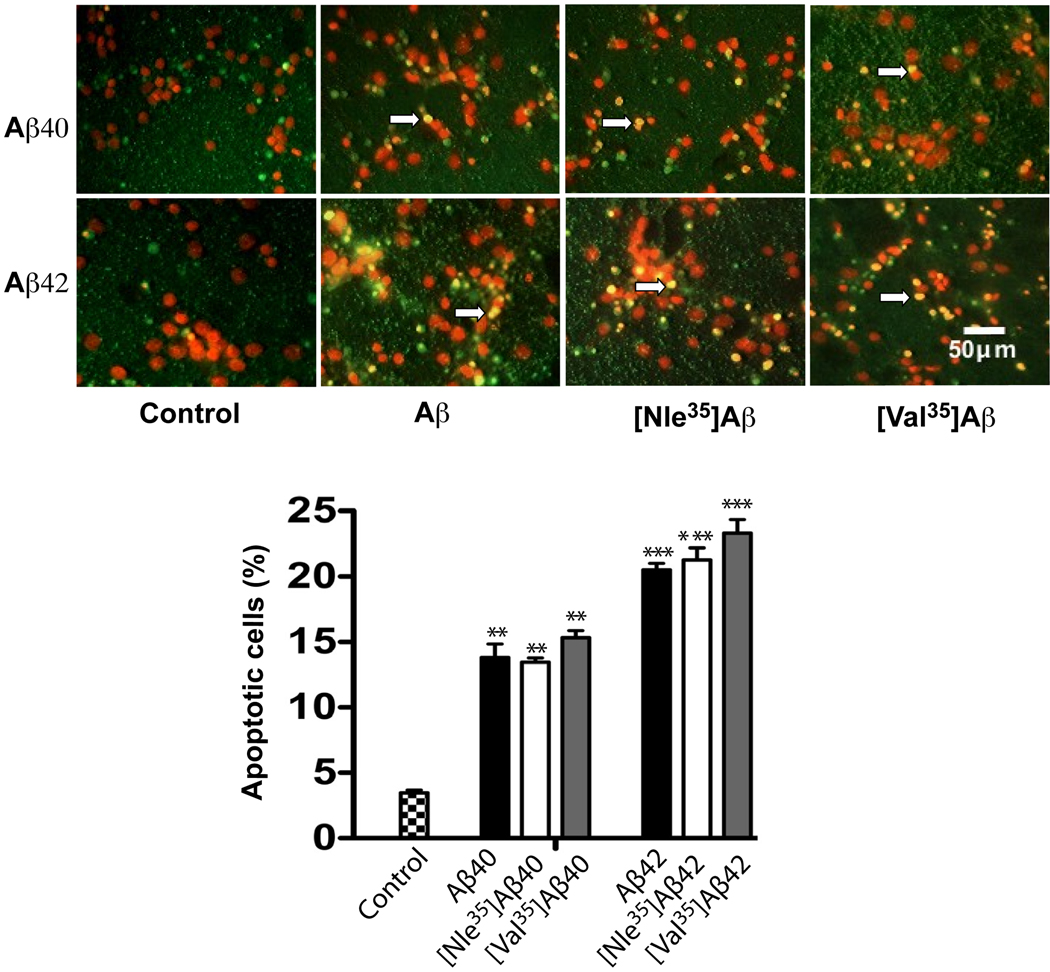
Similar results were obtained using the TUNEL assay, which showed that all the Aβ analogues induced apoptosis. Aβ42 analogues were significantly more toxic that Aβ40 analogues yet no significant differences were found between the Nle- or Val-substituted analogues and the WT peptides (Fig 6).
Fig. 6. Aβ-induced neuronal apoptosis.
Rat primary cortical neurons were grown for 6 days on poly-D-lysine-coated cover slips and then treated with 10 µM of each Aβ analogue for 48 h. DNA fragmentation was probed using APO-BrdU staining. Micrographs are representative of 10–15 different fields of each experiment. Arrows indicate TUNEL-positive cells. The number of apoptotic cells divided by the total number of cells (counted manually) is expressed as % apoptotic death. The data are representative of three independent experiments. **p<0.01 and ***p<0.001 vs control.
DISCUSSION
Substitution of Met35 in Aβ by redox-unreactive, aliphatic groups has been reported to both decrease, in the case of Nle (Varadarajan et al. 1999, Butterfield & Boyd-Kimball 2005, Clementi et al. 2006, Piacentini et al. 2008), and increase, in case of Val (Ciccotosto et al. 2004) Aβ neurotoxicity, with no satisfactory explanation for these contradictory observations. The toxicity findings also were not correlated directly with the assembly properties of the substituted analogues. To re-evaluate these data, we compared the Nle- and Val-substituted and WT analogues of both Aβ40 and Aβ42 using multiple biophysical and cytotoxicity assays.
Using four different biophysical assays, we found that substituting Met35 by Nle or Val tended to increase Aβ assembly. The substitutions effect on the oligomer size distribution (Fig. 1A) or conformational change kinetics (Fig. 2) of Aβ40 was modest relative to the effect on Aβ42. In particular, substitution of Met35 by Val was found to accelerate the aggregation kinetics of Aβ42 (Figs. 2 and 3), which correlated with a shift in the oligomer size distribution towards higher order oligomers (Fig. 1B, C). A similar, yet more subtle effect on acceleration of aggregation (Fig. 3) and a shift in oligomer size distribution (Fig. 1C) was observed for [Nle35]Aβ42. Overall, the tendency to increase self-assembly was more pronounced in Aβ42 than in Aβ40 and correlated with the somewhat higher hydrophobicity of Nle and Val relative to Met. These data support a central role for the C-terminus in the assembly of Aβ42 and less so for Aβ40, in agreement with previous reports (Bitan et al. 2003a, Urbanc et al. 2004, Yun et al. 2007, Yang & Teplow 2008, Bernstein et al. 2009). We note that the DLS data obtained for the Aβ42 analogues are comparable qualitatively but not quantitatively with the CD results due to the differences in samples preparation. Using different preparation protocols was necessary because of the fundamental differences between the two methods. Unlike CD, which is an averaging technique, DLS overemphasizes large particles, as discussed above. Therefore, DLS samples must be filtered prior to measurement to exclude dust particles and cannot be agitated because formation of even a few large particles due to agitation would lead to skewed results. Nevertheless, the capability of DLS to measure assembly dynamics non-invasively and with high sensitivity provides useful, complementary information to the CD and EM measurements.
The correlation we observed between formation of higher order oligomers and increased β-sheet formation concords with recent data showing a similar correlation in isolated Aβ40 oligomers (Ono et al. 2009). These data also are in agreement with the acceleration of β-sheet formation by [Val35]Aβ42 compared to WT Aβ42 in the presence of lipid vesicles and Cu2+ ions observed by Ciccotosto et al. using CD (Ciccotosto et al. 2004). Compared to the latter study, our data reveal that the faster conformational transition of the Val-substituted peptide likely results from the substitution itself rather than from presence of vesicles or Cu2+ ions.
Whereas Ciccotosto et al., (2004) reported that at 5 µM both Aβ42 and [Val35]Aβ42 showed non-fibrillar morphology both initially and after 72 h of incubation at 37°C (Ciccotosto et al. 2004), we found fibrillar morphology for all three Aβ42 analogues after 24 h of incubation. These morphological variations likely result from the difference in experimental setup. We used 10 µM concentration and incubated the peptides at 25°C with continuous agitation, whereas Ciccotosto et al. used 5 µM and incubated at 37°C without agitation.
Despite the differences in assembly kinetics and in contrast to previous reports, we found very similar levels of toxicity for the WT and substituted analogues of both Aβ40 and Aβ42 using three different assays, MTT, LDH, and TUNEL, in both cortical and hippocampal primary neurons.
Dose-response analysis for Aβ42 and its analogues showed nearly identical values in both the MTT and LDH assays (Fig. 5). When the toxicity of Aβ40 and Aβ42 analogues measured by all three assays was compared at 10 µM, the same concentration used in the biophysical studies, the only significant difference found was between the Aβ40 and Aβ42 groups, but not among the WT and substituted analogues within each group (Figs. 5 and 6).
Differences in experimental settings likely explain the difference between our data and those reported by other groups. One obvious possibility is the source of the peptides used. However, in our experience, the differences in biophysical and biological behavior between Aβ analogues prepared by the UCLA Biopolymers Laboratory and those from commercial sources are not bigger than typical batch-to-batch variation. Moreover the toxicity of WT Aβ in our study was comparable that observed by the other laboratories (Ciccotosto et al. 2004, Yatin et al. 1999, Varadarajan et al. 1999), suggesting that the peptide source did not contribute significantly to the observed results.
More likely explanations include differences in the peptide preparation methods and cell types used. For example, Varadarajan et al., used [Nle35]Aβ40 that was pre-incubated for 24 h before addition to culture and measured the effect of this peptide after 6 h of incubation with cells (Varadarajan et al. 1999). Similarly, Yatin et al., used [Nle35]Aβ42, which was dissolved in water at 1 mg/mL, and incubated for 24 h before addition to cells (Yatin et al. 1999). In neither case were the peptides treated to remove aggregates prior to their dissolution. In contrast, we treated all the peptides with HFIP to obtain aggregate-free starting conditions, prepared solutions in cell culture media, and added these solutions to the cells immediately after preparation. It is therefore plausible that we observed toxicity of [Nle35]Aβ oligomers, which likely were not present in the former studies because of the 24 h pre-incubation step.
In different studies, Clementi et al. (Clementi et al. 2006) and Piacentini et al. (Piacentini et al. 2008) found little or no toxicity for [Nle35]Aβ42 using IMR-32 cells. Though neither group used a pre-incubation step, they diluted the peptides from stock solutions prepared in DMSO directly into the cell culture medium. Thus, the differences between their results and the ones presented here may stem both from the higher sensitivity of primary neurons to Aβ-induced toxicity relative to IMR-32 cells and from the differences in the preparation protocols used.
The higher toxicity of [Val35]Aβ42 relative to WT Aβ42 reported by Ciccotosto et al. (Ciccotosto et al. 2004), which we did not observe, also may be due to differences in experimental protocols. They prepared their peptides without HFIP treatment and adjusted the concentration based on measurement of absorbance at 214 nm, a method we found to produce results that are inconsistent with amino acid analysis data (G. Bitan, unpublished results).
Ciccotosto et al. suggested that [Val35]Aβ42 has higher affinity for lipid membranes than Aβ42, which may be linked to increased toxicity, although both Aβ42 and [Val35]Aβ42 were reported to produce similar amount of H2O2 (Ciccotosto et al. 2004). A different view was suggested by Murray et al., who found that substitution of Met35 by Val decreased lipid peroxidation (Murray et al. 2005). Because we did not study lipid peroxidation, our data are not directly comparable with those of Murray et al. Nevertheless, our results suggest that the presence of Met35 in Aβ is not an important factor in Aβ-induced toxicity.
Our conclusion is consistent with a recent elegant study by Butterfield et al. (Butterfield et al. 2009) who used a variant of the PDAPP transgenic mouse model of AD (Masliah et al. 1996), containing an M631L mutation in the amyloid β-protein precursor-encoding gene, which leads to substitution of Met35 in Aβ by Leu. The study showed that Met35 was required for observation of markers of oxidative stress, such as protein carbonylation and lipid peroxidation in the brains of the mice, yet the substitution of Met35 by Leu had no effect on the learning and memory impairment of the mice assessed using the Morris Water Maze (Butterfield et al. 2009). The reasons for this apparent discrepancy are not understood and may be related to differences in deposition patterns between WT and [Leu35]Aβ or to involvement of toxic APP fragments other than Aβ, as suggested by the authors (Butterfield et al. 2009). As is the case with most mouse models of AD, the impairment of learning and memory observed by Butterfield et al. was not associated with neuronal loss (Morrissette et al. 2009). Nonetheless, if one accepts the assumption that the results of the learning and memory tests measured in vivo correlate with our neurotoxicity measurements in primary cultures, these results support our conclusion that the presence of Met35 is not important for Aβ toxicity.
ACKNOWLEDGMENTS
This work was supported by Alzheimer’s Association Grant IIRG-07-5833 (GB) and NIH/NIA grant AG027818 (GB and GBB). We thank Margaret M. Condron for peptide synthesis and amino acid analysis, Dr. David Teplow for the use of his CD spectrometer, and Drs. Farid Rahimi and Huiyuan Li for valuable advice and critical discussion of the manuscript.
Abbreviations
- AD
Alzheimer’s disease
- Aβ
amyloid β-protein
- APS
ammonium persulfate
- BrdU
5-bromo-2-deoxyuridine
- CD
circular dichroism spectroscopy
- DMEM
Dulbecco’s modified Eagle’s medium
- DLS
dynamic light scattering
- EM
electron microscopy
- HFIP
1,1,1,3,3,3-hexafluoroisopropanol
- LDH
lactate dehydrogenase
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- OD
optical density
- PICUP
photo-induced cross-linking of unmodified proteins
- ROS
reactive oxygen species
- Ru(Bpy)
tris(2,2’-bipyridyl)ruthenium dichloride
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- TUNEL
terminal deoxyribonucleotidyl transferase dUTP nick end labeling
- WT
wild-type
REFERENCES
- Barnham KJ, Ciccotosto GD, Tickler AK, et al. Neurotoxic, redox-competent Alzheimer's β-amyloid is released from lipid membrane by methionine oxidation. J. Biol. Chem. 2003;278:42959–42965. doi: 10.1074/jbc.M305494200. [DOI] [PubMed] [Google Scholar]
- Bernstein SL, Dupuis NF, Lazo ND, et al. Amyloid-β protein oligomerization and the importance of tetramers and dodecamers in the aetiology of Alzheimer's disease. Nat. Chem. 2009;1:326–331. doi: 10.1038/nchem.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitan G. Structural study of metastable amyloidogenic protein oligomers by photo-induced cross-linking of unmodified proteins. Methods Enzymol. 2006;413:217–236. doi: 10.1016/S0076-6879(06)13012-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitan G, Kirkitadze MD, Lomakin A, Vollers SS, Benedek GB, Teplow DB. Amyloid β-protein (Aβ) assembly: Aβ40 and Aβ42 oligomerize through distinct pathways. Proc. Natl. Acad. Sci. USA. 2003a;100:330–335. doi: 10.1073/pnas.222681699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitan G, Lomakin A, Teplow DB. Amyloid β-protein oligomerization: prenucleation interactions revealed by photo-induced cross-linking of unmodified proteins. J. Biol. Chem. 2001;276:35176–35184. doi: 10.1074/jbc.M102223200. [DOI] [PubMed] [Google Scholar]
- Bitan G, Tarus B, Vollers SS, Lashuel HA, Condron MM, Straub JE, Teplow DB. A molecular switch in amyloid assembly: Met35 and amyloid β-protein oligomerization. J. Am. Chem. Soc. 2003b;125:15359–15365. doi: 10.1021/ja0349296. [DOI] [PubMed] [Google Scholar]
- Bitan G, Vollers SS, Teplow DB. Elucidation of Primary Structure Elements Controlling Early Amyloid β-Protein Oligomerization. J. Biol. Chem. 2003c;278:34882–34889. doi: 10.1074/jbc.M300825200. [DOI] [PubMed] [Google Scholar]
- Bravo R, Arimon M, Valle-Delgado JJ, Garcia R, Durany N, Castel S, Cruz M, Ventura S, Fernandez-Busquets X. Sulfated polysaccharides promote the assembly of amyloid β(1–42) peptide into stable fibrils of reduced cytotoxicity. J. Biol. Chem. 2008;283:32471–32483. doi: 10.1074/jbc.M709870200. [DOI] [PubMed] [Google Scholar]
- Butterfield DA. Amyloid b-peptide (1–42)-induced oxidative stress and neurotoxicity: Implications for neurodegeneration in Alzheimer's disease brain. Free Radical Res. 2002;36:1307–1313. doi: 10.1080/1071576021000049890. [DOI] [PubMed] [Google Scholar]
- Butterfield DA, Boyd-Kimball D. The critical role of methionine 35 in Alzheimer's amyloid β-peptide (1–42)-induced oxidative stress and neurotoxicity. Biochim. Biophys. Acta. 2005;1703:149–156. doi: 10.1016/j.bbapap.2004.10.014. [DOI] [PubMed] [Google Scholar]
- Butterfield DA, Galvan V, Lange MB, et al. In vivo oxidative stress in brain of Alzheimer disease transgenic mice: Requirement for methionine 35 in amyloid β-peptide of APP. Free Radic. Biol. Med. 2009;48:136–144. doi: 10.1016/j.freeradbiomed.2009.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterfield DA, Kanski J. Methionine residue 35 is critical for the oxidative stress and neurotoxic properties of Alzheimer's amyloid β-peptide 1–42. Peptides. 2002;23:1299–1309. doi: 10.1016/s0196-9781(02)00066-9. [DOI] [PubMed] [Google Scholar]
- Butterfield DA, Reed T, Newman SF, Sultana R. Roles of amyloid β-peptide-associated oxidative stress and brain protein modifications in the pathogenesis of Alzheimer's disease and mild cognitive impairment. Free Radic. Biol. Med. 2007;43:658–677. doi: 10.1016/j.freeradbiomed.2007.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccotosto GD, Tew D, Curtain CC, et al. Enhanced toxicity and cellular binding of a modified amyloid β peptide with a methionine to valine substitution. J. Biol. Chem. 2004;279:42528–42534. doi: 10.1074/jbc.M406465200. [DOI] [PubMed] [Google Scholar]
- Clementi ME, Pezzotti M, Orsini F, Sampaolese B, Mezzogori D, Grassi C, Giardina B, Misiti F. Alzheimer's amyloid β-peptide (1–42) induces cell death in human neuroblastoma via bax/bcl-2 ratio increase: an intriguing role for methionine 35. Biochem. Biophys. Res. Commun. 2006;342:206–213. doi: 10.1016/j.bbrc.2006.01.137. [DOI] [PubMed] [Google Scholar]
- Crouch PJ, Harding SM, White AR, Camakaris J, Bush AI, Masters CL. Mechanisms of Aβ mediated neurodegeneration in Alzheimer's disease. Int. J. Biochem. Cell Biol. 2008;40:181–198. doi: 10.1016/j.biocel.2007.07.013. [DOI] [PubMed] [Google Scholar]
- Cummings JL. Alzheimer's disease. N. Engl. J. Med. 2004;351:56–67. doi: 10.1056/NEJMra040223. [DOI] [PubMed] [Google Scholar]
- Dahlgren KN, Manelli AM, Stine WB, Jr, Baker LK, Krafft GA, LaDu MJ. Oligomeric and fibrillar species of amyloid-β peptides differentially affect neuronal viability. J. Biol. Chem. 2002;277:32046–32053. doi: 10.1074/jbc.M201750200. [DOI] [PubMed] [Google Scholar]
- Decker T, Lohmann-Matthes ML. A quick and simple method for the quantitation of lactate dehydrogenase release in measurements of cellular cytotoxicity and tumor necrosis factor (TNF) activity. J. Immunol. Methods. 1988;115:61–69. doi: 10.1016/0022-1759(88)90310-9. [DOI] [PubMed] [Google Scholar]
- Fancy DA, Kodadek T. Chemistry for the analysis of protein-protein interactions: Rapid and efficient cross-linking triggered by long wavelength light. Proc. Natl. Acad. Sci. USA. 1999;96:6020–6024. doi: 10.1073/pnas.96.11.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradinger EA, Monien BH, Urbanc B, et al. C-terminal peptides coassemble into Aβ42 oligomers and protect neurons against Aβ42-induced neurotoxicity. Proc. Natl. Acad. Sci. USA. 2008;105:14175–14180. doi: 10.1073/pnas.0807163105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haass C, Selkoe DJ. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer's amyloid β-peptide. Nat. Rev. Mol. Cell Biol. 2007;8:101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- Halliwell B. Free radicals, reactive oxygen species and human disease: a critical evaluation with special reference to atherosclerosis. Br. J. Exp. Pathol. 1989;70:737–757. [PMC free article] [PubMed] [Google Scholar]
- Halliwell B. Oxidative stress and neurodegeneration: where are we now? J Neurochem. 2006;97:1634–1658. doi: 10.1111/j.1471-4159.2006.03907.x. [DOI] [PubMed] [Google Scholar]
- Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- Hou L, Kang I, Marchant RE, Zagorski MG. Methionine 35 oxidation reduces fibril assembly of the amyloid Aβ-(1–42) peptide of Alzheimer's disease. J. Biol. Chem. 2002;277:40173–40176. doi: 10.1074/jbc.C200338200. [DOI] [PubMed] [Google Scholar]
- Hou L, Shao H, Zhang Y, et al. Solution NMR studies of the Aβ(1–40) and Aβ(1–42) peptides establish that the Met35 oxidation state affects the mechanism of amyloid formation. J. Am. Chem. Soc. 2004;126:1992–2005. doi: 10.1021/ja036813f. [DOI] [PubMed] [Google Scholar]
- Jarrett JT, Berger EP, Lansbury PT., Jr The carboxy terminus of the β amyloid protein is critical for the seeding of amyloid formation: Implications for the pathogenesis of Alzheimer's disease. Biochemistry. 1993;32:4693–4697. doi: 10.1021/bi00069a001. [DOI] [PubMed] [Google Scholar]
- Josephs KA, Whitwell JL, Ahmed Z, et al. β-Amyloid burden is not associated with rates of brain atrophy. Ann. Neurol. 2008;63:204–212. doi: 10.1002/ana.21223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkitadze MD, Condron MM, Teplow DB. Identification and characterization of key kinetic intermediates in amyloid β-protein fibrillogenesis. J. Mol. Biol. 2001;312:1103–1119. doi: 10.1006/jmbi.2001.4970. [DOI] [PubMed] [Google Scholar]
- Koffie RM, Meyer-Luehmann M, Hashimoto T, et al. Oligomeric amyloid β associates with postsynaptic densities and correlates with excitatory synapse loss near senile plaques. Proc. Natl. Acad. Sci. USA. 2009;106:4012–4017. doi: 10.1073/pnas.0811698106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo YM, Kokjohn TA, Beach TG, et al. Comparative analysis of amyloid-β chemical structure and amyloid plaque morphology of transgenic mouse and Alzheimer's disease brains. J. Biol. Chem. 2001;276:12991–12998. doi: 10.1074/jbc.M007859200. [DOI] [PubMed] [Google Scholar]
- Lomakin A, Chung DS, Benedek GB, Kirschner DA, Teplow DB. On the nucleation and growth of amyloid β-protein fibrils: detection of nuclei and quantitation of rate constants. Proc. Natl. Acad. Sci. USA. 1996;93:1125–1129. doi: 10.1073/pnas.93.3.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markesbery WR, Lovell MA. Four-hydroxynonenal, a product of lipid peroxidation, is increased in the brain in alzheimers-disease. Neurobiol. Aging. 1998;19:33–36. doi: 10.1016/s0197-4580(98)00009-8. [DOI] [PubMed] [Google Scholar]
- Masliah E, Sisk A, Mallory M, Mucke L, Schenk D, Games D. Comparison of neurodegenerative pathology in transgenic mice overexpressing V717F β-amyloid precursor protein and Alzheimer's disease. J. Neurosci. 1996;16:5795–5811. doi: 10.1523/JNEUROSCI.16-18-05795.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissette DA, Parachikova A, Green KN, LaFerla FM. Relevance of transgenic mouse models to human Alzheimer disease. J. Biol. Chem. 2009;284:6033–6037. doi: 10.1074/jbc.R800030200. [DOI] [PubMed] [Google Scholar]
- Murray IV, Sindoni ME, Axelsen PH. Promotion of oxidative lipid membrane damage by amyloid β proteins. Biochemistry. 2005;44:12606–12613. doi: 10.1021/bi050926p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Näslund J, Schierhorn A, Hellman U, et al. Relative abundance of Alzheimer Aβ amyloid peptide variants in Alzheimer disease and normal aging. Proc. Natl. Acad. Sci. USA. 1994;91:8378–8382. doi: 10.1073/pnas.91.18.8378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono K, Condron MM, Teplow DB. Structure-neurotoxicity relationships of amyloid β-protein oligomers. Proc. Natl. Acad. Sci. USA. 2009;106:14745–14750. doi: 10.1073/pnas.0905127106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmblad M, Westlind-Danielsson A, Bergquist J. Oxidation of methionine 35 attenuates formation of amyloid β -peptide 1–40 oligomers. J. Biol. Chem. 2002;277:19506–19510. doi: 10.1074/jbc.M112218200. [DOI] [PubMed] [Google Scholar]
- Piacentini R, Ripoli C, Leone L, Misiti F, Clementi ME, D'Ascenzo M, Giardina B, Azzena GB, Grassi C. Role of methionine 35 in the intracellular Ca2+ homeostasis dysregulation and Ca2+-dependent apoptosis induced by amyloid β-peptide in human neuroblastoma IMR32 cells. J. Neurochem. 2008;107:1070–1082. doi: 10.1111/j.1471-4159.2008.05680.x. [DOI] [PubMed] [Google Scholar]
- Rahimi F, Maiti P, Bitan G. Photo-induced cross-linking of unmodified proteins (PICUP) applied to amyloidogenic peptides. J Vis Exp. 2009 doi: 10.3791/1071. DOI: 10.3791/1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiman EM, Chen K, Liu X, et al. Fibrillar amyloid-β burden in cognitively normal people at 3 levels of genetic risk for Alzheimer's disease. Proc. Natl. Acad. Sci. USA. 2009;106:6820–6825. doi: 10.1073/pnas.0900345106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roher AE, Palmer KC, Yurewicz EC, Ball MJ, Greenberg BD. Morphological and biochemical analyses of amyloid plaque core proteins purified from Alzheimer disease brain tissue. J. Neurochem. 1993;61:1916–1926. doi: 10.1111/j.1471-4159.1993.tb09834.x. [DOI] [PubMed] [Google Scholar]
- Roychaudhuri R, Yang M, Hoshi MM, Teplow DB. Amyloid β-protein assembly and Alzheimer disease. J. Biol. Chem. 2009;284:4749–4753. doi: 10.1074/jbc.R800036200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal M, Manor D. Confocal microscopic imaging of [Ca2+]i in cultured rat hippocampal neurons following exposure to N-methyl-D-aspartate. J. Physiol. 1992;448:655–676. doi: 10.1113/jphysiol.1992.sp019063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkoe DJ. Alzheimer's disease: Genes, proteins, and therapy. Physiol. Rev. 2001;81:741–766. doi: 10.1152/physrev.2001.81.2.741. [DOI] [PubMed] [Google Scholar]
- Tomic JL, Pensalfini A, Head E, Glabe CG. Soluble fibrillar oligomer levels are elevated in Alzheimer's disease brain and correlate with cognitive dysfunction. Neurobiol. Dis. 2009;35:352–358. doi: 10.1016/j.nbd.2009.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanc B, Cruz L, Yun S, Buldyrev SV, Bitan G, Teplow DB, Stanley HE. In silico study of amyloid β-protein folding and oligomerization. Proc. Natl. Acad. Sci. USA. 2004;101:17345–17350. doi: 10.1073/pnas.0408153101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadarajan S, Kanski J, Aksenova M, Lauderback C, Butterfield DA. Different mechanisms of oxidative stress and neurotoxicity for Alzheimer's Aβ(1–42) and Aβ(25–35) J. Am. Chem. Soc. 2001;123:5625–5631. doi: 10.1021/ja010452r. [DOI] [PubMed] [Google Scholar]
- Varadarajan S, Yatin S, Aksenova M, Butterfield DA. Review: Alzheimer's amyloid β-peptide-associated free radical oxidative stress and neurotoxicity. J. Struct. Biol. 2000;130:184–208. doi: 10.1006/jsbi.2000.4274. [DOI] [PubMed] [Google Scholar]
- Varadarajan S, Yatin S, Kanski J, Jahanshahi F, Butterfield DA. Methionine residue 35 is important in amyloid β-peptide-associated free radical oxidative stress. Brain Res. Bull. 1999;50:133–141. doi: 10.1016/s0361-9230(99)00093-3. [DOI] [PubMed] [Google Scholar]
- Vollers SS, Teplow DB, Bitan G. Determination of Peptide oligomerization state using rapid photochemical crosslinking. Methods Mol. Biol. 2005;299:11–18. doi: 10.1385/1-59259-874-9:011. [DOI] [PubMed] [Google Scholar]
- Wang X, Ge J, Wang K, Qian J, Zou Y. Evaluation of MTT assay for measurement of emodin-induced cytotoxicity. Assay Drug Dev. Technol. 2006;4:203–207. doi: 10.1089/adt.2006.4.203. [DOI] [PubMed] [Google Scholar]
- White JA, Manelli AM, Holmberg KH, Van Eldik LJ, Ladu MJ. Differential effects of oligomeric and fibrillar amyloid-β 1–42 on astrocyte-mediated inflammation. Neurobiol. Dis. 2005;18:459–465. doi: 10.1016/j.nbd.2004.12.013. [DOI] [PubMed] [Google Scholar]
- Yang M, Teplow DB. Amyloid β-protein monomer folding: free-energy surfaces reveal alloform-specific differences. J. Mol. Biol. 2008;384:450–464. doi: 10.1016/j.jmb.2008.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatin SM, Varadarajan S, Link CD, Butterfield DA. In vitro and in vivo oxidative stress associated with Alzheimer's amyloid β-peptide (1–42) Neurobiol. Aging. 1999;20:325–330. doi: 10.1016/s0197-4580(99)00056-1. [DOI] [PubMed] [Google Scholar]
- Yun S, Urbanc B, Cruz L, Bitan G, Teplow DB, Stanley HE. Role of Electrostatic Interactions in Amyloid β-Protein (Aβ) Oligomer Formation: A Discrete Molecular Dynamics Study. Biophys. J. 2007;92:4064–4077. doi: 10.1529/biophysj.106.097766. [DOI] [PMC free article] [PubMed] [Google Scholar]