SUMMARY
When to form flowers is a developmental decision that profoundly impacts the fitness of flowering plants. In Arabidopsis this decision is ultimately controlled by the induction and subsequent activity of the transcription factors LEAFY (LFY), FRUITFULL (FUL) and APETALA1 (AP1). Despite their central importance, our current understanding of the regulation of LFY, FUL and AP1 expression is still incomplete. We show here that all three genes are directly activated by the microRNA targeted transcription factor SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 3 (SPL3). Our findings suggest that SPL3 acts together with other microRNA-regulated SPL transcription factors to control the timing of flower formation. Moreover, the identified SPL activity defines a distinct pathway in control of this vital developmental decision.
INTRODUCTION
Flowering plants are sessile organisms that use environmental as well as endogenous cues to optimize the progression through developmental phases. During each developmental phase distinct structures form from the primordia at the flanks of the shoot apical meristem (Araki, 2001; Poethig, 2003; Steeves and Sussex, 1989), which differ in morphology and function. Developmental phases and the transitions between them have been well studied in the reference plant Arabidopsis thaliana. In this species three main phase transitions can be distinguished: the vegetative phase change, the reproductive or floral transition and the meristem identity (MI) transition. After germination, the first primordia formed give rise to juvenile leaves. Following the vegetative phase change, adult leaves are formed instead, which differ from juvenile leaves in many traits (Poethig, 2003). Production of these leaves, collectively referred to as rosette leaves, ceases following the reproductive transition, which is regulated by several independent flowering time pathways (reviewed in Kobayashi and Weigel, 2007; Turck et al., 2008). After this transition, the primary inflorescence grows upward (bolts) and primordia give rise to secondary inflorescences branches subtended by cauline leaves (Araki, 2001; Steeves and Sussex, 1989). Finally, the MI transition controls the onset of the last developmental phase, in which the primordia give rise to the reproductive structures, the flowers (Blazquez et al., 2006).
Proper timing of the MI transition is important for fitness of flowering plants (Roux et al., 2006). This transition is controlled by two types of positive regulators: the plant specific transcription factor LFY and members of the AP1/FUL clade of MADS-box transcription factors (Benlloch et al., 2007; Litt and Irish, 2003). The role of these regulators is conserved in different plant species (Benlloch et al., 2007). Studies in Arabidopsis have shown that LFY, FUL and AP1 expression increases just prior to the MI transition, with LFY and FUL (formerly AGL8) activated very early, and AP1 upregulation occurring later (for example see Hempel et al., 1997). The expression of the three MI genes overlaps at the sites where flowers are specified, the inflorescence meristem and associated primordia (reviewed in Kobayashi and Weigel, 2007; Turck et al., 2008). LFY is considered a master regulator of the MI transition since loss of LFY function causes the most dramatic delay in flower formation (Weigel et al., 1992). ful single mutants exhibit significant delays in the MI transition (Ferrandiz et al., 2000; Melzer et al., 2008). FUL acts together with other MADS-box transcription factors including AP1 to regulate this process (Ferrandiz et al., 2000). ap1 single mutants also cause a delay of the MI transition, and AP1 plays an important role in this process both downstream of and together with LFY (Bowman et al., 1993). Finally, elevated levels of either LFY, AP1, or FUL cause a precocious MI transition, suggesting that upregulation of any one of these regulators is sufficient to trigger this developmental switch (Ferrandiz et al., 2000; Mandel and Yanofsky, 1995; Weigel and Nilsson, 1995). Proper control of the LFY, FUL and AP1 accumulation is therefore key for the correct timing of the onset of flower formation.
Despite their importance in regulation of a vital developmental transition, our understanding of the regulation of LFY, FUL and AP1 expression is incomplete. Recently microRNA regulated SQUAMOSA PROMOTER BINDING PROTEIN (SBP)-box transcription factors have been implicated in regulation of multiple developmental transitions in Arabidopsis and other plant species (Cardon et al., 1997; Chuck et al., 2007; Gandikota et al., 2007; Schwarz et al., 2008; Wu and Poethig, 2006; Xie et al., 2006). Consistent with their role in developmental timing, transcript accumulation of several members of this family strongly increases during development (Cardon et al., 1999; Schmid et al., 2003; Wu and Poethig, 2006). Concomitantly, levels of the microRNA miR156 that specifically targets these genes decrease (Schwab et al., 2005; Wu and Poethig, 2006). How the SBP transcription factors exert their roles in developmental transitions is an area of intense investigation. We show here that one well-studied member of this family, SPL3, directly activates LFY, FUL, and AP1 expression in Arabidopsis identifying a direct molecular link between SPL transcription factors and one of the developmental transitions they regulate. We further show that SPL3, together with additional microRNA regulated SPL transcription factors, is required for LFY, FUL and AP1 upregulation during the MI transition, implicating SPL transcription factors as a central node in the regulatory network that controls flower formation.
RESULTS
SPL3 upregulates LFY, FUL, and AP1
Mutations in the microRNA regulated transcription factor SPL3 cause no visible phenotype, most likely because of functional redundancy with other related SPL proteins in Arabidopsis (Wang et al., 2008; Wu and Poethig, 2006). To examine the role of SPL3 in the MI transition, we constitutively expressed the SPL3 mRNA without the microRNA target site in the 3’UTR (35S:SPL3Δ; Wu and Poethig, 2006) using a gain-of-function approach. 35S:SPL3Δ plants exhibited a precocious reproductive transition (fewer rosette leaves formed) and MI transition (fewer secondary inflorescences and cauline leaves formed) compared to the wildtype in both long-day and short-day conditions (Table S1).
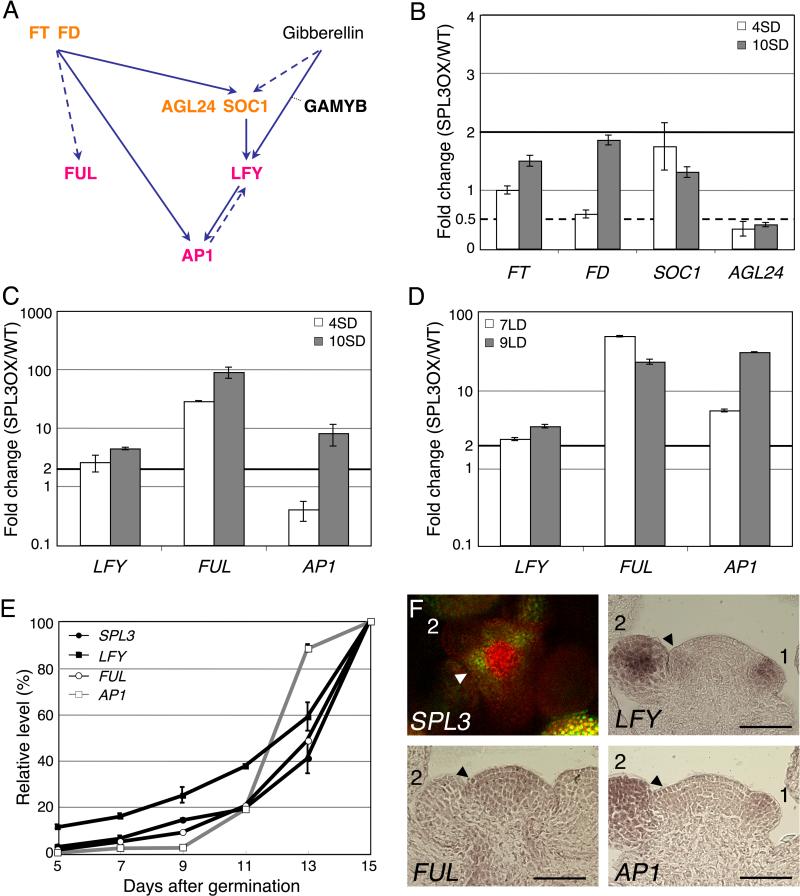
The rapid MI transition in 35S:SPL3Δ could be due to upregulation of upstream regulators of LFY, FUL or AP1 (Fig. 1A). To test this possibility, we examined expression of known activators of the MI genes. We scored two time-points (day four and day ten) in short-day growth conditions to assess the temporal upregulation of genes in 35S:SPLΔ compared to the wildtype. Both time-points precede the MI transition, which occurs at day 25 in short day (not shown). The MADS box transcription factors and flowering time regulators SOC1 and AGAMOUS-LIKE 24 (AGL24) are direct upstream transcriptional activators of LFY (Lee et al., 2008; Liu et al., 2008). Neither AGL24 nor SOC1 expression was strongly elevated in short-day grown 35S:SPL3Δ seedlings (Fig. 1B). We note that AGL24 levels are reduced in 35S:SPL3Δ; the reason for this reduction is currently not clear. The plant hormone gibberellin induces LFY expression through GAMYB transcription factors in short day (Achard et al., 2004; Blazquez and Weigel, 2000). Expression of MYB33, which encodes one of the GAMYB transcription factors known to bind the LFY promoter in vitro (Gocal et al., 2001), was unaltered in 35S:SPL3Δ (Fig. S1). FT is a known photoperiod pathway flowering time regulator that acts upstream of AP1 and FUL (Abe et al., 2005; Teper-Bamnolker and Samach, 2005; Wigge et al., 2005). FT expression was not much altered in 35S:SPL3Δ compared to the wildtype (Fig. 1B). Expression of FD, which encodes a bZIP transcription factor that acts together with FT upstream of AP1 and FUL, was slightly increased (1.8 fold) at the later time-point, in ten-day-old short-day-grown 35S:SPL3Δ plants. Hence, expression of the flowering time regulators was not strongly upregulated in 35S:SPL3Δ with the possible exception of FD.
Fig. 1. SPL3 activates meristem identity gene expression.
(A) Schematic representation of the known interactions involved in the meristem identity (MI) switch (see text for details). Flowering time regulators are indicated in orange, MI regulators in pink. Solid arrows denote direct interactions, dashed arrows denote direct or indirect interactions. (B-D) qRT-PCR analysis of flowering time gene (B) or MI gene expression (C, D) in 35S:SPL3Δ (SPL3OX) compared to wild-type plants. Plants were grown in short-day (SD) for 4 and 10 days (B, C) or in long-day (LD) for 7 and 9 days (D). (E) Temporal expression of SPL3, LFY, FUL and AP1 in wild-type plants grown in LD. The ratio of the expression at each time-point over the final expression level is graphed. (B-E) Values are mean +/- SEM. Black solid horizontal line: two fold increase, black dotted line: two fold decrease. (F) Confocal image of pSPL3:GFP-SPL3 (top view of inflorescence meristem) and in situ hybridization of LFY, FUL and AP1 (longitudinal section through inflorescence meristem). Arrowheads point to the incipient flower primordium, numbers denote stages of flower development. Bar: 50μm.
By contrast, when we examined expression of LFY, FUL and AP1 in the same conditions, we observed a strong increase in LFY levels (2.6 fold at day four and 4.4 fold at day ten) and FUL expression (greater than 25 fold at both time-points) in 35S:SPL3Δ compared to wild-type seedlings (Fig. 1C). AP1 upregulation was only observed at day ten (8-fold increase, Fig. 1C). The combined data suggest that SPL3 activates LFY, FUL, and AP1. To further test whether LFY, FUL and AP1 were also upregulated by SPL3 in different growth conditions, we examined their expression in inductive photoperiod (continuous light) in 35S:SPL3Δ compared to the wildtype (Fig. 1D). Again, the two time-points chosen (day seven and day nine) precede the MI transition, which occurs at day eleven under these growth conditions (Fig 1E, first upregulation of AP1). We observed a very similar increase in LFY and FUL expression in inductive photoperiod to that observed in short day conditions (compared Fig. 1D to Fig. 1C). In addition, AP1 levels were strongly increased at both time-points tested (Fig. 1D). Thus SPL3 may regulate the MI transition via upregulation of LFY, FUL, and AP1.
If SPL3 induces LFY, FUL and AP1, we expect temporal and spatial overlap in the expression of these regulators. In long-day grown wild-type seedlings SPL3, LFY and FUL levels showed very similar temporal upregulation, increasing gradually from day seven onward up to the MI transition at day eleven (first accumulation of AP1; Fig. 1E). The expression of the three genes increased at a similar rate after the MI transition and peaked during late reproductive development (day thirteen to fifteen; Fig. 1E). By contrast, AP1 levels started to increase later (at day nine) and increased at a slow rate up to the MI transition. A similar delay in AP1 induction has been observed in short-day grown plants in response to photoinduction (Hempel et al., 1997; Schmid et al., 2003). After the MI transition, AP1 expression increased very rapidly until day 13, likely because of the elaboration of the first flower primordia, which strongly express AP1 (Fig. 1E; Hempel et al., 1997). The temporal expression of SPL3, and FUL, LFY and AP1 is thus consistent with a role for SPL3 in the regulation of the MI genes.
Previous mRNA in situ hybridization data suggests that SPL3, and FUL, LFY and AP1 are expressed in the inflorescence meristem and associated young flower primordia (Cardon et al., 1997; Hempel et al., 1997; Mandel and Yanofsky, 1995; Weigel et al., 1992). To test for spatial expression overlap between SPL3 and FUL, LFY, and AP1, we generated a translational fusion protein of SPL3 with GFP (pSPL3: GFP-SPL3). This construct contains the microRNA binding site in the 3’UTR of SPL3. Two independent transgenic pSPL3:GFP-SPL3 lines showed GFP fluorescence in adult leaves and in the inflorescence meristem when grown in long-day conditions and did not exhibit early phase transitions (Fig. 1F, Fig. 3C, and data not shown), suggesting that the transgene recapitulates endogenous SPL3 expression and activity. When we compared SPL3 protein accumulation and mRNA accumulation of FUL, LFY, and AP1 in young inflorescences (Fig. 1F), we observed co-expression at the sites of the MI transition in the inflorescence: in incipient flower primordia (SPL3, FUL and LFY) and very young flowers (SPL3, AP1, LFY and, to a lesser extent, FUL; Fig. 1F). The observed spatial expression patterns support a role for SPL3 upstream of FUL, LFY, and AP1 in flower formation.
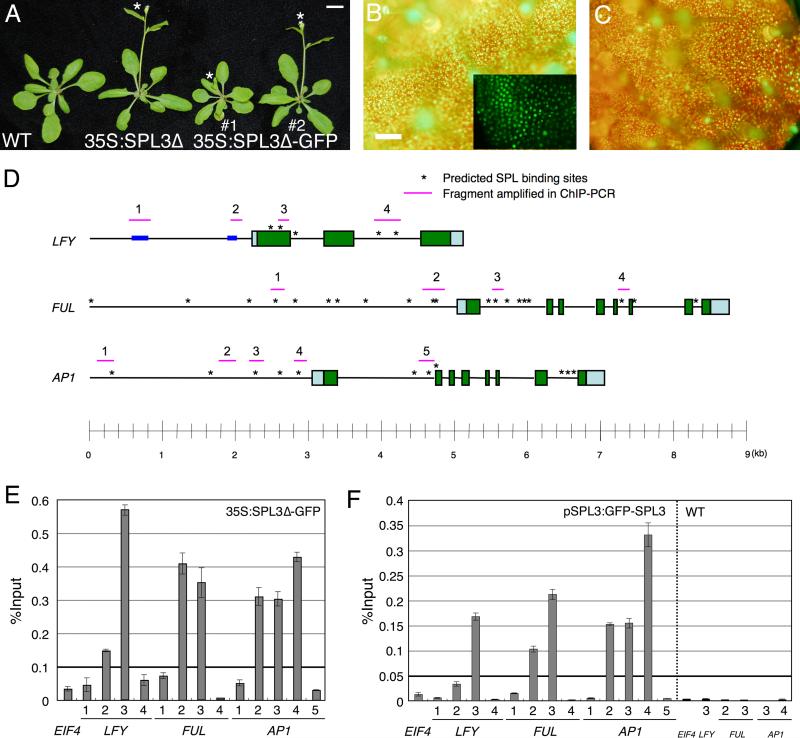
Fig. 3. SPL3 binds regulatory regions of meristem identity genes in vivo.
(A) Early flowering phenotype of LD-grown SPL3 overexpressing plants compared to wildtype. The asterisk marks the primary inflorescence bolt. Bar; 1 cm. Fluorescence image of 35S:SPL3Δ-GFP in leaf one (B) and pSPL3:GFP-SPL3 in leaf four (C). Inset in (B); close-up confocal microscopy image. Bar; 1 mm. (D) Schematic of the LFY, FUL and AP1 loci. Pale blue and green boxes represent untranslated regions and exons, respectively. Asterisks indicate computationally identified SPL consensus motifs. Pink horizontal lines: fragments amplified in by qPCR after ChIP. Blue boxes: regulatory regions in the LFY promoter previously identified (Blazquez and Weigel, 2000). (E, F) qPCR of anti-GFP ChIP in 35S:SPL3-GFPΔ (E), pSPL3:GFP-SPL3 and wildtype (F). Plants were grown in SD for 10 days (E) or LD for 7 days (F). Immunoprecipitated DNA enrichment is presented as percent input DNA. Shown is the mean +/- SEM.
LFY, AP1 and FUL act genetically downstream of SPL3
Since 35S:SPL3Δ plants exhibit a precocious MI transition and elevated LFY, FUL and AP1 levels, this raises the question whether the MI regulators are required for the precocious MI transition of 35S:SPL3Δ plants. We first examined the requirement for LFY by crossing 35S:SPL3Δ to the lfy-1 null mutant (Schultz and Haughn, 1991). lfy-1 was epistatic to 35S:SPL3Δ with regard to cauline leaf formation (Table 1). 35S:SPL3Δ lfy-1 plants formed the same number of cauline leaves as lfy-1, despite the strong reduction in cauline leaf number observed in 35S:SPL3Δ compared to the wildtype (Table 1). In addition, a strong increase was observed in the number of secondary inflorescences in 35S:SPL3Δ lfy-1 compared to 35S:SPL3Δ (Table 1, Fig. 2A). By contrast, lfy-1 had no effect on the number of rosette leaves formed in 35S:SPL3Δ (Table 2). 35S:SPL3Δ plants exhibit a precocious vegetative phase change (Wu and Poethig, 2006), which was not altered in the absence of LFY (Table S2). Thus, consistent with its known role (Blazquez et al., 1997; Weigel et al., 1992), LFY is specifically required for the precocious MI transition observed in 35S:SPL3Δ.
Table 1.
Effect of loss-of-function of meristem identity genes on phenotype of 35S:SPL3Δ phenotype
| Genotype | Number of Rosette Leaves | Number of Cauline Leaves | Number of Secondary Inflorescence |
|---|---|---|---|
| Wild type (Col) | 12.9 ± 0.2 (30) | 3.2 ± 0.1 (30) | 3.2 ± 0.1 (30) |
| 35S:SPL3Δ | 5.6 ± 0.2 (30) | 1.6 ± 0.1 (30) | 1.6 ± 0.1 (30) |
| 35S:SPL3Δ lfy-1 | 5.5 ± 0.2 (ll) | 11.6 ± 0.9 (11)† | 13.4 ± 0.1 (11) |
| lfy-1 | 13.5 ± 0.4 (12) | 10.9 ± 0.5 (16)† | 21.3 ± 0.6 (16) |
| Wild type (Col) | 12.5 ± 0.3 (12)† | 2.7 ± 0.1 (12) | 2.7 ± 0.1 (12) |
| 35S:SPL3Δ | 5.5 ± 0.2 (42) | 1.5 ± 0.1 (42) | 1.5 ± 0.1 (42) |
| 35S:SPL3Δ ful-2 | 9.6 ± 0.4 (12) | 3.5 ± 0.2 (12)† | 3.5 ± 0.2 (12)† |
| ful-2 | 11.8 ± 0.3 (11)† | 3.7 ± 0.2 (11)† | 3.7 ± 0.2 (11)† |
| Wild type (Col) | 12.6 ± 0.5 (7) | 3.4 ± 0.3 (7) | 3.4 ± 0.3 (7) |
| 35S:SPL3Δ | 5.7 ± 0.2 (30) | 1.4 ± 0.1 (30)‡ | 1.4 ± 0.1 (30) |
| 35S:SPL3Δ ap1-10 | 5.4 ± 0.2 (19) | 1.8 ± 0.3 (19)‡ | 8.4 ± 1.6 (9)* 42.1%& |
| ap1-10 | 10.9 ± 0.5 (12) | 3.2 ± 0.1 (12) | 5.8 ± 1.0 (5)* 41.7%& |
Shown is the mean ± SEM (n).
No statistically significant difference (Student's t test, P > 0.4) was detected between the marked genotypes.
Statistically significant difference (Student's t test, P < 0.05) was detected between marked genotypes.
Number of secondary inflorescences in plants that ceased secondary inflorescence and branched flower production prior to node 20.
% of plants that formed more than 20 secondary inflorescences and branched flowers. Plants were grown on soil at 22°C in long-day conditions (LDs; 16h light/8h dark, ~ 110 μmol/m2/s).
Fig. 2. The precocious meristem identity phenotype of 35S:SPL3Δ requires a functional copy of LFY, FUL and AP1.
Phenotype of 35S:SPL3Δ double mutants with lfy-1 (A), ful-2 (B), and ap1-10 (C). Top: side view of same age plants. Note the increased number of secondary inflorescences in the double mutants (left) compared to 35S:SPL3Δ (right). Bottom: top view of shoot apex close-ups from 5 cm tall primary inflorescences. Note the leafy appearance of 35S:SPL3Δ lfy-1 and 35S:SPL3 ap1-10 due to the presence of additional cauline leaves. Plants were grown in LD. Upper panel bar: 1 cm; lower panel bar: 1mm.
Table 2.
Phenotype of 35S:FT-GFP alone and in combination with 35S:miR156a or 35S:SPL3Δ
| Genotype | Number of Rosette Leaves | Number of Cauline Leaves | Number of Secondary Inflorescence |
|---|---|---|---|
| Wild type (Col) | 11.9 ± 0.3 (12) | 2.9 ± 0.1 (12) | 2.9 ± 0.1 (12) |
| 35S:FT-GFP | 4.2 ± 0.2 (16) | 2.0 ± 0.1(16) | 2.1 ± 0.1 (16) |
| 35S:FT-GFP x 35S:miR156a | 3.9 ± 0.2 (24) | 2.2 ± 0.1 (24) | 1.8 ± 0.1 (24) |
| 35S:miR156a x Col | 39.1 ± 1.9 (13) | ND | ND |
| 35S:FT-GFP x Col | 3.3 ± 0.1 (57)‡ | 1.3 ± 0.1 (57) | 1.3 ± 0.1 (57)‡ |
| 35S:FT-GFP x 35S:SPL3 | 2.0 ± 0 (29)‡ | 1.4 ± 0.1 (29) | 1.0 ± 0.1 (29)‡ |
| 35S:SPL3 x Col | 5.1 ± 0.2 (15) | 1.5 ± 0.1 (15) | 1.5 ± 0.1 (15) |
Shown is the mean ± SEM (n). ND denotes not determined.
Statistically significant difference (Student's t test, P < 0.005) was detected between marked genotypes.
Plants were grown on soil at 22°C in LDs
35S:SPL3Δ ful-2 plants had MI phenotypes identical to those of strong loss-of-function ful-2 single mutants (Ferrandiz et al., 2000), both with respect to the number of secondary inflorescences and of cauline leaves formed (Table 1, Fig. 2 B). Thus, ful-2 is epistatic to 35S:SPL3Δ in regulation of the MI transition. ful-2 also partly suppressed the precocious reproductive transition observed in 35S:SPL3Δ (Table 1), suggesting that upregulation of FUL expression contributes to the early flowering of 35S:SPL3Δ. These findings are consistent with the previously described strong and subtle roles of FUL in regulation of the MI and the reproductive transition, respectively (Ferrandiz et al., 2000; Melzer et al., 2008). In addition, ful-2 partially suppressed the precocious vegetative phase transition of 35S:SPL3Δ (Table S2), suggesting a potential novel role for FUL in vegetative phase change. Hence FUL may act downstream of SPL3 in all three developmental transitions.
AP1 is also required for the precocious MI transition in 35S:SPL3Δ (Table 1, Fig. 2C). Like the strong ap1-10 mutant (Schultz and Haughn, 1993), 35S:SPL3Δ ap1-10 plants often continued to produce secondary inflorescences or branched flowers after the 20th node, and the remaining plants showed a strong increase in the number of secondary inflorescences formed (8.4 in 35S:SPL3Δ ap1-10 compared to 5.8 in ap1-10 plants; Table 1). In addition, a slight increase in the number of cauline leaves was observed in 35S:SPL3Δ ap1-10 (Table 1). As expected based on the role of AP1 as an MI regulator (Bowman et al., 1993), ap1-10 had no effect on the vegetative transition (Table S2) or reproductive transition (Table1; Cardon et al., 1997) of 35S:SPL3Δ. Taken together these analyses provide strong support for the hypothesis that SPL3 acts upstream of LFY, AP1 and FUL in the same genetic pathway to control flower formation.
SPL3 directly regulates expression of MI genes
Our combined findings suggest a possible direct role of SPL3 in the upregulation of LFY, FUL, and AP1. Therefore, we computationally identified candidate SPL transcription factor binding sites in the LFY, FUL and AP1 loci (Fig. 3D) using a matrix-based approach (http://www.athamap.de/index.php; Galuschka et al., 2007). To test whether SPL3 directly regulates MI gene expression by binding to any of these cis elements, we probed SPL3 occupancy on genomic DNA in vivo by chromatin immunoprecipitation (ChIP). Towards this end, we generated GFP-tagged 35S:SPL3Δ (35S:SPL3Δ-GFP and 35S:GFP-SPL3Δ). Both transgenes recapitulated the phenotype of 35S:SPL3Δ (Table 1, Fig. 3A and data not shown). The tagged SPL3 protein was constitutively expressed (Fig. 3B, and data not shown) and localized to the nucleus (inset in Fig. 3B). Using two independent transgenic lines and two different polyclonal anti-GFP antibodies, the same genomic regions showed selective SPL3 occupancy in 35S:SPL3Δ-GFP plants (Fig. S2), suggesting that the assay accurately detects SPL3 binding.
No candidate SPL binding sites were identified in the LFY promoter, but consensus binding sites were present in the first exon/intron as well as in the second intron of LFY (region three and region four, respectively, Fig. 3D). Using ChIP-qPCR, we detected strong and selective occupancy of SPL3 at region 3 of the LFY locus, which contains three consensus SPL binding motifs (Fig. 3E). By contrast, the predicted SPL binding sites in the second intron of LFY (region 4) were not bound (Fig. 3E). The close proximity of the three predicted SPL3 binding sites in region 3 of the LFY locus (less than 200 bp apart) is below the resolution limit of ChIP-qPCR since average size of the sonicated DNA is 500 bp. Thus, we were unable to determine which of the three consensus binding sites in region 3 are occupied by SPL3. We also tested for SPL3Δ-GFP occupancy at LFY promoter regions previously shown to be important for photoperiodic induction or gibberellin dependent induction (regions 1 and 2 in the diagram in Fig. 3D; Blazquez and Weigel, 2000). No strong SPL3 binding was observed for either region (Fig. 3E). Hence SPL3 binds regulatory regions outside of the LFY promoter in vivo.
A large number of predicted SPL binding sites are present in the FUL distal and proximal promoter as well as in FUL introns (Fig. 3D). We selected four regions to test for SPL3 occupancy by ChIP-qPCR. SPL3 did not bind to the distal FUL promoter (region 1, Fig. 3E). By contrast, strong SPL3 binding was observed in region 2, which contains a cluster of promoter proximal candidate SPL binding sites (Fig. 3E, Fig. S2; Lannenpaa et al., 2004). In addition, we observed strong SPL3 occupancy in region 3 in the first FUL intron (Fig. 3E). Again the ChIP resolution does not allow us to determine which of the predicted SPL binding sites in region 2 or 3 of FUL are occupied by SPL3. Region 4 of FUL, which contains a putative SPL binding site in the fifth intron, was not bound by SPL3. Hence SPL3 binds regulatory regions in the FUL locus.
AP1 is a direct target of LFY (Busch et al., 1999; Wagner et al., 1999; William et al., 2004), therefore the upregulation of AP1 observed in 35S:SPL3Δ may be an indirect consequence of the elevated LFY levels in these plants. However, prior in vitro studies in both Antirrhinum and Arabidopsis instead point to a direct role of SPL3 in AP1 activation (Klein et al., 1996, Cardon et al. 1997). To examine these possibilities we tested for SPL3Δ-GFP occupancy at AP1 regulatory regions using ChIP. A total of five regions were tested (Fig. 3D), including the SPL3 binding site previously identified by electrophoretic mobility shift assays in the AP1 promoter (region 4; Cardon et al., 1997). We detected strong in vivo SPL3 binding to regions 2, 3 and 4 of the AP1 promoter (Fig. 3E, S2), suggesting that SPL3 occupies multiple binding sites in the AP1 intergenic region. A more distal predicted SPL3 binding motif in the intergenic region upstream of the AP1 transcription start site (region 1) as well as a predicted SPL binding site at the end of the first intron of AP1 (region 5; Fig. 3D) were not occupied by SPL3Δ-GFP (Fig. 3E). Hence SPL3 can bind AP1 regulatory regions in vivo.
These experiments suggest that SPL3 may directly upregulate the expression of LFY, FUL, and AP1 to cause a precocious MI transition in 35S:SPL3Δ, strongly implicating SPL3 in control of this vital phase transition. To further test this hypothesis, we examined whether SPL also binds to regulatory regions of MI genes in wild-type plants expressing pSPL3:GFP-SPL3 (described above). ChIP-qPCR in two independent pSPL3:GFP-SPL3 transgenic lines yielded very similar results (data not shown), suggesting that this construct likely accurately reflects endogenous SPL3 occupancy on the genomic DNA. Representative ChIP-qPCR results for seven-day old long-day grown pSPL3:GFP-SPL3 seedlings are shown in Fig. 3F, with the same regions tested for occupancy as in Fig. 3E. A total of six regions were strongly and selectively occupied by SPL3 in SPL3:GFP-SPL3 (Fig. 3F), including region 3 of the LFY locus; regions 2 and 3 in the FUL locus; and regions 2, 3 and 4 of the AP1 locus. The regions occupied by SPL3 in SPL3:GFP-SPL3 are identical to those bound by SPL3 in 35S:SPL3Δ-GFP lines. We noted some quantitative differences in relative binding strength in 35S:SPL3Δ-GFP compared to pSPL3:GFP-SPL3. These differences may be due to the different SPL3 protein levels in the two lines. Amplification of SPL3-occupied regions after ChIP of non-transgenic wild-type plants yielded no detectable signal under identical conditions (Fig. 3F), indicating the observed enrichment is highly specific. Since SPL3 binds to regulatory regions in the LFY, FUL, and AP1 loci in seedlings that exhibit wild-type growth and development, our combined data indicate that LFY, FUL, and AP1 are direct in vivo targets of SPL3.
SPL3 acts redundantly with other SPL proteins to control the MI transition
Although SPL3 regulates MI gene expression and directly binds their regulatory regions in vivo under physiological conditions, spl3 single mutants do not exhibit any morphological defects, likely due to functional redundancy with other SPL proteins (Wang et al., 2008; Wu and Poethig, 2006). SPL3 is a member of a family of 16 SBP-box transcription factors in Arabidopsis, 11 of which (including SPL3) are regulated post-transcriptionally by the miR156 family of microRNAs (Gandikota et al., 2007; Schwab et al., 2005; Wu and Poethig, 2006). SPL3 has two close paralogs - SPL4 and SPL5; the three genes are grouped together into one SBP subfamily (Guo et al., 2008). Members of this SBP subfamily differ from the remaining seven miR156 targets in two important ways: the genes and cDNAs for SPL3, SPL4, and SPL5 are much smaller than the other SPL genes and encode primarily the DNA binding domain. In addition, the miR156 recognition motif is located in the 3’UTR and not in the coding region in these three SPLs (Gandikota et al., 2007; Guo et al., 2008; Schmid et al., 2003; Wu and Poethig, 2006). Because of these observations, we hypothesized that SPL4 and SPL5 are most likely to have overlapping roles with SPL3.
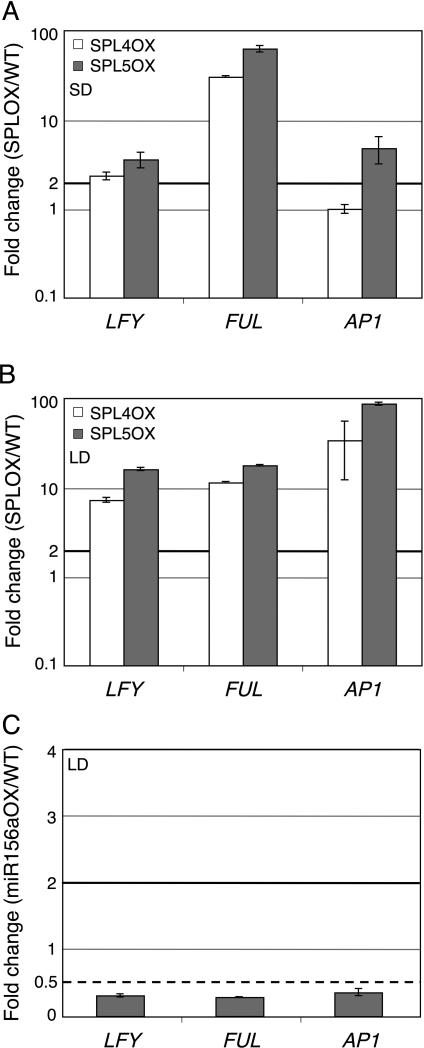
To test for a role of SPL4 and SPL5 in MI gene expression, we examined expression of LFY, FUL, and AP1 in plants overexpressing SPL4 or SPL5 without the miR156 target site in the 3’UTR (35S:SPL4Δ or 35S:SPL5Δ; Wu and Poethig, 2006). 35S:SPL4Δ and 35S:SPL5Δ plants exhibit similar strength phenotypes as 35S:SPL3Δ (Table S1; Wu and Poethig, 2006) and had similar transgene expression levels (SPL4Δ expression was slightly lower than that of SPL3Δ and SPL5Δ; Fig. S3A). We observed a similar increase in LFY and FUL levels in ten-day-old 35S:SPL4Δ and 35S:SPL5Δ plants grown in short-day conditions as we had for 35S:SPL3Δ (compare Fig. 1C and Fig. 4A). AP1 levels were only increased in 35S:SPL5Δ and not 35S:SPL4Δ plants, perhaps owing to the slightly lower transgene levels in the latter. Consistent with this, expression of all three MI genes was increased in 35S:SPL4Δ and 35S:SPL5Δ plants grown in inductive photoperiod (Fig. 4B). Since SPL3 levels are not elevated in 35S:SPL4Δ and 35S:SPL5Δ (Fig. S3B), our results suggest that SPL4 and SPL5 act redundantly with SPL3 in upregulation of LFY, FUL, and AP1 expression. However, in contrast to SPL3 overexpression, which does not result in strong upregulation of any of the flowering time genes tested (Fig. 1E), overexpression of SPL4 or SPL5 resulted in increased expression of several flowering time genes (Fig. S4A). Hence although SPL3, SPL4 and SPL5 are very similar to one another based on primary sequence and regulate MI gene expression, they do not have identical roles and activities, even when constitutively expressed.
Fig. 4. SPL4 and SPL5 upregulate meristem identity genes.
qRT-PCR analysis of meristem identity gene expression in 35S:SPL4Δ and 35S:SPL5Δ compared to wild-type plants (A, B) and in 35S:miR156a compared to wild-type plants (C). Plants were grown in SD for 10 days (A) and in LD for 9 days (B) or 10 days (C). Shown is the mean +/- SEM relative to the wildtype. Black solid horizontal line: two fold increase, black dotted line: two fold decrease.
It is difficult to test whether these SPL proteins are necessary for LFY, FUL, and AP1 regulation since spl3 mutants have no morphological defect and no knockout alleles are available for SPL4 and SPL5 (Wang et al., 2008; Wu and Poethig, 2006). We therefore instead used transgenic plants overexpressing miR156, a microRNA that targets these and additional SPL genes. The Arabidopsis genome encodes for six expressed miR156 genes, which are conserved in different plant species (Xie et al., 2005). Constitutive overexpression of each of these microRNAs causes a delay in the reproductive transition (Schwab et al., 2005; Schwarz et al., 2008; Wu and Poethig, 2006), a phenotype opposite to that observed in plants with elevated levels of SPL3, SPL4, or SPL5 (Table1; Wu and Poethig, 2006). Furthermore, miR156b overexpression reduces the expression level of all SPL genes with a miR156 recognition motif (Schwab et al., 2005). Indeed, we detected a greater than two-fold reduction of SPL3, SPL4, and SPL5 (Fig. S3C) in a transgenic line overexpressing miR156a (Wu and Poethig, 2006).
We assayed expression of the MI genes in ten-day-old 35S:miR156a compared to wild-type seedlings grown in inductive photoperiod. Compared to the wildtype, LFY, FUL, and AP1 expression was markedly reduced in 35S:miR156a seedlings (Fig. 4C). This reduction was specific, because expression of other genes regulating developmental transitions such as FT and AGL24 was not reduced in 35S:miR156a (Fig. S4B). These results strongly suggest that miR156-regulated SBP transcription factors, including SPL3, SPL4, and SPL5, are required for upregulation of LFY, FUL, and AP1 in Arabidopsis.
We next wanted to test whether, consistent with the observed molecular phenotype, 35S:miR156a plants exhibit a delayed MI transition. Unfortunately, we were unable to score the MI phenotype of these plants. As previously described, 35S:miR156a plants were late flowering in long day conditions (Wu and Poethig, 2006). In addition, 35S:miR156a plants exhibit severely reduced apical dominance (Fig. S5; Chuck et al., 2007; Schwarz et al., 2008; Xie et al., 2005). This phenotype, while potentially highly interesting as discussed below, disrupts the normal progression of developmental transitions. Hence it was impossible to assess the timing of the MI transition in 35S:miR156a plants.
Interactions between FT and SPL3
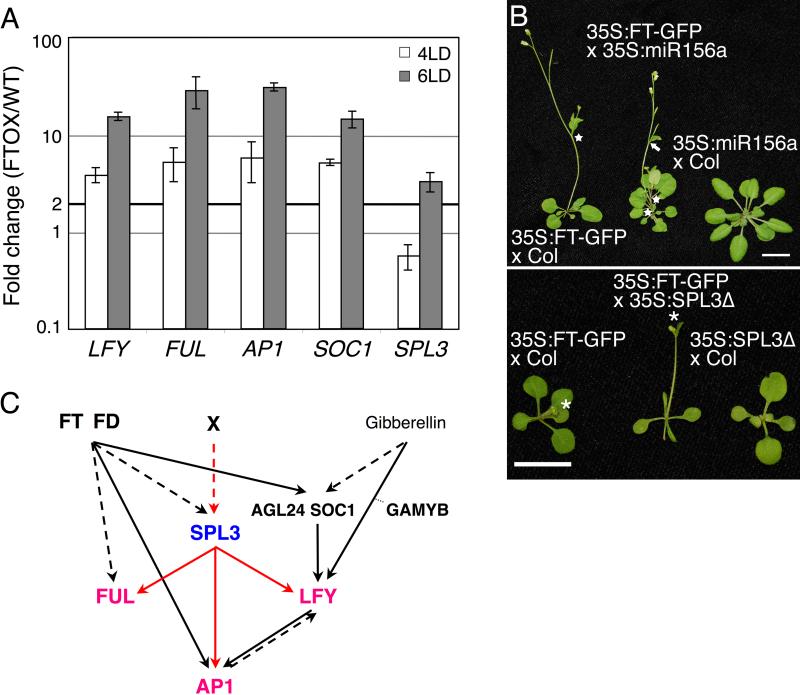
We have identified SPL3 as a new direct upstream regulator of LFY, FUL, and AP1. Interestingly, the flowering time regulator FT also upregulates these three MI genes. Together with the bZIP transcription factor FD, FT directly activates AP1 (Abe et al., 2005; Wigge et al., 2005) and FT/FD either directly or indirectly activate FUL (Teper-Bamnolker and Samach, 2005). FT is thought to indirectly activate LFY via upregulation of SOC1 (Searle et al., 2006). This raises the possibility that FT may upregulate FUL or LFY via SPL3 induction. To test whether FT is sufficient to induce SPL3 expression, we generated transgenic plants overexpressing FT (35S:FT-GFP), which flowered precociously and had a more rapid MI transition (fewer cauline leaves and secondary inflorescences formed than in the wildtype) consistent with previous reports for FT overexpression (Table 2; Kobayashi and Weigel, 2007). No elevated SPL3 expression was observed in four-day-old 35S:FT-GFP plants grown in inductive photoperiod. However, SPL3 levels were increased more than two-fold at day six in this line (Fig. 5A). Since none of the six expressed miR156 precursors showed a reduced expression in six-day-old 35S:FT-GFP compared to wild-type plants (data not shown), the observed increase in SPL3 expression is not likely to be mediated by miR156. Together with previous findings (Schmid et al., 2003), the data suggest that FT acts at least in part upstream of SPL3. Interestingly, LFY, FUL, AP1, as well as SOC1 levels were already elevated by day four in 35S:FT-GFP (Fig. 5A). Since upregulation of FUL and LFY precedes that of SPL3, FT is not likely to activate FUL and LFY via SPL3 induction.
Fig. 5. Regulatory interactions between flowering time and meristem identity regulators.
(A) qRT-PCR analysis of the known FT targets LFY, FUL, AP1 and SOC1 as well as SPL3 in 35S:FT-GFP plants grown in LD. Plants were harvested at day 4 and day 6. Shown is the expression mean +/- SEM relative to the wild type. Black solid horizontal line: two fold increase. (B) Phenotypes of F1 progeny of 35S:FT-GFP x 35S:miR156a (top) and 35S:SPL3 × 35S:FT-GFP (bottom). Star: secondary inflorescences, arrow: flower subtended by cauline leaf, asterisk: primary inflorescence. Bar; 1cm. (C) Role of SPL3 in meristem identity transition. Solid arrows denote direct interactions, dashed arrows denote direct or indirect interactions. SPL3 directly regulates LFY, FUL and AP1 transcription (red arrows) and acts in pathway that is partly in parallel with the FT pathway (red dashed arrow).
To further test the regulatory interaction between FT and SPL3, we examined the timing of developmental transitions in 35S:FT-GFP plants that had strongly reduced SPL levels. F1 progeny of a cross between 35S:FT-GFP and 35S:miR156a still exhibited a precocious reproductive and MI transition compared to the wildtype (Table 2). Overall the phenotype of the F1 progeny was additive with plants forming slightly more rosette leaves, cauline leaves, and secondary inflorescences than the F1 progeny of the control cross (35S:FT-GFP x wildtype; Table 2). As expected, the F1 progeny of 35S:miR156a crossed to wildtype flowered much later than wildtype like the 35S:miR156a parental line (Table 2 and data not shown). The phenotype of 35S:FT-GFP 35S:miR156a plants is shown in Fig. 5B (top). In a parallel approach, we examined developmental transitions in the F1 progeny of the cross of 35S:SPL3 to 35S:FT-GFP. These plants displayed a more precocious reproductive and MI transition than the parental lines (Fig. 5B, bottom). Indeed, 35S:FT-GFP 35S:SPL3Δ plants exhibited a strong significant decrease (P<0.0001) in the number of rosette leaves and a subtle but statistically significant (P<0.001) decrease in the number of secondary inflorescences formed compared to the control cross (35S:FT-GFP x wildtype; Table 2). Because 35S:miR156a is not epistatic to 35S:FT-GFP with respect to the reproductive and MI transition, while 35S:SPL3 enhances 35S:FT-GFP, our genetic data suggest that SPL3 acts at least in part in a pathway parallel to the FT pathway to regulate developmental timing in Arabidopsis.
DISCUSSION
Proper timing of the onset of reproduction is of central importance for fitness across species (Roux et al., 2006). Recently, the SBP-box family of transcription factors has been implicated in regulating this process (Cardon et al., 1997; Schwarz et al., 2008; Wu and Poethig, 2006). Here we show that the SBP-box transcription factor SPL3 directly activates the three key MI genes LFY, FUL and AP1 during the MI transition (Fig. 5C). SPL3 and related proteins form an independent regulatory pathway in the network that controls the onset of reproduction in Arabidopsis. These findings provide a major advance in our understanding of the regulation of this important life history trait. Because of the molecular and functional conservation of the MI regulators (Benlloch et al., 2007; Litt and Irish, 2003; Maizel et al., 2005) and the SPL proteins (Guo et al., 2008; Xie et al., 2006), the identified regulatory interactions may also play a role in the timing of flower formation in other plant species.
SPL3, a new direct upstream activator of LFY, FUL, and AP1
The timing of the MI transition is exquisitely sensitive to LFY levels (Blazquez et al., 1997) and precise spatial and temporal LFY upregulation is likely also important for additional life history traits such as species-specific inflorescence architecture (Prusinkiewicz et al., 2007) and irreversibility of the switch to flower formation (Parcy et al., 2002). Thus far very few direct upstream activators of LFY are known (Gocal et al., 2001; Lee et al., 2008; Liu et al., 2008) and loss- or gain-of-function mutations in these cause only modest MI phenotypes, suggesting that additional types of transcriptional regulators are required for LFY upregulation. Here we identify the SBP-box containing transcription factors as additional LFY activators.
The LFY promoter, which consists of the entire 2.3 kb intergenic region upstream of LFY (Blazquez et al., 1997), does not contain any consensus SPL binding motifs (this study; Cardon et al., 1997). In agreement with this, SPL3 overexpressing plants do not upregulate expression of a reporter gene whose expression is driven by the LFY promoter (Figure S6). Hence regulatory elements outside of the LFY promoter are required for upregulation by SPL3. Indeed, we show that SPL3 strongly binds in vivo to a region in the first exon/intron of LFY that contains three consensus SPL binding sites. LFY introns are known to be critical for proper expression of LFY in monocots (Bomblies and Doebley, 2005; Prasad et al., 2003; Rao et al., 2008) and recent genome-wide transcription factor binding studies support the idea that important cis elements may also reside in exons in Arabidopsis (Winter and Wagner, unpublished; Oh et al., 2009).
Our findings combined with previous studies suggest that LFY accumulation is regulated by a surprisingly complex array of partly redundantly acting transcription factor families and regulatory regions (Blazquez and Weigel, 2000; Lee et al., 2008; Liu et al., 2008). This complexity may form the basis for species-specific differences in induction of flower formation and of inflorescence architecture.
The MADS-box transcription factor FUL plays a role in both the reproductive transition and the MI transition together with other MADS box transcription factors (Ferrandiz et al., 2000; Teper-Bamnolker and Samach, 2005). More recently, FUL and SOC1 were shown to control the annual growth habit of Arabidopsis (Melzer et al., 2008). Hence precise regulation of FUL accumulation is of critical importance for the life strategy of flowering plants. Thus far no direct upstream activator of FUL is known, although there is evidence that FUL acts downstream of the photoperiod pathway regulators FT and FD (Abe et al., 2005; Teper-Bamnolker and Samach, 2005; Wigge et al., 2005). Here we identify SPL3 as a direct upstream activator of FUL. In addition, we show that FUL is not only required for the precocious MI transition, but also for the precocious vegetative phase change and reproductive transition observed in 35S:SPL3Δ plants. Moreover, we observed very strong activation of FUL by SPL3 and occupancy of SPL3 at FUL regulatory regions was highly robust. Taken together, these findings suggest that FUL is an important target of SPL transcription factors in control of developmental timing at multiple stages in the plant lifecycle.
Interestingly, in 35S:miR156a plants, axillary inflorescences bolt prior to the outgrowth of the primary inflorescence and plants have a multiple rosette phenotype (Fig. S5 and data not shown; Schwab et al., 2005; Xie et al., 2006). This phenotype is reminiscent of the perennial growth habit observed in plants with simultaneous downregulation of FUL and SOC1 (Melzer et al., 2008). We show that 35S:miR156a plants have reduced levels of FUL and SOC1 (Fig. 4C and Fig. S4A). One possibility suggested by our results that warrants further investigation is that SPL transcription factors may play a role in the regulation of annual versus perennial growth habit in different species of flowering plants via modulation of FUL and SOC1 expression.
We show that AP1 is a direct in vivo target of SPL3, with strong occupancy of SPL3 at multiple regions in the AP1 promoter. In contrast to the upregulation of LFY and FUL by SPL3, AP1 upregulation is only observed late in non-inductive photoperiod, suggesting that SPL3 is not sufficient for AP1 activation. A delay of AP1 induction compared to FUL and LFY upregulation is also observed in wild-type plants in many different growth conditions (this study; Hempel et al., 1997; Schmid et al., 2003) and may be important for proper timing of the commitment to floral fate. FT and FD together directly upregulate AP1 (Abe et al., 2005; Wigge et al., 2005) and we detect a subtle increase in FD levels in 35S:SPL3Δ that coincides with AP1 upregulation. While we cannot rule out that other factors contribute to the delay in AP1 accumulation, our results suggest that FT/FD may be required for AP1 induction by SPL3. This conclusion is supported by the finding that AP1 is more strongly upregulated in 35S:SPL3Δ plants grown in inductive photoperiod when FT is present.
The position of SPL3 in the regulatory network controlling the onset of reproduction
The MI transition is the final step in reproductive development and leads to formation of the first flowers. Recent investigations determined that flowering time regulators directly activate MI genes (Fig. 5C; Abe et al., 2005; Lee et al., 2008; Liu et al., 2008; Wigge et al., 2005). Since SPL3 and related proteins are important activators of the three main MI genes, this raises the question how these proteins fit in the known regulatory network of flowering time and MI genes. SPL3 and parologous genes are strongly upregulated in response to inductive photoperiod in an FT-dependent fashion (Schmid et al., 2003). Moreover, we show that plants overexpressing FT have elevated SPL3 levels. On the other hand, gain-of SPL3 and FT function are additive and reduction of SPL3 function is not epistatic to gain of FT function, suggesting that SPL3 and FT act in parallel pathways, which converge on upregulation of MI genes (Fig. 5C). Similarly, FT and LFY act in parallel pathways that converge on a common target gene (Abe et al., 2005; Wigge et al., 2005). These regulatory interactions likely allow exquisite fine-tuning of important developmental decisions through integration of multiple environmental and endogenous inputs.
Our findings also point to presence of a second SPL3 activation pathway (X in Fig. 5C). Since we did not detect a decrease in miR156 gene expression in 35S:FT plants with elevated SPL3 levels it is tempting to speculate that this second pathway may involve post-transcriptional regulation of SPL3 levels by this family of microRNAs. Future investigation will be needed to test this hypothesis.
EXPERIMENTAL PROCEDURES
Plant materials and growth conditions
Accession numbers of all genes used in this study are listed in Table S3. Columbia (Col) was used as wildtype. lfy-1, ful-2 and ap1-10 mutants have been described (Ferrandiz et al., 2000; Schultz and Haughn, 1991; Schultz and Haughn, 1993). 35S:SPL3Δ, 35S:SPL4Δ and 35S:SPL5Δ as well as 35S:miR156a have been described (Wu and Poethig, 2006). For phenotype analysis, plants were grown on soil at 22°C in long-day (16 h light/8 h dark) conditions using white fluorescent lights (~ 110 μmol/m2 s) or in short-day (10 h light/14 h dark) conditions with a 3:1 mixture of white fluorescent and GrowLux lights (~ 120 μmol/ m2 s). For expression and chromatin immunoprecipitation analysis, plants were grown on half-strength Murashige and Skoog (MS) medium under long-day (continuous light) conditions with white fluorescent lights (~ 90 μmol/m2 s) or short-day (10 h light/14 h dark) as described above. Seeds were stratified at 4°C for 2 to 4 days and then transferred to 22°C at day 0.
Plasmid construction and transgenic plants
To construct the 35S:SPL3Δ-GFP and 35S:FT-GFP lines, the coding region of SPL3 and FT was amplified by PCR with primer sets containing the attB sequence for Gateway-based cloning, and recombined into pDONR221 using BP reaction (Invitrogen, USA). Primers used are listed in Table S4. After sequencing, each coding sequence was recombined into pGWB5 (Nakagawa et al., 2007) using LR reactions (Invitrogen, USA). The resulting binary vectors were introduced into Agrobacterium strain GV3101::pMP90 and transformed into Col plants. Transgenic plants flowering earlier than wild type were further analyzed.
To construct pSPL3:GFP-SPL3, a 2.9 kb genomic fragment upstream of the ATG and a 0.9 kb fragment downstream of the ATG of SPL3 were cloned before and after eGFP respectively in the binary vector pCAMBIA3300 (CAMBIA, Australia) and transformed into Col plants. Primers used are listed in Table S4.
qRT-PCR and in situ hybridization
RNA extraction and reverse transcription was essentially as described (William et al., 2004) except treatment with RNase-free DNase (Qiagen, USA) was included and the Superscript III kit (Invitrogen, USA) was used. Real-time PCR reactions were performed using Power SYBR Green PCR master mix (Applied Biosystems). The relative amount of a given mRNA was determined based on the threshold cycle number required for amplification compared to the standard curve, and then normalized by the expression values of the eukaryotic translation initiation factor 4A-1 (EIF4A) in each sample. Fold change was calculated by dividing the normalized value of the experimental genotype by that of the wildtype. The mean and standard error were determined using one to two biological replicates with three technical replicates each. Primers used are listed in Table S5.
In situ hybridization was performed as in (Long and Barton, 1998) using probes previously described (Hempel et al., 1997).
Chromatin Immunoprecipitation
300 mg of 10-day-old seedlings grown in short day conditions (35S:SPL3Δ-GFP) or 7-day-old seedlings grown in long day conditions (pSPL3:GFP-SPL3) were employed for ChIP following the procedure previously described (Kwon et al., 2005; William et al., 2004). Anti GFP antibodies A6455 (Invitrogen) and ab290 (Abcam) were used. Real-time PCR was performed using Power SYBR Green PCR master mix (Applied Biosystems). To estimate SPL3 occupancy on genomic DNA we computed the ratio of ChIP over input DNA (% Input) by comparing the reaction threshold cycle for each the ChIP sample to a dilution series of the corresponding input sample. Primers used are summarized in Table S6.
Fluorescence microscopy
GFP fluorescence was visualized using a fluorescent microscope (Olympus, MVX10) or a confocal microscope (Leica, LCS SL).
Supplementary Material
ACKNOWLEGDEMENTS
We thank Xuemei Chen, Kim Gallagher, Sarah Liljegren, and John Wagner, as well as Wagner lab members for comments on this manuscript and Koji Koizumi for help with fluorescence microscopy. This work was supported by NSF IBN grant 0516622 to DW, NIH R01 GM051893 to RSP and a JSPS fellowship to AY.
REFERENCES
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science. 2005;309:1052–1056. doi: 10.1126/science.1115983. [DOI] [PubMed] [Google Scholar]
- Achard P, Herr A, Baulcombe DC, Harberd NP. Modulation of floral development by a gibberellin-regulated microRNA. Development. 2004;131:3357–3365. doi: 10.1242/dev.01206. [DOI] [PubMed] [Google Scholar]
- Araki T. Transition from vegetative to reproductive phase. Curr Opin Plant Biol. 2001;4:63–68. doi: 10.1016/s1369-5266(00)00137-0. [DOI] [PubMed] [Google Scholar]
- Benlloch R, Berbel A, Serrano-Mislata A, Madueno F. Floral initiation and inflorescence architecture: a comparative view. Ann Bot (Lond) 2007;100:659–676. doi: 10.1093/aob/mcm146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazquez MA, Ferrandiz C, Madueno F, Parcy F. How floral meristems are built. Plant Mol Biol. 2006;60:855–870. doi: 10.1007/s11103-006-0013-z. [DOI] [PubMed] [Google Scholar]
- Blazquez MA, Soowal LN, Lee I, Weigel D. LEAFY expression and flower initiation in Arabidopsis. Development. 1997;124:3835–3844. doi: 10.1242/dev.124.19.3835. [DOI] [PubMed] [Google Scholar]
- Blazquez MA, Weigel D. Integration of floral inductive signals in Arabidopsis. Nature. 2000;404:889–892. doi: 10.1038/35009125. [DOI] [PubMed] [Google Scholar]
- Bomblies K, Doebley JF. Molecular evolution of FLORICAULA/LEAFY orthologs in the Andropogoneae (Poaceae). Mol Biol Evol. 2005;22:1082–1094. doi: 10.1093/molbev/msi095. [DOI] [PubMed] [Google Scholar]
- Bowman JL, Alvarez J, Weigel D, Meyerowitz EM, Smyth DR. Control Of Flower Development In Arabidopsis Thaliana By Apetala1 and Interacting Genes. Development. 1993;119:721–743. [Google Scholar]
- Busch MA, Bomblies K, Weigel D. Activation of a floral homeotic gene in Arabidopsis. Science. 1999;285:585–587. doi: 10.1126/science.285.5427.585. [DOI] [PubMed] [Google Scholar]
- Cardon G, Hohmann S, Klein J, Nettesheim K, Saedler H, Huijser P. Molecular characterisation of the Arabidopsis SBP-box genes. Gene. 1999;237:91–104. doi: 10.1016/s0378-1119(99)00308-x. [DOI] [PubMed] [Google Scholar]
- Cardon GH, Hohmann S, Nettesheim K, Saedler H, Huijser P. Functional analysis of the Arabidopsis thaliana SBP-box gene SPL3: a novel gene involved in the floral transition. Plant J. 1997;12:367–377. doi: 10.1046/j.1365-313x.1997.12020367.x. [DOI] [PubMed] [Google Scholar]
- Chuck G, Cigan AM, Saeteurn K, Hake S. The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nat Genet. 2007;39:544–549. doi: 10.1038/ng2001. [DOI] [PubMed] [Google Scholar]
- Ferrandiz C, Gu Q, Martienssen R, Yanofsky MF. Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development. 2000;127:725–734. doi: 10.1242/dev.127.4.725. [DOI] [PubMed] [Google Scholar]
- Galuschka C, Schindler M, Bulow L, Hehl R. AthaMap web tools for the analysis and identification of co-regulated genes. Nucleic Acids Res. 2007;35:D857–862. doi: 10.1093/nar/gkl1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandikota M, Birkenbihl RP, Hohmann S, Cardon GH, Saedler H, Huijser P. The miRNA156/157 recognition element in the 3' UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J. 2007;49:683–693. doi: 10.1111/j.1365-313X.2006.02983.x. [DOI] [PubMed] [Google Scholar]
- Gocal GF, Sheldon CC, Gubler F, Moritz T, Bagnall DJ, MacMillan CP, Li SF, Parish RW, Dennis ES, Weigel D, et al. GAMYB-like Genes, Flowering, and Gibberellin Signaling in Arabidopsis. Plant Physiol. 2001;127:1682–1693. [PMC free article] [PubMed] [Google Scholar]
- Guo AY, Zhu QH, Gu X, Ge S, Yang J, Luo J. Genome-wide identification and evolutionary analysis of the plant specific SBP-box transcription factor family. Gene. 2008;418:1–8. doi: 10.1016/j.gene.2008.03.016. [DOI] [PubMed] [Google Scholar]
- Hempel FD, Weigel D, Mandel MA, Ditta G, Zambryski PC, Feldman LJ, Yanofsky MF. Floral determination and expression of floral regulatory genes in Arabidopsis. Development. 1997;124:3845–3853. doi: 10.1242/dev.124.19.3845. [DOI] [PubMed] [Google Scholar]
- Klein J, Saedler H, Huijser P. A new family of DNA binding proteins includes putative transcriptional regulators of the Antirrhinum majus floral meristem identity gene SQUAMOSA. Mol Gen Genet. 1996;250:7–16. doi: 10.1007/BF02191820. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Weigel D. Move on up, it's time for change--mobile signals controlling photoperiod-dependent flowering. Genes Dev. 2007;21:2371–2384. doi: 10.1101/gad.1589007. [DOI] [PubMed] [Google Scholar]
- Kwon CS, Chen C, Wagner D. WUSCHEL is a primary target for transcriptional regulation by SPLAYED in dynamic control of stem cell fate in Arabidopsis. Genes Dev. 2005;19:992–1003. doi: 10.1101/gad.1276305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lannenpaa M, Janonen I, Holtta-Vuori M, Gardemeister M, Porali I, Sopanen T. A new SBP-box gene BpSPL1 in silver birch (Betula pendula). Physiologia plantarum. 2004;120:491–500. doi: 10.1111/j.0031-9317.2004.00254.x. [DOI] [PubMed] [Google Scholar]
- Lee J, Oh M, Park H, Lee I. SOC1 translocated to the nucleus by interaction with AGL24 directly regulates LEAFY. Plant J. 2008;55:832–43. doi: 10.1111/j.1365-313X.2008.03552.x. [DOI] [PubMed] [Google Scholar]
- Litt A, Irish VF. Duplication and diversification in the APETALA1/FRUITFULL floral homeotic gene lineage: implications for the evolution of floral development. Genetics. 2003;165:821–833. doi: 10.1093/genetics/165.2.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Chen H, Er HL, Soo HM, Kumar PP, Han JH, Liou YC, Yu H. Direct interaction of AGL24 and SOC1 integrates flowering signals in Arabidopsis. Development. 2008;135:1481–1491. doi: 10.1242/dev.020255. [DOI] [PubMed] [Google Scholar]
- Long JA, Barton MK. The development of apical embryonic pattern in Arabidopsis. Development. 1998;125:3027–3035. doi: 10.1242/dev.125.16.3027. [DOI] [PubMed] [Google Scholar]
- Maizel A, Busch MA, Tanahashi T, Perkovic J, Kato M, Hasebe M, Weigel D. The floral regulator LEAFY evolves by substitutions in the DNA binding domain. Science. 2005;308:260–263. doi: 10.1126/science.1108229. [DOI] [PubMed] [Google Scholar]
- Mandel MA, Yanofsky MF. A gene triggering flower formation in Arabidopsis. Nature. 1995;377:522–524. doi: 10.1038/377522a0. [DOI] [PubMed] [Google Scholar]
- Melzer S, Lens F, Gennen J, Vanneste S, Rohde A, Beeckman T. Flowering-time genes modulate meristem determinacy and growth form in Arabidopsis thaliana. Nat Genet. 2008;40:1489–1492. doi: 10.1038/ng.253. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Suzuki T, Murata S, Nakamura S, Hino T, Maeo K, Tabata R, Kawai T, Tanaka K, Niwa Y, et al. Improved Gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci Biotechnol Biochem. 2007;71:2095–2100. doi: 10.1271/bbb.70216. [DOI] [PubMed] [Google Scholar]
- Oh E, Kang H, Yamaguchi S, Park J, Lee D, Kamiya Y, Choi G. Genome-Wide Analysis of Genes Targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE5 during Seed Germination in Arabidopsis. The Plant cell. 2009;21:403–419. doi: 10.1105/tpc.108.064691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parcy F, Bomblies K, Weigel D. Interaction of LEAFY, AGAMOUS and TERMINAL FLOWER1 in maintaining floral meristem identity in Arabidopsis. Development. 2002;129:2519–2527. doi: 10.1242/dev.129.10.2519. [DOI] [PubMed] [Google Scholar]
- Poethig RS. Phase change and the regulation of developmental timing in plants. Science. 2003;301:334–336. doi: 10.1126/science.1085328. [DOI] [PubMed] [Google Scholar]
- Prasad K, Kushalappa K, Vijayraghavan U. Mechanism underlying regulated expression of RFL, a conserved transcription factor, in the developing rice inflorescence. Mech Dev. 2003;120:491–502. doi: 10.1016/s0925-4773(02)00457-4. [DOI] [PubMed] [Google Scholar]
- Prusinkiewicz P, Erasmus Y, Lane B, Harder LD, Coen E. Evolution and development of inflorescence architectures. Science. 2007;316:1452–1456. doi: 10.1126/science.1140429. [DOI] [PubMed] [Google Scholar]
- Rao NN, Prasad K, Kumar PR, Vijayraghavan U. Distinct regulatory role for RFL, the rice LFY homolog, in determining flowering time and plant architecture. Proc Natl Acad Sci U S A. 2008;105:3646–3651. doi: 10.1073/pnas.0709059105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux F, Touzet P, Cuguen J, Le Corre V. How to be early flowering: an evolutionary perspective. Trends Plant Sci. 2006;11:375–381. doi: 10.1016/j.tplants.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Schmid M, Uhlenhaut NH, Godard F, Demar M, Bressan R, Weigel D, Lohmann JU. Dissection of floral induction pathways using global expression analysis. Development. 2003;130:6001–6012. doi: 10.1242/dev.00842. [DOI] [PubMed] [Google Scholar]
- Schultz EA, Haughn GW. LEAFY, a Homeotic Gene That Regulates Inflorescence Development in Arabidopsis. The Plant Cell. 1991;3:771–781. doi: 10.1105/tpc.3.8.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz EA, Haughn GW. Genetic analysis of the floral induction process (FLIP) in Arabidopsis. Development. 1993;119:745–765. [Google Scholar]
- Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D. Specific effects of microRNAs on the plant transcriptome. Dev Cell. 2005;8:517–527. doi: 10.1016/j.devcel.2005.01.018. [DOI] [PubMed] [Google Scholar]
- Schwarz S, Grande AV, Bujdoso N, Saedler H, Huijser P. The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol Biol. 2008;67:183–195. doi: 10.1007/s11103-008-9310-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle I, He Y, Turck F, Vincent C, Fornara F, Krober S, Amasino RA, Coupland G. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 2006;20:898–912. doi: 10.1101/gad.373506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeves TA, Sussex IM. Patterns in Plant Development. Cambridge University Press; Cambridge, UK: 1989. [Google Scholar]
- Teper-Bamnolker P, Samach A. The flowering integrator FT regulates SEPALLATA3 and FRUITFULL accumulation in Arabidopsis leaves. The Plant cell. 2005;17:2661–2675. doi: 10.1105/tpc.105.035766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck F, Fornara F, Coupland G. Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu Rev Plant Biol. 2008;59:573–594. doi: 10.1146/annurev.arplant.59.032607.092755. [DOI] [PubMed] [Google Scholar]
- Wagner D, Sablowski RW, Meyerowitz EM. Transcriptional activation of APETALA1 by LEAFY. Science. 1999;285:582–584. doi: 10.1126/science.285.5427.582. [DOI] [PubMed] [Google Scholar]
- Wang JW, Schwab R, Czech B, Mica E, Weigel D. Dual Effects of miR156-Targeted SPL Genes and CYP78A5/KLUH on Plastochron Length and Organ Size in Arabidopsis thaliana. The Plant Cell. 2008;20:1231–1243. doi: 10.1105/tpc.108.058180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM. LEAFY controls floral meristem identity in Arabidopsis. Cell. 1992;69:843–859. doi: 10.1016/0092-8674(92)90295-n. [DOI] [PubMed] [Google Scholar]
- Weigel D, Nilsson O. A developmental switch sufficient for flower initiation in diverse plants. Nature. 1995;377:495–500. doi: 10.1038/377495a0. [DOI] [PubMed] [Google Scholar]
- Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D. Integration of spatial and temporal information during floral induction in Arabidopsis. Science. 2005;309:1056–1059. doi: 10.1126/science.1114358. [DOI] [PubMed] [Google Scholar]
- William DA, Su Y, Smith MR, Lu M, Baldwin DA, Wagner D. Genomic identification of direct target genes of LEAFY. Proc Natl Acad Sci U S A. 2004;101:1775–1780. doi: 10.1073/pnas.0307842100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Poethig RS. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development. 2006;133:3539–3547. doi: 10.1242/dev.02521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K, Wu C, Xiong L. Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant Physiol. 2006;142:280–293. doi: 10.1104/pp.106.084475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Allen E, Fahlgren N, Calamar A, Givan SA, Carrington JC. Expression of Arabidopsis MIRNA genes. Plant Physiol. 2005;138:2145–2154. doi: 10.1104/pp.105.062943. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.