Abstract
Various mutations in the PARK2 gene which encodes the protein, parkin, are causal of a disease entity termed autosomal recessive juvenile parkinsonism. Parkin can function as an E3 ubiquitin protein ligase, mediating the ubiquitination of specific targeted proteins and resulting in proteasomal degradation. Parkin is thought to lead to parkinsonism as a consequence of a loss in its function. In this study, immunoblot analyses of brain extracts from Balb/c, C57Bl/6, C3H, and 129S mouse strains demonstrated significant variations in immunoreactivity with anti-parkin monoclonal antibodies (PRK8, PRK28, and PRK109). This resulted partly from differences in the steady-state levels of parkin protein across mouse strains. There was also a complete loss of immunoreactivity for PRK8 and PRK28 antibodies in C3H mice which was due to a homologous nucleotide substitution resulting in an E398Q amino acid substitution. In cultured cells, parkin harboring this mutation had a greater tendency to aggregate, exhibited reduced interaction with the E2 ubiquitin conjugating enzymes, UbcH7 and UbcH8, and demonstrated loss of function in promoting the proteasomal degradation of a specific putative substrate, synphilin-1. In situ, C3H mice displayed an age-dependent increased levels of brain cortical synphilin-1 compared to C57Bl/6, suggesting that E398Q parkin in these mice is functionally impaired and that C3H mice may be a suitable model of parkin loss-of-function similar to patients with missense mutations.
Keywords: Parkin, C3H mouse, E3 ligase, Parkinson disease, mutation, recessive
INTRODUCTION
Parkinson disease (PD) is an insidious neurodegenerative disorder that is characterized by the selective demise of specific neuronal populations, including dopaminergic neurons of the substantia nigra, and is associated with an impairment of motor functions. Most cases of PD are sporadic, but several disease-causing genes have been identified (Bonifati 2007). In 1998, it was shown that mutations in the parkin gene (also known as PARK2) were responsible for a disease entity termed autosomal recessive juvenile parkinsonism (AR-JP) (Kitada et al. 1998). To date, a variety of nonsense, missense, exonic deletion, splice site, duplication, and triplication mutations in PARK2 have been identified in individuals with both early and late-onset PD (von Coelln et al. 2004a;Tan and Skipper 2007;Mata et al. 2004;Hedrich et al. 2004). Since several community based reports suggest that parkin mutations may contribute to up to 15% of the inherited cases of PD and are the major cause of early-onset PD (Lucking et al. 2000;Periquet et al. 2003;Guo et al. 2008;Wu et al. 2005;Bonifati 2007), much work has been done to understand the role of parkin in the pathogenesis of PD.
The parkin gene is large, spanning over 1.4 Mega bases with 12 exons and large intronic regions (Kitada et al. 1998;Kitada et al. 1999;Asakawa et al. 2001). The gene encodes a ∼52kDa protein that is 465 amino acids in length (Kitada et al. 1998). The protein has an amino terminal ubiquitin-like (Ubl) domain as well as two Really-Interesting-New-Gene (RING) finger domains which are separated by an in-between-RING (IBR) finger domain at the carboxyl terminus (Kitada et al. 1998;Morett and Bork 1999). These structural features are common to E3 ubiquitin-protein ligases (E3 ligases)(Tanaka et al. 2004) and parkin can function in this capacity (Ciechanover 2001;Hampe et al. 2006;Rankin et al. 2001;Sriram et al. 2005;Imai et al. 2000;Shimura et al. 2000). E3 ligases are a class of proteins that work in concert with ubiquitin-conjugating enzymes (E2s) to mediate the transfer of ubiquitin to specific protein substrates. This ubiquitin transfer often targets substrates for proteolytic degradation by the 26S proteasome (Ciechanover 2001;Joazeiro and Weissman 2000). It is known that parkin can interact with the E2 ubiquitin-conjugating enzymes, UbcH7 and UbcH8 (Shimura et al. 2000;Zhang et al. 2000;Imai et al. 2000). Additionally, many groups have shown that under certain experimental paradigms, parkin can facilitate the ubiquitination of a variety of substrates and can also aid in the subsequent degradation of a subset of these substrates (Zhang et al. 2000;Chung et al. 2001;Moore et al. 2008;Corti et al. 2003;Ko et al. 2006;Huynh et al. 2003;Um et al. 2006;Shimura et al. 2001;Imai et al. 2001;Staropoli et al. 2003;Choi et al. 2003;Ren et al. 2003). Thus, it is widely accepted that parkin functions as an E3 ligase; however, it is unclear how this function may be related to PD (Fitzgerald and Plun-Favreau 2008;Li and Guo 2009;Dodson and Guo 2007).
Several of the pathogenic mutations in parkin have been shown to impair its E3 ligase activity. Pathogenic mutations, such as the T240R mutation, have been shown to reduce the interactions between parkin and E2 ubiquitin-conjugating enzymes (Imai et al. 2000;Zhang et al. 2000;Shimura et al. 2000;Gu et al. 2003). Additionally, this disrupted association of parkin with E2 enzymes can result in reduced ubiquitination and degradation of parkin substrates (Chung et al. 2001;Imai et al. 2000;Zhang et al. 2000;Shimura et al. 2000;Sriram et al. 2005). It is also known that parkin can ubiquitinate itself which then leads to its degradation by the proteasome (Zhang et al. 2000;Choi et al. 2000). Pathogenic mutants which do not demonstrate the ability to autoubiquitinate often show altered protein solubility (Sriram et al. 2005). This altered solubility may be related to decreased protein turnover that is specific to the proteasome pathway (Zhang et al. 2000).
It is hypothesized that parkin mutations may lead to parkinsonism through a loss in parkin function since parkin has been shown to play a protective role in a number of studies (Chung et al. 2004;Imai et al. 2000;Kao 2009;Ved et al. 2005). Parkin deficient mice have been generated by several labs in efforts to study the effects of parkin loss-of-function in vivo (Itier et al. 2003;Goldberg et al. 2003;Kitao et al. 2007;von Coelln et al. 2004b;Perez and Palmiter 2005;Sato et al. 2006). Several inconsistencies have been reported regarding the effects of abolishing parkin in mice. However, in general, parkin loss in mice is not associated with an increased morbidity or loss of dopaminergic neurons (Sato et al. 2006;Zhu et al. 2007;Itier et al. 2003;Perez and Palmiter 2005;Goldberg et al. 2003;von Coelln et al. 2004b). Nor has parkin loss in mice been shown to have an effect on many of the putative parkin substrates (Ko et al. 2005).
In this report, the western blot analyses of endogenous brain parkin from four different mouse strains, C57Bl/6, Balb/c, C3H, and 129S, are described. Dramatic variations in the protein levels of endogenous parkin were observed between mouse strains. Additionally, a novel endogenous homozygous missense mutation was discovered in C3H mice which resulted in the parkin amino acid substitution, E398Q. This mutation abolished the PRK8 and PRK28 parkin antibody epitopes in C3H mice while immunoreactivity with the PRK109 antibody was retained. Because of the use of C3H mice in studies that may involve parkin, the impact of the novel endogenous parkin mutation on protein function was examined. Characterization of this mutation indicates that C3H mouse parkin is functionally impaired, suggesting that the C3H mouse strain may be a suitable in vivo model for endogenous loss-of-function parkin studies.
EXPERIMENTAL PROCEDURES
Mouse strains
C57BL/6, C3H, BALB/c, and 129S mice at 10–12 weeks and 8 months of age were used for studies. All mice were purchased from Charles River Laboratories Inc (Wilmington, MA). Mice were sacrificed by CO2 euthanization as approved by the University of Pennsylvania Institutional Animal Care and Use Committee.
Antibodies
PRK8, PRK28, and PRK109 are mouse monoclonal antibodies raised against human parkin but that react with parkin from various species, including murine (Pawlyk et al. 2003). Mouse anti-GST monoclonal antibody clone A00865.01 was obtained from GenScript (Piscataway, NJ). Affinity purified mouse anti-actin (clone C4) monoclonal antibody reacts with all forms of vertebrate actin (Millipore, Billerica, MA). Two c-Myc antibodies were used in these studies: purified anti-c-Myc monoclonal antibody clone 9E10 (M4439, Sigma-Aldrich, Saint Louis, MO) and an affinity purified anti-c-Myc polyclonal antibody (Sigma-Aldrich, Saint Louis, MO). Two HA antibodies were used in these studies: anti-HA monoclonal antibody clone 12CA5 (Roche Diagnostics, Indianapolis, IN) and affinity purified polyclonal antibody, HA.11 (Covance, Emeryville, CA). UPN79 is an affinity purified polyclonal antibody raised against human full-length recombinant synphilin-1 (Murray et al. 2003).
Relative quantification of the mouse parkin mRNA transcript
Cerebral cortical tissues were harvested from 10 week old Balb/c, BL6, C3H, and 129S mice and divided in half. Half of the tissue was frozen on dry ice for subsequent protein analysis and total RNA was extracted from the remaining half using TRIzol reagent (Invitrogen Corporation, Carlsbad, CA) according to the manufacturer’s protocol. Three tissue samples were prepared for each mouse strain. The RNA products were reverse transcribed to cDNA using Taqman RT-PCR reagents (Applied Biosystems Inc, Foster City, CA) with oligo dT primers that were supplied in the kit. The RT reactions were conducted according to the manufacturer’s protocol. Real-time PCR analyses were performed on the cDNA products using 2X SYBR Green Master Mix reagent (Applied Biosystems) and Applied Biosystems 7000 Real-Time PCR System (Applied Biosystems). Reactions were performed in duplicate for each cDNA sample in a 96 well plate. Primers were designed to amplify a 98 base pair product of mouse parkin cDNA that corresponds to nucleotides 396–494. The sequences for the parkin primers were as follows. Forward: 5’-AGCAGCCAGAGGTCCAGTTA-3’ and Reverse: 5’-CACTGAACTCGGAGCTTTCC-3’. Additionally, a mouse β-actin loading control was amplified. Mouse β-actin primers were designed to amplify a 99 base pair product corresponding to cDNA nucleotides 698–797. The sequences of the β-actin primers were as follows. Forward: 5’-CTTCCTCCCTGGAGAAGAGC-3’ and Reverse: 5’-AAGGAAGGCTGGAAAAGAGC-3’. The thermo cycling conditions for the amplification were as follows: 1 cycle for 2 minutes at 50°C, 1 cycle for 10 minutes at 95°C, followed by 40 cycles of 15 seconds at 95°C and 1 minute at 60°C, followed by 1 cycle for 15 seconds at 95°C, 1 cycle for 20 seconds at 60°C, and 1 cycle for 15 seconds at 95°C. The CT values for each sample were obtained. Parkin values were normalized to β-actin by taking the ratio of parkin to β-actin for each sample. The normalized results were averaged for replicate samples and graphed. Error bars represent standard deviation where n=6. After the reaction, equal volumes of the PCR products, one for each sample type, were loaded onto 7–8% non-denaturing polyacrylamide/TBE gels and were resolved by electrophoresis. The gels were then stained with ethidium bromide at 1ug/mL for 20 minutes and the DNA bands were visualized with a UV light and photographed. Additionally, the reaction products were confirmed by DNA sequencing.
RT-PCR analysis and DNA sequencing of the mouse parkin cDNAs
Mouse total RNA was extracted and reverse transcribed from cortical tissue as described above. Polymerase chain reactions were conducted on the cDNA products using AccuPrime™ SuperMix II (Invitrogen) in order to amplify nucleotides 895–1395 of the mouse Parkin cDNA sequence. The primer sequences for the reaction were as follows. Forward: 5’-GAGCTCCATCACTTCAGGATCCTTGGA-3’ and Reverse: 5’-CTACACGTCAAACCAGTG ATC TCC CAT-3’. The PCR products were digested with ExoSAP-IT ® reagent (USB Corporation, Cleveland, OH) and sequenced by the DNA sequencing facility of the University of Pennsylvania.
Mouse protein analysis
Cerebral cortical tissues were harvested from mice as described above. The tissue was sonicated in 3 tissue volumes of either high salt (HS) buffer (50mM Tris [pH 7.5], 750 mM NaCl, 5mM EDTA, with a protease inhibitor cocktail at 1:1000 and PMSF at 1:500) or 2% SDS/8M urea. Tissue extracts were then quantified using the bicinchoninic acid (BCA) assay (Pierce Biotechnology, Rockford, IL) and bovine serum albumin as the standard. Equal amounts of protein extracts were resolved by SDS-PAGE. The samples were analyzed by western blot with PRK8, PRK28, PRK109 and UPN79 antibodies. Additionally, membranes were blotted with anti-actin antibody to assess equal loading. At least 4 independent tissue samples were analyzed for each mouse strain at each respective age.
Parkin antibodies epitope mapping
Using full length human parkin cDNA as a template, primers were designed to generate various truncated parkin constructs by PCR. See Table 1 for the oligonucleotide sequences used for PCRs. AccuPrime SuperMix II (Invitrogen) was used for the reactions. PCR products were cloned by topoisomerase reaction into the shuttling vector pCR8/GW/TOPO (Invitrogen) and confirmed by DNA sequencing. These cDNA fragments were introduced into the Gateway pDEST 15 vector by recombinase reaction using LR Clonase II enzyme (Invitrogen). BL21 cells were then transformed with positive pDEST15 clones. Transformed cultures were subsequently induced with 500uM IPTG for 2 hours at 37°C in a shaking incubator. After induction of protein expression, the cells were lysed by sonication in 3 pellet volumes of 2% SDS/8M urea. Protein extracts were then quantified using the BCA assay and analyzed by immunoblot with anti-GST, PRK8, PRK28, and PRK109 antibodies.
Table I.
Oligonucleotide sequences used to generate N-and C- terminal truncated human parkin mutants for PRK8, PRK28 and PRK109 epitope mapping studies.
| Nucleotides amplified in PCR |
Corresponding Amino Acid Sequence |
Forward Oligonucleotide sequence |
Reverse Oligonucleotide sequence |
|---|---|---|---|
| 1–1398 | 1–465 | 5’-ATG ATA GTG TTT GTC AGG TTC AAC TCC-3’ |
5’- CTA CAC GTC GAA CCA GTG GTC CCC CAT -3’ |
| 1–1260 | 1–420 | 5’-ATG ATA GTG TTT GTC AGG TTC AAC TCC-3’ |
5’-CTA GCG GGG ACA GGG CTT GGT GGT TTT CTT-3’ |
| 1–1224 | 1–408 | 5’-ATG ATA GTG TTT GTC AGG TTC AAC TCC-3’ |
5’-CTA TTT GGA GGC TGC TTC CCA ACG AGC CTG-3’ |
| 1–1197 | 1–399 | 5’-ATG ATA GTG TTT GTC AGG TTC AAC TCC-3’ |
5’-CTA CTC GGC GGC TCT TTC ATC GAC TCT GT-3’ |
| 661–1398 | 221–465 | 5’-GAA ACA CCA GTA GCT TTG CAC CTG AT- 3’ |
5’- CTA CAC GTC GAA CCA GTG GTC CCC CAT -3’ |
| 1141–1398 | 381–465 | 5’-TTT GAA GCC TCA GGA ACA ACT ACT CAG-3’ |
5’- CTA CAC GTC GAA CCA GTG GTC CCC CAT -3’ |
| 1195–1398 | 399–465 | 5’-GAG CAG GCT CGT TGG GAA GCA GCC-3’ |
5’- CTA CAC GTC GAA CCA GTG GTC CCC CAT -3’ |
Generation of E399Q human parkin construct for bacterial expression
For expression in bacteria, using the Gateway pDEST 15 human parkin 381–465 construct as a template (described above), the QuickChange Site Directed Mutagenesis Kit (Stratagene, La Jolla, CA) was used in order to generate the E399Q mutant form of human parkin protein. The oligonucleotide sequence for the mutagenesis reaction was as follows: Forward-5’- GAAAGAGCCGCCCAGCAGGCTCGTTGG-3’ and Reverse-5’-CCAACGAGCCTGCTG GGCGGCTCT TTC-3’. The sequence of the construct was confirmed by DNA sequencing. The plasmid was subsequently expressed in BL21 cells, harvested, and analyzed as described in the previous section.
Generation of human parkin constructs for expression in mammalian cells
The full-length untagged wild-type (“WT”) human parkin cDNA was cloned into the XhoI and Apa I restriction sites on the pcDNA3.1(+) mammalian expression vector (Invitrogen). This WT Parkin construct was used as a template to perform site directed mutagenesis in order to generate the full-length E399Q mutant. The same oligonucleotides were used as in the E399Q mutagenesis reaction as described in the previous section. Additionally, the WT parkin/pcDNA 3.1 construct was used as a template in a site directed mutagenesis reaction to generate the human parkin T240R pathogenic mutant. The oligonucleotide sequences for the T240R mutagenesis reaction were as follows: Forward-5’- ATCACTTGC ATTAGGTGCACAGACGTC-3’ and Reverse-5’- GACGTCTGTGCACCTAATGCAAGTGAT-3’. The sequence for both the E399Q and T240R parkin mutants were verified by DNA sequencing.
Cell culture
Neuro-2A mouse neuroblastoma (N2A) cells and human embryonic kidney (HEK)-293T cells were cultured in Dulbecco-modified Eagle medium (DMEM) (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS) (Sigma, St.Louis, MO), 100 U/mL penicillin and 100 µg/mL streptomycin (Invitrogen, Carlsbad, CA). Cells were incubated at 37°C and 95% air/5% CO2 atmosphere.
Biochemical fractionation of human parkin variants
HEK293T cells were cultured in 10 cm dishes. In duplicate, cells were transfected with either the pcDNA3.1 full-length WT, E399Q, or T240R human parkin constructs using Lipofectamine Reagent (Invitrogen), following the manufacterer’s protocol. At 48 hours post transfection, cells were rinsed and scraped in 1X PBS and sedimented at 13,000 × g. Cells were vortexed vigorously in 2 pellets volumes of ice-cold PBS/0.1% Triton buffer containing protease inhibitors. The cell debris were sedimented at 13,000 × g for 2 minutes and the supernatants were stored on ice. The PBS/0.1% Triton extraction was repeated and following sedimentation, the supernatants from both extractions were pooled together as the TX fraction. The remaining pellet was rinsed with the PBS/0.1% Triton buffer and the rinse was discarded. The cell pellet was then resuspended into 2 pellet volumes of ice-cold RIPA buffer with protease inhibitors, vortexed vigorously, stored on ice for 5–10 minutes, and then vortexed again. The cell debris were sedimented at 13,000 × g for 2 min and the supernatant was collected as the RIPA fraction. The remaining pellet was rinsed with RIPA buffer and the rinse was discarded. Lastly, the pellet was solubilized in 1 pellet volume of 2% SDS/8M Urea which was collected as the SDS fraction. All extracts were quantified by BCA assay and analyzed by western blot analysis with the PRK109 and actin antibodies. Western blot images were scanned using a CanoScan Lide30 scanner (Canon U.S.A., Inc., Lake Success, NY). The levels of PRK109 signal were quantified for each sample using ImageQuant 5.0 analysis software (GE Healthcare, Piscataway, NJ).
Immunofluorescence and Confocal Analyses of Parkin
HEK293T cells were cultured in 35 mm dishes. Cells were transfected in duplicate either with the pcDNA3.1 full-length WT, E399Q, or T240R human parkin constructs using Lipofectamine Reagent (Invitrogen), following the manufacterer’s protocol. At 24 hours post transfection, cells were rinsed in PBS and fixed by incubation in ice-cold acetic-methanol (1:20) at −20 °C for 30 minutes. Cells were rehydrated with water, rinsed with PBS, and blocked in PBS/2% FBS/0.1% Triton. Cells were then incubated with PRK109 antibody diluted into PBS/2% FBS at a concentration of 1:2000 overnight at 4°C, washed 3 times with PBS at 10 minutes each, and then incubated at room temperature with a goat anti-mouse secondary antibody conjugated to Alexa Fluor ® 594 (Invitrogen) diluted into PBS/2% FBS for 1 hour. Cells were rinsed 3 times with PBS and then stained with Hoerscht 33342 trihydrochloride trihydrate ( 0.5ug/mL) (Invitrogen) for 5 minutes at room temperature. Cells were rinsed with water and then coverslipped with Cytoseal™ 60 mounting media (Richard-Allen Scientific, Kalamazoo, MI). The images were visualized with a Zeiss LSM-510 Meta confocal microscope.
Effects of parkin on synphilin-1 steady-state levels
A mammalian expression construct expressing C-terminal myc-tagged full-length human synphilin-1 (synphilin-c-myc/pcDNA 3.1) was kindly provided by Dr. Virginia Lee. In triplicate, HEK293T cells were co-transfected with synphilin-c-myc/pcDNA 3.1 and either pcDNA3.1 mock vector, or pcDNA3.1 human full-length WT parkin, human E399Q mutant parkin, or T240R mutant parkin constructs. Cells were co-transfected with parkin to synphilin-1 cDNA ratio of 4:1 using the calcium phosphate transfection protocol described by Gallagher et al (Gallagher et al. 1997). At 21 hours post transfection, fresh complete media was added to the cells that were cultured for an additional 16 hours with or without 5uM clasto-lactacystin-β-lactone (Omuralide)(EMD Chemicals Inc, Gibbstown, NJ). Cells were then rinsed in PBS and lysed by sonication in 2% SDS/8M urea. Cell extracts were quantified by BCA assay and analyzed by western blot with monoclonal antibodies anti-c-myc clone 9E10, PRK109, and anti-actin. The 9E10 signal was quantified as described in previous sections.
Pulse-chase synphilin-1 turnover analysis
In triplicate, HEK293T cells were cultured in 35mm dishes. Cells were co-transfected with synphilin-c-myc/pcDNA 3.1 and either pcDNA3.1 mock vector, or pcDNA3.1 human WT parkin, E399Q mutant parkin, or T240R mutant parkin constructs. Cells were co-transfected with a parkin to synphilin-1 cDNA ratio of 4:1 using the calcium phosphate transfection protocol described above. At 24 hours post transection, cells were methionine-deprived for 20 minutes by incubation in pre-warmed methionine-free DMEM (Invitrogen, Carlsbad, CA)/10% dialyzed FBS before adding 100µCi [35S]-methionine (Invitrogen, Carlsbad, CA) per ml of methionine free DMEM/10% dialyzed FBS for 1 hour. Chase experiments were conducted with normal DMEM/FBS for 0, 1, 3, and 6 hours. Cells were then rinsed with PBS and harvested in ice-cold CSK buffer (100 mM NaCl, 50 mM Tris, pH 7.5, 2 mM EDTA, 1 % Triton X-100, protease inhibitors). The radiolabelled protein extracts were pre-cleared with a rabbit serum pre-incubated with protein A-agarose (Santa Cruz Biotechnologies, Santa Cruz, CA) for 3 hours at 4°C and radiolabelled extracts were then immunoprecipitated overnight at 4°C with anti c-Myc polyclonal antibody (Sigma) pre-incubated with protein A-agarose (Santa Cruz Biotechnology, Inc). The antibody-protein complexes were washed 3 times with 10 volumes of CSK buffer, resuspended in 2 volumes of 2X SDS sample buffer and boiled at 100 °C for 5 minutes. The beads were removed by centrifugation and the samples were loaded onto 13 % polyacrylamide gels. Following electrophoresis, gels were fixed with 50% methanol/5% glycerol, dried and exposed to a PhosphorImager plate and the signal was quantified using ImageQuant 5.0 analysis software.
Co-immunoprecipitation/Western studies of parkin and synphilin-1
In triplicate, using the calcium phosphate transfection method, HEK293T cells were transfected in 35mm dishes with either synphilin-c-myc/pcDNA 3.1 alone or co-transfected with synphilin-c-myc/pcDNA 3.1 as well as either the pcDNA3.1 untagged full-length human WT, E399Q, or T240R parkin constructs. At 21 hours post transfection, fresh complete media was added to the cells that were cultured for an additional 24 hours. Cells were then rinsed, scraped, and harvested in PBS by centrifugation at 13,000 × g for 2 minutes. The cell pellets were lysed in 500 uL of ice-cold CSK buffer (100 mM NaCl, 50 mM Tris, pH 7.5, 2 mM EDTA, 1 % Triton X-100) and cell debris was sedimented at 13,000 × g for 2 minutes. 30 uL of the supernatant was incubated at 100°C for 5 minutes with SDS sample buffer as the “Input” fraction. The remaining supernatant was incubated overnight at 4°C with anti-c-Myc polyclonal antibody (Sigma-Aldrich) that had been pre-incubated with protein A-agarose beads (Santa Cruz Biotechnologies, Santa Cruz, CA). The antibody-protein complexes were then washed 3 times with 10 volumes of CSK buffer, resuspended in 20uL of 2X SDS sample buffer, and incubated at 100°C for 5 minutes as the “IP” fractions. 30uL of the unbound supernatants were also incubated at 100°C for 5 minutes with SDS sample buffer as the “Unbound” fractions. The Start, IP, and Unbound fractions were resolved by SDS-PAGE and immunoblotted with PRK109 antibody.
Co-immunoprecipitation/Western studies of Parkin with UbcH7 and UbcH8
The mammalian expression vectors expressing N-terminal HA-tagged full-length human UbcH7 or UbcH8 (pRK5-HA-UbcH7 and pRK5-HA-UbcH8, respectively) were kindly donated by Dr. Ted Dawson. In triplicate, using the calcium phosphate transfection method, HEK293T cells were co-transfected in 35mm dishes with either pRK5-HA-UbcH7 or pRK5-HA-UbcH8 and pcDNA 3.1 mock vector or pcDNA3.1 untagged full-length WT, E399Q, or T240R human parkin constructs. Cells were cultured, harvested, and lysed as described in the previous section. 50uL of the lysate was incubated at 100°C for 5 minutes with SDS sample buffer as the “Input” fraction. The remaining lysate was incubated for 3 hours at 4°C with HA.11 polyclonal antibody (Covance) that had been preabsorbed to protein A-agarose beads (Santa Cruz Biotechnologies). The antibody-protein complexes were then washed 3 times with 10 volumes of CSK buffer, resuspended in 20uL of 2X SDS sample buffer, and incubated at 100°C for 5 minutes as the “IP” fractions. The Input and IP fractions were resolved by SDS-PAGE and immunoblotted with the monoclonal antibodies PRK109 and anti-HA (Roche).
Quantification of mouse synphilin-1 protein
Western blot images were obtained (described above) and scanned using a CanoScan Lide30 scanner. The levels of UPN79 and actin were quantified for each mouse using ImageQuant 5.0 analysis software. The levels of UPN79 were normalized between mice by calculating the ratio between the UPN79 value to that for actin. The normalized results for the mice of the same genetic background were averaged together and graphed.
Relative quantification of the mouse synphilin-1 mRNA transcript
Cerebral cortical tissues were harvested from 8 month old BL6 and C3H mice. Half of the tissue was frozen on dry ice for protein analysis and total RNA was extracted from the remaining half using TRIzol reagent (Invitrogen) as described in detail in previous sections. Three tissue samples were prepared for each mouse strain. Reverse transcription reactions and real-time PCR analyses were performed as described above. Primers were designed to amplify a 107 base pair product of mouse synphilin-1 cDNA that corresponds to nucleotides 988–1095. The sequences for the synphilin-1 primers were as follows. Forward: 5’-ATATCGCTCTTGCCACACCTA-3’ and Reverse: 5’-CAGGCATTCTGCATGGCCCTT-3’. Additionally, a mouse β-actin loading control was amplified as described above. The CT values for each sample were obtained. Synphilin-1 values were normalized to β-actin by taking the ratio of synphilin-1 to β-actin for each sample.
RESULTS
Differential immunoreactivity for parkin protein in various mouse strains
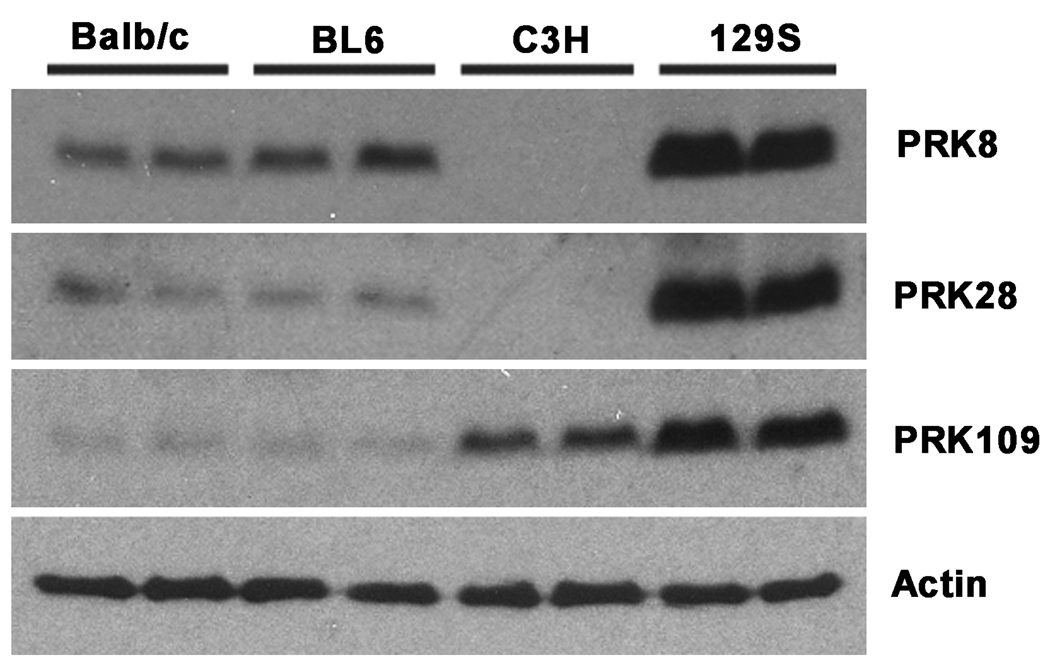
During the course of analyzing parkin expression by immunoblotting in mice from various backgrounds, dramatic differences were observed for immunoreactivity with 3 different monoclonal anti-parkin antibodies (Fig. 1). All three antibodies used here (PRK8, PRK28 and PRK109) were previously shown to specifically recognize human and mouse parkin and to recognize epitopes within the RING2 domain of parkin (amino acids residues 399–465 of human parkin) (Pawlyk et al. 2003). In brain cortex, PRK109 showed the highest level of immunoreactivity in the 129S background, followed by the C3H background and lowest, and about equivalent, levels in the Balb/c and C57BL/6 (BL6) backgrounds. Immunoblotting with antibodies PRK8 and PRK28 also demonstrated equivalent higher immunoreactivity in a 129S background compared to the Balb/c and BL6 background. Surprisingly, the cortical brain extracts from C3H mice were devoid of reactivity with both PRK8 and PRK28 antibodies. However, this result was not due to a complete loss of parkin expression in C3H mice as the PRK109 antibody recognized parkin in C3H tissue extracts. These results suggest that PRK8, PRK28, and PRK109 antibodies recognize different epitopes and that somehow, in C3H mice, the epitope that is specifically recognized by the PRK8 and PRK28 antibodies is altered.
Figure. 1.
Western blot analysis showing differential levels of immunoreactivity with different parkin antibodies in various mouse strains. Cerebral cortical tissues were harvested from Balb/c, C57BL/6 (BL6), C3H, and 129S mice and total protein lysates were extracted. Equal amounts (18 ug) of total protein extracts were loaded on 13% polyacrylamide gels and analyzed by western blot analysis with the anti-parkin monoclonal antibodies, PRK8, PRK28, and PRK109 as well as an actin antibody to confirm equal protein loading. The representative blot above shows extracts from 10 week old mice. Two samples were loaded, each from separate mice of the indicated strains. All experiments were replicated and similar results were obtained for a total of n=4 mice from each strain.
Similar levels of the parkin mRNA transcript are observed in different mice backgrounds
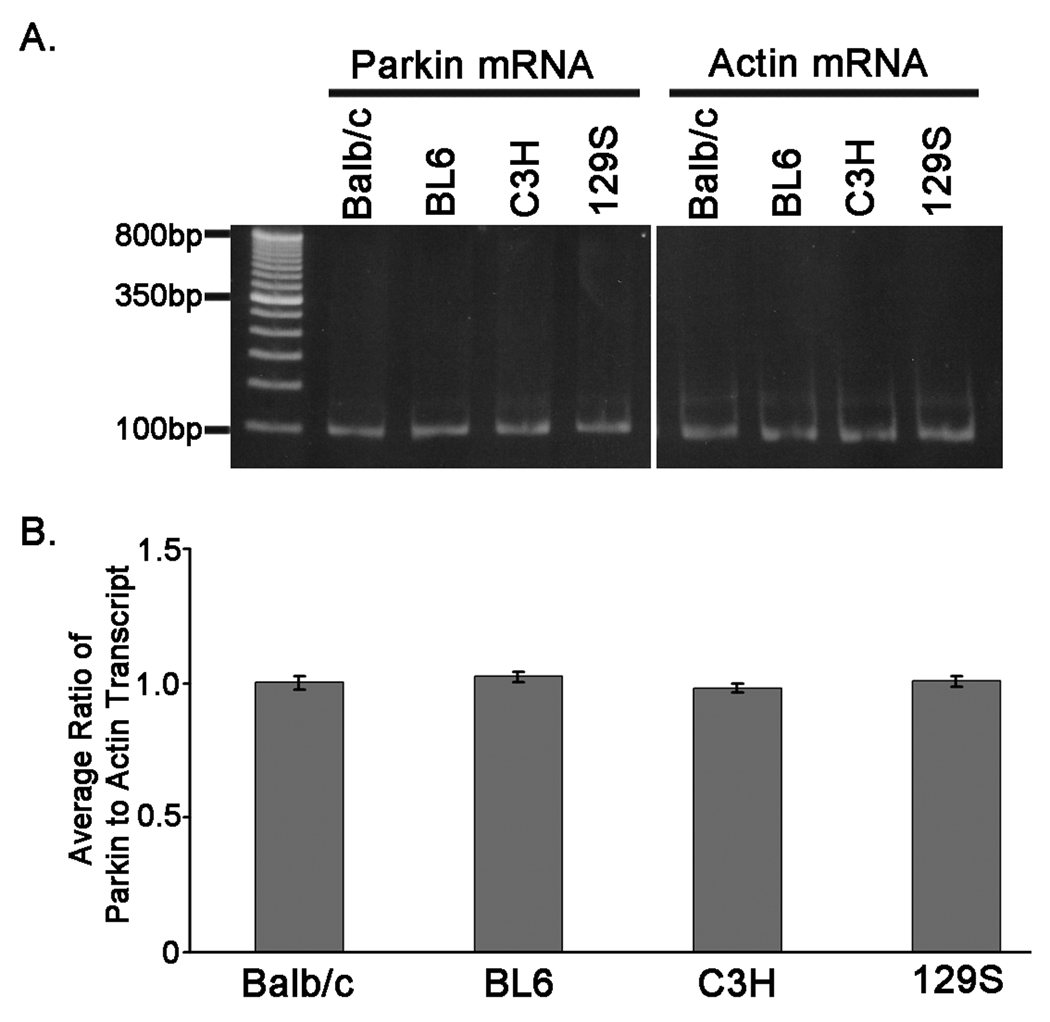
To begin to understand the reasons for the differences in immunoreactivity with parkin antibodies in the Balb/c, BL6, C3H, and 129S mice backgrounds, the levels of parkin mRNA in these mice was determined. Two-step quantitative real-time reverse-transcription polymerase chain reaction (qRT-PCR) analyses were performed using total RNA extracted from cortical tissues of Balb/c, BL6, C3H, and 129S mice. Analyses were performed in duplicate using 3 independent RNA samples for each of the different mouse strains. All mouse strains expressed comparable levels of the parkin mRNA transcript (Fig. 2A and B), indicating that the differences observed in parkin protein immunoreactivity levels are not due to differences in the amount of mRNA.
Figure 2.
Similar levels of the parkin mRNA between different mice stains. Total RNA was extracted from cerebral cortical tissues that were harvested from Balb/c, BL6, C3H, and 129S mice. Parkin and actin mRNA transcripts were quantified using two-step qRT-PCR. Experiments were performed in duplicate and 3 separate samples were analyzed for each mouse strain. A) The parkin and actin qRT-PCR products are shown. The image represents the products from one of the mice analyzed for the indicated strain. B) The ratio of the Parkin to Actin CT value was calculated for each sample. The graph depicts the average of the ratios between replicate samples for each mouse strain. The error bars indicate standard deviation (n=6).
Parkin antibody epitope mapping studies
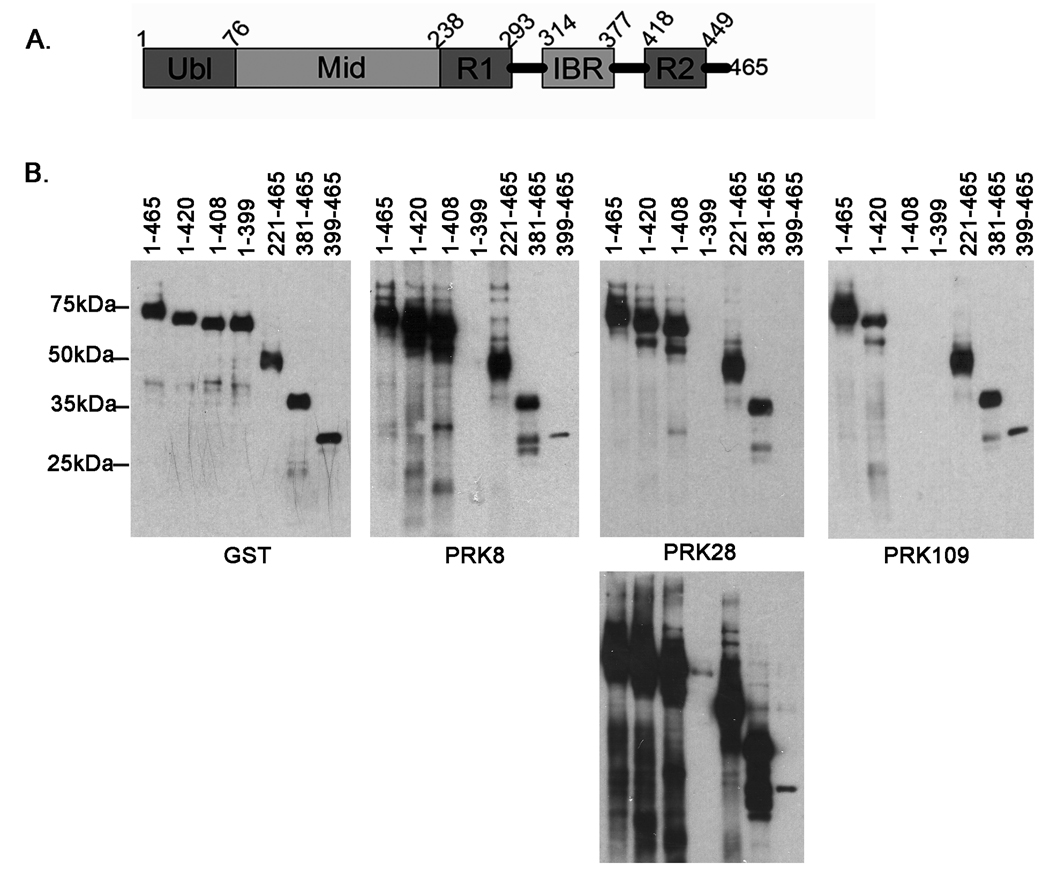
To further explain the differences in immunoreactivity observed in various mouse strains with different parkin antibodies, the epitopes recognized by these antibodies were mapped in greater detail. For these epitope mapping studies, full-length and various N- and C-terminal truncated forms of human parkin were expressed in bacteria as N-terminally GST-tagged recombinant proteins (Fig. 3). Western blotting with an anti-GST antibody demonstrated similar expression for all proteins. The PRK8 antibody recognized GST-parkin(1–408), but not GST-parkin(1–399), indicating that the epitope for this antibody includes residues 400–408. PRK8 reacted with both GST-parkin(381–465) and GST-parkin(399–465), but reactivity for GST-parkin(399–465) was much weaker. These data indicate that the epitope of antibody PRK8 require residues 400–408, but extends further upstream of this sequence. Antibody PRK28 demonstrated similar immunoreactive properties to antibody PRK8, with an additional weak reactivity with GST-parkin(1–399). Therefore, the epitope for PRK28 also requires amino acid residues 400–408 and extends further upstream than PRK8. Antibody PRK109 reacted with GST-parkin(1–420), but not GST-parkin(1–408) indicating that the epitope for this antibody includes residues 409–420.
Figure 3.
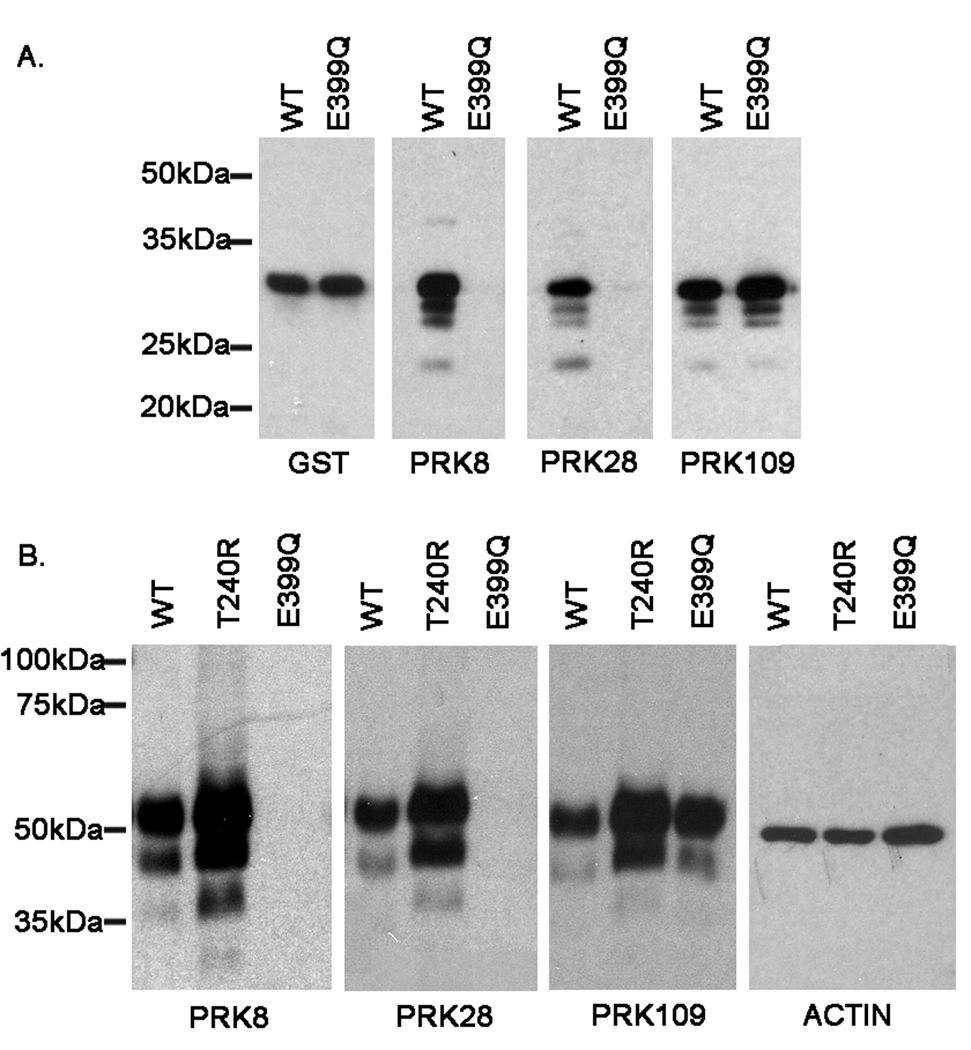
Parkin antibodies epitope mapping studies. A. Schematic of parkin protein. The numbers indicate the amino acid residues that correspond to the respective protein domains. Ubl is the ubiquitin-like domain, Mid is the linker domain, R1 is the first RING finger domain, IBR is the in-between-ring-finger domain, and R2 is the second RING finger domain. B. Full-length human parkin cDNA corresponding to amino acids 1–465 and various truncated cDNA fragments of parkin were cloned in the bacterial expression vector pDEST 15, that expresses N-terminal GST-tagged recombinant proteins. The recombinant proteins were expressed in BL21 E. coli cells. The cells were harvested and equal amounts of total protein lysates were analyzed by western blot with a GST antibody in order to confirm similar expression of the respective proteins. Additionally, equal amounts of the protein extracts were assessed for immunoreactivity with the PRK8, PRK28, and PRK109 antibodies. The experiments were performed in duplicate. Representative blots are shown for each construct. The identical blot for PRK28 is shown at different exposure times in order to reveal the faint immunoreactivity of the 1–399 construct that is not readily detectable at the shorter exposure time. The mobility of molecular mass markers is indicated on the left.
An endogenous mutation in the C3H parkin gene is responsible for the loss of PRK 8 and PRK 28 immunoreactivity
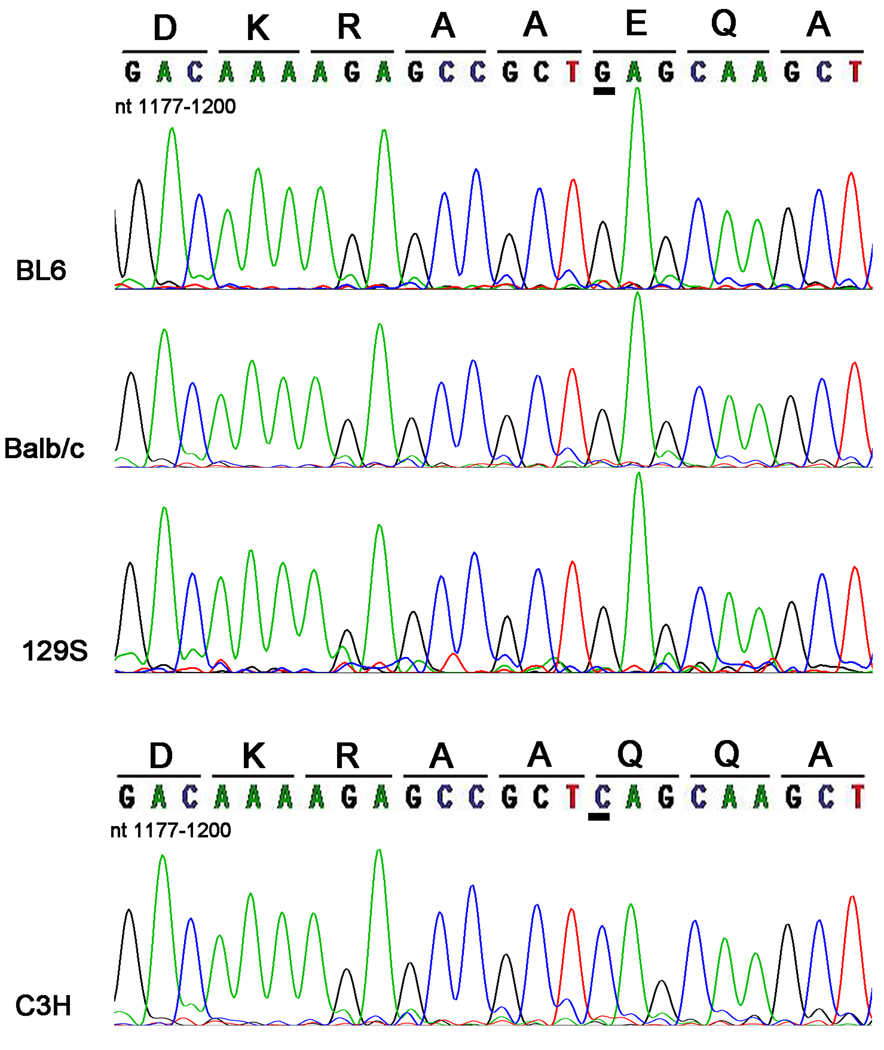
Since genetic alterations can result in differences in the parkin protein that could be responsible for some of the differences in immunoreactivity observed in the different mouse strain background, we proceeded to determine the parkin cDNA sequence (nucleotides 963–1388) around the region comprising the antibody epitopes for each mouse strain. This analysis revealed that the cDNA sequences for Balb/c, BL6 and 129S mice was identical to the previously published sequence for Balb/c (Kitada et al. 2000). However, the cDNA sequence for the C3H strain revealed 2 homologous nucleotide alterations (c. 1140 C>T and c. 1192G>C) (Fig. 4 and data not shown). The nucleotide change at position 1140 is a silent mutation that does not change the amino acid. The 1192G>C substitution results in the missense mutation E398Q. This mutation is equivalent to a E399Q mution in human parkin, since mouse parkin has one less residue (Gly 139) than human parkin (Kitada et al. 2000). DNA sequencing of genomic tail DNA confirmed that the C3H parkin mutation occurred at the DNA level and was not due to post-transcriptional events (see online supporting information).
Figure 4.
Discovery of a novel endogenous parkin mutation in C3H mice. RT-PCR was performed using total RNA that was extracted from cerebral cortical tissues of BL6, Balb/c, C3H, and 129S mice. The amplified PCR products spanned bases 895–1395 of the mouse parkin cDNA sequence. The PCR products were sequenced and analyzed for mutations. The image shows the respective portion (nucleotide bases 1177–1200) of sequencing electropherogram for the various mouse strains. The corresponding amino acid sequences are written above the respective DNA codons. A novel endogenous G1192C parkin cDNA mutation was identified in C3H mice. This mutation translates into a missense E398Q mutation. The 1192 cDNA residue is underlined in order to highlight the location of the novel G1192C mutation in the mouse parkin cDNA sequence.
Since the E399Q mutation is within the epitopes for both PRK8 and PRK28, the effects of this mutation on the reactivity of these antibodies was determined. The E399Q mutation was introduced into the bacterial vector expressing GST-parkin(380–465) and the protein was expressed concurrently with the similar WT protein. Immunoblotting with an anti-GST antibody demonstrated equivalent expression of both proteins (Fig. 5A). Immunoreactivity with PRK109 was not affected by the E399Q mutation, however this mutation abolished the reactivity with both PRK8 and PRK28 (Fig 5A).
Figure 5.
Abolishment of the PRK8 and PRK28 antibody epitopes due to the E399Q human parkin mutant. A) pDEST15/parkin 380–465 and pDEST15/parkin 380–465 E399Q constructs were used to express the respective proteins in BL21 E. coli. Total protein lysates were extracted and equal amounts of protein (600 ng) were resolved on 13% polyacrylamide gels and analyzed by western blot with GST, PRK8, PRK28, and PRK109 antibodies. B) Full-length untagged wild-type and mutant (T240R and E399Q) human parkin cDNA cloned in the mammalian express vector pcDNA3.1 were used to express these proteins in mouse N2A neuroblastoma cells. Following transfection with the respective constructs, equal amounts (4 ug) of total protein lysates were resolved on 13% polyacrylamide gels and assessed by western blot for recognition of the indicated parkin antibodies. Additionally, the blots were probed with an actin antibody to confirm equal protein loading. The mobility of molecular mass markers is indicated on the left.
To confirm these findings in mammalian cells, a construct expressing full-length untagged human WT parkin (parkin/pcDNA3.1) was used to overexpress human parkin in mouse neuroblastoma Neuro-2A (N2A) cells. In addition, similar vectors expressing T240R and E399Q mutant parkin were used. While antibody PRK109 recognized both WT and parkin mutants, antibodies PRK8 and PRK28 did not react with E399Q parkin (Fig. 5B). These data indicate that the E399Q mutation disrupts the epitopes of the PRK8 and PRK28 antibodies and that the equivalent mutation (E398Q) in C3H mice explains the lack of immunoreactivity with these antibodies.
The E399Q human parkin mutant shows reduced solubility without associated inclusion formation
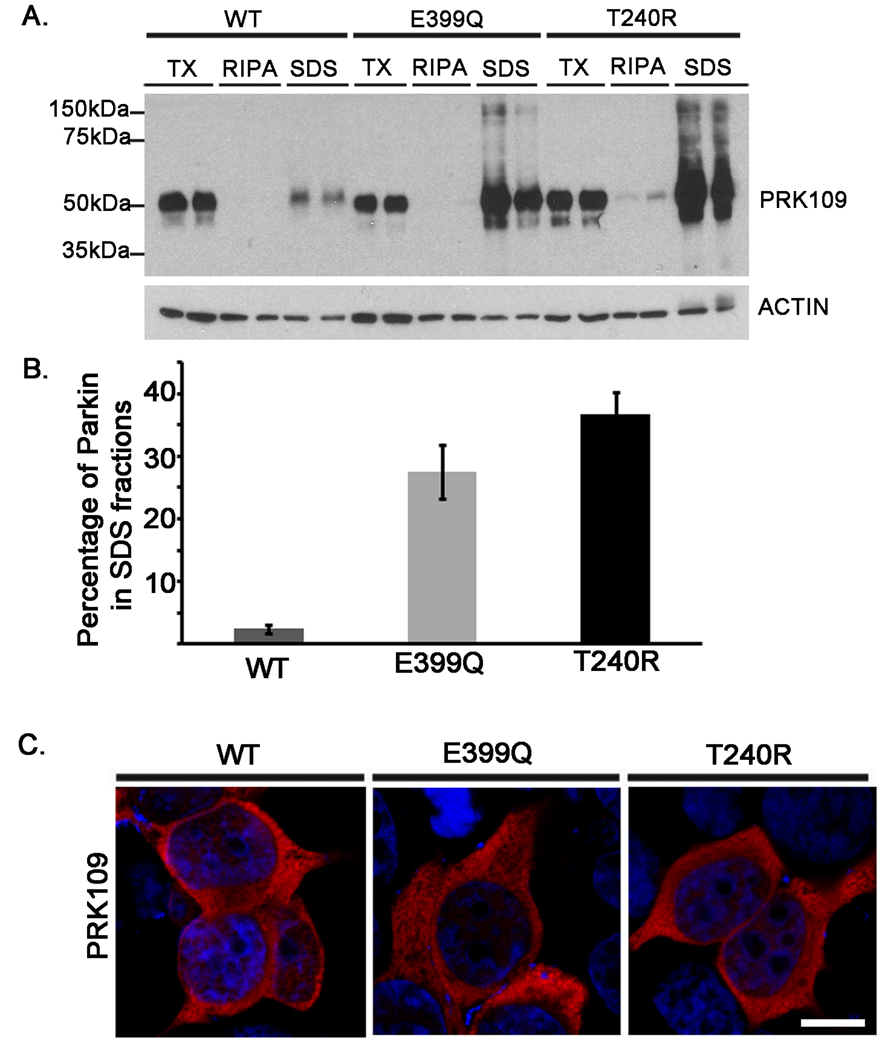
Several of the reported PD-linked parkin mutations, some in close proximity to the E399Q mutation, lead to altered solubility when they are expressed in mammalian cells (Sriram et al. 2005;Wang et al. 2005b;Hampe et al. 2006). To investigate whether the E399Q parkin amino acid substitution alters these biochemical properties of parkin, solubility assays were performed. HEK293T cells were transiently transfected with untagged full-length WT, E399Q, or T240R mutant human parkin constructs. Cell lysates were sequentially extracted in buffers of increasing solubilization strengths. Extracts were then analyzed by western blot with the PRK109 antibody. In comparison to the other fractions, WT Parkin protein was mostly extracted into the soluble TX fractions and to a lesser extent into the SDS insoluble fractions (Fig. 6A and B). A significant fraction of both E399Q and pathogenic T240R human parkin mutants was extracted in the insoluble SDS-fractions (Fig. 6A and B).
Figure 6.
The E399Q human parkin mutant shows reduced solubility in mammalian cells without associated inclusion formation. HEK293T cells were transfected in duplicate with untagged full-length WT, E399Q, or T240R human parkin/pcDNA 3.1 constructs. A) and B) At 48 hours post transfection, cells were harvested and sequentially extracted into PBS/0.1% Triton (TX) buffer, RIPA buffer, and 2% SDS/17mM Tris (SDS) buffer. Equal amounts (16ug) of each sample were loaded onto 10% polyacrylamide gels and analyzed by western blot for parkin distribution with the PRK109 antibody. Blots were also probed with an actin antibody to assess equal protein loading. The mobility of molecular mass markers is indicated on the left. The western blot images were quantified as described in “Materials and Methods”. The total percentage of parkin extracted into SDS fractions was calculated for each sample. Error bars indicate standard deviation (n=4). C) Immunofluorescence analysis of HET393 cells expressing WT or mutant parkin immunostained with PRK109 (red) and counterstained with Hoerscht (blue). The merged images depict the absence of inclusions in cells expressing WT, E399Q, or T240R parkin. Scale bar =10um.
To determine whether this altered extractability of E399Q mutant parkin is associated with the formation of protein inclusions, immunofluorescence studies were conducted. HEK293T cells transiently expressing untagged full-length WT, E399Q, or T240R human parkin were assessed for parkin localization with the PRK109 antibody. Following these analyses, while parkin was found to exhibit similar homogeneous cytoplasmic staining for all of the parkin variants, inclusions were not detected in any of the cells analyzed (Fig. 6C).
The E399Q parkin mutant is functionally impaired
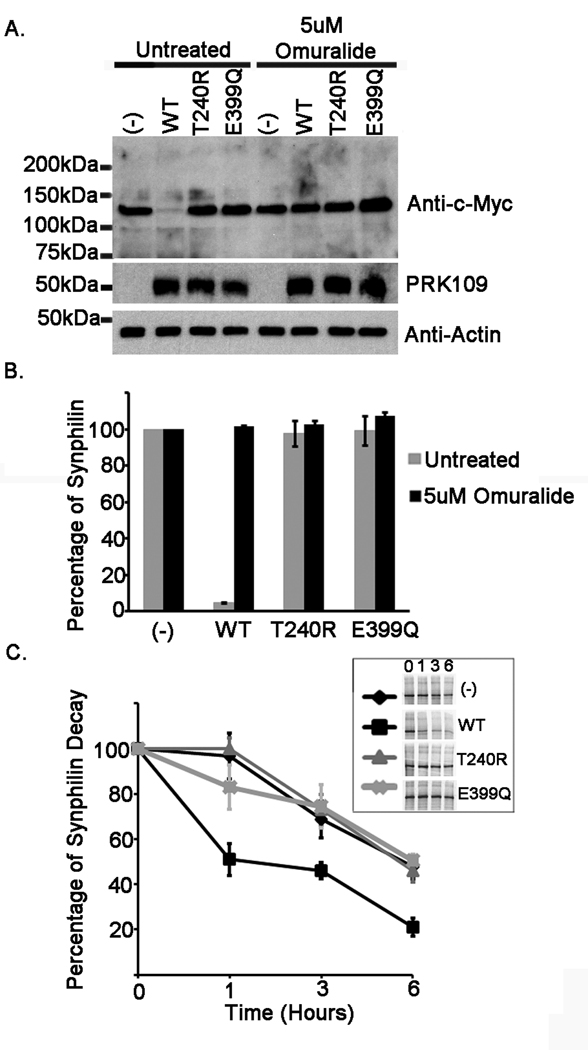
Parkin functions as an E3 ubiquitin-protein ligase, acting to promote the ubiquitination and subsequent degradation of its substrates (Zhang et al. 2000;Imai et al. 2000;Rankin et al. 2001). Synphilin-1 has been identified as one of the putative substrates for parkin (Lim et al. 2005;Chung et al. 2001). Several of the pathogenic mutations in parkin, including the T240R mutation, have been shown to impair the ability for parkin to promote the degradation of synphilin-1 (Chung et al. 2001;Sriram et al. 2005). In order to ascertain whether the E399Q mutation hinders parkin function, steady-state levels of synphilin-1 were analyzed in the presence of WT, T240R, or E399Q parkin. The levels of synphilin-1 were compared between parkin variants. In addition, some transfected cells were treated with the proteasome inhibitor, clasto-lactacystin β-lactone (Omuralide), to assess that the degradation of synphilin-1 promoted by parkin was due to the proteasome. HEK293T cells were co-transfected with myc-tagged synphilin-1 and either pcDNA3.1 mock vector or the respective pcDNA3.1 untagged parkin constructs. Cells were transfected with a 4:1 cDNA ratio of parkin to synphilin-1 as under these conditions, it has been previously shown that parkin promotes the degradation of synphilin-1 (Lim et al. 2005). Total cell lysates were analyzed by immunoblot with anti-c-Myc and PRK109 antibodies. Expression of WT parkin led to a significant decrease in synphilin-1 steady-state levels in comparison to the levels for synphilin-1 when it had been co-expressed with mock vector (Fig. 7A and B). However, neither the T240R nor the E399Q parkin had any effect on the steady-state levels of synphilin-1. Additionally, as it has been previously shown, parkin-mediated degradation of synphilin-1 occurs via the proteasome pathway (Lim et al. 2005), since it was completely blocked in the presence of a specific proteasome inhibitor.
Figure 7.
E399Q mutant parkin is functionally impaired. HEK293T cells were co-transfected either with mock pcDNA3.1 vector “(−)” or full length WT, T240R, or E399Q human parkin pcDNA 3.1 constructs as well as the pcDNA3.1 myc-tagged full-length synphilin-1 construct. The co-transfection experiments were performed using a parkin to synphilin-1 cDNA ratio of 4:1. At 24 hours post transfection, (A and B) cells were incubated for 16 hours with fresh DMEM or with DMEM containing 5uM Omuralide. Cells were then harvested and total protein lysates were extracted. Extracts were resolved by 10 % polyacrylamide gels and immunoblotted with the monoclonal antibodies anti-c-Myc clone 9E10 to assess synphilin-1 levels, PRK109 antibody to confirm parkin expression, and anti-actin antibody to ensure equal protein loading. All experiments were performed in triplicate and repeated at least 3 times. The mobility of molecular mass markers is indicated on the left. The levels of synphilin-1 were quantified as described in “Materials and Methods” and the graph in B depicts the percentage of synphilin-1 protein standardized to that in the mock vector samples. The error bars indicate standard deviation between replicate samples (n=4). C) Cells were pulsed with 35S-methionine for 1 hour and chased for 0, 1, 3, or 6 hours. The inset shows representative pulse-chase experiments. Experiments were conducted in triplicates. The results are plotted as percentage of protein over time standardized to the 0 hrs time point. The error bars show standard deviation (n=3).
To further determine the effects of WT, T240R, and E399Q parkin on the stability of synphilin-1, HEK293T cells were co-transfected with myc-tagged synphilin-1 and either pcDNA3.1 mock vector or the respective pcDNA3.1 untagged human parkin constructs as described above. Synphilin-1 turnover was assessed by pulse-chase analyses with 35 S-methionine and the effects of the respective parkin variants on synphilin-1 turnover were compared. Synphilin-1 exhibited a half-life of ∼6 hours in the absence of parkin (Fig. 7C). WT Parkin enhanced the turn-over of synphilin-1, reducing the half-life to ∼1 hour. However, neither T240R nor E399Q Parkin had an effect on synphilin-1 degradation, suggesting that both mutants are functionally inactive.
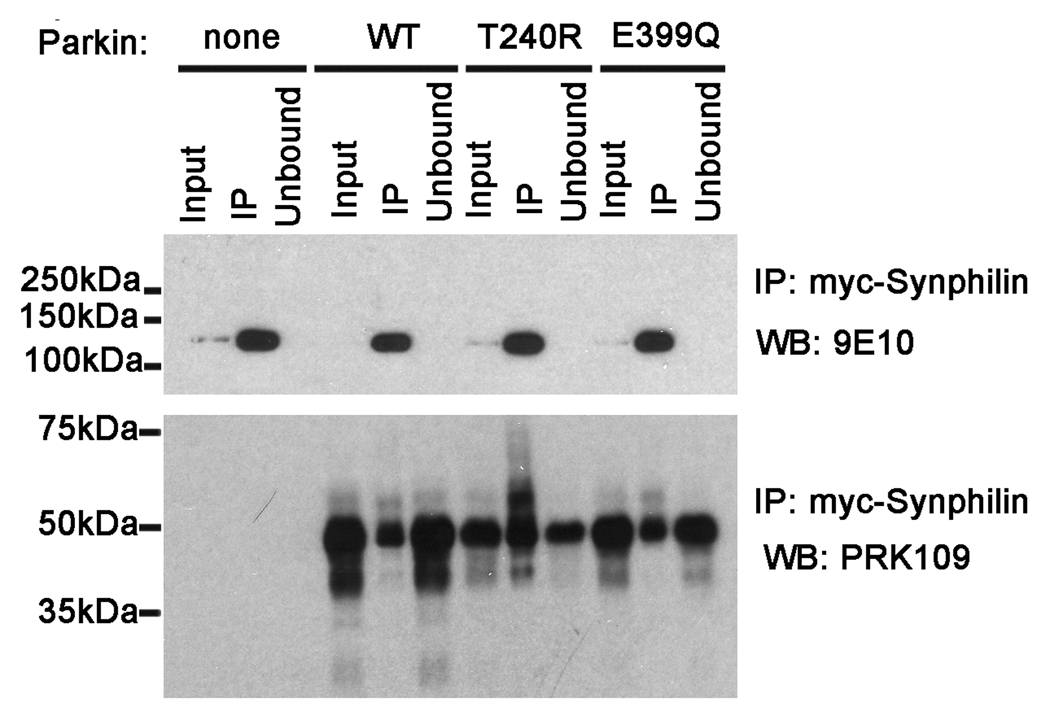
Synphilin-1 was initially identified as a substrate for parkin in studies investigating interacting proteins (Chung et al. 2001). It is possible that the E399Q mutation disrupts the direct interaction between mutant parkin and synphilin-1 that would prevent parkin-mediated degradation. To determine whether there is a loss in the direct interaction between E399Q mutant parkin and synphilin-1, co-transfection and co-immunoprecipitation experiments were conducted. HEK293T cells were co-transfected with myc-tagged synphilin-1 and untagged human WT, E399Q, or T240R mutant parkin constructs. At 48 hours post transfection, soluble cells lysates were immunoprecipated with an anti-c-Myc antibody followed by immunoblot with the PRK109 antibody. All parkin variants, including the E399Q mutant, were co-immunoprecipitated with synphilin-1 (Fig. 8). Thus, the E399Q Parkin mutation does not disrupt the interaction between parkin and synphilin-1.
Figure 8.
E399Q mutant parkin can directly interact with synphilin-1. HEK293T cells were transiently co-transfected with myc-tagged synphilin-1 and pcDNA3.1 mock vector (“none”) or with untagged full-length WT, T240R, or E399Q human parkin pcDNA3.1 constructs. Cells were harvested and soluble protein lysates were extracted (“Input”). The cell lysates were then immunoprecipitated using anti-c-Myc polyclonal antibody (“IP”). Equal amounts of the input, IP, and unbound supernatant (Unbound) fractions were resolved by SDS-PAGE and analyzed by western blot (“WB”) with anti-c-Myc monoclonal antibody 9E10 or with anti-parkin antibody PRK109. Experiments were repeated at least 3 times. The mobility of molecular mass markers is indicated on the left.
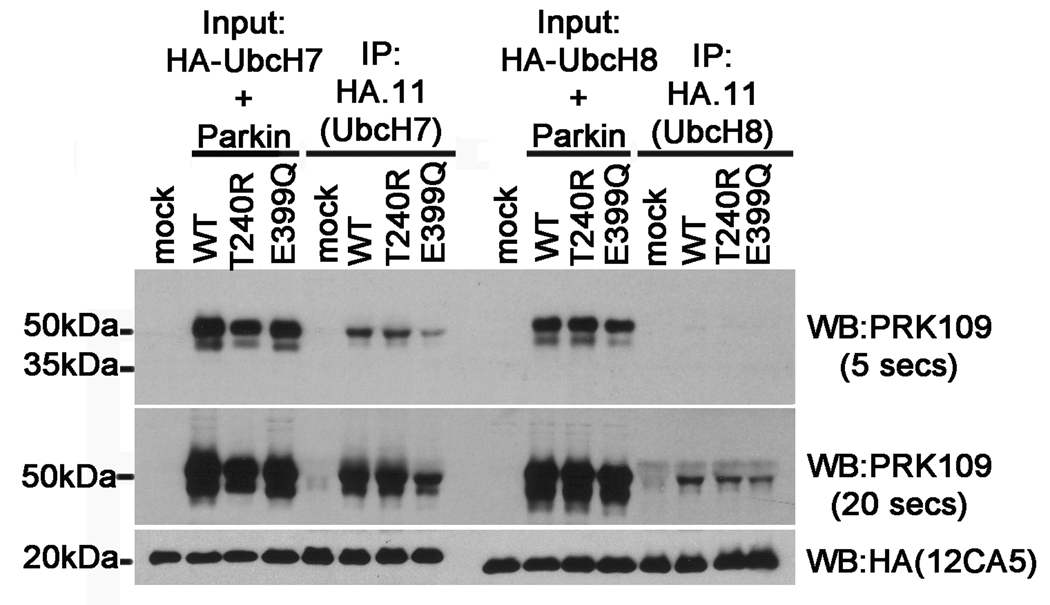
It has been previously shown that parkin specifically interacts with the E2 ubiquitin conjugating enzymes, UbcH7 and UbcH8, in order to exert its function as an E3 ligase (Shimura et al. 2000;Imai et al. 2000;Zhang et al. 2000). One plausible explanation for the impaired function exhibited by the E399Q parkin mutant could be disrupted binding with E2 enzymes. To test this hypothesis, co-transfection and co-immunoprecipitation experiments were conducted. HEK293T cells were co-transfected with HA-tagged UbcH7 or UbcH8 constructs and either mock pcDNA3.1 or untagged pcDNA3.1 full-length WT, T240R, or E399Q human parkin constructs. At 48 hours post transfection, cell lysates were immunoprecipitated with a polyclonal anti-HA antibody, HA.11, followed by immunoblot with the monoclonal antibodies PRK109 and anti-HA (clone 12CA5). These analyses revealed that while UbcH7 recruited similar levels of both WT and T240R parkin, the binding of E399Q parkin with UbcH7 was dramatically reduced (Fig. 9). Interestingly, in comparison to binding with UbcH7, parkin was recruited less effectively by UbcH8 and the levels of T240R and E399Q mutant parkin recruited by UbcH8 were modestly reduced in comparison to WT parkin. Together, these results indicate a loss of function of the E399Q mutant may be due, at least in part, to reduced interactions with E2 ubiquitin conjugating enzymes.
Figure 9.
The E399Q Parkin mutant shows reduced binding to UbcH7 and UbcH8. HEK293T cells were co-transfected with pRK5-HA-UbcH7 or pRK5-HA-UbcH8 and either pcDNA3.1 mock vector (“mock”) or with untagged full-length WT, T240R, or E399Q human parkin pcDNA3.1 constructs. Cells were harvested and soluble protein lysates were extracted (“Input”). The cell lysates were then immunoprecipitated using the anti-HA polyclonal antibody, HA.11 (“IP”). Input and IP fractions were resolved by SDS-PAGE and analyzed by immunoblot with the monoclonal antibodies PRK109 and anti-HA (clone 12CA5). The PRK109 blots are shown at 5 second and 20 second exposure times to highlight the differences in signal intensity reflected by the binding of parkin with UbcH7 versus that with UbcH8.
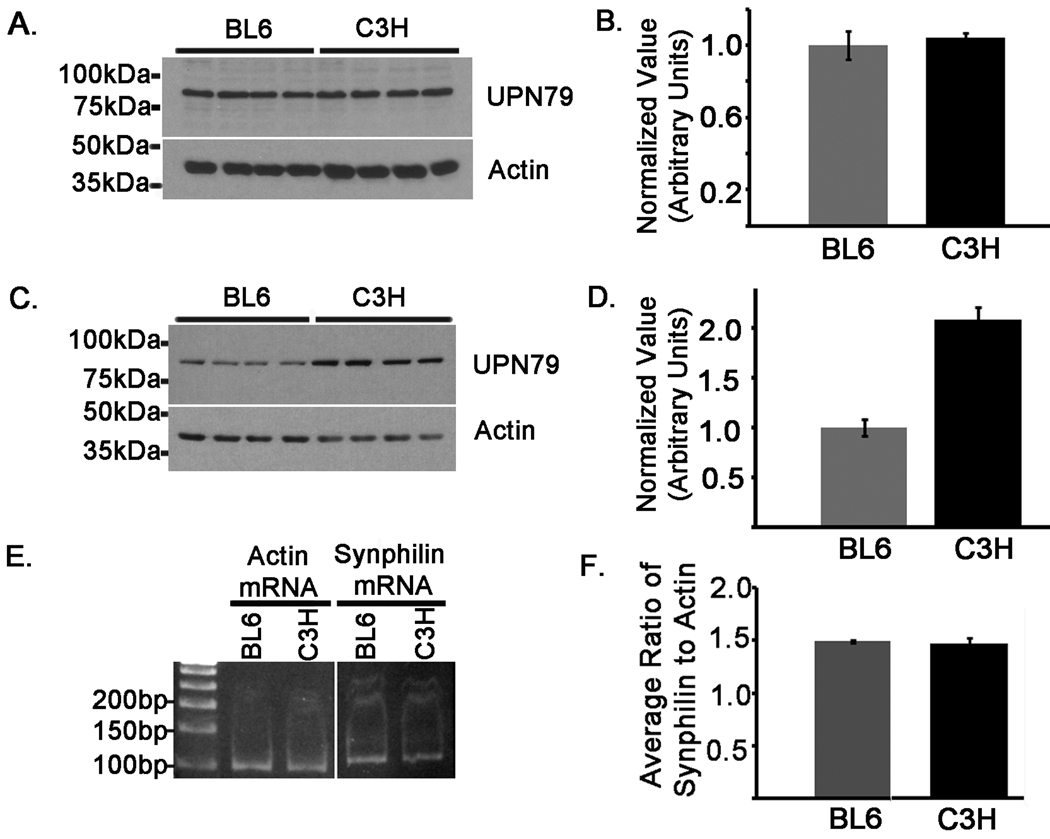
Synphilin-1 levels are increased in C3H mice
Since E399Q mutant parkin was incapable of down-regulating synphilin-1 expression in cultured cells (Fig. 7), it is plausible to hypothesize that the parkin in C3H mice, harboring the homologous E398Q mutation, may show increased levels of the synphilin-1 substrate in situ. To test this hypothesis, total protein extracts from cerebral cortical tissues of 10 week old BL6 and C3H mice were analyzed by western blot with the affinity purified synphilin-1 antibody, UPN79. No differences in synphilin-1 were detected between mice at this age (Fig. 10A and B). However, it is possible that synphilin-1 may accumulate in C3H mice in an age dependent manner. To determine this, cerebral cortical protein extracts from 8 month old BL6 and C3H mice were immunoblotted with antibody UPN79. Remarkably, extracts from 8 month old C3H mice revealed 2.0 ± 0.18 fold higher synphilin-1 levels than BL6 mice of the same age (Fig 10C and D).
Figure 10.
Synphilin-1 protein levels are increased in C3H mice. Cerebral cortical tissues were harvested from BL6 and C3H mice. Protein lysates were extracted from animals at ages 10 weeks (A and B) and 8 months (C and D). Equal amounts (18 ug) of high-salt protein extracts were loaded onto 10% polyacrylamide gels and analyzed by western blot analysis with the affinity purified polyclonal antibody, UPN79 to assess synphilin-1, as well as an actin antibody to confirm equal protein loading. Four samples were loaded, each from separate mice of the indicated strains. The graphs in B and D show the average ratios of the levels of UPN79 to actin normalized to the values in BL6 animals at the respective aforementioned ages. E) Total RNA was extracted from cerebral cortical tissues that were harvested from the same BL6 and C3H mice analyzed in C. Synphilin-1 and actin mRNA transcripts were quantified using two-step qRT-PCR. Experiments were performed in duplicate and 3 separate samples were analyzed for each mouse strain. The synphilin-1 and actin qRT-PCR products are shown. The image shows the products from one of the mice analyzed for the indicated strains. F) The ratio of the synphilin-1 to actin CT value was calculated for each sample. The graph depicts the average of the ratios between replicate samples for each mouse strain. The error bars indicate standard deviation (n=6).
To confirm that these changes were not due to differences at the transcriptional level, qRT-PCR analyses were performed using total RNA extracted from cortical tissues of the 8 month old BL6 and C3H mice used for synphilin-1 protein analysis described above. Both mouse strains expressed comparable levels of the synphilin-1 mRNA transcript (Fig. 10D and E), indicating that the differences observed in synphilin-1 protein immunoreactivity are not due to differences in the amount of mRNA. These findings are consistent with the notion that the E398Q mutation impairs the ability of parkin to mediate the degradation of synphilin-1, leading to an age-dependent accumulation of synphilin-1 in C3H mice.
DISCUSSION
Western blot analyses of brain extracts from Balb/c, C57BL/6, C3H, and 129S mice revealed dramatic variations in immunoreactivity with 3 monoclonal parkin antibodies (PRK8, PRK28, and PRK109). Notably, immunoreactivity for PRK8 and PRK28 was absent in C3H mice. This was especially unexpected since it had been previously reported that the epitopes for the PRK8, PRK28, and PRK109 antibodies were similarly located around the second RING domain of parkin (Pawlyk et al. 2003). In efforts to gain insights into how epitope differences could account for some of these changes in mouse parkin immunoreactivity, thorough epitope mapping studies were conducted. The analyses of a variety of recombinant N-terminal and C-terminal truncated parkin proteins allowed for the PRK8, PRK28 and PRK109 antibody epitopes to be more discretely mapped. The PRK8 antibody epitope requires residues 400–408 of human parkin but also extends upstream of these residues. The PRK28 epitope also requires residues 400–408 but extends slightly further upstream of the PRK8 epitope. Lastly, the PRK109 epitope is further downstream and includes residues 409–420 of human parkin protein.
Genetic analysis revealed a homologous nucleotide substitution (c. 1192G>C) in C3H mice that results in an E398Q missense amino acid change. This alteration is responsible for the disruption of the PRK8 and PRK28 epitopes in C3H mice. These findings and the epitope mapping are particularly important due to the prevalent use of these parkin antibodies in several studies (LaVoie et al. 2005;LaVoie et al. 2007;Henn et al. 2007;Cairns et al. 2004;Pawlyk et al. 2003;Fournier et al. 2009).
Immunoblotting with antibody PRK109 demonstrated variations in the levels of parkin in different mouse strains with the highest level being observed in 129S mice followed by C3H and lowest, and about equivalent levels, in the Balb/c and BL6 mice. Similar results were obtained when comparing parkin levels in 129S, Balb/c and BL6 mice with antibodies PRK8 and PRK28. Genetic studies revealed no alterations that would affect the PRK109 epitope in 129S, C3H, Balb/c and BL6 mice. Nor were any genetic mutations identified that would affect the PRK8 or PRK28 epitopes in 129S, Balb/c and BL6 mice. Additionally, the levels of brain parkin mRNA were found to be similar between strains, indicating post-transcriptional effects. These findings are likely a reflection of differences in parkin protein levels that may result from differences in translation efficiencies or turnover rates in the different mouse strains. Interestingly, no differences in the steady-state levels of other PD relevant proteins including α-synuclein, DJ-1, or tau were observed between mouse strains (data not shown). Neither were there differences in the levels of the E3 ligase, ARIH1 (data not shown), which shares close functional homology to parkin (Marin et al. 2004). Thus, the differences that were observed in parkin between mice strains appear to be specific and not due to the consequence of more generalized effects.
There has been no report of a naturally occurring human E399Q parkin polymorphism or pathogenic mutation, however there are several reported pathogenic mutations that are in close proximity to the this residue [R396G (Wu et al. 2005), A398T(Mata et al. 2004;von Coelln et al. 2004a), R402C (Macedo et al. 2009;Bardien et al. 2009), R402H (Sun et al. 2006), R402W(Poorkaj et al. 2004)], suggesting that it may affect protein structure or function. In addition, this glutamic acid residue at the 398 is conserved in other species including rat, pig, and macaque (Kitada et al. 2000;Kitada et al. 1998); NCBI protein database accession numbers NP_064478, NP_001038068, and ACL68652). Directly supporting this hypothesis, it was shown in this study that the E399Q mutant parkin exhibited reduced solubility when it was overexpressed in mammalian cells. However, this did not lead to the formation of aggregated protein that could be observed at the microscopic level. This suggests that the E399Q parkin mutation may result in protein misfolding without the accumulation into large aggregates. Changes in parkin solubility have been associated with a variety of stimuli including oxidative stress, heat shock, as well as a battery of other types of stress and may have a consequence on parkin function (Winklhofer et al. 2003;LaVoie et al. 2007;Wang et al. 2005a). For instance, changes which induce a shift in parkin solubility could thereby deplete soluble pools of parkin and may result in the impairment of parkin’s function toward substrates.
It was further shown in this report that the E399Q mutant parkin is deficient in the ability to mediate the degradation of the parkin substrate, synphilin-1, similar to the T240R parkin pathological mutant. Neither the steady-state levels nor the turn-over rate of synphilin-1 were affected by co-expression of the E399Q or T240R parkin mutants in HEK293T cells, while expression of WT parkin dramatically reduced the levels of synphilin-1 in these studies. These data supported previous findings which showed a marked decrease in synphilin-1 steady-state levels with the co-expression of WT parkin at a parkin to synphilin-1 cDNA ratio of 4:1, findings which were not observed under the same conditions for the T240R parkin mutant (Lim et al. 2005). It is of interest to note that high molecular weight forms of polyubiquitylated synphilin were not detected in any of the parkin co-expression experiments performed in the studies herein (Fig. 7). This could be because typical ubiquitination “smears” are below detectable levels under the experimental conditions used and/or these observations may be related to the finding by Lim et al which suggests that parkin can mediate different types of ubiquitin linkage (Lim et al. 2005). Nevertheless, the data herein demonstrates impaired function of E399Q and T240R parkin mutants towards down-regulating synphilin protein levels in comparison to WT parkin protein. This was not due to the inability for these mutants to interact with synphilin since WT, E399Q, and T240R parkin all showed similar interactions with synphilin by co-immunoprecipitation studies, consistent with previous findings (Chung et al. 2001).
It is interesting that the novel E399Q parkin mutation lies between the IBR and RING2 domains of the parkin protein. This feature is also common to the previously characterized T415N pathogenic parkin mutant. The T415N parkin mutation interfered with the proper recruitment of E2 enzymes to parkin (Zhang et al. 2000) and resulted in impairments in parkin-mediated degradation (Sriram et al. 2005). However, this was not associated with a loss in substrate binding activity (Chung et al. 2001;Sriram et al. 2005). Similarly, in the current study, in comparison to the other parkin variants analyzed, the E399Q parkin mutant showed dramatically reduced binding with the E2 enzyme, UbcH7. This aberration likely partially contributes to the deficiencies exhibited by the E399Q parkin mutant related to degradation of the synphilin-1 substrate.
The novel E399Q parkin mutant showed reduced solubility and functional impairments when it was analyzed in mammalian cells indicating that the homologous E398Q mutation in C3H mice may result in mice that are deficient or impaired in parkin activity. To support this hypothesis, there was a 2 fold increase in the cortical levels of synphilin-1 in 8 month old C3H mouse brains in comparison to BL6 mice. This difference was not observed in mice at 10 weeks of age which suggests that other E3 ligases or other modes of degradation play a more prominent role in the turnover of synphilin-1 at this age. While C3H mice do not show any overt behavioral characteristics that may be attributed to a functional parkin deficiency, the attempts to identify in vivo effects of parkin’s loss-of-function in mice have proven to be very difficult. Genetically generated parkin null mice fail to display any major differences in most of the behavioral, morphological, and biochemical analyses that have been previously conducted (von Coelln et al. 2006;Perez et al. 2005;Perez and Palmiter 2005;Thomas et al. 2007). Subtle differences in parkin null mice have been identified in a few studies, but many inconsistencies have also been reported (Palacino et al. 2004;Sato et al. 2006;Itier et al. 2003;Zhu et al. 2007;Frank-Cannon et al. 2008;Goldberg et al. 2003). Interestingly, no differences in synphilin-1 levels were detected in brain tissues of parkin null animals in comparison to wild-type animals in previous studies (Goldberg et al. 2003;Ko et al. 2005). However, this does not discredit the finding of the increased steady-state levels of synphilin-1 observed in C3H mice in the current study. It is possible that this discrepancy could be due to a number of factors including antibody specificity, tissue extraction method, or tissue-specific differences in parkin regulation of synphilin-1. Further, since it is likely that synphilin-1 may also be regulated by parkin independent mechanisms, it is unknown how this may be affected in a model system that is completely devoid of parkin versus a system where parkin is available, albeit impaired. Analysis of other putative parkin substrates such as Pael-R or p38/JTV-1 (Ko et al. 2005) could also indirectly help to confirm a loss of parkin function in C3H mice. However, while specific antibodies are not readily available, future studies to assess for alterations in these putative substrates would be informative.
In summary, these studies demonstrate that parkin protein levels differ in various mouse background strains. Additionally, the C3H mouse harbors a previously unreported mutation (E398Q) in parkin that abolishes immunoreactivity with antibodies PRK8 and PRK28. Characterization of this novel mutation revealed that it promotes the misfolding of parkin, impairs its ability to interact with E2 conjugating enzymes, and disrupts its function towards the synphilin-1 substrate. These findings indicate that C3H mice are carriers of a missense parkin mutation and that they may be a suitable model of parkin loss-of-function similar to human patients that have missense mutations. In addition, the difference in the level of parkin expression demonstrated here in various widely-used mouse strains is an important confound that should be taken into account in mouse studies.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the National Institute on Aging [AG09215] and the National Institute of Neurological Disorders and Stroke [NS053488]. C.P. Ramsey is supported by a pre-doctoral NRSA fellowship from the National Institute on General Medicine Sciences [GM082026].
The abbreviations used are
- AR-JP
autosomal recessive juvenile parkinsonism
- BCA
bicinchoninic acid
- BL6
C57BL/6
- Ct
cycle threshold
- DMEM
Dulbecco-modified Eagle medium
- E2
ubiquitin-conjugating enzyme
- E3 ligase
E3 ubiquitin-protein ligase
- HA
hemagglutinin
- HEK
human embryonic kidney
- HS
high-salt
- IBR
in-between-RING
- N2A
Neuro-2A
- PD
Parkinson disease
- PMSF
phenylmethanesulphonylfluoride
- qRT-PCR
quantitative real-time RT-PCR
- RING
Really-Interesting-New-Gene
- RIPA
radio immunoprecipitation assay
- RT-PCR
reverse-transcription polymerase chain reaction
- TX
triton
- Ubl
ubiquitin-like
- WT
wild-type.
REFERENCES
- Asakawa S, Tsunematsu K, Takayanagi A, et al. The genomic structure and promoter region of the human parkin gene. Biochem Biophys Res Commun. 2001;286:863–868. doi: 10.1006/bbrc.2001.5490. [DOI] [PubMed] [Google Scholar]
- Bardien S, Keyser R, Yako Y, Lombard D, Carr J. Molecular analysis of the parkin gene in South African patients diagnosed with Parkinson’s disease. Parkinsonism Relat Disord. 2009;15:116–121. doi: 10.1016/j.parkreldis.2008.04.005. [DOI] [PubMed] [Google Scholar]
- Bonifati V. Genetics of parkinsonism. Parkinsonism Relat Disord. 2007;13 Suppl 3:S233–S241. doi: 10.1016/S1353-8020(08)70008-7. [DOI] [PubMed] [Google Scholar]
- Cairns NJ, Uryu K, Bigio EH, et al. alpha-Internexin aggregates are abundant in neuronal intermediate filament inclusion disease (NIFID) but rare in other neurodegenerative diseases. Acta Neuropathol. 2004;108:213–223. doi: 10.1007/s00401-004-0882-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi P, Ostrerova-Golts N, Sparkman D, Cochran E, Lee JM, Wolozin B. Parkin is metabolized by the ubiquitin/proteosome system. Neuroreport. 2000;11:2635–2638. doi: 10.1097/00001756-200008210-00006. [DOI] [PubMed] [Google Scholar]
- Choi P, Snyder H, Petrucelli L, et al. SEPT5_v2 is a parkin-binding protein. Brain Res Mol Brain Res. 2003;117:179–189. doi: 10.1016/s0169-328x(03)00318-8. [DOI] [PubMed] [Google Scholar]
- Chung KK, Thomas B, Li X, Pletnikova O, Troncoso JC, Marsh L, Dawson VL, Dawson TM. S-nitrosylation of parkin regulates ubiquitination and compromises parkin’s protective function. Science. 2004;304:1328–1331. doi: 10.1126/science.1093891. [DOI] [PubMed] [Google Scholar]
- Chung KK, Zhang Y, Lim KL, Tanaka Y, Huang H, Gao J, Ross CA, Dawson VL, Dawson TM. Parkin ubiquitinates the alpha-synuclein-interacting protein, synphilin- 1: implications for Lewy-body formation in Parkinson disease. Nat Med. 2001;7:1144–1150. doi: 10.1038/nm1001-1144. [DOI] [PubMed] [Google Scholar]
- Ciechanover A. Linking ubiquitin, parkin and synphilin-1. Nat Med. 2001;7:1108–1109. doi: 10.1038/nm1001-1108. [DOI] [PubMed] [Google Scholar]
- Corti O, Hampe C, Koutnikova H, et al. The p38 subunit of the aminoacyl-tRNA synthetase complex is a Parkin substrate: linking protein biosynthesis and neurodegeneration. Hum Mol Genet. 2003;12:1427–1437. doi: 10.1093/hmg/ddg159. [DOI] [PubMed] [Google Scholar]
- Dodson MW, Guo M. Pink1, Parkin, DJ-1 and mitochondrial dysfunction in Parkinson’s disease. Curr Opin Neurobiol. 2007;17:331–337. doi: 10.1016/j.conb.2007.04.010. [DOI] [PubMed] [Google Scholar]
- Fitzgerald JC, Plun-Favreau H. Emerging pathways in genetic Parkinson’s disease: autosomal-recessive genes in Parkinson’s disease--a common pathway? FEBS J. 2008;275:5758–5766. doi: 10.1111/j.1742-4658.2008.06708.x. [DOI] [PubMed] [Google Scholar]
- Fournier M, Vitte J, Garrigue J, et al. Parkin deficiency delays motor decline and disease manifestation in a mouse model of synucleinopathy. PLoS ONE. 2009;4:e6629. doi: 10.1371/journal.pone.0006629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank-Cannon TC, Tran T, Ruhn KA, et al. Parkin deficiency increases vulnerability to inflammation-related nigral degeneration. J Neurosci. 2008;28:10825–10834. doi: 10.1523/JNEUROSCI.3001-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher MJ, Huang H, Grant ER, Lynch DR. The NR2B–specific interactions of polyamines and protons with the N-methyl-D-aspartate receptor. J Biol Chem. 1997;272:24971–24979. doi: 10.1074/jbc.272.40.24971. [DOI] [PubMed] [Google Scholar]
- Goldberg MS, Fleming SM, Palacino JJ, et al. Parkin-deficient mice exhibit nigrostriatal deficits but not loss of dopaminergic neurons. J Biol Chem. 2003;278:43628–43635. doi: 10.1074/jbc.M308947200. [DOI] [PubMed] [Google Scholar]
- Gu WJ, Corti O, Araujo F, et al. The C289G and C418R missense mutations cause rapid sequestration of human Parkin into insoluble aggregates. Neurobiol Dis. 2003;14:357–364. doi: 10.1016/j.nbd.2003.08.011. [DOI] [PubMed] [Google Scholar]
- Guo JF, Xiao B, Liao B, et al. Mutation analysis of Parkin, PINK1, DJ-1 and ATP13A2 genes in Chinese patients with autosomal recessive early-onset Parkinsonism. Mov Disord. 2008;23:2074–2079. doi: 10.1002/mds.22156. [DOI] [PubMed] [Google Scholar]
- Hampe C, Ardila-Osorio H, Fournier M, Brice A, Corti O. Biochemical analysis of Parkinson’s disease-causing variants of Parkin, an E3 ubiquitin-protein ligase with monoubiquitylation capacity. Hum Mol Genet. 2006;13:2059–2075. doi: 10.1093/hmg/ddl131. [DOI] [PubMed] [Google Scholar]
- Hedrich K, Eskelson C, Wilmot B, et al. Distribution, type, and origin of Parkin mutations: review and case studies. Mov Disord. 2004;19:1146–1157. doi: 10.1002/mds.20234. [DOI] [PubMed] [Google Scholar]
- Henn IH, Bouman L, Schlehe JS, et al. Parkin mediates neuroprotection through activation of IkappaB kinase/nuclear factor-kappaB signaling. J Neurosci. 2007;27:1868–1878. doi: 10.1523/JNEUROSCI.5537-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh DP, Scoles DR, Nguyen D, Pulst SM. The autosomal recessive juvenile Parkinson disease gene product, parkin, interacts with and ubiquitinates synaptotagmin XI. Hum Mol Genet. 2003;12:2587–2597. doi: 10.1093/hmg/ddg269. [DOI] [PubMed] [Google Scholar]
- Imai Y, Soda M, Inoue H, Hattori N, Mizuno Y, Takahashi R. An unfolded putative transmembrane polypeptide, which can lead to endoplasmic reticulum stress, is a substrate of Parkin. Cell. 2001;105:891–902. doi: 10.1016/s0092-8674(01)00407-x. [DOI] [PubMed] [Google Scholar]
- Imai Y, Soda M, Takahashi R. Parkin suppresses unfolded protein stress-induced cell death through its E3 ubiquitin-protein ligase activity. J Biol Chem. 2000;275:35661–35664. doi: 10.1074/jbc.C000447200. [DOI] [PubMed] [Google Scholar]
- Itier JM, Ibanez P, Mena MA, et al. Parkin gene inactivation alters behaviour and dopamine neurotransmission in the mouse. Hum Mol Genet. 2003;12:2277–2291. doi: 10.1093/hmg/ddg239. [DOI] [PubMed] [Google Scholar]
- Joazeiro CA, Weissman AM. RING finger proteins: mediators of ubiquitin ligase activity. Cell. 2000;102:549–552. doi: 10.1016/s0092-8674(00)00077-5. [DOI] [PubMed] [Google Scholar]
- Kao SY. Regulation of DNA repair by parkin. Biochem Biophys Res Commun. 2009;382:321–325. doi: 10.1016/j.bbrc.2009.03.048. [DOI] [PubMed] [Google Scholar]
- Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- Kitada T, Asakawa S, Matsumine H, Hattori N, Minoshima S, Shimizu N, Mizuno Y. Positional cloning of the autosomal recessive juvenile parkinsonism (AR-JP) gene and its diversity in deletion mutations. Parkinsonism Relat Disord. 1999;5:163–168. doi: 10.1016/s1353-8020(99)00032-2. [DOI] [PubMed] [Google Scholar]
- Kitada T, Asakawa S, Minoshima S, Mizuno Y, Shimizu N. Molecular cloning, gene expression, and identification of a splicing variant of the mouse parkin gene. Mamm Genome. 2000;11:417–421. doi: 10.1007/s003350010080. [DOI] [PubMed] [Google Scholar]
- Kitao Y, Imai Y, Ozawa K, et al. Pael receptor induces death of dopaminergic neurons in the substantia nigra via endoplasmic reticulum stress and dopamine toxicity, which is enhanced under condition of parkin inactivation. Hum Mol Genet. 2007;16:50–60. doi: 10.1093/hmg/ddl439. [DOI] [PubMed] [Google Scholar]
- Ko HS, Kim SW, Sriram SR, Dawson VL, Dawson TM. Identification of far up stream element binding protein-1 as an authentic parkin substrate. J Biol Chem. 2006;281:16193–16196. doi: 10.1074/jbc.C600041200. [DOI] [PubMed] [Google Scholar]
- Ko HS, von Coelln R, Sriram SR, et al. Accumulation of the authentic parkin substrate aminoacyl-tRNA synthetase cofactor, p38/JTV-1, leads to catecholaminergic cell death. J Neurosci. 2005;25:7968–7978. doi: 10.1523/JNEUROSCI.2172-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaVoie MJ, Cortese GP, Ostaszewski BL, Schlossmacher MG. The effects of oxidative stress on parkin and other E3 ligases. J Neurochem. 2007;103:2354–2368. doi: 10.1111/j.1471-4159.2007.04911.x. [DOI] [PubMed] [Google Scholar]
- LaVoie MJ, Ostaszewski BL, Weihofen A, Schlossmacher MG, Selkoe DJ. Dopamine covalently modifies and functionally inactivates parkin. Nat Med. 2005;11:1214–1221. doi: 10.1038/nm1314. [DOI] [PubMed] [Google Scholar]
- Li H, Guo M. Protein degradation in Parkinson disease revisited: it’s complex. J Clin Invest. 2009;119:442–445. doi: 10.1172/JCI38619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KL, Chew KC, Tan JM, et al. Parkin mediates nonclassical, proteasomal-independent ubiquitination of synphilin-1: implications for Lewy body formation. J Neurosci. 2005;25:2002–2009. doi: 10.1523/JNEUROSCI.4474-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucking CB, Durr A, Bonifati V, et al. Association between early-onset Parkinson’s disease and mutations in the parkin gene. French Parkinson’s Disease Genetics Study Group. N Engl J Med. 2000;342:1560–1567. doi: 10.1056/NEJM200005253422103. [DOI] [PubMed] [Google Scholar]
- Macedo MG, Verbaan D, Fang Y, et al. Genotypic and phenotypic characteristics of Dutch patients with early onset Parkinson’s disease. Mov Disord. 2009;24:196–203. doi: 10.1002/mds.22287. [DOI] [PubMed] [Google Scholar]
- Marin I, Lucas JI, Gradilla AC, Ferrus A. Parkin and relatives: the RBR family of ubiquitin ligases. Physiol Genomics. 2004;17:253–263. doi: 10.1152/physiolgenomics.00226.2003. [DOI] [PubMed] [Google Scholar]
- Mata IF, Lockhart PJ, Farrer MJ. Parkin genetics: one model for Parkinson’s disease. Hum Mol Genet. 2004;13:R127–R133. doi: 10.1093/hmg/ddh089. Spec No 1. [DOI] [PubMed] [Google Scholar]
- Moore DJ, West AB, Dikeman DA, Dawson VL, Dawson TM. Parkin mediates the degradation-independent ubiquitination of Hsp70. J Neurochem. 2008;105:1806–1819. doi: 10.1111/j.1471-4159.2008.05261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morett E, Bork P. A novel transactivation domain in parkin. Trends Biochem Sci. 1999;24:229–231. doi: 10.1016/s0968-0004(99)01381-x. [DOI] [PubMed] [Google Scholar]
- Murray IJ, Medford MA, Guan HP, Rueter SM, Trojanowski JQ, Lee VM. Synphilin in normal human brains and in synucleinopathies: studies with new antibodies. Acta Neuropathol (Berl) 2003;105:177–184. doi: 10.1007/s00401-002-0629-2. [DOI] [PubMed] [Google Scholar]
- Palacino JJ, Sagi D, Goldberg MS, Krauss S, Motz C, Wacker M, Klose J, Shen J. Mitochondrial dysfunction and oxidative damage in parkin-deficient mice. J Biol Chem. 2004;279:18614–18622. doi: 10.1074/jbc.M401135200. [DOI] [PubMed] [Google Scholar]
- Pawlyk AC, Giasson BI, Sampathu DM, et al. Novel monoclonal antibodies demonstrate biochemical variation of brain parkin with age. J Biol Chem. 2003;278:48120–48128. doi: 10.1074/jbc.M306889200. [DOI] [PubMed] [Google Scholar]
- Perez FA, Curtis WR, Palmiter RD. Parkin-deficient mice are not more sensitive to 6-hydroxydopamine or methamphetamine neurotoxicity. BMC Neurosci. 2005;6:71. doi: 10.1186/1471-2202-6-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez FA, Palmiter RD. Parkin-deficient mice are not a robust model of parkinsonism. Proc Natl Acad Sci U S A. 2005;102:2174–2179. doi: 10.1073/pnas.0409598102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Periquet M, Latouche M, Lohmann E, et al. Parkin mutations are frequent in patients with isolated early-onset parkinsonism. Brain. 2003;126:1271–1278. doi: 10.1093/brain/awg136. [DOI] [PubMed] [Google Scholar]
- Poorkaj P, Nutt JG, James D, Gancher S, Bird TD, Steinbart E, Schellenberg GD, Payami H. parkin mutation analysis in clinic patients with early-onset Parkinson disease. Am J Med Genet A. 2004;129A:44–50. doi: 10.1002/ajmg.a.30157. [DOI] [PubMed] [Google Scholar]
- Rankin CA, Joazeiro CA, Floor E, Hunter T. E3 ubiquitin-protein ligase activity of Parkin is dependent on cooperative interaction of RING finger (TRIAD) elements. J Biomed Sci. 2001;8:421–429. doi: 10.1007/BF02255952. [DOI] [PubMed] [Google Scholar]
- Ren Y, Zhao J, Feng J. Parkin binds to alpha/beta tubulin and increases their ubiquitination and degradation. J Neurosci. 2003;23:3316–3324. doi: 10.1523/JNEUROSCI.23-08-03316.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Chiba T, Nishiyama S, et al. Decline of striatal dopamine release in parkin-deficient mice shown by ex vivo autoradiography. J Neurosci Res. 2006;84:1350–1357. doi: 10.1002/jnr.21032. [DOI] [PubMed] [Google Scholar]
- Shimura H, Hattori N, Kubo S, et al. Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat Genet. 2000;25:302–305. doi: 10.1038/77060. [DOI] [PubMed] [Google Scholar]
- Shimura H, Schlossmacher MG, Hattori N, Frosch MP, Trockenbacher A, Schneider R, Mizuno Y, Kosik KS, Selkoe DJ. Ubiquitination of a new form of alpha-synuclein by parkin from human brain: implications for Parkinson’s disease. Science. 2001;293:263–269. doi: 10.1126/science.1060627. [DOI] [PubMed] [Google Scholar]
- Sriram SR, Li X, Ko HS, Chung KK, Wong E, Lim KL, Dawson VL, Dawson TM. Familial-associated mutations differentially disrupt the solubility, localization, binding and ubiquitination properties of parkin. Hum Mol Genet. 2005;14:2571–2586. doi: 10.1093/hmg/ddi292. [DOI] [PubMed] [Google Scholar]
- Staropoli JF, McDermott C, Martinat C, Schulman B, Demireva E, Abeliovich A. Parkin is a component of an SCF-like ubiquitin ligase complex and protects postmitotic neurons from kainate excitotoxicity. Neuron. 2003;37:735–749. doi: 10.1016/s0896-6273(03)00084-9. [DOI] [PubMed] [Google Scholar]
- Sun M, Latourelle JC, Wooten GF, et al. Influence of heterozygosity for parkin mutation on onset age in familial Parkinson disease: the GenePD study. Arch Neurol. 2006;63:826–832. doi: 10.1001/archneur.63.6.826. [DOI] [PubMed] [Google Scholar]
- Tan EK, Skipper LM. Pathogenic mutations in Parkinson disease. Hum Mutat. 2007;28:641–653. doi: 10.1002/humu.20507. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Suzuki T, Hattori N, Mizuno Y. Ubiquitin, proteasome and parkin. Biochim Biophys Acta. 2004;1695:235–247. doi: 10.1016/j.bbamcr.2004.09.026. [DOI] [PubMed] [Google Scholar]
- Thomas B, von Coelln R, Mandir AS, et al. MPTP and DSP-4 susceptibility of substantia nigra and locus coeruleus catecholaminergic neurons in mice is independent of parkin activity. Neurobiol Dis. 2007;26:312–322. doi: 10.1016/j.nbd.2006.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Um JW, Min DS, Rhim H, Kim J, Paik SR, Chung KC. Parkin ubiquitinates and promotes the degradation of RanBP2. J Biol Chem. 2006;281:3595–3603. doi: 10.1074/jbc.M504994200. [DOI] [PubMed] [Google Scholar]
- Ved R, Saha S, Westlund B, et al. Similar patterns of mitochondrial vulnerability and rescue induced by genetic modification of alpha-synuclein, parkin, and DJ-1 in Caenorhabditis elegans. J Biol Chem. 2005;280:42655–42668. doi: 10.1074/jbc.M505910200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Coelln R, Dawson VL, Dawson TM. Parkin-associated Parkinson’s disease. Cell Tissue Res. 2004a;318:175–184. doi: 10.1007/s00441-004-0924-4. [DOI] [PubMed] [Google Scholar]
- von Coelln R, Thomas B, Andrabi SA, et al. Inclusion body formation and neurodegeneration are parkin independent in a mouse model of alpha-synucleinopathy. J Neurosci. 2006;26:3685–3696. doi: 10.1523/JNEUROSCI.0414-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Coelln R, Thomas B, Savitt JM, Lim KL, Sasaki M, Hess EJ, Dawson VL, Dawson TM. Loss of locus coeruleus neurons and reduced startle in parkin null mice. Proc Natl Acad Sci U S A. 2004b;101:10744–10749. doi: 10.1073/pnas.0401297101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Ko HS, Thomas B, et al. Stress-induced alterations in parkin solubility promote parkin aggregation and compromise parkin’s protective function. Hum Mol Genet. 2005a;14:3885–3897. doi: 10.1093/hmg/ddi413. [DOI] [PubMed] [Google Scholar]
- Wang C, Tan JM, Ho MW, et al. Alterations in the solubility and intracellular localization of parkin by several familial Parkinson’s disease-linked point mutations. J Neurochem. 2005b;93:422–431. doi: 10.1111/j.1471-4159.2005.03023.x. [DOI] [PubMed] [Google Scholar]
- Winklhofer KF, Henn IH, Kay-Jackson PC, Heller U, Tatzelt J. Inactivation of parkin by oxidative stress and C-terminal truncations: a protective role of molecular chaperones. J Biol Chem. 2003;278:47199–47208. doi: 10.1074/jbc.M306769200. [DOI] [PubMed] [Google Scholar]
- Wu RM, Bounds R, Lincoln S, Hulihan M, Lin CH, Hwu WL, Chen J, Gwinn-Hardy K, Farrer M. Parkin mutations and early-onset parkinsonism in a Taiwanese cohort. Arch Neurol. 2005;62:82–87. doi: 10.1001/archneur.62.1.82. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Gao J, Chung KK, Huang H, Dawson VL, Dawson TM. Parkin functions as an E2-dependent ubiquitin- protein ligase and promotes the degradation of the synaptic vesicle-associated protein, CDCrel-1. Proc Natl Acad Sci U S A. 2000;97:13354–13359. doi: 10.1073/pnas.240347797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu XR, Maskri L, Herold C, Bader V, Stichel CC, Gunturkun O, Lubbert H. Non-motor behavioural impairments in parkin-deficient mice. Eur J Neurosci. 2007;26:1902–1911. doi: 10.1111/j.1460-9568.2007.05812.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.