Mechanisms that regulate microtubule length are thought to play crucial roles in a number of cellular processes including nuclear positioning, the establishment and maintenance of mitotic spindles, the regulation of mitotic chromosome alignment, process extension, and directed motility. However, the molecular control of microtubule length in cells is not well understood. Members of the kinesin-8 family of motors have been implicated in the direct regulation of microtubule length in cells (Gupta et al., 2006; Stumpff et al., 2008; Tischer et al., 2009; Varga et al., 2006), and a recent paper from Varga et al. (2009) indicates that the budding yeast kinesin-8, Kip3p, uses a surprising cooperative mechanism to monitor microtubule lengths and shorten them “as needed.”
Kip3p uses the energy of ATP to “walk” toward the plus-ends of microtubules for unusually long distances without releasing from the polymer. This ensures that a motor that lands anywhere on the microtubule lattice, even on long cellular microtubules, is likely to eventually reach the plus end, leading to an accumulation of motors there. Surprisingly, once a sufficient number of Kip3p molecules reach the plus-end of the microtubule, disassembly commences at a rate that is proportional to the concentration of Kip3p molecules. Due to the motor’s inherently high processivity, the depolymerization rate, by extension, is proportional to the length of the microtubule (Varga et al., 2006). Microtubule length-dependent activity is proposed as a mechanism by which the kinetic properties of the motor could be used to measure and “trim” microtubules to a length that is appropriate to the needs of the cell.
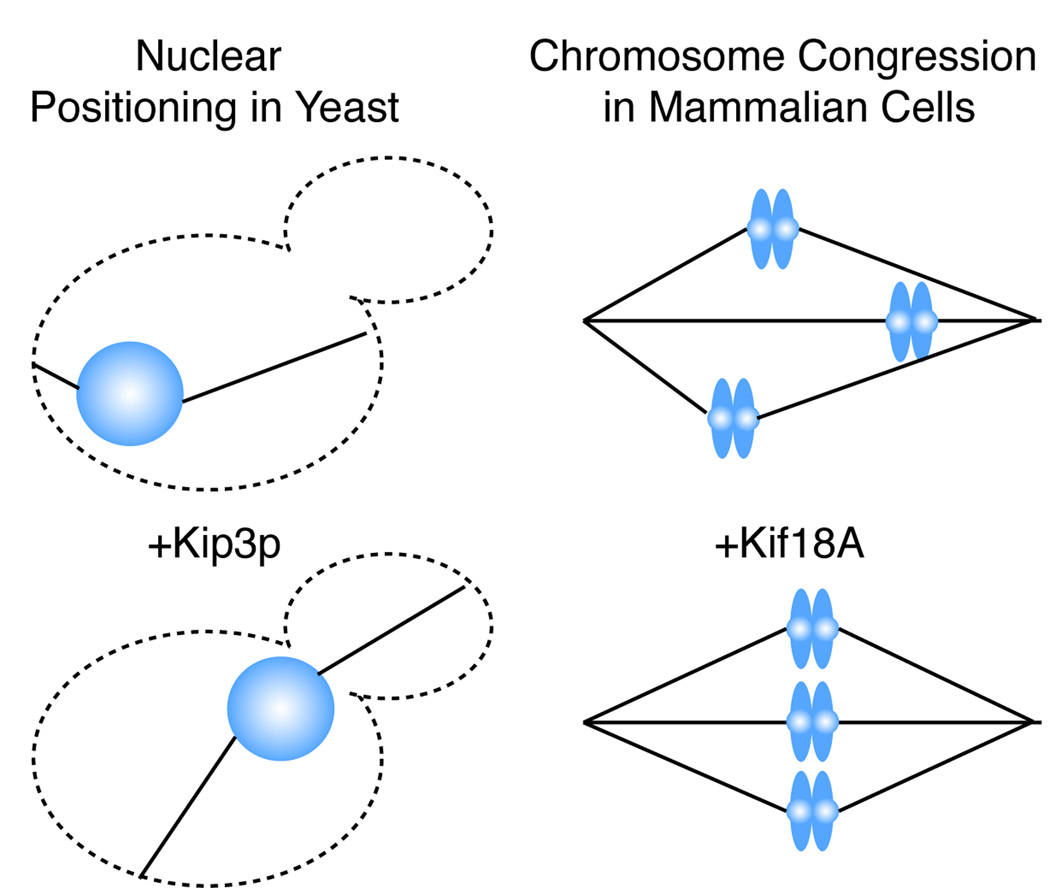
Regulation of microtubule length is consistent with many of the described cellular activities of Kip3p (Figure 1). The molecule is found associated with both astral (cytoplasmic) and spindle (nuclear) microtubules in S. cerevesiae. It is required to transport the preanaphase nucleus to the daughter bud site: a function dependent on the adjustment of astral microtubule length. Loss of Kip3p leads to excessively long, bent, anaphase spindles suggesting that length-dependent microtubule depolymerization activity is utilized at the plus ends of spindle midzone microtubules to limit the length of anaphase spindles (Straight et al., 1998). The mechanism by which Kip3p “knows” how to disassemble a microtubule, but only so far, to set spindle position and length is puzzling. An impressive study by Varga and colleagues (Varga, 2009) using TIRF microscopic assays in conjunction with purified GFP-Kip3p showed that once a motor reaches the end of the microtubule it will linger there until another motor arrives knocking off the terminal motor in conjunction with between 1–2 tubulin dimers. One motor molecule, consisting most likely of a dimer of two motor domains, removes only one or two tubulin dimers and then recycles rather slowly to rebind to the microtubule.
Figure 1.
Kinesin-8 motors are implicated in processes that lead to equalization of microtubule length. Both preanaphase nuclear positioning in S. cerevesiae and chromosome congression in mammalian cells tend to equalize cytoplasmic or kinetochore microtubule (solid black lines) lengths, respectively.
By comparison to kinesin-13 microtubule depolymerases, this is an extraordinarily weak microtubule depolymerizer. High processivity combined with exceptionally weak, cooperative removal of terminal tubulin dimers, are features that could enable Kip3p to establish a gradient of motors at the microtubule tip that serves as a readout of microtubule length. If, for example, the processive motors translocated to the end of the microtubule and then halted benignly, the gradient would disappear over time as the lattice became saturated and the positional information along the microtubule would be lost.
An important question to consider is what, structurally, the kinesin-8 family of motors does at the end of the microtubule? Why do kinesin-8 family members halt at the end of the microtubule while other processive, motile kinesins do not under physiological conditions. Significant insight into the mechanism by which the robust microtubule depolymerizers of the kinesin-13 family disassemble microtubules is obtained from electron microscopic studies showing that kinesin-13s stabilize a curled from of the microtubule protofilament in the ATP-bound state (Moores et al., 2002). These curls are clearly evident in the electron microscope. Mutational analysis of kinesin-13 motors demonstrates that removal of a tubulin dimer from the microtubule and the subsequent release of this dimer from the motor is tightly coupled to the ATP hydrolysis cycle (Wagenbach et al., 2008). In contrast, electron microscopic studies of kinesin-8 family molecules (klp5/6 from S. pombe) liberally decorating the microtubule lattice and protofilament extensions showed no evidence of protofilament bending (Grissom et al., 2009). In the unusual case of Kip3p, it is unknown whether loss of the terminal tubulin dimer is coupled to any part of the ATP hydrolysis cycle of the tubulin-associated motor. However, given the extraordinarily efficient means by which some kinesins (those of the kinesin-13 family for example) remove tubulin dimers from microtubule polymer, this haphazard tubulin removal begs the question: does this activity completely explain the mechanism used by Kip3p to modulate microtubule polymer length in cells?
To begin to answer this question one must revisit the effects that kinesin-8 motors have on “live”, dynamic microtubules. In S. cerevesiae, loss of Kip3p reduces both the catastrophe frequency and the rescue frequency of dynamic microtubules (Gupta et al., 2006). The catastrophe frequency is the frequency with which a microtubule will transition to depolymerization (one would expect a microtubule depolymerizer to increase this frequency). The rescue frequency is the frequency with which the microtubule will transition to polymerization. Kip3p can promote both catastrophes and, unexpectedly, rescues of microtubules in cells. Even more unexpectedly for a depolymerizer, Kip3p slows the disassembly rate of live microtubules in S. cerevesiae. This activity contrasts with the kinesin-13 depolymerase, MCAK, which promotes catastrophes and, not unexpectedly, reduces rescues in pure solutions of dynamic, unstabilized microtubules (Newton et al., 2004). By simultaneously increasing both rescues and catastrophes over time the kinesin-8 motor Kip3p has the potential to limit microtubules to a median steady state length. This activity is compatible with promoting the transport of the preanaphase nucleus to the bud site (DeZwaan et al., 1997). In this case long astral microtubules extending toward the bud will shorten and short astral microtubules in the mother cell will lengthen in order to center the nucleus near the bud neck. Similarly, in mammalian cells, the kinesin-8 motor Kif18A is restricted to kinetochore fibers during mitosis. Loss of the motor unexpectedly increases both the speed of chromosome movement and reduces the frequency with which chromosomes switch directions (an activity that simultaneously requires rescue and catastrophe of kinetochore fiber microtubules in the sister-kinetochores) (Stumpff et al., 2008). In contrast, adding excess Kif18A motor to the kinetochore fibers strongly restricts the movement of chromosomes to the spindle midline. Progressive restriction of the movement of the chromosomes, in effect, normalizes the length of all the kinetochore fiber bundles to the same median length, in turn promoting the congregation of chromosomes at the metaphase plate. A potent kinetochore-coupled depolymerase, such as the kinesin-13 MCAK, would be expected to increase the speed of chromosome movement (Wordeman et al., 2007) and increase the dynamics of live microtubules (Newton et al., 2004) as published studies have indicated. The fact that Kif18A seems to have the opposite effect is puzzling. However, if kinesin-8s can promote both catastrophe and rescue then this would be consistent with the observed suppression of chromosome movement (Stumpff et al., 2008) and suppression of live microtubule dynamics associated with these motors in vivo (Gupta et al., 2006). Presently, all signs point toward kinesin-13s and kinesin-8s as having opposite, even antagonistic roles at the kinetochore in mammalian cells. This is counterintuitive if both motors are depolymerizers. The Varga et al. (2009) study illustrates beautifully the likely source and mechanism of the catastrophe promoting activity of kinesin-8s. However, published work strongly suggests that a key component of kinesin-8 activity remains to be explored: the rescue promoting activity. Both of these activities are required to explain the role of kinesin-8s in living cells.
Acknowledgements
We thank Mike Wagenbach for helpful discussions and comments on the manuscript. This work was supported by the National Institutes of Health (grant GM69429 to L. Wordeman).
Literature Cited
- DeZwaan TM, Ellingson E, Pellman D, Roof DM. Kinesin-related KIP3 of Saccharomyces cerevisiae is required for a distinct step in nuclear migration. The Journal of cell biology. 1997;138:1023–1040. doi: 10.1083/jcb.138.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grissom PM, Fiedler T, Grishchuk EL, Nicastro D, West RR, McIntosh JR. Kinesin-8 from fission yeast: a heterodimeric, plus-end-directed motor that can couple microtubule depolymerization to cargo movement. Molecular biology of the cell. 2009;20:963–972. doi: 10.1091/mbc.E08-09-0979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta ML, Jr, Carvalho P, Roof DM, Pellman D. Plus end-specific depolymerase activity of Kip3, a kinesin-8 protein, explains its role in positioning the yeast mitotic spindle. Nature cell biology. 2006;8:913–923. doi: 10.1038/ncb1457. [DOI] [PubMed] [Google Scholar]
- Moores CA, Yu M, Guo J, Beraud C, Sakowicz R, Milligan RA. A mechanism for microtubule depolymerization by KinI kinesins. Mol Cell. 2002;9:903–909. doi: 10.1016/s1097-2765(02)00503-8. [DOI] [PubMed] [Google Scholar]
- Newton CN, Wagenbach M, Ovechkina Y, Wordeman L, Wilson L. MCAK, a Kin I kinesin, increases the catastrophe frequency of steady-state HeLa cell microtubules in an ATP-dependent manner in vitro. FEBS Lett. 2004;572:80–84. doi: 10.1016/j.febslet.2004.06.093. [DOI] [PubMed] [Google Scholar]
- Straight AF, Sedat JW, Murray AW. Time-lapse microscopy reveals unique roles for kinesins during anaphase in budding yeast. The Journal of cell biology. 1998;143:687–694. doi: 10.1083/jcb.143.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpff J, von Dassow G, Wagenbach M, Asbury C, Wordeman L. The kinesin-8 motor Kif18A suppresses kinetochore movements to control mitotic chromosome alignment. Dev Cell. 2008;14:252–262. doi: 10.1016/j.devcel.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischer C, Brunner D, Dogterom M. Force- and kinesin-8-dependent effects in the spatial regulation of fission yeast microtubule dynamics. Mol Syst Biol. 2009;5:250. doi: 10.1038/msb.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga V, Helenius J, Tanaka K, Hyman AA, Tanaka TU, Howard J. Yeast kinesin-8 depolymerizes microtubules in a length-dependent manner. Nature cell biology. 2006;8:957–962. doi: 10.1038/ncb1462. [DOI] [PubMed] [Google Scholar]
- Varga V, Leduc C, Bormuth V, Diez S, Howard J. Cooperative removal of the terminal tubulin dimer underlies length-dependent depolymerization by yeast kinesin-8. Dev Cell. 2009 doi: 10.1016/j.cell.2009.07.032. In press. [DOI] [PubMed] [Google Scholar]
- Wagenbach M, Domnitz S, Wordeman L, Cooper J. A kinesin-13 mutant catalytically depolymerizes microtubules in ADP. The Journal of cell biology. 2008;183:617–623. doi: 10.1083/jcb.200805145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wordeman L, Wagenbach M, von Dassow G. MCAK facilitates chromosome movement by promoting kinetochore microtubule turnover. The Journal of cell biology. 2007;179:869–879. doi: 10.1083/jcb.200707120. [DOI] [PMC free article] [PubMed] [Google Scholar]