Abstract
Cellular microtubules are rigid in comparison to other cytoskeletal elements [1, 2]. To facilitate cytoplasmic remodeling and timely responses to cell signaling events, microtubules depolymerize and repolymerize rapidly at their ends [3]. These dynamic properties are critically important for many cellular functions, such as spindle assembly, the capture and segregation of chromosomes during cell division and cell motility. Microtubule dynamics are spatially and temporally controlled in the cell by accessory proteins. Molecular motor proteins of the kinesin superfamily that act to destabilize microtubules play important roles in this regulation [4].
Keywords: kinesin, microtubule, MCAK, cytoskeleton
1 Introduction
The kinesin superfamily of proteins has recently been classified into fourteen subfamilies based on primary protein structure [5]. The proteins that comprise many of these subfamilies utilize the energy from ATP hydrolysis to transport along microtubules. However, at least two kinesin subfamilies, the kinesin-13s and kinesin-14s, contain motors with microtubule destabilizing activity [6–8]. Detailed mechanistic studies of the microtubule destabilizing activity exhibited by these motors have significantly contributed to our understanding of their cellular functions.
In this chapter, we discuss assays for measuring the microtubule destabilizing activity of kinesins in both a purified system and within the context of mammalian cells. We first describe a turbidity assay that can be used to determine whether a kinesin is a bona fide microtubule destabilizer and to quantitatively compare the activities of different kinesin preparations. In addition, we report a fluorescence microscopy assay for analyzing the effect of kinesin expression on microtubule stability in cells. We have applied these approaches in combination with site-directed mutagenesis to study the microtubule destabilizing activity of MCAK, one of the founding members of the kinesin-13 family [9–12]. For many of these assays MCAK protein or constructs provides an admirable positive control for microtubule destabilizing activity.
2 Materials
2.1 Materials for Microtubule Turbidity Assay
Purified kinesin: Recombinant kinesin should be expressed in bacteria or insect cells and purified using standard chromatography techniques. Elute the kinesin from the final purification column in motor storage buffer and store frozen in working aliquots.
Motor storage buffer: 300 mM KCl, 1 mM MgCl2, 200 mM imidazole-HCl pH 7.0, 5 µM ATP and 20% glycerol in ddH2O.
Tubulin: Purified bovine brain tubulin can be purchased from Cytoskeleton, Inc (catalog number T237) as a 10 mg/ml (100 µM) stock solution.
GMP-CPP: GMP-CPP can be obtained from Jena Biosciences as a 10 mM stock.
BRB80 buffer: 80 mM piperazine-N,N-bis[2-ethanesulfonic acid] (PIPES), 1 mM MgCl2, 1mM EGTA (pH 6.8 with potassium hydroxide). Sterile filter and store at 4°C. BRB80 can also be made and stored as a 5× stock.
Poly-L-lysine coated glass coverslips: Coat glass coverslips in bulk by soaking in a 0.1% solution of poly-L-lysine (Sigma) for 15 minutes. Remove coverslips and allow them to dry individually on a clean Kimwipe or paper towel. Store in a small covered dish or beaker at room temperature until use.
Antibody dilution buffer (Abdil): 1% BSA (IgG-free, protease-free), 0.1% Triton ×-100 and 0.02% sodium azide in TBS (20 mM Tris-HCL pH 7.4, 150 mM NaCl).
Nitrocellulose membranes: Use 25 mm diameter nitrocellulose membranes with 0.45 µm pores.
A screen or frit support that allows suction to be applied to filters while collecting flow-through for radioactive waste disposal will be needed.
Mg-ATP: A stock solution of 100 mM ATP and 100 mM MgCl2 in water should be sterile filtered and stored at −20°C in working aliquots.
α32P-ATP: α32P-ATP stock should be purchased from a reputable source as a 10mCi/ml stock.
Turbidity Buffer: BRB80 buffer supplemented with 250 µM Mg-ATP, 75 mM KCl, 1 mM DTT and 200 µg/ml BSA
2.2 Materials for In Vivo Microtubule Depolymerization Assay
CHO cell culture: CHO cells are cultured using standard sterile tissue culture techniques in Minimal Essential Medium Alpha (MEMα) medium supplemented with 10% fetal bovine serum and grown in a 37°C, 5% CO2 incubator.
Kinesin expression plasmid: Choose a well-characterized vector that expresses the fluorescent protein of choice (see Note 1).
Transfection reagent: Any commercially available transfection reagent or procedure suitable for transfecting plasmid DNA into mammalian cells will work.
Glass coverslip preparation: Wash 12 mm diameter glass coverslips in 1N HCl at room temperature for 2 hours, stirring occasionally. Rinse 5 times with ddH2O. Wash 5 times with 95% ethanol. Store in 70% ethanol. Flame coverslips in laminar flow hood prior to use.
Phosphate-buffered saline (PBS): Prepare a 1× stock solution of 0.2 M monobasic sodium phosphate, 0.2 M dibasic sodium phosphate and 150 mM NaCl in ddH2O. PBS can be prepared as a 10× stock and stored at room temperature after autoclaving.
Mounting Media: Use an anti-fade cell mounting media containing a DNA stain such as DAPI.
Image analysis software: An image analysis software package that allows quantification of fluorescence intensity in user-defined regions of an image will be needed. We routinely use ImageJ, which is available for free download at http://rsb.info.nih.gov/ij. The specific commands given for quantifying fluorescence in section 3.2.3 are for the Macintosh version of ImageJ 1.33.
3 Methods
3.1 In Vitro Microtubule Turbidity Assay
Assembled microtubules cause detectable light scattering at 350 nm. This characteristic turbidity can be utilized to assay kinesin-mediated microtubule disassembly [12]. The turbidity assay is best performed using GMP-CPP stabilized microtubules. Longer taxol stabilized microtubules tend to be bundled by added protein, and bundling of microtubules will artificially contribute to light scattering. We describe here how to prepare GMP-CPP microtubules and carry out the turbidity assay. To facilitate accurate comparisons between different preparations or variants of depolymerizing kinesins using the turbidity assay, we also describe a method used to determine the concentration of active motors competent to release ADP and bind ATP.
3.1.1 Preparation of GMP-CPP Microtubules
Tubulin has a higher affinity for GTP than it does for GMP-CPP [13]. Thus, in order to grow microtubules with an uninterrupted GMP-CPP lattice, a method is required for removing the GTP that is commonly present in solution with purchased tubulin. To accomplish this, we recommend cycling the tubulin once in GMPCPP as noted in steps 1–4.
Prepare a 200 µl reaction on ice containing 20 µM tubulin and 1 mM GMP-CPP diluted in BRB80. Incubate on ice for 10 min. Polymerize reaction at 37°C for 30 min.
Sediment the reaction at 150,000 × g in a small volume ultracentrifuge at room temperature for 10 min.
Resuspend the pellet in BRB80 and disassemble microtubules on ice for 20 min. Sediment at 13,000 × g for 2 minutes at 2–4°C to remove insoluble protein.
Remove the supernatant and dilute with BRB80 to achieve a final concentration of 20 µM tubulin. Add GMP-CPP to 1 mM final concentration and incubate on ice for 10 min.
Polymerize the reaction at 37°C for 30 min. Sediment the microtubules at 150,000 × g for 10 minutes at 25°C.
Resuspend the resulting pellet to a tubulin concentration of 89 µM. The original tubulin concentration prior to assembly can be determined by measuring the absorbance (A) at 280nm (see Note 2).
Aliquots of these cycled GMP-CPP assembled microtubules can be frozen in liquid nitrogen and stored at −80°C. A fresh aliquot must be thawed for each experiment because assays for depolymerase activity are sensitive to the concentration of microtubule ends and aliquots of assembled GMP-CPP microtubules will undergo end-to-end annealing over time. Thaw one aliquot and calibrate it for microtubule length and end concentration as described in (Section 3.1.2).
3.1.2 Determination of Microtubule-End Concentration
Spin down 100 µL of GMP-CPP microtubule solution in a small volume ultracentrifuge at 150,000 × g onto poly-L-lysine coated glass coverslips (see Note 3).
Fix coverslips in methanol, rehydrate by rinsing in PBS and block with 20% donkey serum
Label with anti-tubulin primary antibody and appropriate fluorescent secondary antibody.
Mount coverslips microtubule side down in a 1–2 µl drop of anti-fade mounting media.
Record images of fluorescently labeled microtubules from multiple locations on the coverslip using a widefield fluorescent microscope equipped with a CCD camera.
Measure the average microtubule lengths using an image analysis program. Scale by the magnification factor of the microscope and lens used to convert the microtubule lengths to microns. We find that GMP-CPP microtubules prepared as described in section 3.1.1 are approximately 2.5 microns in length.
Calculate the microtubule end concentration in the GMP-CPP stock solution (see Note 4).
3.1.3 Determination of Kinesin ATP-Binding Site Concentration
Wash nitrocellulose filters in 0.4 N KOH for 10 minutes at room temperature, rinse 5 times with ddH20 and 2 times with BRB80 (see Note 5). Let stand at room temperature for at least 60 minutes.
Supplement motor storage buffer with 5 µM cold ATP and 110 nM α32P-ATP, correcting for 32P decay (see Note 6).
Thaw protein to be assayed on ice. Mix well 2 µl protein with 18 µl solution from step 2. This solution is the assay mixture. Also prepare a buffer blank with 3 µl motor storage buffer + 27 µl solution from step 2. Incubate at room temperature for10–20 minutes and then place on ice (see Note 7).
Apply suction to a washed nitrocellulose filter. Spot 5 µl of assay mixture from step 3 on the filter and immediately wash with 0.5 ml ice-cold BRB80. Remove filter with forceps and place in a scintillation vial (see Note 8).
Apply 1 µl of the buffer blank solution from step 3 to each of three dry filters and without filtering or washing, place these in scintillation vials. These filters will provide the total signal for 1 µl of 100 nM α32P-ATP.
Add scintillation cocktail to all filters and count each for 1 minute. Calculate mean values for each reaction.
Calculate ATP-binding activity as follows: Subtract the counts per minute (cpm) value for the washed buffer blank from the protein assay’s value to get blank-subtracted cpm. Multiply this value by the dilution factor used when setting up the binding reaction (20 µl / 2 µl = 10×) and any intermediate dilution made of the protein stock. Next divide by the number of microliters spotted on each filter (5 µl) to give the calculated signal per microliter of protein stock solution. Then multiply by the ratio of cold: hot ATP in step 4 (50×), divide by the mean cpm from the unwashed filters prepared in step 5 and multiply by the concentration of hot ATP in step 5 (100 nM). For an example calculation, see Note 9.
3.1.4 Microtubule Turbidity Assay
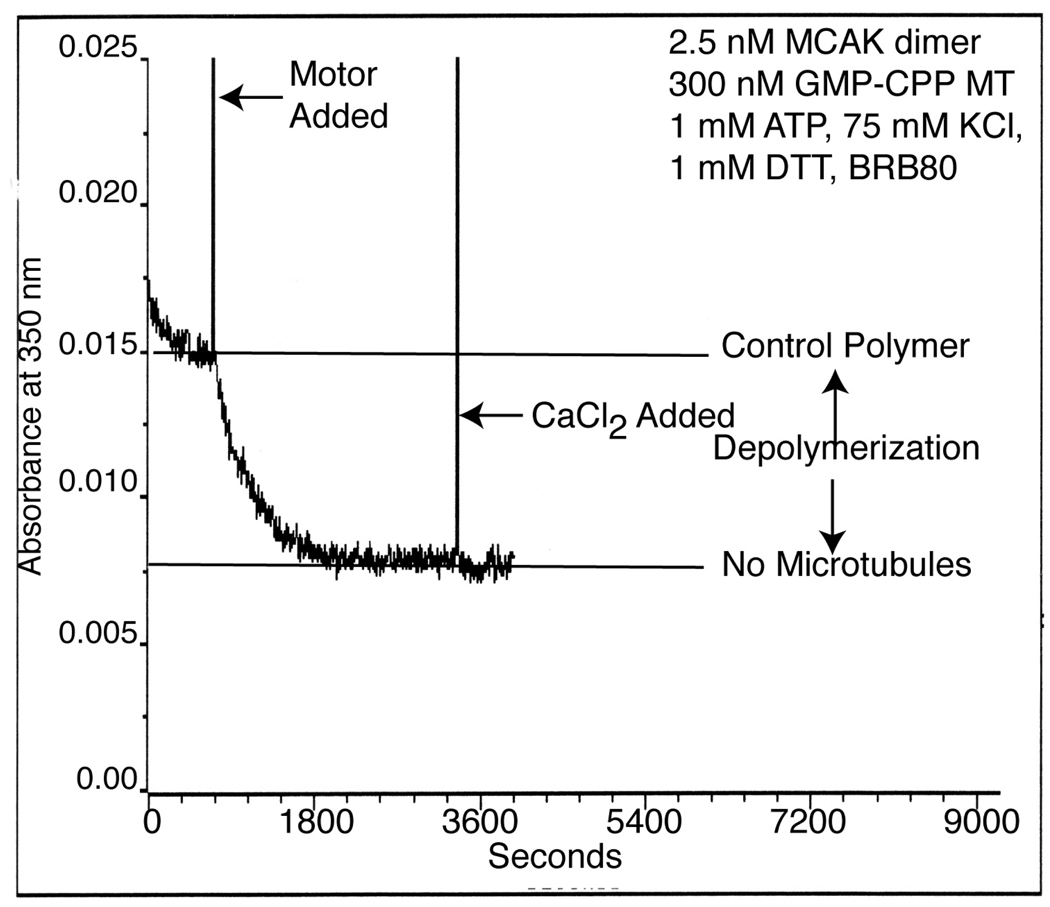
Thaw an aliquot of GMP-CPP tubulin and dilute it to a concentration of 300 nM in Turbidity Buffer. The ionic strength of this solution is optimal for microtubule depolymerization assays using MCAK. Equilibrate the solution to 23°C in a quartz cuvette prior to the addition of motor.
Add purified motor that has been calibrated for ATP binding (section 3.1.3) and mix rapidly using a P1000 pipettor (see Note 10).
Monitor turbidity in real time by measuring the absorbance at 350 nm at 5 second intervals using a spectrophotometer.
Measure the final extent of depolymerization by the addition of 5 mM CaCl2 to the reaction (see Note 11).
Normalize turbidity traces from absorbance units to tubulin polymer concentration using a standard curve (see Note 12 and example in Figure 1).
Figure 1.
Example of a typical raw data trace of MCAK-dependent microtubule depolymerization as assayed by turbidity (or absorbance at 350nm).
3.2 In Vivo Assay for Microtubule Depolymerization
In addition to measuring microtubule depolymerization in vitro, it is important to determine the ability of a kinesin to depolymerize microtubules in vivo. This in vivo depolymerization assay depends in large part on the tubulin autoregulatory system. When microtubules are depolymerized by nocodazole or transiently transfected MCAK, the decrease of microtubule polymer and increase of tubulin dimer causes the degradation of β-tubulin mRNA [10, 14, 15]. Over a period of 24 hours, this causes an overall reduction of tubulin in the cell, which can be detected by a decrease in tubulin immunofluorescence. To measure microtubule depolymerization of a known or suspected depolymerizing kinesin in vivo, tubulin immunofluorescence intensity can be measured after kinesin expression and compared to tubulin immunofluorescence in control cells [10]. We utilize Chinese hamster ovarian (CHO) cells for this assay because they consistently express transfected DNA quickly (i.e. within 12–24 hours).
3.2.1 Preparation of Cultured Cells
CHO cells should be grown using standard sterile cell culture techniques to 70–90% confluency (see Note 13) and then plated onto glass coverslips (see Note 14).
Transfect cells with a recombinant expression plasmid that will result in a high level of GFP-kinesin fusion protein expression (see Note 1). As a negative control, an empty EGFP vector should be transfected into cells on another coverslip in parallel. Transfection can be carried out with any procedure suited for transfection of mammalian cells.
Incubate cells in a 37°C, 5% CO2 incubator for 24 hours before fixation.
3.2.2 Fixing and Staining Cells for Tubulin
Fix cells with 1% para-formaldehyde in −20°C methanol for 10 minutes (see Note 15).
Wash cells twice with PBS for 5 minutes each at room temperature.
Block for 30 minutes in Abdil supplemented with 20% donkey serum (see Note 16).
Briefly rinse coverslips in PBS.
Stain cells with primary antibodies against alpha-tubulin (mouse anti-DM1α, Sigma) diluted 1:100 in Abdil. Incubate with rotation for 1 hour.
Wash coverslips for 5 minutes in PBS.
Incubate cells with Texas Red conjugated donkey anti-mouse secondary antibody (Jackson Immunoresearch Laboratories, Inc.) diluted 1:75 in Abdil. Incubate with rotation for 1 hour.
Wash coverslips for 5 minutes in PBS.
On a slide, lay coverslips cell-side down in 4 µl anti-fade mounting media containing a DNA stain such as DAPI.
Seal coverslip edges with nail polish.
Once the nail polish is dry (10–15 minutes), slides can be viewed immediately or stored at −20°C.
3.2.3 Quantification of Fluorescence
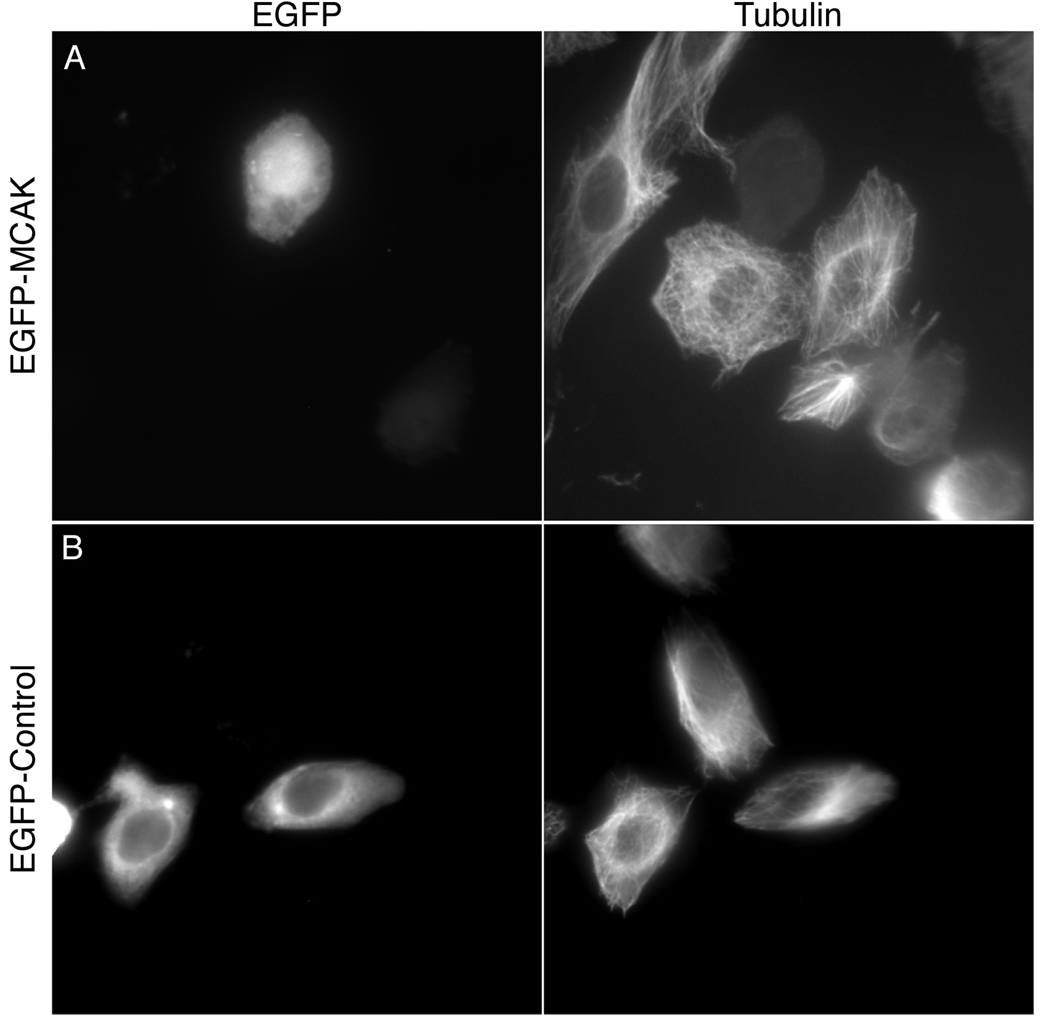
Once the coverslips are fixed, stained and mounted, the cells are ready to be assayed using a widefield fluorescent microscope equipped with a CCD camera. For this analysis, choose only interphase cells that are well spread on the coverslip, and that have similar levels of GFP expression (examples in Figure 2).
Starting with untransfected cells, determine an exposure length for tubulin in a normal cell (see Note 17). Use this exposure length for all tubulin images analyzed. (With our antibodies and fixation/staining procedure typical exposure times are between 200–400 ms).
Determine the expression level of kinesin that gives complete microtubule depolymerization without protein aggregation (clumps of GFP). Use this exposure length for all GFP images.
Acquire images of at least 50 transfected cells per experiment for each construct being tested. Make sure that for each transfected cell imaged, there is an untransfected cell in the same frame, which has similar size and shape. Depending on transfection efficiency and cell density, one to three coverslips per condition may be needed to find enough useable cells. Organize image files in a way that is optimized for analysis later (see Note 18).
Once all the images are acquired, open them in ImageJ or similar image analysis software. (File-Import-Image Sequence-Image Folder, then select the first image in the folder and click Open.) Select Reduce to 8-bit and open all files in the folder (see Note 19). Set-up measurement parameters to collect the following data: area of the cell, average fluorescence intensity and maximum fluorescence intensity (see Note 20).
For the first cell, select the area of the cell (use the freehand tool in ImageJ) while viewing either the GFP image or the tubulin image, whichever gives a better outline of the cell. Go to the GFP image and measure the area of the cell and GFP fluorescence intensity, then measure the same parameters in the tubulin image (see Note 21).
Repeat step 7 for an untransfected cell in the same frame.
Copy the mean GFP and tubulin pixel intensity for both the transfected and untransfected cell and paste these values into a spreadsheet.
Divide the tubulin fluorescence of the transfected cell by the tubulin fluorescence of the untransfected cell to get a tubulin fluorescence ratio.
Repeat steps 8– 11 for the rest of the photos for a given construct and repeat the entire procedure for each expression construct being analyzed.
Calculate the average and standard deviation of the GFP intensities. Determine the average and standard deviation of the tubulin fluorescent ratios calculated in step 9. Comparison of the data obtained from GFP expressing and GFP-kinesin expressing cells will indicate the effectiveness of microtubule depolymerization (see Note 22).
Figure 2.
Example of in vivo microtubule depolymerization by MCAK. CHO cells were fixed and stained for tubulin 24 hours after transfection with either EGFP-MCAK (A) or EGFP-control (B) plasmids. (A) The average pixel intensity for the tubulin immunofluorescence of the MCAK transfected cell (TF) is 24.0 arbitrary units. The average pixel intensity for the tubulin immunofluorescence of a neighboring untransfected cell (UTF) is 48.9. The tubulin intensity ratio (TF/UTF) is 0.49. The average GFP intensity is 137.3. (B) The average pixel intensity for the tubulin immunofluorescence of the control transfected cell is 102.5. The average pixel intensity of a neighboring untransfected cell is 114.4. The tubulin intensity ratio is 0.90. The average GFP intensity is 100.8.
Acknowledgements
We gratefully acknowledge the excellent work of Dave Coy, Jo Howard, Andy Hunter, Yulia Ovechkina, and Todd Maney in the development of these assays. This work was supported by a National Science Foundation NCI IGERT grant to J. Cooper, a National Institutes of Health Predoctoral training grant (GM07270) to K. Rankin and a National Institutes of Health grant (GM69429) to L. Wordeman.
Footnotes
We routinely use pEGFP-C1 (Clontech). This vector drives strong, quick expression of EGFP fusion proteins under the control of a CMV promoter. The EGFP coded in the vector folds quickly and bleaches slowly, which allows for quick expression and stable photographing, respectively. Additionally, this vector has an f1 single strand DNA origin for replication that is useful for fast production of multiple mutant transgenes. N-terminal versus C-terminal placement of the transgene relative to the fluorescent protein should be considered and tested to determine whether placement has an effect on expression and function.
The original tubulin concentration prior to assembly can be measured by measuring the absorbance (A) at 280nm assuming an extinction coefficient for tubulin of 115,000 mol−1cm−1. Using this value, a 1 mM solution of tubulin would possess an A280=1.15. This extinction coefficient is noted in Hyman et al. 1991 [16].
Coverslips are mounted inside the centrifuge tube. Depending on the type of centrifuge tubes used, it may be necessary to machine a small fitting for mounting the coverslips securely at the bottom of the tube. It is important that the coverslip fits securely into the tube to ensure all microtubules are pelleted onto the glass. We routinely spin microtubules onto 5 mm diameter coverslips in 8 mm (outer diameter) centrifuge tubes using a Beckman airfuge. As an alternative to a machined fitting for the centrifuge tubes, a small amount of epoxy can be placed in the bottom of the tube and then spun while the epoxy is curing. This will generate a flat angled surface on which to lay the coverslip.
Measure the average number of microtubules per frame, multiply this number by the total coverslip surface area, and divide by the surface area of one frame. This gives the total number of microtubules on the coverslip. Next, divide by the initial starting volume to determine the initial microtubule concentration. Multiply this number by two to determine the microtubule end concentration.
For a typical ATP binding assay, prepare three nitrocellulose filters per protein prep being assayed and three for the buffer blank control. Additionally, we routinely wash a few spare membranes in case some break during the procedure.
α32P-ATP, 800 Ci/mmol, 10 mCi/ml or 12.5 µM α32P-ATP on reference date. Concentration on date of assay is A(t) = Ao * e−lt where Ao = concentration on reference date, t = number of days after reference date, and l = ln2(1/half-life) or 0.0485 day−1 for 32P.
Note that the hot: cold ATP ratio in this solution is assumed to be 1:50. If the stock concentration of active protein much exceeds 1 µM, the cold ATP carried by the protein will be significant, and a dilution of protein to 1–2 µM in motor storage buffer + 5 µM ATP should be made prior to the assay
Each step in this process should be done as quickly and consistently as possible. Process 3 filters for each assay mixture or buffer blank.
- 2 µL MCAK, average of 3 × 5 µL samples: 20,667 cpm
- blank reaction, average of 3 × 5 µL samples: 1587 cpm
- 100 nM hot ATP, average of 3 × 1 µL samples: 198,690 cpm
- 20,667 cpm – 1587 cpm = 19,080 cpm × (20 µL / 2 µL) × (1 / 5 µL) × 50 cold:hot ATP = 1.91 × 106 cpm / µL corrected for sample volumes and dilution
- 1.91 × 106 cpm / µL × (1 × 10−7 M × 1 µL / 198,690 cpm) = 0.96 µM active ATP binding sites.
A typical test reaction would employ 300 nM GMP-CPP microtubules and 3 nM full-length MCAK dimmer, but optimal motor concentration of other kinesins should be determined empirically.
Addition of CaCl2 to a final concentration of 5 mM will disassemble all GMP-CPP microtubule polymer [17]. This is a useful control to determine the A350 of fully disassembled microtubule polymer. When modest concentrations of active motor are added to microtubules, destabilizers like MCAK will disassemble the microtubules to a new steady state polymer concentration. It is unwise to assume that the lowest A350 reading represents fully disassembled microtubules.
Turbidity traces can be converted to tubulin polymer concentration using a standard curve in which the zero time point corresponds to the concentration of tubulin in the assembled microtubules (i.e. 300 nM) and the turbidity after complete disassembly in CaCl2 is equal to 0 nM assembled tubulin.
Confluency refers to the density of the cells adhered to the plate. For example, 100% confluency means cells that have adhered to and spread out on the plate are dense enough that they are touching each other and taking up 100% of the plate surface.
One day prior to transfection, we plate CHO cells in a 24-well plate containing one acid-washed 12 mm coverslip in each well. Under these conditions, seed cells at a density of 3.5 × 104 cells per well in 500 µl culturing media. If cells are seeded too densely, analysis of tubulin fluorescence will be difficult because cells will not spread properly (section 3.2.3).
We fix and wash our cells in 250 ml plastic beakers containing 100 ml of fix solution or PBS. The coverslips are held vertically in a coverglass staining rack and the cell containing side of each coverslip is carefully tracked through the procedure.
We block and stain cells by placing coverslips cell-side up on a piece of parafilm laid flat inside a 25 mm Petri dish. Damp paper towels are rolled and placed around the inside of the dish to provide humidity and prevent drying during incubation periods.
Choose cells that do not touch any other cells for this analysis because fluorescence from contacting cells can complicate the quantification procedure. Also, choose cells that provide a good dynamic range with respect to fluorescence intensity and avoid cells that contain areas of fluorescence saturation (at or above the maximal intensity value).
We routinely take separate images for microtubules and GFP, and number the files sequentially (e.g. Kif2C_01_GFP; Kif2C__01_MT, where “Kif2C” is the name of the kinesin being analyzed). We keep all the files for each construct in a single folder. This organization is optimal for importing the files as a sequence into Image J for analysis.
All the photos in the folder will open as a stack, with the GFP channel first and the microtubule channel second. To go forward within the stack, use the period key (.), and to go backward use the comma key (,).
In ImageJ, set parameters by opening Analyze-Set measurements. Click the boxes for “Area”, “Mean Gray Value”, “Min and Max Gray Value” and “Display Label.”
In ImageJ, take measurements with the Analyze-Measure command. Toggle between the GFP and tubulin images to take measurements using the same cell outline.
The average GFP intensities should be statistically the same for all constructs in the assay by a student’s t-test analysis. Cells transfected with GFP should give a microtubule fluorescence ratio of approximately one, while cells expressing a microtubule depolymerizing kinesin should give a tubulin fluorescence ratio that is less than one (for example see Figure 2).
References
- 1.Felgner H, Frank R, Schliwa M. Flexural rigidity of microtubules measured with the use of optical tweezers. J Cell Sci. 1996;109(Pt 2):509–516. doi: 10.1242/jcs.109.2.509. [DOI] [PubMed] [Google Scholar]
- 2.Gittes F, Mickey B, Nettleton J, Howard J. Flexural rigidity of microtubules and actin filaments measured from thermal fluctuations in shape. J Cell Biol. 1993;120:923–934. doi: 10.1083/jcb.120.4.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Desai A, Mitchison TJ. Microtubule polymerization dynamics. Annu Rev Cell Dev Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- 4.Wordeman L. Microtubule-depolymerizing kinesins. Curr Opin Cell Biol. 2005;17:82–88. doi: 10.1016/j.ceb.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 5.Lawrence CJ, Dawe RK, Christie KR, Cleveland DW, Dawson SC, Endow SA, Goldstein LS, Goodson HV, Hirokawa N, Howard J, Malmberg RL, McIntosh JR, Miki H, Mitchison TJ, Okada Y, Reddy AS, Saxton WM, Schliwa M, Scholey JM, Vale RD, Walczak CE, Wordeman L. A standardized kinesin nomenclature. J Cell Biol. 2004;167:19–22. doi: 10.1083/jcb.200408113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chu HM, Yun M, Anderson DE, Sage H, Park HW, Endow SA. Kar3 interaction with Cik1 alters motor structure and function. Embo J. 2005;24:3214–3223. doi: 10.1038/sj.emboj.7600790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sproul LR, Anderson DJ, Mackey AT, Saunders WS, Gilbert SP. Cik1 targets the minus-end kinesin depolymerase kar3 to microtubule plus ends. Curr Biol. 2005;15:1420–1427. doi: 10.1016/j.cub.2005.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desai A, Verma S, Mitchison TJ, Walczak CE. Kin I kinesins are microtubule-destabilizing enzymes. Cell. 1999;96:69–78. doi: 10.1016/s0092-8674(00)80960-5. [DOI] [PubMed] [Google Scholar]
- 9.Wordeman L, Mitchison TJ. Identification and partial characterization of mitotic centromere-associated kinesin, a kinesin-related protein that associates with centromeres during mitosis. J Cell Biol. 1995;128:95–104. doi: 10.1083/jcb.128.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ovechkina Y, Wagenbach M, Wordeman L. K-loop insertion restores microtubule depolymerizing activity of a "neckless" MCAK mutant. J Cell Biol. 2002;159:557–562. doi: 10.1083/jcb.200205089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moore A, Wordeman L. C-terminus of mitotic centromere-associated kinesin (MCAK) inhibits its lattice-stimulated ATPase activity. Biochem J. 2004;383:227–235. doi: 10.1042/BJ20040736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hunter AW, Caplow M, Coy DL, Hancock WO, Diez S, Wordeman L, Howard J. The kinesin-related protein MCAK is a microtubule depolymerase that forms an ATP-hydrolyzing complex at microtubule ends. Mol Cell. 2003;11:445–457. doi: 10.1016/s1097-2765(03)00049-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hyman AA, Salser S, Drechsel DN, Unwin N, Mitchison TJ. Role of GTP hydrolysis in microtubule dynamics: information from a slowly hydrolyzable analogue, GMPCPP. Mol Biol Cell. 1992;3:1155–1167. doi: 10.1091/mbc.3.10.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonzalez-Garay ML, Cabral F. alpha-Tubulin limits its own synthesis: evidence for a mechanism involving translational repression. J Cell Biol. 1996;135:1525–1534. doi: 10.1083/jcb.135.6.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cleveland DW, Lopata MA, Sherline P, Kirschner MW. Unpolymerized tubulin modulates the level of tubulin mRNAs. Cell. 1981;25:537–546. doi: 10.1016/0092-8674(81)90072-6. [DOI] [PubMed] [Google Scholar]
- 16.Hyman A, Drechsel D, Kellogg D, Salser S, Sawin K, Steffen P, Wordeman L, Mitchison T. Preparation of modified tubulins. Methods Enzymol. 1991;196:478–485. doi: 10.1016/0076-6879(91)96041-o. [DOI] [PubMed] [Google Scholar]
- 17.O'Brien ET, Salmon ED, Erickson HP. How calcium causes microtubule depolymerization. Cell Motil Cytoskeleton. 1997;36:125–135. doi: 10.1002/(SICI)1097-0169(1997)36:2<125::AID-CM3>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]