Abstract
Objectives
Clinical studies show that Asians (ASN) are more susceptible to toxicities associated with platinum-containing regimens. We hypothesized that studying ASN as an `enriched phenotype' population could enable the discovery of novel genetic determinants of platinum susceptibility.
Methods
Using well-genotyped lymphoblastoid cell lines from the HapMap, we determined cisplatin and carboplatin cytotoxicity phenotypes (IC50s) for ASN, Caucasians (CEU), and Africans (YRI). IC50s were used in genome-wide association studies.
Results
ASN were most sensitive to platinums, corroborating clinical findings. ASN genome-wide association studies produced 479 single-nucleotide polymorphisms (SNPs) associating with cisplatin susceptibility and 199 with carboplatin susceptibility (P<10−4). Considering only the most significant variants (P< 9.99 × 10−6), backwards elimination was then used to identify reduced-model SNPs, which robustly described the drug phenotypes within ASN. These SNPs comprised highly descriptive genetic signatures of susceptibility, with 12 SNPs explaining more than 95% of the susceptibility phenotype variation for cisplatin, and eight SNPs approximately 75% for carboplatin. To determine the possible function of these variants in ASN, the SNPs were tested for association with differential expression of target genes. SNPs were highly associated with the expression of multiple target genes, and notably, the histone H3 family was implicated for both drugs, suggesting a platinum-class mechanism. Histone H3 has repeatedly been described as regulating the formation of platinum-DNA adducts, but this is the first evidence that specific genetic variants might mediate these interactions in a pharmacogenetic manner. Finally, to determine whether any ASN-identified SNPs might also be important in other human populations, we interrogated all 479/199 SNPs for association with platinum susceptibility in an independent combined CEU/YRI population. Three unique SNPs for cisplatin and 10 for carboplatin replicated in CEU/YRI.
Conclusion
Enriched `platinum susceptible' populations can be used to discover novel genetic determinants governing interindividual platinum chemotherapy susceptibility.
Keywords: interethnic differences, pharmacogenomic susceptibility, platinum chemotherapy
Introduction
Interindividual variation in susceptibility to chemotherapy toxicity poses a significant therapeutic obstacle for clinicians, as prediction of individuals at greatest risk of experiencing toxicity remains difficult. Cisplatin and carboplatin are two widely used chemotherapy agents whose toxicities – predominantly nephrotoxicity and ototoxicity for cisplatin [1,2] and myelosuppression for carboplatin [3] – are common causes of treatment-related morbidity and mortality. Although multiple factors can influence toxicity susceptibility and response to these agents, genetic factors likely explain a portion of the interindividual heterogeneity. In fact, our laboratory has shown earlier that up to 47 and 36%, respectively, of cisplatin-induced and carboplatin-induced cytotoxicity can be explained by genetic factors [4,5].
One means by which to illuminate the genetic causes underlying a heritable trait is to examine a population enriched for the given phenotype of interest. There is some clinical evidence that individuals from East Asia are particularly sensitive to the effects of platinating agents and therefore may be an especially informative cohort in which to begin a search for genetic determinants of platinum susceptibility. As an example, Millward et al. [6] studied 68 Caucasian and Asian lung cancer patients treated with carboplatin and paclitaxel and found that although the incidence of febrile neutropenia for the overall study population was 26%, 50% of an initially treated cohort of Asians experienced this toxicity and carboplatin was subsequently dose reduced in all Asian patients. However, the overall disease response rate for Asians was 65%, compared with only 31% for Caucasians (P = 0.01). As a second example, Watanabe et al. [7] tested the tolerability of a US-published, cisplatin-containing induction chemotherapy regimen in Asian head and neck cancer patients and found that although the overall doses of the various agents, including cisplatin, were 80% of the total doses given in the US study, Asians still had a 19% incidence of grade 3 or 4 neutropenia, compared with 8% in the US population. Notwithstanding potential differences in environmental factors, local treatment practice differences, or unique drug–drug interactions, these and other clinical observations [8,9] suggest that there may be underlying genetic causes of the unique susceptibility of Asians to platinum chemotherapies.
On the basis of these data and the principle that a population enriched for a given phenotype is ideal for the study of genetic variants responsible for that phenotype, we have chosen to use a cell-based model previously refined by our laboratory and others [5,10–12] using the International HapMap collection [13] but which specifically examines individuals from East Asia as a `phenotype-enriched' discovery cohort. Using these cell lines, and comparing them to lines from individuals of other ethnic backgrounds within the HapMap collection, we take advantage of the abundance of existing genetic information [>2 000 000 common single-nucleotide polymorphisms (SNPs)] on HapMap individuals to use a genome-wide approach to elucidate possible genetic determinants of susceptibility to cisplatin and carboplatin. We believe that approaches that are unbiased to known genes in the metabolic or mechanistic pathways of the platinum agents are necessary because even previously identified platinum pharmacogenomic candidates (e.g. ERCC1 for cisplatin) [14,15] have been unable to completely explain the clinical heterogeneity in susceptibility to toxicity or response. Our genome-wide approach permits the possibility that platinum susceptibility is likely to be a multigenic trait [16].
This study proposed to test the following three hypotheses: (i) cell lines from Asian individuals recapitulate clinical evidence of increased platinum sensitivity, thereby comprising an especially informative cohort in which to conduct a search for platinum susceptibility determinants; (ii) novel germline genetic polymorphisms – and their related genes – are identifiable as important determinants of cisplatin and carboplatin susceptibility in Asians; and (iii) although Asian-identified susceptibility determinants may be population-specific, some variants will be also important in individuals from other ethnic populations.
Materials and methods
Cell lines
We utilized the well-genotyped, Epstein–Barr virus-transformed lymphoblastoid cell lines (LCLs) from apparently healthy individuals in the International HapMap project collection [13,17]: Asian (ASN) cell lines (total n = 90) represented those combined unrelated individuals from Tokyo, Japan (n = 45) and Beijing, China (n = 45); unrelated Caucasians (CEU) of northern and western European ancestry were from Utah, US (n = 57); unrelated Africans (YRI) were from the Yoruba people in Ibadan, Nigeria (n = 59).
Phenotyping
Cell culture methods and cytotoxicity assays were performed as described earlier [10] except that ASN cell lines were passaged to a dilution concentration of 200 000 cells/ml three times weekly. Plating density was identical to the earlier method. Alamar-blue reagent [10] was used to determine cell growth inhibition at increasing concentrations of drug. IC50s (concentration at which 50% cell growth inhibition occurred) were calculated as described [10]. As all individuals in the ASN population were unrelated, for comparison purposes we included a subset of the previously reported phenotype data [11,18] on CEU and YRI so as to only include unrelated individuals within these populations in all analyses.
Genome-wide association studies in Asians
ASN IC50s were used along with genotypes for each ASN cell line in genome-wide association studies (GWAS) to identify SNPs associating (individually) with cisplatin and carboplatin susceptibility. Approximately 2.4 × 106 HapMap SNPs included in HapMap release 23a (March, 2008, http://www.hapmap.org) having a minor allele frequency (MAF) of more than 5% and a genotyping rate of greater than 90% were tested. The HapMap database was accessed to determine the risk allele frequencies for all SNPs. An additive model incorporating sex as a covariate [18] was employed using the PLINK software (v1.06, http://pngu.mgh.harvard.edu/purcell/plink/) [19]. As we intended to be as inclusive as possible at the discovery stage to avoid eliminating potentially important candidates, we chose a relatively loose initial-step cut-off of P ≤ 10−4. False discovery rates (FDRs) [20] were calculated at this raw P value cut-off for both drugs and were based on the number of SNPs tested.
Backwards elimination analysis to define `genetic signature' of susceptibility
As a means of filtering the initial ASN GWAS SNPs to identify those most likely to be true positives, a method of backwards elimination was used [11]. Using the ASN population, linear models were constructed with IC50 values after log2 transforming. The independent variables included sex and the most significant ASN SNPs, which were defined as SNPs associating at a raw P value cut-off of less than 9.99×10−6 in the initial GWAS, and any SNPs from the full initial GWAS (P<10−4) which were uniquely polymorphic in ASN (monomorphic in CEU and YRI). Linkage disequilibrium (LD) patterns were considered and SNPs in complete LD were not doubly included. Additive genetic effects were assumed for each SNP. Models were reduced using a backwards elimination approach to identify only SNPs statistically significant at the α=0.05 level. These SNPs were included in the final model for each drug. R-squared estimates were reported from the final (`reduced-SNP') models to quantify the genetic contribution of the reduced-SNP genetic signature on drug susceptibility.
Association of SNPs of interest with the expression of target genes in ASN
SNPs comprising the backwards elimination-filtered genetic signatures were tested for association with differential baseline expression of any target genes using the publicly available Illumina gene expression array data in the 90 ASN cell lines [21]. Linear regression was performed evaluating genotypes against all 47 293 probes, although it is understood that nearly 50% of the interrogated genes are not expressed in LCLs and only approximately 25% have reasonable variability. Sex was included as a covariate in the association analysis [22]. We chose a statistical cut-off for association significance at this step of P<10−4. As a final verification step, genes surviving this SNP-expression analysis were subjected to linear regression against the original IC50 phenotypes (for each drug) to determine which genes also associated with cytotoxicity.
Exploratory testing in independent, secondary populations
Significant SNPs from the initial ASN GWAS were tested for phenotype–genotype association in an independent secondary population, the combined CEU/YRI samples. To do this, we developed a computer program for the analysis of genotype–phenotype data using linear regression and chose an additive model (e.g. AA=0, AB=1, BB=2). IC50s, which were log2 transformed for normalization, were taken as dependent variables in the regression analysis. For each genotype–phenotype association, the correlation coefficient and associated P value were reported. ASN-derived SNPs significant in the CEU/YRI population at a raw P value of less than 0.05 were termed possible `cross-population' susceptibility SNPs. Adjusted P values were calculated and reported using the QVALUE software (http://genomics.princeton.edu/storeylab/qvalue/) and the Benjamini–Hochberg correction [20,23]. The potential cross-population SNPs were then subjected to backwards elimination analysis, as described above, in the CEU and YRI populations, respectively, to determine the SNPs most likely to be true positives in these different populations.
Results
Population differences in platinum susceptibility
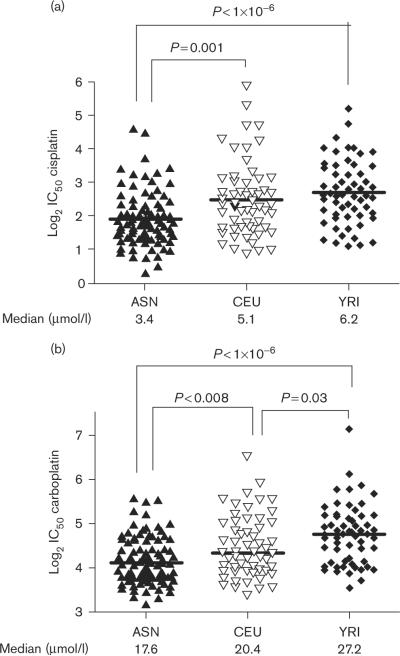
Cytotoxicity phenotypes (IC50s) for the population of ASN cell lines were compared with the previously determined platinum IC50s of unrelated CEU and YRI individuals from the original HapMap release [18]. For cisplatin, ASN had a median IC50 of 3.4 μmol/l, which was significantly lower than the median for CEU (5.1 μmol/l; P=0.001) and YRI (6.2 μmol/l; P<1×10−6). There was no statistical difference between the cisplatin medians for the unrelated CEU and YRI (Fig. 1a), in agreement with our similar previous report using all CEU and YRI trios [18].
Fig. 1.
Comparative population cytotoxicities upon treatment of lymphoblastoid cell lines with (a) cisplatin and (b) carboplatin for Asians (ASN), Caucasians (CEU), and Africans (YRI). Each individual cell line is represented by a unique dot. Population medians are shown by horizontal bars through the population clusters. Values corresponding to these medians are shown horizontally across the bottom of the graph.
Similarly for carboplatin, ASN were the most sensitive with a population median IC50 of 17.6 μmol/l, significantly lower than that for CEU (20.4 μmol/l; P<0.008) and YRI (27.2 μmol/l; P<1×10−6). For carboplatin, the difference between YRI and CEU was also significant (P=0.03), indicating that YRI were the most resistant of all three populations to carboplatin-induced cytotoxicity (Fig. 1b).
Genome-wide association studies in Asians
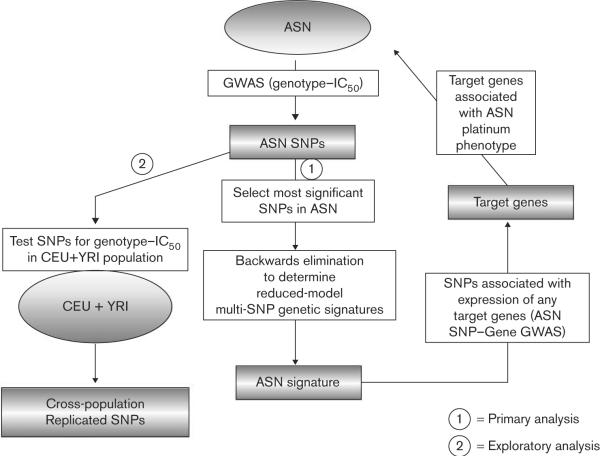
As the population `enriched' for the platinum susceptibility phenotype, we first performed GWAS in 90 ASN individuals as a means to identify potential SNPs governing platinum susceptibility. As we planned to secondarily test the ASN GWAS SNPs using a backwards elimination model, and because we also planned testing in a separate exploratory population (the combined CEU/YRI) (see Fig. 2 for overall schema), we chose a liberal P value of 10−4 as a cut-off for association at this stage.
Fig. 2.
Flow diagram of the single-nucleotide polymorphism (SNP)-discovery/replication methods used. Asian (ASN) cells were phenotyped by exposure to cisplatin and carboplatin, separately. Genome-wide association studies (GWAS) were performed in ASN cell lines associating genotype with IC50, and SNPs of interest which associated at P<10−4 for each drug were retained. Next (analysis pathway 1), the most significant of these ASN SNPs (P<9.99 × 10−6) were included in a backwards elimination approach to identify SNPs comprising genetic signatures which robustly describe the ASN drug phenotype variation. These final reduced-model SNPs were then further validated by testing their association with the differential expression of any target genes (P<10−4), and any identified target genes were then interrogated against the original ASN platinum phenotypes to provide final, summary evidence of the importance of these genes and SNPs. In analysis pathway 2, the initial ASN GWAS SNPs were also tested for genotype–phenotype association in an independent set of combined Caucasian (CEU) and African (YRI) cell lines. SNPs replicating (as single SNPs) for each drug at P<0.05 were identified as `cross-population' platinum variants.
Of 2 034 708 HapMap SNPs tested, 479 were strongly associated with cisplatin susceptibility in the ASN population. The corresponding FDR associated with this result was 0.42. For carboplatin, 199 SNPs were strongly associated with susceptibility in ASN (corresponding FDR for this result was 0.90). To decrease the FDR within ASN, only the most significant SNPs (those associated at P<9.99×10−6, totaling 70 SNPs for cisplatin and 18 SNPs for carboplatin) were included in the backwards elimination analysis (see schema Fig. 2, analysis pathway 1). In contrast, all of the ASN GWAS SNPs identified at P<10−4 were included in the cross-population analysis described later, as that analysis was considered more exploratory.
`Genetic signatures' of susceptibility using a backwards elimination analysis
As a means of further filtering, the most significant ASN GWAS SNPs to identify those more likely to be true positives, a method of backwards elimination was used that identified the SNPs that remained significant with IC50 after reduction of a linear model. r2 was used to assess the degree to which the SNP signature model described the drug susceptibility phenotype. For cisplatin, the 70 `most significant' SNPs mentioned above plus three additional SNPs from the ASN GWAS that were uniquely polymorphic in ASN were included in the full linear model. After removing SNPs in LD, 34 unique SNPs were tested using backwards elimination reduction. Twelve of these 34 SNPs were significant in the reduced (final) model: rs10964552, rs11179377, rs12119783, rs1520896, rs2051396, rs224786, rs224788, rs2309997, rs509620, rs6558058, rs7022486, and rs904106 (P<0.05 for all 12 SNPs). The 12-SNP final model gives a robust estimate of r2=0.96 for predicting the phenotype of cisplatin susceptibility. The full model (including 34 SNPs) gives an estimate of r2 = 0.99, suggesting a relatively small incremental contribution from the 22 eliminated SNPs. We therefore defined the refined, final model list as a novel ASN-derived `genetic signature' of cisplatin susceptibility (Table 1).
Table 1.
Genetic signatures of cisplatin and carboplatin susceptibility in Asians
| Drug | SNP | Chromosome | Gene | Location | r2 |
|---|---|---|---|---|---|
| Cisplatin | rs12119783 | 1 | NA | NA | 0.96 |
| rs904106 | 1 | NA | NA | ||
| rs2309997 | 2 | TBC1D8 | Intron | ||
| rs6558058 | 8 | FBXO16 | Intron | ||
| rs10964552 | 9 | MLLT3 | Intron | ||
| rs7022486 | 9 | NA | NA | ||
| rs1520896 | 11 | NA | NA | ||
| rs11179377 | 12 | NA | NA | ||
| rs509620 | 18 | MBP | Intron | ||
| rs224786 | 20 | NA | NA | ||
| rs224788 | 20 | NA | NA | ||
| rs2051396 | 21 | NA | NA | ||
| Carboplatin | rs13241797 | 7 | NA | NA | 0.76 |
| rs955980 | 8 | C8ORF68 | Intron | ||
| rs3789911 | 9 | KIAA0649 | Intron | ||
| rs4740928 | 9 | NA | NA | ||
| rs7102746 | 11 | NA | NA | ||
| rs4792990 | 17 | NA | NA | ||
| rs16939986 | 18 | NA | NA | ||
| rs1321391 | X | NA | NA |
The multi-SNP signatures describe the most significant SNPs (all P <9.99×10−6) from initial genome-wide association study in Asians which also then remained significant after backwards elimination analyses. Significance in the backwards elimination step (Preduced < 0.05) for all cisplatin SNPs shown in the final signature. Preduced < 0.007 for all carboplatin SNPs comprising the final signature.
r2 describes the degree to which each genetic signature explains the drug susceptibility variability within ASN.
NA, not applicable; SNP, single-nucleotide polymorphism.
Similarly for carboplatin, the 18 `most significant' SNPs mentioned above and one additional SNP from the ASN GWAS that was uniquely polymorphic in ASN were included in the full linear carboplatin model. After removing SNPs in LD, 13 unique SNPs were tested using backwards elimination. Eight of these 13 SNPs were significant in the reduced (final) model: rs1321391, rs13241797, rs16939986, rs3789911, rs4740928, rs4792990, rs7102746, and rs955980 (P<0.007 for all eight SNPs). The eight-SNP final model gives an estimate of r2=0.76 for predicting the carboplatin susceptibility phenotype. The full model (including all 13 SNPs) gives an estimate of r2=0.78, again suggesting a relatively small contribution from the five eliminated SNPs. The final model list was therefore defined as a novel ASN-derived genetic signature of carboplatin susceptibility (Table 1).
Potential functions of the implicated SNPs in Asians - associations with the expression of target genes
As the genetic signature SNPs were located either in intronic regions of their host genes or in unknown regions of the genome, we hypothesized that these SNPs might exert their function through regulation of target genes located either cis or trans to the SNPs themselves. We chose to explore this possibility using Illumina gene expression array data for the ASN samples [21]. We interrogated whether the 12 cisplatin and eight carboplatin SNPs of interest (those included in the final reduced-SNP models) were associated with the differential expression of any target genes.
For cisplatin, the 12 reduced-model SNPs were surprisingly strongly associated with the differential expression of 82 different target genes (P<10−4). As a means of identifying the associations most likely to be true positives and validating the importance of the genes (and the related SNPs), we then tested these 82 genes for significant linear relationship with the original phenotype, cisplatin IC50, in the 90 ASN cell lines. We believe this integrative, three-step method (wherein phenotype-SNP, SNP-expression, and then expression-back-to-phenotype associations are sequentially performed) [10] increases the likelihood that findings surviving all association steps are true positives. Sixty-one genes survived the rigorous gene–phenotype verification association at P<0.05 (TMSL2, HIST1H3A, HIST2H2AB, TMSL4, TMSL1, FLJ10565, FKBP1A, TMSL3, NFYC, KREMEN2, FTO, SRR, HIST2H3C, ING1, SSBP2, LY86, P15RS, EEF1A1, TH1L, AAMP, LRRTM4, RPS13, MAPK10, AP1M1, LIF, MTMR1, CNNM4, FLJ14981, C14ORF109, YPEL1, RPS14, COX6C, CDH15, HMGN2, EVA1, FLJ21290, STX10, ZHX3, RRP46, RBBP6, ACF, ASNS, MOAP1, RPS23, RPS21, C12ORF10, FOXF2, ARHGAP15, SOX30, ARID1B, C1QTNF1, C21ORF128, RAB20, DDX4, ABLIM1, CACNB1, PAN3, FLJ10496, KCNJ16, UNQ431, and TFDP2). For 10 of these 61 genes, the gene–phenotype level of association was significant even if a strict Bonferroni correction was applied (Pc<0.05): TMSL2, HIST1H3A, HIST2H2AB, TMSL4, TMSL1, FLJ10565, FKBP1A, TMSL3, and KREMEN2. As a final observation suggesting the importance of these gene–SNP–phenotype findings, four of the 12 cisplatin genetic signature SNPs were noted to be `master regulators' [24] – SNPs associated with the expression of 10 or more different target genes such as rs2309997 (26 target genes), rs12119783 (24 target genes), rs224786 (12 target genes), and rs6558058 (11 target genes).
For carboplatin, SNP–expression association analysis identified 15 target genes strongly associated with the eight reduced-model SNPs of interest (P<10 −4). Eight of these 15 target genes were also independently associated (P<0.05) with carboplatin IC50 in the 90 ASN cell lines: CCR3, FBXO32, FLJ21736, FN1, HIST1H3I, KHK, MGC71745, and SORBS1.
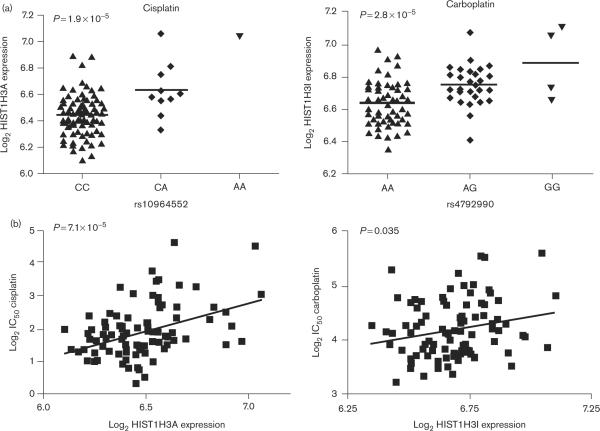
It is particularly interesting to note that two members of the H3 histone family (of histone cluster 1) were commonly implicated by both drugs (HIST1H3A for cisplatin, and HIST1H3I for carboplatin), suggesting a possible platinum drug class effect for this gene family. To illustrate this as an example of the above findings for cisplatin and carboplatin, the SNP–expression and expression–phenotype relationships involving HIST1H3A/I are illustrated in Fig. 3.
Fig. 3.
Platinum susceptibility single-nucleotide polymorphisms (SNPs) of interest may function through regulation of a common target gene family. (a) In Asians (ASN), an example SNP of interest derived from the cisplatin analysis (left panel) and one derived from the carboplatin analysis (right panel) are each associated with the differential expression of a member of the histone H3 family of genes (HIST1H3A and HIST1H3I). Baseline histone H3 expression is plotted as a function of genotype for each SNP. (b) ASN cisplatin and carboplatin IC50s are plotted separately as a function of expression for the respective target histone H3 genes. The cytotoxicity phenotypes for both drugs were correlated with histone H3-member expression levels.
Identifying cross-population susceptibility SNPs
As an exploratory aim of our study, we desired to determine whether ASN-derived (`the enriched population') SNP candidates also conferred platinum susceptibility in other ethnic populations (i.e. as `cross-population' platinum susceptibility determinants) or whether the ASN genetic signatures were unique to ASN and therefore perhaps explain a portion of the population susceptibility differences that are seen. To do this, we interrogated the broad ASN GWAS-significant SNPs for genotype–phenotype relationships in the combined CEU and YRI populations (Fig. 2, analysis pathway 2).
For cisplatin, we first excluded SNPs that could simply not be tested in CEU and YRI because they were not genotyped in these populations. This reduced the number of tested SNPs from 479 to 466. An additional 13 SNPs were excluded from testing because the combined MAF in CEU/YRI was less than 0.05. Ten SNPs of interest showed genotype–IC50 susceptibility associations in CEU/YRI (P<0.05); however, none were individually significant after correction for multiple testing. The directionality of the association in CEU/YRI was opposite to that in ASN for seven of these 10 SNPs, so these seven were discarded from further consideration. Three SNPs remained: rs293331, rs7937567, and rs16946737 (Table 2). rs293331 and rs7937567 were also significant upon backwards elimination (Preduced<0.05; Preduced = 0.002) in CEU and YRI, respectively, although the magnitude of their effects (r2 = 0.07; r2 = 0.16) in predicting cisplatin susceptibility variation in these populations was limited, as might be expected for single SNPs.
Table 2.
`Cross-population' SNPs: some ASN-derived SNPs also may be important in determining cisplatin and carboplatin susceptibility phenotypes in other populations
| Drug | SNP | Chromosome | Gene | Location | P (ASN GWAS) | Pcross (CEU+YRI population) |
|---|---|---|---|---|---|---|
| Cisplatin | rs293331 | 10 | PRKG1 | Intron | 8.7×(10−5) | 0.014 |
| rs7937567 | 11 | GALNTL4 | Intron | 8.9×(10−5) | 0.013 | |
| rs16946737 | 16 | KIAA1576 | Intron | 9.4×(10−5) | 0.015 | |
| Carboplatin | rs11120986 | 1 | CAMTA1 | Intron | 1.6×(10−5) | 0.00080a |
| rs11120989 | 1 | CAMTA1 | Intron | 3.0×(10−5) | 0.0077a | |
| rs11587056 | 1 | CAMTA1 | Intron | 2.1×(10−5) | 0.0066a | |
| rs11590447 | 1 | CAMTA1 | Intron | 1.6×(10−5) | 0.0037a | |
| rs6691275 | 1 | CAMTA1 | Intron | 1.6×(10-5) | 0.0092a | |
| rs7519687 | 1 | CAMTA1 | Intron | 7.2×(10−5) | 0.0024a | |
| rs2463460 | 2 | NA | NA | 3.0×(10−5) | 0.049 | |
| rs7867305 | 9 | C9ORF150 | Intron | 7.9×(10−5) | 0.0020a | |
| rs7134205 | 12 | NA | NA | 4.5×(10−5) | 0.023 | |
| rs1426897 | 15 | NA | NA | 9.7×(10−5) | 0.020 |
For all listed SNPs, the direction of the association in CEU/YRI is the same direction as is observed in ASN.
Pcross describes the raw significance of each SNP when tested in isolation in the combined CEU+YRI replication set.
ASN, Asian; CEU, Caucasian; GWAS, genome-wide association study; SNP, single-nucleotide polymorphism; YRI, Yoruban.
When tested as single SNPs in the replication population (combined CEU + YRI), denotes those SNPs having corrected P values ≤0.15 using the Benjamini-Hochberg false-discovery rate correction.
For carboplatin, an identical process for examining ASN-derived SNPs in the CEU/YRI populations was performed. Of the 199 SNPs from the initial carboplatin ASN GWAS, one SNP was not genotyped in either CEU or YRI, and four additional SNPs had combined MAF of less than 0.05, leaving 194 SNPs for replication testing. Genotype–phenotype associations (P<0.05) were found for 12 unique SNPs, and 10 of these SNPs (rs11120986, rs11120989, rs11587056, rs11590447, rs6691275, rs7519687, rs2463460, rs7867305, rs7134205, rs1426897) showed the association in the same direction as seen in ASN (Table 2). In addition, seven of these 10 SNP associations were significant even after a correction for multiple testing. None of the SNPs were in significant LD with each other across all three populations (although some SNPs were in LD within a single population). rs7867305 was significant upon backwards elimination in CEU (Preduced = 0.0003; r2 = 0.25), whereas rs11120986 (Preduced = 0.0009) and rs2463460 (Preduced = 0.02) were significant upon backwards elimination in YRI (r2 for the pair = 0.38). The exploratory nature of these specific SNP results in CEU and YRI – especially considering the large differences in the SNP LD patterns between the populations – needs to be emphasized. Note that all of the SNPs replicating in CEU/YRI had raw P values from the initial ASN GWAS that fell below the adopted P value cut-off for the ASN-specific analysis (Fig. 2, analysis pathway 1), explaining why none of the SNPs themselves were part of the prior ASN-specific signatures. Furthermore, because the `cross-population' SNPs were first derived using the enriched-phenotype ASN population, there are likely additional SNPs that are important especially in CEU and YRI, respectively, which are not captured in the cross-population analyses.
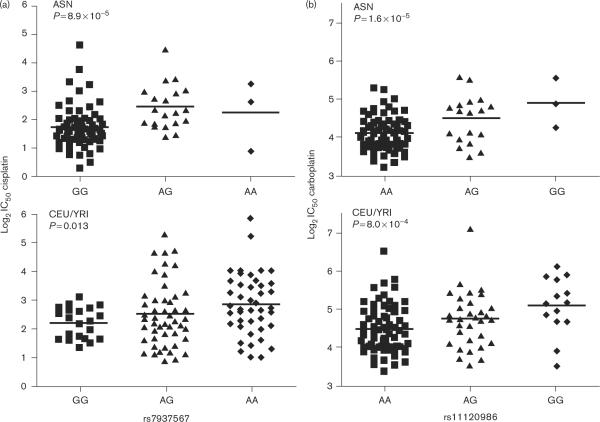
The genotype–phenotype relationships for a representative `cross-population' SNP for both cisplatin and carboplatin are shown in Fig. 4 in both the ASN and CEU/YRI populations.
Fig. 4.
Representative graphical depiction of the genotype–phenotype relationships for one of the SNPs (rs7937567) identified as associated with cisplatin susceptibility (a) and a SNP (rs11120986) associated with carboplatin susceptibility (b) in Asians (ASN; top panels) and replicated in the combined population of Caucasians/Africans (CEU/YRI; bottom panels). Each individual cell line is represented by a unique dot. Genotype groups are shown on the horizontal axis. Genotype group medians are shown by horizontal bars through the respective clusters.
As a final analysis of the cross-population SNPs, we examined whether these SNPs were associated with any target genes in ASN using the Illumina platform. Intriguingly, when the `cross-population' cisplatin-identified SNP rs7937567 – located in GALNTL4 (a host gene previously implicated in a genome-wide analysis of cisplatin susceptibility in YRI [11]) – was analyzed using the ASN Illumina gene expression array data, we found that it was strongly associated with the differential expression of the target gene LY86 (P = 3.0 × 10 −5), which was one of the genes that survived the rigorous three-step association analysis with rs2309997 of the original ASN-derived cisplatin genetic signature.
Perhaps most interestingly, for carboplatin the cross-population SNP rs7134205 is strongly associated with the differential expression of HIST1H3A (P = 1.8 × 10−5), a gene in the same histone family that was implicated by ASN-derived SNPs for both cisplatin and carboplatin and a gene which is independently and strongly associated with susceptibility to both drugs (P = 7.1 × 10−5 and P = 1.1 × 10−3, respectively). These findings further suggest the importance of this gene in platinum susceptibility and show possible coordinated control or biological redundancy in the genetic variants governing the effect.
Discussion
Importantly, our lymphoblastoid cell model findings recapitulated clinical observations that Asian individuals are particularly sensitive to platinum agents compared with their international counterparts. Using the principle, then, that a population enriched for a given phenotype is ideal for the study of genetic variants responsible for that phenotype, we were able to identify a novel list of SNPs and related genes associated with platinum susceptibility in ASN. The SNPs comprised highly descriptive genetic signatures of susceptibility in ASN, explaining more than 95% of the observed variability in cisplatin susceptibility, and more than 75% of the variation in carboplatin susceptibility. The final-model SNPs were also further validated by demonstrating their association with the differential expression of several target genes in ASN (for each drug) which themselves were shown to be linearly correlated with platinum-induced cytotoxicity in ASN. Finally, several ASN-derived SNPs were also shown to be associated with platinum susceptibility in an independent Caucasian/African population, indicating that, in addition to population-specific signatures, our method was able to identify variants that may be important across populations.
In considering the means by which the SNPs of interest might govern platinum susceptibility, our analysis of genes related to these SNPs produced several interesting findings. From the standpoint of target genes whose expression was associated with the SNPs of interest, the most interesting finding was that one gene family, histone H3, was identified as a key target for both cisplatin and carboplatin, suggesting a platinum drug class effect. Given the function of histones as essential parts of the nucleosome, the evidence that histone-free DNA favors platinum adduct formation [25], and the fact that nucleosome disassembly must occur for platinum lesions to be repaired [26], it is especially interesting that our genome-wide approach identified histone H3 as a consistent, strong finding. Others have previously shown that platinums also form adducts on histones – specifically with histone H3 for cisplatin – and have suggested that such histone adducts could act as a nuclear `reservoir' of platinum compounds for further DNA adduct formation [27]. Wang and Lippard [28] showed that cisplatin treatment results in histone H3 phosphorylation in cancer cells. To our knowledge, our findings are the first to suggest that specific genetic variants might mediate these mechanisms in a pharmacogenetic manner.
From the standpoint of `host genes' for the SNPs of interest, six unique SNPs within CAMTA1 were associated with carboplatin susceptibility in the cross-population analysis of our study. CAMTA1 encodes Homo sapiens calmodulin binding transcription activator 1, a transcription factor that may participate in the induction of cell differentiation and cell cycle regulation [29]. In the arena of oncology, it is noteworthy that CAMTA1 is frequently deleted in human neuroblastomas, and low expression of CAMTA1 is related to worse outcomes [30]. For cisplatin, it was interesting to find a significant SNP in the gene GALNTL4, as this gene was earlier identified as associated with cisplatin susceptibility by our laboratory using LCLs derived from YRI samples and a modified method of analysis [11]. Even more intriguingly, the cross-population cisplatin-identified SNP in GALNTL4 – rs7937567 – was associated with the differential expression of the target gene LY86, which was one of the genes that survived the rigorous three-step association analysis with rs2309997 in ASN. These two SNPs are not in LD. While not only heightening the potential importance of LY86 (lymphocyte antigen 86, which itself has been found to be uniquely upregulated in tumor-associated macrophages compared to normal macrophages [31]), this finding suggests that an SNP like rs7937567, which is apparently important across multiple populations, may, in ASN, coregulate the common target gene LY86 along with another SNP (rs2309997) thereby providing biological pliability and a potential basis for allelic dose regulation of the phenotype.
We compared our current SNP findings to the results we reported earlier using linkage-directed association studies of these two drugs [5,32]. Although the prior studies included only Caucasians, because this is the only population that LCLs from pedigrees are available, there is some interesting overlap. For carboplatin, rs7102746 (Table 1) falls under the previously identified linkage peak on chromosome 11 with the 1-logarithm of odds (LOD) confidence interval of 11q21-q23.1 (95–105 cm, peak at 99 cm) having a peak LOD score of genome-wide significance of 3.36 (for the IC50 phenotype) [5]. This SNP was not interrogated in subsequent linkage-directed association studies because those studies were limited to SNPs within genes. In addition, all of the currently identified SNPs in CAMTA1 (Table 2) fall under the previously identified linkage peak on chromosome 1 with the 1-LOD confidence interval 1p36.32-36.22 (6–23 cm, peak at 15 cm) with a peak at LOD 1.64 for the 10 μmol/l carboplatin phenotype [5]. The current novel carboplatin SNPs in CAMTA1 fall very close to the peak of this region. In our earlier publication, we performed linkage analysis on all drug concentrations but followed up with association studies on only linkage regions associated with the IC50 phenotype [5].
For cisplatin, one current SNP (rs1520896) (Table 1) interestingly falls under two previously identified cisplatin linkage peaks (also on chromosome 11, 11p15.4-q13.2), which had peak LOD scores of 2.15 (5 μmol/l cisplatin phenotype, 1-LOD confidence interval 11–67 cm, peak at 22 cm) and 2.50 (10 μmol/l cisplatin phenotype, 1-LOD confidence interval 13–55 cm, peak at 22 cm) [32]. This SNP did not meet the P<1×10−4 significance threshold in a linkage-directed association study of cisplatin IC50 phenotype in 86 CEU samples [32].
It is important to note that the individual SNPs identified in our genome-wide approach may not themselves be the causative SNPs governing the phenotype relationship. As the identified SNPs were all genotyped as part of the HapMap project, it is equally possible that any SNP in LD with the identified SNP is the causative locus. This does not, however, affect the translational applicability of our findings, as clinical genotyping of any SNP in perfect LD with the causative SNP should be adequate; however, the possibility exists that a rarer SNP in LD may have an even larger effect. In addition, each individual's specific platinum susceptibility phenotype is likely to be a composite of the risk allele effects for some or all of the identified SNPs (in addition to other undiscovered SNPs), explaining why, upon inspection of the data from Figs 3 and 4, some individuals homozygous for one apparent `risk' allele, for example, can have relatively lower platinum susceptibility (higher IC50s) than other individuals who are heterozygous or absent for that same given allele. In other words, rather than functioning alone to determine the phenotype, each SNP of interest acts in concert with the other genetic determinants to determine phenotype in a given individual. This concept has been advanced since other studies of genetic susceptibility that assess or test the effects of one SNP in isolation often reveal a very low odds-ratios of effect on the phenotype, or no replicable effect at all, but studies which instead consider complex trait genetics as determined by a genetic signature (or haplotype) of multiple interacting SNPs have found that relatively large odds ratios of effect can be found, even when any one SNP by itself is not found to be significant [33]. We believe that this concept is likely also true of the multiple SNPs of interest implicated by this work. Our backwards elimination predictive model supports this statement, showing the robust ability with which a group of SNPs (a genetic signature) can describe the overall phenotypic variation.
Several candidate genes implicated by other earlier studies of platinum toxicity susceptibility, such as ERCC1, ABCC2, and EPHA2 [4,15,34], were not identified using our model, but this may be because of characteristics of the gene expression profile of lympho-blasts, limited HapMap genotyping coverage of previously-implicated candidate genes, relatively modest (yet still important) statistical association in the initial ASN GWAS, or because these genes are not important in Asians, from which all findings in our model originated. These considerations emphasize the paramount relevance of combining our genome-wide method's results with those of candidate gene methods to arrive at a composite picture of platinum susceptibility. In addition, it should be emphasized that there may be critical rare variants in the ASN population (having a MAF <5%) that would not have been detected by this study because of the method we used. Such SNPs, in fact, may be most important toward explaining interethnic susceptibility differences [35]. Our cross-population SNP-identifying method, however, offers the advantage of identifying relatively common SNPs testable in individuals of diverse ethnicities. Restriction of our analysis to identify the uncommon variants might also be fruitful and could represent a novel model for investigating genetic variation underlying ethnic differences in response to chemotherapy drugs [36,37].
One limitation of our work is that, on account of the relatively small sizes of our population cohorts, the chosen P value cut-offs for the initial ASN GWAS and for cross-population significance testing, when corrected for multiple testing, would not meet the most stringent levels of significance [38]. Although it remains possible that some or all of the SNP associations are produced by chance alone, we believe that this possibility is highly unlikely given the facts that (a) the SNPs of interest, when tested in multi-SNP genetic signatures (which more closely approximate the biology) rather than in isolation, are highly significant in backwards elimination models which show that the genetic contribution of these signatures on predicting platinum susceptibility is robust; (b) the finding of multiple, independently identified SNPs within the same host gene (as with the SNPs identified in CAMTA1 for carboplatin) suggests that this location is much less likely to be identified by chance, but rather, adds import to this region as a true region of significance for susceptibility to the drug; (c) several of our SNPs fall under previously identified linkage regions associated with susceptibility to these drugs; and (d) target gene expression analysis for the SNPs shows that several are strong master regulators of multiple genes, that these genes themselves are independently correlated to platinum sensitivity, and that one gene is implicated for both cisplatin and carboplatin showing an expected commonality within the platinum family. Separately, our approach defined ASN as the combined Chinese and Japanese HapMap populations – a method proposed by the plan of the International HapMap project [13], supported by genetic ancestry/migration data on these populations [39], and consistent with earlier data suggesting significant genetic homogeneity [40] – but it is still likely that some genetic differences exist between these two populations [41]. Finally, these cell-based studies of human LCLs are intended to allow discovery of SNPs which can then be tested in a formal clinical trial, following the paradigm set forth by Hirschhorn and Daly [42]. Such clinical testing is currently ongoing.
In summary, we used a cell-based, genome-wide approach centered on a phenotypically enriched population to identify novel genetic variants governing cisplatin and carboplatin susceptibility. The composite list of novel SNPs and related genes, along with SNPs in previously identified platinum pathway genes, deserve study in clinical settings toward the goal of identifying individuals at risk of toxicity from, and perhaps response to, these commonly used platinum agents.
Acknowledgements
This Pharmacogenetics of Anticancer Agents Research Group (http://pharmacogenetics.org) study was supported by NIH/NIGMS grant UO1GM61393 and data deposits are supported by UO1GM61374 (http://pharmgkb.org) and P50 CA125183, The University of Chicago Breast Cancer SPORE grant. In addition, P.H.O. was supported by NIH/NCI T32 CA09566.
References
- 1.Rabik CA, Dolan ME. Molecular mechanisms of resistance and toxicity associated with platinating agents. Cancer Treat Rev. 2007;33:9–23. doi: 10.1016/j.ctrv.2006.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Celik I, Kars A, Ozyar E, Tekuzman G, Atahan L, Firat D. Major toxicity of cisplatin, fluorouracil, and leucovorin following chemoradiotherapy in patients with nasopharyngeal carcinoma [comment] J Clin Oncol. 1996;14:1043–1044. doi: 10.1200/JCO.1996.14.3.1043. [DOI] [PubMed] [Google Scholar]
- 3.Van Glabbeke M, Renard J, Pinedo HM, Cavalli F, Vermorken J, Sessa C, et al. Iproplatin and carboplatin induced toxicities: overview of phase II clinical trial conducted by the EORTC Early Clinical Trials Cooperative Group (ECTG) Eur J Cancer Clin Oncol. 1988;24:255–262. doi: 10.1016/0277-5379(88)90262-3. [DOI] [PubMed] [Google Scholar]
- 4.Dolan ME, Newbold KG, Nagasubramanian R, Wu X, Ratain MJ, Cook EH, Jr, Badner JA. Heritability and linkage analysis of sensitivity to cisplatin-induced cytotoxicity. Cancer Res. 2004;64:4353–4356. doi: 10.1158/0008-5472.CAN-04-0340. [DOI] [PubMed] [Google Scholar]
- 5.Shukla SJ, Duan S, Wu X, Badner JA, Kasza K, Dolan ME. Whole-genome approach implicates CD44 in cellular resistance to carboplatin. Human Genomics. 2009;3:128–142. doi: 10.1186/1479-7364-3-2-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Millward MJ, Boyer MJ, Lehnert M, Clarke S, Rischin D, Goh BC, et al. Docetaxel and carboplatin is an active regimen in advanced non-small-cell lung cancer: a phase II study in Caucasian and Asian patients. Ann Oncol. 2003;14:449–454. doi: 10.1093/annonc/mdg118. [DOI] [PubMed] [Google Scholar]
- 7.Watanabe A, Taniguchi M, Sasaki S. Induction chemotherapy with docetaxel, cisplatin, fluorouracil and l-leucovorin for locally advanced head and neck cancers: a modified regimen for Japanese patients. Anti-Cancer Drugs. 2003;14:801–807. doi: 10.1097/00001813-200311000-00005. [DOI] [PubMed] [Google Scholar]
- 8.Ogawara M, Kawahara M, Hosoe S, Atagi S, Kawaguchi T, Okishio K, et al. A feasibility study of paclitaxel 225 mg/m(2) and carboplatin AUC= 6 in untreated advanced non-small cell lung cancer patients in Japan. Jpn J Clin Oncol. 2002;32:48–53. doi: 10.1093/jjco/hyf014. [DOI] [PubMed] [Google Scholar]
- 9.Takei Y, Suzuki M, Ohwada M, Saga Y, Kohno T, Machida S, Sato I. A feasibility study of paclitaxel and carboplatin therapy in Japanese patients with epithelial ovarian cancer. Oncol Reports. 2003;10:951–955. [PubMed] [Google Scholar]
- 10.Huang RS, Duan S, Bleibel WK, Kistner EO, Zhang W, Clark TA, et al. A genome-wide approach to identify genetic variants that contribute to etoposide-induced cytotoxicity. Proc Natl Acad Sci U S A. 2007;104:9758–9763. doi: 10.1073/pnas.0703736104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang RS, Duan S, Shukla SJ, Kistner EO, Clark TA, Chen TX, et al. Identification of genetic variants contributing to cisplatin-induced cytotoxicity by use of a genomewide approach. Am J Hum Genet. 2007;81:427–437. doi: 10.1086/519850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duan S, Bleibel WK, Huang RS, Shukla SJ, Wu X, Badner JA, Dolan ME. Mapping genes that contribute to daunorubicin-induced cytotoxicity. Cancer Res. 2007;67:5425–5433. doi: 10.1158/0008-5472.CAN-06-4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.International HapMap C: a haplotype map of the human genome [see comment] Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gossage L, Madhusudan S. Current status of excision repair cross complementing-group 1 (ERCC1) in cancer. Cancer Treatment Rev. 2007;33:565–577. doi: 10.1016/j.ctrv.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 15.Suk R, Gurubhagavatula S, Park S, Zhou W, Su L, Lynch TJ, et al. Polymorphisms in ERCC1 and grade 3 or 4 toxicity in non-small cell lung cancer patients. Clin Cancer Res. 2005;11:1534–1538. doi: 10.1158/1078-0432.CCR-04-1953. [DOI] [PubMed] [Google Scholar]
- 16.Kotti S, Bickeboller H, Clerget-Darpoux F. Strategy for detecting susceptibility genes with weak or no marginal effect [erratum appears in Hum Hered 2008; 65:119] Hum Heredity. 2007;63:85–92. doi: 10.1159/000099180. [DOI] [PubMed] [Google Scholar]
- 17.Shukla SJ, Dolan ME. Use of CEPH and non-CEPH lymphoblast cell lines in pharmacogenetic studies. Pharmacogenomics. 2005;6:303–310. doi: 10.1517/14622416.6.3.303. [DOI] [PubMed] [Google Scholar]
- 18.Huang RS, Kistner EO, Bleibel WK, Shukla SJ, Dolan ME. Effect of population and gender on chemotherapeutic agent-induced cytotoxicity. Mol Cancer Ther. 2007;6:31–36. doi: 10.1158/1535-7163.MCT-06-0591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc. 1995;B(57):289–300. [Google Scholar]
- 21.Stranger BE, Nica AC, Forrest MS, Dimas A, Bird CP, Beazley C, et al. Population genomics of human gene expression. Nat Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dixon AL, Liang L, Moffatt MF, Chen W, Heath S, Wong KC, et al. A genome-wide association study of global gene expression. Nat Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 23.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ. Trait associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010 doi: 10.1371/journal.pgen.1000888. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galea AM, Murray V. The interaction of cisplatin and analogues with DNA in reconstituted chromatin. Biochim Biophys Acta. 2002;1579:142–152. doi: 10.1016/s0167-4781(02)00535-3. [DOI] [PubMed] [Google Scholar]
- 26.Ransom M, Dennehey BK, Tyler JK. Chaperoning histones during DNA replication and repair. Cell. 140:183–195. doi: 10.1016/j.cell.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu B, Droge P, Davey CA. Site selectivity of platinum anticancer therapeutics. Nat Chem Biol. 2008;4:110–112. doi: 10.1038/nchembio.2007.58. [DOI] [PubMed] [Google Scholar]
- 28.Wang D, Lippard SJ. Cisplatin-induced post-translational modification of histones H3 and H4. J Biol Chem. 2004;279:20622–20625. doi: 10.1074/jbc.M402547200. [DOI] [PubMed] [Google Scholar]
- 29.Nakatani K, Nishioka J, Itakura T, Nakanishi Y, Horinouchi J, Abe Y, et al. Cell cycle-dependent transcriptional regulation of calmodulin-binding transcription activator 1 in neuroblastoma cells. Intl J Oncol. 2004;24:1407–1412. [PubMed] [Google Scholar]
- 30.Henrich KO, Fischer M, Mertens D, Benner A, Wiedemeyer R, Brors B, et al. Reduced expression of CAMTA1 correlates with adverse outcome in neuroblastoma patients. Clin Cancer Res. 2006;12:131–138. doi: 10.1158/1078-0432.CCR-05-1431. [DOI] [PubMed] [Google Scholar]
- 31.Gottfried E, Faust S, Fritsche J, Kunz-Schughart LA, Andreesen R, Miyake K, Kreutz M. Identification of genes expressed in tumor-associated macrophages. Immunobiology. 2003;207:351–359. doi: 10.1078/0171-2985-00246. [DOI] [PubMed] [Google Scholar]
- 32.Shukla SJ, Duan S, Badner JA, Wu X, Dolan ME. Susceptibility loci involved in cisplatin-induced cytotoxicity and apoptosis. Pharmacogenet Genomics. 2008;18:253–262. doi: 10.1097/FPC.0b013e3282f5e605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang L, Lu J, An J, Shi Q, Spitz MR, Wei Q. Polymorphisms of cytosolic serine hydroxymethyltransferase and risk of lung cancer: a case-control analysis. Lung Cancer. 2007;57:143–151. doi: 10.1016/j.lungcan.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Filipski KK, Loos WJ, Marsh S, Verweij J, Sparreboom A. Effect of genetic variation in ABCC2 on the renal clearance of cisplatin. Clin Pharmacol Ther. 2009;85(Suppl 1):S66. [Google Scholar]
- 35.Torgerson DG, Boyko AR, Hernandez RD, Indap A, Hu X, White TJ, et al. Evolutionary processes acting on candidate cis-regulatory regions in humans inferred from patterns of polymorphism and divergence. PLoS Genet. 2009;5:e1000592. doi: 10.1371/journal.pgen.1000592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang SM, Temple R. Is this the drug or dose for you? Impact and consideration of ethnic factors in global drug development, regulatory review, and clinical practice. Clin Pharmacol Ther. 2008;84:287–294. doi: 10.1038/clpt.2008.144. [DOI] [PubMed] [Google Scholar]
- 37.Yasuda SU, Zhang L, Huang SM. The role of ethnicity in variability in response to drugs: focus on clinical pharmacology studies. Clin Pharmacol Ther. 2008;84:417–423. doi: 10.1038/clpt.2008.141. [DOI] [PubMed] [Google Scholar]
- 38.Maitland ML, Ratain MJ, Cox NJ. Interpreting P values in pharmacogenetic studies: a call for process and perspective [comment] J Clin Oncol. 2007;25:4513–4515. doi: 10.1200/JCO.2007.12.7803. [DOI] [PubMed] [Google Scholar]
- 39.Su B, Xiao J, Underhill P, Deka R, Zhang W, Akey J, et al. Y-Chromosome evidence for a northward migration of modern humans into Eastern Asia during the last ice age. Am J Hum Genet. 1999;65:1718–1724. doi: 10.1086/302680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spielman RS, Bastone LA, Burdick JT, Morley M, Ewens WJ, Cheung VG. Common genetic variants account for differences in gene expression among ethnic groups [see comment] Nat Genet. 2007;39:226–231. doi: 10.1038/ng1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He M, Gitschier J, Zerjal T, de Knijff P, Tyler-Smith C, Xue Y. Geographical affinities of the HapMap samples. PLoS ONE [Electronic Resource] 2009;4:e4684. doi: 10.1371/journal.pone.0004684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat Rev Genet. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]