SUMMARY
Proteins of the death domain (DD) superfamily mediate assembly of oligomeric signaling complexes for the activation of caspases and kinases via unknown mechanisms. Here we report the crystal structure of the PIDD DD and RAIDD DD complex, which forms the core of the caspase-2 activating complex PIDDosome. While RAIDD DD and PIDD DD are monomers, they assemble into a complex that comprises seven RAIDD DDs and five PIDD DDs. Despite the use of an asymmetric assembly mechanism, all DDs in the complex are in quasi-equivalent environments. The structure provided eight unique asymmetric interfaces, which can be classified into three types. These three types of interactions together cover a majority of the DD surface. Mutagenesis on almost all interfaces leads to disruption of the assembly resulting in defective caspase-2 activation. The three types of interactions may represent most, if not all, modes of interactions in the DD superfamily for assembling complexes of different stoichiometry.
INTRODUCTION
The death domain (DD) superfamily comprises the death domain (DD) subfamily, the death effector domain (DED) subfamily, the caspase recruitment domain (CARD) subfamily and the pyrin domain (PYD) subfamily. It is one of the largest protein domain superfamilies (Kohl and Grutter, 2004; Park et al., 2007; Reed et al., 2004). These domains mediate homotypic interactions within each subfamily and play critical roles in the formation of oligomeric signaling complexes such as the death inducing signaling complex (DISC) assembled by some members of the TNF receptor family for caspase-8 and caspase-10 activation, the apoptosome for caspase-9 activation, the inflammasome for caspase-1 activation and the PIDDosome for caspase-2 activation (Kohl and Grutter, 2004; Park et al., 2007; Reed et al., 2004). These domains also participate in the assembly of signaling complexes for kinase and NF-κB activation in TNF signaling, T-cell and B-cell receptor signaling, intracellular pathogen sensing and defense and response to DNA damage (Kohl and Grutter, 2004; Park et al., 2007; Reed et al., 2004).
The DD superfamily domains appear to mediate two types of functions in these oligomeric signaling complexes for caspase and kinase activation. One function is to mediate the assembly of oligomeric platforms for these complexes and the other is to recruit downstream effectors. In a simplified view, these molecular complexes activate their effectors via proximity induced auto-activation such as dimerization, proteolytic processing and trans-phosphorylation. For caspases, proximity induced dimerization is sufficient for their activation (Baliga et al., 2004; Pop et al., 2006; Yin et al., 2006).
The unifying feature of the DD superfamily is the six-helical bundle structural fold as first revealed by NMR structures of Fas DD, FADD DED, RAIDD CARD, and NALP1 PYD (Kohl and Grutter, 2004; Park et al., 2007; Reed et al., 2004). There are currently two complex structures in the DD superfamily involved in effector recruitment, the Pelle DD: Tube DD complex involved in Drosophila Toll signaling (Xiao et al., 1999) and the Apaf-1 CARD: procaspase-9 CARD complex involved in caspase-9 activation (Qin et al., 1999). Despite the fundamental importance of the DD superfamily in apoptotic and immune signaling pathways, no structures of any oligomeric DD superfamily complexes are currently available.
Caspase-2 is an initiator caspase and the most evolutionarily conserved caspase (Lassus et al., 2002; Wang et al., 1994). Caspase-2 deficient germ cells and oocytes are resistant to cell death after treatment with chemotherapeutic agents (Bergeron et al., 1998). In response to DNA damage, caspase-2 acts upstream of the mitochondria by inducing Bid cleavage, Bax translocation and cytochrome c release (Guo et al., 2002; Lassus et al., 2002; Robertson et al., 2002). When added to purified mitochondria, caspase-2 leads to cytochrome c release (Guo et al., 2002; Robertson et al., 2002).
The PIDDosome for caspase-2 activation is composed of 3 components, PIDD (Lin et al., 2000; Telliez et al., 2000a), RAIDD (Duan and Dixit, 1997) and caspase-2 (Tinel and Tschopp, 2004) (Fig. 1A). It is assembled via a DD: DD interaction between RAIDD and PIDD and a CARD: CARD interaction between RAIDD and caspase-2. PIDD DD is not only essential for the activation of caspase-2, it can also interact with the DD of RIP1, a kinase implicated in the activation of NF-κB (Janssens et al., 2005). PIDD appears to act as a molecular switch, controlling the balance between life and death upon DNA damage (Janssens et al., 2005).
Figure 1.
Characterization of the PIDD DD: RAIDD DD complex. A. Domain organizations of the PIDDosome components, PIDD, RAIDD and caspase-2. The cleavage fragments of PIDD are shown. B. Gel filtration profiles of PIDD DD alone (cyan), RAIDD DD alone (blue) and the complex (red). C. Determination of the molar mass of the complex by multi-angle light scattering. D. Representative class averages obtained with negatively stained sample. Each class contains 50–170 particles. Scale bar is 10 nm.
Full length PIDD contains 910 residues with seven leucine rich repeats (LRRs), two ZU-5 domains and a C-terminal DD (Fig. 1A). It is often auto-processed via an intein-like mechanism into shorter fragments of 51kD, 48kD and 37kD (Pick et al., 2006; Tinel et al., 2006; Tinel and Tschopp, 2004). The cleavage sites have been mapped to S446 and S588 (Tinel et al., 2006). Cleavage at S446, which locates in between the two ZU-5 domains, generates a PIDD-N fragment of 48kD (residues 1–446) and a PIDD-C fragment of 51kD (residues 447–910). Further cleavage at S588, which locates in between the second ZU-5 domain and the C-terminal DD, generates a PIDD-CC fragment of 37kD (residues 589–910). Auto-cleavage of PIDD determines the outcome of the downstream signaling events. The initially formed PIDD-C fragment mediates the activation of NF-κB via the recruitment of RIP1 and NEMO and the subsequent formation of PIDD-CC causes caspase-2 activation and cell death (Tinel et al., 2006) (Fig. 1A). Full-length PIDD is inactive in either NF-κB or caspase-2 activation.
PIDD-CC containing the C-terminal DD is sufficient for PIDDosome formation and caspase-2 activation (Tinel et al., 2006). In addition, PIDD DD alone is sufficient in sensitizing a colon carcinoma cell line for UV-induced apoptosis (Pick et al., 2006) and in formation of a ternary complex with RAIDD and caspase-2 CARD upon overexpression in 293T cells (Tinel and Tschopp, 2004). The PIDD DD: RAIDD DD complex forms the core oligomeric platform in PIDDosome, while the RAIDD CARD: caspase-2 CARD interaction is responsible for caspase-2 recruitment.
To elucidate the molecular basis of caspase-2 activation and of the assembly mechanisms of the DD superfamily, we determined the crystal structure of the PIDD DD: RAIDD DD complex, which comprises 7 RAIDD DD and 5 PIDD DD molecules. Despite the use of an asymmetric assembly mechanism, all DDs in the complex are in quasi-equivalent environments. The structure provided multiple observations of 8 unique asymmetric interfaces, which can be further classified into 3 types. These interactions can co-exist on a single DD and together cover a majority of the DD surface. Structure-based mutagenesis on almost all interfaces leads to disruption of the assembly resulting in defective caspase-2 activation. In contrast to the concept that DD superfamily interactions may involve any available surfaces and may be very diverse, we show here that the 3 types of interactions in this complex may represent most, if not all, modes of interactions in the DD superfamily and may be used to assemble oligomeric complexes of different stoichiometry.
RESULTS
Overall Structure of the PIDD DD: RAIDD DD Complex, the Core Oligomerization Platform of the PIDDosome
As a first step towards elucidating the molecular basis of PIDDosome formation, we expressed and purified the DDs of PIDD and RAIDD. Although PIDD DD and RAIDD DD are both monomeric in solution, when mixed together, the complex containing both DDs eluted at around 150 kD from a Superdex 200 gel filtration column (Fig. 1B). Because both mass and shape affect gel filtration positions, we further used multi-angle light scattering (MALS) with refractive index to accurately measure its molecular mass. MALS measurement gave a molecular mass of 152.4 kD (0.8% fitting error) for the complex with a polydispersity of 1.001 (Fig. 1C). These data suggest that PIDD DD and RAIDD DD assemble into an oligomeric complex.
Electron microscopy (EM) of negatively stained PIDD DD: RAIDD DD complex revealed a monodisperse and homogeneous particle population (Supplemental Fig. S1). Classification of 3,708 particle images into 25 groups produced class averages that depicted molecules of similar size, about 9 nm in diameter, but with varying structural features (Fig. 1D, Supplemental Fig. S1). The differences in the projections most likely arise from different orientations, in which the complex had adsorbed to the carbon support film.
We crystallized the complex and determined its structure at 3.2Å resolution using single wavelength anomalous diffraction of a mercury derivative (Table 1). The structure revealed that the PIDD DD: RAIDD DD complex contains 5 PIDD DD and 7 RAIDD DD molecules. It forms a compact globular structure of approximately 90 Å in diameter (Fig. 2A 2B). The globular shape of the structure is consistent with the normal elution behavior of the complex in gel filtration. This size agrees well with the EM images. In addition, despite the strong contrast between the individual domains in the EM projection averages due to stain accumulation, comparison of the experimental class averages with projections calculated from the atomic model and resolution-filtered to 30 Å clearly showed that the class averages depict the same complex. The differences between the class averages arise from different orientations in which the complex had adsorbed to the carbon support film (Supplemental Fig. S1). As the calculated molecular weights of monomeric PIDD DD and RAIDD DD are 13,036 Dalton and 13,075 Dalton, respectively, the calculated molecular mass of a 5:7 PIDD DD: RAIDD DD complex is 156.7 kD, which agrees well with the molecular mass measured by MALS.
Table 1.
Crystallographic Statistics
| Mercury derivative | Native | |
|---|---|---|
| Data collection | ||
| Beamline | X4A of NSLS | NE-CAT of APS |
| Space group | P65 | P65 |
| Cell dimensions | ||
| a, b, c | 138.9Å, 138.9Å, 208.3Å | 138.4Å, 138.4Å, 207.5Å |
| Resolution | 30 – 4.0Å | 30 – 3.2Å |
| Rsym | 7.5% (42.8%) | 7.4% (36.4%) |
| I/σI | 29.7 (3.2) | 40.5 (2.1) |
| Completeness | 100% (100%) | 97.7% (79.4%) |
| Redundancy | 5.7 (5.7) | 10.3 (5.9) |
| Refinement | ||
| Resolution | 30 – 3.2Å | |
| No. reflections | 34,580 | |
| Rwork/Rfree | 23.6%/27.5% | |
| No. atoms | ||
| Protein/water and other small molecules | 9,082/19 | |
| Average B-factors | ||
| Protein/water and other small molecules | 84.5Å2/34.3Å2 | |
| Root mean square deviations | ||
| Bond lengths/angles | 0.01Å/1.7° | |
| Ramachandran Plot | ||
| Most favored/additionally allowed | 85.8%/13.6% | |
Highest resolution shell is shown in parenthesis.
Figure 2.
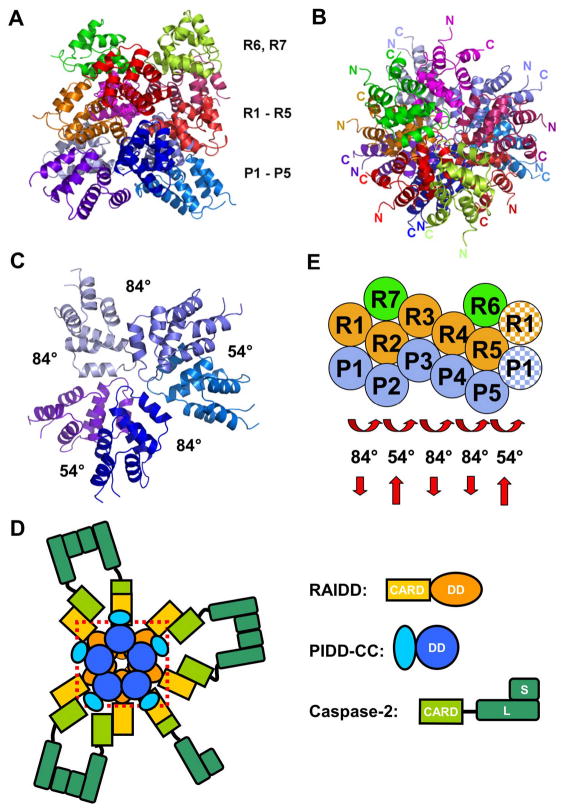
Overview of the PIDD DD: RAIDD DD complex. A. Side view of the complex. The top layer contains 2 RAIDD DD molecules (green and yellow). The middle layer contains 5 RAIDD DD molecules (red, purple, orange, magenta and pink). The bottom layer contains 5 PIDD DD molecules (different shades of blue). B. Top view of the complex. C. Top view of the 5 PIDD DDs at the bottom layer, showing the rotational relationships. D. A model of the PIDDosome for caspase-2 activation, showing the PIDD DD: RAIDD DD complex inside a dotted red box and the rest of the domains and molecules. Caspase-2 molecules are schematically dimerized to illustrate proximity induced dimerization in the PIDDosome. E. A schematic planar diagram for the construction of the complex, showing the successive screw rotations of a hypothetical PR sub-complex. The shaded R1 and P1 molecules at the right indicate that the rotations have brought the PR sub-complex back to the beginning and completed the ring.
PIDDosome for caspase-2 activation contains the PIDD auto-processing fragment PIDD-CC, RAIDD and caspase-2 (Tinel et al., 2006; Tinel and Tschopp, 2004), with calculated molecular weights of 36,473 Dalton, 22,745 Dalton and 50,685 Dalton, respectively. If the same PIDD DD: RAIDD DD stoichiometry is present in the PIDDosome for caspase-2 activation, the calculated molecular mass of the PIDDosome with 5 PIDD-CC, 7 RAIDD and 7 casapse-2 molecules would be 696.4 kD. This molecular mass is in striking agreement with gel filtration analysis of the PIDDosome, which showed a molecular mass of around 670 kD (Read et al., 2002; Tinel and Tschopp, 2004).
The structure of the PIDD DD: RAIDD DD complex may be divided into 3 layers viewing from the side of the complex, 2 RAIDD DDs at the top layer (R6 and R7), 5 RAIDD DDs in the middle layer (R1-R5) and 5 PIDD DDs at the bottom layer (P1-P5) (Fig. 2A). Viewing from the top of the complex, the middle and the bottom layers form two stacked closed rings (Fig. 2B, 2C). The termini of the DDs point to the periphery of the complex (Fig. 2B, 2C). The peripheral locations of the N-termini allow PIDD DD to connect to the N-terminal region of PIDD-CC and RAIDD DD to connect to its CARD domain (Fig. 2D). Therefore, one could envision that the PIDD DD: RAIDD DD complex localizes in the center of the PIDDosome to mediate oligomerization while the N-terminal region of PIDD-CC, RAIDD CARD and caspase-2 occupy the outer part of the PIDDosome. In this scenario, the 7 caspase-2 molecules in the complex are brought into proximity for their dimerization and activation (Fig. 2D).
The PIDD DD: RAIDD DD Complex is Constructed by Successive Screw Rotations
Strikingly, the core complex of 5 PIDD DDs and 5 RAIDD DDs does not possess a recognizable symmetry. Looking down from the top, the RAIDD DDs and the PIDD DDs around the two stacked rings are related by rotations around a common central axis (Fig. 2C, Supplemental Fig. S2). Pair wise superposition showed that the molecules are related by two different rotation angles at different locations (Fig. 2C). In addition, viewing from the side of the complex, the DDs in each layer are not localized on the exact same plane within the complex, suggesting that the rotations are screw rotations (Fig. 2A, Supplemental Fig. S2).
If we cut open the structure from the side and lay the molecules flat, the locations of the DDs form a staggered hexagonal pattern (Fig. 2E). If we take a hypothetical sub-complex of 1 PIDD DD and 1 RAIDD DD (PR sub-complex) as a central building block of this pattern, the complex may be described as 5 successive screw rotations of the PR sub-complex around the central vertical axis. There are two types of such screw rotations in the assembly of the complex, one rotating approximately 84° and translating down the axis and the other rotating approximately 54° and translating up the axis (Fig. 2C, 2E). Among the 5 screw rotations in the complex, 3 are of 84° and 2 are of 54°. This gives a total rotation of 84×3+54×2 = 360° and a zero net translation to close the rings. The net translation is zero because each upward translation is 1.5-fold of the downward translation. The top layer molecules R6 and R7 are built above R5 and R2 respectively and the relationships of R6-R5 and R7-R2 are similar to the R1-P1 relationship.
Massive surface areas in a total of 17,207 Å2 are buried upon complex formation, which correspond to an average of 1,434 Å2 per DD in the complex. With the exception of the surface from which the N- and C-termini protrude out, virtually all other surfaces of each DD are used in complex formation.
Despite Lack of Conventional Symmetry, Each DD in the Complex has Quasi-equivalent Environment
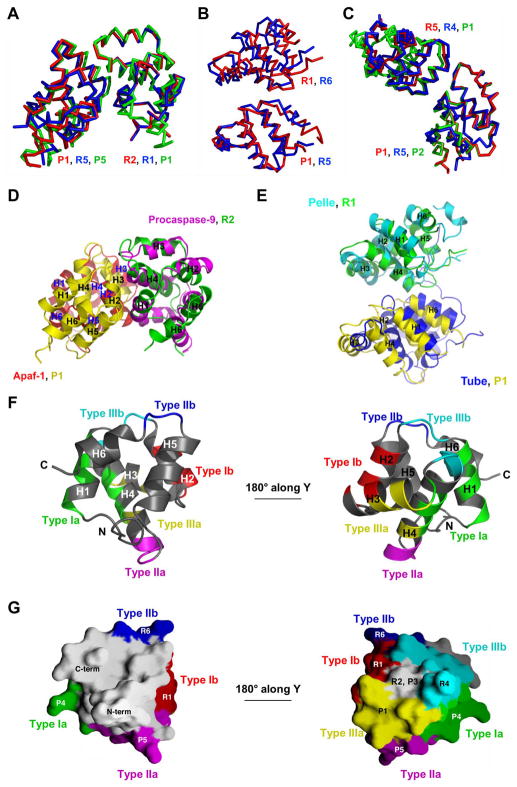
Given this unusual assembly mechanism, we investigated whether the different DDs in the complex are surrounded by different environments. We first superimposed the 5 pairs of PR sub-complexes and found that they align well with pair wise root mean square deviation (RMSD) of below 0.4Å (Fig. 3A). In addition, there is a high degree of structural similarity between RAIDD DD and PIDD DD (Fig. 3B). They superimpose to an RMSD of 1.1Å among 64 aligned Cα atoms. Although it is known that both RAIDD DD and PIDD DD exhibit the same six-helical bundle structure characteristic of the DD superfamily, this level of structural similarity is unexpected as the sequence identity between the two domains is only 15% (Fig. 3D).
Figure 3.
Quasi-equivalent environment. A. Superposition of the 5 PIDD DD: RAIDD DD PR sub-complexes. B. Superposition of RAIDD DD (purple) and PIDD DD (green). C. Quasi-equivalence of the contacts on the plane of the staggered hexagonal construction. R5 and its neighboring DDs (gray) are superimposed with P1 and its neighboring DDs (different colors). D. Structure-based sequence alignment between RAIDD DD and PIDD DD. Residues of RAIDD DD and PIDD DD involved in the 8 different interfaces, which are classified into 3 types of interactions, are highlighted in yellow and marked. P: PIDD. R: RAIDD. Type Ia, IIa and IIIa residues are marked in red and type Ib, IIb and IIIb residues are marked in blue.
As seen from the construction model of the complex, each DD has maximally 6 immediate neighboring DDs (Fig. 2E). Some DDs such as R2 and R5 have all 6 neighboring DDs. Other DDs have 3 to 5 neighboring DDs. Strikingly, all DDs in the complex form similar contacts with their respective neighboring DDs. As an example, Fig. 3C shows the superposition of R5 and its 6 neighboring DDs with P1 and its 5 neighboring DDs. If we take R5 and its neighboring DDs as a standard, all other DDs can be superimposed to this standard along with their respective neighboring DDs (Supplemental Fig. S3), demonstrating that all DDs have an equivalent environment in the complex. Because at the same relative positions around a central DD there may either be a RAIDD DD or a PIDD DD, this equivalent environment is a quasi-equivalent environment.
There are Eight Kinds of Interfaces in the PIDD DD: RAIDD DD Complex, Which may be Classified into Three Types of Interactions
The different interfaces in the PIDD DD: RAIDD DD complex may be classified into 3 types based on the regions involved in these interactions (Fig. 4A). Because it has been proposed earlier that DDs may use diverse mechanisms of interactions (Xiao et al., 1999), we were surprised to find that these 3 types of interactions are similar to the procaspase-9 CARD: Apaf-1 CARD interaction (Qin et al., 1999), the Pelle DD: Tube DD interaction (Xiao et al., 1999) and an interaction proposed to exist in the Fas DD: FADD DD complex (Weber and Vincenz, 2001). Because these interactions have been previously named type I, II and III, respectively (Weber and Vincenz, 2001), we followed the same convention in our description of the interactions. Depending whether the interactions are between a RAIDD DD and a PIDD DD (R:P), between 2 RAIDD DDs (R:R) or between 2 PIDD DDs (P:P), the type I, II, and III interactions contain 3, 2 and 3 subtypes, respectively, making a total of 8 kinds of interfaces.
Figure 4.
The 3 types of interactions and their subtypes, a total of 8 interactions. A. A schematic diagram for the locations of the 3 types of contacts in the PIDD DD: RAIDD DD complex. B. The 3 different subtypes of the type I interaction. C. The 2 different subtypes of the type II interaction. D. The 3 different subtypes of the type III interaction. Important residues and hydrogen bonding interactions are labeled. E. A hypothetical Fas DD: FADD DD complex constructed from the same 3 types of interactions. The same view is used as in A and the Fas DD: FADD DD complex may be considered as a portion of the PIDD DD: RAIDD DD complex composed of R7, R2, P2, R3, P3 and R4.
In the type I interaction, residues at H1 and H4 of the first DD (type Ia surface) interact with residues at H2 and H3 of the second DD (type Ib surface) (Fig. 3D). In the first subtype of this type, the interaction is between a RAIDD DD and a PIDD DD molecule (R:P interaction) (Fig. 4B). Many hydrophobic and polar interactions form the interface. Residues D117, V161, E162 and I165 of RAIDD pack against Y814 of PIDD. The side chains of R118 and N121 of RAIDD make hydrogen bonds respectively with the carbonyl oxygen and the side chain of H809 of PIDD. The side chain of Q125 of RAIDD forms a hydrogen bond with the main chain carbonyl oxygen of Q859 of PIDD. H154 and N155 of RAIDD interact with L801 of PIDD and the side chain of N155 of RAIDD makes a hydrogen bond with the carbonyl oxygen of L801 of PIDD (Fig. 4B). In the second subtype of this type, the PIDD DD is replaced by a RAIDD DD to generate the R:R interaction and in the third subtype of this type of interaction, the RAIDD DD is replaced by a PIDD DD to generate the P:P interaction (Fig. 4B).
In the type II interaction, residues at the H4 helix and the H4-H5 loop of the first DD (type IIa surface) and residues at the H5-H6 loop and H6 helix of the second DD (type IIb surface) mediate this interaction (Fig. 3D). In the first subtype of this type, the interaction is between the RAIDD DD and the PIDD DD in the PR sub-complexes (R:P interaction) (Fig. 4C). The interface appears to be mostly polar and charged. Especially, there is a salt bridge interaction between D861 of PIDD and R170 of RAIDD, a hydrogen bonding network between R862, N863, D864 of PIDD and Q169 of RAIDD and a hydrogen bond between the main chain of K173 of RAIDD and E867 of PIDD (Fig. 4C). In the second subtype of this type, the PIDD DD is replaced by a RAIDD DD and the interaction is between 2 RAIDD DDs such as those between R6 and R5 and between R7 and R2 (R:R interaction) (Fig. 4C). This R:R interaction appears to be much less extensive than the corresponding R:P interaction.
In the type III interaction, residues at H3 of the first DD (type IIIa) interact with residues near the H1-H2 and the H3-H4 loops of the second DD (type IIIb) (Fig. 3D). In the first subtype of this type, the interaction is between a RAIDD DD and a PIDD DD (R:P interaction) (Fig. 4D). A mixture of hydrophobic, polar and charged interactions occur at this interface, including the hydrophobic interaction between L801 of PIDD and Y146 of RAIDD, the salt bridge between D829 of PIDD and R147 of RAIDD and a hydrogen bond between the main chain of L828 of PIDD and the side chain of N151 of RAIDD (Fig. 4D). In the second subtype of this type, the PIDD DD is replaced by a RAIDD DD molecule (R:R interaction) (Fig. 4D). In the third subtype of this type, the RAIDD DD is replaced by a PIDD DD molecule (P:P interaction) (Fig. 4D).
Conservation, Plasticity and Coverage of the Type I, II and III Interactions in the DD Superfamily
Comparison among the different observations within each type revealed conservation, variation and plasticity in these interactions. First, different observations within each subtype of interactions in the PIDD DD: RAIDD DD complex are completely conserved. In type I interactions, there are 3 observations of the R:P subtype, 4 observations of the R:R subtype and 2 observations of the P:P subtype (Fig. 4A). These are all conserved and align well to within RMSD of 0.4Å. Similar well conserved alignment statistics are also observed within the 5 observations of the type II interaction R:P subtype, the 2 observations of the type II interaction R:R subtype, the 2 observations of the type III interaction R:P subtype, the 5 observations of the type III interaction R:R subtype and the 3 observations of the type III interaction P:P subtype. These data suggest that each observed interaction is specific to the particular partners in the interaction.
Second, among the different interactions within each type, variations in orientations are observed. The different subtypes within each type of interactions in the PIDD DD: RAIDD DD complex show small adjustment in orientation (Fig. 5A, 5B, 5C). More significant adjustments are observed when the type I interaction in the PIDD DD: RAIDD DD complex is compared with the procaspase-9 CARD: Apaf-1 CARD interaction (Fig. 5D) and when the type II interaction in the PIDD DD: RAIDD DD complex is compared with the Pelle DD: Tube DD complex (Fig. 5E). Nonetheless, in all type I interactions, it is the H1 and H4 region of the first molecule (type Ia) interacting with the H2 and H3 region of the second molecule (type Ib). In the type II interaction, however, the regions of contact are somewhat different. In addition to the common interaction between the H4-H5 region of the first DD and the H5-H6 region of the second DD, in the Pelle DD: Tube DD complex, the adjacent H2 region of the Pelle DD and the adjacent H1-H2 region of the Tube DD also participate in the interaction. In comparison with the type I interaction, the type II interaction buries a smaller surface area. In the Pelle DD: Tube DD complex, this interaction is strengthened by an additional interaction between a long tail of Tube and the H2-H3 and H4-H5 region of the Pelle DD. Therefore, depending on the exact partners in the complex, there is adjustment in orientation within each type of interactions. This structural plasticity may be important for accommodating the different sequences at these interfaces and for achieving specificity of different interaction pairs.
Figure 5.
Conservation, plasticity and coverage of the type I, II and III interactions. A. Comparison of the R:P (red), R:R (blue) and P:P (green) subtypes of the type I interaction. One molecule in each subtype is superimposed. B. Comparison of the R:P (red) and R:R (blue) subtypes in the type II interaction. One molecule in each subtype is superimposed. C. Comparison of the R:P (red), R:R (blue) and P:P (green) subtypes in the type III interaction. One molecule in each subtype is superimposed. D. Comparison of the type I interaction (R2:P1) with the procaspase-9 CARD: Apaf-1 CARD interaction. R2 is superimposed with procaspase-9 CARD. E. Comparison of the type II interaction (R1:P1) with the Pelle DD: Tube DD interaction. R1 is superimposed with Pelle DD. F. The 6 types of regions of R5 in its interaction with neighboring DDs in the complex. Two views of R5 are shown. Green and red: type Ia and Ib regions. Magenta and blue: type IIa and IIb regions. Yellow and cyan: type IIIa and IIIb regions. G. Surface representation of R5, showing the same 6 surfaces of contacts. Same color coding is used as in F. The small gray area of surface at the 180° rotated view of R5 that does not contact any of the 6 immediate neighboring molecules interact with R2 and P3 in the three-dimensional assembly.
Not only the regions of contacts are relatively conserved, the surface shape complementarity appears to be preserved within each type of interaction. In type I interaction, the type Ia surface is concave that receives the convex surface of the type Ib surface. In both type II and type III interactions, the IIa and IIIa surfaces are convex and the IIb and IIIb surfaces are concave. However, the nature of contacts is not conserved within each type of interaction. For example, in the procaspase-9 CARD: Apaf-1 CARD complex, the interacting surfaces are complementary in charge. In the analogous type I interactions in the PIDD DD: RAIDD DD complex, a complex network of hydrophobic contacts and hydrogen bonds mediate the interfaces.
The 3 types of interactions can coexist on a single DD and each DD in the PIDD DD: RAIDD DD complex uses all types of surfaces to interact with neighboring DDs. Strikingly, when these interactions are mapped onto a particular DD (e.g. R5), the DD surface is almost all covered by these interactions, with the exception of the surface from which the termini protrude out (Fig. 5F, 5G). In the standard orientation we use in this report, the DDs use type IIa and IIb surfaces to interact with other DDs above or below and use other types of surfaces for lateral interactions. The full coverage of these interactions on a DD and their conservation suggest that these 3 types of interaction may likely represent the major, if not all, modes of interactions in the DD superfamily.
Mutations that Disrupt the PIDD DD: RAIDD DD Interaction Prevent PIDDosome Formation and Caspase-2 Activation
To correlate the PIDD DD: RAIDD DD structure with PIDDosome function, we generated extensive structure-based mutations on all 8 potential interfaces of the 3 types and assayed complex formation (Fig. 6A). We were able to obtain mutations on 7 of the 8 subtypes of interfaces that disrupted formation of the PIDD DD: RAIDD DD complex, as judged by native gel electrophoresis and gel filtration chromatography (Supplemental Fig. S4). No intermediate complexes were observed for any of the mutations. These data suggest that complex assembly may require the simultaneous presence of most, if not all, interfaces. We have so far not been able to obtain disruptive mutations on the R:R subtype of the type II interaction (Fig. 6A), which mediates the assembly of R6 and R7 on the top layer.
Figure 6.
Mutational analysis of the PIDD DD: RAIDD DD interaction. A. Structure-based mutations and their effects on assembly of the PIDD DD: RAIDD DD complex in vitro. ** and * show mutations that completely and partially disrupted complex formation, respectively. B. HEK293T cells were transiently transfected with expression vectors encoding wild type or mutant Flag-PIDD and wild type VSV-RAIDD. PIDD or RAIDD was immunoprecipitated from the lysates with anti-Flag or anti-VSV antibodies respectively, and co-immunoprecipitated proteins were revealed by Western blotting (WB). * corresponds to the p20 subunit of caspase-2, which is due the presence of overexpressed RAIDD (PIDD independent). C. Instead of PIDD mutants, the activity of RAIDD mutants was analyzed. D. as B, but PIDD proteins were expressed in the absence of overexpressed RAIDD.
We next investigated whether mutations at the PIDD DD: RAIDD DD interfaces would impact on PIDDosome formation and caspase-2 activation. To this end, several PIDD mutants were overexpressed in HEK293T cells along with wild type RAIDD and complex formation and caspase-2 activation were assessed by co-immunoprecipitation experiments after transient co-transfection (Fig. 6B). While combined expression of the wild type version resulted in formation of a complex containing PIDD (most likely the PIDD-CC form), RAIDD and active caspase-2, complex formation and caspase-2 activation were either completely absent or attenuated with PIDD mutants. Several RAIDD mutants were also examined for their capacity to form the PIDDosome and shown to be defective (Fig. 6C). Partial complex formation and caspase-2 activation were observed with several PIDD and RAIDD mutants, most of which also exhibited less drastic effects on PIDD DD: RAIDD DD interaction in vitro (Fig. 6A). In addition, a complete absence of caspase-2 processing was seen with all PIDD mutants in the absence of RAIDD overexpression (using endogenous RAIDD) (Fig. 6D). This indicates that caspase-2 recruitment and activation in the PIDDosome are critically dependent on the PIDD DD: RAIDD DD interaction we observe in the structure.
DISCUSSION
Molecular Mechanism of Caspase-2 Activation in the PIDDosome
Caspase activation is a hallmark of apoptotic cell death (Riedl and Shi, 2004; Salvesen, 2002). According to their sequence of activation, caspases may be divided into two groups: initiator caspases such as caspase-2, 8, 9 and 10, and effector caspases such as caspase-3 and 7 (Riedl and Shi, 2004; Salvesen, 2002). Unlike effector caspases, initiator caspases possess a domain of the DD superfamily at their N-terminal region for recruitment to oligomeric adapter protein complexes upon apoptosis induction. Caspases are synthesized as single chain pro-caspases, which undergo intra-chain cleavage to generate the large and small subunits.
Caspases need to form specific dimers to be active. Because effector caspases are constitutive dimers, their activation is strictly a consequence of intra-chain cleavage by initiator caspases. In contrast, intra-chain cleavage does not appear to be the crucial factor for initiator caspase activation due to the relative longer lengths of the inter-subunit linker regions. A proximity induced dimerization model was proposed for initiator caspase activation because initiator caspases such as caspase-2, 8 and 9 are not constitutive dimers in solution and specific homo-dimerization appears to be crucial for their activation (Baliga et al., 2004; Pop et al., 2006; Yin et al., 2006).
In agreement with this analysis, the different caspase activating platforms are in different oligomerization states and may recruit caspases with different stoichiometry. While the mammalian apoptosome is a heptamer (Yu et al., 2005a), the drosophila apoptosome is octameric (Yu et al., 2005b). CED4, the Apaf-1 homolog in C. elegans, is a tetramer (Yan et al., 2005). The DISC contains a trimer or likely multiple trimers, which may recruit 3 or more caspase-8 or caspase-10 molecules (Yang et al., 2005). Regardless of the stoichiometry, the key common event is oligomerization, which allows neighboring caspases to form specific activating dimers. This appears to be the case even when the oligomeric platform recruits odd numbers of caspases so that not all recruited caspases have dimeric partners. In this scenario, understanding caspase activation is reduced to understanding the oligomerization mechanisms of their activating complexes.
Therefore it is not surprising that the PIDDosome brings 7 caspase-2 molecules into proximity for their activation. One might ask why such an apparently unusual PIDD DD: RAIDD DD oligomerization platform is used to induce the proximity of caspase-2. Perhaps nature has evolved many different oligomerization platforms for caspase activation and the PIDD DD: RAIDD DD complex is simply one of such examples. Upon recruitment to the PIDDosome, caspase-2 is able to form dimers and be activated, even in the absence of processing. The 50 residue linker region between the CARD and the catalytic region of caspase-2 likely facilitates dimerization in the correct orientations for caspase activation. Auto-processing then proceeds and caspase-2 dimerization and activity are both enhanced to induce mitochondrial events and cell death (Baliga et al., 2004; Read et al., 2002).
General Mechanisms of Interactions in the DD Superfamily
The DD superfamily is one of the largest and most widely distributed domain superfamilies. Evolutionarily, it seems that the ever-expanding DD superfamily may have evolved by inserting its domains into various signal transduction proteins such as caspases, kinases and adapter proteins. In this regard, it is amazing that almost all oligomeric signaling complexes in apoptosis and inflammation contain domains of the DD superfamily. For caspase activating complexes, the DD superfamily domains may either be the major oligomerization platforms or the major mediators in recruiting the caspases. The DDs in the PIDDosome for caspase-2 activation and in the DISC for caspase-8 and caspase-10 activation fall into the former category while the CARDs in the PIDDosome, DEDs in the DISC and CARDs in the inflammasome fall into the latter category.
Our structure of the PIDD DD: RAIDD DD complex provides the first glimpse of an oligomeric complex of the DD superfamily and forms a template for other interactions in this superfamily. Because the RIP1 kinase DD is homologous to RAIDD DD, it is likely that the PIDD DD: RIP1 DD complex uses a similar assembly mechanism. In a much broader scenario, in contrast to the concept that the DD superfamily interactions may involve any available surfaces and may be very diverse, our study suggests that the observed 3 types of asymmetric interactions may represent preferred modes of interactions for DDs, and likely for the entire DD superfamily. This may be shown by the conservation between the type I interactions in the PIDD DD: RAIDD DD complex and the procaspase-9 CARD: Apaf-1 CARD interaction. In addition, in the DED1: DED2 interaction in the tandem DED-containing viral FLIP MC159, the H1 and H4 of DED2 interacts with H2 and H5 of DED1, which is somewhat similar to the type I interaction as well (Li et al., 2006; Yang et al., 2005).
Curiously, all these known interactions in the DD superfamily are asymmetric despite what might have been expected of homotypic interactions. This appears to be true for both effector recruitment as in the procaspase-9 CARD: Apaf-1 CARD complex and the Pelle DD: Tube DD complex and for oligomerization as in the PIDD DD: RAIDD DD complex. This is not true, however, for secondary self-associations of DD superfamily domains that are involved in effector recruitment and are linked to oligomerization domains outside the DD superfamily. For example, Apaf-1 CARD is linked to a nucleotide binding oligomerization domain (NOD). In the heptameric apoptosome, Apaf-1 NOD confers a 7 fold symmetry and the Apaf-1 CARD also self-contacts with this symmetry in the presence of the NOD (Yu et al., 2005a).
On the subject of asymmetric interactions, an asymmetric trimeric model has also been proposed for the Fas DD: FADD DD complex (Weber and Vincenz, 2001). This model was generated from the structures of the Apaf-1 CARD: caspase-9 CARD complex (type I interaction) and the Pelle DD: Tube DD complex (type II interaction). When Pelle DD is superimposed with procaspase-9 CARD, the associated Tube DD and the Apaf-1 CARD pack against Pelle DD (or procaspase-9 CARD) in a well organized trimer. The newly formed interaction between Tube DD and Apaf-1 CARD forms a different interface, which strikingly, is very similar to the type III interaction observed in the PIDD DD: RAIDD DD complex. By continuing building interactions using these structures, a hexamer of DDs can be formed, in which the 3 central DDs may either be Fas DD or FADD DD (Fig. 4E). Therefore, similar to the PIDD DD: RAIDD DD complex, the Fas DD: FADD DD complex may also be constructed from these 3 types of interfaces. In an independent experiment, type I, II and III interactions were also found in docking models between Fas DD and FADD DD (Thakar et al., 2006).
The involvement of all 3 types of interactions in the assembly of various caspase activating complexes is consistent with and explains the existing mutagenesis data on Fas (Huang et al., 1996; Martin et al., 1999), FADD (Hill et al., 2004), TNFR1 (Tartaglia et al., 1993; Telliez et al., 2000b) and TRADD (Park and Baichwal, 1996). In each case, residues affecting the binding and/or function of the DD spread throughout its entire sequence. Similarly, our structure-based mutagenesis also identified important residues throughout the PIDD DD and RAIDD DD sequences (Fig. 6A).
Asymmetry and the apparent preference for the 3 types of interactions may represent a unique feature of the homotypic interactions in the DD superfamily. One potentially strong rationale is that the conserved common fold of the DD superfamily members determines, or at least contributes, to the surface shape complementarity seen in all 3 types of asymmetric interactions. However, the nature of contacts may not be conserved within each type of interaction as exemplified by the different surface hydrophobicity, hydrophilicity and charge features of the different DDs (Park and Wu, 2006). In addition, as the major function of these domains is homotypic interaction, the interactions may have evolved from several primordial interaction pairs and be preserved through co-evolution. Therefore, the preservation of these preferred interactions may reflect both fold conservation and evolutionary circumstances.
As the 3 types of interactions essentially cover a majority of the available surface of a DD (Fig. 5G), it is likely that these 3 types of interactions represent the major, if not all, modes of interactions of the DD superfamily. For effector recruitment, only one of the 3 types of interactions is required. For oligomerization, it is likely that all 3 types of interactions are needed. The oligomerization stoichiometry is probably dictated by both the structural plasticity of the exact interaction pairs and how the interactions could be terminated. For the PIDD DD: RAIDD DD complex, the structure terminates within the layers as it forms rings. The top layer has only 2 RAIDD DD molecules, likely because of the 2 available spaces for interactions and the less extensive R:R subtype of the type II interaction (Fig. 2E, 4A). The lower surfaces of the bottom layer of the PIDD DD molecules apparently have no affinity for either RAIDD DDs or PIDD DDs, therefore terminating the buildup of further layers. In the Fas DD: FADD DD complex, the inability of either FADD DD or Fas DD to associate with itself in the complex may leave the complex in a trimeric form. Given some plasticity at the interfaces, it is likely that by choosing among the 3 types of interactions around each DD, a wide number of oligomeric complexes with different stoichiometry may be built. Selective usage of a certain type of interaction may even switch the binding to alternative DD adapters (Sandu et al., 2005). Therefore, these conserved asymmetric interactions may underlie the unique, but elegant common assembly mechanism of the DD superfamily.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The PIDD DD (residues 778–883) and RAIDD DD (residues 94–199) with C-terminal His-tags were expressed in E. coli and purified using the Ni-NTA affinity resin (Qiagen). They were then mixed and incubated at room temperature for 1 hour before subjecting to gel filtration chromatography using Superdex 200 HR 10/30 (GE Healthcare). The complex eluted at around 12 ml and was concentrated to 10–12 mg/ml.
Multi-angle Light Scattering (MALS)
Molar mass of the PIDD DD: RAIDD DD complex was determined by MALS. The complex was injected onto a Superdex 200 HR 10/30 gel filtration column (GE Healthcare) equilibrated in a buffer containing 20 mM Tris at pH 8.0 and 50 mM NaCl. The chromatography system was coupled to a three-angle light scattering detector (mini-DAWN EOS) and a refractive index detector (Optilab DSP) (Wyatt Technology). Data were collected every 0.5 s at a flow rate of 0.2 ml/min. Data analysis was carried out using the program ASTRA.
Electron Microscopy and Image Processing
PIDD DD: RAIDD DD complex was negatively stained with uranyl formate (Ohi et al., 2004). Images were recorded using low-dose conditions at a magnification of 52,000x and a defocus of −1.5 μm with an FEI Tecnai T12 electron microscope operated at 120 kV. 2 × 2 pixels were averaged yielding a pixel size of 2.69 Å. Using the SPIDER software (Frank et al., 1996), 3,708 particles (from 16 images) were windowed into 80 × 80 pixel images and subjected to ten cycles of multireference alignment and K-means classification specifying 25 classes. For comparison with the crystal structure, the atomic model was resolution-filtered to 30 Å and projections were calculated at 2-degree angular intervals. The re-projections were cross-correlated to the class averages and the re-projections with the highest correlation coefficient were selected.
Structure Determination and Analysis
The assembled complex was crystallized at 20°C using 5.5% PEG 3350, 200 mM NaCl, and 100 mM Na/K phosphate at pH 6.5. To obtain a heavy atom derivative, the crystals were soaked with 1 mM 1,4-diacetoxymercuri-2,3-dimethoxybutane for 30 minutes. One crystal diffracted to 4.0Å resolution and a complete anomalous data set was collected at a wavelength of 1.0079Å at the X4A beamline of NSLS. A 3.2Å native data set was collected at the NE-CAT beamline of APS. Both data sets were processed using HKL2000 (Otwinowski and Minor, 1997).
The structure was determined by single-wavelength anomalous diffraction. There is a single partially surface exposed free cysteine in the RAIDD DD structure (Park and Wu, 2006) and no cysteine in PIDD DD. Five strong mercury sites (15–20σ) were found and the structure was phased using the program SOLVE/RESOLVE (Terwilliger, 2004). A six-dimensional search of the electron density map using the RAIDD DD structure found ten DD molecules in the crystallographic asymmetric unit. The locations of the mercury sites and crystallographic refinement in CNS (Brunger et al., 1998) using the native data confirmed that 5 of the DD molecules were RAIDD DDs and that the 5 remaining molecules were PIDD DDs. Two additional RAIDD DD molecules were also found. The final atomic model contains 7 RAIDD DDs and 5 PIDD DDs (Table 1). The structure was analyzed using O (Jones et al., 1991) and Pymol (DeLano Scientific).
Mutational Analysis of Complex Formation in vitro
Site-directed mutagenesis was performed using the Quikchange kit (Stratagene) and confirmed by sequencing. Purified wild type or mutant PIDD DD and RAIDD DD proteins were first mixed and incubated at room temperature for 1 hour. The mixed solutions were subjected to electrophoresis under native conditions on pre-made 8–25% acrylamide gradient gels using the PhastSystem (GE Healthcare). The gels were stained with Coomassie Blue and complex formation was determined by the appearance of shifted bands. Mutational effects were also characterized by gel filtration chromatography using the Superdex 200 HR 10/30 column (GE Healthcare).
PIDDosome Formation and Caspase-2 Activation
PIDDosome formation was revealed by co-immunoprecipitation experiments after transient co-transfection of PIDD wild type and RAIDD mutants or RAIDD wild type and PIDD mutants. After 48h transfection, cells were lysed in lysis buffer containing 1% NP-40, 20 mM Tris at pH 7.4, 250 mM NaCl, 5% glycerol and a protease inhibitor cocktail. After lysis, the extracts were incubated with anti-Flag or anti-VSV beads for 2 h. After incubation the beads were washed 4 times with lysis buffer, and analyzed by immunoblotting. The antibodies used for Western blotting were: anti-caspase-2 11B4 (Apotech), mouse anti-VSV and rabbit anti-Flag (Sigma).
Supplementary Material
Acknowledgments
We thank Drs. David Eliezer, Olga Boudhka and Fred Maxfield for helpful discussions, Randy Abramowitz and John Schwanof for data collection at X4A of NSLS, Kanagalaghatta Rajashankar and Igor Kourinov for data collection at NE-CAT of APS, Dr. Sally Kornbluth for providing the cDNA of human RAIDD, Dr. Jin Kuk Yang for help with data collection, Jin Wu for maintaining X-ray equipment and computers, Su-Chang Lin for help with the light scattering experiment and the Wu lab members for discussions. The molecular EM facility at Harvard Medical School was established with a generous donation from the Giovanni Armenise Harvard Center for Structural Biology and is maintained with funds from NIH.
Footnotes
Accession Numbers
The Coordinates have been deposited in the RCSB Protein Data Bank with the PDB code 2OF5.
References
- Baliga BC, Read SH, Kumar S. The biochemical mechanism of caspase-2 activation. Cell Death Differ. 2004;11:1234–1241. doi: 10.1038/sj.cdd.4401492. [DOI] [PubMed] [Google Scholar]
- Bergeron L, Perez GI, Macdonald G, Shi L, Sun Y, Jurisicova A, Varmuza S, Latham KE, Flaws JA, Salter JC, et al. Defects in regulation of apoptosis in caspase-2-deficient mice. Genes Dev. 1998;12:1304–1314. doi: 10.1101/gad.12.9.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Duan H, Dixit VM. RAIDD is a new ‘death’ adaptor molecule. Nature. 1997;385:86–89. doi: 10.1038/385086a0. [DOI] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Guo Y, Srinivasula SM, Druilhe A, Fernandes-Alnemri T, Alnemri ES. Caspase-2 induces apoptosis by releasing proapoptotic proteins from mitochondria. J Biol Chem. 2002;277:13430–13437. doi: 10.1074/jbc.M108029200. [DOI] [PubMed] [Google Scholar]
- Hill JM, Morisawa G, Kim T, Huang T, Wei Y, Werner MH. Identification of an expanded binding surface on the FADD death domain responsible for interaction with CD95/Fas. J Biol Chem. 2004;279:1474–1481. doi: 10.1074/jbc.M304996200. [DOI] [PubMed] [Google Scholar]
- Huang B, Eberstadt M, Olejniczak ET, Meadows RP, Fesik SW. NMR structure and mutagenesis of the Fas (APO-1/CD95) death domain. Nature. 1996;384:638–641. doi: 10.1038/384638a0. [DOI] [PubMed] [Google Scholar]
- Janssens S, Tinel A, Lippens S, Tschopp J. PIDD mediates NF-kappaB activation in response to DNA damage. Cell. 2005;123:1079–1092. doi: 10.1016/j.cell.2005.09.036. [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou J-Y, Cowan SW, Kjeldgaard M. Improved methods for building models in electron density maps and the location of errors in those models. Acta Crystallgr. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kohl A, Grutter MG. Fire and death: the pyrin domain joins the death-domain superfamily. C R Biol. 2004;327:1077–1086. doi: 10.1016/j.crvi.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Lassus P, Opitz-Araya X, Lazebnik Y. Requirement for caspase-2 in stress-induced apoptosis before mitochondrial permeabilization. Science. 2002;297:1352–1354. doi: 10.1126/science.1074721. [DOI] [PubMed] [Google Scholar]
- Li FY, Jeffrey PD, Yu JW, Shi Y. Crystal structure of a viral FLIP: insights into FLIP-mediated inhibition of death receptor signaling. J Biol Chem. 2006;281:2960–2968. doi: 10.1074/jbc.M511074200. [DOI] [PubMed] [Google Scholar]
- Lin Y, Ma W, Benchimol S. Pidd, a new death-domain-containing protein, is induced by p53 and promotes apoptosis. Nat Genet. 2000;26:122–127. doi: 10.1038/79102. [DOI] [PubMed] [Google Scholar]
- Martin DA, Zheng L, Siegel RM, Huang B, Fisher GH, Wang J, Jackson CE, Puck JM, Dale J, Straus SE, et al. Defective CD95/APO-1/Fas signal complex formation in the human autoimmune lymphoproliferative syndrome, type Ia. Proc Natl Acad Sci U S A. 1999;96:4552–4557. doi: 10.1073/pnas.96.8.4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi M, Li Y, Cheng Y, Walz T. Negative Staining and Image Classification - Powerful Tools in Modern Electron Microscopy. Biol Proced Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Park A, Baichwal VR. Systematic mutational analysis of the death domain of the tumor necrosis factor receptor-1-associated protein TRADD. J Biol Chem. 1996;271:9858–9862. doi: 10.1074/jbc.271.16.9858. [DOI] [PubMed] [Google Scholar]
- Park HH, Lo YC, Lin SC, Wang L, Yang JK, Wu H. The Death Domain Superfamily in Intracellular Signaling of Apoptosis and Inflammation. Ann Rev Immunology. 2007;25:561–586. doi: 10.1146/annurev.immunol.25.022106.141656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HH, Wu H. Crystal structure of RAIDD death domain implicates potential mechanism of PIDDosome assembly. J Mol Biol. 2006;357:358–364. doi: 10.1016/j.jmb.2005.12.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pick R, Badura S, Bosser S, Zornig M. Upon intracellular processing, the C-terminal death domain-containing fragment of the p53-inducible PIDD/LRDD protein translocates to the nucleoli and interacts with nucleolin. Biochem Biophys Res Commun. 2006;349:1329–1338. doi: 10.1016/j.bbrc.2006.08.176. [DOI] [PubMed] [Google Scholar]
- Pop C, Timmer J, Sperandio S, Salvesen GS. The apoptosome activates caspase-9 by dimerization. Mol Cell. 2006;22:269–275. doi: 10.1016/j.molcel.2006.03.009. [DOI] [PubMed] [Google Scholar]
- Qin H, Srinivasula SM, Wu G, Fernandes-Alnemri T, Alnemri ES, Shi Y. Structural basis of procaspase-9 recruitment by the apoptotic protease-activating factor 1. Nature. 1999;399:549–557. doi: 10.1038/21124. [DOI] [PubMed] [Google Scholar]
- Read SH, Baliga BC, Ekert PG, Vaux DL, Kumar S. A novel Apaf-1-independent putative caspase-2 activation complex. J Cell Biol. 2002;159:739–745. doi: 10.1083/jcb.200209004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JC, Doctor KS, Godzik A. The domains of apoptosis: a genomics perspective. Sci STKE 2004. 2004:re9. doi: 10.1126/stke.2392004re9. [DOI] [PubMed] [Google Scholar]
- Riedl SJ, Shi Y. Molecular mechanisms of caspase regulation during apoptosis. Nat Rev Mol Cell Biol. 2004;5:897–907. doi: 10.1038/nrm1496. [DOI] [PubMed] [Google Scholar]
- Robertson JD, Enoksson M, Suomela M, Zhivotovsky B, Orrenius S. Caspase-2 acts upstream of mitochondria to promote cytochrome c release during etoposide-induced apoptosis. J Biol Chem. 2002;277:29803–29809. doi: 10.1074/jbc.M204185200. [DOI] [PubMed] [Google Scholar]
- Salvesen GS. Caspases and apoptosis. Essays Biochem. 2002;38:9–19. doi: 10.1042/bse0380009. [DOI] [PubMed] [Google Scholar]
- Sandu C, Gavathiotis E, Huang T, Wegorzewska I, Werner MH. A mechanism for death receptor discrimination by death adaptors. J Biol Chem. 2005;280:31974–31980. doi: 10.1074/jbc.M506938200. [DOI] [PubMed] [Google Scholar]
- Tartaglia LA, Ayres TM, Wong GH, Goeddel DV. A novel domain within the 55 kd TNF receptor signals cell death. Cell. 1993;74:845–853. doi: 10.1016/0092-8674(93)90464-2. [DOI] [PubMed] [Google Scholar]
- Telliez JB, Bean KM, Lin LL. LRDD, a novel leucine rich repeat and death domain containing protein. Biochim Biophys Acta. 2000a;1478:280–288. doi: 10.1016/s0167-4838(00)00029-7. [DOI] [PubMed] [Google Scholar]
- Telliez JB, Xu GY, Woronicz JD, Hsu S, Wu JL, Lin L, Sukits SF, Powers R, Lin LL. Mutational analysis and NMR studies of the death domain of the tumor necrosis factor receptor-1. J Mol Biol. 2000b;300:1323–1333. doi: 10.1006/jmbi.2000.3899. [DOI] [PubMed] [Google Scholar]
- Terwilliger T. SOLVE and RESOLVE: automated structure solution, density modification and model building. J Synchrotron Radiat. 2004;11:49–52. doi: 10.1107/s0909049503023938. [DOI] [PubMed] [Google Scholar]
- Thakar J, Schleinkofer K, Borner C, Dandekar T. RIP death domain structural interactions implicated in TNF-mediated proliferation and survival. Proteins. 2006;63:413–423. doi: 10.1002/prot.20895. [DOI] [PubMed] [Google Scholar]
- Tinel A, Janssens S, Lippens S, Cuenin S, Logette E, Jaccard B, Quadroni M, Tschopp J. Autoproteolysis of PIDD marks the bifurcation between pro-death caspase-2 and pro-survival NF-kappaB pathway. Embo J. 2006 doi: 10.1038/sj.emboj.7601473. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinel A, Tschopp J. The PIDDosome, a protein complex implicated in activation of caspase-2 in response to genotoxic stress. Science. 2004;304:843–846. doi: 10.1126/science.1095432. [DOI] [PubMed] [Google Scholar]
- Wang L, Miura M, Bergeron L, Zhu H, Yuan J. Ich-1, an Ice/ced-3-related gene, encodes both positive and negative regulators of programmed cell death. Cell. 1994;78:739–750. doi: 10.1016/s0092-8674(94)90422-7. [DOI] [PubMed] [Google Scholar]
- Weber CH, Vincenz C. A docking model of key components of the DISC complex: death domain superfamily interactions redefined. FEBS Lett. 2001;492:171–176. doi: 10.1016/s0014-5793(01)02162-7. [DOI] [PubMed] [Google Scholar]
- Xiao T, Towb P, Wasserman SA, Sprang SR. Three-dimensional structure of a complex between the death domains of Pelle and Tube. Cell. 1999;99:545–555. doi: 10.1016/s0092-8674(00)81542-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan N, Chai J, Lee ES, Gu L, Liu Q, He J, Wu JW, Kokel D, Li H, Hao Q, et al. Structure of the CED-4-CED-9 complex provides insights into programmed cell death in Caenorhabditis elegans. Nature. 2005;437:831–837. doi: 10.1038/nature04002. [DOI] [PubMed] [Google Scholar]
- Yang JK, Wang L, Zheng L, Wan F, Ahmed M, Lenardo MJ, Wu H. Crystal structure of MC159 reveals molecular mechanism of DISC assembly and FLIP inhibition. Mol Cell. 2005;20:939–949. doi: 10.1016/j.molcel.2005.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Q, Park HH, Chung JY, Lin SC, Lo YC, da Graca LS, Jiang X, Wu H. Caspase-9 holoenzyme is a specific and optimal procaspase-3 processing machine. Mol Cell. 2006;22:259–268. doi: 10.1016/j.molcel.2006.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Acehan D, Menetret JF, Booth CR, Ludtke SJ, Riedl SJ, Shi Y, Wang X, Akey CW. A structure of the human apoptosome at 12.8 A resolution provides insights into this cell death platform. Structure (Camb) 2005a;13:1725–1735. doi: 10.1016/j.str.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Yu X, Wang L, Acehan D, Wang X, Akey CW. Three-dimensional Structure of a Double Apoptosome Formed by the Drosophila Apaf-1 Related Killer. J Mol Biol. 2005b;355:577–589. doi: 10.1016/j.jmb.2005.10.040. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.