Abstract
By screening C. elegans mutants for severe defects in germline proliferation, we isolated a new loss-of-function allele of cdc-25.1, bn115. bn115 and another previously identified loss-of-function allele nr2036 do not exhibit noticeable cell division defects in the somatic tissues but have reduced numbers of germ cells and are sterile, indicating that cdc-25.1 functions predominantly in the germ line during postembryonic development, and that cdc-25.1 activity is probably not required in somatic lineages during larval development. We analyzed cell division of germ cells and somatic tissues in bn115 homozygotes with germline-specific anti-PGL-1 immunofluorescence and GFP transgenes that express in intestinal cells, in distal tip cells, and in gonadal sheath cells, respectively. We also analyzed the expression pattern of cdc-25.1 with conventional and quantitative RT-PCR. In the presence of three other family members of cdc-25 in C. elegans, defects are observed only in the germ line but not in the somatic tissues in cdc-25.1 single mutants, and cdc-25.1 is expressed predominantly, if not exclusively, in the germ line during postembryonic stages. Our findings indicate that the function of cdc-25.1 is unique in the germ line but likely redundant with other members in the soma.
Keywords: bn115, Caenorhabditis elegans, CDC-25.1, germline proliferation, somatic gonadal tissues
INTRODUCTION
Germline development is essential for genomic transmission from generation to generation. Despite the availability of advanced technologies, the molecular mechanisms of germline development are still not fully elucidated. C. elegans is an ideal model system to study germline development because all the developmental stages of germline progression can be observed sequentially in the adult hermaphrodite gonad. It contains distal to proximal distribution of mitotic germline stem cells, early meiotic prophase germ cells, differentiating oocytes, and sperm (Hirsh et al., 1976; Kimble and Hirsh, 1979).
In C. elegans, germ cells have incomplete plasma membranes: they proliferate and differentiate in a syncytium, sharing common cytoplasm within the core of the gonad (Crittenden et al., 1994; Hirsh et al., 1976). All germ cells except mature sperm contain germline-specific cytoplasmic organelles called P granules (Kawasaki et al., 1998; 2004; Strome and Wood, 1983). In the first larval stage (L1), the gonad primordium contains four cells including two somatic gonadal precursor cells (Z1 and Z4) and two primordial germ cells (PGCs, Z2 and Z3). During larval development, Z1 and Z4 give rise to all of the somatic gonadal cells and structures including the distal tip cells (DTCs), the gonad sheath, the spermatheca, and the uterus (Kimble and White, 1981; Sulston et al., 1983; Sulston and Horvitz, 1977). By contrast, Z2 and Z3 give rise to all the germ cells (Kimble and Hirsh, 1979). The first postembryonic germ cell division occurs during the L1 stage, and is followed by germline proliferation that continues throughout the life of the worm to produce as many as 1,000 germ cells in each of the two gonadal arms of the hermaphrodite (Hirsh et al., 1976; Kimble and Crittenden, 2005). Thus, all but two (Z2 and Z3) of the germ cells are produced in postembryonic stages, while more than half (558 out of 959) of the somatic cells are produced during embryogenesis in hermaphrodite development.
In order to understand the onset of postembryonic germline proliferation at the molecular level, we performed mutagenesis and screened for mutants that had severe defects in early germline proliferation. We identified a new loss-of-function (lf) mutation, bn115, in the cdc-25.1 gene. cdc-25.1 is one of four C. elegans orthologs of cdc25, a key cell cycle promoting factor originally identified in S. pombe fission yeast (Russell and Nurse, 1986).
The function of cdc-25.1 in C. elegans development was first reported by Ashcroft et al. (1999). Worms treated with cdc-25.1 RNAi produced aberrantly fertilized eggs with the mispositioned meiotic spindles, failed chromosome segregation, and abnormal cleavage furrows. This RNAi study suggested that cdc-25.1 might be required for completion of meiosis prior to the onset of embryogenesis. Another function of cdc-25.1 was demonstrated by the study of gain-of-function (gf) alleles of cdc-25.1 (Clucas et al., 2002; Kostic and Roy, 2002). Two cdc-25.1(gf) mutations, rr31 and ij48, were shown to induce tissue-specific hyperplasia in the intestinal E lineage during embryogenesis. The gf mutations render the mutant CDC-25.1 proteins more stable than the wild-type CDC-25.1 protein (Hebeisen and Roy, 2008). Finally, we and others identified a role for cdc-25.1 in postembryonic germline proliferation by studying the loss-of-function mutations nr2036 (Ashcroft and Golden, 2002) and bn115. Ashcroft and Golden (2002) found that the nr2036 deletion mutant worms developed into sterile adults, but they did not make detailed analyses for the somatic development. In this study, we analyzed the somatic tissues at the cellular levels using GFP transgenes in bn115 and have demonstrated that cdc-25.1(lf) mutants do not exhibit obvious cell division defects in the somatic tissues which develop postembryonically. We also analyzed the expression pattern of cdc-25.1 during developmental processes and the expression levels in different genetic backgrounds, and demonstrated that cdc-25.1 is expressed predominantly, if not exclusively, in the germ line during postembryonic stages. Our findings indicate that cdc-25.1 functions predominantly in the germ line in postembryonic development and that cdc-25.1 activity is probably not required or functionally redundant with other cdc25 family members in some somatic tissues during larval development.
MATERIALS AND METHODS
Strains and general methods
Caenorhabditis elegans strains were maintained and manipulated as described (Brenner, 1974). C. elegans variety Bristol, strain N2 was used as wild type for all experiments. Most strains were maintained at 20°C on Nematode Growth Medium (NGM) agar plates containing Escherichia coli strain OP50, unless noted otherwise. Mutant strains used in this study were as follows: SS671: cdc-25.1(bn115)/+ I, YHS2: cdc-25.1(bn115)/hT2g I, AG151: cdc-25.1(nr2036)/dpy-5(e61) unc-13(e450) I (Ashcroft and Golden, 2002), MR142: cdc-25.1(rr31) I; rrls 1 (Kostic and Roy, 2002), IA123: cdc-25.1(ij48) I; unc-76(e911) ijlS10 V (Clucas et al., 2002), BA17: fem-1(hc17) IV (Nelson et al., 1978), JK816: fem-3(q20) IV, JK509: glp-1(q231) III (Austin and Kimble, 1987). To construct a strain carrying cdc-25.1(bn115) along with LAG-2::GFP or LIM-7::GFP transgene, L4 hermaphrodites of strain XA6226: mrg-1(qa6200)/qC1 dpy-19(e1259) glp-1(q339) [qls26] III or strain DG1575: tnls6 [lim-7::GFP + rol-6(su1006)] were mated with YHS2: cdc25.1 (bn115)/hT2g I males. Expression of the LAG-2::GFP and LIM-7::GFP fusion proteins were observed with a fluorescence microscope (Axioskop 2 MOT, ZEISS).
Sterile mutant screening and maintenance
L4-stage N2 hermaphrodites were treated with 40 mM ethyl methanesulfonate (EMS) for 6 h at 20°C for mutagenesis as described (Brenner, 1974). Healthy-looking F1 hermaphrodite progeny were picked to individual plates and allowed to self-fertilize. From any plates on which about one fourth of F2 progeny showed sterility, some F2 fertile heterozygous siblings were transferred to new plates to maintain the sterile mutation. Heterozygous mutants that mapped to LGI were eventually mated with hT2g/+ males to produce mutation/hT2g strains. hT2g, hT2 [qls48; myo-2::GFP], is a GFP-marked variant of hT2(I;III), a well characterized effective balancer for the left portion of chromosome I and the right portion of chromosome III. Worms containing hT2g as heterozygotes develop normally and generate green fluorescence in their pharynx after hatching, while hT2g homozygotes are embryonic lethal. Homozygous sterile mutant worms produced from hT2g heterozygous mothers were distinguished from their heterozygous fertile siblings by the absence of green fluorescence from the L1 stage onward using a dissecting fluorescence microscope (SMZ1500, Nikon).
Genetic mapping and positional cloning of bn115
The triply marked strain, MT465: dpy-5(e61) I; bli-2(e768) II; unc-32(e189) III, was used for linkage group analysis. For sterile mutations linked to chromosome I, the genetic location was determined by two-factor mapping with dpy-5(e61) and/or by three-factor mapping with dpy-5(e61) unc-13(e51). bn115, which mapped 0.8 cM to the right of dpy-5 on chromosome I, was identified as a new allele of cdc-25.1 by its failure to complement nr2036, a loss-of-function allele of cdc-25.1. Sequencing was performed to confirm the presence of a mutation in cdc-25.1 in genomic DNA prepared from bn115 homozygous mutant worms. Genomic DNA was prepared as follows. Worm lysate was prepared from 50 young adult bn115 homozygous animals by digesting them in 25 μl of lysis buffer (10 mM Tris-HCl, pH 8.3, 50 mM KCl, 2.5 mM MgCl2, 0.45% Tween-20, 0.45% NP-40, and 0.01% gelatin) containing 100 μg/ml proteinase K at 60°C for 1 h. DNA of the cdc-25.1 locus was PCR-amplified from the worm lysate as four overlapping DNA segments using the following primer pairs: 5′-ATG GCT ACC ACC GGG GAA AAA GCA A-3′ and 5′-GGA GCA ACA AGA GAC AAA TTC CGG C-3′, 5′-CGG TGT TTG ATT TGG ATT CTC TCG A-3′ and 5′-TGT ATT TCT TAT CGA AAT CAT CTC C-3′, 5′-AAG TAC ACT CTT CCC GGT GTT GAT A-3′ and 5′-GTT CAA AGA AGA ATT ATT TCT TCG-3′, 5′-ATG GAA TTT AAA TCG GCT CGT CTC G-3′ and 5′-TTA TTC GGC GTC GTC AGA AAT CGA T-3′ (GenBank accession No. NC_003279). The PCR products were purified with QIAEX II Gel Extraction Kit (QIAGEN) and cloned into the pCR2.1-TOPO vector (Invitrogen). Plasmid DNA was prepared from individual E. coli transformant colonies by alkaline lysis method with QIAprep Spin Miniprep Kit (QIAGEN). For each of the four PCR fragments covering the cdc-25.1 genomic locus, two independent plasmid-inserted clones were sequenced with M13 forward and M13 reverse primers, using an ABI310 DNA sequencer (PE Biosystem).
Immunofluorescence methods and microscopy
Immunostaining of germ lines was performed as previously described with minor adaptations (Kawasaki et al., 1998; Strome and Wood, 1983). Staged animals were placed in 10 μl of M9 buffer on a poly-lysine coated slide glass, dissected with a 26 1/2-gauge needle to extrude their gonads, covered with a cover slip, and quickly frozen in liquid nitrogen. After 3 min, the slide glass was recovered from liquid nitrogen, the cover slip quickly removed, and the specimen fixed in methanol for 10 min, post-fixed in acetone for 10 min at 4°C, and air dried for 5 min. For immunostaining, dried specimens were blocked with 30 μl of 3% BSA (bovine serum albumin) in PBS (phosphate-buffered saline) for 1 h at RT in a humidity chamber. After wicking off the BSA, 30 μl of rabbit polyclonal anti-PGL-1 (diluted 1:5,000 in PBS (Kawasaki et al., 1998) or mouse monoclonal antibody K76 (Strome, 1986a) was applied. The specimens were incubated in a humidity chamber for more than 6 h at 4°C, washed twice in PBS for 20 min at 16°C, and treated with 30 μl of 3% BSA for 15 min at 4°C for the second blocking. After wicking off the BSA, goat anti-rabbit IgG-FITC (Santa Cruz Biotechnology, diluted 1:200 in PBS) or goat anti-mouse IgG-FITC (Santa Cruz Biotechnology, diluted 1:100 in PBS) was applied and incubated for more than 4 h at 4°C in a humidity chamber in the dark. The specimens were washed 3 times in PBS for 10 min, with 0.5 μg/ml Hoechst 33342 included in the second PBS wash, rinsed with distilled water, mounted in Sigma Mounting Medium (M1289) beneath a cover slip, and sealed with nail polish. For nuclear staining without immunostaining with antibodies, 20 young adult animals were dissected in 10 μl of M9 buffer on a poly-lysine coated slide glass, stained with 10 μl of 5 μg/ml Hoechst 33342 dye, fixed in 10 μl of 5% glutaraldehyde in M9 buffer, covered with a cover slip, sealed with nail polish, and left for more than 2 h in a dark chamber at 4°C before observation. The mounted specimens were observed under a fluorescence microscope (Axioplan 2 or Axioskop 2 MOT, ZEISS). Images were acquired using an Orca ERG digital camera (Hamamatsu) and processed with Openlab (Improvision) and Photoshop (Adobe) softwares.
Counting numbers of intestinal nuclei
cdc-25.1(bn115) homozygotes containing elt-2::GFP transgene rrls 1, which was introduced by crossing with MR142, were examined for numbers of intestinal nuclei by fluorescence microscopy. Synchronized worms were mounted on 4% agar pad with a droplet of 10 mM tetramisole, covered with a coverslip, and observed.
RT-PCR
Synchronized young adult worms (100) were collected into 300 μl of Trizol (Invitrogen) and frozen in liquid nitrogen. Total RNA was extracted using phase lock gel (MaXtract High Density, QIAGEN). cDNA was synthesized using oligo-dT primer and amplified for 25 cycles in the PCR reactions as previously described (Kim et al., 2008). Primers for PCR of cdc-25.1 were 5′-ATG GCT ACC ACC GGG GAA AAA GCA A-3′ and 5 ′-TTA TTC GGC GTC GTC AGA AAT CGA T-3′ (GenBank accession No. NM_059461); primers for PCR of act-3 were 5′-GAG GCC CCA TCC AAG AGA-3′ and 5′-TGT TGG AAG GTG GAG AGG-3′ (GenBank accession No. Z81584).
Quantitative real-time RT-PCR
Total RNA was prepared from synchronized populations of L1, L2, L3, L4, and young adult worms of wild-type N2 grown at 20°C, and from fem-1 and fem-3 mutants grown at 25°C, and collected into 300 μl of Trizol (Invitrogen). Total RNA and cDNA were prepared as described for conventional RT-PCR. PCR reactions were conducted using Power SYBR®Green PCR Master Mix (Applied Biosystems) in a 96-well plate with 25 μl reaction volume. Primers for act-1, which served as the internal control, were 5′-CCA GGA ATT GCT GAT CGT ATG CAG AA-3′ and 5′-TGG AGA GGG AAG CGA GGA TAG A-3′ (GenBank accession No. NM_073418). Primers for cdc-25.1 were 5′-ATG TTA TCA AGG TCG TCT AGT G-3′ and 5′-CTA AAT CAT TCT CCG ATT CCG T-3′ (GenBank accession No. NM_059461). The mRNA level of cdc-25.1 at each developmental stage or in each mutant was averaged from triplicate experiments and normalized to that of act-1, the internal control, for comparison.
RESULTS
Identification of bn115 as a new loss-of-function allele of cdc-25.1
To gain new insights into the molecular mechanism of postembryonic germline proliferation in C. elegans, we screened for sterile mutants that have severe defects in early larval stage germ cell proliferation. One of the isolated alleles, bn115, was mapped 0.8 cM to the right of dpy-5 on chromosome I. This map position was close to that of cdc-25.1, a mutation of which was previously shown to cause sterility due to defects in germ-line proliferation (Ashcroft and Golden, 2002). Therefore we performed a complementation test between bn115 and nr2036, a loss-of-function allele of cdc-25.1. bn115 and nr2036 did not complement each other. Sequencing analysis revealed that the cytosine at position 105 in the coding sequence of cdc-25.1 was deleted in bn115, which would cause premature termination of translation before the amino acid 50 (Figs. 1A and 1B). Thus, bn115 was identified as a new loss-of-function allele of cdc-25.1.
Fig. 1.

bn115 mutation in cdc-25.1. (A) bn115 has a single base-pair deletion at cytosine 105 in the first exon of the cdc-25.1 coding sequence (1,815 bp), which is transcribed to a 2,566 bp transcript. Grey box, black boxes, carets, and a grey box with arrowhead indicate the 5′UTR, exons, introns, and 3′UTR, respectively. (B) The cytosine deletion causes a frame shift after the 35th amino acid and creates a premature stop codon at amino acid 50. Bold and boxed amino acid residues indicate frame shifted amino acids.
bn115 is defective in germline proliferation
The mutant phenotype of bn115 was observed through postembryonic development with nuclear staining and immunostaining for PGL-1, a germline-specific marker (Kawasaki et al., 1998; 2004; Fig. 2). Although bn115 homozygous mutant worms contained morphologically normal Z2 and Z3 primordial germ cells (PGCs) at the L1 stage (Fig. 2D), they divided more slowly than wild-type PGCs, and underwent only a few rounds of division to generate approximately 14 germ nuclei per gonad arm at the L4 stage (Figs. 2P and 2U). The number of germ cells has never increased more than that, and they even gradually degenerated during adult stage with only a few germ cells remaining by aged adulthood in most bn115 mutant worms (Figs. 2T and 2U).
Fig. 2.
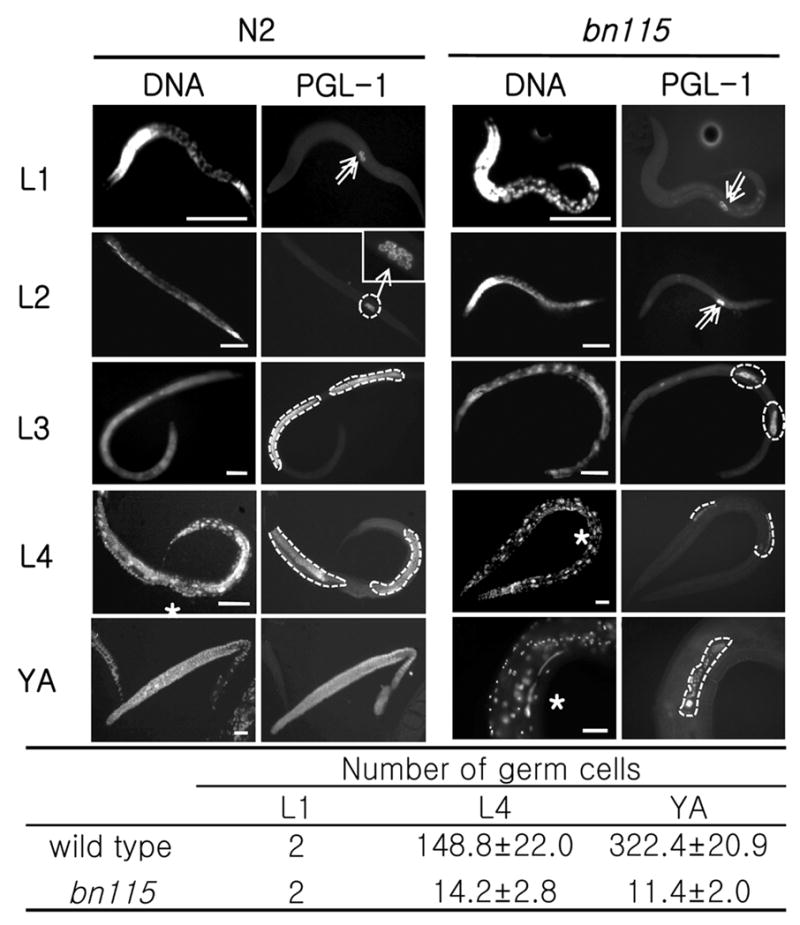
Loss of function of cdc-25.1 causes a germ cell proliferation defect. Postembryonic germline development was observed by immunostaining with anti-PGL-1, a germline-specific marker, along with Hoechst nuclear staining. Left: Wild-type N2, Right: cdc-25.1 (bn115) mutant. (A–D) At L1 stage, both N2 and bn115 worms have two primordial germ cells (PGCs), Z2 and Z3 (arrows). (E–H) At L2 stage, N2 contains around 10–20 germ cells (see magnified view in F), but bn115 still contains only 2 germ cells because of delayed cell divisions. (I–L and M–P) At L3 and L4 stages, germline mitotic proliferation continues in N2, producing hundreds of germ cells, which are circled with dotted lines, while the numbers of germ cells are much smaller in bn115 gonads. (Q–T) At the adult stage, N2 gonads display a progression of germline development from distal to proximal, including mitotic region, transition zone, pachytene region, oocytes in diakinesis, and sperm, while bn115 gonads contain only small numbers of undifferentiated germ cells. The N2 gonad shown (Q and R) was dissected out of the body. (U) Numbers of germ cells per gonad arm in cdc-25.1(bn115) hermaphrodites at different postembryonic stages. Numbers of PGL-1 positive germ nuclei per gonad arm in hermaphrodite worms of wild-type N2 and cdc-25.1 (bn115) mutant at each developmental stage are shown. Only for L1 stage, numbers are given per worm and not per gonad arm, as the two gonad arms in a hermaphrodite are not separated until the L3 stage. For each value, 10 hermaphrodite gonad arms were examined by fluorescence microscopy. L1, first larval stage; L4, fourth larval stage; YA, young adult stage, in which worms had already finished L4-to-adult molting but had not started laying eggs yet. Numbers are presented as mean ± SD. In each panel, anterior is left and dorsal is up. Asterisks indicate the position of vulva. Scale bars, 50 μm.
cdc-25.1 expression level correlates with the number of germ cells
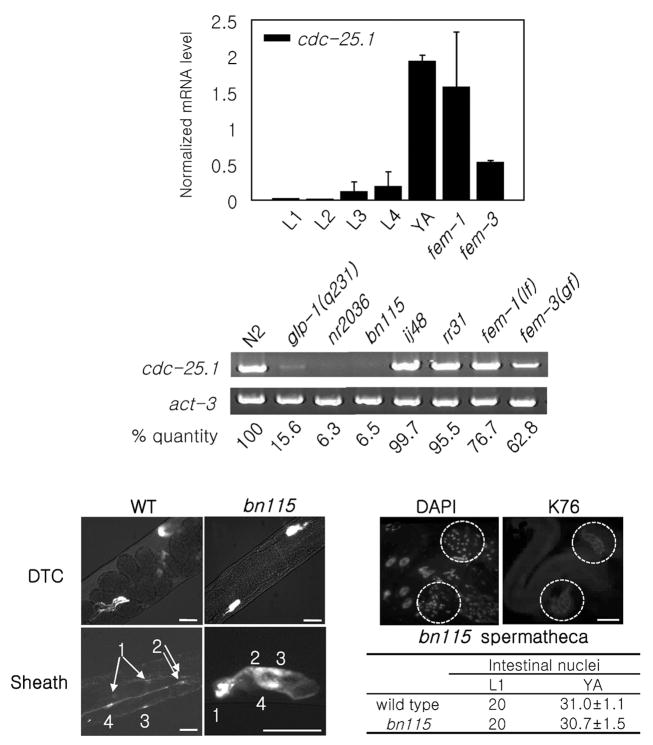
If cdc-25.1 is essential in the germ line as evidence suggests, then cdc-25.1 might well be expressed specifically in the germ line during postembryonic development. To test this hypothesis, the relative expression levels of cdc-25.1 during postembryonic development and in different genetic backgrounds were examined by two RT-PCR methods (Figs. 3A and 3B). First, we demonstrated that cdc-25.1 expression levels increase during postembryonic stages using quantitative real-time RT-PCR (qPCR) (Fig. 3A). The number of germ cells is also known to increase from two (Z2 and Z3) to over 1,000 in wild-type N2 hermaphrodites during this period of development. Second, we used conventional RT-PCR to demonstrate that the cdc-25.1 expression level is very low in adult hermaphrodites of two cdc-25.1 loss-of-function mutants, bn115 and nr2036, and one germ cell proliferation defective mutant, glp-1(q231) (Fig. 3B). In contrast, two cdc-25.1 gain-of-function mutants which contain normal numbers of germ cells, ij48 and rr31, had cdc-25.1 expression levels comparable to wild type levels (Fig. 3B). These results indicate that the expression level of cdc-25.1 correlates with the number of germ cells, and it is therefore likely that cdc-25.1 is expressed predominantly, if not exclusively, in the germ line during postembryonic stages. We also examined the sex dependence of cdc-25.1 expression. The cdc-25.1 expression level was comparable to wild type in fem-1(hc17) loss-of-function mutants which contain oocytes but no sperm, whereas the expression level was reduced in fem-3(q20) gain-of-function mutant worms which contain sperm but no oocytes (Figs. 3A and 3B). These results are consistent with the expression of cdc-25.1 occurring during oogenesis to provide a maternal load of the gene product functioning during embryogenesis. Abundant expression of CDC-25.1 protein in oocytes was also reported (Ashcroft et al., 1999).
Fig. 3.
Analyses of cdc-25.1 expression and somatic tissues in bn115. (A) Real-time qPCR results showing relative expression levels of cdc-25.1 during five postembryonic developmental stages (from L1 to L4 larval, and young adult (YA) stages), in fem-1(hc17lf) mutants, and in fem-3(q20gf) mutants. The average values from three independent experiments, normalized to act-1 control, are shown. Error bars represent SD. (B) RT-PCR was performed with the following strains; young adult worms of wild-type N2, a germline proliferation defective mutant, glp-1(q231), a complete deletion mutant of cdc-25.1, nr2036, a loss-of-function mutant of cdc-25.1, bn115, gain-of-function mutants of cdc-25.1, ij48 and rr31, which contain normal numbers of germ cells, as well as fem-1(hc17lf) and fem-3(q20gf) mutants. Relative mRNA levels of cdc-25.1 in each mutant compared to that in N2 after normalized to act-3 internal control are shown. (C–H) Somatic gonad development in wild type and bn115. (C, D) Wild type (WT) and bn115 mutant containing LAG-2::GFP. Two DTCs were observed in both strains. (E, F) Wild type and bn115 mutant containing LIM-7::GFP. Four pairs of gonadal sheath cells were observed in both strains. Arrows and numbers indicate sheath cell nuclei and order of the nuclear pairs relative to the orientation of the gonad (most distal pair is numbered 1). (G, H) DAPI nuclear staining and monoclonal antibody K76 immunostaining of bn115 mutant gonad showed no aberration in the spermatheca. Dashed circles indicate two spermatheca structures that have normal numbers of nuclei (about 20 nuclei). Scale bars, 30 μm. (I) Numbers of intestinal nuclei in wild-type and bn115 mutant worms at L1 larval and young adult (YA) stages. Numbers are presented as mean ± SD. Ten worms were examined per stage.
bn115 has normal somatic gonad structures and normal number of intestinal nuclei
Since CDC-25.1 is a C. elegans ortholog of the evolutionally conserved cell cycle regulator CDC25 phosphatase, we predicted it to have functions in postembryonic cell division of other tissues as well. Therefore, we examined the somatic gonad structures of bn115 mutant hermaphrodites to assess the development of the distal tip cells (DTCs), gonadal sheath cells, vulva and spermatheca, which develop postembryonically presumably without the influences of maternally loaded gene products. LAG-2::GFP and LIM-7::GFP were introduced into bn115 mutants to label the DTCs and gonadal sheath cells, respectively (Figs. 3D and 3F). LAG-2::GFP and LIM-7::GFP are known to be expressed specifically in the two DTCs and in four pairs out of five pairs of gonadal sheath cells, respectively, in wild-type worms (Figs. 3C and 3E). Although bn115 gonads were small because of the reduced number of germ cells, all bn115 gonads examined contained two DTCs and the normal number of gonadal sheath cells (Figs. 3D and 3F). Spermathecae were examined by DAPI nuclear staining and K76 monoclonal antibody immunostaining, which specifically recognizes spermathecae in addition to germline-specific P granules. We found that all bn115 gonads contained a spermatheca structure that was indistinguishable from that of wild type (Figs. 3G and 3H). We also examined vulvas and intestinal nuclei for abnormalities. First, we observed no morphological changes in the vulvas of either bn115 or nr2036 mutants (data not shown). Second, we found that the average number of bn115 intestinal nuclei was 20 at L1 and around 30 at the young adult stage, which were comparable to wild type numbers (Fig. 3I). In addition, the bn115 mutant worms did not exhibit any obvious “uncoordinated” movement, which would have been expected if the postembryonic cell divisions of neuronal or muscle tissues were affected. These results indicate that postembryonic cell divisions of at least some, if not all, somatic lineages are normal in bn115 mutants. Our finding that some somatic tissues are unaffected by the single depletion of cdc-25.1 suggests that cdc-25.1 is either not required or functionally redundant with other cdc25 family members in these somatic tissues.
DISCUSSION
In this study, we showed that CDC-25.1 is essential in the germ line for postembryonic germ cell proliferation while it is dispensable in some somatic tissues. These results suggest that the function of cdc-25.1 is unique in the germ line, but is not required or can be compensated by other family members in some somatic tissues. CDC-25.1 protein was previously reported to be expressed in Z2 and Z3 primordial germ cells (Ashcroft and Golden, 2002). In our study, the mRNA levels of cdc-25.1 increased during postembryonic stages in parallel with the increasing number of germ cells. It was also expressed in both fem-1(lf) and fem-3(gf) mutant backgrounds, in which only oocytes and sperm are generated, respectively. These expression patterns of cdc-25.1 support the idea that it functions at all stages for germline proliferation in both hermaphrodites and males. There are four cdc25 family members in C. elegans (Ashcroft et al., 1998). None of the three other members of the C. elegans cdc25 family, cdc-25.2, cdc-25.3, and cdc-25.4 appeared to be capable of compensating for the role of cdc-25.1 in germline proliferation. According to the previously reported microarray analysis (Reinke et al., 2004), transcripts from all four cdc25 genes were enriched in the germ line during postembryonic development. These results may suggest that all four cdc25 family members have unique roles in the germ line, and their uniqueness is likely rendered by specific regulatory mechanisms for each gene in the germ line. In contrast, we did not observe any obvious abnormalities in some somatic tissues of cdc-25.1(bn115) mutants including the gonad structures, intestine, and the vulva. The cdc25 family members may function redundantly in these somatic tissues, while in other untested somatic tissues, any of the members may execute its unique and indispensable function. Multiple genetic mutants or multiple RNAi of the cdc25 family members are required to see if they function redundantly in the somatic tissues. Examination of specific functions of each cdc25 family member as well as observation of developmental expression patterns of each member remain to be elucidated.
Acknowledgments
We thank colleagues at Indiana University for helping us initiate and pursue this project. We also thank Young-Ki Paik for comments on the manuscript. This work was supported by Forest Science & Technology Projects (No. S110707L0501101) provided by Korea Forest Service, the second Brain Korea 21 projects, and NIH grant GM34059 to S.S. Worm strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health National Center for Research Resources.
References
- Ashcroft N, Golden A. CDC-25.1 regulates germline proliferation in Caenorhabditis elegans. Genesis. 2002;33:1–7. doi: 10.1002/gene.10083. [DOI] [PubMed] [Google Scholar]
- Ashcroft NR, Kosinski ME, Wickramasinghe D, Donovan PJ, Golden A. The four cdc25 genes from the nematode Caenorhabditis elegans. Gene. 1998;214:59–66. doi: 10.1016/s0378-1119(98)00228-5. [DOI] [PubMed] [Google Scholar]
- Ashcroft NR, Srayko M, Kosinski ME, Mains PE, Golden A. RNA-Mediated interference of a cdc25 homolog in Caenorhabditis elegans results in defects in the embryonic cortical membrane, meiosis, and mitosis. Dev Biol. 1999;206:15–32. doi: 10.1006/dbio.1998.9135. [DOI] [PubMed] [Google Scholar]
- Austin J, Kimble J. glp-1 is required in the germ line for regulation of the decision between mitosis and meiosis in C. elegans. Cell. 1987;51:589–599. doi: 10.1016/0092-8674(87)90128-0. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clucas C, Cabello J, Bussing I, Schnabel R, Johnstone IL. Oncogenic potential of a C. elegans cdc25 gene is demonstrated by a gain-of-function allele. EMBO J. 2002;21:665–674. doi: 10.1093/emboj/21.4.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crittenden SL, Troemel ER, Evans TC, Kimble J. GLP-1 is localized to the mitotic region of the C. elegans germ line. Development. 1994;120:2901–2911. doi: 10.1242/dev.120.10.2901. [DOI] [PubMed] [Google Scholar]
- Hebeisen M, Roy R. CDC-25.1 stability is regulated by distinct domains to restrict cell division during embryogenesis in C. elegans. Development. 2008;135:1259–1269. doi: 10.1242/dev.014969. [DOI] [PubMed] [Google Scholar]
- Hirsh D, Oppenheim D, Klass M. Development of the reproductive system of Caenorhabditis elegans. Dev Biol. 1976;49:200–219. doi: 10.1016/0012-1606(76)90267-0. [DOI] [PubMed] [Google Scholar]
- Kawasaki I, Amiri A, Fan Y, Meyer N, Dunkelbarger S, Motohashi T, Karashima T, Bossinger O, Strome S. The PGL family proteins associate with germ granules and function redundantly in Caenorhabditis elegans germline development. Genetics. 2004;167:645–661. doi: 10.1534/genetics.103.023093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki I, Shim YH, Kirchner J, Kaminker J, Wood WB, Strome S. pGL-1, a predicted RNA-binding component of germ granules, is essential for fertility in C. elegans. Cell. 1998;94:635–645. doi: 10.1016/s0092-8674(00)81605-0. [DOI] [PubMed] [Google Scholar]
- Kim S, Park DH, Shim J. Thymidylate synthase and dihydropyrimidine dehydrogenase levels are associated with response to 5-fluorouracil in Caenorhabditis elegans. Mol Cells. 2008;26:344–349. [PubMed] [Google Scholar]
- Kimble J, Hirsh D. The postembryonic cell lineages of the hermaphrodite and male gonads in Caenorhabditis elegans. Dev Biol. 1979;70:396–417. doi: 10.1016/0012-1606(79)90035-6. [DOI] [PubMed] [Google Scholar]
- Kimble JE, White JG. On the control of germ cell development in Caenorhabditis elegans. Dev Biol. 1981;81:208–219. doi: 10.1016/0012-1606(81)90284-0. [DOI] [PubMed] [Google Scholar]
- Kimble J, Crittenden SL. Germline proliferation and its control. WormBook. 2005:1–14. doi: 10.1895/wormbook.1.13.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic I, Roy R. Organ-specific cell division abnormalities caused by mutation in a general cell cycle regulator in C. elegans. Development. 2002;129:2155–2165. doi: 10.1242/dev.129.9.2155. [DOI] [PubMed] [Google Scholar]
- Nelson GA, Lew KK, Ward S. Intersex, a temperature-sensitive mutant of the nematode Caenorhabditis elegans. Dev Biol. 1978;66:386–409. doi: 10.1016/0012-1606(78)90247-6. [DOI] [PubMed] [Google Scholar]
- Reinke V, Gil IS, Ward S, Kazmer K. Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development. 2004;131:311–323. doi: 10.1242/dev.00914. [DOI] [PubMed] [Google Scholar]
- Russell P, Nurse P. cdc25+ functions as an inducer in the mitotic control of fission yeast. Cell. 1986;45:145–153. doi: 10.1016/0092-8674(86)90546-5. [DOI] [PubMed] [Google Scholar]
- Strome S. Asymmetric movements of cytoplasmic components in Caenorhabditis elegans zygotes J. Embryol. Exp Morph. 1986a;97:15–29. [PubMed] [Google Scholar]
- Strome S, Wood WB. Generation of asymmetry and segregation of germ-line granules in early C. elegans embryos. Cell. 1983;35:15–25. doi: 10.1016/0092-8674(83)90203-9. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]