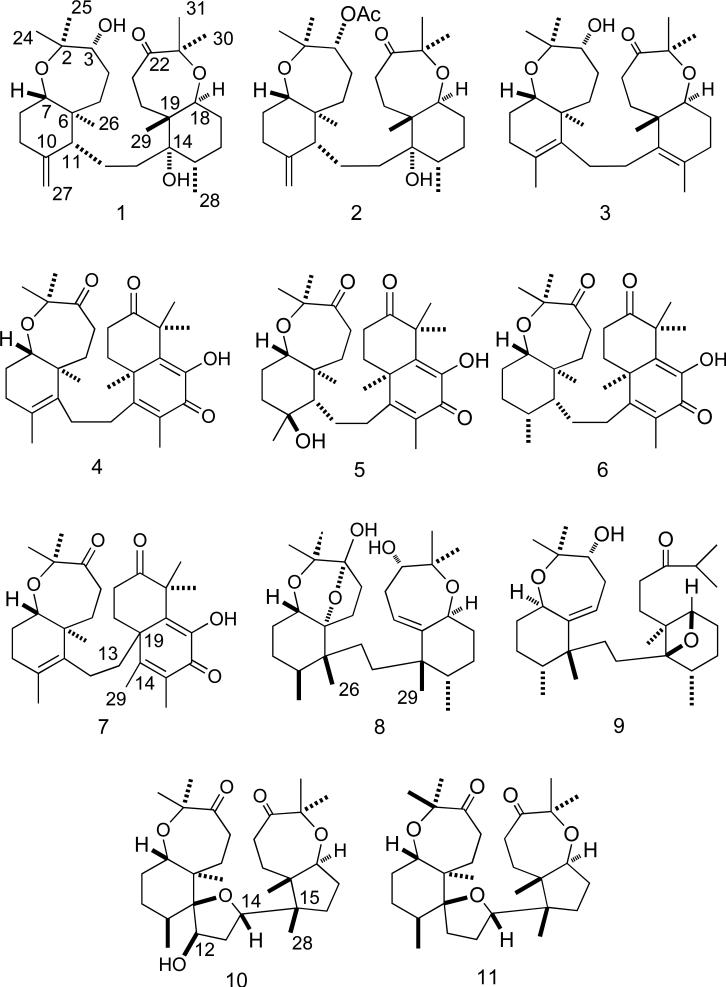
Abstract
Hypoxia-inducible factor-1 (HIF-1) is a transcription factor that promotes tumor cell adaptation and survival under hypoxic conditions. HIF-1 is currently recognized as an important molecular target for anti-cancer drug discovery. A T47D breast tumor cell-based reporter assay was used to evaluate the NCI Open Repository of marine invertebrates and algae lipid extracts for HIF-1 inhibitory activity. Bioassay-guided fractionation and isolation of an active extract from Axinella sp. yielded seven new sodwanone triterpenoids [3-epi-sodwanone K (1), 3-epi-sodwanone K 3-acetate (2), 10,11-dihydrosodwanone B (4), sodwanones T–W (3, 7, 8, 9), the new yardenone triterpene 12R-hydroxyyardenone (10), and the previously reported compounds sodwanone A (5), sodwanone B (6), and yardenone (11). The structures and relative configurations of these Axinella metabolites were determined spectroscopically. The absolute configuration of 1 was determined by the modified Mosher ester procedure. Sodwanone V (8) inhibited both hypoxia-induced and iron chelator (1,10-phenanthroline)-induced HIF-1 activation in T47D breast tumor cells (IC50 15 μM) and 8 was the only sodwanone that inhibited HIF-1 activation in PC-3 prostate tumor cells (IC50 15 μM). Compounds 1, 3, 4, and 5 inhibited hypoxia-induced HIF-1 activation in T47D cells (IC50 values 20-25 μM). Compound 2 was cytotoxic to T47D cells (IC50 22 μM) and 8 showed cytotoxicity to MDA-MB-231 breast tumor cells (IC50 23 μM).
Intratumoral hypoxia (a reduced state of oxygen tension) is a common feature of solid tumors.1,2 The extent of tumor hypoxia correlates with advanced disease stages, malignant progression, poor prognosis, and is a significant contributor to radiation and chemotherapy treatment resistance.1-3 Current approaches to overcome tumor hypoxia include those that increase oxygenation during radiotherapy and those that develop hypoxia-activated prodrugs such as radiation sensitizers and hypoxia-activated cytotoxins.1-3 No small molecule drug that specifically targets tumor hypoxia is in clinical use and there are only two hypoxic cytotoxins (tirapazamine and AQ4N) in clinical trials.1 The promising results from tirapazamine clinical trials (phase II and III) have validated the feasibility of small molecule drugs that target tumor hypoxia.4 However, the application of tirapazamine for cancer treatment is limited by toxicity.
To discover potential drug leads that target tumor hypoxia, our research efforts focus on the discovery of natural product-derived inhibitors of hypoxia-inducible factor-1 (HIF-1). First identified as a transcription factor that is activated by hypoxia,5 HIF-1 has been shown to be a major regulator of oxygen homeostasis.2,6-8 Extensive studies indicate that HIF-1 plays an important role in cancer biology by regulating the expression of hundreds of genes.2,6-8 The processes that are regulated by HIF-1 target genes range from tumorigenesis, angiogenesis, glycolysis, survival, growth, invasion, metastasis, to treatment resistance.2,6-8 In addition to hypoxia, the activation of oncogenes and/or inactivation of tumor suppressor genes can also lead to HIF-1 activation.6-8 Over-expression of the oxygen-regulated HIF-1α subunit is associated with advanced disease stages and poor prognosis in many forms of cancer.9 In animal models, HIF-1 inhibition suppresses tumor angiogenesis and retards tumor growth.10 Since HIF-1 inhibitors represent potential tumor-selective drugs with low side effects on the well-oxygenated normal tissues, intensive research efforts are currently underway to discover HIF-1 inhibitors.7,8,11 The majority of the known small molecule HIF-1 inhibitors are natural products or derived from natural products.11 Most of these compounds are also known to regulate other molecular targets at the concentrations required to inhibit HIF-1.
Using established human breast carcinoma cells as in vitro models, we have developed a T47D cell-based reporter assay for HIF-1 inhibitors and examined over 10,000 natural product-rich plant, marine invertebrate, and microbe extracts.12 Potent HIF-1 inhibitors such as manassantin B were subsequently identified from these studies.12,13 Iron chelators like desferrioxamine and 1,10-phenanthroline activate HIF-1 by inhibiting the hydroxylases that promote the degradation and inactivation of the oxygen-regulated HIF-1α subunit.12,14 Manassantin B and related compounds selectively inhibited HIF-1 activation by hypoxia, in contrast to their relatively weak ability to inhibit 1,10-phenanthroline-induced HIF-1 activation.12,13 Herein, we report the isolation and molecular characterization of eleven sodwanone/yardenone triterpenoids from an active marine sponge Axinella sp. extract, that inhibited HIF-1 activation.
Results and Discussion
In a T47D breast tumor cell-based reporter assay,12 the crude extract from a South African collection of the marine sponge Axinella sp. (at 5 μg mL-1) inhibited hypoxia (1% O2)-induced HIF-1 activation by 90% without pronounced cytotoxicity (<50%). At 5 μg mL-1, this extract also inhibited an iron chelator (1,10-phenanthroline at 10 μM)-induced HIF-1 activation by 50% in T47D cells. Five grams of the active extract were obtained from the NCI open repository. Examination of a freshly prepared sample of the extract revealed that it inhibited hypoxia-induced HIF-1 activation by 67% and 82% at the concentrations of 1 and 5 μg mL-1, respectively. Bioassay-guided fractionation yielded eight new sodwanone/yardenone triterpenoids and three previously reported sodwanones/yardenones.
The first compound isolated was a colorless oil with the molecular formula C30H50O5, as deduced from HRESIMS spectrometric and 13C NMR spectroscopic data. The IR spectrum showed a strong absorption (3412 cm-1) that indicated the presence of hydroxyl groups. The 1H NMR spectrum (Table 1) exhibited the presence of seven methyl groups (δ = 0.65, 0.80, 1.02, 1.04, 1.14, 1.18 and 1.21 ppm) and three protons attached to oxygenated carbons at δ = 3.39, 3.60, and δ = 3.73 ppm, respectively. The 13C NMR spectrum (Table 2) contained 30 carbon resonances, and the 13C DEPT spectrum indicated the presence of seven methyl groups, eleven methylenes, five methines and seven quaternary carbon atoms.
Table 1.
1H NMR Spectral Data for 1 – 4 and 7 – 10 (400 MHz, CDCl3).
| Proton | 1 | 2 | 3 | 4 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|
| 3 | 3.73 (d, 6.0) | 4.94 (d, 6.0) | 3.79 (d, 6.0) | 3.72 (d, 6.0) | ||||
| 4a | 1.86 m | 2.05 m | 1.95 m | 3.23 (t, 11.6) | 3.14 (t, 12.0) | 1.88 m | 3.01 m | 3.05 m |
| b | 1.68 m | 1.81 m | 1.72 m | 2.18 m | 2.15 m | 1.50 m | 2.31 m | 2.20 m |
| 5a | 1.55 m | 1.65 m | 1.70 m | 1.97 m | 1.94 m | 2.36 m | 5.21 brs | 1.77 m |
| b | 1.35 m | 1.50 m | 1.50 m | 1.42 m | 1.35 m | 1.12 m | 1.32 m | |
| 7 | 3.60 (dd, 11.3, 4.9) | 3.55 (dd, 11.3, 4.9) | 3.69 (dd, 11.3, 4.9) | 3.09 (dd, 12.0, 4.1) | 3.01 (dd, 11.2, 4.1) | 4.19 brs | 4.13 brs | 3.05 (dd, 11.3, 5.4) |
| 8a | 1.56 m | 1.56 m | 1.70 m | 1.75 m | 1.66 m | 1.80 m | 1.83 m | 2.00 m |
| b | 1.24 m | 1.24 m | 1.65 m | 153 m | 1.49 m | 1.40 m | 1.70 m | |
| 9a | 2.14 m | 2.11 m | 1.90 m | 2.16 m | 2.07 m | 1.32 m | 1.40 m | 1.48 m |
| b | 1.85 m | 1.88 m | 1.97 m | 1.86 m | ||||
| 10 | 1.75 m | 1.37 m | 1.91 m | |||||
| 11 | 1.49 m | 1.49 m | ||||||
| 12a | 1.45 m | 1.45 m | 2.03 m | 2.20 m | 2.20 m | 2.38 m | 1.50 m | 4.41 |
| b | 1.35 m | 1.35 m | 1.88 m | 1.90 m | 1.70 m | 1.88 m | 1.22 m | |
| 13a | 1.80 m | 1.80 m | 2.03 m | 2.64 m | 2.65 m | 1.50 m | 1.72 m | 1.93 m |
| b | 1.03 m | 1.10 m | 2.35 m | 2.35 m | 1.22 m | 1.20 m | 1.65 m | |
| 15 | 1.72 m | 1.72 m | 1.39 m | 1.35 m | 4.07 | |||
| 16 | 1.26 m | 1.26 m | 1.90 m | 1.50 m | 1.84 m | 1.55 m | ||
| 16 | 1.69 m | 1.45 m | ||||||
| 17 | 1.43 m | 1.43 m | 1.70 m | 1.83 m | 1.50 m | 1.62 m | ||
| 17 | 1.61 m | 1.40 m | 1.48 m | |||||
| 18 | 3.39 (dd, 10.4, 5.2) | 3.45 (dd, 10.4, 5.2) | 3.06 (dd, 9.6, 4.9) | 4.01 m | 3.96 brs | 3.84 (dd, 9.6, 4.9) | ||
| 20a | 1.87 m | 1.87 m | 2.03 m | 2.47 m | 2.63 m | 5.23 brs | 2.48 m | 1.74 m |
| b | 1.50 m | 1.55 m | 1.48 m | 1.77 m | 1.70 m | 2.30 m | 1.30 m | |
| 21a | 3.08 m | 3.11 m | 3.19 m | 2.80 m | 2.68 m | 3.04 m | 1.50 m | 3.04 m |
| b | 2.05 m | 2.05 m | 2.18 m | 2.62 m | 2.59 m | 2.25 m | 2.22 m | |
| 22 | 3.74 (d, 5.6) | |||||||
| 23 | ||||||||
| 24 | 1.18 s | 1.17 s | 1.25 s | 1.34 s | 1.28 s | 1.45 s | 1.26 s | 1.20 s |
| 25 | 1.02 s | 1.03 s | 1.16 s | 1.30 s | 1.24 s | 1.30 s | 1.19 s | 1.19 s |
| 26 | 0.65 s | 0.70 s | 1.02 s | 1.24 s | 1.05 s | 0.86 s | 1.02 s | 0.95 s |
| 27 | 4.80 s, 4.46 s | 4.90 s, 4.48 s | 1.63 s | 1.73 s | 1.49 s | 1.18 (d, 6.5) | 0.86 s | 1.08 (d, 6.5) |
| 28 | 0.80 (d, 6.5) | 0.80 (d, 6.5) | 1.65 s | 2.05 s | 1.94 s | 0.88 (d, 6.5) | 0.88 s | 0.70 s |
| 29 | 1.04 s | 1.04 s | 1.12 s | 1.17 s | 1.24 s | 1.02 s | 1.27 s | 1.09 s |
| 30 | 1.21 s | 1.21 s | 1.29 s | 1.56 s | 1.53 s | 1.24 s | 1.12 s | 1.20 s |
| 31 | 1.14 s | 1.13 s | 1.25 s | 1.50 s | 1.48 s | 1.10 s | 1.10 s | 1.20 s |
Table 2.
13C NMR Spectral Data for 1 – 4 and 7 – 10 (100 MHz, CDCl3).
| Carbon | 1 | 2 | 3 | 4 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|
| 2 | 76.5 | 79.1 | 77.9 | 82.7 | 82.7 | 77.8 | 77.5 | 81.6 |
| 3 | 76.2 | 78.3 | 77.0 | 218.0 | 217.8 | 104.2 | 77.8 | 217.9 |
| 4 | 25.8 | 23.5 | 25.9 | 35.6 | 35.6 | 23.8 | 30.4 | 34.8 |
| 5 | 33.5 | 33.5 | 32.6 | 38.1 | 37.7 | 32.9 | 115.3 | 31.8 |
| 6 | 43.2 | 43.3 | 43.0 | 42.3 | 42.3 | 88.7 | 149.0 | 45.3 |
| 7 | 75.4 | 76.1 | 74.9 | 79.7 | 79.5 | 71.3 | 71.9 | 77.5 |
| 8 | 32.6 | 32.8 | 27.4 | 31.1 | 31.7 | 26.8 | 36.1 | 30.2 |
| 9 | 35.3 | 35.3 | 32.1 | 26.9 | 26.9 | 28.8 | 28.7 | 29.7 |
| 10 | 147.0 | 146.9 | 126.8 | 129.0 | 129.0 | 33.5 | 42.2 | 32.7 |
| 11 | 54.7 | 55.0 | 137.9 | 135.5 | 135.0 | 41.4 | 42.9 | 92.5 |
| 12 | 19.7 | 19.7 | 29.2 | 28.6 | 23.7 | 26.4 | 25.8 | 74.4 |
| 13 | 35.9 | 35.6 | 29.2 | 32.2 | 35.4 | 30.5 | 29.6 | 39.6 |
| 14 | 77.8 | 77.8 | 137.0 | 163.4 | 160.0 | 43.7 | 86.5 | 82.8 |
| 15 | 33.2 | 33.2 | 127.2 | 129.5 | 127.8 | 42.1 | 51.1 | 48.2 |
| 16 | 27.9 | 28.2 | 32.0 | 181.4 | 179.5 | 29.4 | 26.2 | 26.1 |
| 17 | 29.8 | 30.1 | 27.1 | 141.9 | 145.0 | 36.0 | 38.0 | 28.5 |
| 18 | 75.9 | 76.2 | 79.9 | 137.6 | 137.5 | 71.7 | 82.0 | 81.9 |
| 19 | 45.0 | 45.3 | 42.2 | 43.0 | 46.1 | 149.0 | 49.5 | 48.8 |
| 20 | 32.4 | 32.4 | 37.9 | 27.0 | 26.7 | 116.0 | 36.1 | 31.6 |
| 21 | 34.9 | 34.9 | 35.8 | 32.5 | 37.6 | 30.1 | 24.8 | 35.5 |
| 22 | 218.0 | 218.0 | 218.0 | 214.0 | 213.0 | 78.7 | 215.0 | 216.0 |
| 23 | 81.4 | 81.8 | 82.6 | 48.1 | 48.1 | 78.7 | 40.6 | 82.6 |
| 24 | 21.0 | 21.0 | 21.7 | 20.8 | 20.7 | 18.7 | 19.5 | 20.2 |
| 25 | 28.7 | 28.9 | 29.0 | 26.4 | 26.9 | 24.5 | 29.8 | 26.9 |
| 26 | 11.8 | 12.0 | 18.3 | 17.9 | 17.6 | 17.5 | 22.7 | 15.2 |
| 27 | 106.9 | 107.1 | 20.5 | 20.8 | 19.7 | 15.8 | 16.4 | 21.4 |
| 28 | 15.8 | 15.8 | 20.5 | 12.5 | 11.9 | 16.5 | 13.5 | 20.2 |
| 29 | 14.1 | 14.0 | 17.8 | 21.6 | 24.2 | 20.2 | 18.1 | 15.8 |
| 30 | 20.0 | 20.0 | 20.8 | 20.5 | 22.1 | 20.2 | 18.3 | 22.1 |
| 31 | 26.5 | 26.9 | 26.7 | 24.3 | 24.8 | 30.5 | 18.3 | 26.1 |
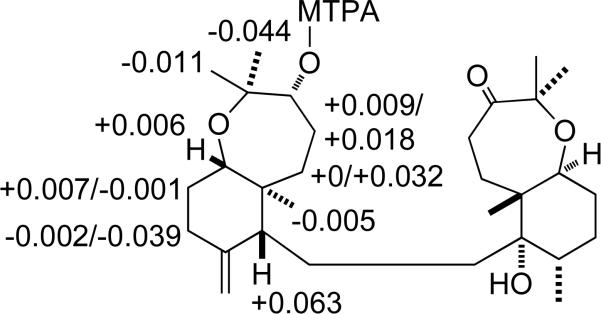
Analysis of the 1H–1H COSY and 1H–13C HMQC spectra suggested the presence of five clear 1H–1H spin systems: -CH(3)-CH2(4)-CH2(5)-, -CH(7)-CH2(8)-CH2(9)-, -CH(11)-CH2(12)-CH2(13)-, -CH3(28)-CH(15)-CH2(16)-CH2(17)-CH(18)-, and a -CH2(20)-CH2(21)- system. Long-range 1H-13C correlations were observed by 1H–13C HMBC (Figure 1) between C-2 and H-3, H-4, H-24, H-25; from C-6 to H-4, H-5, H-7, H-11, H-26; from C-10 to H-9, H-11, H-27; C-14, H-13, H-29, H-15, H-28; from C-19 to H-20, H-18, H-29; from C-22 to H-21, H-30, H-31; as well as between C-23 and H-21, H-30, H-31, to establish the carbon skeleton and the substitution pattern of functional groups that was identical to that of sodwanone K, a triterpenoid previously isolated from the marine sponge Axinella weltneri.15 Comparison of the 13C NMR spectroscopic data of 1 with those of sodwanone K, showed nearly complete analogy of the chemical shifts, except for C-2 and C-3. Clear NOESY correlations between H-24 and H-7; and between H-25 and H-26 suggested that the relative configuration of this portion of 1 was similar to that of sodwanone K, except that a NOE between H-3 and H-24 indicated that 1 was the C-3 epimer of sodwanone K. The absolute configuration of C-3 was determined using the modified Mosher ester method.16 Two separate samples of 1 were each treated with (R)-(+)- and (S)-(–)-α-methoxy-α-(trifluoromethyl)phenylacetyl chloride (MTPA) in anhydrous pyridine, respectively, to yield the (S)- and (R)-MTPA ester derivatives 1s and 1r. The 1H NMR chemical shift values of the (R)-MTPA ester derivative (1r) were subtracted from the values of the (S)-MTPA ester (1s) [Δδ= δ (S)-MTPA – δ (R)-MTPA]. The distribution of positive and negative Δδ values around the MTPA esters (Figure 2) indicated an R-configuration for C-3. Therefore, 1 was deduced to be 3-epi-sodwanone K (1).
Figure 1.
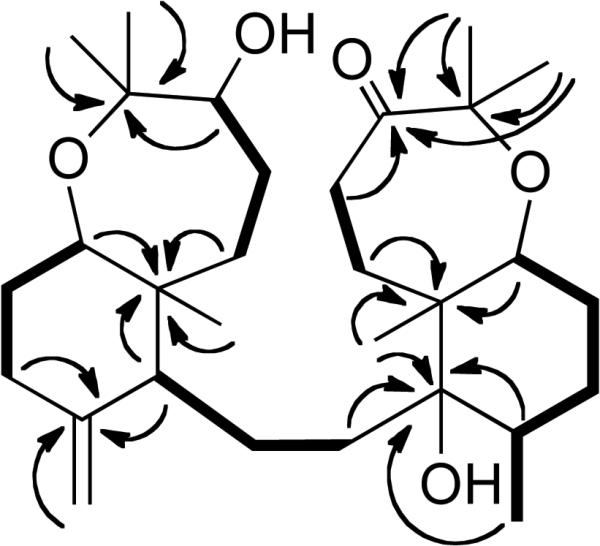
Selected 1H-1H COSY (bold solid bars) and 1H-13C HMBC (arrows) correlations of 1.
Figure 2.
Selected Δδ values [Δδ (ppm) = δS - δR] for (R)- and (S)-MTPA esters of 1.
In contrast to 1, compound 2 contained one acetate ester. The 1D (1H, 13C and 13C DEPT) and 2D (COSY, HMQC and HMBC) NMR spectra of 2 were similar to those of 1, except for the presence of the acetate ester and the H-3 proton resonance. The chemical shift observed for the H-3 resonance in 1 was shifted downfield from δ = 3.73 to δ = 4.94 ppm with respect to the resonance observed for H-3 in 2 and were consistent with a proposed structure with a C-3-substituted acetate ester. Therefore, compound 2 was deduced to be 3-epi-sodwanone K 3-acetate (2).
Compound 3 was obtained as a colorless resin with the molecular formula C30H48O4, as deduced from HRESIMS data. Its IR spectrum possessed a strong hydroxyl absorption at 3425 cm-1. Comparison of the 1H and 13C NMR spectra of 3 (Tables 1, 2) with those of 1 revealed that 3 was a structurally related sodwanone triterpene. However, the presence of olefinic resonances in the 13C NMR spectrum of 3, the downfield chemical shift of the H3-28 methyl group at δ 1.65 ppm in the 1H NMR spectrum, and the additional methyl group (at δ 1.63 ppm), indicated that 3 was consistent with a proposed structure with double bonds located at C-10,11 and C-14,15. Long-range 1H-13C HMBC couplings between both the H-9 and H-27 resonances and C-10 (δ 126.8) further supported the location of the double bonds. In addition, the proton resonances at H-12, H-26 and H-27 were coupled to C-11 (δ 137.9); H-13, H-29 and H-28 were coupled to C-14 (δ 137.0); and the H-16 and H-28 proton resonances were coupled to C-15 (δ 127.2). Comparison of the 13C NMR spectroscopic data with those of sodwanone O17 revealed that the chemical shifts of the bicyclic C2 – C11 portions of the structures were analogous. This suggested that 3 is a structurally related new Axinella triterpene, herein referred to as sodwanone T.
The molecular formula of 4 was determined to be C30H42O5 by HRESIMS and 13C NMR data. Analysis of the 13C NMR spectrum (Table 2) revealed the presence of 30 carbons and the 13C DEPT experiment indicated the presence of eight methyl groups, eight methylenes, one methine and 13 quaternary carbons atoms. Portions of the 1H NMR spectrum (Table 1) were similar to those of the previously reported compound sodwanone A (5).18 The major differences in the spectra were that 5 contained two additional olefinic resonances in the 13C NMR. The remaining 13C resonances, the proton chemical shifts, and coupling constants were consistent with most of the spectrum of 5. Long-range 1H-13C HMBC couplings were observed in 4 (Figure 3) between the H-9 and H-27 proton resonances and C-10 (δ 129.0), and the H-12, H-26 and H-27 proton resonances were coupled to C-11 (δ 135.5). The 1H NMR resonances and the 1H-1H coupling patterns observed for the bicyclic C-14 – C-23 portions of 4 were identical to that of 5. This indicated that 4 is a new unsaturated version of the previously identified compound sodwanone B (6)18 that can be referred to as 10,11-dihydrosodwanone B (4).
Figure 3.
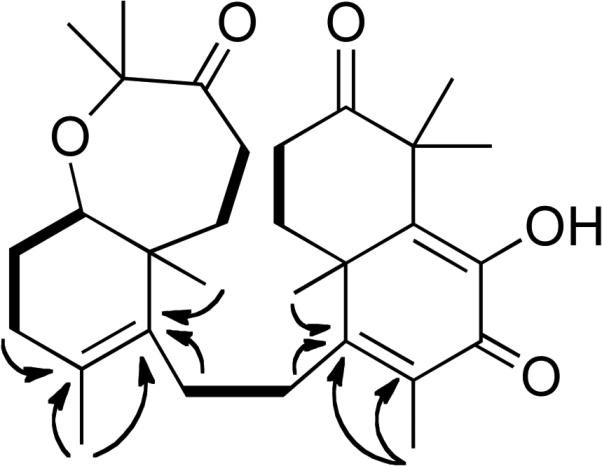
Selected 1H-1H COSY (bold solid bars) and 1H-13C HMBC (arrows) correlations of 4.
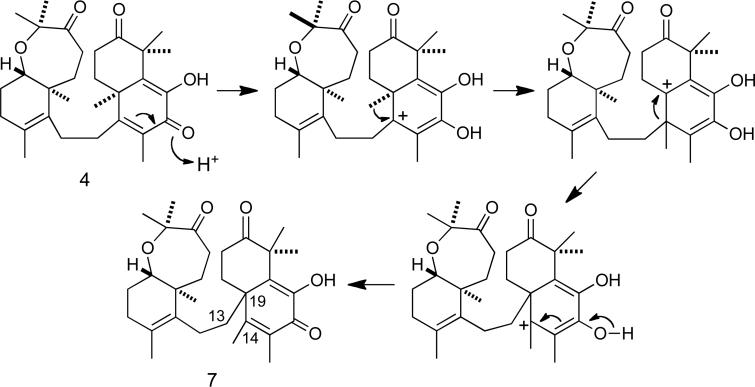
The molecular formula of 7 was determined to be the same as that of 10,11-dehydrosodwanone B (4) (C30H42O5) by HRESIMS and analysis of the 13C NMR spectrum. Comparison of the 1H and 13C NMR spectra of 7 (Tables 1 and 2) with those of 4 indicated that the bicyclic C-2–C-11 portion of the structures were analogous in both compounds. Long-range 1H-13C HMBC couplings (Figure 4) were observed between the H-20 and H-13 proton resonances and the C-19 resonance at δ 46.1; the H-28 and H-29 proton resonances were coupled to C-15 (δ 127.8); and H-28, H-29 and OH-17 were coupled to C-16 (δ 179.5). Thus 7 possess a new carbon skeleton where the two separate bicyclic portions of the structure are connected at the bridgehead carbon C-19 by the C-12 – C-13 linker. This new sodwanone analog was assigned the trivial name sodwanone U (7). The occurrence of 7 presents a biogenetic problem. If a divergence in the biosynthetic pathways for the formation of most sodwanones is responsible for the production of 7, it would appear likely that alternate ring formation would result in a significantly different functional group substitution pattern. Therefore, the occurrence of 7 appears most likely to result from an acid-catalyzed rearrangement of 4, as in Figure 5.
Figure 4.
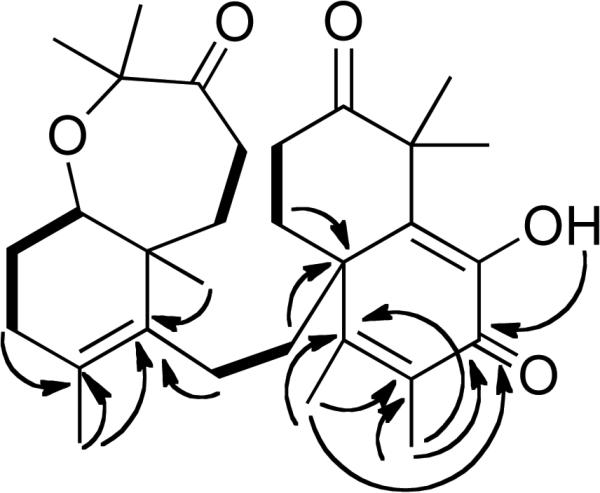
Selected 1H-1H COSY (bold solid bars) and 1H-13C HMBC (arrows) correlations of 7.
Figure 5.
Proposed mechanism for the formation of 7.
The molecular formula of 8 was determined to be C30H50O5 by a combination of HRESIMS and 13C NMR data. Comparison of the NMR spectra of 8 (Table 1 and 2) with those of the known sodwanones suggested that 8 possessed the same tricyclic C-2 – C-11 system as previously reported for sodwanone F19 and the same bicyclic C-14 – C-23 system as reported in the structure of sodwanone S.20 This was confirmed by evaluation of the COSY, HMQC, and HMBC spectra. Additional analysis of the COSY and HMQC spectra of 8 suggested the presence of five partial structures: [-CH2(4)-CH2(5)-, -CH(7)-CH2(8)-CH2(9)-CH(10)-CH3(27), -CH2(12)-CH2(13)-, -CH3(28)-CH(15)-CH2(16)-CH2(17)-CH(18)- and -CH(20)-CH2(21)-CH(22)-]. Connection of the partial structures was facilitated by interpretation of the 1H-13C HMBC spectrum. Specifically, long-range 1H-13C couplings were observed between C-3 and H-4, H-5, H-24, H-25; from C-6 to H-4, H-5, H-7, H-8, H-26; from C-11 to H-5, H-7, H-10, H-12, H-26; from C-14 to H-13, H-15, H-18, H-20, H-28, H-29, as well as between C-19 and H-18, H-20, H-29. Therefore, compound 8 was deduced to be sodwanone V.
The molecular formula of 9 was deduced to be C30H50O4 by interpretation of the HRESIMS and 13C NMR data. Comparison of the 1H and 13C NMR spectra of 9 (Table 1 and 2) with those of known sodwanones suggested that 9 had the same tricyclic C-2 – C-11 system as in sodwanone S20 and the C14-C18 epoxide-bridged C14-C19 cyclohexane ring and C-19 substituted side chain (C20 – C31) that were previously reported for the structure of sodwanone I.21 Analysis of the COSY and HMQC spectra of 9 confirmed these partial structures and facilitated the deduction of the six partial structures: [-CH(3)-CH2(4)-CH(5)-, -CH(7)-CH2(8)-CH2(9)-CH(10)-CH3(27), -CH2(12)-CH2(13)-, -CH3(28)-CH(15)-CH2(16)-CH2(17)-CH(18)-, -CH2(20)-CH2(21)- and -CH3(30)-CH(23)-CH3(31)-]. As in 8, the partial structures of 9 were readily connected by interpretation of long-range 1H-13C HMBC couplings from C-6 to H-4, H-5, H-7, H-8, H-26; from C-11 to H-5, H-7, H-10, H-12, H-26, H-27; from C-14 to H-13, H-15, H-18, H-20, H-28, H-29, and between C-23 and H-21, H-23, H-30, H-31. In this new sodwanone, trivially named sodwanone W (9), the ether link of the seven-membered cyclic ether (C18 – C23) was opened and an additional ether bridge was formed between C14 and C18. The relative configuration of 9 was deduced from the NOESY spectrum, where NOE interactions were observed between H-7/H-25, H-3/H-24, H-10/H-26, H-18/H-29 and H-28/H-29.
Compound 10 was obtained as colorless needles with the molecular formula C30H48O6, as deduced by HRESIMS. The IR spectrum suggested the presence of hydroxyl (3438 cm-1) and carbonyl (1712 cm-1) groups. The 13C NMR spectrum of 10 (Table 2) showed carbon resonances for two keto-carbonyls (δC 217.9 and 216.0) and seven other oxygen-substituted carbon resonances, three of which were quaternary carbons (δC 92.5, 82.6, 81.6) and four were methines (δC 82.8, 81.9, 77.5, 74.4). The 1H NMR spectrum of 10 (Table 1) showed the presence of seven tertiary methyl groups and one methyl resonance at δH 1.08 (d, J = 6.5 Hz). The 13C NMR resonances observed in 10 were very similar to those of the previously reported compound yardenone22 (11) except for the chemical shift of C-12 (Table 2). As the major difference, the C-12 methylene resonance observed in 11 (δC 28.5) was replaced with a downfield methine resonance at δC 74.4, typical for an oxygen-bearing carbon. Analysis of the 1H–1H COSY and 1H-13C HMQC spectra suggested the presence of five distinct spin systems: [-CH2(4)-CH2(5)-, -CH(7)-CH2(8)-CH2(9)-CH (10)-CH3(27)-, -CH (12)-CH2(13)-CH(14)-, -CH2(16)-CH2(17)-CH(18)- and -CH2(20)-CH2(21)-]. Long-range couplings were observed in the 1H-13C HMBC spectrum (Figure 6) between C-11 and H-10, H-12, H-26, H-27, and from C-15 to H-14, H-16, H-28, H-29, suggesting that the structure contained a hydroxyl group at C-12. The relative configuration of 10 was assigned to be the same as in 11 since both have nearly identical NOESY correlations and JH-H coupling constants. However, a clear NOE was observed in the NOESY spectrum between H-12 and H-26, while there was no NOE observed between H-12 and H-27. Therefore, this metabolite is a C-12 hydroxylated form of the previously reported compound yardenone22 (11) and is trivially named 12R-hydroxyyardenone (10).
Figure 6.
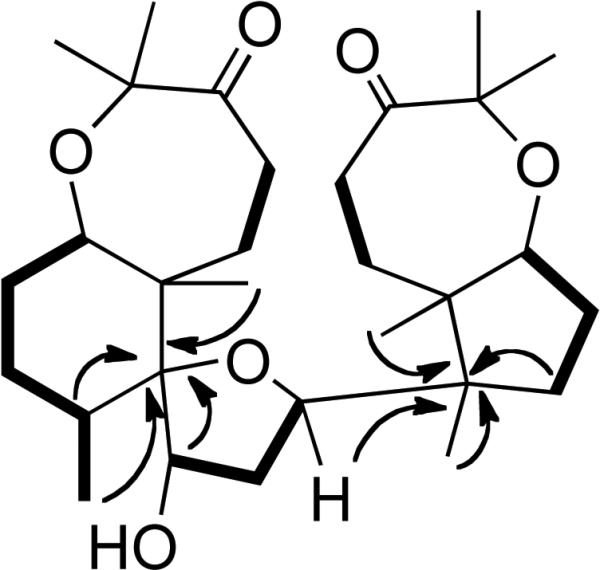
Selected 1H-1H COSY (bold solid bars) and 1H-13C HMBC (arrows) correlations of 10.
Three previously reported Axinella triterpenes were also isolated and identified as sodwanone A (4), sodwanone B (6), and yardenone (11) by comparison of their 1H and 13C NMR spectra with those reported in literature.18,22
Concentration-response studies were performed to determine the effects of 1-11 on HIF-1 activation and the data are shown in the Supporting Information. In T47D breast tumor cells, the most active compound 8 inhibited both hypoxia (1% O2)- and an iron chelator (10 μM 1,10-phenanthroline)-induced HIF-1 activation with an IC50 of 15 μM. The IC50 values for 1-5 to inhibit hypoxia-induced HIF-1 activation range from 20 to 25 μM in a T47D cell-based reporter assay. Compounds 3 and 4 also inhibited 1,10-phenanthroline-induced HIF-1 activation with comparable IC50 values (20 and 30 μM, respectively). The remaining compounds inhibited by less than 50% at the highest concentration tested (30 μM). Among the compounds examined, only 8 inhibited hypoxia (1% O2)-induced HIF-1 activation with an IC50 of 15 μM in PC-3 prostate tumor cells. Under experimental conditions (hypoxia for 16 h), no statistically significant cytotoxicity was observed in T47D cells. In the T47D cell-based primary screening assay, the crude extract from this Axinella sp. inhibited hypoxia-induced HIF-1 activation by 90% at 5 μg mL-1. If we assume that the compounds responsible for the HIF-1 inhibitory activity in the crude extract had a molecular weight of 500 and the yield was 1%, then the anticipated IC50 value of the purified active compound should be sub-micromolar. However, none of the compounds (1-11) displayed this level of expected potency. One possible explanation is that the potent HIF-1 inhibitory activity exhibited by the crude extract was caused by a synergistic effect of the compounds identified. To test this hypothesis, a mixture of 1-8 and 10 was prepared and the effect of the mixture on HIF-1 activation examined. Compounds 1-8 and 10 were mixed at the ratio of 10:1:5:2.5:7:1:2:1:5, based on the yield of each respective compound from the extract. In a T47D cell-based reporter assay, the mixture (5 μg mL-1) inhibited hypoxia-induced HIF-1 activation by 41 ± 6% and 1,10-phenanthroline-induced HIF-1 activation by 37 ± 3% (n = 3). Therefore, significant synergistic HIF-1 inhibitory activity does not appear to exist between these compounds. The most active constituents may have been lost in the separation. It is noted that considerable mass from each of the active fractions irreversibly bound to the CC Si gel during the isolation process. Certain sodwanones have been proposed to be generated via cyclization of unstable epoxide intermediates.20 It remains possible that a such structurally related unstable analog(s) is responsible for the potent HIF-1 inhibitory activity and that, many if not most, of the sodwanone triterpenes isolated from various collections of Axinella species actually represent degradation artifacts.
Cell line-dependent cytotoxicity has been reported for sodwanones A, G, H, and S.15,20-22 The effect of 1-11 on tumor cell proliferation/viability was examined in T47D and MDA-MB-231 breast tumor cells and in PC-3 and DU145 prostate tumor cells. Following 48 hr incubation under normoxic conditions (95% Air:5% CO2), 2 inhibited T47D cell proliferation/viability with an IC50 of 22 μM and 8 inhibited MDA-MB-231 cells with an IC50 of 23 μM. None of the other compounds inhibited by greater than 50% at the highest concentration tested (30 μM). A previous study indicated that sodwanone A is cytotoxic to DU-145 cells (IC50 3.6 μM).20 In our study, the IC50 for sodwanone A (5) to inhibit DU-145 cell proliferation/viability was greater than 30 μM. This variation in cytotoxic activities may be caused by the extremely poor solubility of sodwanone A in aqueous solution. In fact, crystals were observed in the tissue culture medium that contained sodwanone A at 30 μM.
Experimental Section
General Experimental Procedures
Optical rotation was measured on an Autopol IV Automatic Polarimeter. The IR spectrum was obtained using a Genesis Series FTIR. The NMR spectra were recorded in CDCl3 on Bruker AMX-NMR spectrometers operating at either 400 or 500 MHz for 1H and either 100 or 125 MHz for 13C, respectively. The NMR spectra were recorded running gradients and using residual solvent peaks (δ 7.27 for 1H) and (δ 77.0 for 13C) as internal references. The HRESIMS spectra were measured using a Daltonic micro TOF with electrospray ionization. Silica gel (200-400 mesh) was used for column chromatography. TLCs were run on Merck Si60F254 or Si60RP18F254 plates and visualized under UV at 254 nm or by heating after spraying with a 1% anisaldehyde solution in acetic acid:H2SO4 (50:1).
Sponge Material
The sponge material was obtained from the National Cancer Institute's Open Repository Program. A new species of Axinella (Axinellidae) was collected (collection C020287) at a depth of -16 M off the southeastern coast of South Africa on March 22, 2000 (photograph in Supporting Information). While the complete taxonomic characterization of this species has not been fully completed, the sample was identified to be a new Axinella species by Dr. Michele Kelly (National Institute of Water and Atmospheric Research Limited, Auckland, New Zealand). The sample was frozen at -20° C and ground in a meat grinder. A voucher sample was deposited in the Natural History Museum, Washington, D.C. in the Department of Worms under the collection number 0CDN7239.
Extraction and Isolation
Ground sponge material was extracted with H2O. The residual sample was then lyophilized and extracted with CH2Cl2:MeOH (1:1), residual solvents were removed under vacuum, and the crude extract stored at -20° C in the NCI repository at the Frederick Cancer Research and Development Center (Frederick, Maryland). The crude extract of the marine sponge Axinella sp. was subjected to a process of bioassay-guided fractionation. The crude extract (5 g) was separated into five fractions by CC on Si gel (50 g), using gradients of hexanes and EtOH (85:15, 50:50, 0:100). The active fractions 1-5 (0.5 mg mL-1 HIF-1 assay inhibition values 46%, 74%, 86%, 44%, and 81%, respectively) were further separated by a second level of CC to produce compounds 1 (10.0 mg, 0.2% yield), 2 (1.0 mg, 0.02% yield), 3 (5.0 mg, 0.1% yield), 4 (2.5 mg, 0.05% yield), 5 (7.0 mg, 0.14% yield), 6 (1.0 mg, 0.02% yield), 7 (2.0 mg, 0.04% yield), 8 (1.0 mg, 0.02% yield), 9 (5.0 mg, 0.1% yield), 10 (5.0 mg, 0.1% yield), and 11 (10.0 mg, 0.2% yield).
3-epi-Sodwanone K (1)
Colorless oil: [α]D24 +6.5 (c 0.20, CH2Cl2); IR (film) νmax 3412, 2935, 1709 cm-1; 1H and 13C NMR data in Tables 1 and 2; HREIMS: m/z 490.3636 (calcd for C30H50O5 490.3658).
Preparation of (R)- and (S)-MTPA esters of 3-epi-sodwanone K (1)
Compound 1 (2.0 mg, 4.0 μmol) was dissolved in pyridine-d5 (180 μL) and treated with 10 μL (53.4 μmol) of (R)-(-)-MTPA chloride at room temperature in a NMR tube for 12 h to yield the (S)-MTPA ester 1s. The (R)-MTPA ester (1r) was prepared from 1 (2.0 mg, 4.0 μmol) and (S)-(+)-MTPA chloride (53.4 μmol) using the same procedure as described for 1s.
3-epi-Sodwanone K, (S)-MTPA ester (1s)
1H NMR (pyridine-d5, 400 MHz) 0.69 (s, H-26), 1.17 (s, H-24), 1.04 (s, H-25), 2.00 (m, H-4β), 1.88 (m, H-4α), 1.54 (m, H-5β), 1.41 (m, H-5α), 1.50 (m, H-11), 1.537 (m, H-8β), 1.23 (m, 8α), 2.202 (m, H-9β), 1.88 (m, 9α), 3.53 (dd, 11.2, 4.9, H-7).
3-epi-Sodwanone K, (R)-MTPA ester (1r)
1H NMR (pyridine-d5, 400 MHz) δ 0.70 (s, H-26), 1.18 (s, H-24), 1.08 (s, H-25), 1.99 (m, H-4β), 1.87 (m, H-4α), 1.54 (m, H-5β), 1.38 (m, H-5α), 1.44 (m, H-11), 1.538 (m, H-8β), 1.22 (m, 8α), 2.204 (m, H-9β), 1.93 (m, 9α), 3.52 (dd, 11.2, 4.9, H-7).
3-epi-Sodwanone K 3-acetate (2)
Colorless oil: [α]D24 +5.2 (c 0.15, CH2Cl2); IR (film): νmax 3410, 2089, 1644 cm-1; 1H and 13C NMR data in Tables 1 and 2; HRESIMS: m/z 532.3731 (calcd for C32H52O6 532.3764).
Sodwanone T (3)
Colorless oil: [α]D24 -78.7 (c 0.14, CH2Cl2); IR (film) νmax 3425, 2976, 2939, 1713, 1645 cm-1; 1H and 13C NMR data in Tables 1 and 2; HRESIMS: m/z 472.3559 (calcd for C30H48O4 472.3553).
10,11-Dihydrosodwanone B (4)
White powder: [α]D24 -8.9 (c 0.06, CH2Cl2); IR (film) νmax 3413, 1713, 1625 cm-1; UV (CH2Cl2) λmax (log ε) 262 (3.58), 300 (3.23); 1H and 13C NMR data in Tables 1 and 2; HRESIMS: m/z 482.3053 (calcd for C30H42O5 482.3032).
Sodwanone U (7)
White powder: [α]D24 +6.0 (c 0.13, CH2Cl2); IR (film) νmax 3436, 1641 cm-1; UV (CH2Cl2) λmax (log ε) 256 (3.66), 300 (3.33); 1H and 13C NMR data in Tables 1 and 2; HREIMS: m/z 482.3031 (calcd for C30H42O5 482.3032).
Sodwanone V (8)
Colorless oil: [α]D24 +1.3 (c 0.30, CH2Cl2); IR (film) νmax 3442, 2360, 1642 cm-1; 1H and 13C NMR data in Tables 1 and 2; HREIMS: m/z 490.3676 (calcd for C30H50O5 490.3658).
Sodwanone W (9)
Colorless oil: [α]D24 -9.8 (c 0.30, CH2Cl2); IR (film) νmax 3411, 2974, 2360, 1633, 1585 cm-1; 1H and 13C NMR data in Tables 1 and 2; HREIMS: m/z 474.3707 (calcd for C30H50O4 474.3709).
12R-Hydroxyyardenone (10)
White powder: [α]D24 +74.7 (c 0.35, CH2Cl2); IR (film) νmax 3438, 2976, 1712, 1442, 1031 cm-1; 1H and 13C NMR data in Tables 1 and 2; HRESIMS: m/z 504.3442 (calcd for C30H48O6 504.3451).
Cell-Based Reporter Assay for HIF-1 Activity
The transfection, compound treatment, exposure to hypoxic conditions (1% O2:5% CO2:94% N2), normoxic conditions (5% CO2:95% Air), and a hypoxia mimetic (10 μM 1,10-phenanthroline), and luciferase activity determination were performed as previously described.12 The compounds were prepared as 10 mM stock solutions in isopropanol and stored at -80° C.
Neutral Red Assay for Cell Proliferation/Viability
The T47D, MDA-MB-231, PC-3, and DU145 tumor cells were purchased from ATCC and maintained in DMEM/F12 medium (JRH Biosciences) supplemented with 10% (v/v) FBS (Hyclone) and antibiotics (50 U mL-1 penicillin G sodium, and 50 μg mL-1 streptomycin, GIBCO) under humidified atmosphere (5% CO2:95% Air) at 37° C. Exponentially grown cells were plated at the density of 30,000 cells per well in a volume of 100 μL DMEM/F12 medium with 10% FBS and antibiotics into 96-well plates (Corning). After 24 h, test compounds were diluted in serum-free DMEM/F12 with antibiotics and added to the wells in a volume of 100 μL (n = 3). The incubation continued for another 48 h at 37° C. Cell viability was determined by the Neutral Red method24 with modifications described earlier.25 The following formula was used to calculate % inhibition of cell proliferation/viability: % Inhibition = 1 - OD540(treated)/OD540(control).
Supplementary Material
Supporting Info
S1. IC50 values of 1-11 on HIF-1 activation in T47D and PC-3 cells, and cell proliferation/viability in T47D, MDA-MB-231, PC-3, and DU145 cells.
S2. New species of Axinella (Axinellidae) collected (collection C020287) at a depth of -16 M off the southeastern coast of South Africa on March 22, 2000. Photograph provided by Dr. M.T. Davies-Coleman (Dept. of Chemistry, Rhodes University, Grahamstown, South Africa).
Acknowledgment
The authors thank the Natural Products Branch Repository Program at the National Cancer Institute for providing marine extracts from the NCI Open Repository used in these studies, T. Smillie (NCNPR, University of Mississippi) for coordinating sample acquisition from the NCI, D.K. Jones (NCNPR, University of Mississippi) for screening NCI samples in the HIF-1 assay, S.L. McKnight (University of Texas Southwestern Medical Center at Dallas) for providing the pTK-HRE3-luc construct, M.T. Davies-Coleman (Dept. of Chemistry, Rhodes University, Grahamstown, South Africa) for providing a photograph of the Axinella sp., and F.T. Wiggers (NCNPR, University of Mississippi) for obtaining some NMR spectra. This work was supported by the National Institutes of Health-NCI CA 98787-03 (DGN/YDZ), the DOD-Prostate Cancer Research Program PC040931 (DGN), and NOAA NURP/NIUST NA16RU1496. This investigation was conducted in a facility constructed with support from Research Facilities Improvement Grant No. C06 RR-14503-01 from the National Institutes of Health. The U.S. Army Medical Research Acquisition Activity, 820 Chandler Street, and Fort Detrick MD 21702-5014 is the awarding and administering acquisition office for the DOD support. The content herein reported does not necessarily reflect the position or the policy of the Government, and no official endorsement should be inferred.
Footnotes
Supporting Information Available: Table with the IC50 values of 1-11 on HIF-1 activation in T47D and PC-3 cells, and cell proliferation/viability in T47D, MDA-MB-231, PC-3, and DU145 cells. Photograph of the Axinella sponge collection C020287. The material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Brown JM, Wilson WR. Nat. Rev. Cancer. 2004;4:437–447. doi: 10.1038/nrc1367. [DOI] [PubMed] [Google Scholar]
- 2.Harris AL. Nat. Rev. Cancer. 2002;2:38–47. doi: 10.1038/nrc704. [DOI] [PubMed] [Google Scholar]
- 3.Weinmann M, Welz S, Bamberg M. Curr. Med. Chem. - Anti-Cancer Agents. 2003;3:364–374. doi: 10.2174/1568011033482350. [DOI] [PubMed] [Google Scholar]
- 4.a Rischin D, Peters L, Fisher R, Macann A, Denham J, Poulsen M, Jackson M, Kenny L, Penniment M, Corry J, Lamb D, McClure B. J. Clin. Oncol. 2005;23:79–87. doi: 10.1200/JCO.2005.01.072. [DOI] [PubMed] [Google Scholar]; b Williamson SK, Crowley JJ, Lara PN, Jr, McCoy J, Lau DH, Tucker RW, Mills GM, Gandara DR. J. Clin. Oncol. 2005;23:9097–9104. doi: 10.1200/JCO.2005.01.3771. [DOI] [PubMed] [Google Scholar]; c Rischin D, Hicks RJ, Fisher R, Binns D, Corry J, Porceddu S, Peters LJ. J. Clin. Oncol. 2006;24:2098–2104. doi: 10.1200/JCO.2005.05.2878. [DOI] [PubMed] [Google Scholar]
- 5.Semenza GL, Wang GL. Mol. Cell. Biol. 1992;12:5447–5454. doi: 10.1128/mcb.12.12.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Semenza GL. Nat. Rev. Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 7.Giaccia A, Siim BG, Johnson RS. Nat. Rev. Drug Discov. 2003;2:1–9. doi: 10.1038/nrd1199. [DOI] [PubMed] [Google Scholar]
- 8.Semenza GL. Expert Opin. Ther. Targets. 2006;10:267–280. doi: 10.1517/14728222.10.2.267. [DOI] [PubMed] [Google Scholar]
- 9.Zhong H, De Marzo AM, Laughner E, Lim M, Hilton DA, Zagzag D, Buechler P, Isaacs WB, Semenza GL, Simons JW. Cancer Res. 1998;59:5830–5835. [PubMed] [Google Scholar]
- 10.a Maxwell PH, Dachs GU, Gleadle JM, Nicholls LG, Harris AL, Stratford IJ, Hankinson O, Pugh CW, Ratcliffe PJ. Proc. Natl. Acad. Sci. U.S.A. 1997;94:8104–8109. doi: 10.1073/pnas.94.15.8104. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Ryan HE, Poloni M, McNulty W, Elson D, Gassmann M, Arbeit JM, Johnson RS. Cancer Res. 2000;60:4010–4015. [PubMed] [Google Scholar]; c Kung AL, Wang S, Klco JM, Kaelin WG, Livingston DM. Nat. Med. 2000;6:1335–1340. doi: 10.1038/82146. [DOI] [PubMed] [Google Scholar]
- 11.Nagle DG, Zhou Y-D. Curr. Drug Target. 2006;7:355–369. doi: 10.2174/138945006776054979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hodges TW, Hossain CF, Kim Y-P, Zhou Y-D, Nagle DG. J. Nat. Prod. 2004;67:767–771. doi: 10.1021/np030514m. [DOI] [PubMed] [Google Scholar]
- 13.Hossain CF, Kim Y-P, Baerson SR, Zhang L, Bruick RK, Mohammed KA, Agarwal AK, Nagle DG, Zhou Y-D. Biochem. Biophys. Res. Comm. 2005;333:1026–1033. doi: 10.1016/j.bbrc.2005.05.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang GL, Semenza GL. Blood. 1993;82:3610–3615. [PubMed] [Google Scholar]
- 15.Rudi A, Aknin M, Gaydou EM, Kashman Y. J. Nat. Prod. 1997;60:700–703. doi: 10.1021/np960727u. [DOI] [PubMed] [Google Scholar]
- 16.Ohtani I, Kusumi T, Kashman Y, Kakisawa H. J. Am. Chem. Soc. 1991;113:4902–4906. [Google Scholar]
- 17.Rudi A, Yosief T, Schleyer M, Kashman Y. Tetrahedron. 1999;55:5555–5566. [Google Scholar]
- 18.Rudi A, Goldberg I, Stein Z, Benayahu Y, Schleyer M, Kashman Y. Tetrahedron Lett. 1993;34:3943–3944. [Google Scholar]
- 19.Rudi A, Kashman Y, Benayahu Y, Schleyer M. J. Nat. Prod. 1994;57:1416–1423. doi: 10.1021/np50108a023. [DOI] [PubMed] [Google Scholar]
- 20.Funel-Le Bon C, Berrué F, Thomas OP, Reyes F, Amade P. J. Nat. Prod. 2005;68:1284–1287. doi: 10.1021/np050100o. [DOI] [PubMed] [Google Scholar]
- 21.Rudi A, Goldberg I, Stein Z, Kashman Y, Benayahu Y, Schleyer M, Gravalos MDG. J. Nat. Prod. 1995;58:1702–1712. doi: 10.1021/np50125a009. [DOI] [PubMed] [Google Scholar]
- 22.Carletti I, Long C, Funel C, Amade P. J. Nat. Prod. 2003;66:25–29. doi: 10.1021/np020208t. [DOI] [PubMed] [Google Scholar]
- 24.Borenfreund E, Puerner JA. Toxicol. Lett. 1985;24:119–124. doi: 10.1016/0378-4274(85)90046-3. [DOI] [PubMed] [Google Scholar]
- 25.Zhou Y-D, Kim Y-P, Mohammed KA, Jones DK, Muhammad I, Dunbar DC, Nagle DG. J. Nat. Prod. 2005;68:947–950. doi: 10.1021/np050029m. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Info
S1. IC50 values of 1-11 on HIF-1 activation in T47D and PC-3 cells, and cell proliferation/viability in T47D, MDA-MB-231, PC-3, and DU145 cells.
S2. New species of Axinella (Axinellidae) collected (collection C020287) at a depth of -16 M off the southeastern coast of South Africa on March 22, 2000. Photograph provided by Dr. M.T. Davies-Coleman (Dept. of Chemistry, Rhodes University, Grahamstown, South Africa).