Abstract
The Forkhead family of transcription factors mediates many aspects of physiology, including stress response, metabolism, commitment to apoptosis, and development. The Forkhead box subfamily O (FoxO) proteins have garnered particular interest due to their involvement in the modulation of cardiovascular biology. In this review, we discuss the mechanisms of FoxO regulation and outcomes of FoxO signaling under normal and pathological cardiovascular contexts.
Keywords: Forkhead, Akt signaling, stress response, cardiac hypertrophy
INTRODUCTION
The Forkhead/winged helix family of transcription factors, characterized by a conserved DNA binding domain known as the Forkhead box (Fox), is essential in both development and adult physiology. First described in 1989 (1), further identification of related Forkhead proteins has led to the characterization of 19 subfamilies, designated A–S (reviewed in 2, 3). Several Forkhead proteins, including FoxC1, FoxC2, FoxH1, FoxM1, FoxO1, and FoxP1, appear to be essential for normal cardiac development, as their deletion in mice results in severe arterial and cardiac defects and early lethality (reviewed in 4). In addition, an abundance of evidence suggests that three members of the FoxO subfamily—FoxO1, FoxO3, and FoxO4—are critical in maintaining cardiac function and mediating cardiac stresses in the adult (the remaining FoxO family member, FoxO6, is localized in the brain). Within the setting of cardiac function, the FoxO family is believed to be involved in diverse activities, including response to oxidative stress (5), regulation of metabolism (6), cell cycle control (7), and commitment to apoptosis (8). This review focuses primarily on the regulation and outcomes of FoxO signaling in the heart.
FOXO TRANSCRIPTIONAL ACTIVITY
In the nucleus, FoxO proteins target a conserved DNA binding sequence, TTGTTTAC (9), and can associate with coactivators such as Smad (10), Notch (11), β-catenin (12), and PGC-1α (6). The end result of FoxO transcriptional activity is generally associated with counteracting oxidative stress and promoting cell cycle arrest and apoptosis. Early experiments studying FoxO function demonstrated that overexpression of constitutively active FoxO1 causes apoptosis, and this outcome is dependent on FoxO-DNA binding (13). This proapoptotic function is mediated through transcription of several target genes, including Fas (14), Bim (15), TRAIL (16), and Puma (17). In addition, FoxO causes cell cycle arrest via transcription of p27kip1 (8), p21cip1 (10), p130, and the inhibitory cyclin G2 (18). Conversely, FoxO proteins are also involved in the promotion of DNA repair through increased expression of several genes, including Gadd45a (19). In addition, the antioxidant enzymes MnSOD (manganese superoxide dismutase) and catalase are FoxO target genes (5), and transcription of these enzymes is induced by FoxO to counteract oxidative stress. FoxO can activate the synthesis of PEPCK (phosphoenolpyruvate carboxykinase) and glucose-6-phophatase, two enzymes necessary for gluconeogenesis (20). Lastly, FoxO can mediate the stress response by interfering with other transcription factors to inhibit transcription (21-23).
MECHANISMS OF FOXO REGULATION
FoxO and Akt Signaling
The FoxO subfamily members were originally identified by their involvement in chromosomal translocations associated with leukemias and rhabdomyosarcomas (24, 25). Concurrent work in Caenorhabditis elegans demonstrated a link between hormonal inputs, Akt signaling, and FoxO. In C. elegans, inactivating mutations in daf-2 (homolog of the insulin receptor) or age-1 [homolog of phosphoinositide-3 kinase (PI3K)] cause growth arrest and life span extension; however, these effects are dependent on the activity of the FoxO homolog daf-16 (26, 27). This observation led to the discovery that FoxO transcriptional activity is inhibited by PI3K-Akt signaling (28) whereby PI3K phosphorylation causes Akt to translocate to the nucleus (29), where it phosphorylates FoxO (Thr24, Ser256, Ser319 of FoxO1; Thr32, Ser253, Ser315 of FoxO3). FoxO contains a nuclear export sequence and a nuclear localization sequence, the latter of which is masked following phosphorylation by Akt (30, 31). Phosphorylation by Akt promotes FoxO interaction with a subset of 14-3-3 proteins localized in the nucleus, causing translocation of the FoxO-14-3-3 complex to the cytoplasm via the nuclear export protein Crm1. Once in the cytosol, FoxO is then sequestered by 14-3-3 proteins and is rendered inactive (14, 32). Mutation of the Akt phosphorylation sites on FoxO proteins results in exclusive FoxO nuclear localization and insusceptibility to Akt-mediated inhibition (33). Cumulatively, PI3K-Akt signaling is a major regulator of FoxO activity.
Additional FoxO Regulation
In addition to phosphorylation and inhibition by Akt, recent work suggests that FoxO is also regulated by several posttranslational modifications, including phosphorylation by multiple kinases, glycosylation, acetylation, and ubiquitination. The details of these regulatory pathways are described below.
Phosphorylation and inhibition
In addition to Akt, SGK1 (serum and glucocorticoid-induced kinase 1) can phosphorylate and inhibit FoxO3 activity through nuclear exclusion (34). Although Akt and SGK1 share FoxO phosphorylation sites, SGK1 favors phosphorylation of Ser315, whereas Akt most actively phosphorylates Ser253. Under some stimuli, coordinate activities of these proteins may synergistically inhibit FoxO3 function. Like Akt, SGK1 is activated by the PI3K signaling cascade and has many overlapping functions with Akt. However, in the case of FoxO3 at least, the regulation by SGK1 and Akt appears to be a nonredundant, complementary relationship. For example, radiation-induced DNA damage stimulates p53 expression, which inhibits FoxO3 through an SGK1-dependent, but not an Akt-dependent, manner (35). In addition, Wnt activation and β-catenin accumulation lead to an upregulation of SGK1 and subsequent inhibition of FoxO3 activity (36).
CDK2 (cyclin-dependent kinase 2) promotes G1/S cell cycle progression by associating with cyclin E and cyclin A. CDK2 can phosphorylate Ser249 and Ser298 of FoxO1 to cause cytoplasmic retention and inhibit FoxO1 activity (37). However, under DNA-damaging conditions, CDK2 activity is inhibited by the FoxO target genes p21cip1 and p27kip1 (38), allowing FoxO1 to activate target gene expression, thereby forming a positive feedback loop.
IκB kinase (IκK) can also phosphorylate FoxO3 at Ser644, inhibiting FoxO3 function independently of Akt (39). In addition to promoting cytoplasmic retention of FoxO3, IκK mediates the polyubiquitination and degradation of FoxO3 via the proteosome system. Similarly, ERK (extracellular signal–regulated protein kinase) phosphorylation of FoxO3 (at Ser 294, Ser 344, and Ser 425) in response to EGF stimulation also increases cytosolic FoxO3 levels and promotes interaction and degradation by the E3 ubiquitin ligase MDM2 (mouse double minute 2) (40).
FoxO1, FoxO3, and FoxO4 share a highly conserved serine-rich domain (Ser319, Ser322, Ser325, Ser329 of FoxO1), suggestive of regulation by kinase signaling pathways. The phosphorylation of Ser319 by Akt or SGK1 is required for CK1 (casein kinase 1)-dependent Ser322 and Ser325 phosphorylation (41). Furthermore, Ser329 is phosphorylated by DYRK1A (dual-specificity tyrosine phosphorylated and regulated kinase 1A) (42). Together, phosphorylation of these residues provides a strong signal promoting nuclear exclusion.
Phosphorylation and activation
FoxO3 can be directly phosphorylated by AMPK (AMP-activated protein kinase), an energy-sensing protein activated when ATP levels are low (43). However, unlike Akt phosphorylation, which results in FoxO cytoplasmic retention and inhibition of FoxO target gene transcription, AMPK phosphorylation increases FoxO3 target gene transcription without affecting FoxO3 subcellular localization. The observation that Akt phosphorylation and AMPK phosphorylation produce opposite outcomes on FoxO activity can be attributed to the fact that Akt does not share any of the six unique AMPK phosphorylation sites identified in vitro on FoxO3, of which three (Ser413, Ser588, Ser626) are functional in vivo. Furthermore, the signals leading to Akt or AMPK activation generally occur under the opposite circumstances of energy abundance versus energy conservation, respectively. Although it is not currently known whether FoxO1 is also activated by AMPK, in vitro assays have demonstrated that FoxO1 is phosphorylated by AMPK (44).
A second kinase capable of promoting FoxO activation is JNK (c-Jun-N-terminal kinase). Oxidative stress induced by hydrogen peroxide exposure leads to activation of the GTPase Ral and subsequent activation of JNK, which can in turn phosphorylate FoxO4 on two C-terminal residues (Thr447 and Thr451) to promote nuclear translocation and activation of FoxO4 gene transcription (45). JNK-dependent activation of FoxO in response to reactive oxygen species induces expression of the FoxO target genes encoding MnSOD and catalase to form an efficient feedback loop. Interestingly, although JNK-dependent FoxO activation in response to stress is conserved in Drosophila and C. elegans, the sites of JNK phosphorylation are not conserved in either species. Instead, the Drosophila homolog dFOXO (46), and the C. elegans homolog daf-16 are phosphorylated by JNK in their N termini (47). The JNK target sites on FoxO4 are also not conserved in FoxO1 or FoxO3, but evidence suggests that FoxO1 and FoxO3 are also activated, either by direct phosphorylation or by inhibition of the FoxO-retaining protein 14-3-3, in a JNK-dependent manner (48, 49).
Oxidative stress also induces autophosphorylation and activation of MST1 (mammalian sterile twenty 1), a member of a kinase family involved in stress response and JNK activation. Activated MST1 interacts with the forkhead domain of FoxO3 to directly phosphorylate a conserved serine residue (Ser207), which reduces FoxO interaction with 14-3-3 proteins and initiates nuclear translocation and target gene transcription (50). Overexpression of the C. elegans MST1 homolog cst-1 increases longevity in a daf-16-dependent manner. Although MST1 and JNK participate in similar signaling pathways, JNK is unable to phosphorylate the forkhead domain, suggesting that MST1 and JNK may act in a complementary fashion to activate FoxOs following oxidative stress. Although these studies demonstrate the capability of MST1 to phosphorylate FoxO1 in vitro, subsequent in vitro work has shown that MST1 does not phosphorylate DNA-bound FoxO1 (51). In fact, MST1-phosphorylated FoxO1 has greatly diminished DNA binding affinity (52). Although it appears that MST1-mediated FoxO phosphorylation promotes nuclear translocation, the subsequent dephosphorylation of FoxO may be required for transcriptional activity. Incidentally, Akt suppresses MST1-mediated FoxO3 phosphorylation, demonstrating an indirect mechanism of FoxO suppression by Akt (53).
Glycosylation
Another mechanism of FoxO regulation is mediated through addition of N-acetylglucosamine (O-GlcNAc) by O-GlcNAc transferase (OGT), a dynamic modification of serine/threonine residues similar to phosphorylation. Generally, this modification is highly sensitive to glucose concentrations and cell stress, and several proteins involved in glucose metabolism, oxidative stress, and mitosis, including transcription factors, are O-GlcNAcylated (reviewed in 54). FoxO1 O-GlcNAcylation by OGT increases transcription of some FoxO target genes independent of Akt phosphorylation (55, 56). In addition, PGC-1α, previously shown to be a FoxO coactivator and now shown to also be O-GlcNAcylated, recruits OGT to FoxO1 to increase FoxO activation (57).
Acetylation
Early studies suggested acetylation-dependent FoxO regulation when it was demonstrated that the p300/CBP coactivator complex, which possesses histone acetyltransferase (HAT) activity, interacts with FoxO1, and inhibition of this interaction reduces FoxO transcriptional activity (58). However, subsequent examinations of the effects of p300/CBP acetylation have been conflicting. A current model suggests that p300-induced FoxO1 acetylation promotes FoxO1-mediated gene transcription in the absence of insulin (59). Conversely, insulin receptor signaling increases FoxO1 acetylation dependent on Akt-mediated phosphorylation but does not induce FoxO1 transcriptional activity. In contrast, CBP acetylates a region of the DNA-binding domains of FoxO1, FoxO3, and FoxO4 (Lys242, Lys245, Lys262 on FoxO1), resulting in decreased DNA binding affinity, reduced transcriptional activity, and increased susceptibility to Akt-mediated phosphorylation at Ser253 (60, 61).
The NAD-dependent histone deacetylase SirT1 (silent mating type information regulation 2 homolog 1) targets many substrates, particularly proteins involved in metabolism and stress response (reviewed in 62). In C. elegans, caloric restriction–dependent life span extension via overexpression of the SirT1 homolog sir-2 is dependent on daf-16 (63), suggesting a relationship between the human homologs SirT1 and FoxO. FoxO3 and SirT1 loosely interact under unstressed conditions, but following hydrogen peroxide exposure or heat shock, FoxO3 is highly acetylated and tightly interacts with SirT1 (64). The SirT1-FoxO interaction represents a complicated biology that remains to be elucidated. SirT1 deacetylates p300 and inhibits p300-dependent increases in FoxO transcription of Bim (65). However, SirT1 enhances transcription of some FoxO target genes (66), suggesting an overall model in which SirT1 increases the ability of FoxO to respond to stress through cell cycle arrest and other adaptations but inhibits FoxO transcription of apoptotic genes. In addition, evidence suggests that SirT1 deacetylation increases FoxO polyubiquitination and degradation (67).
Ubiquitination
Following activation of Akt, both FoxO1 and FoxO3 are degraded by the proteosome (68); however, the ubiquitination of FoxO1 and FoxO3 is mediated through different pathways. FoxO1 is degraded by Skp2, an F-box protein and subunit of Skp1/Cul1/F-box ubiquitin ligase complex (69). FoxO1 ubiquitination by Skp2 is dependent on Akt phosphorylation of Ser256, providing an additional level of FoxO regulation by Akt. In addition, Skp2 also ubiquitinates and degrades p27kip1 (7). However, Skp2 does not appear to interact with FoxO3. Instead, FoxO3 appears to be polyubiquitinated and degraded via MDM2, which is promoted by phosphorylation of FoxO3 by ERK (40), as discussed above. FoxO3 may also be degraded through cleavage by caspase-3-like proteases (70).
Ubiquitin-dependent degradation of FoxO4 has not been identified; however, following hydrogen peroxide treatment, MDM2 monoubiquitinates FoxO4 to increase nuclear localization and transcriptional activity (71, 72). This modification is reversed by the ubiquitin-specific protease USP7, which deubiquitinates FoxO4 and causes nuclear export.
The FoxO target gene product atrogin-1 ubiquitinates and degrades calcineurin, preventing Akt-mediated hypertrophic signaling. In addition, atrogin-1 also acts as a FoxO coactivator through noncanonical polyubiquitination, which serves to increase FoxO activity and oppose Akt-mediated signaling (73). Atrogin-1 does not tag FoxO with canonical polyubiquitin chains that lead to proteosomal degradation and does not appear to directly affect FoxO stability. The consequences of atrogin-1-dependent ubiquitination are discussed in more detail below.
FOXO AND CARDIAC BIOLOGY
Cardiac Development
Despite some of the overlapping effects of the FoxO proteins, studies involving the deletion of individual FoxO family members suggest that each of the FoxOs has very different roles in vascular and cardiac development. For instance, whereas FoxO3−/− and FoxO4−/− embryos develop normally (74), FoxO1−/− embryos die by day E10.5–E11 and exhibit grossly deficient vascular and cardiac growth (74, 75). FoxO1 is expressed in embryonic vessels, so it is not surprising that FoxO1−/− embryos exhibit impaired yolk sac vasculature, underdeveloped dorsal aorta, and impaired cardiac looping. Interestingly, though, FoxO1+/− mice develop normally. Conversely, mice overexpressing a cardiac-specific FoxO1 transgene also die by E10.5 due to abnormal p21cip1 and p27kip1 expression, resulting in impaired cardiomyocyte proliferation, reduced heart size and myocardium thickness, and subsequent heart failure (76).
FoxO proteins are also involved in the regulation of vascular development. FoxO3 represses the expression of BMPER (bone morphogenetic protein [BMP]-binding endothelial cell precursor-derived regulator), which is involved in endothelial cell migration and sprouting during development (77, 78). Consistent with these data, FoxO1 or FoxO3 overexpression in HUVEC cells decreases endothelial cell sprouting and migration; conversely, silencing FoxO1 or FoxO3 promotes angiogenesis (79). Additionally, negative regulation of FoxO1 by SirT1 appears to be important for supporting sprout formation (80).
The enhancer region of myocardin, which acts as a coactivator to induce smooth muscle cell and cardiomyocyte differentiation, contains FoxO binding sites and requires FoxO for its activity (81). FoxOs also maintain normal endothelial cell physiology in the adult, as demonstrated by mice engineered with floxed FoxO alleles that develop hemangiomas with advancing age (82).
Cardiac Hypertrophy
Maintaining FoxO activity is essential in preventing Akt-mediated cardiac hypertrophy. The IGF-Akt signaling cascade promotes cardiac hypertrophy by increasing cell size and protein translation via activation of S6K and mTOR and by decreasing antihypertrophic transcriptional activity via inhibition of GSK3 (reviewed in 83). By inhibiting FoxO activity, Akt also suppresses transcription of two key FoxO target gene products, the muscle-specific ubiquitin ligases atrogin-1/MAFbx and MuRF1 (84). The cumulative effect of Akt activation in the heart is an increase in progrowth pathways and the inhibition of FoxO-mediated catabolism, resulting in hypertrophy. The CIRKO (cardiac insulin receptor knockout) mouse, which displays a reduction in cardiac mass due to reduced cardiomyocyte size (85), demonstrates the necessity of Akt signaling in cardiac hypertrophy. The phenotype of the CIRKO mouse is mimicked in mice with heart-specific FoxO3 expression (86). Furthermore, adenovirus-mediated FoxO3 overexpression reduces growth factor– and stretch-induced hypertrophy in cultured cardiomyocytes (86). In contrast, heart size is doubled in the heart-specific Akt transgenic mouse (87), consistent with evidence that FoxO3−/− mice demonstrate an increase in cardiac mass (88).
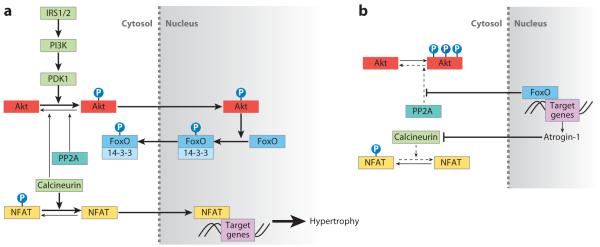
In addition to Akt signaling, a second major pathway involved in cardiac hypertrophy is mediated by the calcium-regulated protein phosphatase calcineurin (or PP2B). Calcineurin dephosphorylates the transcription factor NFAT (nuclear factor of activated T cells), which promotes NFAT nuclear translocation and induces transcription of genes characteristic of the hypertrophic response such as those encoding α-skeletal actin and β-myosin heavy chain (89). Akt, FoxO, atrogin-1, and calcineurin were first connected in cardiomyocytes with the discovery that atrogin-1 targets calcineurin for degradation, decreases NFAT-dependent transcription, and prevents calcineurin-dependent cardiac hypertrophy (90), suggesting that FoxO might regulate calcineurin-NFAT signaling. Accordingly, overexpression of FoxO in cultured cardiomyocytes increases atrogin-1 expression, inhibits calcineurin activity, and decreases angiotensin II–induced cardiac hypertrophy (88). Atrogin-1 overexpression decreases Akt-mediated FoxO1/3 phosphorylation, and furthermore, transgenic mice with cardiomyocyte-restricted overexpression of atrogin-1 are resistant to Akt-dependent hypertrophy (73). Although it was previously shown that the phosphatase activity of PP2A inhibits Akt (91), it has since been shown that calcineurin also dephosphorylates and inactivates Akt and that FoxO attenuates the interaction between PP2A and Akt (92). In summary, when FoxO is activated, FoxO-dependent atrogin-1 transcription leads to decreased calcineurin levels and, accordingly, decreased phosphatase activity directed toward Akt. Furthermore, because FoxO inhibits the interaction between Akt and PP2A, the net result is hyperphosphorylation of Akt (Figure 1). Surprisingly, the FoxO-induced increase in Akt phosphorylation appears to preferentially cause FoxO phosphorylation, and not phosphorylation of the Akt targets mTOR and GSK3. Thus, FoxO activity is controlled through a negative feedback loop.
Figure 1.
Akt and FoxO signaling pathways in cardiac hypertrophy. (a) Insulin receptor signaling activates the PI3K (phosphoinositide-3 kinase) signaling cascade, leading to Akt phosphorylation and translocation into the nucleus. Akt phosphorylates three conserved FoxO residues to promote FoxO association with 14-3-3 proteins and nuclear export to the cytosol, where the complex remains transcriptionally inactive. Calcineurin dephosphorylates NFAT (nuclear factor of activated T cells), allowing NFAT nuclear translocation and subsequent transcription of NFAT target genes, such as those encoding α-skeletal actin and β-myosin heavy chain. PP2A and calcineurin provide negative feedback by dephosphorylating Akt. (b) In the absence of PI3K signaling, FoxO is able to transcribe target genes such as atrogin-1, which encodes a ubiquitin ligase that promotes calcineurin degradation and prevents NFAT activation. The decrease in calcineurin levels reduces Akt-associated phosphatase activity. FoxO also reduces PP2A phosphatase activity on Akt. The reduction in calcineurin and PP2A-associated phosphatase activity leads to Akt hyperphosphorylation, which eventually inhibits FoxO activity.
Cardiac Stress Response and Aging
The outcome of FoxO signaling in different contexts can appear contradictory. For example, although FoxOs increase antioxidant enzymes to counter oxidative stress, FoxOs also promote apoptotic signaling pathways under other circumstances. The effects of FoxOs on aging and stress response in the heart and vasculature involve a complex relationship between many signaling pathways.
As mentioned above, an antiaging role of forkhead proteins is supported by the evidence of increased longevity in C. elegans with active daf-16. FoxOs also have an antiaging function in the heart, first exhibited by the prevention of cardiac decline by transgenic expression in Drosophila (93). One major pathway through which FoxO signaling prevents cardiac aging is via activation of SirT1, which has a conserved role in preventing an aging phenotype in many tissues (reviewed in 62). SirT1 is upregulated by pressure overload, and cardiac-specific overexpression of SirT1 helps prevent oxidative stress and aging via FoxO1-dependent expression of catalase (94). Additionally, moderate SirT1 transgenic expression diminishes age-related heart hypertrophy; in contrast, a transgenic mouse line demonstrating ~12-fold SirT1 overexpression demonstrates left ventricular fibrosis, apoptosis, and hypertrophy-associated fetal gene expression, suggestive of dose-dependent SirT1 effects. Finally, activation of SirT1 via resveratrol treatment in a cardiomyocyte cell line inhibits hypoxia-induced apoptosis (95). These effects appear to be mediated through decreased expression of the proapoptotic FoxO target Bim and increased expression of the FoxO1 target p27kip1, which causes cell cycle arrest. Incidentally, it has been demonstrated that the inhibition of hypertrophy by resveratrol treatment is mediated through activation of AMPK, which, as described above, can also promote FoxO activity (96).
In addition to stimulating the antioxidant activities of MnSOD and catalase, FoxO3 also regulates transcription of a mitochondrial hydrogen peroxide scavenger, peroxiredoxin III, in cardiac fibroblasts (97). Peroxiredoxin III expression prevents serum withdrawal–induced apoptosis and may have an important role as a cardioprotectant in vivo.
The endothelial cell stress response is partially mediated by VEGF (vascular endothelial growth factor), which promotes angiogenesis through endothelial cell survival, migration, and proliferation. VEGF and FoxO have a complicated relationship in endothelial cells. VEGF induces NADPH oxidase activity, which activates a redox-sensitive Akt signaling pathway and mediates some effects of VEGF. VEGF also causes transient phosphorylation and nuclear exclusion of FoxO1, FoxO3, and FoxO4 via NADPH oxidase–dependent Akt signaling, which prevents apoptosis, promotes cell cycle progression, and downregulates expression of most FoxO1 target genes (98, 99). However, a small subset of FoxO target genes (including those encoding MnSOD, BMP2, VCAM-1, and MMP10) is upregulated by VEGF in a FoxO1-dependent manner (100). Unlike most FoxO1 target genes, expression of this subset is not immediately inhibited by VEGF, and transcription of these genes may be aided by NF-κB or NFAT. Despite the fact that VEGF generally inhibits FoxO activity, the VEGF-dependent activation of a subset of FoxO1 target genes helps explain why FoxO1−/− mice demonstrate an impaired vasculature phenotype.
In the vasculature, Akt is the major antagonist of FoxO signaling. Under some stress conditions, FoxO inhibition is beneficial to maintaining vessel viability. Shear stress, heat shock, and anoxia induce Akt signaling and subsequently inhibit FoxO, which promotes cell survival, evasion of apoptosis, and early senescence (101-103).
Cardiac Metabolism and Autophagy
Through a variety of transcriptional targets, FoxOs can respond to a changing metabolic environment via regulation of metabolic enzymes and energy-dependent proteins. The direct metabolic effects of FoxO signaling are not entirely clear; however, because FoxOs may be involved in transcription of many metabolic genes, a bioinformatics approach may help uncover FoxO-regulated genes. In Drosophila, a significant portion of genes upregulated by fasting-refeeding are also activated in cells expressing constitutively active dFOXO (104). In humans, mice, and rats, FoxO binding sites are found in the promoters of several metabolic genes, including those encoding acetyl CoA carboxylase 2, the fatty acid transporter CD36, lipoprotein lipase, and Glut4 (105). FoxO proteins may also indirectly exert metabolic effects through transcription of PGC-1α, which associates with PPARγ to activate genes involved in mitochondrial biogenesis, fatty acid oxidation, and gluconeogenesis (66, 106). Additionally, PGC-1α can serve as a FoxO coactivator to regulate expression of metabolic genes. For example, in hepatocytes, PGC-1α and FoxO1 associate with estrogen-related receptors to activate transcription of pyruvate dehydrogenase kinase 4, which limits the production of acetyl CoA from pyruvate and suppresses glucose oxidation in the absence of insulin (107).
FoxOs also regulate expression of genes that sense changes in the metabolic environment. For example, FoxO1 and FoxO3 activate transcription of Kir6.2, an ATP-dependent potassium channel (KATP) subunit (105). In the presence of ATP, KATP channels close, causing membrane depolarization and calcium influx, thereby maintaining contractile function. Under metabolically stressful conditions, ATP levels are low and KATP channels remain open, such that the cell is hyperpolarized and calcium demand is limited. Transgenic mice lacking Kir6.2 demonstrate severe heart failure within hours of transaortic banding (108).
FoxO also provides a balance between metabolic pathways and protein degradation. In addition to regulating the transcription of the E3 ligases atrogin-1 and MuRF1, FoxO can promote proteolysis through autophagy. Autophagy recaptures amino acids and fatty acids for ATP production and new protein translation (reviewed in 109). In cardiomyocytes, autophagy is activated during ischemia-reperfusion and transaortic constriction–induced heart failure. Accordingly, increased autophagy reduces apoptosis following ischemia-reperfusion (110), whereas acute cardiac-specific inhibition of autophagy causes left ventricular dilation, decreased contractile function, and hypertrophy (111). In myocytes, FoxO3 regulates transcription of autophagy genes, such as those encoding Gabarapl1, Atg12l, and LCB3, and promotes autophagosome formation (112). Thus, FoxOs promote proteolysis through activation of the ubiquitin ligases MuRF1 and atrogin-1 and through increased transcription of genes necessary for autophagy.
CONCLUDING REMARKS
The Forkhead subfamily O is an important mediator of the stress response. First characterized for its necessity for longevity in C. elegans, it has since been demonstrated that FoxOs have unexpected and diverse roles in countering stress, determining cell fate, and regulating energy availability. Because the heart is constantly adapting to different stresses and metabolic conditions, FoxOs appear to play an extremely important role in cardiac physiology. As we learn more precise information about FoxO regulation, direct transcriptional targets, and dynamic signaling responses, a therapeutic role for FoxOs may be elucidated. The rapidly growing field of forkhead biology will surely provide information and models of how cells grow, change, and adapt.
SUMMARY POINTS.
FoxO proteins mediate stress response through transcription of genes involved in oxidative stress, metabolism, autophagy, aging, and apoptosis.
FoxO proteins are regulated by a variety of mechanisms, including phosphorylation, glycosylation, acetylation, and ubiquitination. These modifications can activate or inhibit FoxO activity.
In cardiomyocytes, regulation of FoxO signaling is complex and inhibited by negative feedback. Akt inhibits FoxO activity while promoting NFAT-mediated hypertrophy, whereas overexpression of FoxO leads to eventual Akt hyperphosphorylation and FoxO inhibition. FoxO overexpression prevents Akt-dependent cardiac hypertrophy.
FoxO helps mediate the antiaging effects of SirT1 through increased transcription of antioxidant and cell cycle arrest proteins while inhibiting transcription of FoxO proapoptotic targets.
ACKNOWLEDGMENTS
We thank Andrea Portbury for helpful discussion. This work was supported by National Institutes of Health grants GM61728, HL65619, and AG024282 to C.P.
Glossary
- FoxO
Forkhead box subfamily O
- MnSOD
manganese superoxide dismutase
- PI3K
phosphoinositide-3 kinase
- PI3K-Akt signaling
activated by insulin receptor. Akt activity leads to increased glucose uptake, cell cycle progression, evasion of apoptosis, and protein translation
- SirT1
silent mating type information regulation 2 homolog 1
- Cardiac hypertrophy
increased cell size in cardiomyocytes in response to stress or growth factor stimulation
- MuRF1
muscle ring finger 1
- NFAT
nuclear factor of activated T cells
- Autophagy
catabolic lysosome-mediated protein degradation involved in energy and protein reallocation
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Contributor Information
Sarah M. Ronnebaum, McAllister Heart Institute, University of North Carolina, Chapel Hill, North Carolina 27599; sarah.ronnebaum@unc.edu
Cam Patterson, McAllister Heart Institute, University of North Carolina, Chapel Hill, North Carolina 27599; cpatters@med.unc.edu.
LITERATURE CITED
- 1.Weigel D, Jurgens G, Kuttner F, Seifert E, Jackle H. The homeotic gene fork head encodes a nuclear protein and is expressed in the terminal regions of the Drosophila embryo. Cell. 1989;57:645–58. doi: 10.1016/0092-8674(89)90133-5. [DOI] [PubMed] [Google Scholar]
- 2.Tuteja G, Kaestner KH. Forkhead transcription factors II. Cell. 2007;131:192. doi: 10.1016/j.cell.2007.09.016. [DOI] [PubMed] [Google Scholar]
- 3.Tuteja G, Kaestner KH. SnapShot: forkhead transcription factors I. Cell. 2007;130:1160. doi: 10.1016/j.cell.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 4.Papanicolaou KN, Izumiya Y, Walsh K. Forkhead transcription factors and cardiovascular biology. Circ. Res. 2008;102:16–31. doi: 10.1161/CIRCRESAHA.107.164186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kops GJ, Dansen TB, Polderman PE, Saarloos I, Wirtz KW, et al. Forkhead transcription factor FOXO3a protects quiescent cells from oxidative stress. Nature. 2002;419:316–21. doi: 10.1038/nature01036. [DOI] [PubMed] [Google Scholar]
- 6.Puigserver P, Rhee J, Donovan J, Walkey CJ, Yoon JC, et al. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1α interaction. Nature. 2003;423:550–55. doi: 10.1038/nature01667. [DOI] [PubMed] [Google Scholar]
- 7.Medema RH, Kops GJ, Bos JL, Burgering BM. AFX-like Forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. Nature. 2000;404:782–87. doi: 10.1038/35008115. [DOI] [PubMed] [Google Scholar]
- 8.Stahl M, Dijkers PF, Kops GJ, Lens SM, Coffer PJ, et al. The forkhead transcription factor FoxO regulates transcription of p27Kip1 and Bim in response to IL-2. J. Immunol. 2002;168:5024–31. doi: 10.4049/jimmunol.168.10.5024. [DOI] [PubMed] [Google Scholar]
- 9.Furuyama T, Nakazawa T, Nakano I, Mori N. Identification of the differential distribution patterns of mRNAs and consensus binding sequences for mouse DAF-16 homologues. Biochem. J. 2000;349:629–34. doi: 10.1042/0264-6021:3490629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seoane J, Le HV, Shen L, Anderson SA, Massagué J. Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell. 2004;117:211–23. doi: 10.1016/s0092-8674(04)00298-3. [DOI] [PubMed] [Google Scholar]
- 11.Kitamura T, Kitamura YI, Funahashi Y, Shawber CJ, Castrillon DH, et al. A Foxo/Notch pathway controls myogenic differentiation and fiber type specification. J. Clin. Investig. 2007;117:2477–85. doi: 10.1172/JCI32054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Essers MA, de Vries-Smits LM, Barker N, Polderman PE, Burgering BM, Korswagen HC. Functional interaction between β-catenin and FOXO in oxidative stress signaling. Science. 2005;308:1181–84. doi: 10.1126/science.1109083. [DOI] [PubMed] [Google Scholar]
- 13.Tang ED, Nuñez G, Barr FG, Guan KL. Negative regulation of the forkhead transcription factor FKHR by Akt. J. Biol. Chem. 1999;274:16741–46. doi: 10.1074/jbc.274.24.16741. [DOI] [PubMed] [Google Scholar]
- 14.Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–68. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 15.Dijkers PF, Medema RH, Lammers JW, Koenderman L, Coffer PJ. Expression of the proapoptotic Bcl-2 family member Bim is regulated by the forkhead transcription factor FKHR-L1. Curr. Biol. 2000;10:1201–4. doi: 10.1016/s0960-9822(00)00728-4. [DOI] [PubMed] [Google Scholar]
- 16.Modur V, Nagarajan R, Evers BM, Milbrandt J. FOXO proteins regulate tumor necrosis factor-related apoptosis inducing ligand expression. Implications for PTEN mutation in prostate cancer. J. Biol. Chem. 2002;277:47928–37. doi: 10.1074/jbc.M207509200. [DOI] [PubMed] [Google Scholar]
- 17.You H, Pellegrini M, Tsuchihara K, Yamamoto K, Hacker G, et al. FOXO3a-dependent regulation of Puma in response to cytokine/growth factor withdrawal. J. Exp. Med. 2006;203:1657–63. doi: 10.1084/jem.20060353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen J, Yusuf I, Andersen HM, Fruman DA. FOXO transcription factors cooperate with δ EF1 to activate growth suppressive genes in B lymphocytes. J. Immunol. 2006;176:2711–21. doi: 10.4049/jimmunol.176.5.2711. [DOI] [PubMed] [Google Scholar]
- 19.Tran H, Brunet A, Grenier JM, Datta SR, Fornace AJ, Jr, et al. DNA repair pathway stimulated by the forkhead transcription factor FOXO3a through the Gadd45 protein. Science. 2002;296:530–34. doi: 10.1126/science.1068712. [DOI] [PubMed] [Google Scholar]
- 20.Nakae J, Kitamura T, Silver DL, Accili D. The forkhead transcription factor Foxo1 (Fkhr) confers insulin sensitivity onto glucose-6-phosphatase expression. J. Clin. Investig. 2001;108:1359–67. doi: 10.1172/JCI12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dowell P, Otto TC, Adi S, Lane MD. Convergence of peroxisome proliferator-activated receptor γ and Foxo1 signaling pathways. J. Biol. Chem. 2003;278:45485–91. doi: 10.1074/jbc.M309069200. [DOI] [PubMed] [Google Scholar]
- 22.Hirota K, Daitoku H, Matsuzaki H, Araya N, Yamagata K, et al. Hepatocyte nuclear factor-4 is a novel downstream target of insulin via FKHR as a signal-regulated transcriptional inhibitor. J. Biol. Chem. 2003;278:13056–60. doi: 10.1074/jbc.C200553200. [DOI] [PubMed] [Google Scholar]
- 23.Zhao HH, Herrera RE, Coronado-Heinsohn E, Yang MC, Ludes-Meyers JH, et al. Forkhead homologue in rhabdomyosarcoma functions as a bifunctional nuclear receptor-interacting protein with both coactivator and corepressor functions. J. Biol. Chem. 2001;276:27907–12. doi: 10.1074/jbc.M104278200. [DOI] [PubMed] [Google Scholar]
- 24.Borkhardt A, Repp R, Haas OA, Leis T, Harbott J, et al. Cloning and characterization of AFX, the gene that fuses to MLL in acute leukemias with a t(X;11)(q13;q23) Oncogene. 1997;14:195–202. doi: 10.1038/sj.onc.1200814. [DOI] [PubMed] [Google Scholar]
- 25.Galili N, Davis RJ, Fredericks WJ, Mukhopadhyay S, Rauscher FJ, 3rd, et al. Fusion of a fork head domain gene to PAX3 in the solid tumour alveolar rhabdomyosarcoma. Nat. Genet. 1993;5:230–35. doi: 10.1038/ng1193-230. [DOI] [PubMed] [Google Scholar]
- 26.Lin K, Dorman JB, Rodan A, Kenyon C. daf-16: an HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 1997;278:1319–22. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- 27.Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, et al. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–99. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 28.Kops GJ, de Ruiter ND, de Vries-Smits AM, Powell DR, Bos JL, Burgering BM. Direct control of the Forkhead transcription factor AFX by protein kinase B. Nature. 1999;398:630–34. doi: 10.1038/19328. [DOI] [PubMed] [Google Scholar]
- 29.Meier R, Alessi DR, Cron P, Andjelkovic M, Hemmings BA. Mitogenic activation, phosphorylation, and nuclear translocation of protein kinase Bβ. J. Biol. Chem. 1997;272:30491–97. doi: 10.1074/jbc.272.48.30491. [DOI] [PubMed] [Google Scholar]
- 30.Brownawell AM, Kops GJ, Macara IG, Burgering BM. Inhibition of nuclear import by protein kinase B (Akt) regulates the subcellular distribution and activity of the forkhead transcription factor AFX. Mol. Cell. Biol. 2001;21:3534–46. doi: 10.1128/MCB.21.10.3534-3546.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brunet A, Kanai F, Stehn J, Xu J, Sarbassova D, et al. 14-3-3 transits to the nucleus and participates in dynamic nucleocytoplasmic transport. J. Cell Biol. 2002;156:817–28. doi: 10.1083/jcb.200112059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Biggs WH, 3rd, Meisenhelder J, Hunter T, Cavenee WK, Arden KC. Protein kinase B/Akt-mediated phosphorylation promotes nuclear exclusion of the winged helix transcription factor FKHR1. Proc. Natl. Acad. Sci. USA. 1999;96:7421–26. doi: 10.1073/pnas.96.13.7421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takaishi H, Konishi H, Matsuzaki H, Ono Y, Shirai Y, et al. Regulation of nuclear translocation of forkhead transcription factor AFX by protein kinase B. Proc. Natl. Acad. Sci. USA. 1999;96:11836–41. doi: 10.1073/pnas.96.21.11836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brunet A, Park J, Tran H, Hu LS, Hemmings BA, Greenberg ME. Protein kinase SGK mediates survival signals by phosphorylating the forkhead transcription factor FKHRL1 (FOXO3a) Mol. Cell. Biol. 2001;21:952–65. doi: 10.1128/MCB.21.3.952-965.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.You H, Jang Y, You-Ten AI, Okada H, Liepa J, et al. p53-dependent inhibition of FKHRL1 in response to DNA damage through protein kinase SGK1. Proc. Natl. Acad. Sci. USA. 2004;101:14057–62. doi: 10.1073/pnas.0406286101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dehner M, Hadjihannas M, Weiske J, Huber O, Behrens J. Wnt signaling inhibits Forkhead box O3a-induced transcription and apoptosis through up-regulation of serum- and glucocorticoid-inducible kinase 1. J. Biol. Chem. 2008;283:19201–10. doi: 10.1074/jbc.M710366200. [DOI] [PubMed] [Google Scholar]
- 37.Huang H, Regan KM, Lou Z, Chen J, Tindall DJ. CDK2-dependent phosphorylation of FOXO1 as an apoptotic response to DNA damage. Science. 2006;314:294–97. doi: 10.1126/science.1130512. [DOI] [PubMed] [Google Scholar]
- 38.Harper JW, Elledge SJ, Keyomarsi K, Dynlacht B, Tsai LH, et al. Inhibition of cyclin-dependent kinases by p21. Mol. Biol. Cell. 1995;6:387–400. doi: 10.1091/mbc.6.4.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu MC, Lee DF, Xia W, Golfman LS, Ou-Yang F, et al. IκB kinase promotes tumorigenesis through inhibition of forkhead FOXO3a. Cell. 2004;117:225–37. doi: 10.1016/s0092-8674(04)00302-2. [DOI] [PubMed] [Google Scholar]
- 40.Yang JY, Zong CS, Xia W, Yamaguchi H, Ding Q, et al. ERK promotes tumorigenesis by inhibiting FOXO3a via MDM2-mediated degradation. Nat. Cell Biol. 2008;10:138–48. doi: 10.1038/ncb1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rena G, Woods YL, Prescott AR, Peggie M, Unterman TG, et al. Two novel phosphorylation sites on FKHR that are critical for its nuclear exclusion. EMBO J. 2002;21:2263–71. doi: 10.1093/emboj/21.9.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Woods YL, Rena G, Morrice N, Barthel A, Becker W, et al. The kinase DYRK1A phosphorylates the transcription factor FKHR at Ser329 in vitro, a novel in vivo phosphorylation site. Biochem. J. 2001;355:597–607. doi: 10.1042/bj3550597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Greer EL, Oskoui PR, Banko MR, Maniar JM, Gygi MP, et al. The energy sensor AMP-activated protein kinase directly regulates the mammalian FOXO3 transcription factor. J. Biol. Chem. 2007;282:30107–19. doi: 10.1074/jbc.M705325200. [DOI] [PubMed] [Google Scholar]
- 44.Dixit M, Bess E, Fisslthaler B, Hartel FV, Noll T, et al. Shear stress-induced activation of the AMP-activated protein kinase regulates FoxO1a and angiopoietin-2 in endothelial cells. Cardiovasc. Res. 2008;77:160–68. doi: 10.1093/cvr/cvm017. [DOI] [PubMed] [Google Scholar]
- 45.Essers MA, Weijzen S, de Vries-Smits AM, Saarloos I, de Ruiter ND, et al. FOXO transcription factor activation by oxidative stress mediated by the small GTPase Ral and JNK. EMBO J. 2004;23:4802–12. doi: 10.1038/sj.emboj.7600476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang MC, Bohmann D, Jasper H. JNK extends life span and limits growth by antagonizing cellular and organism-wide responses to insulin signaling. Cell. 2005;121:115–25. doi: 10.1016/j.cell.2005.02.030. [DOI] [PubMed] [Google Scholar]
- 47.Oh SW, Mukhopadhyay A, Svrzikapa N, Jiang F, Davis RJ, Tissenbaum HA. JNK regulates lifespan in Caenorhabditis elegans by modulating nuclear translocation of forkhead transcription factor/DAF-16. Proc. Natl. Acad. Sci. USA. 2005;102:4494–99. doi: 10.1073/pnas.0500749102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kawamori D, Kaneto H, Nakatani Y, Matsuoka TA, Matsuhisa M, et al. The forkhead transcription factor Foxo1 bridges the JNK pathway and the transcription factor PDX-1 through its intracellular translocation. J. Biol. Chem. 2006;281:1091–98. doi: 10.1074/jbc.M508510200. [DOI] [PubMed] [Google Scholar]
- 49.Sunayama J, Tsuruta F, Masuyama N, Gotoh Y. JNK antagonizes Akt-mediated survival signals by phosphorylating 14-3-3. J. Cell Biol. 2005;170:295–304. doi: 10.1083/jcb.200409117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lehtinen MK, Yuan Z, Boag PR, Yang Y, Villen J, et al. A conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell. 2006;125:987–1001. doi: 10.1016/j.cell.2006.03.046. [DOI] [PubMed] [Google Scholar]
- 51.Anand R, Kim AY, Brent M, Marmorstein R. Biochemical analysis of MST1 kinase: elucidation of a C-terminal regulatory region. Biochemistry. 2008;42:6719–26. doi: 10.1021/bi800309m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brent MM, Anand R, Marmorstein R. Structural basis for DNA recognition by FoxO1 and its regulation by posttranslational modification. Structure. 2008;16:1407–16. doi: 10.1016/j.str.2008.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jang SW, Yang SJ, Srinivasan S, Ye K. Akt phosphorylates MstI and prevents its proteolytic activation, blocking FOXO3 phosphorylation and nuclear translocation. J. Biol. Chem. 2007;282:30836–44. doi: 10.1074/jbc.M704542200. [DOI] [PubMed] [Google Scholar]
- 54.Slawson C, Housley MP, Hart GW. O-GlcNAc cycling: how a single sugar post-translational modification is changing the way we think about signaling networks. J. Cell. Biochem. 2006;97:71–83. doi: 10.1002/jcb.20676. [DOI] [PubMed] [Google Scholar]
- 55.Housley MP, Rodgers JT, Udeshi ND, Kelly TJ, Shabanowitz J, et al. O-GlcNAc regulates FoxO activation in response to glucose. J. Biol. Chem. 2008;283:16283–92. doi: 10.1074/jbc.M802240200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kuo M, Zilberfarb V, Gangneux N, Christeff N, Issad T. O-glycosylation of FoxO1 increases its transcriptional activity towards the glucose 6-phosphatase gene. FEBS Lett. 2008;582:829–34. doi: 10.1016/j.febslet.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 57.Housley MP, Udeshi ND, Rodgers JT, Shabanowitz J, Puigserver P, et al. A PGC-1α:O-GlcNAc transferase complex regulates FoxO transcription factor activity in response to glucose. J. Biol. Chem. 2008;284:5148–57. doi: 10.1074/jbc.M808890200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nasrin N, Ogg S, Cahill CM, Biggs W, Nui S, et al. DAF-16 recruits the CREB-binding protein coactivator complex to the insulin-like growth factor binding protein 1 promoter in HepG2 cells. Proc. Natl. Acad. Sci. USA. 2000;97:10412–17. doi: 10.1073/pnas.190326997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Perrot V, Rechler MM. The coactivator p300 directly acetylates the forkhead transcription factor Foxo1 and stimulates Foxo1-induced transcription. Mol. Endocrinol. 2005;19:2283–98. doi: 10.1210/me.2004-0292. [DOI] [PubMed] [Google Scholar]
- 60.Matsuzaki H, Daitoku H, Hatta M, Aoyama H, Yoshimochi K, Fukamizu A. Acetylation of Foxo1 alters its DNA-binding ability and sensitivity to phosphorylation. Proc. Natl. Acad. Sci. USA. 2005;102:11278–83. doi: 10.1073/pnas.0502738102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van der Horst A, Tertoolen LG, de Vries-Smits LM, Frye RA, Medema RH, Burgering BM. FOXO4 is acetylated upon peroxide stress and deacetylated by the longevity protein hSir2(SIRT1) J. Biol. Chem. 2004;279:28873–79. doi: 10.1074/jbc.M401138200. [DOI] [PubMed] [Google Scholar]
- 62.Ghosh HS. The anti-aging, metabolism potential of SIRT1. Curr. Opin. Investig. Drugs. 2008;9:1095–102. [PubMed] [Google Scholar]
- 63.Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–30. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 64.Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–15. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 65.Motta MC, Divecha N, Lemieux M, Kamel C, Chen D, et al. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116:551–63. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- 66.Daitoku H, Hatta M, Matsuzaki H, Aratani S, Ohshima T, et al. Silent information regulator 2 potentiates Foxo1-mediated transcription through its deacetylase activity. Proc. Natl. Acad. Sci. USA. 2004;101:10042–47. doi: 10.1073/pnas.0400593101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kitamura YI, Kitamura T, Kruse JP, Raum JC, Stein R, et al. FoxO1 protects against pancreatic β cell failure through NeuroD and MafA induction. Cell Metab. 2005;2:153–63. doi: 10.1016/j.cmet.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 68.Plas DR, Thompson CB. Akt activation promotes degradation of tuberin and FOXO3a via the proteasome. J. Biol. Chem. 2003;278:12361–66. doi: 10.1074/jbc.M213069200. [DOI] [PubMed] [Google Scholar]
- 69.Huang H, Regan KM, Wang F, Wang D, Smith DI, et al. Skp2 inhibits FOXO1 in tumor suppression through ubiquitin-mediated degradation. Proc. Natl. Acad. Sci. USA. 2005;102:1649–54. doi: 10.1073/pnas.0406789102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Charvet C, Alberti I, Luciano F, Jacquel A, Bernard A, et al. Proteolytic regulation of Forkhead transcription factor FOXO3a by caspase-3-like proteases. Oncogene. 2003;22:4557–68. doi: 10.1038/sj.onc.1206778. [DOI] [PubMed] [Google Scholar]
- 71.Brenkman AB, de Keizer PL, van den Broek NJ, Jochemsen AG, Burgering BM. Mdm2 induces mono-ubiquitination of FOXO4. PLoS ONE. 2008;3:e2819. doi: 10.1371/journal.pone.0002819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van der Horst A, de Vries-Smits AM, Brenkman AB, van Triest MH, van den Broek N, et al. FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat. Cell Biol. 2006;8:1064–73. doi: 10.1038/ncb1469. [DOI] [PubMed] [Google Scholar]
- 73.Li HH, Willis MS, Lockyer P, Miller N, McDonough H, et al. Atrogin-1 inhibits Akt-dependent cardiac hypertrophy in mice via ubiquitin-dependent coactivation of Forkhead proteins. J. Clin. Investig. 2007;117:3211–23. doi: 10.1172/JCI31757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hosaka T, Biggs WH, 3rd, Tieu D, Boyer AD, Varki NM, et al. Disruption of forkhead transcription factor (FOXO) family members in mice reveals their functional diversification. Proc. Natl. Acad. Sci. USA. 2004;101:2975–80. doi: 10.1073/pnas.0400093101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Furuyama T, Kitayama K, Shimoda Y, Ogawa M, Sone K, et al. Abnormal angiogenesis in Foxo1 (Fkhr)-deficient mice. J. Biol. Chem. 2004;279:34741–49. doi: 10.1074/jbc.M314214200. [DOI] [PubMed] [Google Scholar]
- 76.Evans-Anderson HJ, Alfieri CM, Yutzey KE. Regulation of cardiomyocyte proliferation and myocardial growth during development by FOXO transcription factors. Circ. Res. 2008;102:686–94. doi: 10.1161/CIRCRESAHA.107.163428. [DOI] [PubMed] [Google Scholar]
- 77.Heinke J, Wehofsits L, Zhou Q, Zoeller C, Baar KM, et al. BMPER is an endothelial cell regulator and controls bone morphogenetic protein-4-dependent angiogenesis. Circ. Res. 2008;103:804–12. doi: 10.1161/CIRCRESAHA.108.178434. [DOI] [PubMed] [Google Scholar]
- 78.Moser M, Binder O, Wu Y, Aitsebaomo J, Ren R, et al. BMPER, a novel endothelial cell precursor-derived protein, antagonizes bone morphogenetic protein signaling and endothelial cell differentiation. Mol. Cell. Biol. 2003;23:5664–79. doi: 10.1128/MCB.23.16.5664-5679.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Potente M, Urbich C, Sasaki K, Hofmann WK, Heeschen C, et al. Involvement of Foxo transcription factors in angiogenesis and postnatal neovascularization. J. Clin. Investig. 2005;115:2382–92. doi: 10.1172/JCI23126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Potente M, Ghaeni L, Baldessari D, Mostoslavsky R, Rossig L, et al. SIRT1 controls endothelial angiogenic functions during vascular growth. Genes Dev. 2007;21:2644–58. doi: 10.1101/gad.435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Creemers EE, Sutherland LB, McAnally J, Richardson JA, Olson EN. Myocardin is a direct transcriptional target of Mef2, Tead and Foxo proteins during cardiovascular development. Development. 2006;133:4245–56. doi: 10.1242/dev.02610. [DOI] [PubMed] [Google Scholar]
- 82.Paik JH, Kollipara R, Chu G, Ji H, Xiao Y, et al. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128:309–23. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shiojima I, Walsh K. Regulation of cardiac growth and coronary angiogenesis by the Akt/PKB signaling pathway. Genes Dev. 2006;20:3347–65. doi: 10.1101/gad.1492806. [DOI] [PubMed] [Google Scholar]
- 84.Stitt TN, Drujan D, Clarke BA, Panaro F, Timofeyva Y, et al. The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol. Cell. 2004;14:395–403. doi: 10.1016/s1097-2765(04)00211-4. [DOI] [PubMed] [Google Scholar]
- 85.Belke DD, Betuing S, Tuttle MJ, Graveleau C, Young ME, et al. Insulin signaling coordinately regulates cardiac size, metabolism, and contractile protein isoform expression. J. Clin. Investig. 2002;109:629–39. doi: 10.1172/JCI13946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Skurk C, Izumiya Y, Maatz H, Razeghi P, Shiojima I, et al. The FOXO3a transcription factor regulates cardiac myocyte size downstream of AKT signaling. J. Biol. Chem. 2005;280:20814–23. doi: 10.1074/jbc.M500528200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shioi T, McMullen JR, Kang PM, Douglas PS, Obata T, et al. Akt/protein kinase B promotes organ growth in transgenic mice. Mol. Cell. Biol. 2002;22:2799–809. doi: 10.1128/MCB.22.8.2799-2809.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ni YG, Berenji K, Wang N, Oh M, Sachan N, et al. Foxo transcription factors blunt cardiac hypertrophy by inhibiting calcineurin signaling. Circulation. 2006;114:1159–68. doi: 10.1161/CIRCULATIONAHA.106.637124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Molkentin JD, Lu JR, Antos CL, Markham B, Richardson J, et al. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell. 1998;93:215–28. doi: 10.1016/s0092-8674(00)81573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li HH, Kedar V, Zhang C, McDonough H, Arya R, et al. Atrogin-1/muscle atrophy F-box inhibits calcineurin-dependent cardiac hypertrophy by participating in an SCF ubiquitin ligase complex. J. Clin. Investig. 2004;114:1058–71. doi: 10.1172/JCI22220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ivaska J, Nissinen L, Immonen N, Eriksson JE, Kähäri VM, Heino J. Integrin α2 β1 promotes activation of protein phosphatase 2A and dephosphorylation of Akt and glycogen synthase kinase 3 β. Mol. Cell. Biol. 2002;22:1352–59. doi: 10.1128/mcb.22.5.1352-1359.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ni YG, Wang N, Cao DJ, Sachan N, Morris DJ, et al. FoxO transcription factors activate Akt and attenuate insulin signaling in heart by inhibiting protein phosphatases. Proc. Natl. Acad. Sci. USA. 2007;104:20517–22. doi: 10.1073/pnas.0610290104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wessells RJ, Fitzgerald E, Cypser JR, Tatar M, Bodmer R. Insulin regulation of heart function in aging fruit flies. Nat. Genet. 2004;36:1275–81. doi: 10.1038/ng1476. [DOI] [PubMed] [Google Scholar]
- 94.Alcendor RR, Gao S, Zhai P, Zablocki D, Holle E, et al. Sirt1 regulates aging and resistance to oxidative stress in the heart. Circ. Res. 2007;100:1512–21. doi: 10.1161/01.RES.0000267723.65696.4a. [DOI] [PubMed] [Google Scholar]
- 95.Chen CJ, Yu W, Fu YC, Wang X, Li JL, Wang W. Resveratrol protects cardiomyocytes from hypoxia-induced apoptosis through the SIRT1-FoxO1 pathway. Biochem. Biophys. Res. Commun. 2009;378:389–93. doi: 10.1016/j.bbrc.2008.11.110. [DOI] [PubMed] [Google Scholar]
- 96.Chan AY, Dolinsky VW, Soltys CL, Viollet B, Baksh S, et al. Resveratrol inhibits cardiac hypertrophy via AMP-activated protein kinase and Akt. J. Biol. Chem. 2008;283:24194–201. doi: 10.1074/jbc.M802869200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chiribau CB, Cheng L, Cucoranu IC, Yu YS, Clempus RE, Sorescu D. FOXO3A regulates peroxiredoxin III expression in human cardiac fibroblasts. J. Biol. Chem. 2008;283:8211–17. doi: 10.1074/jbc.M710610200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Abid MR, Guo S, Minami T, Spokes KC, Ueki K, et al. Vascular endothelial growth factor activates PI3K/Akt/forkhead signaling in endothelial cells. Arterioscler. Thromb. Vasc. Biol. 2004;24:294–300. doi: 10.1161/01.ATV.0000110502.10593.06. [DOI] [PubMed] [Google Scholar]
- 99.Abid MR, Spokes KC, Shih SC, Aird WC. NADPH oxidase activity selectively modulates vascular endothelial growth factor signaling pathways. J. Biol. Chem. 2007;282:35373–85. doi: 10.1074/jbc.M702175200. [DOI] [PubMed] [Google Scholar]
- 100.Abid MR, Shih SC, Otu HH, Spokes KC, Okada Y, et al. A novel class of vascular endothelial growth factor-responsive genes that require forkhead activity for expression. J. Biol. Chem. 2006;281:35544–53. doi: 10.1074/jbc.M608620200. [DOI] [PubMed] [Google Scholar]
- 101.Allard D, Figg N, Bennett MR, Littlewood TD. Akt regulates the survival of vascular smooth muscle cells via inhibition of FoxO3a and GSK3. J. Biol. Chem. 2008;283:19739–47. doi: 10.1074/jbc.M710098200. [DOI] [PubMed] [Google Scholar]
- 102.Chlench S, Mecha Disassa N, Hohberg M, Hoffmann C, Pohlkamp T, et al. Regulation of Foxo-1 and the angiopoietin-2/Tie2 system by shear stress. FEBS Lett. 2007;581:673–80. doi: 10.1016/j.febslet.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 103.Kim HS, Skurk C, Maatz H, Shiojima I, Ivashchenko Y, et al. Akt/FOXO3a signaling modulates the endothelial stress response through regulation of heat shock protein 70 expression. FASEB J. 2005;19:1042–44. doi: 10.1096/fj.04-2841fje. [DOI] [PubMed] [Google Scholar]
- 104.Gershman B, Puig O, Hang L, Peitzsch RM, Tatar M, Garofalo RS. High-resolution dynamics of the transcriptional response to nutrition in Drosophila: a key role for dFOXO. Physiol. Genomics. 2007;29:24–34. doi: 10.1152/physiolgenomics.00061.2006. [DOI] [PubMed] [Google Scholar]
- 105.Philip-Couderc P, Tavares NI, Roatti A, Lerch R, Montessuit C, Baertschi AJ. Forkhead transcription factors coordinate expression of myocardial KATP channel subunits and energy metabolism. Circ. Res. 2008;102:e20–35. doi: 10.1161/CIRCRESAHA.107.166744. [DOI] [PubMed] [Google Scholar]
- 106.Daitoku H, Yamagata K, Matsuzaki H, Hatta M, Fukamizu A. Regulation of PGC-1 promoter activity by protein kinase B and the forkhead transcription factor FKHR. Diabetes. 2003;52:642–49. doi: 10.2337/diabetes.52.3.642. [DOI] [PubMed] [Google Scholar]
- 107.Zhang Y, Ma K, Sadana P, Chowdhury F, Gaillard S, et al. Estrogen-related receptors stimulate pyruvate dehydrogenase kinase isoform 4 gene expression. J. Biol. Chem. 2006;281:39897–906. doi: 10.1074/jbc.M608657200. [DOI] [PubMed] [Google Scholar]
- 108.Yamada S, Kane GC, Behfar A, Liu XK, Dyer RB, et al. Protection conferred by myocardial ATP-sensitive K+ channels in pressure overload-induced congestive heart failure revealed in KCNJ11 Kir6.2-null mutant. J. Physiol. 2006;577:1053–65. doi: 10.1113/jphysiol.2006.119511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Matsui Y, Kyoi S, Takagi H, Hsu CP, Hariharan N, et al. Molecular mechanisms and physiological significance of autophagy during myocardial ischemia and reperfusion. Autophagy. 2008;4:409–15. doi: 10.4161/auto.5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hamacher-Brady A, Brady NR, Gottlieb RA. Enhancing macroautophagy protects against ischemia/reperfusion injury in cardiac myocytes. J. Biol. Chem. 2006;281:29776–87. doi: 10.1074/jbc.M603783200. [DOI] [PubMed] [Google Scholar]
- 111.Nakai A, Yamaguchi O, Takeda T, Higuchi Y, Hikoso S, et al. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat. Med. 2007;13:619–24. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- 112.Zhao J, Brault JJ, Schild A, Cao P, Sandri M, et al. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6:472–83. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]