Abstract
Phagosome maturation is the process by which internalized particles (such as bacteria and apoptotic cells) are trafficked into a series of increasingly acidified membrane-bound structures, ultimately leading to particle degradation. Studies in model organisms and mammals, along with characterization of the phagosomal proteome have led to the identification of numerous candidate proteins that cooperate to control the maturation of phagosomes containing different particles. A subset of these candidates make up the first pathway that has been identified for maturation of apoptotic cell-containing phagosomes. suggesting the use of a machinery that is distinct from receptor-mediated endocytosis.
Eukaryotic cells internalize a variety of particles during their lifetime. The uptake of particles >0.5 μm in size by cells is considered phagocytosis, whereas particles <0.5 μm are taken up by receptor-mediated endocytosis or pinocytosis. There are distinct types of phagocytosis, which tend to be ligand specific: bacteria (~0.5–3 μm) or yeast (~3–4 μm) are internalized by macrophages via scavenger receptors; microorganisms can also be coated with serum components (eg. complement) or antibodies and then taken up via complement1, 2 or Fc receptors3, 4, respectively. Cells undergoing apoptosis, which can range in size from 5–50 μm, must also be removed (Box 1): in fact, we turn over ~200 billion cells each day of our life, perhaps making apoptotic cell removal one of the most common type of phagocytosis occurring throughout life5. Thus, understanding how apoptotic cells are phagocytosed and processed is a fundamentally important biological problem. Apoptotic cell turnover begins with the induction of an apoptotic programme or other cellular changes that mark them for removal6, 7. The subsequent recognition of altered features by phagocytes leads to their highly efficient and immunologically silent removal8. Apoptotic cells can be taken up either by neighbouring cells or by professional phagocytes such as macrophages9–11 and dendritic cells12, 13; the contribution of each type of phagocyte to clearance in vivo has not been extensively addressed.
Phagosome maturation can be viewed as the end of the phagocytic process, when internalized apoptotic cells or bacteria must be efficiently degraded. Once a particle is internalized, it “matures” through a series of increasingly acidified membrane-bound structures called phagosomes14. These phagosomes sequentially acquire different proteins (eg. the Rab GTPases15) during maturation, ultimately leading to fusion with an acidic lysosome structure. Defects in degradation have a number of consequences, depending on the type of particle that must be removed: defects in lysosome-associated membrane protein 2 (LAMP-2; see below) block phagosome maturation and have been linked to periodontal disease due to a decreased neutrophil-mediated bacterial killing16. Defects in apoptotic cell degradation can result in autoimmune disease17: apoptotic cell removal by dendritic cells has been implicated in the establishment of tolerance, the process by which the immune system is educated to not respond to self-derived antigens9, 12, 13, 18, 19. Phagosome maturation is therefore of great importance to normal homeostasis of the immune system, to the response to bacterial pathogens and in influencing the immune response to our normal bacterial flora. Moreover, since certain pathogens seem to use the phagocytic machinery to enter and/or live within the host, understanding the various steps of cargo handling in the context of phagocytosis could be key in therapeutic design.
In this review, we focus on how apoptotic cell-containing phagosomes mature within the phagocyte, and on the recent developments in our understanding of the genetic, molecular and functional aspects of phagosome maturation in a variety of contexts. Extensive studies on receptor-mediated endocytosis over the past few decades have given us a general paradigm for this process: after binding of a soluble ligand (such as a growth factor) to a receptor, there is invagination of the membrane and scission of the vesicle that contains the receptor and the ligand/cargo from the plasma membrane20–22 (Figure 1). Time-lapse studies have shown that this vesicle moves within the cell through a series of stages (coated with the GTPases Rab5 and Rab723) leading to progressive acidification and, eventually, fusion with lysosomes24, 25. During these acidification steps, dissociation of cargo from receptor occurs, and the receptor may be recycled back to the membrane, while the cargo may be trafficked for degradation or use within the cell26, 27. While this general paradigm has been extremely useful for our current knowledge of vesicular transport within cells and membrane biogenesis/fusion events, the phagocytic process poses several unique challenges.
Figure 1. Endocytosis as a paradigm for phagocytosis.
Phagocytosis and receptor mediated endocytosis are similar processes, but also have distinct features.
(a) During receptor-mediated endocytosis, activated receptor sinks into the cell, in this case into a clathrin-coated pit21. Following scission of the vesicle from the membrane mediated by dynamin, clathrin is removed20; receptors are then trafficked to the `recycling endosome,' where they are eventually sorted back to the cell surface22. Activated receptors are also removed from phagosomes and presumably are also targeted to recycling endosomes. Endocytosed vesicles acquire the GTPase Rab5, which is activated by GEFs (such as RabEx-5 and GapEx-5)150 which allows homotypic fusion with other Rab5-positive structures23, 57, as well as heterotypic fusion with Rab4-positive recycling endosomes70. Rab5 is then exchanged for Rab7 via the HOPS complex23, which contains the Rab7 GEF Vps39, resulting in recruitment of the Rab7 effector RILP and ultimately fusion with acidic lysosomes151 and acquisition of the lysosomal markers LAMP-1 and LAMP-2.
(b) In contrast, during phagocytosis the plasma membrane is extended around the apoptotic cell, forming the phagosome8, 14. Phagosomes also `mature' through Rab5(+) and Rab7(+) stages, ultimately resulting in fusion with lysosome structures28, 33, 41, 52, 57, 58, however phagosomes appear to utilize a distinct mechanism for GTPase regulation. In the nematode, known RAB-5 GEFs are not required for phagosome maturation), and the HOPS complex is not required for the recruitment of Rab7 to the phagosome. This may reflect the observations that phagosomes rarely fuse together and tend to mature individually. Other events, such as trafficking of proteins from the Golgi to the phagosome have been described43, although whether this occurs during endocytosis remains to be determined. A number of GTPases have also been found associated with the phagosome during phagosome maturation (see Box 3), many of which have not been described to play a role on the endosome.
First, when a cell engulfs an apoptotic neighbour, the plasma membrane must be temporarily extended over the entire surface area of the apoptotic cell, which in many cases is as large or larger than the phagocyte14. This is accomplished by the mobilization of intracellular vesicles to the plasma membrane, which also potentially transport signalling proteins to the site of internalization14, 28, 29. Second, the contents of the phagocytic cargo brought into the cell (for example, bacteria or apoptotic cells) needs to be `unpacked' and processed in a way that allows for either an immunological response (in the case of microorganisms) or immunological tolerance (to self-antigens derived from apoptotic cells; Box 2)30, 31. Third, the engulfment of larger particles, such as apoptotic cells, often involves the simultaneous activation of multiple types of receptors that are distinct from those involved in receptor-mediated endocytosis or phagocytosis of bacteria (see Box 1)8, 32. Even though the general paradigm that phagocytosed particles mature through progressively acidic structures appears to be true33, defining which molecules regulate phagosome maturation, and our knowledge of how signalling events overlap or differ from endocytosis and other phagocytic events have become active areas of investigation.
Insights from proteomics approaches
Early studies attempted to identify proteins involved in phagosome maturation through biochemical and proteomic approaches using targets such as whole bacteria or latex beads33. Although this resulted in the identification of a vast number of candidate proteins localized to phagosomes, these studies largely did not address the functional requirements for these proteins in phagosome maturation. What these studies did accomplish, however, was an excellent descriptive analysis of the different stages of phagosome maturation and the development of a basic model describing protein recruitment and release from the nascent phagosome33–35. A variety of proteins must be delivered to the phagosome, including V-ATPases (which acidify the phagosome), acidic proteases (such as cathepsins) and major histocompatibility complex (MHC) class II molecules (for presentation of phagosome-derived antigens on the cell surface).
The acid test and its consequences
Acidification of the phagosome is perhaps the most common readout used for addressing protein function in phagosome maturation. Acidification is essential during phagosome maturation: it is only when the phagosome hits a sufficiently low pH that cathepsins (a family of acidic proteases) become active and the phagocytosed particle can begin to be degraded36. In addition to degrading proteins into peptides for loading onto class II and class I major histocompatibility (MHC) molecules, cathepsin D also plays a role in the activation of MHC class II proteins, without which peptides cannot be loaded37, 38. Thus, acidification is a marker of a `functional' phagosome.
Acidification of the phagosome occurs in two stages: first, an `early' acidification step that results in a relatively small drop in pH and is not well understood39; subsequent to this, V-type (vacuolar) ATPases, proton pumps driven by the hydrolysis of ATP40, are trafficked to the phagosome at relatively late stages. In the nematode Caenorhabditis elegans, phagosome acidification begins quite early, with Rab5(+) phagosomes staining weakly with acridine orange, a marker for acidic organelles41. However, these phagosomes do not stain as strongly as late-stage apoptotic cells, consistent with multiple modes of acidification dependent on the stage of maturation.
Vacuolar ATPases
The V-type (vacuolar) ATPase is a multi-subunit complex composed of 14 subunits in two separate structures: the V1 complex mediates ATP hydrolysis, whereas the V0 complex transports H+ across the endosomal membrane40. V-type ATPases are trafficked to the phagosome and function to acidify its contents, but where do these ATPases (and other proteins trafficked onto the phagosome) come from?
In Dictyostelium discoideum, yeast-containing phagosomes acquire V-type ATPases by fusion with V-type ATPase-containing endosomes42. In multicellular organisms, trafficking of V-type ATPases is more complex: rather than being delivered by fusing with endosomes, these proteins appear to be trafficked from the trans-Golgi to the phagosomealong with cathepsins43. Proteomic studies have identified a large number of components that localize to phagosomes33–35, 44–48; however, the mechanism by which many of these components arrive at the phagosomes has yet to be addressed. Intriguingly, the COPI complex, which is required for budding of vesicles from the Golgi, is also required for phagosome maturation49, although a role for COPI in vesicle budding from the phagosome (or other organelles) is possible.
Studies in zebrafish have suggested that V-type ATPases may have roles in phagosome maturation in addition to phagosome acidification: the V-ATPase A1 subunit is required for fusion of apoptotic cell-containing phagosomes with lysosomes, although the exact stage at which phagosomes are arrested is not known50. C. elegans vha-16 (the D subunit of the V0 ATPase) was identified in a screen for genes that are required for the early stages of phagosome maturation41. A direct role for these ATPases in vesicle fusion seems unlikely; however, it is possible that low pH may induce conformational changes in proteins required for phagosome maturation. Alternatively, acidification may result in cleavage of a target protein by activated cathepsins, which would then induce maturation. However, at this time, these are speculations, and the mechanism by which V-type ATPases regulate maturation remains uncertain.
Proteomics and contamination
In fairness, one must sound a note of caution when analysing proteomic data: one recent study has suggested that proteomic purification schemes suffer from relatively high contamination with other cellular membranes, which could be misleading51. While transient localization of a protein to the phagosome may provide an argument for a role in this process, further studies need to be undertaken to validate such candidates, either by RNA interference or dominant-negative approaches. In this respect, recent studies in model organisms like the nematode C. elegans have for the first time identified a functional pathway for phagosome maturation41, 52, 53.
Defining signalling pathways
While studies in cultured cells have been instrumental in identifying the protein components of the phagosome, a comprehensive pathway for how phagosome maturation is regulated has been lacking. Recent genetic studies, combined with cell biological approaches in the context of model organisms, have begun to define the basic pathway for the acidification of apoptotic cell-containing phagosomes.
C. elegans has been a powerful genetic tool for the identification of genes involved in programmed cell death, and genes identified in this model have also been shown to play a role in mammalian phagocytic uptake (see Box 1). However, the events leading to corpse degradation following phagocytosis have only recently begun to be addressed. One of the major impediments to these studies has been the requirement for many endocytic proteins during life, requiring the identification of rare temperature-sensitive mutations41 or time-consuming clonal screens28, 54. However, RNA interference (RNAi), which is remarkably efficient in the nematode, has bypassed this issue41, although it should be emphasized that complementary studies using mutant alleles are preferable, whenever such mutants are available. RNAi-based approaches, combined with genetics and fluorescent markers defining specific steps in phagosome maturation, have allowed unbiased screens to identify players that regulate various steps in phagosome maturation (Table 1). Several groups have made great strides towards developing the nematode as an in vivo model for phagosome maturation41, 52, 53.
Table 1.
Genes and proteins required for phagosome maturation during apoptotic cell removal
| C. elegans | ||||
|---|---|---|---|---|
| Gene name | Mammalian Orthologue | Function | Loss-of-function phenotypes | Ref. |
| Phagocytic genes | ||||
|
| ||||
| ced-1 | LRP/MEGF10, GULP/hCED-6, ABCA1/ABCA7 | Recognition of the apoptotic cell and intracellular signaling | Delayed phagocytosis and accumulation of RAB-7 and FYVE marker | 53, 104, 134, 136, 137, 153 |
| ced-6 | ||||
| ced-7 | ||||
| ced-5 | Dock180 | GTPase exchange factor (GEF) for CED-10/Rac1 | Delayed phagocytosis and accumulation of FYVE marker | 53, 131, 154, 155 |
|
| ||||
| GTPases | ||||
|
| ||||
| rab-2 | Rab2 | ER to Golgi transport and potentially ER/Golgi to phagosome | Delay (or possibly abnormal) maturation | 52, 54 |
| rab-5 | Rab5 | Early endosome maturation | Phagosome arrest, block in acidification | 41 |
| rab-7 | Rab7 | Late endosome maturation | Arrest of phagosomes at the RAB-5(+) stage | 41, 53 |
| dyn-1 | Dynamin | Recruitment of Rab5 | Enlarged phagosomes, pre-RAB-5 arrest | 41 |
|
| ||||
| RAB-5 effectors | ||||
|
| ||||
| vps-34 | Vps34 | PtdIns(3)-kinase | Block in RAB-5 recruitment, no acidification | 41 |
|
| ||||
| RAB-7 effectors | ||||
|
| ||||
| vps-11 | Vps11 | RING-motif containing protein | Phagosome arrest at RAB-7(+) stage | |
| vps-16 | Vps16 | unknown | 41 | |
| vps-18 | Vps18 | SNARE binding? | ||
| vps-33 | Vps33 | Sec1 homologue, SNARE binding? | ||
| vps-39 | Vps39 | GEF for RAB-7 | ||
|
| ||||
| Genes with `unknown' arrest | ||||
|
| ||||
| vps-41 | Vps41 | Adaptor protein, vacuolar biogenesis | No gross effect on recruitment of RAB-5 or RAB-7 to phagosomes | 41 |
| vps-45 | Vps45 | Sec1 homologue, vacuolar biogenesis | ||
| Mammals | ||||
|---|---|---|---|---|
| Protein | Nematode Orthologue | Function | Reduction of function phenotype | Ref. |
| Dynamin | DYN-1 | Required for Rab5 recruitment | Arrest of nascent phagosome, block in acidification | 28, 41 |
| Rab5 | RAB-5 | Required for maturation from nascent phagosome to Rab7(+) stage | Block in acidificaiton | 41, 61 |
| Rab7 | RAB-7 | Maturation from Rab5(+) to LAMP-1(+) phagosome | Not addressed | 89 |
| GapEx-5 | RME-6 | Rab5 activation | Block in Rab5 activation during phagosome maturation | 61 |
| RhoA | RHO-1 | Regulate early steps of phagosome maturation | Blocks Rab7 recruitment, phagosome acidification, and protease delivery | 89 |
| ERM proteins | ERM-1 | |||
Recent studies in the nematode C. elegans and mammalian systems have helped identify proteins involved in various steps of phagosome maturation during apoptotic cell removal. The genes in the nematode were identified in genetic screens, while those in the mammalian systems were identified using a candidate approach targeting individual molecules.
Phagosome maturation: a “Rab”-id obsession
Rab GTPases have been implicated in controlling transport between membrane-bound organelles. These proteins function as `molecular switches': in the inactive conformation, the Rab protein is GDP bound and resides in the cytoplasm, complexed with a RabGDI (Rab GDP dissociation inhibitor), which keeps the protein inactive55, 56. Following a signalling event, the RabGDI releases the GTPase, exposing the prenylated tail and allowing it to bind membranous organelles. Rab proteins will then bind to a GTPase exchange factor (GEF), resulting in the exchange of GDP for GTP55; the now active RabGTP will bind effector proteins57, resulting in the recruitment of machinery for further steps of maturation.
Recent genetic studies have shown that the GTPases RAB-541 and RAB-741, 53 are required for removal of apoptotic cells in the nematode, with RAB-5 and RAB-7 marking distinct (but partially overlapping) stages of maturation41. Similar to our current knowledge of maturation of internalized particles in mammalian systems15, 58, RAB-5 functions upstream of RAB-7, as phagosomes are arrested at the RAB-5(+) stage in rab-7-deficient worms. Based on the role of Rab5 and Rab7 on endosomes, Rab GTPases appear to function as part of a fusion pore59, which ultimately determines the types of interactions vesicles have with other membrane-bound organelles. In this respect, RAB-5 and RAB-7 potentially serve as nodes for recycling proteins back to the plasma membrane and as entry points for vesicles to deposit lysosomal proteases, with `effector' proteins binding to the activated Rab protein to mediate this function (see below).
A novel recruitment strategy for Rab5
One of the earliest known maturation events in a number of systems is the recruitment of Rab5 to phagosomes41, 58, 60, 61; however, a detailed mechanism for how this is accomplished during phagosome maturation is lacking. Recent studies41 have implied an unexpected role for dynamin and the Rab5 effector Vps34 in Rab5 recruitment to the nascent phagosomes (Figure 2), suggesting that recruitment of GTPases to the phagosome is a complex regulated process41, 62.
Figure 2. Activation of Rab5 during phagosome maturation.
During internalization of apoptotic cells Dynamin and Vps34 are recruited to the phagosome by an unknown mechanism. Vps34 can interact with inactive (GDP-bound) or active (GTP-bound) Rab5; the recruitment of Dynamin to the forming phagosome results in the recruitment of Vps34 and RAB-5GDP to the phagosome41. Dynamin (and DYN-1) has also been shown to play a role in the maintenance of the actin cytoskeleton, which has implications for phagocytosis as well41, 152. Once on the phagosome, an unidentified GEF likely activates RAB-5 (converting it to the GTP-bound state), activating Vps34 kinase activity61, which generates PtdIns(3)P (indicated as yellow dots) on the phagosome. It is noteworthy that GapEx-5 has been identified as a possible GEF for Rab5 in one mammalian cell context61. Effector proteins bind to active Rab5 in the context of PtdIns(3)P on the phagosome57; the assembly of effectors on the phagosome (which have yet to be identified) results in the exchange of Rab5 for Rab7, leading to the continued maturation of the phagosome.
The GTPase DYN-1, the nematode homologue of dynamin, was identified in forward and reverse genetic screens for genes involved in programmed cell death28, 41. Although initial reports linked DYN-1 to internalization of apoptotic cells (playing a role in focal exocytosis28, this view has now been revised41, 53. The current view based on studies in mammalian cells and nematodes41, 62 is that DYN-1 (and mammalian dynamin) play a role not in exocytosis or internalization of corpses, but rather is essential for phagosome maturation, namely the recruitment of RAB-5 to the nascent phagosome. Phagosomes in dyn-1(lf) mutant worms appear arrested, without recycling of the phagocytic receptor CED-1 back to the plasma membrane41. Further genetic and cell biological analysis identified the PtdIns(3)-kinase VPS-34 (a homologue of human PI3KC3/hVPS34) as another protein that regulates RAB-5 recruitment41. Biochemical analysis revealed a novel protein complex, where DYN-1/dynamin mediates the recruitment of Rab5GDP via interaction with Vps34, which serves as a bridging protein. To make matters more complex, RAB-5 also appears to regulate VPS-34 activity63 (also shown during removal of IgG-opsonized RBCs in mammals58, 63), and knockdown of RAB-5 blocks accumulation of PtdIns(3)P on the phagosome (see below).
While this novel complex suggests how Rab5 is recruited to apoptotic cell-containing phagosomes, it does not suggest a mode of regulation, as Rab5 must still be converted to the GTP-bound form in order for phagosomes to mature41, 64. There are at least three canonical Rab5 GEFs in the nematode genome (identified as VPS9-domain-containing proteins)41: RME-6, RABX-5 and TAG-333 (homologues of mammalian GapEx-5, RabEx-5 and RIN1, respectively). However, none of these are required (either singly or redundantly) for the removal of apoptotic cells, suggesting a novel GEF may play this role. This is distinct from other endocytic events (such as the internalization of proteins from the extracellular space), which jointly require RME-6 and RABX-5, suggesting that regulation of phagosome maturation is different from receptor-mediated endocytosis.
Intriguingly, a recent study, using a FRET (fluorescence resonance energy transfer) probe to monitor Rab5 activation in Swiss 3T3 cells suggested that the GEF GapEx-5 may play a role in regulating Rab5GTP loading during removal of apoptotic cells61. One caveat to this is that the experimental system utilizes overexpression of integrin proteins (and opsonization with the integrin ligand MFG-E8), rather than assaying basal engulfment, and the importance of GapEx-5 in the context of endogenous corpse removal (which does not require the opsonin MFG-E8 in most cases65) therefore remains to be determined.
Phosphoinositides and RAB-5 effectors
The enrichment of PtdIns(3)P on the surface of the phagosome can be regarded as a sign of a phagosome that is maturing from the Rab5(+) to the Rab7(+) stage, in both mammalian and nematode models (see below). The recruitment of Rab5 effectors (such as Rabenosyn-566) by interaction with Rab5GTP [in conjunction with PtdIns(3)P57], results in the recruitment of Rab7 and continued maturation of the phagosome (see below). To date, there are many known Rab5 effectors in cultured cells that serve diverse functions57. Effectors typically mediate homotypic fusion events during endocytosis, either among Rab5(+) vesicles, as in the case of EEA159, 67, 68, or they mediate heterotypic fusion between structures coated with different Rab proteins (potentially between Rab5- and Rab7-staining structures, as in the case of Rabenosyn66, 69) or between Rab5- and Rab4-coated structures (for Rabaptin-5)70. Typically, Rab5 effectors share a similar architecture, containing a FYVE domain for binding PtdIns(3)P generated on the phagosome by the PtdIns-3-kinase Vps3457.
VPS-34 is required for accumulation of PtdIns(3)-P on the phagosome41, and GFP-tagged constructs consisting of the FYVE domain from EEA-1 (a component of the Rab5 fusion pore) or HGRS1 (required for formation of multivesicular endosomes71) have been successfully used to monitor this process41, 53, 54. VPS-34 is currently the only RAB-5 effector to be linked to phagosome maturation in the nematode, and worms deficient in eea-1 or hgrs-1 show no defect in corpse removal41, although EEA1 is required for maturation of Mycobacterium-containing phagosomes in mammalian macrophages72. Knockdown of another known Rab5 effector, F22G12.4 (Rabankyrin; required for maturation of pinocytosed particles73) suggested that this protein is dispensable for maturation41. Moreover, a targeted screen of all FYVE-domain-containing proteins in the genome failed to identify other candidate effectors. Other phospholipid-binding proteins, such as those containing the Phox-homology (PX) domain, have been described to function in endocytic events and can bind PtdIns(3)P. Targeted screening of these factors identified two previously uncharacterized genes with weak defects in corpse removal, but could not place these genes within the pathway for phagosome maturation41.
In summary, it appears that RAB-5 function and regulation during phagosome maturation is distinct from other endocytic events. Intriguingly, studies in mammalian macrophages have suggested that Mycobacterium target EEA1 as a means of arresting phagosome maturation72, suggesting that the requirements for some proteins may vary by phagosome cargo. Generation of PtdIns(3)P on phagosome surfaces must play a function in maturation given that knockdown of VPS-34 generates a potent inhibition of phagosome maturation41, but the identity of other PtdIns(3)P-binding proteins and RAB-5 effectors, and their roles in phagosome maturation, remain to be determined.
Control of Rab7 activation and function
To date, we have little idea of how RAB-7 is recruited to the phagosome, or how recruitment is related to the release of RAB-5. Recent studies in cultured cells suggest Rab5 is simultaneously exchanged for Rab7 during endocytosis23, a process called Rab conversion, rather than by a series of fusion events between Rab5- and Rab7-positive structures, as was previously proposed74. Recruitment of RAB-7 independent of vesicle fusion events has also been observed during time-lapse studies during apoptotic cell removal in the nematode53. One potential candidate to regulate this process, the HOPS (homotypic fusion and vacuole protein sorting) complex, has been implicated in Rab7 recruitment during endocytosis in mammals23; however, this complex appears to serve a different function during phagosome maturation (Figure 3).
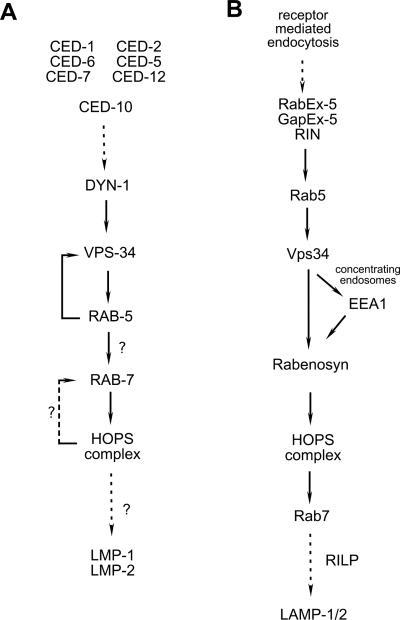
Figure 3. Pathways for phagosome maturation versus endocytosis.
Pathways for phagosome maturation and endosome maturation derived from publications describing the requirements of each protein in the different endocytic processes. (a) In the nematode, recruitment of RAB-5 to the phagosome (via interaction with DYN-1 and VPS-3441) begins the process of acidification, leading to VPS-34 activation and recruitment of as-yet unidentified effectors. Rab7 is then recruited to the phagosome, resulting in recruitment of the HOPS complex, which activates Rab7, promoting further maturation events and eventual fusion with lysosome structures.
(b) Activation of Rab5 on endocytic vesicles (by the GEFs RabEx-5, GapEx-5, or RIN1) begins a concentration step, where vesicles merge and ligand is concentrated23, potentially through the formation of a fusion pore by the Rab5 effector EEA1, which binds PtdIns(3)P on the phagosome generated by Vps34. Another Rab5 effector, Rabenosyn, binds Vps45, a component of the Rab7 GEF/effector HOPS complex, potentially mediating Rab5 recruitment to the maturing vesicle23, 66. The HOPS complex member Vps39 activated Rab7, resulting in recruitment of the Rab7 effector RILP, potentially resulting in lysosome fusion151. While some of the players have been shown to play a role in mammalian engulfment, a step-wise pathway for apoptotic cell-containing phagosome maturation in mammalian systems is just beginning to be defined.
The HOPS complex was first identified in the yeast S. cerevisiae75, and is composed of the proteins VPS-11, VPS-16, VPS-18, VPS-33, VPS-39, VPS-41 and VPS-45 (see Table 1). A subset of these proteins are also involved in transport from the Golgi to endosomal structures as the CORVET complex76. Vps39 has been shown to be a GEF for Ypt7 (Rab7) in S. cerevisae65, and VPS-33 and VPS-45 are Sec1 homologues, which have been proposed to function in vesicular tethering during fusion events77, 78. While the function of the HOPS complex has been tied to Rab5 function during endocytosis23, genetic studies in the nematode suggest that this complex instead functions downstream of RAB-7 recruitment during phagosome maturation41. Worms mutant in vps-11, vps-16, vps-18, vps-33 or vps-39 show phagosomes arrested at the RAB-7(+) stage, suggesting these proteins may be required for maturation of phagosomes to phagolysosomes (and/or fusion with existing lysosome structures)41.
Not all HOPS complex members function downstream of RAB-7 recruitment. One member of the complex, VPS-45, appears to function after loss of RAB-5 from the phagosome and prior to RAB-7 recruitment41. VPS-45 interacts with the Rab5 effector Rabenosyn (RABS-5 in C. elegans) during endocytosis in worms23 and mammals69, providing a mechanism for HOPS complex recruitment to Rab5(+) vesicles. However, RABS-5 is not required for phagosome maturation in C. elegans41, suggesting that VPS-45 may interact with another RAB-5 effector (or may play an alternative, unidentified role) during phagosome maturation. VPS-41, on the other hand, appears to be required downstream of RAB-7 function, as phagosomes can recruit (and lose) both RAB-5 and RAB-7 staining in vps-41-depleted worms41. The molecular function of VPS-41 has not been described, although like most HOPS complex members it is required for vesicle docking41, and further studies are needed to address how this protein functions both during phagosome maturation and endocytosis.
Signalling required for maturation (or fusion) of Rab7(+) phagosomes into acidic lysosomes has not been studied in great depth; to date, the HOPS complex has not been implicated in maturation of bacteria- or bead-containing phagosomes. One Rab7 effector, RILP (Rab7 interacting lysosomal protein) is required following FcR-mediated uptake of opsonized RBCs; Brucella have been shown to require RILP for conversion of the phagosome into an ER-derived replicative organelle, suggesting that this protein is relevant for phagosome maturation79. Similarly, M. bovis secretes a protein that interferes with Rab7 activation, blocking recruitment of RILP to the phagosome80. The nematode possesses a homologue of RILP (JMK and KSR, unpublished observations), but the function of this gene has yet to be linked to phagosome maturation or to endocytosis in the nematode.
Other Rab GTPases and the phagosome
While the roles of RAB-5 and RAB-7 in phagosome maturation can be inferred from mammalian studies, the role of RAB-2 is something of an enigma. A number of different Rab GTPases (including Rab2) have been identified in a proteomics studies34, 48, 81–84 (Box 3); genetic studies have shown that loss of RAB-2 delays lysosome fusion54, but how RAB-2 mediates this has not been addressed. Markers of maturation such as PtdIns(3)P-binding proteins are recruited to the phagosome with normal kinetics54, suggesting that the defect lies after RAB-5 recruitment. Consistent with this, knockdown of rab-5 blocks the recruitment of RAB-2 to phagosomes (JMK and KSR, unpublished observations).
RAB-2 co-localizes with RAB-5, RAB-7 and LAMP-152 and PtdIns(3)P markers54 on the phagosome, suggesting that rather than marking a distinct stage, RAB-2 localizes to phagosomes throughout their lifespan. Indeed, depletion of other phagosome maturation markers invariably results in RAB-2(+)-arrested phagosomes (JMK and KSR, unpublished observations). RAB-2 is also not required for phagosome acidification per se: recent studies have shown that apoptotic cell-containing phagosomes in rab-2-deficient worms are acidified and stain with acridine orange52. However, another group has suggested that in rab-2-deficient worms, phagosomes may be less acidified compared to wild-type54.
What is the function of RAB-2 on phagosomes, if it does not mark a particular stage of maturation? Studies in mammalian cells have shown that Rab2 is specifically localized to the ER and Golgi, and may play a role in retrograde transport, or the movement of proteins from the ER to the Golgi81, 82. It is thus conceivable that RAB-2's function may be to direct vesicles from the ER/Golgi to the nascent phagosome, delivering cargo-containing degradatory proteases (and, potentially, MHC molecules in mammals). In this context, it should be mentioned that many Rab GTPases associated with the ER/Golgi complex have been identified on phagosomes using proteomics approaches (see Box 3)15, 34, 35, 45–48, 60. RAB-10, another GTPase associated withGolgi transport85 and recycling endocytosis86, has also been identified in screens for factors involved in corpse removal41. However, at this time we have no clear understanding of what molecular process RAB-2 and its mammalian orthologue regulates during phagosome maturation.
Efficient targeting of vesicles to the phagosome (and appropriate acidification) also has important implications for antigen presentation. Dendritic cells isolated from Rab27a-deficient mice show defects in antigen presentation related to overacidification of the phagosome87. Another factor, NOX2, is an NADPH oxidase that generates reactive oxygen species, contributing to the alkalinization of the phagosome88. Fusion of latex bead-containing phagosomes with NOX2(+) structures slows acidification, allowing the controlled generation of antigenic peptides and loading onto MHC molecules. Loss of Rab27a, which is required for this fusion event, leads to over-acidified latex bead-containing phagosomes, greatly reducing efficiency of antigen presentation87. This work emphasizes the complex sorting requirements of phagosomes, and the importance of Rab proteins to phagosome maturation (Box 3). Other studies have suggested that this method of delayed acidification may also be relevant in apoptotic cell removal, as apoptotic cell-containing phagosomes are acidified more slowly in dendritic cells as compared to macrophages89.
LAMP proteins and lysosome fusion
LAMP-1 and LAMP-2 (and the nematode orthologues LMP-1 and LMP-2) are glycoproteins that are specifically localized to acidic lysosome structures90; the precise molecular function of these proteins is unknown. LAMP-1 and LAMP-2 (and nematode LMP-1) are recruited to the phagosome; intriguingly, in lmp1−/−lmp2−/− mutant mice, phagosomes containing IgG-opsonized beads appear arrested at the Rab5(+), PtdIns(3)P(+) stage91, suggesting that these two proteins play a role in the recruitment of Rab7 to the phagosome. Rab7 and its effector RILP can be found on LAMP(+) structures91, however, a molecular mechanism for LAMP function during phagosome maturation remains to be determined.
Phagocytic receptors and maturation
Conceptually, recognition of the target during phagocytosis represents a desirable signalling mechanism for distinguishing cargo. Therefore, the possibility that phagocytic receptors could play a key role in determining how a given target is processed for degradation is enticing. In endocytosis, ligand-bound receptors (eg. EGFR) have been shown to control Rab5 recruitment, although the mechanisms by which this occurs are sketchy92–94, and the NGF (nerve growth factor) receptor actively signals from the endosome to activate the Ras-MAPK signalling pathway95. Signalling during internalization may be particularly important for generating an immune response, whether tolerogenic (apoptotic cells) or inflammatory (bacteria). Recent studies in both nematode53 and mouse models96, 97 have led to the proposal of a `phagosome autonomous' model, according to which signalling proteins enriched on the phagosome during internalization determine how the phagosome will mature. This becomes especially relevant in the case of internalized pathogens, which can have complex secretion systems by which they inject factors outside of the phagosome, potentially altering signalling and disrupting maturation. However, our knowledge of how early steps in phagocytosis dictate later events of phagosome maturation is minimal.
Do phagocytic receptors control phagosome maturation?
Recently, the contribution of proteins involved in apoptotic cell phagocytosis to phagosome maturation was addressed. In engulfment-deficient worms (e.g. a ced-1 or ced-10/rac1 mutant), the efficiency of phagocytosis is greatly decreased, although many cells are eventually engulfed53,98. Using time-lapse microscopy of embryogenesis as a model, it was shown that CED-1, a transmembrane protein thought to function as a phagocytic receptor (see Box 1), was required for the recruitment of a FYVE domain marker and of RAB-7 to the phagosome53. This work also suggested that another key engulfment protein, CED-5/Dock180 (a RacGEF that functions in an alternate engulfment pathway downstream of an unknown receptor), also showed reduced phagosomal PtdIns(3)P and a mild delay in RAB-7 recruitment28. Studies addressing apoptotic cell removal in mammalian macrophages have also shown signalling from RhoA (which negatively regulates apoptotic cell removal99) and ERM (ezrin-radixin-moesin) proteins also play a role in the timely recruitment of Rab7 to the phagosome89, suggesting that there may be feedback between phagosome maturation and apoptotic cell uptake. Indeed, studies using non-digestible latex beads have shown that the inability to degrade a target can result in decreased uptake100.
Many of the works that have studied this process focus on localization of Rab7 or other lysosomal markers (rather than on earlier markers like Rab5 or Vps34), studying a stage once removed from where the action is. Studies on endocytosis have revealed that internalized receptors can modulate Rab5 activity on vesicles93, 94, and one tantalizing possibility is that a receptor (or co-receptor) would recruit Rab5 or a Rab5-regulatory machinery (such as a GEF or GAP, GTPase Activating Protein) to the phagosome. It is intriguing to note that the punctate localization of dynamin on the phagosome appears similar to the localization pattern of PtdSer on the phagosome and LRP1 (a phagocytic receptor required for apoptotic cell removal) on the phagocytic cup41, 101, suggesting recruitment of proteins involved in phagosome maturation may indeed be mediated by receptors. Studies in mammalian systems have also suggested that GULP, a homologue of the adaptor protein CED-6 that binds CED-1 (LRP1) (see Box 1), can interact with the endocytic machinery102, 103. Whether this represents a mechanism for recycling of CED-1 to the cell surface or a mechanism to recruit maturation proteins remains to be addressed.
One caveat with these studies is that they do not address how delays in phagocytosis may ultimately affect phagosome maturation: for example, one recent study has shown that microtubules are required for delivery of GapEx-5 (a GEF for Rab5) to the phagosome61. In this respect, a delay in microtubule attachment to the phagosome could affect timing of maturation without a `direct' role in signalling. Addressing this issue is likely to be extremely difficult, and may require the identification of mutant proteins that can affect phagosome maturation while leaving phagocytosis intact. In the case of CED-1 (and LRP1), one study has addressed functional domains of the protein that are required for corpse removal104. Further clarification of whether increased numbers of apoptotic cells in this case represent defects in uptake vs. maturation would greatly strengthen the hypothesis that these proteins direct phagosome maturation.
Mammalian TLRs and maturation of phagosomes containing bacteria
Toll-like receptors (TLRs) have many functions and can recognize multiple bacterial antigens to mediate their effects. During phagocytosis, TLRs serve as co-receptors for bacteria (in conjunction with a primary receptor, such as CD14 or CD36)105; TLRs typically do not function as phagocytic receptors, but instead induce macrophage activation in response to bacterial antigens such as lipopolysaccharide (LPS), resulting in the induction of an inflammatory response106. Most importantly in this context, TLR2 and TLR4 appear to control phagosome maturation, and steer the bacterial cargo into the `fast lane' for antigen presentation and induction of an inflammatory immune response96. When bacteria and apoptotic cells (which do not engage TLRs) are co-incubated with phagocytes, the different targets are not present within the same phagosome, and each target matures at a different rate. It is important to note that fast-tracking a phagosome may require multifactorial signalling events, as studies have shown that addition of TLR agonists alone are not sufficient to enhance the maturation of IgG-opsonized beads and other particles107, though this needs to be addressed in greater detail in vivo.
One of the end consequences of phagosome maturation is the generation of peptide antigens that are subsequently loaded onto MHC molecules and targeted to the cell surface38, 108. The differential maturation of phagosomes appears to be an important regulatory mechanism to exclude apoptotic cell antigens from class II presentation (and avoid subsequent stimulation of a self-reactive response). MHC class II molecules are typically activated by a proteolyic event on the phagosome (as a result of cathepsin activation37); engagement of TLR signalling and `fast-track' maturation results in steps that favour MHC class II loading, leading to phagocyte activation and induction of an immune response. Apoptotic cell-containing phagosomes do not possess peptide-loaded MHC class II, and apoptotic cell antigens are not presented on the cell surface on class II molecules97.
Normally, peptides derived from antigens that enter an antigen-presenting cell (e.g. macrophages or dendritic cells) get presented on class II MHC molecules. However, it has now been well documented that antigens derived from apoptotic cells are presented by a process referred to as “antigen cross-presentation” on class I MHC molecules that generally present ER-derived or endogenously produced proteins11. This again suggests that early steps in recognition of targets entering the phagocyte and/or phagosome maturation dictate how the antigens derived from the cargo would be processed. This influence on phagosome maturation could have important implications for the immune responses to antigens derived from the target.
Similar to mammals, the fly has developed primitive innate and cellular immune systems to target and remove both foreign and self particles109. A number of phagocytic receptors have been identified in the fly, including croquemort (apoptotic cells, Staphylococcus aureus)110, 111, draper (apoptotic cells, injured axons)112, 113, eater (S. aureus, E. coli and Serratia marcescans)114, nimrod C1 (S. aureus)115 and peste (Mycobacterium fortuitum)116. These receptors have yet to be linked to phagosome maturation in the fly, but homologues of croquemort in mammals (CD36) have been linked to TLR2 and TLR6 signalling, suggesting these receptors may also modulate phagosome maturation in the fly. In addition, Toll-like receptors have been implicated in the innate immune response to bacteria in the nematode: tol-1-deficient worms show significant invasion of Salmonella into the pharynx117. Whether this is true `invasion' or due to deficiencies in phagosome maturation and bacterial degradation remains to be determined.
Removal of bacteria and apoptotic cells: similarities and differences
The outcomes of recognition of bacteria (immunogenic) and apoptotic cells (tolerogenic) are very different; the molecular basis of this difference is perhaps the differential activation of signalling during both phagocytosis and phagosome maturation. Little is known regarding how apoptotic cells generate a tolerogenic state, but much work has gone into the identification of signalling pathways regulating the inflammatory response to bacteria (reviewed in105, 118). The activation of MHC class II molecules on bacterial phagosomes (but not phagosomes containing apoptotic cells) supports distinct signalling during each process97. The molecular basis for this difference is difficult to understand: since MHC class II molecules are present on phagosomes containing apoptotic cells97, but apoptotic cell-derived antigens are not loaded, this suggests a negative regulatory event. Other signalling events on the phagosome appear to be conserved: the GTPases Rab5 and Rab7 are present on both bacteria- and apoptotic cell-containing phagosomes; further, both types of phagosomes accumulate and lose PtdIns(3)P during maturation and require Vps3441, 89 15, 58. How these Rab proteins are regulated during bacterial phagocytosis has yet to be addressed.
Escaping a timely death
The immune system is incredibly efficient in seeking out and destroying potential pathogens; however, certain pathogens have developed mechanisms to escape degradation, persisting (and often replicating) within phagosomes by co-opting the phagosome maturation machinery (Table 2). Legionella pneumophila, for example, changes the composition of the phagosomal membrane so that it contains many features of the endoplasmic reticulum, allowing it to persist and replicate within the cell119. To achieve this, L. pneumophila has developed proteins that act specifically on Rab1, although how this leads to phagosome transformation is not well understood120–122. Many other bacteria similarly modify signalling to block their degradation within the cell (Table 2). Furthermore, the yeast (Cryptococcus neoformans), rather than blocking phagosome maturation, is able to induce expulsion of the phagosome from the cell. One can speculate that this yeast may convert the phagosome into an exocytic vesicle, though the mechanism of how this would be accomplished is unknown.
Table 2.
Bacteria have evolved mechanisms to modify the phagosome
| Bacteria | Mode of Action | References |
|---|---|---|
| Brucella | Converts phagosome into autophagosome/ER-like vesicle | 156, 157 |
| Chlamydia | Form inclusions enriched in sphingomyelin | 158, 159 |
| Coxiella burnetii | Converts phagosome into hybrid autophagosome-Rab7(+) phagosome | 160, 161 |
| Cryptococcus neoformans | Phagosome is extruded from living cells by unknown mechanism | 162, 163 |
| Helicobacter pylori | Sequesters Rab7 to block delivery of proteases | 164, 165 |
| Legionella pneumophila | Converts phagosome into rough ER | 120 – 122 |
| Leishmania sp. | Inhibits phagosome maturation | 166, 167 |
| Mycobacterium tuberculosis | Block at the Rab5(+) stage | 168 |
| Salmonella | Blocks maturation at the Rab5(+) phagosome, potentially by influencing Rab7 function | 166, 169 |
| Toxoplasma | Phagosome has characteristics of host membrane (non-receptor-mediated uptake) | 164, 165 |
Intriguingly, recent work has shown that one pathogen, Shigella, appears to utilize some of the genes required for corpse removal, ELMO1 and Dock180, for entry into the mammalian cells (see Box 1)123. Since phagocytic receptors/co-receptors have been linked to maturation events in other contexts, and ced-5 is required for aspects of phagosome maturation in the nematode53, it is possible that some bacteria may use apoptotic cell uptake machinery to disguise itself and avoid detection. A recent study showed that vaccinia virus may disguise itself in a phosphatidylserine (PtdSer) coat to mimic an apoptotic cell124. The consequence of such PtdSer-dependent entry to phagosome maturation, and presentation of bacterial/viral epitopes is not yet defined.
One of the major problems designing drugs to treat diseases associated with these pathogens is poor understanding of how they adapt the phagosome to their life cycle. A more thorough evaluation of the functional role of different proteins on the phagosome, and an assembly of these proteins into a coherent pathway, is of key importance to the development of therapeutics that can restore `normal' phagosome maturation, rather than inhibiting bacterial growth or replication.
Where do we go from here?
The recent identification of genes and proteins involved in phagosome maturation is of key importance, not only to basic science but also to the understanding of bacterial pathogenesis and as targets for drug design. Unbiased genetic screening approaches have identified >60 proteins that are potentially involved in phagosome maturation in C. elegans. Further characterization of proteins identified by genetic and proteomic approaches and testing them in mammalian systems would likely shed light on the control of phagosome maturation.
So far, most studies have focused on the acidification of the phagosomes as the primary readout. However, there are a number of other steps of phagosome maturation, such as accumulation of specific proteins on the phagosome (e.g. ATPases involved in phagosome acidification, myosin motors, acidic proteases, etc89, 125, 126) or other steps in processing (see Box 2) which could provide new insights. Indeed, a recent study in zebrafish in which V0 ATPase function was monitored revealed a novel role in phagosome fusion50, though the molecular details (or the stage at which the phagosome is arrested) remain to be defined. Moreover, while significant attention has been paid to the Rab family GTPases in phagosome maturation (justifiably so, given the significance of the Rab GTPases in vesicular traffic in other systems, see Box 3), the role of Arf family GTPases, another group linked to vesicular traffic in cells, is less well defined. Since one of the engulfment proteins, GULP, has been linked to Arf6-mediated signalling103, this may be worthy of a future avenue of investigation. Similarly, the role of microtubules, which have recently been suggested to play a role in delivery of a Rab5 GEF to the phagosome61, in phagosome maturation needs more detailed investigation.
Another important topic is whether phagosome maturation and autophagy might share a common mechanism. Both processes share a similar aetiology: the degradation of a target (internalized particles or organelles, respectively) within a double-membraned vesicle. Markers for autophagy (such as LC3 and Beclin) are recruited to mammalian phagosomes127, and loss of Atg5 or Atg7 impairs phagosome acidification. How these proteins are recruited to the phagosome (and whether they overlap with Rab5 or Rab7/LAMP-1) has not been addressed; however, these studies suggest that a simplistic view of phagosome maturation as movement from a phagosome into a lysosomal structure is simplistic, and the signalling may be more complex than envisioned.
Current knowledge of how bacteria can adapt phagocytic and phagosomal signalling to their own designs is sketchy (see Box 3), and further research on this topic is important for the generation of novel therapeutic targets. Removal of apoptotic cells is important for innate immunity, and defects in apoptotic cell phagocytosis and degradation has been shown to result in autoimmune disease. Comparative studies on how apoptotic cells and bacteria are degraded is thus of great clinical import, and further studies into the basic biology of phagosome maturation are crucial to the identification of relevant therapeutic targets for persistent bacterial diseases. How signalling proteins required for phagosome maturation (Table 1) can be co-opted by bacterial pathogens (Table 2) is a topic of great interest. In particular, Rab GTPases (Box 3) may represent the best target, and the identification of diverse functions of these proteins in phagosome maturation is of tantamount importance to the field.
Online Summary.
Eukaryotic cells phagocytose a variety of particles during their lifetime, including potentially pathogenic microorganisms and apoptotic cells. Approximately 200 billion cells must be cleared each day of our life, making removal of apoptotic cells one of the most common type of phagocytosis. Understanding how bacteria and apoptotic cells are phagocytosed and processed is a fundamentally important biological problem, both for normal homeostasis and disease.
Internalized particles are present in membrane organelles termed “phagosomes.” The phagosome functions as more than just as an organelle for `garbage disposal'. Proteins from the ingested target are degraded into peptides and presented on MHC class II molecules (in the case of bacteria) for the generation of an immune response; apoptotic cell-derived antigens are typically cross-presented on MHC class I molecules and are tolerogenic.
Phagosome maturation is the process by which a particle-containing phagosome “matures” through a series of increasingly acidic membrane bound structures, ultimately becoming an acidic phagolysosome before fusion with lysosomes. Proteomic approaches have identified a number of candidates localized to the phagosome, including the GTPases Rab5 and Rab7.
Recent studies in model systems such as Drosophila, Dictyostelium and C. elegans have developed genetic models for the identification and characterization of proteins required for maturation. Studies in the nematode has led to the development of a pathway for maturation of apoptotic cell-containing phagosomes.
Following phagocytosis, apoptotic cells (and other particles) exist within a membrane-bound organelle termed the phagosome. The proteins bound to the intracellular face of the phagosome membrane change as the phagosome matures. Soon after uptake, the phagosome is coated with the GTPase Rab5, which is then subsequently exchanged for Rab7 and ultimately for lysosomal markers, such as LAMP-1.
The regulation of phagosome maturation is quite complex, requiring a series of GEFs (Guanine Nucleotide Exchange Proteins), GAPs (GTPase Activating Proteins) and effectors. How Rab5 is regulated on the phagosome in vivo is just beginning to be described. The HOPS complex, a Rab7 activator and effector, is required for maturation of phagosome from the Rab7(+) stage.
A number of different bacterial pathogens have evolved mechanisms for co-opting phagosome maturation as a means of immune evasion or as a replicative niche. These bacteria target the machinery regulating maturation, in some cases converting the phagosome into other types of organelles.
Future studies are expected to focus on signalling pathways that determine whether the immune response to an internalized particle would be immunogenic (bacteria) or tolerogenic (apoptotic cells). The identification of novel players, and their placement within a pathway for phagosome maturation, may be important in the future development of new therapeutics targeting intracellular pathogens (such as M. tuberculosis).
Box 1. Removal of apoptotic cells.
While this review mainly deals with phagosome maturation, the recognition and subsequent internalization of the target precedes processing of the corpse. Signalling during internalization appears to regulate phagosome maturation and the specific post-engulfment responses of the phagocyte, and in turn, important biological consequences.
In the nematode and mammalian models, numerous receptors (e.g. CED-1, LRP1, CD36, TIM4, BAI1, and Mer, reviewed in8, 128) and adhesion molecules (e.g. CD14129) have been linked to removal of apoptotic cells, however, the specific ligands recognized and the intracellular signalling leading to corpse uptake have only been defined in some cases (see Box 2). Based initially on the simpler nematode model followed by studies in mammals, two partially redundant pathways have been identified to play an evolutionarily conserved role in apoptotic cell removal. In the first pathway, the proteins CED-2 (CrkII), CED-5 (Dock180) and CED-12 (ELMO) function to activate CED-10 (Rac1)130, 131 downstream of BAI1 (a G-protein-coupled receptor)132 and potentially integrins133 in mammals. In the second pathway, the candidate receptor CED-1 (MEGF10/LRP1) binds an unknown ligand on the apoptotic cell104 and signals via its cytoplasmic tail to the adaptor protein CED-6 (hCED-6/GULP)104, 134–136, whereas CED-7 (ABCA1) is thought to play a role in membrane dynamics137. This pathway has also been linked to Rac activation in both nematode and mammalian models98, 101, and both pathways appear to coordinately regulate the actin cytoskeleton41. Studies on phagocytic signalling in other models (such as Drosophila and mammalian cells) have underlined the importance of this pathway for the evolutionarily conserved removal of apoptotic cell corpses138.
Box 2. Consequences of apoptotic cell removal.
There are a number of consequences following internalization of apoptotic cell corpses, which loosely fall into three classes, which may also be affected by signalling from the phagosome. First, the removal of apoptotic cells has been linked to enhanced secretion of “pro-healing” cytokines (such as TGF-β10 and IL-10139), which serve to reduce inflammation in the extracellular environs and promote wound healing. The signalling pathways that alter cytokine secretion are not well characterized, but many phagocytic receptors (e.g. Stabilin-2140 and CD36139) have been implicated in this process. Intriguingly, endothelial cells have been reported to induce pro-inflammatory signalling (increased secretion of TNF-α) following recognition of apoptotic cells141, which has implications for atheroschlerotic disease.
Second, phagocytes must also dispose of internalized components of the apoptotic cell. Few studies have tracked the ultimate fate of apoptotic cell components, but two recent studies have addressed signalling pathways leading to efflux of cholesterol from the cell, suggesting that internalization of apoptotic cells (or targets) activates the nuclear receptor LXR, resulting in enhanced transcription of the transporter ABCA1103, 142.
Third, protein components of the cell are degraded and cross-presented via MHC class I molecules11, 143 and tend to be excluded from class II presentation97. Processing of apoptotic cell-associated antigens appears to be especially important, and has been linked to the maintenance of self-tolerance12, 19, 144. Potential pathways for DNA degradation have been described in both the nematode145–147 and mouse models17; defects in this process, like defects in corpse removal, have been linked to autoimmune disease, emphasizing that efficient phagosome maturation is required for health of the organism.
Box 3. Rab-family GTPases implicated in phagosome maturation.
Proteomics approaches have identified a number of different Rab GTPases that can be found localized to the phagosome at different times. However, the majority of these GTPases appear to be localized to the ER or Golgi bodies, observations which may have biased previous reports of phagosome-ER fusion51, 148.
What are the functions of these GTPases on the phagosome? Rab-family GTPases typically mediate transport between membrane-bound organelles, so the most likely role would be delivery of cargo (eg from the Golgi and ER to the phagosome), potentially delivering lysosomal proteases and other degradatory components. With respect to Rab5, potentially the most well-characterized Rab GTPase, recruitment appears to be related to the carefully timed acidification of the phagosome, which may have consequences for the generation of peptide antigens. Another tantalizing possibility is that one (or multiple) of these GTPases mediates transport from the phagosome to the ER, carrying processed antigens to ultimately be presented on the cell surface (via MHC class I proteins). Regardless of function, these GTPases are an important target of bacterial pathogens, and several have been described to require the function of these proteins (eg Rab14 and Rab22 by Mycobacterium tuberculosis60, 149, Rab1 by Legionella pneumophila119) for phagosome arrest.
| Protein | Function | Reference |
|---|---|---|
| Rab1 | ER to Golgi transport | 15, 35, 46–48 |
| Rab2 | ER to Golgi transport | 34, 48, 52, 54, 81–84 |
| Rab3 | Exocytosis | 34 |
| Rab5 | Intermediate between plasma membrane and early endosome and/or recycling endosomes, etc | 15, 34, 35, 41, 44, 47, 48 |
| Rab6 | Retrograde transport, Golgi to ER | 35 |
| Rab7 | trafficking from early to late endosome/lysosome | 15, 34, 35, 41, 44, 46–48 |
| Rab8 | Trafficking between Golgi, endosomes, and plasma membrane | 35, 47 |
| Rab9 | Lysosomal enzyme trafficking, late endosome to trans-Golgi transport | 15, 44, 48 |
| Rab10 | Transport form Golgi to polarized membrane (basolateral face, may cooperate with Rab8) | 15, 34, 35, 48 |
| Rab11 | Transport from Golgi to polarized membrane (apical face), v-ATPase transport | 15, 34, 35, 44, 47 |
| Rab14 | Phagosome and early endosome fusion, trafficking between early endosomes and Golgi | 15, 34, 48 |
| Rab20 | apical transport, Golgi, v-ATPase trafficking | 15 |
| Rab22 | Trans-Golgi-endosome transport | 15 |
| Rab23 | Endosomes and plasma membrane | 15 |
| Rab32 | Mitochondria? | 15, 48 |
| Rab33 | Retrograde transport from Golgi to ER | 15 |
| Rab35 | Cytokinesis | 15 |
Glossary terms
- Receptor-mediated endocytosis
the process by which a ligand-bound receptor is internalized in a membrane-bound vesicle that sequentially acquires different Rab GTPases
- Pinocytosis
the process by which liquids and small particles are internalized by the cell
- Complement
a protein complex component of the innate immune system that can bind to foreign particles and initiate their phagocytosis
- Fc-Receptor
a group of proteins which bind to IgG-opsonized particles, leading to activation of phagocytosis
- prenylation
the process by which a hydrocarbon moiety is attached to a conserved CAAX motif a the C-terminus of GTPases
- Programmed cell death
the process by which a healthy cell is induced to die, undergoes apoptosis, and then is cleared and degraded by a phagocyte
- Rab GTPases
a family of proteins which cycle between GDP-bound inactive and GTP-bound active forms and play a role in targeting transport to membrane-bound organelles
- Dynamin
a large GTPases that plays roles in membrane scission, actin cytoskeletal dynamics, and phagosome maturation
- Focal exocytosis
during phagocytosis, small vesicles are targeted to the plasma membrane; this, in turn, is thought to produce a local increase in plasma membrane volume allowing the cell to extend its membrane around large particles.
- FYVE and Phox Homology (PX) domains
domains within proteins which bind to phosphatidylinositol on membranes
- LAMP-1
a membrane glycoprotein enriched on lysosomes and phagolysosomes
- Retrograde transport
Transport from the Golgi to the endoplasmic reticulum, in `reverse' from the `normal' mode of ER to Golgi transport
- Recycling endosomes
a membrane-bound organelle for the recycling of receptors following ligand dissociation
- Exocyst
a protein complex required for the exocytosis of proteins
- Toll-like receptors
transmembrane proteins which recognize bacterial pathogens and induce an inflammatory response
- Arf family GTPases
small GTPases which regulate aspects of membrane trafficking related to the budding of vesicles from membranes
References
- 1.van Lookeren Campagne M, Wiesmann C, Brown EJ. Macrophage complement receptors and pathogen clearance. Cell Microbiol. 2007;9:2095–102. doi: 10.1111/j.1462-5822.2007.00981.x. [DOI] [PubMed] [Google Scholar]
- 2.Underhill DM, Ozinsky A. Phagocytosis of microbes: complexity in action. Annu Rev Immunol. 2002;20:825–52. doi: 10.1146/annurev.immunol.20.103001.114744. [DOI] [PubMed] [Google Scholar]
- 3.Swanson JA, Hoppe AD. The coordination of signaling during Fc receptor-mediated phagocytosis. J Leukoc Biol. 2004;76:1093–103. doi: 10.1189/jlb.0804439. [DOI] [PubMed] [Google Scholar]
- 4.Nimmerjahn F, Ravetch JV. Fcgamma receptors: old friends and new family members. Immunity. 2006;24:19–28. doi: 10.1016/j.immuni.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 5.Metzstein MM, Stanfield GM, Horvitz HR. Genetics of programmed cell death in C. elegans: past, present and future. Trends Genet. 1998;14:410–6. doi: 10.1016/s0168-9525(98)01573-x. [DOI] [PubMed] [Google Scholar]
- 6.Lettre G, Hengartner MO. Developmental apoptosis in C. elegans: a complex CEDnario. Nat Rev Mol Cell Biol. 2006;7:97–108. doi: 10.1038/nrm1836. [DOI] [PubMed] [Google Scholar]; These two reviews are an excellent introduction to the field of apoptosis in the nematode. The review by Metzstein et al is a classic in the field, and is an excellent introduction to the genetics of programmed cell death. The review by Lettre and Hengartner extends and updates these findings.
- 7.Taylor RC, Cullen SP, Martin SJ. Apoptosis: controlled demolition at the cellular level. Nat Rev Mol Cell Biol. 2008;9:231–41. doi: 10.1038/nrm2312. [DOI] [PubMed] [Google Scholar]
- 8.Ravichandran KS, Lorenz U. Engulfment of apoptotic cells: signals for a good meal. Nat Rev Immunol. 2007;7:964–74. doi: 10.1038/nri2214. [DOI] [PubMed] [Google Scholar]
- 9.Miyake Y, et al. Critical role of macrophages in the marginal zone in the suppression of immune responses to apoptotic cell-associated antigens. J Clin Invest. 2007;117:2268–78. doi: 10.1172/JCI31990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fadok VA, et al. Macrophages that have ingested apoptotic cells in vitro inhibit proinflammatory cytokine production through autocrine/paracrine mechanisms involving TGF-beta, PGE2, and PAF. J Clin Invest. 1998;101:890–8. doi: 10.1172/JCI1112. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article is the first characterization of the “anti-inflammatory” cytotkine response elicited by recognition of apoptotic cells by phagocytes. It should be noted that this response occurs in both professional and nonprofessional phagocytes
- 11.Bellone M, et al. Processing of engulfed apoptotic bodies yields T cell epitopes. J Immunol. 1997;159:5391–9. [PubMed] [Google Scholar]
- 12.Huang FP, et al. A discrete subpopulation of dendritic cells transports apoptotic intestinal epithelial cells to T cell areas of mesenteric lymph nodes. J Exp Med. 2000;191:435–44. doi: 10.1084/jem.191.3.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Albert ML, et al. Immature dendritic cells phagocytose apoptotic cells via alphavbeta5 and CD36, and cross-present antigens to cytotoxic T lymphocytes. J Exp Med. 1998;188:1359–68. doi: 10.1084/jem.188.7.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]; Defects in apoptotic cell removal have long been associated with autoimmune disease; previous thought was that release of cellular contents led to the generation of an immune response. This article suggests that defects are in the establishment or maintenance of tolerance rather than faulty induction of an immune response.
- 14.Swanson JA. Shaping cups into phagosomes and macropinosomes. Nat Rev Mol Cell Biol. 2008;9:639–49. doi: 10.1038/nrm2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith AC, et al. A network of Rab GTPases controls phagosome maturation and is modulated by Salmonella enterica serovar Typhimurium. J Cell Biol. 2007;176:263–8. doi: 10.1083/jcb.200611056. [DOI] [PMC free article] [PubMed] [Google Scholar]; A number of GTPases can be found physically on the phagosome; this article uses dominant negative approaches to identify which GTPases are functionally required for maturation of Salmonella-containing phagosomes.
- 16.Beertsen W, et al. Impaired phagosomal maturation in neutrophils leads to periodontitis in lysosomal-associated membrane protein-2 knockout mice. J Immunol. 2008;180:475–82. doi: 10.4049/jimmunol.180.1.475. [DOI] [PubMed] [Google Scholar]
- 17.Kawane K, et al. Chronic polyarthritis caused by mammalian DNA that escapes from degradation in macrophages. Nature. 2006;443:998–1002. doi: 10.1038/nature05245. [DOI] [PubMed] [Google Scholar]
- 18.Wallet MA, et al. MerTK is required for apoptotic cell-induced T cell tolerance. J Exp Med. 2008;205:219–32. doi: 10.1084/jem.20062293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Albert ML, Jegathesan M, Darnell RB. Dendritic cell maturation is required for the cross-tolerization of CD8+ T cells. Nat Immunol. 2001;2:1010–7. doi: 10.1038/ni722. [DOI] [PubMed] [Google Scholar]
- 20.Ungewickell EJ, Hinrichsen L. Endocytosis: clathrin-mediated membrane budding. Curr Opin Cell Biol. 2007;19:417–25. doi: 10.1016/j.ceb.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 21.Young A. Structural insights into the clathrin coat. Semin Cell Dev Biol. 2007;18:448–58. doi: 10.1016/j.semcdb.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 22.Hanyaloglu AC, von Zastrow M. Regulation of GPCRs by endocytic membrane trafficking and its potential implications. Annu Rev Pharmacol Toxicol. 2008;48:537–68. doi: 10.1146/annurev.pharmtox.48.113006.094830. [DOI] [PubMed] [Google Scholar]
- 23.Rink J, Ghigo E, Kalaidzidis Y, Zerial M. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 2005;122:735–49. doi: 10.1016/j.cell.2005.06.043. [DOI] [PubMed] [Google Scholar]; This groundbreaking study used time-lapse microscopy to track endocytic vesicles as they acquired the GTPases Rab5 and Rab7. This work provides support for the direct exchange of Rab5 for Rab7 on the endosome; studies in the nematode have also observed this `Rab conversion' during phagosome maturation.
- 24.Russell MR, Nickerson DP, Odorizzi G. Molecular mechanisms of late endosome morphology, identity and sorting. Curr Opin Cell Biol. 2006;18:422–8. doi: 10.1016/j.ceb.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 25.Luzio JP, et al. Lysosome-endosome fusion and lysosome biogenesis. J Cell Sci. 2000;113(Pt 9):1515–24. doi: 10.1242/jcs.113.9.1515. [DOI] [PubMed] [Google Scholar]
- 26.van Ijzendoorn SC. Recycling endosomes. J Cell Sci. 2006;119:1679–81. doi: 10.1242/jcs.02948. [DOI] [PubMed] [Google Scholar]
- 27.Maxfield FR, McGraw TE. Endocytic recycling. Nat Rev Mol Cell Biol. 2004;5:121–32. doi: 10.1038/nrm1315. [DOI] [PubMed] [Google Scholar]
- 28.Yu X, Odera S, Chuang CH, Lu N, Zhou ZC. elegans Dynamin mediates the signaling of phagocytic receptor CED-1 for the engulfment and degradation of apoptotic cells. Dev Cell. 2006;10:743–57. doi: 10.1016/j.devcel.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 29.Lee WL, Mason D, Schreiber AD, Grinstein S. Quantitative analysis of membrane remodeling at the phagocytic cup. Mol Biol Cell. 2007;18:2883–92. doi: 10.1091/mbc.E06-05-0450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Albert ML. Death-defying immunity: do apoptotic cells influence antigen processing and presentation? Nat Rev Immunol. 2004;4:223–31. doi: 10.1038/nri11308. [DOI] [PubMed] [Google Scholar]
- 31.Blander JM. Signalling and phagocytosis in the orchestration of host defence. Cell Microbiol. 2007;9:290–9. doi: 10.1111/j.1462-5822.2006.00864.x. [DOI] [PubMed] [Google Scholar]
- 32.Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 33.Desjardins M, Huber LA, Parton RG, Griffiths G. Biogenesis of phagolysosomes proceeds through a sequential series of interactions with the endocytic apparatus. J Cell Biol. 1994;124:677–88. doi: 10.1083/jcb.124.5.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garin J, et al. The phagosome proteome: insight into phagosome functions. J Cell Biol. 2001;152:165–80. doi: 10.1083/jcb.152.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stuart LM, et al. A systems biology analysis of the Drosophila phagosome. Nature. 2007;445:95–101. doi: 10.1038/nature05380. [DOI] [PubMed] [Google Scholar]
- 36.Lennon-Dumenil AM, et al. Analysis of protease activity in live antigen-presenting cells shows regulation of the phagosomal proteolytic contents during dendritic cell activation. J Exp Med. 2002;196:529–40. doi: 10.1084/jem.20020327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bryant PW, Lennon-Dumenil AM, Fiebiger E, Lagaudriere-Gesbert C, Ploegh HL. Proteolysis and antigen presentation by MHC class II molecules. Adv Immunol. 2002;80:71–114. doi: 10.1016/S0065-2776(02)80013-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramachandra L, Boom WH, Harding CV. Class II MHC antigen processing in phagosomes. Methods Mol Biol. 2008;445:353–77. doi: 10.1007/978-1-59745-157-4_23. [DOI] [PubMed] [Google Scholar]
- 39.Hackam DJ, et al. Regulation of phagosomal acidification. Differential targeting of Na+/H+ exchangers, Na+/K+-ATPases, and vacuolar-type H+-atpases. J Biol Chem. 1997;272:29810–20. doi: 10.1074/jbc.272.47.29810. [DOI] [PubMed] [Google Scholar]
- 40.Beyenbach KW, Wieczorek H. The V-type H+ ATPase: molecular structure and function, physiological roles and regulation. J Exp Biol. 2006;209:577–89. doi: 10.1242/jeb.02014. [DOI] [PubMed] [Google Scholar]
- 41.Kinchen JM, et al. A pathway for phagosome maturation during engulfment of apoptotic cells. Nat Cell Biol. 2008;10:556–66. doi: 10.1038/ncb1718. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article uses the nematode C. elegans as a model to screen for regulators of phagosome maturation, identifies a genetic pathway for phagosome maturation, and shows evolutionary conservation of this pathway in mammals. This work also identifies an unexpected role for Dynamin in the recruitment of Vps34 and Rab5 to the phagosome.
- 42.Clarke M, et al. Dynamics of the vacuolar H(+)-ATPase in the contractile vacuole complex and the endosomal pathway of Dictyostelium cells. J Cell Sci. 2002;115:2893–905. doi: 10.1242/jcs.115.14.2893. [DOI] [PubMed] [Google Scholar]
- 43.Fratti RA, Chua J, Vergne I, Deretic V. Mycobacterium tuberculosis glycosylated phosphatidylinositol causes phagosome maturation arrest. Proc Natl Acad Sci U S A. 2003;100:5437–42. doi: 10.1073/pnas.0737613100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fratti RA, Vergne I, Chua J, Skidmore J, Deretic V. Regulators of membrane trafficking and Mycobacterium tuberculosis phagosome maturation block. Electrophoresis. 2000;21:3378–85. doi: 10.1002/1522-2683(20001001)21:16<3378::AID-ELPS3378>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 45.McCoy JJ, Mann BJ. Proteomic analysis of Gal/GalNAc lectin-associated proteins in Entamoeba histolytica. Exp Parasitol. 2005;110:220–5. doi: 10.1016/j.exppara.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 46.Okada M, et al. Proteomic analysis of phagocytosis in the enteric protozoan parasite Entamoeba histolytica. Eukaryot Cell. 2005;4:827–31. doi: 10.1128/EC.4.4.827-831.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Okada M, et al. Kinetics and strain variation of phagosome proteins of Entamoeba histolytica by proteomic analysis. Mol Biochem Parasitol. 2006;145:171–83. doi: 10.1016/j.molbiopara.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 48.Jutras I, et al. Modulation of the phagosome proteome by interferon-gamma. Mol Cell Proteomics. 2008;7:697–715. doi: 10.1074/mcp.M700267-MCP200. [DOI] [PubMed] [Google Scholar]
- 49.Botelho RJ, Hackam DJ, Schreiber AD, Grinstein S. Role of COPI in phagosome maturation. J Biol Chem. 2000;275:15717–27. doi: 10.1074/jbc.M910068199. [DOI] [PubMed] [Google Scholar]
- 50.Peri F, Nusslein-Volhard C. Live imaging of neuronal degradation by microglia reveals a role for v0-ATPase a1 in phagosomal fusion in vivo. Cell. 2008;133:916–27. doi: 10.1016/j.cell.2008.04.037. [DOI] [PubMed] [Google Scholar]
- 51.Touret N, et al. Quantitative and dynamic assessment of the contribution of the ER to phagosome formation. Cell. 2005;123:157–70. doi: 10.1016/j.cell.2005.08.018. [DOI] [PubMed] [Google Scholar]
- 52.Lu Q, et al. C. elegans Rab GTPase 2 is required for the degradation of apoptotic cells. Development. 2008;135:1069–80. doi: 10.1242/dev.016063. [DOI] [PubMed] [Google Scholar]
- 53.Yu X, Lu N, Zhou Z. Phagocytic receptor CED-1 initiates a signaling pathway for degrading engulfed apoptotic cells. PLoS Biol. 2008;6:e61. doi: 10.1371/journal.pbio.0060061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mangahas PM, Yu X, Miller KG, Zhou Z. The small GTPase Rab2 functions in the removal of apoptotic cells in Caenorhabditis elegans. J Cell Biol. 2008;180:357–73. doi: 10.1083/jcb.200708130. [DOI] [PMC free article] [PubMed] [Google Scholar]; References 52 – 54 introduce time-lapse microscopy to the study of phagosome maturation in the whole organism and identifies UNC-108/RAB-2 (Ref 52, 54) and phagocytic signalling (Ref 53) as important for phagosome maturation.
- 55.Ullrich O, Horiuchi H, Bucci C, Zerial M. Membrane association of Rab5 mediated by GDP-dissociation inhibitor and accompanied by GDP/GTP exchange. Nature. 1994;368:157–60. doi: 10.1038/368157a0. [DOI] [PubMed] [Google Scholar]
- 56.Seabra MC, Wasmeier C. Controlling the location and activation of Rab GTPases. Curr Opin Cell Biol. 2004;16:451–7. doi: 10.1016/j.ceb.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 57.Grosshans BL, Ortiz D, Novick P. Rabs and their effectors: achieving specificity in membrane traffic. Proc Natl Acad Sci U S A. 2006;103:11821–7. doi: 10.1073/pnas.0601617103. [DOI] [PMC free article] [PubMed] [Google Scholar]; This is an excellent review of Rab effectors and their functions.
- 58.Vieira OV, et al. Modulation of Rab5 and Rab7 recruitment to phagosomes by phosphatidylinositol 3-kinase. Mol Cell Biol. 2003;23:2501–14. doi: 10.1128/MCB.23.7.2501-2514.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article shows the requirements for kinase function (as inhibited by wortmannin) for fusion of phagosomes with late endosomes/lysosomes. Intriguingly, Rab7 can still be recruited to the phagosome, supporting a model for `Rab conversion' on the phagosome.
- 59.McBride HM, et al. Oligomeric complexes link Rab5 effectors with NSF and drive membrane fusion via interactions between EEA1 and syntaxin 13. Cell. 1999;98:377–86. doi: 10.1016/s0092-8674(00)81966-2. [DOI] [PubMed] [Google Scholar]
- 60.Roberts EA, Chua J, Kyei GB, Deretic V. Higher order Rab programming in phagolysosome biogenesis. J Cell Biol. 2006;174:923–9. doi: 10.1083/jcb.200603026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kitano M, Nakaya M, Nakamura T, Nagata S, Matsuda M. Imaging of Rab5 activity identifies essential regulators for phagosome maturation. Nature. 2008;453:241–5. doi: 10.1038/nature06857. [DOI] [PubMed] [Google Scholar]; This article uses a Raichu-Rab5 biosensor probe to monitor Rab5 activation during apoptotic cell removal, and suggests that GapEx-5 is a major player in Rab5 activation during lactadherin/MFGE-8-mediated uptake.
- 62.Huynh KK, Grinstein S. Phagocytosis: Dynamin's Dual Role in Phagosome Biogenesis. Curr Biol. 2008;18:R563–5. doi: 10.1016/j.cub.2008.05.032. [DOI] [PubMed] [Google Scholar]
- 63.Vieira OV, et al. Distinct roles of class I and class III phosphatidylinositol 3-kinases in phagosome formation and maturation. J Cell Biol. 2001;155:19–25. doi: 10.1083/jcb.200107069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bucci C, et al. The small GTPase rab5 functions as a regulatory factor in the early endocytic pathway. Cell. 1992;70:715–28. doi: 10.1016/0092-8674(92)90306-w. [DOI] [PubMed] [Google Scholar]
- 65.Hanayama R, et al. Autoimmune disease and impaired uptake of apoptotic cells in MFGE8-deficient mice. Science. 2004;304:1147–50. doi: 10.1126/science.1094359. [DOI] [PubMed] [Google Scholar]
- 66.Nielsen E, et al. Rabenosyn-5, a novel Rab5 effector, is complexed with hVPS45 and recruited to endosomes through a FYVE finger domain. J Cell Biol. 2000;151:601–12. doi: 10.1083/jcb.151.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Christoforidis S, McBride HM, Burgoyne RD, Zerial M. The Rab5 effector EEA1 is a core component of endosome docking. Nature. 1999;397:621–5. doi: 10.1038/17618. [DOI] [PubMed] [Google Scholar]
- 68.Simonsen A, et al. EEA1 links PI(3)K function to Rab5 regulation of endosome fusion. Nature. 1998;394:494–8. doi: 10.1038/28879. [DOI] [PubMed] [Google Scholar]
- 69.Gengyo-Ando K, et al. The SM protein VPS-45 is required for RAB-5-dependent endocytic transport in Caenorhabditis elegans. EMBO Rep. 2007;8:152–7. doi: 10.1038/sj.embor.7400882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vitale G, et al. Distinct Rab-binding domains mediate the interaction of Rabaptin-5 with GTP-bound Rab4 and Rab5. Embo J. 1998;17:1941–51. doi: 10.1093/emboj/17.7.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Williams RL, Urbe S. The emerging shape of the ESCRT machinery. Nat Rev Mol Cell Biol. 2007;8:355–68. doi: 10.1038/nrm2162. [DOI] [PubMed] [Google Scholar]
- 72.Fratti RA, Backer JM, Gruenberg J, Corvera S, Deretic V. Role of phosphatidylinositol 3-kinase and Rab5 effectors in phagosomal biogenesis and mycobacterial phagosome maturation arrest. J Cell Biol. 2001;154:631–44. doi: 10.1083/jcb.200106049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schnatwinkel C, et al. The Rab5 effector Rabankyrin-5 regulates and coordinates different endocytic mechanisms. PLoS Biol. 2004;2:E261. doi: 10.1371/journal.pbio.0020261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Storrie B, Desjardins M. The biogenesis of lysosomes: is it a kiss and run, continuous fusion and fission process? Bioessays. 1996;18:895–903. doi: 10.1002/bies.950181108. [DOI] [PubMed] [Google Scholar]
- 75.Rothman JH, Stevens TH. Protein sorting in yeast: mutants defective in vacuole biogenesis mislocalize vacuolar proteins into the late secretory pathway. Cell. 1986;47:1041–51. doi: 10.1016/0092-8674(86)90819-6. [DOI] [PubMed] [Google Scholar]
- 76.Peplowska K, Markgraf DF, Ostrowicz CW, Bange G, Ungermann C. The CORVET tethering complex interacts with the yeast Rab5 homolog Vps21 and is involved in endo-lysosomal biogenesis. Dev Cell. 2007;12:739–50. doi: 10.1016/j.devcel.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 77.Seals DF, Eitzen G, Margolis N, Wickner WT, Price A. A Ypt/Rab effector complex containing the Sec1 homolog Vps33p is required for homotypic vacuole fusion. Proc Natl Acad Sci U S A. 2000;97:9402–7. doi: 10.1073/pnas.97.17.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Peterson MR, Burd CG, Emr SD. Vac1p coordinates Rab and phosphatidylinositol 3-kinase signaling in Vps45p-dependent vesicle docking/fusion at the endosome. Curr Biol. 1999;9:159–62. doi: 10.1016/s0960-9822(99)80071-2. [DOI] [PubMed] [Google Scholar]
- 79.Starr T, Ng TW, Wehrly TD, Knodler LA, Celli J. Brucella intracellular replication requires trafficking through the late endosomal/lysosomal compartment. Traffic. 2008;9:678–94. doi: 10.1111/j.1600-0854.2008.00718.x. [DOI] [PubMed] [Google Scholar]
- 80.Sun J, et al. Mycobacterium bovis BCG disrupts the interaction of Rab7 with RILP contributing to inhibition of phagosome maturation. J Leukoc Biol. 2007;82:1437–45. doi: 10.1189/jlb.0507289. [DOI] [PubMed] [Google Scholar]
- 81.Tisdale EJ. A Rab2 mutant with impaired GTPase activity stimulates vesicle formation from pre-Golgi intermediates. Mol Biol Cell. 1999;10:1837–49. doi: 10.1091/mbc.10.6.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tisdale EJ, Balch WE. Rab2 is essential for the maturation of pre-Golgi intermediates. J Biol Chem. 1996;271:29372–9. doi: 10.1074/jbc.271.46.29372. [DOI] [PubMed] [Google Scholar]
- 83.Tisdale EJ, Bourne JR, Khosravi-Far R, Der CJ, Balch WE. GTP-binding mutants of rab1 and rab2 are potent inhibitors of vesicular transport from the endoplasmic reticulum to the Golgi complex. J Cell Biol. 1992;119:749–61. doi: 10.1083/jcb.119.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chun DK, McEwen JM, Burbea M, Kaplan JM. UNC-108/Rab2 Regulates Post-endocytic Trafficking in C. elegans. Mol Biol Cell. 2008 doi: 10.1091/mbc.E07-11-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schuck S, et al. Rab10 is involved in basolateral transport in polarized Madin-Darby canine kidney cells. Traffic. 2007;8:47–60. doi: 10.1111/j.1600-0854.2006.00506.x. [DOI] [PubMed] [Google Scholar]
- 86.Chen CC, et al. RAB-10 is required for endocytic recycling in the Caenorhabditis elegans intestine. Mol Biol Cell. 2006;17:1286–97. doi: 10.1091/mbc.E05-08-0787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jancic C, et al. Rab27a regulates phagosomal pH and NADPH oxidase recruitment to dendritic cell phagosomes. Nat Cell Biol. 2007;9:367–78. doi: 10.1038/ncb1552. [DOI] [PubMed] [Google Scholar]; This intriguing paper highlights the importance of trafficking events to processing of internalized cargo: trafficking of Rab27a(+) vesicles to the phagosome slows acidification, allowing efficient proteolysis of proteins and loading of antigens for presentation.
- 88.Savina A, et al. NOX2 controls phagosomal pH to regulate antigen processing during crosspresentation by dendritic cells. Cell. 2006;126:205–18. doi: 10.1016/j.cell.2006.05.035. [DOI] [PubMed] [Google Scholar]
- 89.Erwig LP, et al. Differential regulation of phagosome maturation in macrophages and dendritic cells mediated by Rho GTPases and ezrin-radixin-moesin (ERM) proteins. Proc Natl Acad Sci U S A. 2006;103:12825–30. doi: 10.1073/pnas.0605331103. [DOI] [PMC free article] [PubMed] [Google Scholar]; This is the first characterization of maturation relating to apoptotic cell removal, and suggests a link between RhoA (whose activity is downregulated during apoptotic cell engulfment), and ERM family proteins for phagosome maturation.
- 90.Eskelinen EL, Tanaka Y, Saftig P. At the acidic edge: emerging functions for lysosomal membrane proteins. Trends Cell Biol. 2003;13:137–45. doi: 10.1016/s0962-8924(03)00005-9. [DOI] [PubMed] [Google Scholar]
- 91.Huynh KK, et al. LAMP proteins are required for fusion of lysosomes with phagosomes. Embo J. 2007;26:313–24. doi: 10.1038/sj.emboj.7601511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Barbieri MA, Fernandez-Pol S, Hunker C, Horazdovsky BH, Stahl PD. Role of rab5 in EGF receptor-mediated signal transduction. Eur J Cell Biol. 2004;83:305–14. doi: 10.1078/0171-9335-00381. [DOI] [PubMed] [Google Scholar]
- 93.Barbieri MA, et al. Epidermal growth factor and membrane trafficking. EGF receptor activation of endocytosis requires Rab5a. J Cell Biol. 2000;151:539–50. doi: 10.1083/jcb.151.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lanzetti L, et al. The Eps8 protein coordinates EGF receptor signalling through Rac and trafficking through Rab5. Nature. 2000;408:374–7. doi: 10.1038/35042605. [DOI] [PubMed] [Google Scholar]
- 95.Howe CL, Valletta JS, Rusnak AS, Mobley WC. NGF signaling from clathrin-coated vesicles: evidence that signaling endosomes serve as a platform for the Ras-MAPK pathway. Neuron. 2001;32:801–14. doi: 10.1016/s0896-6273(01)00526-8. [DOI] [PubMed] [Google Scholar]
- 96.Blander JM, Medzhitov R. Regulation of phagosome maturation by signals from toll-like receptors. Science. 2004;304:1014–8. doi: 10.1126/science.1096158. [DOI] [PubMed] [Google Scholar]
- 97.Blander JM, Medzhitov R. Toll-dependent selection of microbial antigens for presentation by dendritic cells. Nature. 2006;440:808–12. doi: 10.1038/nature04596. [DOI] [PubMed] [Google Scholar]; Refs 96, 97 present evidence suggesting a phagosome-autonomous model for maturation, where receptor ligation during internalization determines the ultimate fate of the cargo and whether antigens will be presented on MHC class II molecules at the cell surface.
- 98.Kinchen JM, et al. Two pathways converge at CED-10 to mediate actin rearrangement and corpse removal in C. elegans. Nature. 2005;434:93–9. doi: 10.1038/nature03263. [DOI] [PubMed] [Google Scholar]
- 99.Tosello-Trampont AC, Nakada-Tsukui K, Ravichandran KS. Engulfment of apoptotic cells is negatively regulated by Rho-mediated signaling. J Biol Chem. 2003;278:49911–9. doi: 10.1074/jbc.M306079200. [DOI] [PubMed] [Google Scholar]
- 100.Schrijvers DM, De Meyer GR, Kockx MM, Herman AG, Martinet W. Phagocytosis of apoptotic cells by macrophages is impaired in atherosclerosis. Arterioscler Thromb Vasc Biol. 2005;25:1256–61. doi: 10.1161/01.ATV.0000166517.18801.a7. [DOI] [PubMed] [Google Scholar]
- 101.Gardai SJ, et al. Cell-surface calreticulin initiates clearance of viable or apoptotic cells through trans-activation of LRP on the phagocyte. Cell. 2005;123:321–34. doi: 10.1016/j.cell.2005.08.032. [DOI] [PubMed] [Google Scholar]
- 102.Martins-Silva C, et al. A rat homologue of CED-6 is expressed in neurons and interacts with clathrin. Brain Res. 2006;1119:1–12. doi: 10.1016/j.brainres.2006.08.064. [DOI] [PubMed] [Google Scholar]
- 103.Ma Z, Nie Z, Luo R, Casanova JE, Ravichandran KS. Regulation of Arf6 and ACAP1 signaling by the PTB-domain-containing adaptor protein GULP. Curr Biol. 2007;17:722–7. doi: 10.1016/j.cub.2007.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhou Z, Hartwieg E, Horvitz HR. CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans. Cell. 2001;104:43–56. doi: 10.1016/s0092-8674(01)00190-8. [DOI] [PubMed] [Google Scholar]
- 105.Underhill DM, Gantner B. Integration of Toll-like receptor and phagocytic signaling for tailored immunity. Microbes Infect. 2004;6:1368–73. doi: 10.1016/j.micinf.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 106.Taylor PR, et al. Macrophage receptors and immune recognition. Annu Rev Immunol. 2005;23:901–44. doi: 10.1146/annurev.immunol.23.021704.115816. [DOI] [PubMed] [Google Scholar]
- 107.Yates RM, Russell DG. Phagosome maturation proceeds independently of stimulation of toll-like receptors 2 and 4. Immunity. 2005;23:409–17. doi: 10.1016/j.immuni.2005.09.007. [DOI] [PubMed] [Google Scholar]; This article questions the phagosome-autonomous model and instead suggests that maturation does not depend on signalling from Toll-like receptors.
- 108.Houde M, et al. Phagosomes are competent organelles for antigen cross-presentation. Nature. 2003;425:402–6. doi: 10.1038/nature01912. [DOI] [PubMed] [Google Scholar]
- 109.Stuart LM, Ezekowitz RA. Phagocytosis and comparative innate immunity: learning on the fly. Nat Rev Immunol. 2008;8:131–41. doi: 10.1038/nri2240. [DOI] [PubMed] [Google Scholar]
- 110.Franc NC, Heitzler P, Ezekowitz RA, White K. Requirement for croquemort in phagocytosis of apoptotic cells in Drosophila. Science. 1999;284:1991–4. doi: 10.1126/science.284.5422.1991. [DOI] [PubMed] [Google Scholar]
- 111.Franc NC, Dimarcq JL, Lagueux M, Hoffmann J, Ezekowitz RA. Croquemort, a novel Drosophila hemocyte/macrophage receptor that recognizes apoptotic cells. Immunity. 1996;4:431–43. doi: 10.1016/s1074-7613(00)80410-0. [DOI] [PubMed] [Google Scholar]
- 112.Awasaki T, et al. Essential role of the apoptotic cell engulfment genes draper and ced-6 in programmed axon pruning during Drosophila metamorphosis. Neuron. 2006;50:855–67. doi: 10.1016/j.neuron.2006.04.027. [DOI] [PubMed] [Google Scholar]
- 113.Freeman MR, Delrow J, Kim J, Johnson E, Doe CQ. Unwrapping glial biology: Gcm target genes regulating glial development, diversification, and function. Neuron. 2003;38:567–80. doi: 10.1016/s0896-6273(03)00289-7. [DOI] [PubMed] [Google Scholar]
- 114.Kocks C, et al. Eater, a transmembrane protein mediating phagocytosis of bacterial pathogens in Drosophila. Cell. 2005;123:335–46. doi: 10.1016/j.cell.2005.08.034. [DOI] [PubMed] [Google Scholar]
- 115.Kurucz E, et al. Nimrod, a putative phagocytosis receptor with EGF repeats in Drosophila plasmatocytes. Curr Biol. 2007;17:649–54. doi: 10.1016/j.cub.2007.02.041. [DOI] [PubMed] [Google Scholar]
- 116.Philips JA, Rubin EJ, Perrimon N. Drosophila RNAi screen reveals CD36 family member required for mycobacterial infection. Science. 2005;309:1251–3. doi: 10.1126/science.1116006. [DOI] [PubMed] [Google Scholar]
- 117.Tenor JL, Aballay A. A conserved Toll-like receptor is required for Caenorhabditis elegans innate immunity. EMBO Rep. 2008;9:103–9. doi: 10.1038/sj.embor.7401104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.O'Neill LA. When signaling pathways collide: positive and negative regulation of toll-like receptor signal transduction. Immunity. 2008;29:12–20. doi: 10.1016/j.immuni.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 119.Kagan JC, Stein MP, Pypaert M, Roy CR. Legionella subvert the functions of Rab1 and Sec22b to create a replicative organelle. J Exp Med. 2004;199:1201–11. doi: 10.1084/jem.20031706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Machner MP, Isberg RR. Targeting of host Rab GTPase function by the intravacuolar pathogen Legionella pneumophila. Dev Cell. 2006;11:47–56. doi: 10.1016/j.devcel.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 121.Machner MP, Isberg RR. A bifunctional bacterial protein links GDI displacement to Rab1 activation. Science. 2007;318:974–7. doi: 10.1126/science.1149121. [DOI] [PubMed] [Google Scholar]
- 122.Ingmundson A, Delprato A, Lambright DG, Roy CR. Legionella pneumophila proteins that regulate Rab1 membrane cycling. Nature. 2007;450:365–9. doi: 10.1038/nature06336. [DOI] [PubMed] [Google Scholar]
- 123.Handa Y, et al. Shigella IpgB1 promotes bacterial entry through the ELMO-Dock180 machinery. Nat Cell Biol. 2007;9:121–8. doi: 10.1038/ncb1526. [DOI] [PubMed] [Google Scholar]
- 124.Mercer J, Helenius A. Vaccinia virus uses macropinocytosis and apoptotic mimicry to enter host cells. Science. 2008;320:531–5. doi: 10.1126/science.1155164. [DOI] [PubMed] [Google Scholar]; References 123 and 124 suggest that pathogens can co-opt phagocytic receptors for apoptotic cells to gain entry into the cell. Whether such mode of entry also alters phagosome maturation remains to be determined.
- 125.Santic M, Asare R, Skrobonja I, Jones S, Abu Kwaik Y. Acquisition of the vacuolar ATPase proton pump and phagosome acidification are essential for escape of Francisella tularensis into the macrophage cytosol. Infect Immun. 2008;76:2671–7. doi: 10.1128/IAI.00185-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Araki N. Role of microtubules and myosins in Fc gamma receptor-mediated phagocytosis. Front Biosci. 2006;11:1479–90. doi: 10.2741/1897. [DOI] [PubMed] [Google Scholar]
- 127.Sanjuan MA, et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450:1253–7. doi: 10.1038/nature06421. [DOI] [PubMed] [Google Scholar]
- 128.Gardai SJ, Bratton DL, Ogden CA, Henson PM. Recognition ligands on apoptotic cells: a perspective. J Leukoc Biol. 2006;79:896–903. doi: 10.1189/jlb.1005550. [DOI] [PubMed] [Google Scholar]
- 129.Devitt A, et al. Persistence of apoptotic cells without autoimmune disease or inflammation in CD14−/− mice. J Cell Biol. 2004;167:1161–70. doi: 10.1083/jcb.200410057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Reddien PW, Horvitz HR. CED-2/CrkII and CED-10/Rac control phagocytosis and cell migration in Caenorhabditis elegans. Nat Cell Biol. 2000;2:131–6. doi: 10.1038/35004000. [DOI] [PubMed] [Google Scholar]
- 131.Gumienny TL, et al. CED-12/ELMO, a novel member of the CrkII/Dock180/Rac pathway, is required for phagocytosis and cell migration. Cell. 2001;107:27–41. doi: 10.1016/s0092-8674(01)00520-7. [DOI] [PubMed] [Google Scholar]
- 132.Park D, et al. BAI1 is an engulfment receptor for apoptotic cells upstream of the ELMO/Dock180/Rac module. Nature. 2007;450:430–4. doi: 10.1038/nature06329. [DOI] [PubMed] [Google Scholar]
- 133.Albert ML, Kim JI, Birge RB. alphavbeta5 integrin recruits the CrkII-Dock180-rac1 complex for phagocytosis of apoptotic cells. Nat Cell Biol. 2000;2:899–905. doi: 10.1038/35046549. [DOI] [PubMed] [Google Scholar]
- 134.Su HP, et al. Interaction of CED-6/GULP, an adapter protein involved in engulfment of apoptotic cells with CED-1 and CD91/low density lipoprotein receptor-related protein (LRP) J Biol Chem. 2002;277:11772–9. doi: 10.1074/jbc.M109336200. [DOI] [PubMed] [Google Scholar]
- 135.Su HP, et al. Identification and characterization of a dimerization domain in CED-6, an adapter protein involved in engulfment of apoptotic cells. J Biol Chem. 2000;275:9542–9. doi: 10.1074/jbc.275.13.9542. [DOI] [PubMed] [Google Scholar]
- 136.Liu QA, Hengartner MO. Candidate adaptor protein CED-6 promotes the engulfment of apoptotic cells in C. elegans. Cell. 1998;93:961–72. doi: 10.1016/s0092-8674(00)81202-7. [DOI] [PubMed] [Google Scholar]; References 130 – 136 identify intracellular signaling molecules that function during apoptotic cell removal in the nematode and mammals, and also provide insights into their mechanism of action.
- 137.Hamon Y, Chambenoit O, Chimini G. ABCA1 and the engulfment of apoptotic cells. Biochim Biophys Acta. 2002;1585:64–71. doi: 10.1016/s1388-1981(02)00325-6. [DOI] [PubMed] [Google Scholar]
- 138.Kinchen JM, Ravichandran KS. Journey to the grave: signaling events regulating removal of apoptotic cells. J Cell Sci. 2007;120:2143–9. doi: 10.1242/jcs.03463. [DOI] [PubMed] [Google Scholar]
- 139.Voll RE, et al. Immunosuppressive effects of apoptotic cells. Nature. 1997;390:350–1. doi: 10.1038/37022. [DOI] [PubMed] [Google Scholar]
- 140.Park SY, et al. Rapid cell corpse clearance by stabilin-2, a membrane phosphatidylserine receptor. Cell Death Differ. 2008;15:192–201. doi: 10.1038/sj.cdd.4402242. [DOI] [PubMed] [Google Scholar]
- 141.Kirsch T, et al. Engulfment of apoptotic cells by microvascular endothelial cells induces proinflammatory responses. Blood. 2007;109:2854–62. doi: 10.1182/blood-2006-06-026187. [DOI] [PubMed] [Google Scholar]; The cytokine response of phagocytes to apoptotic cells is typically tolerogenic, however this paper suggests that in certain circumstances, the presence of apoptotic cells can elicit an inflammatory response.
- 142.Gerbod-Giannone MC, et al. TNFalpha induces ABCA1 through NF-kappaB in macrophages and in phagocytes ingesting apoptotic cells. Proc Natl Acad Sci U S A. 2006;103:3112–7. doi: 10.1073/pnas.0510345103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Blachere NE, Darnell RB, Albert ML. Apoptotic cells deliver processed antigen to dendritic cells for cross-presentation. PLoS Biol. 2005;3:e185. doi: 10.1371/journal.pbio.0030185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Perruche S, et al. CD3-specific antibody-induced immune tolerance involves transforming growth factor-beta from phagocytes digesting apoptotic T cells. Nat Med. 2008;14:528–35. doi: 10.1038/nm1749. [DOI] [PubMed] [Google Scholar]
- 145.Wu YC, Stanfield GM, Horvitz HR. NUC-1, a caenorhabditis elegans DNase II homolog, functions in an intermediate step of DNA degradation during apoptosis. Genes Dev. 2000;14:536–48. [PMC free article] [PubMed] [Google Scholar]
- 146.Parrish JZ, Yang C, Shen B, Xue D. CRN-1, a Caenorhabditis elegans FEN-1 homologue, cooperates with CPS-6/EndoG to promote apoptotic DNA degradation. Embo J. 2003;22:3451–60. doi: 10.1093/emboj/cdg320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Parrish JZ, Xue D. Functional genomic analysis of apoptotic DNA degradation in C. elegans. Mol Cell. 2003;11:987–96. doi: 10.1016/s1097-2765(03)00095-9. [DOI] [PubMed] [Google Scholar]; References 145 – 147 characterize DNAses that play a role during the degradation of apoptotic cells in the nematode.
- 148.Gagnon E, et al. Endoplasmic reticulum-mediated phagocytosis is a mechanism of entry into macrophages. Cell. 2002;110:119–31. doi: 10.1016/s0092-8674(02)00797-3. [DOI] [PubMed] [Google Scholar]
- 149.Kyei GB, et al. Rab14 is critical for maintenance of Mycobacterium tuberculosis phagosome maturation arrest. Embo J. 2006;25:5250–9. doi: 10.1038/sj.emboj.7601407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Delprato A, Lambright DG. Structural basis for Rab GTPase activation by VPS9 domain exchange factors. Nat Struct Mol Biol. 2007;14:406–12. doi: 10.1038/nsmb1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Cantalupo G, Alifano P, Roberti V, Bruni CB, Bucci C. Rab-interacting lysosomal protein (RILP): the Rab7 effector required for transport to lysosomes. Embo J. 2001;20:683–93. doi: 10.1093/emboj/20.4.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Orth JD, McNiven MA. Dynamin at the actin-membrane interface. Curr Opin Cell Biol. 2003;15:31–9. doi: 10.1016/s0955-0674(02)00010-8. [DOI] [PubMed] [Google Scholar]; This excellent review characterizes the role of dynamin-2 in actin-related processes such as maintenance of the actin cytoskeleton.
- 153.Wu YC, Horvitz HR. The C. elegans cell corpse engulfment gene ced-7 encodes a protein similar to ABC transporters. Cell. 1998;93:951–60. doi: 10.1016/s0092-8674(00)81201-5. [DOI] [PubMed] [Google Scholar]
- 154.Wu YC, Horvitz HRC. elegans phagocytosis and cell-migration protein CED-5 is similar to human DOCK180. Nature. 1998;392:501–4. doi: 10.1038/33163. [DOI] [PubMed] [Google Scholar]
- 155.Brugnera E, et al. Unconventional Rac-GEF activity is mediated through the Dock180-ELMO complex. Nat Cell Biol. 2002;4:574–82. doi: 10.1038/ncb824. [DOI] [PubMed] [Google Scholar]
- 156.Pizarro-Cerda J, et al. Brucella abortus transits through the autophagic pathway and replicates in the endoplasmic reticulum of nonprofessional phagocytes. Infect Immun. 1998;66:5711–24. doi: 10.1128/iai.66.12.5711-5724.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Pizarro-Cerda J, Moreno E, Sanguedolce V, Mege JL, Gorvel JP. Virulent Brucella abortus prevents lysosome fusion and is distributed within autophagosome-like compartments. Infect Immun. 1998;66:2387–92. doi: 10.1128/iai.66.5.2387-2392.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Scidmore MA, Fischer ER, Hackstadt T. Sphingolipids and glycoproteins are differentially trafficked to the Chlamydia trachomatis inclusion. J Cell Biol. 1996;134:363–74. doi: 10.1083/jcb.134.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Heinzen RA, Scidmore MA, Rockey DD, Hackstadt T. Differential interaction with endocytic and exocytic pathways distinguish parasitophorous vacuoles of Coxiella burnetii and Chlamydia trachomatis. Infect Immun. 1996;64:796–809. doi: 10.1128/iai.64.3.796-809.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Romano PS, Gutierrez MG, Beron W, Rabinovitch M, Colombo MI. The autophagic pathway is actively modulated by phase II Coxiella burnetii to efficiently replicate in the host cell. Cell Microbiol. 2007;9:891–909. doi: 10.1111/j.1462-5822.2006.00838.x. [DOI] [PubMed] [Google Scholar]
- 161.Beron W, Gutierrez MG, Rabinovitch M, Colombo MI. Coxiella burnetii localizes in a Rab7-labeled compartment with autophagic characteristics. Infect Immun. 2002;70:5816–21. doi: 10.1128/IAI.70.10.5816-5821.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ma H, Croudace JE, Lammas DA, May RC. Expulsion of live pathogenic yeast by macrophages. Curr Biol. 2006;16:2156–60. doi: 10.1016/j.cub.2006.09.032. [DOI] [PubMed] [Google Scholar]
- 163.Alvarez M, Casadevall A. Phagosome extrusion and host-cell survival after Cryptococcus neoformans phagocytosis by macrophages. Curr Biol. 2006;16:2161–5. doi: 10.1016/j.cub.2006.09.061. [DOI] [PubMed] [Google Scholar]
- 164.Terebiznik MR, et al. Helicobacter pylori VacA toxin promotes bacterial intracellular survival in gastric epithelial cells. Infect Immun. 2006;74:6599–614. doi: 10.1128/IAI.01085-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Allen LA. Intracellular niches for extracellular bacteria: lessons from Helicobacter pylori. J Leukoc Biol. 1999;66:753–6. doi: 10.1002/jlb.66.5.753. [DOI] [PubMed] [Google Scholar]
- 166.Scianimanico S, et al. Impaired recruitment of the small GTPase rab7 correlates with the inhibition of phagosome maturation by Leishmania donovani promastigotes. Cell Microbiol. 1999;1:19–32. doi: 10.1046/j.1462-5822.1999.00002.x. [DOI] [PubMed] [Google Scholar]
- 167.Dermine JF, Goyette G, Houde M, Turco SJ, Desjardins M. Leishmania donovani lipophosphoglycan disrupts phagosome microdomains in J774 macrophages. Cell Microbiol. 2005;7:1263–70. doi: 10.1111/j.1462-5822.2005.00550.x. [DOI] [PubMed] [Google Scholar]
- 168.Chua J, Vergne I, Master S, Deretic V. A tale of two lipids: Mycobacterium tuberculosis phagosome maturation arrest. Curr Opin Microbiol. 2004;7:71–7. doi: 10.1016/j.mib.2003.12.011. [DOI] [PubMed] [Google Scholar]
- 169.Meresse S, Steele-Mortimer O, Finlay BB, Gorvel JP. The rab7 GTPase controls the maturation of Salmonella typhimurium-containing vacuoles in HeLa cells. Embo J. 1999;18:4394–403. doi: 10.1093/emboj/18.16.4394. [DOI] [PMC free article] [PubMed] [Google Scholar]