Abstract
Background
A growing number of laboratories are using the mouse as a model system in developmental biology as well as in molecular biology. Surprisingly, most of these laboratories do not have reliable computerized systems to track these animals, and the few commercial solutions available are expensive. We thus developed MICE (Mouse Information and Classification Entity), a program aimed at facilitating the monitoring of animals in animal facilities.
Results
This program consists of a virtual facility in which scientists can perform all the tasks done in the real world (i.e., receiving animals, breeding them, preparing cage labels, etc.). Recording of each animal (birth date, cage number, ID number, tail analysis number, parents, genetic status, genetic background, etc.) enables reliable tracking. According to any parameter of interest, animals can then be identified, grouped, sorted, moved, and so forth. Crossings are automatically processed by the program. For example, new genetic backgrounds, generation number, and anticipated due dates are determined. The program also reminds the user when new births are expected and entering newborn animals only requires a few clicks. The genealogy of each animal can be determined in two different ways, one being the visualization of a genealogical tree from which information of ancestors can be retrieved.
Conclusion
This standalone program, that will be distributed free of charge to academic laboratories requesting a license, represents a new and valuable tool for all animal facility users, and permits simple and reliable tracking and retrieving of animals.
Background
A survey of laboratories working with mice revealed that, except for the very recent "Mousebase" [1], which runs only on Macintoshes and requires the purchase of a license for multiple user configuration, and expensive commercial programs, no convenient system exists to handle recording information of mouse in animal facilities in a reliable manner. When this information is conventionally stored in notebooks, the tracking of different animals becomes quite tedious. Some laboratories have developed computerized lists using word processing applications, allowing animal searches with the word processor "find" function. But accidental modifications are possible and animal tracking, sorting and genealogy determination are basically impossible to perform. In order to sanitize this situation plaguing the majority of laboratories, we developed MICE (Mouse Information and Classification Entity), a standalone application which simulates an animal facility, and runs on both Windows and Macintosh computers. This allows laboratory members to virtually receive, breed and sort their animals, and to automatically record their history and genealogy, together with any other associated information. This program relies on the same strategy as Frozen Cell Stock Monitor (FCSM), another standalone program that we developed previously in order to simplify the monitoring of cell lines stored in liquid nitrogen containers [2].
Results
The MICE program, mimicking an animal facility, consists of 5 main sections:
1- receiving animals,
2- crossing animals,
3- searching and sorting animals,
4- information on animals,
5- moving animals.
These sections, which are described in more detail below, are accessible via the use of self explanatory buttons. Once entered, modifications are automatically saved, thus limiting the risk of data loss in case of a computer crash.
Receiving animals
Upon entering the application, the user is asked to identify himself through the use of a pop-up list that has been customized to identify all laboratory members using the animal facilities. The user is then offered several choices. Among them is the option "Enter animals" (Fig. 1). This opens a screen on which the origin of the animals entered has to be specified.
Figure 1.
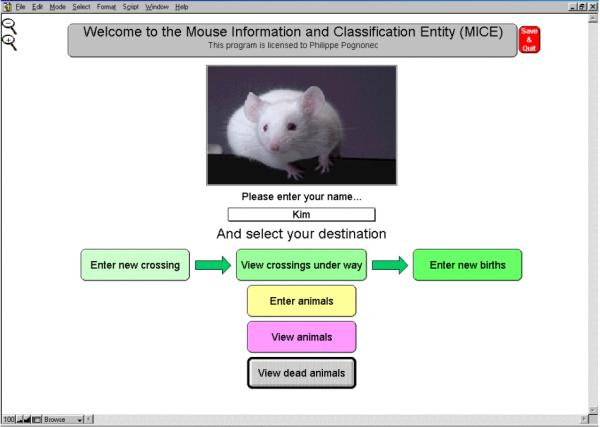
Opening screen.This opening screen displays the different choices available when entering MICE. First, a user name has to been entered from the pop up and customizable list. Then, clicking the buttons will allow either displaying, crossing, or importing animals.
If these animals come from another MICE-monitored facility, the process of transfer is automatic: one simply has to select the file containing the imported animals (by default, named "Exported MICE", see below), and to open it. All the animals will automatically be added to the local MICE database. The date and the importing process will be recorded in the field "Comments" of each imported animal, and their parental description will include the note: "Not from that facility". To create the file containing the imported animals, the sender had to select the animals to be exported (using "mark") from his own MICE database, search all marked animals, and click on "export these animals" (see below).
If the received animals come without MICE information, they have to be manually entered into the database. To this end, select the "Enter non MICE animals". The genetic background, sex, cage location, genetic status, and so forth can then be specified. When several identical animals are received, they can be entered in a single step by simply indicating the number of animals received (Fig. 2).
Figure 2.
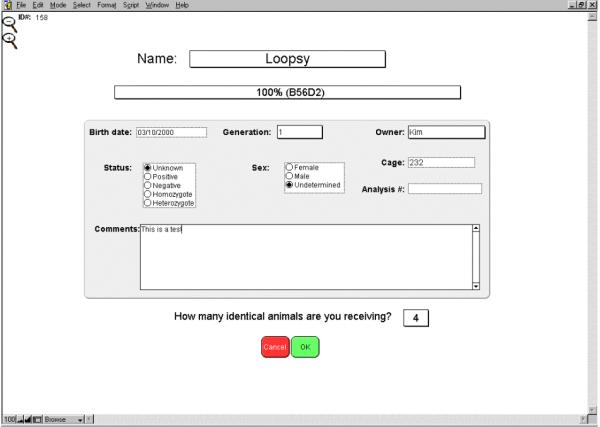
Entering new animals. This screen displays the different parameters that can be entered when a new animal is received in the facility. By default, the date is set to the date that the program is opened, and the owner identified as the user of the program (name selected from the opening screen when opening the application). Number of animals received allows to enter in one single step all the identical animal received. The other parameters can be entered later on.
It should be noted that a unique and definitive ID number is automatically attributed to each animal in the MICE database. A customized number, usually corresponding to the tail analysis, can be entered manually by the user in the "Analysis #" field. This latter number does not necessarily have to be unique and may be replaced later on if another analysis is available (see below).
Crossing animals
To cross animals, the user must select "Enter new crossing". The user is then prompted to select the parent names from two distinct male and female lists present in MICE. To facilitate selection of the parents, typing the first letter of the name will bring the animal on top of the displayed list. The crossing date and cage number must also be entered for the crossing to proceed. Crossing of a homozygote animal with a negative one will yield offspring automatically labeled "Heterozygote", a negative with a negative will yield a negative offspring, and so on.... In all other conditions like a crossing between a heterozygote animal with a negative one, "Unknown" will be entered by default. If both parents have the same name (strain, genetic background, transgenic or knock out lines), that name will automatically be attributed to the offspring by default. In addition, if both parents are of the same generation, generation + 1 will automatically be attributed to the offspring, allowing for a smooth and straightforward family expansion. If parents do not share the same name, entering a name for the future offspring will be required. An interesting feature of MICE is to automatically calculate the genetic background of the offspring, as long as the genetic background of each parent is available. This allows the user to know precisely the genetic background of animals he/she is working with, especially during backcrosses where a pure strain is required. The predicted due date will be calculated and displayed. Finally, additional comments can be added to the crossing. These comments will subsequently follow each animal of that litter throughout its virtual existence (Fig. 3).
Figure 3.
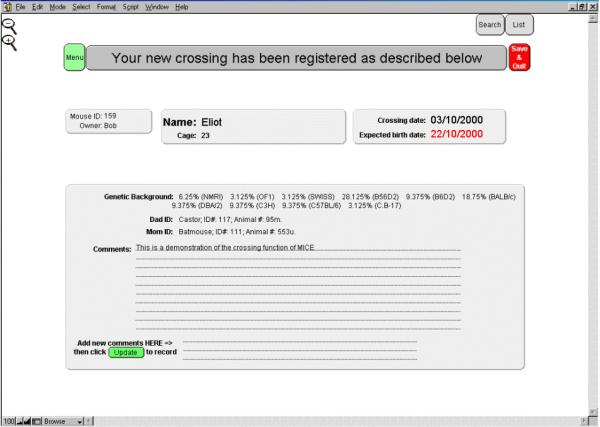
Entering crossings. This screen appears when a crossing has been entered. All the information entered during the crossing procedure are displayed, as well as the calculated due date and the unique ID number attributed by the program. The parents are clearly identified through their names, unique ID#, and analysis #. When one or both parents originate from another facility, it is also specified. If additional comments have been forgotten, it is still possible to enter them on the bottom field, and validate the entry by clicking on the "Update" button. These comments and parameters will follow each of the future animals throughout their life.
Although it is possible to verify the specific details of a crossing by selecting "View crossings under way", MICE will systematically remind the user of crossings as of 2 days prior to the expected due date. Once the litter has arrived, the user will go to "Register births" where he/she will select the offspring number from a pop up list and will need to correct the birth date if it is different from the predicted due date. If no birth is observed, the crossing is erased by selecting "No birth". Once the animals are born and are entered into MICE, they join other animals already present in the database. Each can be sexed by simply clicking on the corresponding box. Genetic analysis results (positive, negative, homo- or heterozygote) can also be entered at any time by a simple click on the different parameters (Fig. 4). If the initial analysis number is later replaced with a newer number, it is safely stored together with the date and the name of the user who performed the new entry.
Figure 4.
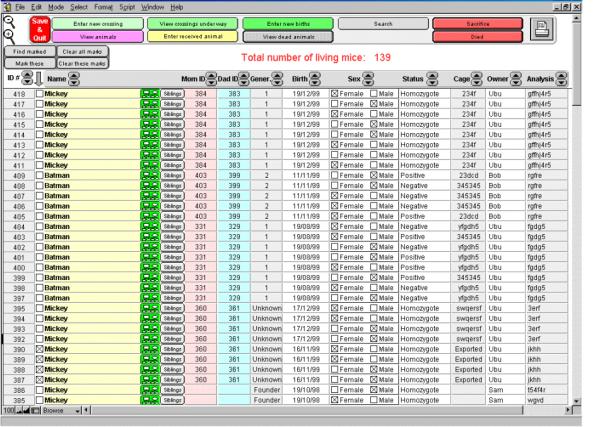
List of animal present in MICE. This is the list of animals present in MICE. The most important information is displayed on one lane per animal: ID#, name, parents ID#, generation, birth date, genetic status, sex, status, cage#, owner and analysis #. Clicking on the name will display all information available for that specific animal (see Fig. 5). The little symbolized tree gives access to the genealogy of the animal chosen, while clicking on the siblings button will post the list of all the animals from the same litter. Clicking on the status, cage, owner or analysis will allow to modify these parameters if necessary, as well as the generation number. Sex can be entered by clicking on the corresponding case. Note that the birth date, for reliability reasons, cannot be modified. Animals of interest can be marked by clicking on their "mark" case to be quickly isolated later on, by clicking on the "Find marked" button. "Mark these" will mark the animals currently displayed, for example those isolated following a search (see Fig. 6). "Clear these marks" is used to remove marks present within the list of animals currently browsed, while "Clear all marks" will erase all marks, even those that are not in the list currently browsed. The top control panel gives access to the other options of MICE, and the print icon controls the printing of the list of animals being browsed.
Searching and sorting animals
To retrieve an animal from a MICE database by list browsing would be too cumbersome especially if thousands of animals are present. For the user's convenience, a powerful search function is available to identify a particular animal, by entering any parameter, alone or in combination with other parameters, including words present in the "Comments" section. Once animals are found, they can then be marked by clicking on "Mark these" or by clicking on the mark box of each animal of interest, to facilitate later retrieval using the "Find marked animals" button. Using the mark parameter, identified animals can be sacrificed individually or as a group, using the corresponding red buttons in the upper control panel. Once animals are recorded as sacrificed ("Dead"), they remain virtually present in the MICE database and are available for genealogy studies. All previous information that was entered also remains accessible after animals have been sacrificed. If the database becomes saturated with information concerning dead animals that are no longer of interest, these animals can be permanently removed by selecting them in the "View dead animal" window and clicking on one of the proposed "Discard" buttons. When a subset of animals is frequently used, it may be useful to use the "Mark" option, so that they can be easily sorted from other animals by clicking on "Find marked".
Living and dead animals are stored separately in the MICE database. They are accessible by clicking on the "View living animals" or "View dead animals" buttons. It is also possible to sort all the displayed lists by clicking on the small up and down arrows present next to each parameter (Fig. 4). One should note that except for ID numbers, up or down sorting are performed on an alphabetical basis. Since analysis numbers or cage locations frequently contain letters as well as numbers, the first character determines the position, then the second, and so on, and priority is given to numbers over letters. (for example, cage "1a" will be displayed before cage "a1", and "a11" before "a2").
It is often necessary to compare animals from the same litter. To that end, clicking on the sibling button in the list view will isolate all siblings of the animal chosen.
Finally, MICE offers the possibility to print cage labels. To this end, select all the animals present in the cage of interest (for example by using the "search" button, and entering the cage number), and click on the print icon. A choice will be offered to either print the list of the selected animals, or to print a cage label, on which the name (if common to all animals in cage), analysis # (if common to all animals in cage), owner (if common to all animals in cage), birth date (if common to all animals in cage), parents (if common to all animals in cage), and cage number are displayed, together with the number of females and males present in the cage.
Information on animals
With MICE, comments and results associated with each animal are always kept together. Additional information can be entered any time, with MICE automatically recording the dates on which new comments are entered together with the names of the users entering them. For reliability reasons, comments that have been entered cannot be erased, however, corrections can always be added. To access these comments, one simply has to click on the name of the animal of interest. A new window, specific for the chosen animal, will be displayed. All information related to a specific animal is present on the screen and can be printed if necessary. The name and sex of the animal, its ID number, and parents are indicated. Birth date together with the animal's present age in days, and death date, when applicable, are also displayed. The genetic background and genetic status (homo/heterozygote, positive/negative) are also indicated, together with cage identification and analysis number. Of course, all comments concerning the specific animal are also displayed (Fig. 5). Finally, the genealogy of the animal can be accessed and presented under two different formats:
Figure 5.
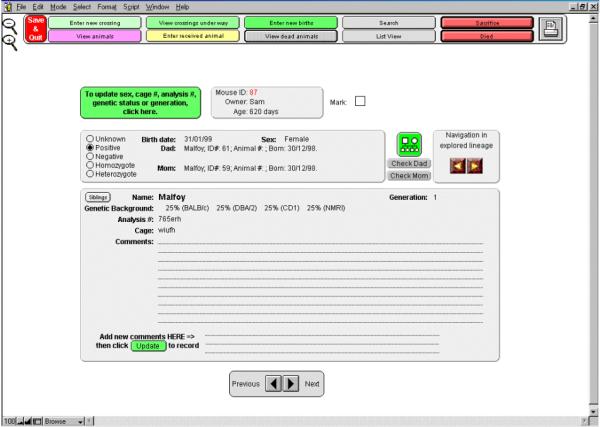
Information on a specific animal. Clicking on the animal name from the list display will open this window, on which all information available for the current animal is presented. As in the list presentation, the animal can be marked, and genealogy can be explored, either through a visual tree (tree icon), or by selecting the "Check Dad" or the "Check Mom" buttons. Information on animals placed before and after in the list display can also be visualized by clicking on either the "Previous" and "Next" button, at the bottom of the screen.
1) Clicking on the "Check Dad" or "Check Mom" buttons will bring up the same window for the corresponding parent, from which the same ancestor browsing can be performed. It is possible to travel back and forth in the explored lineage using the left and right arrows.
2) Clicking on the symbolized tree will display a genealogical tree for the animal analyzed (Fig. 6). From that tree, which goes back 5 generations, any ancestor can be accessed by clicking on it. The same information screen will then be displayed for this specific ancestor. By selecting the tree button again, it is then possible to either display the original tree, or to build a new one corresponding to the chosen ancestor, and thus to go back further in the genealogy if necessary. Those trees can be printed to keep a visual track of the studied lineage.
Figure 6.
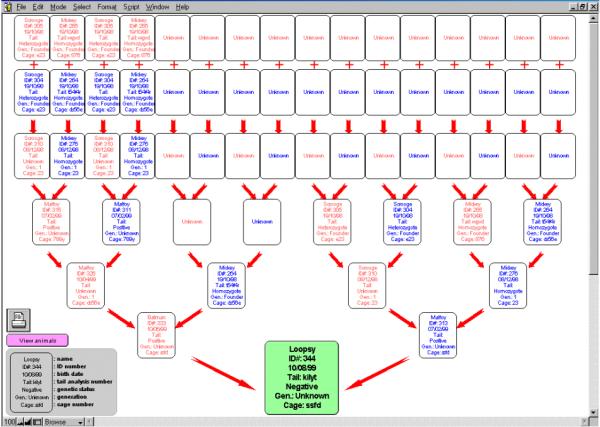
Genealogy display. When the symbolized tree button is selected, this page is displayed. It represents the genealogy of the selected animal, up to 5 generations. Each ancestor is identified by its name and ID#. In addition, birth dates, analyses #, genetic status, generations and cage numbers are presented. When a parent is not available, his box displays "Unknown". When an ancestor is dead, the cage # is replaced by "Dead". Any ancestor presented can be accessed by clicking on its card, in order to get additional information. A new genealogy search can be performed from this ancestor, allowing to go back 5 more generations. This process can be indefinitely repeated. The trees can be printed by clicking on the printer icon.
Moving animals
Animals can be moved either within the MICE facility (i.e., to different cages), or to another facility.
To move animals from cage to cage, two options are possible: if only one mouse is to be moved from the "view animals" list, click on the cage number to be changed, and enter the new cage address. If the name of the animal has been clicked, and the window of information specific to that animal is being displayed, click on the green "update..." button and modify the cage address. In the case where several animals are to be moved, first select the animals and mark them as described above, then using the search function for marked animals select "move all to another cage". It is worth noting that when a crossing is performed, the Dad and Mom are automatically moved to the cage selected for the crossing.
To move animals to another facility, select and mark them as indicated above. Then using the search function for marked animals, select "export these animals". A new file containing these animals together with their corresponding information will be created on the desktop, named "Exported MICE". This file can be e-mailed to the receiving facility. Information on exported animals will still be present in your MICE facility, but in place of their cage addresses, "Exported" will be indicated. These animals will still be available for genealogical searches but will no longer be available for crossing. If one wants to definitively remove them from MICE, it is necessary to mark and select them with the search option, and choose "sacrifice all". Then, from "show dead animals", select "discard marked records". All traces of these animals will then be removed from MICE.
Discussion
MICE is a virtual animal facility. It allows laboratories to efficiently keep track of their animals throughout time and despite the high people turn-over frequently associated with laboratories. It is easy and intuitive to use. This program, developed on FileMaker Pro™, runs indifferently on PC or Mac computers. Its runtime binding engine allows for standalone application, i.e., will run without having to install FileMaker Pro™ on the receiving computer. Printouts of summarized or detailed information corresponding to groups of animals or individuals, respectively, will simplify the manipulation of the real animals. We recommend using a computer with a screen definition of at least 768X1024, G3 or Pentium II processors, and a minimum of 50 Mo of free disk space for smooth operations. For information on the distribution of this program, that will be distributed free of charge to academic laboratories requesting a license, please e-mail to: pognonec@unice.fr and indicate "MICE" in the subject field.
Acknowledgments
Acknowledgements
This work was supported by contract #5231 from ARC (Association pour la Recherche contre le Cancer) to KEB. KEB and PP are supported by CNRS.
Contributor Information
Kim E Boulukos, Email: boulukos@unice.fr.
Philippe Pognonec, Email: pognonec@unice.fr.
References
- R Hopley, A Zimmer. MouseBank: a database application for managing transgenic mouse breeding programs. Biotechniques. 2001;30:130–2. doi: 10.2144/01301bc04. [DOI] [PubMed] [Google Scholar]
- A Zaldumbide, KE Boulukos, P Pognonec. Virtual nitrogen tank to monitor frozen cell stocks. Biotechniques. 2000;29:122–6. doi: 10.2144/00291bc04. [DOI] [PubMed] [Google Scholar]


