Abstract
The tcpRXABCYD operon of Cupriavidus necator JMP134 is involved in the degradation of 2,4,6-trichlorophenol (TCP). All of the gene products except TcpY have assigned functions in TCP metabolism. Sequence comparison identified TcpY as a member of COG4313, a group of hypothetical proteins. TcpY has a signal peptide, indicating it is a membrane or secreted protein. Secondary structure and topology analysis indicated TcpY as a β-barrel outer membrane protein, similar to the Escherichia coli outer membrane protein FadL that transports hydrophobic long-chain fatty acids. Constitutive expression of tcpY in two C. necator strains rendered the cells more sensitive to TCP and other polychlorophenols. Further, C. necator JMP134 expressing cloned tcpY transported more TCP into the cell than a control with the cloning vector. Thus, TcpY is an outer membrane protein that facilitates the passing of polychlorophenols across the outer membrane of C. necator. Similarly, other COG4313 proteins are possibly outer membrane transporters of hydrophobic aromatic compounds.
Keywords: Gram negative bacteria, Outer membrane transporter, β-Barrel protein, Substrate uptake, Trichlorophenol biodegradation
Introduction
2,4,6-Trichlorophenol (TCP), a major environmental pollutant, has been used extensively as a preservative and a biocide (Czaplicka 2004). The gram negative bacterium Cupriavidus necator JMP134, a versatile degrader of aromatic compounds, can use TCP as a carbon and energy source (Clement et al. 1995; Padilla et al. 2000). A gene cluster, tcpRXABCYD, is involved in TCP degradation (Louie et al. 2002; Matus et al. 2003; Sanchez and Gonzalez 2007), and all the encoded proteins have been characterized except TcpY (Fig. 1). First, TcpA (TCP 4-monooxygenase) oxidizes TCP to 2,6-dichloro-p-benzoquinone and then hydrolyzes the latter to 6-chloro-2-hydroxy-p-benzoquinone (Xun and Webster 2004). Since TcpA is a reduced flavin adenine dinucleotide (FADH2)-dependent monooxygenase, TcpX (NADH:FAD oxidoreductase) supplies FADH2 to TcpA in coupled reactions (Belchik and Xun 2008). Second, TcpB (NADH:quinone oxidoreductase) reduces the quinone to 6-chloro-2-hydroxy-p-hydroquinone (Belchik and Xun 2008). Third, TcpC (dioxygenase) catalyzes the ring-cleavage of the latter to produce 2-chloromaleylacetate (Louie et al. 2002). Fourth, TcpD (maleylacetate reductase) removes the last chlorine and then converts maleylacetate to 3-ketoadipate, a common metabolic intermediate of microbial degradation of aromatic compounds (Harwood and Parales 1996). The tcpR gene codes for a regulatory protein controlling the expression of tcpXABCYD (Sanchez and Gonzalez 2007).
Fig. 1.
a Degradation pathway of C. necator JMP134. TcpX and TcpA catalyze the conversion of TCP to 6-chloro-2-hydroxy-p-benzoquinone. TcpB reduces 6-chloro-2-hydroxy-p-benzoquinone to 6-chloro-2-hydroxy-p-hydroquinone. TcpC converts the latter to 2-chloromaleylacetate, which is reduced by TcpD to maleylacetate and then to 3-ketoadipate, b the tcp operon in C. necator JMP134. The bar represents 1 kb
The tcpY gene is located in the middle of the tcp gene cluster, but the function of TcpY remains unknown. TcpY does not share significant sequence similarity to any known proteins, and a tcpY mutant is normal in TCP degradation (Sanchez and Gonzalez 2007). However, a structural homology search described below indicated TcpY displayed significant homology with FadL, an Escherichia coli outer membrane transporter protein required for long-chain fatty acid (LCFA) transport (van den Berg et al. 2004). Gram-negative bacteria have hydrophilic outer membranes that act as a barrier against hydrophobic molecules and thus require transporters to facilitate diffusion into the cytoplasm (van den Berg 2005). Like LCFAs, TCP is a hydrophobic molecule. Therefore, TcpY may function as a transporter to facilitate TCP diffusion into the cell. Here, we report evidence supporting TcpY as an outer membrane transporter that facilitates the diffusion of polychlorophenols into C. necator.
Materials and methods
Chemicals and enzymes
Reagents used were purchased from Sigma Chemical Co. (St. Louis, MO) or Aldrich Chemical Co. (Milwaukee, WI). Enzymes were purchased from Invitrogen (Carlsbad, CA) or New England Biolabs (Beverly, MA).
Bacterial strains and culture conditions
Cupriavidus necator JMP134 and C. necator H16 were grown in a mineral salt medium with 0.2% glutamate as the carbon source at 30°C (Louie et al. 2002). E. coli strains were grown in Luria–Bertani (LB) medium at 37°C. Kanamycin was used at a final concentration of 30 μg ml−1.
Sequence analysis
The BLASTP program (Altschul et al. 1997) was used to search sequence similarity. Signal sequence of membrane proteins was predicted by using SignalP http://www.cbs.dtu.dk/services/SignalP/) (Emanuelsson et al. 2007).
TcpY modeling
The likelihood of proteins to form β-strands in the outer membrane of gram-negative bacteria was calculated using the PRED-TMBB (http://bioinformatics.biol.uoa.gr/PREDTMBB) (Bagos et al. 2004). The three-dimensional model of TcpY was constructed using Phyre version 0.2 (http://www.sbg.bio.ic.ac.uk/phyre/) (Bennett-Lovsey et al. 2008). Phyre calculates the best fit towards a solved crystal structure with estimated precision predicting likelihood of fit.
Phylogenetic analysis
Protein sequences were aligned by using CLUSTALW (Larkin et al. 2007), and a phylogenetic tree was generated by using MEGA version 4.0.2 (Tamura et al. 2007). The evolutionary distances were computed using the Jones et al. (1992) method and are in units of the number of amino acid substitutions per site.
Chromosomal disruption of tcpY in C. necator JMP134
A 493-bp internal fragment of tcpY was amplified from C. necator JMP134 DNA by PCR using primer pair TcpY-internalF (gtggtaccgatcgtgtctgc) and TcpY-internalR (actgatactgggtggcttcg). The PCR products were cloned into pCR2.1-TOPO, forming plasmid pKOY. The plasmid was amplified in E. coli and isolated. The purified plasmid DNA (50 ng) was electroporated into C. necator JMP134 cells (Louie et al. 2002). Recombinant strains were selected on LB agar plates containing kanamycin.
Constitutive expression of TcpY in C. necator
The full length tcpY gene was cloned in the plasmid pTrc99a (Pharmacia, Piscataway, NJ), where tcpY was under the control of the trc promoter that contains a LacI binding site (Amann et al. 1988). The tcpY gene was PCR amplified with primers TcpYR (gctttcatcgaagcttctcccgtc) and TcpYF-pTrc99 (gttcgacagatgccatggaggtcc), digested with HindIII and NcoI, and ligated into pTrc99a to generate pTrc99a-tcpY. As the pTrc99a-tcpY plasmid cannot replicate in C. necator JMP134, the cloned tcpY is moved to pBBR1MCS2 that can replicate in a broad host range (Kovach et al. 1994, 1995). The pTrc99a-tcpY plasmid was used as a template for PCR using pTrc99-F (gttctggataagcttttttgcgcc) with a HindIII site and pTrc99-R (atcttctctcatccgcca). This PCR product was digested with HindIII and ligated into pBBR1MCS2 to create pTcpY, in which tcpY is under the control of the trc promoter from pTrc99a. Electroporation was utilized to transfer pTcpY into C. necator JMP134 and C. necator H16. In C. necator, the trc promoter is constitutively expressed due to the lack of repressor, LacI. pBBR1MCS2 was also electroporated into these strains as the empty vector control.
Agar plate toxicity assay for TCP
To determine the effects of constitutive tcpY expression when TCP is present, we utilized mineral salt medium agar plates with 0.2% glutamate and varying amounts of TCP (from 0 to 400 μM). Glutamate represses the tcp operon in C. necator JMP134, so no degradation occurs (Louie et al. 2002). Briefly, cells were cultured overnight in LB medium, harvested and resuspended in the mineral salt medium without carbon source at a turbidity of 1.0 at 600 nm (~1 × 107 cells ml−1). Serial dilutions were performed in the medium, and 3 μl was spotted onto the agar plates. The plates were incubated for 3 days at 30°C and pictures were taken. If the cells are sensitive to the TCP concentrations, the cells at higher dilutions would have small or no colonies due to slower growth.
Liquid culture toxicity assay for polychlorophenols
Several polychlorophenols were tested to determine if TcpY was specific to TCP. C. necator JMP134 was cultured overnight in LB, harvested, and resuspended in the mineral salt medium with no carbon source to a turbidity of 1.0 at 600 nm. A 1% inoculum was then transferred to the mineral salt medium containing 0.2% glutamate and 100 μM 2,6-dichlorophenol (DiCP), TCP, 2,3,4,6-tetrachlorophenol (TeCP), 2,3,5,6-TeCP, or pentachlorophenol (PCP). Cultures were shaken for 18 h at 30°C and a final turbidity of 600 nm was measured.
TCP uptake
Cells of C. necator JMP134 carrying pTcpY or pBBR1MCS2 were cultured for 5 h to a turbidity of 2 at 600 nm in LB medium, harvested and washed with mineral salt medium containing 0.2% glutamate, and resuspended in the same medium to a turbidity of 2.0 at 600 nm. Protein concentration was determined to be about 0.4 mg per ml by using modified Lowry protein assay (Pierce Biotechnology, Rockford, IL). One ml of culture was transferred to 1.7-ml micro-fuge tube, to which varying amounts of TCP (12.5–100 μM TCP) was added. The tubes were shaken for 1 min at room temperature, and then were centrifuged at 12,000×g for 1 min. First, the pellets were washed with 1 ml of 50 mM KPi (pH 7.0) without suspending the cells. Then, they were washed twice with 1 ml of 50 mM KPi (pH 7.0) by suspension and centrifugation. After the washing, each pellet was resuspended in 100 μl of the KPi buffer with 1% sodium dodecyl sulfate. The sample was vortexed and incubated at 50°C for 5 min. About 100 μl of hexane:1-propanol (vol/vol, 4:1) was added, and the mixture was vortexed for 1 min. After centrifugation, TCP in the organic phase was analyzed with high performance liquid chromatography equipped with a C-18 column and photodiode array detector, as previously described (Louie et al. 2002). The volume of the organic phase was assumed to be 100 μl for calculations.
Results and discussion
Sequence analysis of the TcpY revealed a β-barrel outer membrane protein
BLASTP search with TcpY sequence returned only hypothetical proteins that are grouped together in the cluster of orthologous groups (COG) 4313 (Marchler-Bauer et al. 2007). Although no experimental data exists for this particular COG, members are thought to be involved in the meta-pathway of phenol degradation due to their gene location with genes responsible for phenol degradation (Xu et al. 2003). SignalP indicated that TcpY had a signal peptide and a predicted cleavage site between amino acid residues 38 and 39. The estimated molecular mass of the mature polypeptide was 31,715.24 Da. The signal peptide suggested that TcpY was a membrane or secreted protein. C. necator has both cytoplasmic and outer membranes. Integral membrane proteins of cytoplasmic membranes usually consist of α-helices for the transmembrane portions; whereas, the outer membrane proteins are composed of antiparallel β-strands for transmembrane segments that form a barrel in the outer membrane, e.g., porins (Schulz 2002). PRED-TMBB, a web-server capable of predicting and discriminating β-barrel outer membrane proteins, was utilized to analyze TcpY (Bagos et al. 2004). PRED-TMBB scored TcpY with a value of 2.877 that is less than the threshold value of 2.965. Values below this threshold correctly predict β-barrel membrane proteins with 95% accuracy using a database of known β-barrel outer membrane proteins (Bagos et al. 2004). PRED-TMBB predicted the topology of TcpY with fourteen transmembrane β-strands. Structural homology modeling using Phyre predicted TcpY structure with an estimated 95% precision related to the three-dimensional structure of FadL from E. coli. FadL is a transporter that facilitates the diffusion of hydrophobic LCFAs across hydrophilic outer membranes of E. coli and related gram-negative bacteria (van den Berg et al. 2004). Thus, bioinformatic analysis indicated TcpY as a β-barrel outer membrane protein, possibly involved in TCP uptake.
Phylogenetic analysis of TcpY and structural homologs
Other structural homologs of FadL have been linked to the uptake of hydrophobic molecules for biodegradation (van den Berg 2005). XylN is identified as an outer membrane transporter for the uptake of xylene (Kasai et al. 2001); TodX and TbuX are for toluene uptake (Wang et al. 1995; Kahng et al. 2000); StyE is for styrene uptake (Mooney et al. 2006). The resolved structures of TodX and TbuX are similar to FadL (Hearn et al. 2008). To determine how TcpY is related to these proteins, a multiple alignment was utilized. The proteins are listed in Table 1 and the phylogenetic tree created from the alignment is shown in Fig. 2. Bootstrap values of 100 indicated the proteins fell into three separate clades: LCFA transporters, xenobiotic transporters, and COG4313 hypothetical proteins. TcpY was grouped with the COG4313 proteins. The substrates for the three groups are also different: hydrophobic LCFA, aromatic compounds without hydrophilic groups, and aromatic compounds with hydroxyl groups, respectively.
Table 1.
TcpY and related proteins used in the phylogenetic analysis
| Protein | Organism | Accession no. | Pairwise score (%)a | Proposed function |
|---|---|---|---|---|
| TcpY | Cupriavidus necator JMP134 | YP_295798 | 100 | Conserved hypothetical protein |
| 383-Hypo | Burkholderia sp. 383 | YP_373710 | 38 | Meta-pathway phenol degradation-like proteinb |
| GB-1-Hypo | Pseudomonas putida GB-1 | YP_001669533 | 35 | Meta-pathway phenol degradation-like proteinb |
| SB-Hypo | Syntrophus aciditrophicus SB | YP_461556 | 32 | Conserved hypothetical protein |
| Orf8 | Acinetobacter calcoaceticus PHEA-2 | CAD92317 | 31 | Meta-pathway phenol degradation-like proteinb |
| TbuX | Ralstonia pickettii PKO1 | AAF03168 | 12 | Transport of toluene across the cell membrane |
| XylN | Pseudomonas putida KT2440 | BAA09665 | 9 | Transport of xylene across the cell membrane |
| StyE | P. putida CA-3 | AAR24508 | 9 | Transport of styrene across the cell membrane |
| FadL-ET1/99 | Erwinia tasmaniensis Et1/99 | YP_001907087 | 9 | LCFA outer membrane transporter |
| FadL-TTO1 | Photorhabdus luminescens subsp. laumondii TTO1 | NP_930431 | 9 | LCFA outer membrane transporter |
| TodX | P. putida F1 | AAC43318 | 7 | Transport of toluene across the cell membrane |
| FadL | Escherichia coli W3110 | AP_002944 | 6 | LCFA outer membrane transporters |
| FadL-SCRI1043 | Pectobacterium atrosepticum SCRI1043 | YP_051171 | 6 | LCFA outer membrane transporter |
Percentage sequence identity to TcpY divided by the number of residues in the alignment by MEGA4 (Tamura et al. 2007)
No specific information was given
Fig. 2.
Phylogenetic relationship of β-barrel outer membrane transporters as determined using MEGA4. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (2,000 replicates) is shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The proteins, species, GenBank accession number, pairwise score, and proposed function are listed in Table 1
The COG4313 proteins used for phylogenetic analysis (Fig. 2) were further analyzed (Table 2). Like TcpY, the four hypothetical proteins all contained signal peptides, scored as β-barrel outer membrane proteins, and shared high structural homology with FadL. COG4313 proteins are within gene clusters responsible for biodegradation of phenolic compounds, which are likely the substrates of COG4313 proteins. The orf8 gene is located near meta-pathway of phenol degradation genes in Acinetobacter calcoaceticus PHEA-2 (Xu et al. 2003). The P. putida GB-1-Hypo protein gene is adjacent to phenol degradation genes.
Table 2.
COG4313 proteins computational analysis
| Proteina | Signal peptide cleavage sitea,b | PRED-TMBB scorec | Estimated precision with FadL structure (%)d |
|---|---|---|---|
| Orf8 | 24 | 2.876 | 90 |
| 383-Hypo | 27 | 2.904 | 95 |
| GB-1-Hypo | 27 | 2.871 | 95 |
| SB-Hypo | 34 | 2.883 | 95 |
Accession numbers are given in Table 1
As determined by SignalP 3.0 Server (Emanuelsson et al. 2007)
A score lower than the cutoff of 2.965 indicates a β-barrel outer membrane protein with 95% confidence (Bagos et al. 2004)
As determined by Phyre server (Bennett-Lovsey et al. 2008)
Knockout of tcpY caused no apparent difference in degradation or sensitivity to TCP
The tcpY gene was disrupted via homologous integration of a suicidal plasmid that carried an internal fragment of the tcpY gene in C. necator JMP134. The integration of pKOY resulted in a kanamycin resistant mutant that contained two truncated copies of tcpY on the chromosome. Integration of pKOY was confirmed by PCR using one primer located on the chromosome TcpYKOF (gtattccggcctgatgttgc) and one located on the plasmid pCR-R (gttttcccagtcacgacgtt) (data not shown). The tcpY mutant and C. necator JMP134 wild type degraded TCP at comparable rates using TCP concentrations from 10 to 400 μM. Also, the mutant and wild type showed similar sensitivity to TCP (100–400 μM) as well as other polychlorophenols both in the mineral salt liquid medium and on agar plates (data not shown). Sanchez and Gonzalez have also reported that a tcpY knockout mutant can grow at the same rate as the wild type with TCP as the sole carbon source (Sanchez and Gonzalez 2007). Thus, TCP is able to pass the outer membrane of C. necator JMP134 without the aid of TcpY under the culturing conditions. However, it is unknown if TcpY affects TCP transport in the environment, as different growth conditions alter the permeability of the outer membrane of gram-negative bacteria (Nikaido 2003).
Constitutively expressed tcpY was toxic to C. necator JMP134
To evaluate whether TcpY facilitated the diffusion of TCP into C. necator JMP134 cells, we compared the sensitivity to TCP with and without the presence of constitutively expressed tcpY. If TcpY functions as an outer membrane transporter for TCP, constitutive expression of tcpY should produce TcpY at higher concentrations than in the wild type where the gene is repressed in the presence of glutamate (Louie et al. 2002). Without the operon expressing the genes required to degrade TCP, TCP would slow down cell growth. The decreased growth would produce small or no colonies on agar plates inoculated with serially diluted cells. On agar plates, C. necator JMP134 containing pBBR1MCS2 (empty vector) displayed decreased growth starting at 200 μM TCP but was still able to grow at 400 μM TCP (Fig. 3). On the other hand, the strain containing pTcpY, which constitutively expressed tcpY, exhibited decreased growth at 100 μM and almost a complete abolishment of growth at 400 μM TCP (Fig. 3). The results indicated that TcpY allowed more TCP to enter the cells.
Fig. 3.

Sensitivity of C. necator JMP134 on agar plates with TCP. Cultures were grown overnight, harvested, washed, and resuspended to a turbidity of 1.0 in the mineral salt medium. Three micro litres of serial diluted cells were dropped onto the mineral salt agar plates with 0.2% glutamate and various concentrations of TCP. The plates were incubated at 30°C for 3 days. Ctl refers to clone containing pBBR1MCS2, and pTcpY refers to the clones containing constitutively expressed tcpY
Constitutive expression of tcpY in C. necator H16 also increased TCP toxicity
To study the function of TcpY in a closely related bacterium, pBBR1MCS2 and pTcpY were electroporated into C. necator H16, a bacterium unable to degrade TCP. Overall, C. necator H16 had a higher sensitivity to TCP in comparison to C. necator JMP134. On agar plates, C. necator H16 control did not display decreased growth until 100 μM TCP while the strain carrying pTcpY started to exhibit decreased growth at 50 μM TCP and almost a complete inhibition of growth at 200 μM (data not shown). The results indicated that TcpY also brought more TCP into the cells and made them more sensitive to TCP in comparison to the control carrying the empty vector.
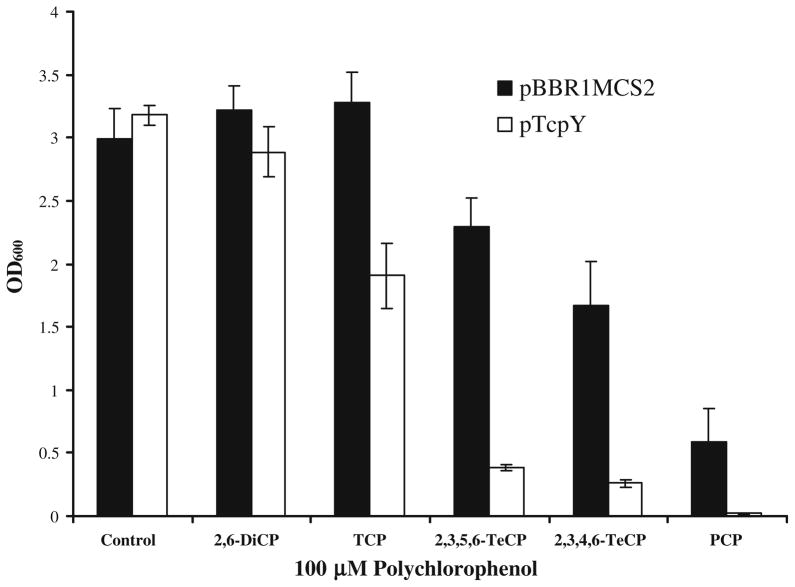
TcpY increased toxicity towards other polychlorophenols
Since FadL transports various LCFAs, we tested the ability of tcpY to transport other polychlorophenols. The experiments were done by growing the cells with glutamate that suppresses the expression of the tcp operon. Further, the tcp operon is only induced by TCP and not by other polychlorophenols (Sanchez and Gonzalez 2007). We determined the sensitivity of C. necator JMP134 towards DiCP, TCP, 2,3,4,6-TeCP, 2,3,5,6-TeCP, and PCP. DiCP did not have any apparent effect on the growth of C. necator JMP134 carrying pTcpY compared to the empty vector control (Fig. 4), indicating it is likely not a substrate for TcpY. The two TeCPs and PCP all reduced growth when compared to without polychlorophenols in the medium for cells carrying the empty vector, demonstrating toxicity of the TeCPs and PCP. Moreover, C. necator JMP134 cells carrying pTcpY displayed more sensitivity to TCP, 2,3,4,6-TeCP, 2,3,5,6-TeCP, and PCP than the cells with the empty vector (Fig. 4).
Fig. 4.
Toxicity of chlorophenols to C. necator JMP134. A 1% inoculum from overnight cultures was added to the minimal salt medium with 0.2% glutamate containing 100 μM of DiCP, TCP, 2,3,4,6-TeCP, 2,3,5,6-TeCP, or PCP. Cultures were incubated with shaking at 30°C for 18 h before turbidities were taken at 600 nm. C. necator JMP134 containing pBBR1MCS2 or pTcpY was tested
TcpY facilitated the uptake of TCP into C. necator JMP134 cells
2,4,6-Trichlorophenol uptake by JMP134 cells carrying pTcpY or pBBR1MCS2 was determined after 1 min incubation in the presence of glutamate. JMP134 cells constitutively expressing tcpY from the plasmid pTcpY transported more TCP into the cells than the cells carrying the vector pBBR1MCS2 at various TCP concentrations (Fig. 5). The method was taking advantage of TCP accumulation in lipid membranes so that quick washes did not result in any significant loss of the transported TCP. Further, glutamate inhibits TCP metabolism (Louie et al. 2002). Because centrifugation and wash both took time, the calculated rate may not be accurate. However, the measured rates reflected the difference mediated by TcpY. The data provided direct evidence that TcpY facilitated TCP transportation into C. necator cells.
Fig. 5.
TCP uptake by JMP134 cells carrying pTcpY (filled square) or pBBR1MCS2 (filled triangle). Late log-phase cells were suspended in mineral medium with glutamate and various amounts of TCP. After 1 min shaking, the cells were collected by centrifugation and washed three times with KPi buffer. The TCP transported into the cells were extracted with hexane and determined
Conclusion
β-Barrel outer membrane proteins share similar structures but limited sequence homology due to the lack of evolutionary constraints for loop regions that connect the β-sheets (Jeanteur et al. 1991). Some β-barrel outer membrane proteins can facilitate the transport of hydrophobic substrates across the outer membrane, which is often hydrophilic due to lipopolysaccharides (Nikaido 2003). TcpY is identified as a β-barrel outer membrane protein because of its structural similarity to characterized β-barrel outer membrane proteins, including FadL, TodX and TbuX (Wang et al. 1995; Kahng et al. 2000; van den Berg 2005). The toxicity and uptake studies support that TcpY facilitates the transport of polychlorophenols across the outer membrane of gram negative bacterium C. necator.
Acknowledgments
The research was supported by the US National Science Foundation grant MCB-0323167. Sara Belchik was also supported by an NIH Biotechnology Training Grant.
Contributor Information
Sara Mae Belchik, School of Molecular Biosciences, Washington State University, Life Sciences Building, Room 202, 100 Dairy Road, Pullman, WA 99164-7520, USA.
Scott M. Schaeffer, School of Molecular Biosciences, Washington State University, Life Sciences Building, Room 202, 100 Dairy Road, Pullman, WA 99164-7520, USA
Shelley Hasenoehrl, School of Molecular Biosciences, Washington State University, Life Sciences Building, Room 202, 100 Dairy Road, Pullman, WA 99164-7520, USA.
Luying Xun, Email: xun@mail.wsu.edu, School of Molecular Biosciences, Washington State University, Life Sciences Building, Room 202, 100 Dairy Road, Pullman, WA 99164-7520, USA.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann E, Ochs B, Abel KJ. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene. 1988;69:301–315. doi: 10.1016/0378-1119(88)90440-4. [DOI] [PubMed] [Google Scholar]
- Bagos PG, Liakopoulos TD, Spyropoulos IC, Hamodrakas SJ. PRED-TMBB: a web server for predicting the topology of beta-barrel outer membrane proteins. Nucleic Acids Res. 2004;32:W400–W404. doi: 10.1093/nar/gkh417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belchik SM, Xun L. Functions of flavin reductase and quinone reductase in 2,4,6-trichlorophenol degradation by Cupriavidus necator JMP134. J Bacteriol. 2008;190:1615–1619. doi: 10.1128/JB.01697-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett-Lovsey RM, Herbert AD, Sternberg MJE, Kelley LA. Exploring the extremes of sequence/structure space with ensemble fold recognition in the program Phyre. Proteins. 2008;70:611–625. doi: 10.1002/prot.21688. [DOI] [PubMed] [Google Scholar]
- Clement P, Matus V, Cardenas L, Gonzalez B. Degradation of trichlorophenols by Alcaligenes eutrophus JMP134. FEMS Microbiol Lett. 1995;127:51–55. doi: 10.1111/j.1574-6968.1995.tb07449.x. [DOI] [PubMed] [Google Scholar]
- Czaplicka M. Sources and transformations of chlorophenols in the natural environment. Sci Total Environ. 2004;322:21–39. doi: 10.1016/j.scitotenv.2003.09.015. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP, and related tools. Nat Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- Harwood CS, Parales RE. The beta-ketoadipate pathway and the biology of self-identity. Annu Rev Microbiol. 1996;50:553–590. doi: 10.1146/annurev.micro.50.1.553. [DOI] [PubMed] [Google Scholar]
- Hearn EM, Patel DR, van den Berg B. Outer-membrane transport of aromatic hydrocarbons as a first step in biodegradation. Proc Natl Acad Sci USA. 2008;105:8601–8606. doi: 10.1073/pnas.0801264105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanteur D, Lakey JH, Pattus F. The bacterial porin superfamily—sequence alignment and structure prediction. Mol Microbiol. 1991;5:2153–2164. doi: 10.1111/j.1365-2958.1991.tb02145.x. [DOI] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Kahng HY, Byrne AM, Olsen RH, Kukor JJ. Characterization and role of tbuX in utilization of toluene by Ralstonia pickettii PKO1. J Bacteriol. 2000;182:1232–1242. doi: 10.1128/jb.182.5.1232-1242.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai Y, Inoue J, Harayama S. The TOL plasmid pWWO xylN gene product from Pseudomonas putida is involved in m-xylene uptake. J Bacteriol. 2001;183:6662–6666. doi: 10.1128/JB.183.22.6662-6666.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovach ME, Phillips RW, Elzer PH, Roop RM, Peterson KM. pBBR1MCS: a broad-host-range cloning vector. Biotechniques. 1994;16:800–802. [PubMed] [Google Scholar]
- Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, Peterson KM. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Louie TM, Webster CM, Xun L. Genetic and biochemical characterization of a 2,4,6-trichlorophenol degradation pathway in Ralstonia eutropha JMP134. J Bacteriol. 2002;184:3492–3500. doi: 10.1128/JB.184.13.3492-3500.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Derbyshire MK, DeWeese-Scott C, Gonzales NR, Gwadz M, et al. CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res. 2007;35:D237–D240. doi: 10.1093/nar/gkl951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus V, Sanchez MA, Martinez M, Gonzalez B. Efficient degradation of 2,4,6-trichlorophenol requires a set of catabolic genes related to tcp genes from Ralstonia eutropha JMP134(pJP4) Appl Environ Microbiol. 2003;69:7108–7115. doi: 10.1128/AEM.69.12.7108-7115.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney A, O’Leary ND, Dobson ADW. Cloning and functional characterization of the styE gene, involved in styrene transport in Pseudomonas putida CA-3. Appl Environ Microbiol. 2006;72:1302–1309. doi: 10.1128/AEM.72.2.1302-1309.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla L, Matus V, Zenteno P, Gonzalez B. Degradation of 2,4,6-trichlorophenol via chlorohydroxyquinol in Ralstonia eutropha JMP134 and JMP222. J Basic Microbiol. 2000;40:243–249. doi: 10.1002/1521-4028(200008)40:4<243::AID-JOBM243>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Sanchez AA, Gonzalez B. Genetic characterization of 2,4,6-trichlorophenol degradation in Cupriavidus necator JMP134. Appl Environ Microbiol. 2007;73:2769–2776. doi: 10.1128/AEM.02584-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz GE. The structure of bacterial outer membrane proteins. Biochim Biophys Acta. 2002;1565:308–317. doi: 10.1016/s0005-2736(02)00577-1. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- van den Berg B. The FadL family: unusual transporters for unusual substrates. Curr Opin Struct Biol. 2005;15:401–407. doi: 10.1016/j.sbi.2005.06.003. [DOI] [PubMed] [Google Scholar]
- van den Berg B, Black PN, Clemons WM, Rapoport TA. Crystal structure of the long-chain fatty acid transporter FadL. Science. 2004;304:1506–1509. doi: 10.1126/science.1097524. [DOI] [PubMed] [Google Scholar]
- Wang Y, Rawlings M, Gibson DT, Labbe D, Bergeron H, Brousseau R, Lau PCK. Identification of a membrane-protein and a truncated LysR-type regulator associated with the toluene degradation pathway in Pseudomonas putida F1. Mol Gen Genet. 1995;246:570–579. doi: 10.1007/BF00298963. [DOI] [PubMed] [Google Scholar]
- Xu YQ, Chen M, Zhang W, Lin M. Genetic organization of genes encoding phenol hydroxylase, benzoate 1,2-dioxygenase alpha subunit and its regulatory proteins in Acinetobacter calcoaceticus PHEA-2. Curr Microbiol. 2003;46:235–240. doi: 10.1007/s00284-002-3840-4. [DOI] [PubMed] [Google Scholar]
- Xun L, Webster CM. A monooxygenase catalyzes sequential dechlorinations of 2,4,6-trichlorophenol by oxidative and hydrolytic reactions. J Biol Chem. 2004;279:6696–6700. doi: 10.1074/jbc.M312072200. [DOI] [PubMed] [Google Scholar]