Abstract
N-myristoyltransferases (NMT) add myristate to the NH2 termini of certain proteins, thereby regulating their localization and/or biological function. Using RNA interference, this study functionally characterizes the two NMT isozymes in human cells. Unique small interfering RNAs (siRNA) for each isozyme were designed and shown to decrease NMT1 or NMT2 protein levels by at least 90%. Ablation of NMT1 inhibited cell replication associated with a loss of activation of c-Src and its target FAK as well as reduction of signaling through the c-Raf/mitogen-activated protein kinase/extracellular signal-regulated kinase kinase/extracellular signal-regulated kinase pathway. Terminal deoxynucleotidyl transferase–mediated dUTP nick end labeling assays showed that depletion of either NMT isozyme induced apoptosis, with NMT2 having a 2.5-fold greater effect than NMT1. Western blot analyses revealed that loss of NMT2 shifted the expression of the BCL family of proteins toward apoptosis. Finally, intratumoral injection of siRNA for NMT1 or for both NMT1 and NMT2 inhibited tumor growth in vivo, whereas the same treatment with siRNA for NMT2 or negative control siRNA did not. Overall, the data indicate that NMT1 and NMT2 have only partially overlapping functions and that NMT1 is critical for tumor cell proliferation.
Introduction
The mature forms of many proteins contain various covalent modifications, including lipids, phosphates, acetyl groups, formyl groups, and nucleotides. The structural changes resulting from these modifications have effects on the stability, cellular location, and biological activity of the proteins (1, 2). These processes are becoming increasingly important for the study of cancer, as several key oncoproteins require this type of “post-translational maturation” for their biological active and for their ability to transform cells (3, 4). The Src family of tyrosine kinases (e.g., c-Src, Yes, and Fyn) is an example of oncogenic molecules that require a myristate moiety attached to their NH2 terminus in order for them to function in cells. This was first shown in studies showing that the viral oncogene product v-Src requires myristoylation for membrane binding and cellular transformation (5–7). Mutation of the NH2-terminal myristoylated glycine residue of Src-related tyrosine kinases blocks myristoylation and inhibits the ability of these proteins to transform cells without affecting their intrinsic kinase activity (7, 8).
The roles of the Src family of tyrosine kinases have been explored in several transformed cell lines, and these proteins are central intermediates in conveying both extracellular and intracellular signals to downstream effectors (9, 10). Interestingly, activating mutations in c-src are rare and have only been found in a small subset of advanced colon cancers and in a single endodermal cancer. However, high levels of Src activity are common in cancer as a result of a variety of Src stimulators (11, 12). Once Src activity is increased, it is involved in the migration, proliferation, adhesion, and angiogenesis of the affected cells (11). A large body of evidence has accumulated regarding the expression and activation of Src in several tumor types, including colon, breast, and ovarian tumors. Unfortunately, there is no single activator in these diseases and the transformation of the cells is mediated by an unmutated cellular protein (c-Src), making it difficult to design a therapeutic agent for their treatment. However, the fact that c-Src is a key regulator of cellular transformation and progression also presents an opportunity for therapeutic manipulation.
Recently, it has been shown that the enzyme myristoyl-CoA:protein N-myristoyltransferase (NMT) that is involved in the post-translational modification of c-Src is also overex-pressed in colon tumor cells and human brain tumors (13–17). Further, inhibiting the myristoylation of Src in colon cancer cell lines prevents the localization of the kinase to the plasma membrane and results in decreased colony formation and cell proliferation (18). Because NMTs and Src are overexpressed in colonic tumors (16), NMT inhibitors have the potential to be an important advance in colon cancer therapeutics.
N-myristoylation involves the covalent attachment of myristate, a 14-carbon saturated fatty acid, to the NH2-terminal glycine residue of specific proteins. NMT is responsible for this activity in eukaryotic cells and works by modifying its polypeptide substrate after the removal of the initiator methionine residue by methionyl aminopeptidase (19, 20). This modification occurs primarily as a cotranslational process (21, 22), although myristoylation can also occur post-translationally (23–25). Two isozymes of the mammalian NMT enzymes have been cloned and are designated NMT1 and NMT2. The two human NMT enzymes share ~77% identity (26), with the majority of divergence occurring in the NH2-terminal domains. Splice variants of NMT1 have also been observed in some cells. It is thought that these NH2-terminal differences may allow differential cellular localization of the isozymes, thereby allowing either cotranslational ribosome-based or post-translational cytosol-based protein myristoylation. An in vitro comparison of the activity of NMT1 and NMT2 toward a small panel of substrate peptides indicated that the isozymes have similar, but distinguishable, relative selectivity (26, 27). However, there has been few published demonstration of differential functions of NMT isozymes in mammalian cells. In fact, only as this work was coming to a close was there a report published where both NMT1 and NMT2 were considered (28). In this report, the authors conclude that NMT2 is not active in embryonic stem cells but they also show that NMT2 levels increase during development. This report also shows the distribution of both isozymes in normal tissue. Unfortunately, they do not pursue the function of NMT2 either in the fetal mouse or in the pups.
To elucidate the roles of the NMT isozymes in human cells, small interfering RNAs (siRNA) have been used to selectively knockdown the expression of NMT1, NMT2, or both isozymes in human colon cancer HT-29 cells and ovarian carcinoma SK-OV-3 cells. This study reports siRNA sequences unique to each NMT message that selectively reduce the expression of the NMT1 or NMT2 isozyme by >90% for at least 72 hours. With these reagents, we have shown that NMT1 and NMT2 have both redundant and unique effects on protein processing, apoptosis, and cell proliferation. Consequently, these siRNAs provide novel tools to determine the molecular mechanisms by which the individual NMT enzymes function to produce their cellular effects in mature organisms.
Results
Ablation of NMT1 and NMT2 Using siRNAs
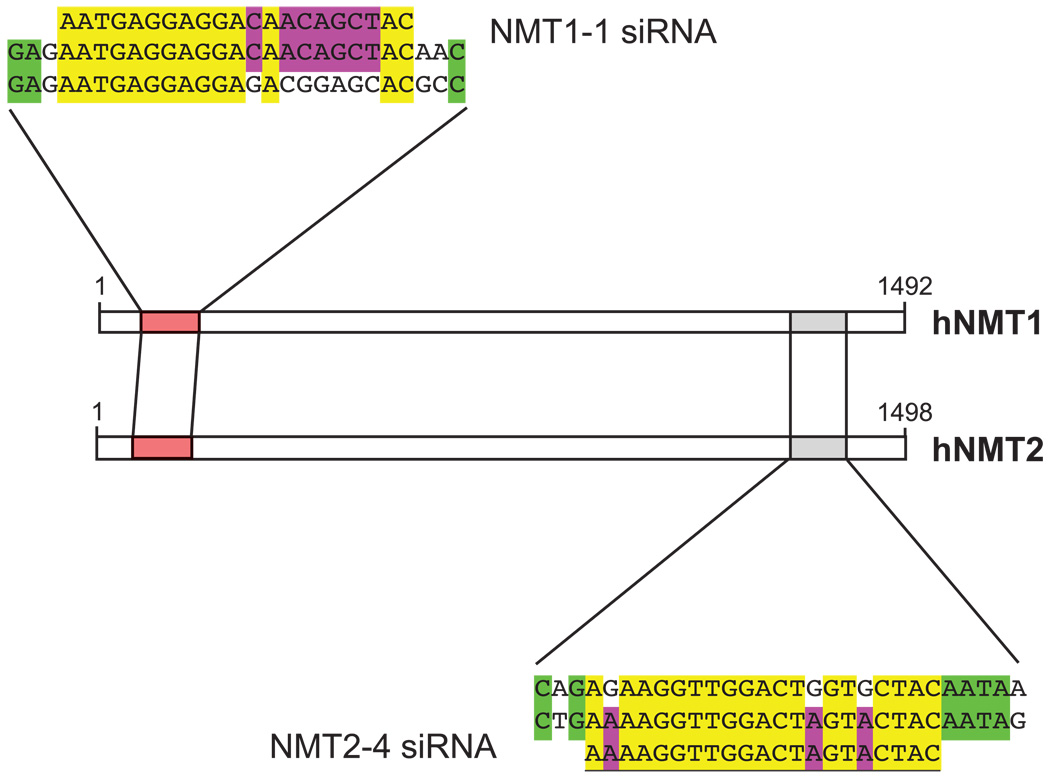
In all fungi and nematodes that have been examined, the single NMT enzyme is essential for viability. However, this question has not been addressed for the two human isozymes. Therefore, we sought to selectively abrogate the expression of the two NMT isozymes using siRNA to assess the roles of NMT1 and NMT2 in human cells. Target sites for siRNA constructs for both NMT1 and NMT2 were located and analyzed using the Ambion (Austin, TX) siRNA Target Finder. Of the numerous potential sites in each NMT gene, four were chosen so that they were distributed relatively evenly throughout the coding sequences and were checked to ensure specificity using BLAST searches against the available public databases. Preliminary experiments identified the siRNA constructs that produced the greatest reduction in NMT protein levels (NMT1-1 and NMT2-4), and these reagents were used for subsequent transfection experiments (Fig. 1).
FIGURE 1.
siRNA constructs. The sense strand sequences for the NMT1-1 and NMT2-4 siRNAs are shown with the alignment of the cDNA of both enzymes. NMT1-1 represents nucleotides 103 to 124 of NMT1 and NMT2-4 represents nucleotides 1,471 to 1,492 of NMT2.
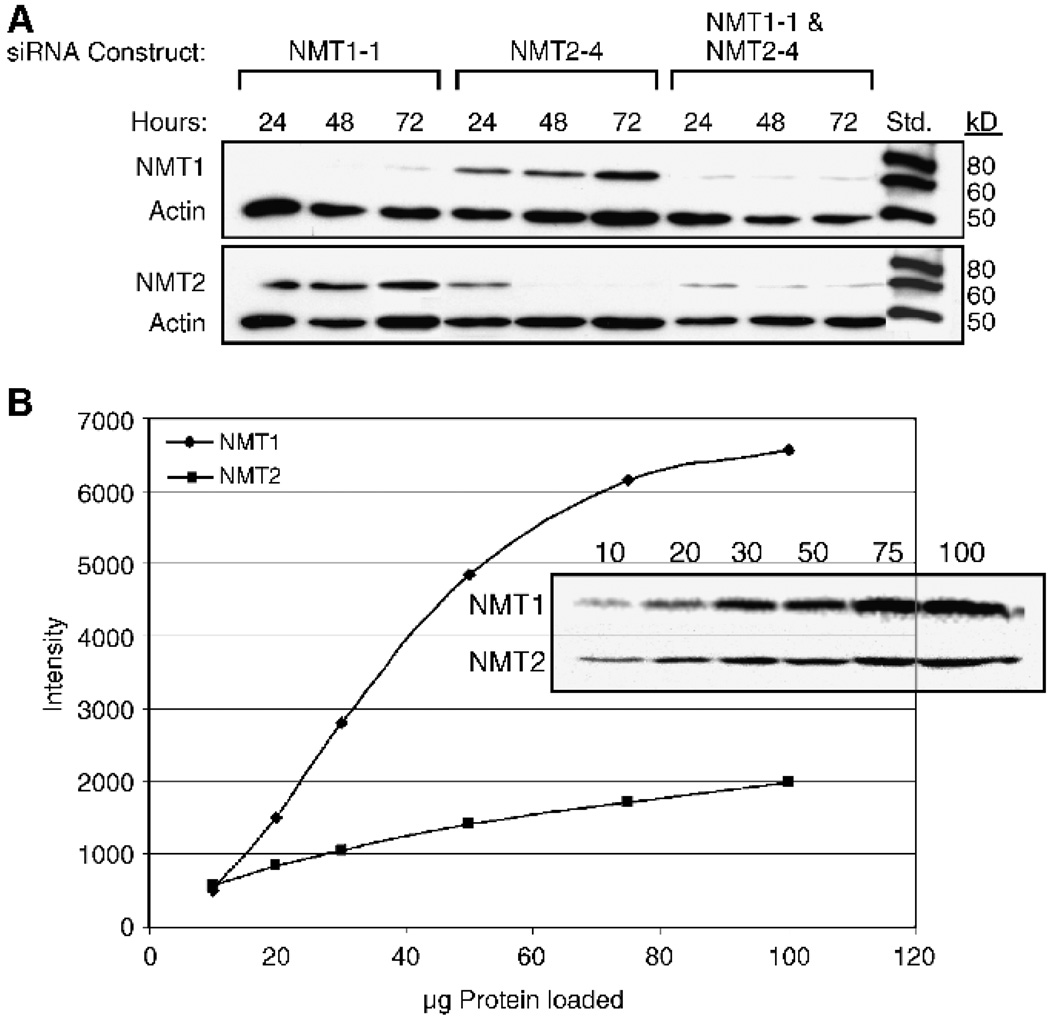
The duration of the knockdown of NMT1 and NMT2 in SK-OV-3 cells is indicated in Fig. 2A. As shown in this Western blot analysis, the NMT1-1 siRNA reduced the levels of NMT1 protein to below detectable limits within 24 hours, and the effect persisted for at least 72 hours. The NMT2-4 siRNA reduced the levels of NMT2 protein by ~75% at 24 hours, and complete ablation was seen at 48 and 72 hours. The double knockdown using both siRNAs significantly reduced the levels of both NMT1 and NMT2 at 24 hours and persisted for at least 72 hours. However, it is interesting to note that levels of NMT2 were not fully ablated when NMT1 was absent, suggesting the need to maintain some level of NMT activity. It was also apparent in these experiments that the cells undergoing the double siRNA treatment did not proliferate as fast as those that were untreated and in fact were dying. Figure 2B shows a titration of whole cell lysates probed with either NMT1 or NMT2 antibodies. A comparison of the signal for each band shows that the quantity of protein (50 µg) used for quantifying the level of NMT1 and NMT2 knockdown is in the linear range of the signal-to-antigen ratio.
FIGURE 2.
Time course of NMT knock-down. A. SK-OV-3 cells were treated with 10 nmol/L NMT1-1 siRNA, 10 nmol/L NMT2-4 siRNA, or 10 nmol/L each of NMT1-1 and NMT2-4 siRNA for 48 hours as described in Materials and Methods. Western blots were prepared using 50 µg total protein harvested from the cells and were first probed with either anti-NMT1 or anti-NMT2 antibodies, stripped, and then reprobed with a pan-actin antibody. The standards are Magic Mark Western standards (Invitrogen) that are visualized by anti-mouse horseradish peroxidase – conjugated secondary antibodies. B. A titration of HT-29 total cell protein (10–100 µg) probed for NMT1 and NMT2 and quantified on the Fujifilm imager. The graph shows the signal intensity of each band versus the concentration of total cell protein loaded for both NMT1 and NMT2 blots.
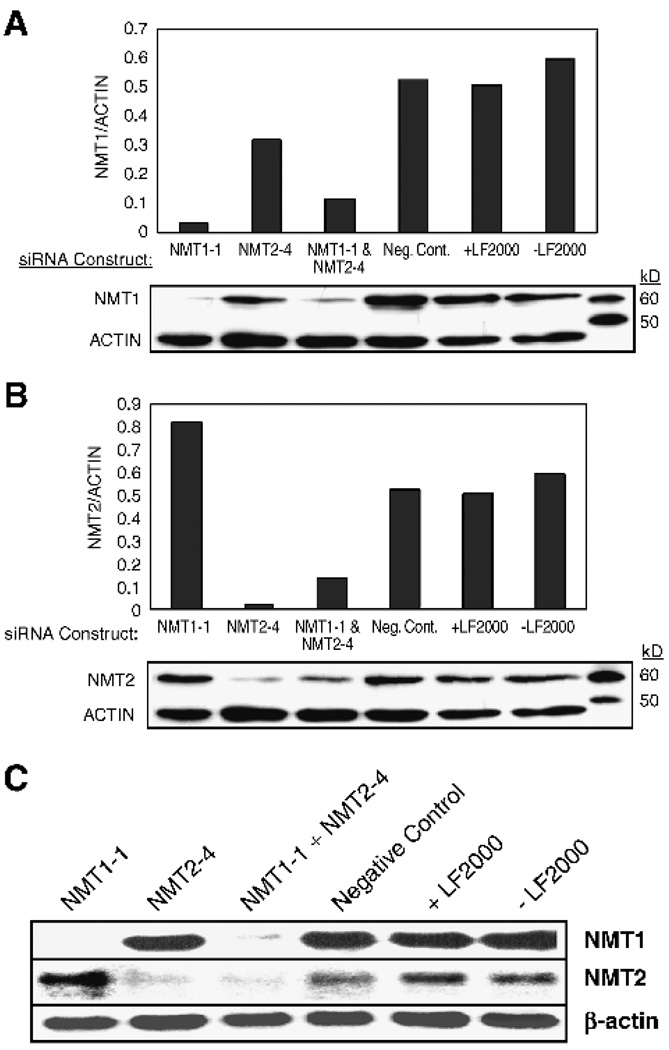
To quantify the effects of the siRNA treatments, additional experiments were done with total cellular protein and RNA being collected at 48 hours after transfection. In these experiments, images were captured using a CCD imaging system. β-Actin protein and mRNA were used to normalize for small variations in gel loading. Figure 3 shows the quantification of Western blots of total protein from cells treated with 10 nmol/L NMT1-1 siRNA, 10 nmol/L NMT2-4 siRNA, 10 nmol/L each of NMT1-1 and NMT2-4, 20 nmol/L negative control siRNA, and cells treated with no siRNA with and without the transfection reagent, LipofectAMINE 2000, and then probed with antibodies against human NMT1 (Fig. 3A) or NMT2 (Fig. 3B). In cells treated with NMT1-1 siRNA, the levels of NMT1 protein were reduced by 93% and levels of NMT2 protein were increased by 60% compared with negative control siRNA-treated cells or cells treated with only Lipofect-AMINE 2000. In cells treated with NMT2-4 siRNA, the levels of NMT2 protein were reduced by 95% and levels of NMT1 protein were reduced by 25% compared with negative controls. Finally, in cells treated with both NMT1-1 and NMT24 siRNA, levels of NMT1 protein were reduced by 80% and levels of NMT2 protein were reduced by 75% compared with controls. Northern blot analyses were conducted on the same cells and are shown in Fig. 3C. There was an excellent correlation between the mRNA levels of NMT1 and NMT2 and the protein levels observed in the Western blot analyses. Interestingly, NMT2 mRNA levels were increased in the NMT1-1 siRNA-treated cells to the same level as the protein, showing that the increase in NMT2 protein is due to an increase in mRNA levels and not to an increase in protein stability. The Northern blot also allows for an estimation of the relative abundance of NMT1 and NMT2 mRNA in the cell. There was approximately five times more NMT1 message than NMT2 message in untreated (no siRNA and no LipofectAMINE 2000) cells. Effective knockdown of NMT1 and NMT2 in HT-29 cells was achieved with NMT1-1 and NMT2-4 siRNAs; however, 40 nmol/L of each siRNA construct were required for efficient transfection of these cells (data not shown).
FIGURE 3.
Quantification of NMT knockdown. SK-OV-3 cells were treated with 10 nmol/L NMT1-1 siRNA, 10 nmol/L NMT2-4 siRNA, 10 nmol/L each of NMT1-1 and NMT2-4 siRNA, 20 nmol/L negative control siRNA (Neg. Cont.), or no siRNA in the absence and presence of LipofectAMINE 2000 (LF2000) for 48 hours as described in Materials and Methods. Western blots were prepared using 50 µg total protein harvested from cells and were first probed with either anti-NMT1 (A) or anti-NMT2 antibodies (B), stripped, and then reprobed with a pan-actin antibody. The standards are Magic Mark Western standards that are visualized by anti-mouse horseradish peroxidase – conjugated secondary antibodies. The graphs represent the ratio of NMT1 or NMT2 to actin in each lane. Northern blots (C) were prepared using 10 µg mRNA per lane from the same cell treatments and were probed with NMT1, NMT2, and β-actin probes as described in Materials and Methods.
Effects of NMT1 and NMT2 Ablation on the Localization of N-Myristoylated Green Fluorescent Protein
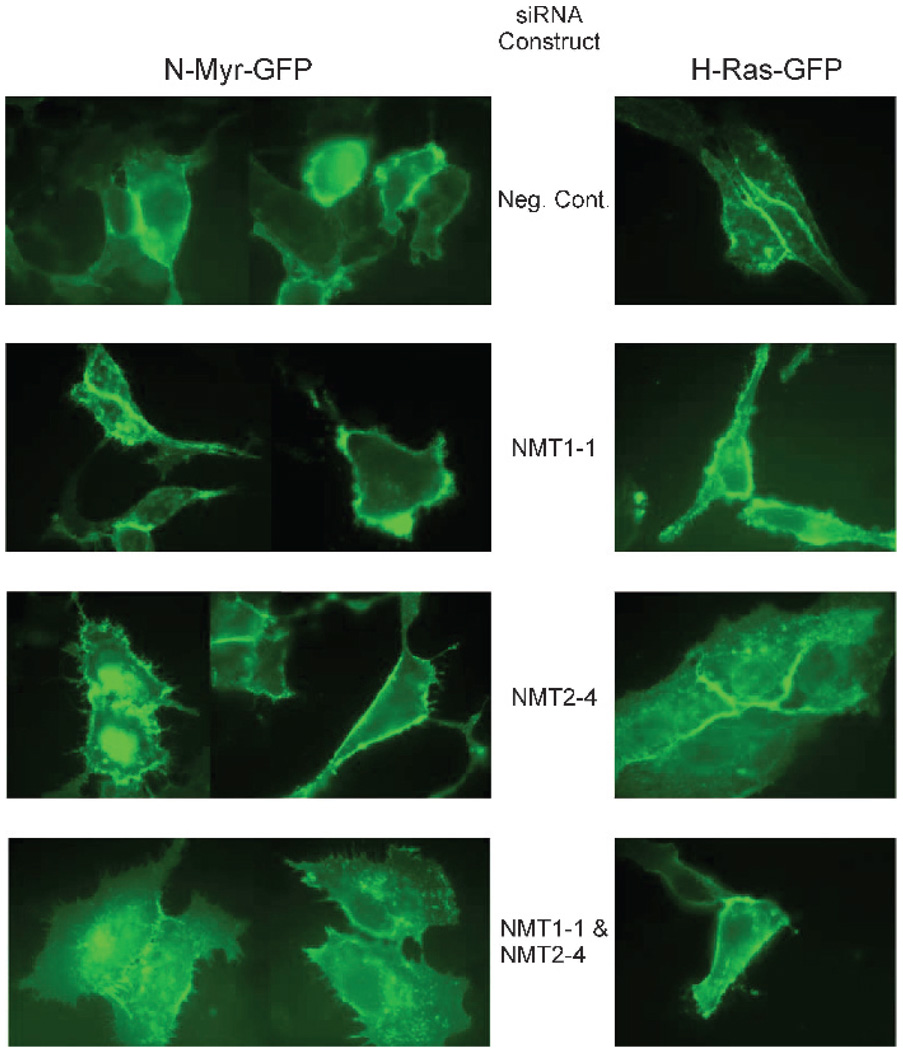
As Src-related tyrosine kinases (e.g., Fyn) are translated, the NH2-terminal methionine is removed by methionyl amino-peptidase and the newly formed NH2-terminal glycine is N-myristoylated by NMT. Members of this family, other than c-Src, are then palmitoylated on a cysteine residue near the myristoylated NH2 terminus. These modifications are necessary for the maturation and functionality of these proteins because blocking N-myristoylation abolishes their proper localization to the inner leaflet of the plasma membrane (29, 30). We have shown previously that fusing the NH2-terminal sequence of Fyn to green fluorescent protein (GFP; N-myr-palm-GFP) directs GFP to be myristoylated and palmitoylated resulting in its localization in the plasma membrane and that relocalization of GFP to the cytosol occurs when the protein is mutated so that myristoylation cannot occur (31). This reporter construct was used to determine if removal of endogenous NMT1 and/or NMT2 affects the subcellular localization of the modified GFP. A second GFP construct, GFP-farn-palm, containing the COOH-terminal sequence of H-Ras thereby directing it to be farnesylated and palmitoylated, was used as a control because this fusion protein localizes to the plasma membrane in a myristoylation-independent manner. Figure 4 shows SK-OV-3 cells transiently transfected with N-myr-palm-GFP or GFP-farn-palm and NMT1-1 siRNA, NMT2-4 siRNA, both NMT1-1 and NMT2-4, or negative control siRNA. As expected, normal localization of both fusion proteins to the plasma membrane occurred in cells treated with the negative control siRNA. The localization of the N-myr-palm-GFP was not affected by treatment of the cells with either NMT1-1 or NMT2-4 siRNA individually. However, the double knockdown clearly resulted in blockage of GFP localization to the plasma membrane. In contrast, the localization of GFP-farn-palm to the plasma membrane was not affected by either the single knockdowns or the double NMT knockdown. This suggests that NMT1 and NMT2 are each capable of processing this NH2-terminal sequence of Fyn fused to this GFP.
FIGURE 4.
Inhibition of N-myristoylated GFP processing by NMT siRNAs. N-myr-palm-GFP and GFP-farn-palm expression constructs were cotransfected into SK-OV-3 cells with the indicated siRNAs as described in Materials and Methods. After 48 hours, the subcellular localization of the GFP was visualized by fluorescence microscopy.
Effects of NMT1 and NMT2 Ablation on Cell Proliferation and Apoptosis
As indicated above, there were always fewer cells in the NMT siRNA-treated cultures than in untreated cultures or cultures treated with the negative control siRNA. To determine the mechanism of the decrease in cell number, cell replication and survival were examined in NMT1-1 and NMT2-4 siRNA-treated cells. First, [3H]thymidine incorporation into DNA was monitored to determine if the knockdowns of NMT1, NMT2, or both had an effect on cell proliferation. In this assay, cells that are actively dividing will incorporate the labeled nucleoside into DNA, whereas cells that are not replicating will not. By measuring the amount of 3H incorporated into DNA, it is possible to assess the number of cells actively undergoing DNA replication. Equal numbers of SK-OV-3 cells were treated with either 10 nmol/L NMT1-1 siRNA, 10 nmol/L NMT2-4 siRNA, 10 nmol/L each of NMT1-1 and NMT2-4, or 20 nmol/L negative control siRNA, incubated for 48 hours, and then pulsed for 2 hours with [3H]thymidine. This experiment was done twice in triplicate with the final numbers representing the mean ± SD of both sets of replicates. [3H]Thymidine incorporation into DNA in SK-OV-3 cells treated with NMT1-1 siRNA was inhibited 27 ± 3%, whereas cells treated with NMT2-4 siRNA were inhibited 2 ± 4% and cells treated with both NMT1-1 and NMT2-4 siRNA were inhibited 29 ± 12%. These results indicate that NMT1, but not NMT2, is involved in the control of cellular replication. Similar results were obtained from experiments done in HT-29 cells using 40 nmol/L of each siRNA construct, indicating that this phenomenon is not unique to the SK-OV-3 cells (data not shown).
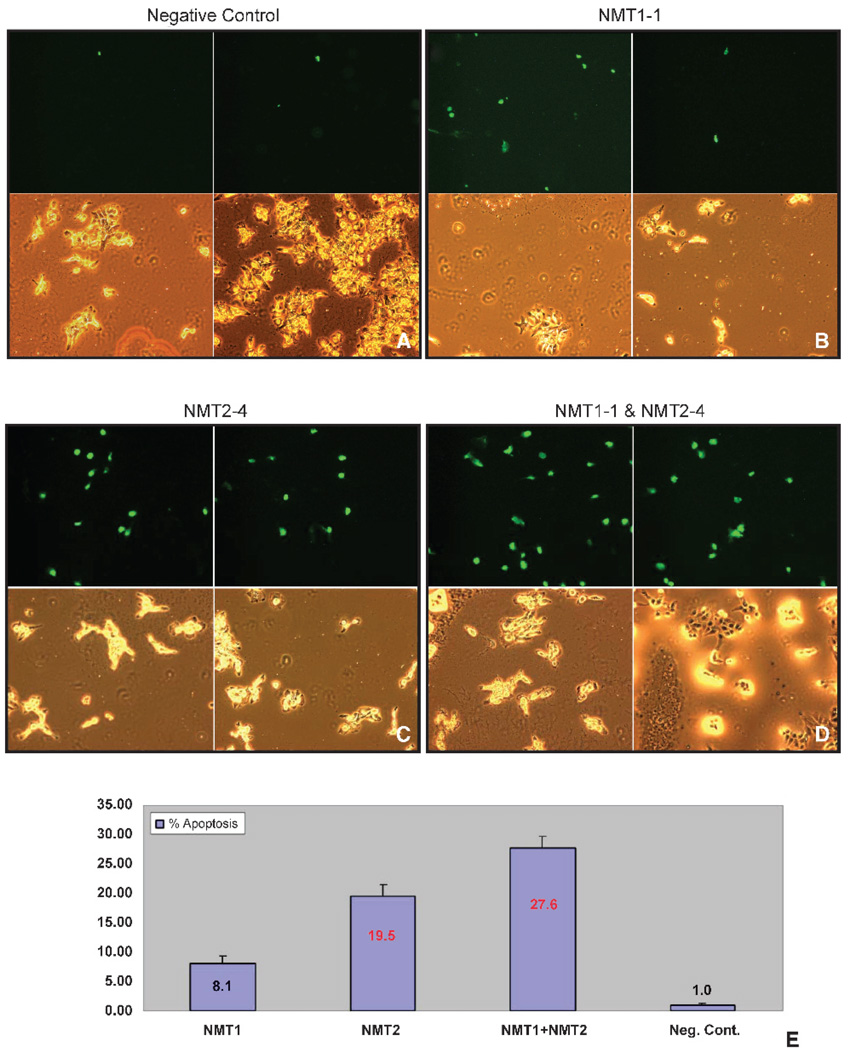
Second, the effects of the siRNAs on the percentage of cells undergoing apoptosis were determined using the terminal deoxynucleotidyl transferase (TdT)–mediated dUTP nick end labeling assay. Cells undergoing apoptosis accumulate double-stranded breaks in their DNA and these “free ends” can be labeled with fluorescein-conjugated dUTP through the action of TdT and visualized by microscopy. As above, SK-OV-3 cells were treated with 10 nmol/L NMT1-1 siRNA, 10 nmol/L NMT2-4 siRNA, 10 nmol/L each of NMT1-1 and NMT2-4, or 20 nmol/L negative control siRNA. After 48 hours, the cells were fixed and double-stranded breaks in the DNA were labeled. The experiment was done in triplicate and 15 fields were counted for each slide of three replicates, with the final fold changes representing all 45 fields ± SD. Figure 5 shows representative matched bright-field and fluorescent images of cells treated with NMT1-1, NMT2-4, both NMT1-1 and NMT2-4, or negative control siRNA. The images clearly show that the double knockdown of NMT1 and NMT2 had the most dramatic effect on the cells, with ~30% of the cells undergoing programmed cell death. Interestingly, both NMT1 and NMT2 seem to contribute to this phenotype because the depletion of NMT1 caused an 8-fold increase in apoptosis and the depletion of NMT2 caused a 19-fold increase in apoptosis compared with the control cells.
FIGURE 5.
Induction of apoptosis by NMT siRNAs. SK-OV-3 cells were treated with 20 nmol/L negative control siRNA (A), 10 nmol/L NMT1-1 siRNA (B), 10 nmol/L NMT2-4 siRNA (C), or 10 nmol/L each of NMT1-1 and NMT2-4 siRNA (D) for 48 hours as described in Materials and Methods and then scored for apoptosis using the TdT-mediated dUTP nick end labeling assay. Panels are matched sets of bright field (bottom) and fluorescent (TdT-mediated dUTP nick end labeling positive; top) images for each treatment. E. Graphical representation of the percentage of apoptotic cells in each treatment condition. Negative control – treated cells had an average of 1.0 ± 0.3% of apoptotic cells. NMT11-treated cells had an average of 7.8 ± 1.1% of apoptotic cells. NMT2-4-treated cells had an average of 19.0 ± 2.0% of apoptotic cells. NMT1-1- and NMT2-4-treated cells had an average of 29.8 ± 2.0% of apoptotic cells.
Effects of NMT1 and NMT2 Ablation on Signaling Proteins
Microarray analyses were done to examine the molecular pathways that NMT1 and NMT2 impact to affect the observed changes in replication and apoptosis. These experiments were done with RNA isolated from HT-29 cells that had been treated with 40 nmol/L NMT1-1, 40 nmol/L NMT2-4, or 40 nmol/L negative control siRNA (data not shown). HT-29 cells were used in these assays because it has been shown that SK-OV-3 cells are not optimal for investigating intracellular signaling due to their constitutive activation of the phosphatidylinositol 3-kinase pathway and their modest response to cellular perturbations (32, 33). The data revealed several interesting findings that were further analyzed in protein expression studies shown in Figs. 6 and 7.
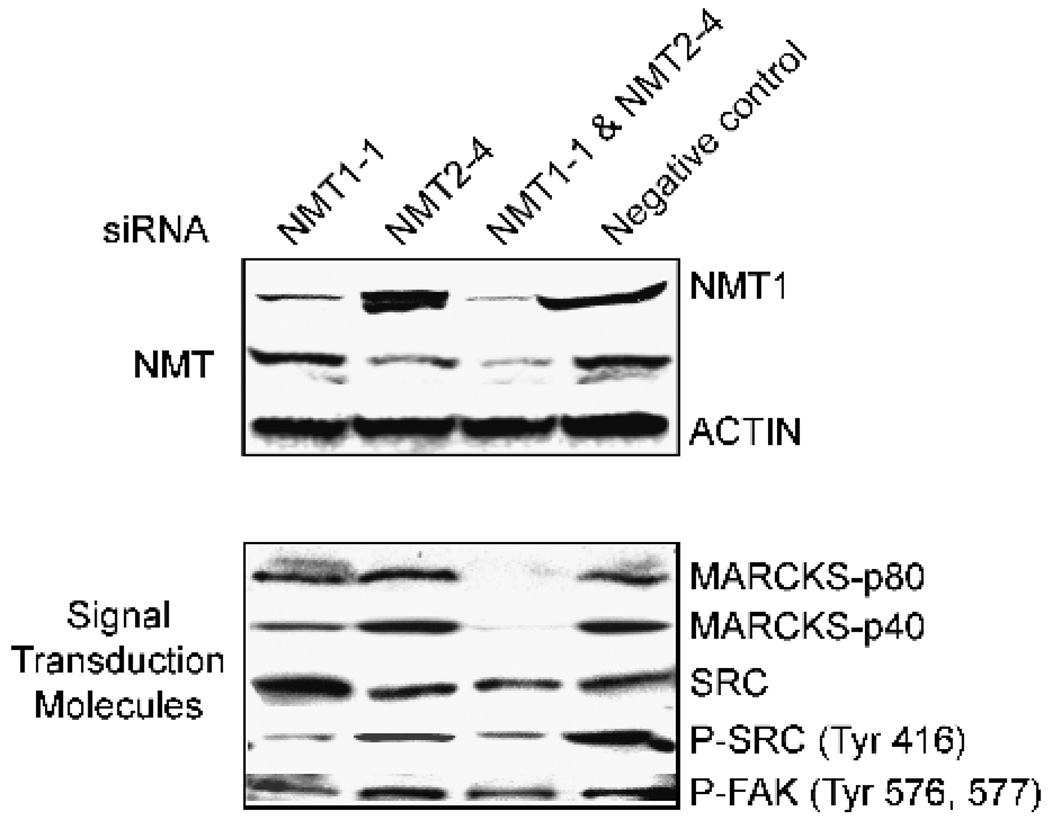
FIGURE 6.
Effects of NMT siRNAs on MARCKS and c-Src. HT-29 cells were treated with 40 nmol/L NMT1-1 siRNA, 40 nmol/L NMT2-4 siRNA, 40 nmol/L each of NMT1-1 and NMT2-4 siRNA, or 80 nmol/L negative control for 48 hours as described in Materials and Methods. Western blots were prepared using 50 µg total cellular protein. The p80 signal in the MARCKS blot is the intact cellular protein, whereas the p40 band is the cleaved COOH terminus of the protein. See Table 1 for a complete list of antibodies used in these studies.
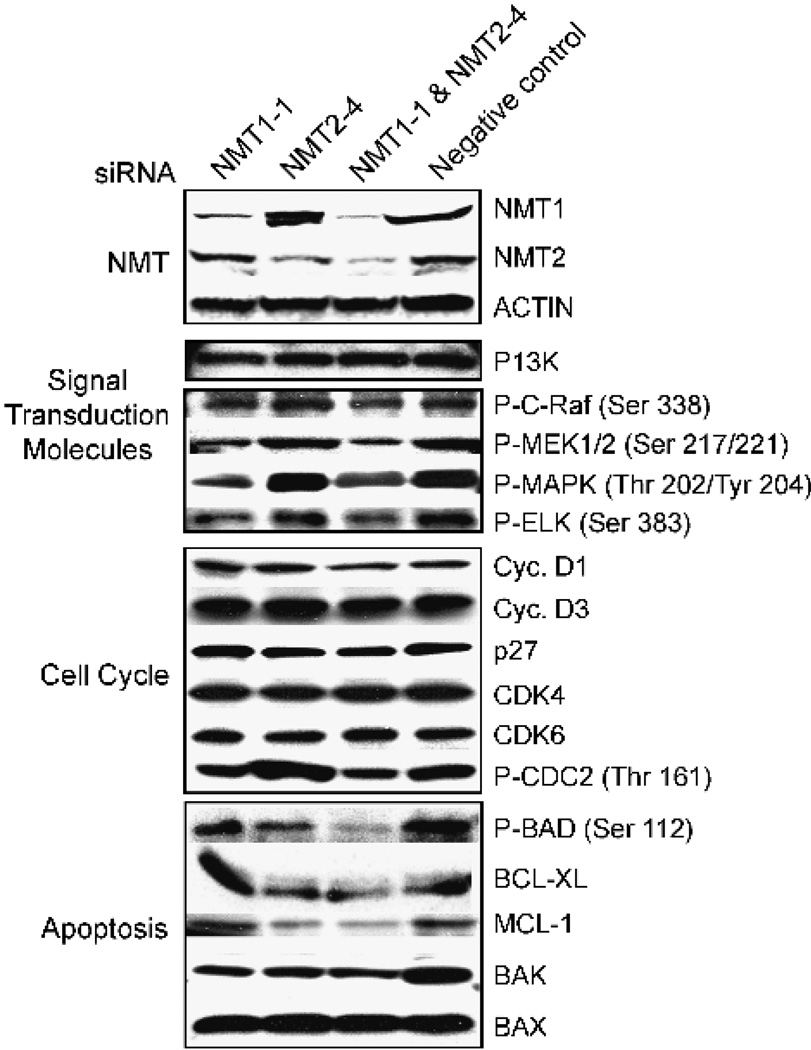
FIGURE 7.
Effects of NMT siRNAs on signal transduction, cell cycle, and apoptosis proteins. HT-29 cells were treated with 40 nmol/L NMT1-1 siRNA, 40 nmol/L NMT2-4 siRNA, 40 nmol/L each of NMT1-1 and NMT2-4 siRNA, or 80 nmol/L negative control for 48 hours as described in Materials and Methods. Western blots were prepared using 50 µg total cellular protein. See Table 1 for a complete list of antibodies used in these studies.
As with the SK-OV-3 cells, the siRNAs NMT1-1 and NMT2-4 promoted selective depletion of NMT1 and NMT2, respectively. The extent of knockdown in the HT-29 cells was slightly lower than in the SK-OV-3 cells; however, the 40 nmol/L doses were sufficient to perturb signaling pathways as described below. In the double knockdown, cellular NMT levels were decreased by >85%. The effects of NMT ablation on the expression of the myristoylated proteins, myristoylated alanine-rich C kinase substrate (MARCKS) and Src, are shown in Fig. 6. Surprisingly, the microarray analyses indicated that c-Src message was up-regulated in the NMT1 knockdown cells compared with the control cells. Western blot analyses of total c-Src protein confirmed that the protein levels are increased in the NMT1 knockdown (Fig. 6). In contrast, levels of activated (i.e., phosphorylated) c-Src were down-regulated in the NMT1 knockdown cells as well as in the double knockdown cells. Because activated c-Src binds to and phosphorylates FAK at residues 576 and 577 (34, 35), the effects of the NMT siRNAs on phospho-FAK levels were also examined. The Western blot in Fig. 6 shows that, when c-Src phosphorylation was reduced by ablation of NMT1, FAK phosphorylation was also reduced. These results suggest that NMT1 is responsible for myristoylation of c-Src and that loss of c-Src myristoylation and activity leads to down-regulation of FAK activity.
A second interesting finding from the microarray analyses was the differential regulation of the MARCKS protein by the knockdown of NMT1 versus NMT2. This protein is of interest for two reasons. First, MARCKS translocation between the plasma membrane and the cytosol (36) has been suggested to mediate destabilization of the cytoskeleton by cross-linking actin filaments at the plasma membrane (37). The activation of MARCKS by phosphorylation and myristoylation has also been shown to be involved in cellular adhesion, cell spreading, and vesicle trafficking, which could play a role in the metastasis of some cancers. Second, the change in total MARCKS protein provides a cellular marker that can be used as a indicator of the inhibition of the NMT1 isozyme. Levels of message for MARCKS were down-regulated in the NMT1 knockdown cells relative to control cells. Western blot analyses of total MARCKS protein confirmed that the protein levels are reduced in the NMT1 knockdown and the double knockdown (Fig. 6). The myristoylated form of MARCKS, which migrates with an apparent molecular weight of 80 kDa, is clearly reduced in the NMT1 knockdown cells and barely visible in the double knockdown cells. MARCKS can be cleaved by calpain to produce a fragment that migrates with an apparent molecular weight of 40 kDa, and this cleavage has been shown to increase the rate of MARCKS-induced polymerization of actin (38). Levels of the p40 MARCKS product were also reduced in the NMT1 knockdown and double knockdowns.
The microarray data also indicated changes in the c-Raf/mitogen-activated protein kinase/extracellular signal-regulated kinase (ERK) kinase (MEK)/ERK/Elk signaling pathway on ablation of the NMTs. In depicted in Fig. 7, Western blot analyses showed that levels of phospho-c-Raf, phospho-MEK, and phospho–mitogen-activated protein kinase were reduced in cells treated with the NMT1-1 siRNA, whereas treatment with the NMT2-4 siRNA had no effect. In the same experiments, phosphatidylinositol 3-kinase protein and phospho-Akt levels were unaltered in any of the siRNA treatments. The maintenance of the c-Raf/MEK/ERK/Elk pathway in the NMT2-ablated cells is consistent with the proliferation data presented above. Interestingly, both c-Src activation and signaling through the c-Raf/MEK/ERK/Elk pathway have been implicated in the promotion of cellular replication (39, 40). The modulation of the c-Src activity profile in Fig. 6 is consistent with its activation of the c-Raf/MEK/ERK/Elk pathway in these cells.
Figure 7 shows Western blots using a panel of antibodies against a selection of proteins that regulate the cell cycle. These data indicate that the cells are not arrested in any particular stage of the cell cycle, as the signals for cyclin D1, cyclin D3, p27, CDK4, CDK6, and phospho-CDC2 were equal in all of the treatments. Fluorescence-activated cell sorting analyses of siRNA-treated cells confirmed the Western blot data in that similar cell cycle distributions were observed under all treatment conditions (data not shown). These data serve to strengthen the argument for the specificity of the siRNA knockdowns, as they are not affecting the expression of multiple “off-target” proteins.
Finally, the microarray studies suggested that levels of the BCL-2 family of proteins are differentially regulated in the NMT knockdowns. This was of interest as the BCL-2 family of proteins is involved in regulating apoptosis by controlling mitochondrial permeability and the release of cytochrome c (41). Consistent with the TdT-mediated dUTP nick end labeling data presented in Fig. 5, the antiapoptotic proteins BCL-XL and MCL-1 were down-regulated in the NMT2 and double knockdown (Fig. 7). Phosphorylation of BAD was also down-regulated in these conditions, with strong suppression occurring in the double knockdown cells. Removal of the phosphorylation signal from BAD allows it to complex with the BCL-XL protein to promote apoptosis, and this is consistent with the marked induction of apoptosis in the double knockdown cells. The levels of the proapoptotic protein BAK were reduced ~70% in all three knockdown conditions, whereas the levels of the proapoptotic protein BAX remained relatively unchanged by the siRNA treatments. As the effects of BAX are regulated by changes in its subcellular localization, it was not necessary to elevate the cellular protein levels to promote apoptosis. These data, in conjunction with the TdT-mediated dUTP nick end labeling results, indicate that NMT2 plays an important role in regulating the BCL-2 family of proteins and their ability to promote apoptosis in tumor cells.
Antitumor Activity of NMT1-1 and NMT2-4 siRNA
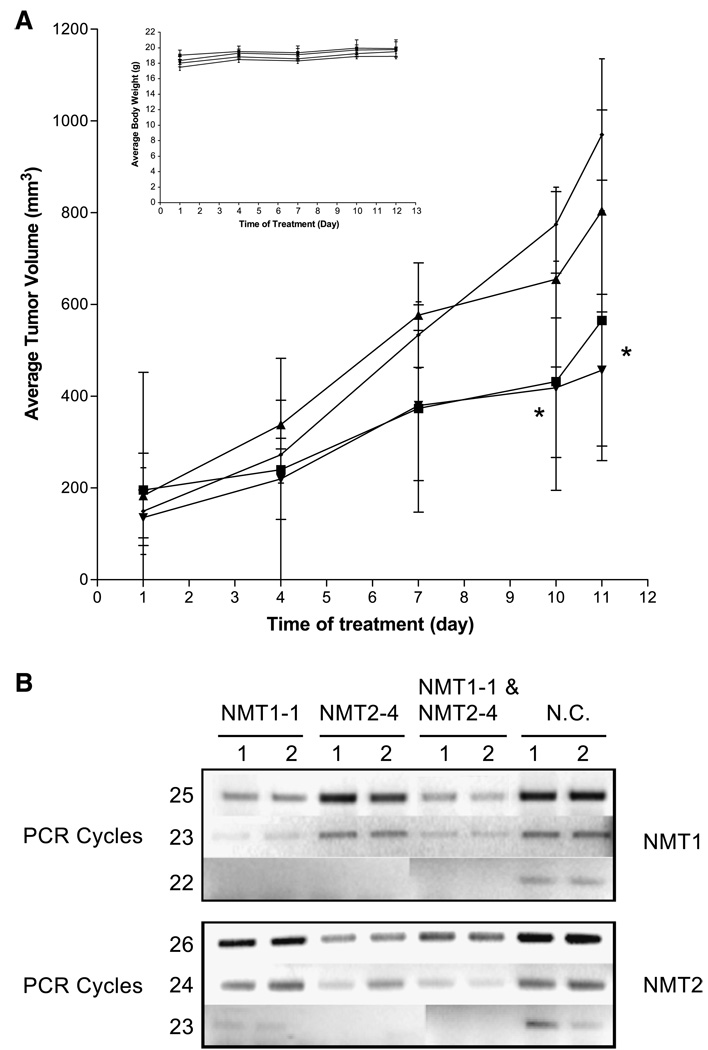
To determine if the ablation of either NMT1 or NMT2 had an effect on tumor progression in vivo, a syngeneic tumor model was used (42). Female BALB/c mice were injected with 1 × 106 JC (mammary adenocarcinoma) cells and palpable tumors were allowed to form. Mice were split into four groups of four mice each. Each group was treated with the designated siRNA with one-site intratumoral injection and tumor volumes were monitored (Fig. 8A). On days 1, 4, 7, and 10, group 1 was given 10 µmol/L NMT1-1 siRNA, group 2 was given 10 µmol/L NMT2-4 siRNA, group 3 was given 10 µmol/L NMT1-1 plus 10 µmol/L NMT2-4 siRNA, and group 4 was given 20 µmol/L negative control siRNA. On day 11, the mice were euthanized and the tumors were measured for a final time and then excised. As indicated in Fig. 8A, tumor growth in animals treated with both NMT1-1 and NMT2-4 siRNA was significantly lower (>50% decrease by day 11) than tumor growth in animals treated with control siRNA. The reduction in tumor growth in animals treated with NMT1-1 was almost as good (42% decrease by day 11) as the double treatment. Tumor growth was decreased by only 17% in animals treated with NMT2-4 and was not statistically significantly different from that of animals treated with the negative control siRNA. The body weights of animals in all groups increased at the same rate, indicating that the siRNA administration was not overtly toxic to the mice.
FIGURE 8.
Antitumor activity of NMT siRNAs. A. Female BALB/c mice received subcutaneous injections of 106 JC cells suspended in PBS. NMT1-1 (■), NMT2-4 (▲), NMT1-1 and NMT2-4 (▼), or negative control (♦) siRNAs were injected intratumorally on days 1, 4, 7, and 10, and tumor volumes were measured. Inset, body weights for the animals during this study. *, P < 0.05, unpaired t tests. B. RT-PCR analyses of NMT mRNA levels in siRNA-treated tumors. RT-PCR analyses were preformed using 1 µg total RNA isolated from the indicated tumors and primers described in Materials and Methods. NMT1 message was amplified 22, 23, or 25 cycles, whereas NMT2 was amplified 23, 24, or 26 cycles.
To determine if the siRNA treatments were knocking down the appropriate target in vivo, reverse transcription-PCR (RT-PCR) was done on total RNA isolated from two tumors from each group. RT-PCR was used in these experiments because the antibodies used in Figs. 2, 3, 6, and 7 react efficiently with human NMT1 or human NMT2 but do not react with the murine proteins in the JC tumor cells. Figure 8B shows that message levels for NMT1 or NMT2 were substantially reduced in tumors treated with NMT1-1 or NMT2-4 siRNAs, respectively. Because the NMT message and protein levels are well coordinated (Fig. 3), it is very likely that NMT protein expression in the tumors was also significantly depleted. Overall, these results suggest that ablating NMT1 is more effective in inhibiting tumor growth than is ablation of NMT2. Additionally, this confirms the potential efficacy of small-molecule inhibitors of NMT as an anticancer therapy.
Discussion
In studies of lower eukaryotes, their single NMT gene is essential for viability (43). Mammals possess two NMT genes; however, it is not presently known if these encode redundant or specialized isozymes. Given the critical cellular proteins that NMTs affect, such as Src-related oncoproteins, it is important to discern the specific functions of these two NMT isozymes. This may be important for the generation of antineoplastic therapies that target a particular NMT isozyme. Therefore, siRNAs have been used to knockdown NMT1, NMT2, or both NMT1 and NMT2 in ovarian and colon carcinoma cells to distinguish the roles of the two isozymes.
Using siRNA-induced selective ablation of NMT1 and NMT2, we have shown that there is cross-talk between the levels of NMT1 and NMT2 expression. Specifically, when NMT1 levels were reduced, NMT2 protein and mRNA levels rose, whereas when NMT2 levels were reduced NMT1 protein and mRNA levels remained unaffected. Although the carcinoma cells normally express more NMT1 message than NMT2 message, the data in Fig. 3 suggest that cells need to maintain a threshold level of NMT expression, because NMT2 could not be fully ablated when NMT1 was depleted. This observation is consistent with expression data in normal mouse tissue showing higher levels of NMT1 in most tissues examined (28). Although it has been shown previously that NMT2 protein levels can be altered by certain xenobiotics (44), this study is the first demonstration that the expression levels of NMT2 protein are linked to NMT1 protein levels. Although it is possible that NMT2 is part of the cell’s general stress response pathway, knocking down β-actin had no effect on NMT2 protein levels (data not shown). The mechanism for coordinated expression of the NMT isozymes is presently unknown, so it will be interesting to examine the NMT2 promoter region to determine if there are uncharacterized elements that mediate this coordinated regulation.
These studies have also shown that NMT1 and NMT2 have both unique and common substrates in cells. A preliminary examination of the abilities of NMT1 and NMT2 to modify a small set of substrate peptides indicated that they may have different specificities (26). The siRNAs described herein provide the tools to examine this issue in intact cells and have revealed an interesting dichotomy regarding the Src family of proteins. The N-myr-palm-GFP reporter used in these studies contains the NH2-terminal 14 – amino acid residues of Fyn (MGCVQCKTKLTEER) fused to GFP. After removal of the methionine residue, it is myristoylated and palmitoylated, allowing its stable association with the plasma membrane (45). Thus, if Fyn is a unique substrate of either NMT1 or NMT2, it was expected that siRNAs targeting these isozymes should reveal the modifying enzyme. However, the experiments clearly showed that individually reducing the expression of NMT1 or NMT2 had no effect on the membrane localization of this modified GFP. In contrast, simultaneously reducing the levels of both NMT1 and NMT2 resulted in redistribution of the Fyn-GFP to the cytosol without affecting the plasma membrane localization of the GFP-farn-palm protein that reaches the membrane in a myristoylationin-dependent manner. These results suggest that both NMT isozymes are able to modify the Fyn myristoylation motif. This finding is significant because it has been proposed that the specificities of these isozymes might result from differential subcellular localization through the divergent NH2 termini (46).
In contrast to the finding with the Fyn myristoylation signal, c-Src, which is myristoylated but not palmitoylated, seems to be primarily processed by NMT1. Although there is not yet evidence for a direct interaction between NMT1 and c-Src, the present data consistently indicate that c-Src is not myristoylated in cells that lack NMT1 and therefore cannot localize to the plasma membrane where it is activated. The increased protein levels of c-Src in the NMT1 knockdown cells may reflect an attempt to compensate for decreased c-Src activity. Further, these results show that loss of c-Src phosphorylation leads to the loss of FAK phosphorylation, and this is consistent with the requirement of c-Src to be myristoylated before its interaction with FAK at the plasma membrane. These results are also consistent with the finding that NMTs are overexpressed in colon cancer, which could underlie the increased Src activity observed in these and other tumors. It is possible that the increase in NMT1 protein levels drives all of the available c-Src to the plasma membrane, thereby increasing the local concentration of c-Src where it is required to activate downstream effectors.
The differential processing of Fyn and c-Src could identify sequence differences that make one a substrate for both NMT enzymes and the other specific for NMT1. This may reflect processing selectivity for proteins that are only myristoylated versus those that are further processed by palmitoylation. Further characterization of a more diverse set of GFP constructs, as well as examination of the myristoylation of endogenous cellular proteins, in the NMT knockdowns will be necessary to fully explain this issue.
Consistent with NMT1 modification of c-Src, data presented in this study indicate that NMT1, but not NMT2, is involved in cellular proliferation, because DNA synthesis was decreased with the NMT1-1 and double siRNA treatment but not with the NMT2-4 siRNA treatment. The Western blot data suggest that a critical NMT1 substrate is c-Src, which requires myristoylation for membrane association and its ability to transform cells (7, 47). Further, Src activation is involved in several aspects of cellular transformation, including proliferation, survival, migration, and invasion (48–50). Additionally, activity of the Src-family of kinases is required for signal transducers and activators of transcription 3 and FAK phosphorylation (51), and this phosphorylation is required for the progression of the cell cycle (52, 53). Furthermore, it has been reported that overexpression of FAK leads to an accelerated G1-S–phase transition, whereas overexpression of a dominant-negative FAK inhibits cell cycle progression at G1 (39, 40). These authors also reported that the dominant-negative FAK competed for binding to c-Src, which resulted in a decrease in ERK (mitogen-activated protein kinase) activation. These results are consistent with our findings that ablation of NMT1 leads to loss of c-Src and FAK activation as well as down-regulation of the c-Raf/MEK/ERK/Elk pathway. Zhao et al. have shown that disrupting FAK activation and down-regulating the c-Raf/MEK/ERK/Elk pathway leads to a slowing of cellular replication (54). The data presented above clearly show that replication is slowed when NMT1 activity is inhibited. Overall, these findings suggest that loss of NMT1 is responsible for slowing of the replication process due to the decreased activation of c-Src.
In contrast to the cellular replication, cell survival seems to be regulated by both NMT1 and NMT2. Consequently, simultaneous ablation of NMT1 and NMT2 was particularly lethal to the cells, producing 29-fold increase in the apoptosis index. This is consistent with experiments with fungi (55), Arabidopsis thaliana (56), and flies (57) that have shown that loss of all NMT activity is lethal to these organisms. Additionally, compounds that inhibit both NMT1 and NMT2 are cytotoxic to tumor cells (31). Western blot analyses of the BCL-2 family of apoptosis regulators showed that there is a shift toward induction of apoptosis when NMT2 is ablated from the cells. At the present time, there is no clear target protein for NMT2 that can be linked to this process. It is of interest to note that SK-OV-3 cells are p53 null and that HT-29 cells have elevated levels of p53, indicating that the effects of loss of both NMT1 and NMT2 are p53 independent and therefore may be useful for inducing apoptosis in a wide range of human tumors. Using the tools established in this study, it is now possible to explore the substrate specificity of the individual NMT isozymes to determine which protein(s) NMT2 regulates to affect this pathway.
Finally, in vivo studies consisting of intratumoral injection of siRNAs against the NMT isozymes indicate that pharmacologically reducing the activity of NMT1 will have antitumor activity. Although clearly not optimized in terms of delivery and schedule, the siRNAs provide preliminary proof-of-principle that inhibitors of NMT1 will be useful as therapeutic agents. This is consistent with the antitumor activity of small-molecule inhibitors of NMT that we have recently described (31). Interestingly, the data indicate that knocking down NMT2, which had the greatest effect on apoptotic induction in vitro, had little effect on tumor progression in vivo. This was not due to lack of in vivo ablation of NMT2, because the NMT1-1 and NMT2-4 siRNAs caused pronounced reductions in the message levels for NMT1 and NMT2, respectively. The rate of in vivo tumor growth reflects the relative rates of cell proliferation and apoptosis. The JC cells growing as xenografts represent an aggressive tumor model, with an in vivo doubling time of ~3 days (Fig. 8). Under these conditions, the rate of tumor growth is dominated by the rate of proliferation because the apoptotic index is very low. Therefore, it is not surprising that depletion of NMT1, which is more closely related to proliferation than is NMT2, has a stronger growth suppressive effect than does depletion of NMT2. It is also possible that the tumors may develop mechanisms to evade the deleterious effects of loss of NMT2. In either case, definition of selective substrates for NMT1 and NMT2 will be important for understanding the mechanisms of the differential antitumor effects of the siRNAs.
Because NMT activity is essential for the growth and survival of a variety of infectious organisms, NMT has been considered as a target for the development of new antibiotics (55, 58–60). For example, several studies have targeted the NMTs of Candida albicans and Cryptococcus neoformans, which are responsible for systemic fungal infections in immunocompromised humans, as treatment of such infections (43, 61–64). NMTs have also been targeted for the development of antiviral drugs due to their essential role in viral replication. For example, the myristoylated glycine residues of several viral Gag proteins have been mutated, and this prevents the assembly of viral particles (65–67). We can now add antineoplastic therapies to the growing list of possible uses for NMT inhibitors. The results presented in this study provide evidence that targeting a specific NMT isozyme could have dramatic effects on tumor cells reliant on these enzymes for survival. It is possible to envision NMT1-specific inhibitors affecting the growth of c-Src-driven colon tumors while having little effect on nonmetastatic tissues in the body.
In conclusion, we have described siRNA sequences that efficiently and selectively ablate the expression of the two NMT isozymes in human cells. These tools have allowed the functional characterization of human NMT1 and NMT2, providing insight into the cellular processes involving these isozymes. The knockdown system will also aid in the development of specific inhibitors for NMT1 and NMT2, which could provide a much needed therapy for diseases, such as colon and ovarian cancer.
Materials and Methods
Cell Culture
SK-OV-3 cells (ATCC-HTB-77) were maintained in DMEM containing 10% fetal bovine serum, 50 µg/mL gentamicin, and 1 mmol/L sodium pyruvate at 37°C in an atmosphere of 5% CO2 and 95% air. HT-29 cells (ATCC-HTB-38) were maintained in RPMI containing 10% fetal bovine serum with 50 µg/mL gentamicin at 37°C in an atmosphere of 5% CO2 and 95% air. DMEM and RPMI, fetal bovine serum, gentamicin, and sodium pyruvate were from Life Technologies (Gaithersburg, MD).
Constructs
siRNA Constructs
Oligonucleotide templates for the generation of siRNAs were designed using the Ambion siRNA Target Finder (http://www.ambion.com). Four sets of oligonucleotides were designed for each gene encoding NMT1 (National Center for Biotechnology Information accession no. NM_021079) and NMT2 (National Center for Biotechnology Information accession no. NM_004808), and the siRNAs were synthesized using the Silencer siRNA construction kit (Ambion). The siRNA used to knockdown NMT1, designated NMT1-1, is 5′-AATGAGGAGGACAACAGCTAC-3′ and was determined to be optimally effective at 10 nmol/L. The siRNA used to knockdown NMT2, designated NMT2-4, is 5′-AAAAGGTTGGACTAGTACTAC-3′ and was also determined to be optimally effective at 10 nmol/L. Transfections using both NMT1 and NMT2 siRNAs were done at a final total concentration of 20 nmol/L. Negative control siRNA templates were obtained from Ambion (Negative Control 1 siRNA Templates) and used to generate the negative control siRNA, which was used at a concentration of 20 nmol/L in all experiments.
GFP Constructs
The GFP-farn-palm construct (also designated as pEGFP-F) was purchased from Clontech (Mountain View, CA) and contains the COOH terminus of H-Ras fused to GFP. The N-myr-palm-GFP construct was generated by the addition of the NH2-terminal myristoylation signal of Fyn to the NH2 terminus of GFP by PCR. The following primers were used in the PCR reaction: 5′-GGGAATTCACCATGGGCTGTGTGCAATGTAAGACAAAACTGACGGAGGAGAGGGTGAGCAAGGGGGAGGAGCTG-3′ and 5′-GGGGATCCTTATCAGGACTTGTACAGCTCGTCCATGCC-3′. The PCR fragment was digested with EcoRI and BamHI restriction enzymes and cloned into the pZEOSV2-expression vector (Invitrogen, Carlsbad, CA). Once clones were generated, the construct was isolated from bacteria and sequenced to ensure its identity. This construct was used in transient transfection assays at 0.66 µg per reaction.
Transfection with siRNAs
Transfection of cells with siRNA was done as follows: cells were plated in six-well or 10 cm dishes in either RPMI or DMEM with 10% fetal bovine serum (without antibiotic) such that cells were 40% to 50% confluent at the time of transfection. Transient transfection of siRNA constructs into both cell types was achieved using 7.5 µg/mL LipofectAMINE 2000 (Invitrogen) for 4 hours at 37°C. Following the 4-hour incubation, the medium was replaced with RPMI or DMEM plus 10% fetal bovine serum (without antibiotic) and the cells were incubated at 37°C for 24 to 72 hours. Cells were harvested at the indicated times, and each sample was divided into two aliquots. One cell aliquot was used for Western blot analysis and the other for Northern blot analysis using standard methods as described below.
Western Blot Analyses
Following the transfection, harvested cells were resuspended in hypotonic lysis buffer [10 mmol/L HEPES (pH 7.4), 10 mmol/L KCl, 1.5 mmol/L MgCl2, and 5 µmol/L phenyl-methylsulfonyl fluoride] containing 1% SDS. The protein concentration in each sample was determined using the fluorescamine assay (68), and samples normalized for equal amounts of protein (50–100 µg/lane) were separated on 10% SDS-PAGE gels and electrotransferred to polyvinylidene difluoride membranes. The membranes were incubated in PBST [100 mmol/L sodium phosphate, 100 mmol/L NaCl (pH 7.6) containing 0.1% Tween 20] or TBST [20 mmol/L Tris, 137 mmol/L NaCl (pH 7.6) containing 0.1% Tween 20] plus 5% (w/v) powdered milk at room temperature for 2 hours. For the phosphospecific blots, the membranes were washed thrice in TBST at room temperature for 5 minutes. Membranes were then incubated with various antibodies (Table 1) in TBST (phosphospecific antibodies) plus 5% bovine serum albumin or PBST on a shaking platform for 18 hours at 4°C. The membranes were washed thrice in TBST (phosphospecific antibodies) or PBST (10 minutes each at room temperature), and anti-mouse antibodies conjugated to horseradish peroxidase (Sigma, St. Louis, MO) were then incubated with the membranes in TBST (phosphospecific antibodies) or PBST containing 3% powdered milk (w/v) on a shaking platform for 1 hour at room temperature. The blots were washed four times in TBST (phosphospecific antibodies) or PBST for 5 minutes each at room temperature. Blots were developed with Super-Signal development reagents (Pierce, Rockford, IL). Filters were stripped of the original antibody by incubation in stripping buffer containing 100 mmol/L 2-mercaptoethanol, 2% SDS, and 62.5 mmol/L Tris-HCl (pH 6.7) for 30 minutes at 50°C and were then reprobed with antibodies against pan-actin. Blots were imaged with a Fujifilm (Bedford, United Kingdom) Intelligent Dark Box II, and the appropriate bands were quantified using Image Gauge 4.0 software. More information on the imager can be found at http://home.fujifilm.com/products/science/si_product/las1000plus/spc.html.
Table 1.
Antibody Panel
| Antibody | Phosphorylation Site | Vender |
|---|---|---|
| NMT1 | — | BD Transduction Laboratories (Mountain View, CA) |
| NMT2 | — | BD Transduction Laboratories |
| Actin | — | Chemicon (Temecula, CA) |
| MARCKS | — | Calbiochem (San Diego, CA) |
| c-Src | — | Santa Cruz Biotechnology (Santa Cruz, CA) |
| Phospho-c-Src | Tyr416 | Cell Signaling (Beverly, MA) |
| Phospho-FAK | Tyr576, Tyr577 | Cell Signaling |
| Phosphatidylinositol 3-kinase | — | Upstate Biotechnology (Charlottesville, VA) |
| Phospho-c-Raf | Ser338 | Cell Signaling |
| Phospho-MEK1/2 | Ser217/Ser221 | Cell Signaling |
| Phospho – mitogen-activated protein kinase p42/p44 | Thr202/Tyr204 | Cell Signaling |
| Phospho-Elk | Ser383 | Cell Signaling |
| Cyclin D1 | — | Cell Signaling |
| Cyclin D3 | — | Cell Signaling |
| CDK4 | — | Cell Signaling |
| CDK6 | — | Cell Signaling |
| p27 | — | Cell Signaling |
| Phospho-CDC2 | Thr161 | Cell Signaling |
| Phospho-BAD | Ser112 | Cell Signaling |
| BCL-XL | — | Cell Signaling |
| MCL-1 | — | Cell Signaling |
| BAK | — | Cell Signaling |
| BAX | — | Cell Signaling |
Northern Blot Analyses
After transfection, total RNA was isolated from the harvested cell aliquots using the RNeasy total RNA isolation kit (Qiagen, Valencia, CA). Total RNA (~10 µg) was loaded onto 1% agarose gels containing formaldehyde, electrophoresed for 2 hours at 70 V, and transferred to a nylon membrane (Immobilon_NY+, Millipore, Billerica, MA) and UV cross-linked. The membrane was sequentially hybridized with the NMT1 cDNA (1,492 bp), a 500-bp PCR fragment of the NMT2 gene (−1 to +503), and a β-actin probe (1,076 bp) obtained from Ambion. All probes were random prime labeled with [α-32P]dCTP to a specific activity of 109 cpm/µg. The membranes were exposed to film, and the densities of appropriate bands were quantified using ImageQuant software.
Subcellular Localization of GFP
Transfections of SK-OV-3 cells for GFP localization analyses were carried out as described above with the following modification. The cells were plated into 24-well culture dishes containing sterile glass coverslips and transfected with siRNAs as indicated above, except that 0.66 µg vector encoding either N-myr-palm-GFP or GFP-farn-palm was included. At 48 hours after transfection, the cells were fixed with 4% paraformaldehyde in PBS for 30 minutes at room temperature. Coverslips were mounted on glass slides with mounting medium (Sigma) and sealed with rubber cement. The cells were visualized by epifluorescence microscopy and images were captured digitally.
Cell Proliferation Assay
SK-OV-3 or HT-29 cells were transfected in six-well plates as described above and incubated at 37°C in an atmosphere of 5% CO2 and 95% air for 48 hours. At 48 hours after transfection, [3H]thymidine (16 µCi) was added to each well and cultures were incubated for an additional 2 hours. The medium was then removed by aspiration and 0.8 mL ice-cold 10% trichloroacetic acid was added to each well. After 10 minutes, the trichloroacetic acid was removed by aspiration and replaced with 0.4 mL of 40 µg/mL type I DNA (Sigma) in 0.2 mol/L NaOH. The samples were incubated at 37°C for 30 minutes and scraped and the amount of 3H in the supernatant containing the recovered genomic DNA was quantified by scintillation counting.
Apoptosis Assay
HT-29 or SK-OV-3 cells were plated on glass coverslips and transfected with siRNAs as indicated above. After 48 hours, the cells were permeabilized and cells with fragmented DNA were labeled following the instructions for the In situ Cell Death Detection kit, Fluorescein (Roche, Indianapolis, IN). Fluorescent and bright-field images were captured digitally and the percentage of cells undergoing apoptosis was calculated as the ratio of fluorescent cells to total cells.
NMT siRNA Tumor Study
Female BALB/c mice (6–8 weeks old) were injected subcutaneously in the right hind flank with 1 × 106 JC (mammary adenocarcinoma) cells suspended in PBS (42). On the appearance of palpable tumors, tumor volumes were determined using calipers measuring the length (L) and width (W) of the tumor and calculated using the formula: (L × W2) / 2. Groups of four mice were treated with single-site intratumoral injections of NMT1-1 (10 µmol/L), NMT2-4 (10 µmol/L), NMT1-1 and NMT2-4 (20 µmol/L total), or negative control (20 µmol/L) siRNA in a total volume of 50 µL PBS on days 1, 4, 7, and 10. Animals were euthanized and tumors were excised and weighed on day 11. Whole body weight and tumor measurements were done on days, 1, 4, 7, 10, and 11. Ps were determined by unpaired t tests using GraphPad InStat (San Diego, CA).
RT-PCR Analysis
Total RNA was isolated from flash-frozen tumors using the RNeasy RNA isolation kit (Qiagen). Total RNA (1 µg) was used in each RT-PCR reaction using the One-Step RT-PCR kit (Qiagen). NMT1 mRNA was amplified using primers that span the intron between exons 10 and 12. The 5′ primer is 5′-GCGACCAATGGAAACAAAGG-3′ and the 3′ primer is 5′-GGTGACTGGTTATTGTAGCACCAG-3. The NMT1 primer pair generated a 487-bp PCR product. NMT2 mRNA was amplified using primers that span the intron between exons 10 and 12. The 5V primer is 5′-CAGGTTTGAGACCAATGGAACC-3′ and the 3′ primer is 5′-TCTTCTTTGGAATGGCAGTTCC-3′. The NMT2 primer pair generated a 566-bp PCR product. RT-PCR products were resolved on a 2% agarose gel and visualized by staining with ethidium bromide.
Acknowledgments
We thank The Penn State Functional Genomics Core Facility for performing the microarray analyses and for their assistance in analyzing the data, Jean Copper for providing the GFP fusion constructs and technical assistance with the fluorescence microscopy, and all of the members of the Smith laboratory for their helpful comments and suggestions in the preparation of this article.
Grant support: NIH grants 1 R43 CA102853 (Yan Zhuang) and 5 R01 CA075248 (C.D. Smith).
References
- 1.Maurer-Stroh S, Eisenhaber B, Eisenhaber F. N-terminal N-myristoylation of proteins: refinement of the sequence motif and its taxon-specific differences. J Mol Biol. 2002;317:523–540. doi: 10.1006/jmbi.2002.5425. [DOI] [PubMed] [Google Scholar]
- 2.Maurer-Stroh S, Eisenhaber B, Eisenhaber F. N-terminal N-myristoylation of proteins: prediction of substrate proteins from amino acid sequence. J Mol Biol. 2002;317:541–557. doi: 10.1006/jmbi.2002.5426. [DOI] [PubMed] [Google Scholar]
- 3.Selvakumar P, Pasha MK, Ashakumary L, Dimmock JR, Sharma RK. Myristoyl-CoA:protein N-myristoyltransferase: a novel molecular approach for cancer therapy [review] Int J Mol Med. 2002;10:493–500. [PubMed] [Google Scholar]
- 4.Boutin JA. Myristoylation. Cell Signal. 1997;9:15–35. doi: 10.1016/s0898-6568(96)00100-3. [DOI] [PubMed] [Google Scholar]
- 5.Krueger JG, Garber EA, Goldberg AR, Hanafusa H. Changes in amino-terminal sequences of pp60src lead to decreased membrane association and decreased in vivo tumorigenicity. Cell. 1982;28:889–896. doi: 10.1016/0092-8674(82)90068-x. [DOI] [PubMed] [Google Scholar]
- 6.Cross FR, Garber EA, Pellman D, Hanafusa H. A short sequence in the p60src N terminus is required for p60src myristylation and membrane association and for cell transformation. Mol Cell Biol. 1984;4:1834–1842. doi: 10.1128/mcb.4.9.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamps MP, Buss JE, Sefton BM. Rous sarcoma virus transforming protein lacking myristic acid phosphorylates known polypeptide substrates without inducing transformation. Cell. 1986;45:105–112. doi: 10.1016/0092-8674(86)90542-8. [DOI] [PubMed] [Google Scholar]
- 8.Kamps MP, Buss JE, Sefton BM. Mutation of NH2-terminal glycine of p60src prevents both myristoylation and morphological transformation. Proc Natl Acad Sci U S A. 1985;82:4625–4628. doi: 10.1073/pnas.82.14.4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Playford MP, Schaller MD. The interplay between Src and integrins in normal and tumor biology. Oncogene. 2004;23:7928–7946. doi: 10.1038/sj.onc.1208080. [DOI] [PubMed] [Google Scholar]
- 10.Parsons SJ, Parsons JT. Src family kinases, key regulators of signal transduction. Oncogene. 2004;23:7906–7909. doi: 10.1038/sj.onc.1208160. [DOI] [PubMed] [Google Scholar]
- 11.Summy JM, Gallick GE. Src family kinases in tumor progression and metastasis. Cancer Metastasis Rev. 2003;22:337–358. doi: 10.1023/a:1023772912750. [DOI] [PubMed] [Google Scholar]
- 12.Ishizawar R, Parsons SJ. c-Src and cooperating partners in human cancer. Cancer Cell. 2004;6:209–214. doi: 10.1016/j.ccr.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 13.Selvakumar P, Lakshmikuttyamma A, Lawman Z, Bonham K, Dimmock JR, Sharma RK. Expression of methionine aminopeptidase 2, N-myristoyltransferase, and N-myristoyltransferase inhibitor protein 71 in HT29. Biochem Biophys Res Commun. 2004;322:1012–1017. doi: 10.1016/j.bbrc.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 14.Rajala RV, Dehm S, Bi X, Bonham K, Sharma RK. Expression of N-myristoyltransferase inhibitor protein and its relationship to c-Src levels in human colon cancer cell lines. Biochem Biophys Res Commun. 2000;273:1116–1120. doi: 10.1006/bbrc.2000.3066. [DOI] [PubMed] [Google Scholar]
- 15.Raju RV, Moyana TN, Sharma RK. N-Myristoyltransferase overexpression in human colorectal adenocarcinomas. Exp Cell Res. 1997;235:145–154. doi: 10.1006/excr.1997.3679. [DOI] [PubMed] [Google Scholar]
- 16.Magnuson BA, Raju RV, Moyana TN, Sharma RK. Increased N-myristoyltransferase activity observed in rat and human colonic tumors. J Natl Cancer Inst. 1995;87:1630–1635. doi: 10.1093/jnci/87.21.1630. [DOI] [PubMed] [Google Scholar]
- 17.Lu Y, Selvakumar P, Ali K, et al. Expression of N-myristoyltransferase in human brain tumors. Neurochem Res. 2005;30:9–13. doi: 10.1007/s11064-004-9680-9. [DOI] [PubMed] [Google Scholar]
- 18.Shoji S, Kurosawa T, Inoue H, Funakoshi T, Kubota Y. Human cellular src gene product: identification of the myristoylated pp60c-src and blockage of its myristoyl acylation with N-fatty acyl compounds resulted in the suppression of colony formation. Biochem Biophys Res Commun. 1990;173:894–901. doi: 10.1016/s0006-291x(05)80870-8. [DOI] [PubMed] [Google Scholar]
- 19.Towler DA, Eubanks SR, Towery DS, Adams SP, Glaser L. Amino-terminal processing of proteins by N-myristoylation. Substrate specificity of N-myristoyl transferase. J Biol Chem. 1987;262:1030–1036. [PubMed] [Google Scholar]
- 20.Wolven A, Okamura H, Rosenblatt Y, Resh MD. Palmitoylation of p59fyn is reversible and sufficient for plasma membrane association. Mol Biol Cell. 1997;8:1159–1173. doi: 10.1091/mbc.8.6.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilcox C, Hu JS, Olson EN. Acylation of proteins with myristic acid occurs cotranslationally. Science. 1987;238:1275–1278. doi: 10.1126/science.3685978. [DOI] [PubMed] [Google Scholar]
- 22.Deichaite I, Casson LP, Ling HP, Resh MD. In vitro synthesis of pp60vsrc: myristylation in a cell-free system. Mol Cell Biol. 1988;8:4295–4301. doi: 10.1128/mcb.8.10.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pillai S, Baltimore D. Myristoylation and the post-translational acquisition of hydrophobicity by the membrane immunoglobulin heavy-chain polypeptide in B lymphocytes. Proc Natl Acad Sci U S A. 1987;84:7654–7658. doi: 10.1073/pnas.84.21.7654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.King MJ, Sharma RK. N-myristoyl transferase assay using phosphocellulose paper binding. Anal Biochem. 1991;199:149–153. doi: 10.1016/0003-2697(91)90082-5. [DOI] [PubMed] [Google Scholar]
- 25.Vijayasarathy C, Bhat NR, Avadhani NG. Intramitochondrial fatty acylation of a cytoplasmic imported protein in animal cells. J Biol Chem. 1989;264:7772–7775. [PubMed] [Google Scholar]
- 26.Giang DK, Cravatt BF. A second mammalian N-myristoyltransferase. J Biol Chem. 1998;273:6595–6598. doi: 10.1074/jbc.273.12.6595. [DOI] [PubMed] [Google Scholar]
- 27.Aitken A. Structure determination of acylated proteins. Biochem Soc Trans. 1989;17:871–875. doi: 10.1042/bst0170871. [DOI] [PubMed] [Google Scholar]
- 28.Yang SH, Shrivastav A, Kosinski C, et al. N-myristoyltransferase 1 is essential in early mouse development. J Biol Chem. 2005;280:18990–18995. doi: 10.1074/jbc.M412917200. [DOI] [PubMed] [Google Scholar]
- 29.van’t Hof W, Resh MD. Dual fatty acylation of p59(Fyn) is required for association with the T cell receptor ζ chain through phosphotyrosine-Src homology domain-2 interactions. J Cell Biol. 1999;145:377–389. doi: 10.1083/jcb.145.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang X, Lu Y, Wilkes M, Neubert TA, Resh MD. The N-terminal SH4 region of the Src family kinase Fyn is modified by methylation and heterogeneous fatty acylation: role in membrane targeting, cell adhesion, and spreading. J Biol Chem. 2004;279:133–139. doi: 10.1074/jbc.M311180200. [DOI] [PubMed] [Google Scholar]
- 31.French KJ, Zhuang Y, Schrecengost RS, Copper JE, Xia Z, Smith CD. Cyclohexyl-octahydro-pyrrolo[1,2-a]pyrazine-based inhibitors of human N-myristoyltransferase. J Pharmacol Exp Ther. 2004;309:340–347. doi: 10.1124/jpet.103.061572. [DOI] [PubMed] [Google Scholar]
- 32.Razzini G, Berrie CP, Vignati S, et al. Novel functional PI 3-kinase antagonists inhibit cell growth and tumorigenicity in human cancer cell lines. FASEB J. 2000;14:1179–1187. doi: 10.1096/fasebj.14.9.1179. [DOI] [PubMed] [Google Scholar]
- 33.Yang G, Thompson JA, Fang B, Liu J. Silencing of H-ras gene expression by retrovirus-mediated siRNA decreases transformation efficiency and tumor growth in a model of human ovarian cancer. Oncogene. 2003;22:5694–5701. doi: 10.1038/sj.onc.1206858. [DOI] [PubMed] [Google Scholar]
- 34.Westhoff MA, Serrels B, Fincham VJ, Frame MC, Carragher NO. SRC-mediated phosphorylation of focal adhesion kinase couples actin and adhesion dynamics to survival signaling. Mol Cell Biol. 2004;24:8113–8133. doi: 10.1128/MCB.24.18.8113-8133.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Calalb MB, Polte TR, Hanks SK. Tyrosine phosphorylation of focal adhesion kinase at sites in the catalytic domain regulates kinase activity: a role for Src family kinases. Mol Cell Biol. 1995;15:954–963. doi: 10.1128/mcb.15.2.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thelen M, Rosen A, Nairn AC, Aderem A. Regulation by phosphorylation of reversible association of a myristoylated protein kinase C substrate with the plasma membrane. Nature. 1991;351:320–322. doi: 10.1038/351320a0. [DOI] [PubMed] [Google Scholar]
- 37.Hartwig JH, Thelen M, Rosen A, Janmey PA, Nairn AC, Aderem A. MARCKS is an actin filament crosslinking protein regulated by protein kinase C and calcium-calmodulin. Nature. 1992;356:618–622. doi: 10.1038/356618a0. [DOI] [PubMed] [Google Scholar]
- 38.Tapp H, Al-Naggar IM, Yarmola EG, et al. MARCKS is a natively unfolded protein with an inaccessible actin-binding site: evidence for long-range intramolecular interactions. J Biol Chem. 2005;280:9946–9956. doi: 10.1074/jbc.M414614200. [DOI] [PubMed] [Google Scholar]
- 39.Reiske HR, Zhao J, Han DC, Cooper LA, Guan JL. Analysis of FAK-associated signaling pathways in the regulation of cell cycle progression. FEBS Lett. 2000;486:275–280. doi: 10.1016/s0014-5793(00)02295-x. [DOI] [PubMed] [Google Scholar]
- 40.Zhao JH, Reiske H, Guan JL. Regulation of the cell cycle by focal adhesion kinase. J Cell Biol. 1998;143:1997–2008. doi: 10.1083/jcb.143.7.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mishra NC, Kumar S. Apoptosis: a mitochondrial perspective on cell death. Indian J Exp Biol. 2005;43:25–34. [PubMed] [Google Scholar]
- 42.Lee BD, French KJ, Zhuang Y, Smith CD. Development of a syngeneic in vivo tumor model and its use in evaluating a novel P-glycoprotein modulator, PGP-4008. Oncol Res. 2004;14:49–60. doi: 10.3727/000000003108748603. [DOI] [PubMed] [Google Scholar]
- 43.Farazi TA, Waksman G, Gordon JI. The biology and enzymology of protein N-myristoylation. J Biol Chem. 2001;276:39501–39504. doi: 10.1074/jbc.R100042200. [DOI] [PubMed] [Google Scholar]
- 44.Kolluri SK, Balduf C, Hofmann M, Gottlicher M. Novel target genes of the Ah (dioxin) receptor: transcriptional induction of N-myristoyltransferase 2. Cancer Res. 2001;61:8534–8539. [PubMed] [Google Scholar]
- 45.Liang X, Lu Y, Neubert TA, Resh MD. The N-terminal SH4 region of the Src family kinase Fyn is modified by methylation and heterogeneous fatty acylation: role in membrane targeting, cell adhesion and spreading. J Biol Chem. 2004;279:8133–8139. doi: 10.1074/jbc.M311180200. [DOI] [PubMed] [Google Scholar]
- 46.Glover CJ, Hartman KD, Felsted RL. Human N-myristoyltransferase amino-terminal domain involved in targeting the enzyme to the ribosomal subcellular fraction. J Biol Chem. 1997;272:28680–28689. doi: 10.1074/jbc.272.45.28680. [DOI] [PubMed] [Google Scholar]
- 47.Buss JE, Kamps MP, Gould K, Sefton BM. The absence of myristic acid decreases membrane binding of p60src but does not affect tyrosine protein kinase activity. J Virol. 1986;58:468–474. doi: 10.1128/jvi.58.2.468-474.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Irby RB, Yeatman TJ. Increased Src activity disrupts cadherin/catenin-mediated homotypic adhesion in human colon cancer and transformed rodent cells. Cancer Res. 2002;62:2669–2674. [PubMed] [Google Scholar]
- 49.Irby RB, Yeatman TJ. Role of Src expression and activation in human cancer. Oncogene. 2000;19:5636–5642. doi: 10.1038/sj.onc.1203912. [DOI] [PubMed] [Google Scholar]
- 50.Susa M, Luong-Nguyen NH, Crespo J, Maier R, Missbach M, McMaster G. Active recombinant human tyrosine kinase c-Yes: expression in baculovirus system, purification, comparison to c-Src, and inhibition by a c-Src inhibitor. Protein Expr Purif. 2000;19:99–106. doi: 10.1006/prep.2000.1221. [DOI] [PubMed] [Google Scholar]
- 51.Laird AD, Li G, Moss KG, et al. Src family kinase activity is required for signal tranducer and activator of transcription 3 and focal adhesion kinase phosphorylation and vascular endothelial growth factor signaling in vivo and for anchorage-dependent and -independent growth of human tumor cells. Mol Cancer Ther. 2003;2:461–469. [PubMed] [Google Scholar]
- 52.Scholz A, Heinze S, Detjen KM, et al. Activated signal transducer and activator of transcription 3 (STAT3) supports the malignant phenotype of human pancreatic cancer. Gastroenterology. 2003;125:891–905. doi: 10.1016/s0016-5085(03)01064-3. [DOI] [PubMed] [Google Scholar]
- 53.Bhunia AK, Piontek K, Boletta A, et al. PKD1 induces p21(waf1) and regulation of the cell cycle via direct activation of the JAK-STAT signaling pathway in a process requiring PKD2. Cell. 2002;109:157–168. doi: 10.1016/s0092-8674(02)00716-x. [DOI] [PubMed] [Google Scholar]
- 54.Zhao J, Zheng C, Guan J. Pyk2 and FAK differentially regulate progression of the cell cycle. J Cell Sci. 2000;113:3063–3072. doi: 10.1242/jcs.113.17.3063. [DOI] [PubMed] [Google Scholar]
- 55.Duronio RJ, Towler DA, Heuckeroth RO, Gordon JI. Disruption of the yeast N-myristoyl transferase gene causes recessive lethality. Science. 1989;243:796–800. doi: 10.1126/science.2644694. [DOI] [PubMed] [Google Scholar]
- 56.Qi Q, Rajala RV, Anderson W, et al. Molecular cloning, genomic organization, and biochemical characterization of myristoyl-CoA:protein N-myristoyltransferase from Arabidopsis thaliana. J Biol Chem. 2000;275:9673–9683. doi: 10.1074/jbc.275.13.9673. [DOI] [PubMed] [Google Scholar]
- 57.Ntwasa M, Aapies S, Schiffmann DA, Gay NJ. Drosophila embryos lacking N-myristoyltransferase have multiple developmental defects. Exp Cell Res. 2001;262:134–144. doi: 10.1006/excr.2000.5086. [DOI] [PubMed] [Google Scholar]
- 58.Weinberg RA, McWherter CA, Freeman SK, Wood DC, Gordon JI, Lee SC. Genetic studies reveal that myristoylCoA:protein N-myristoyltransferase is an essential enzyme in Candida albicans. Mol Microbiol. 1995;16:241–250. doi: 10.1111/j.1365-2958.1995.tb02296.x. [DOI] [PubMed] [Google Scholar]
- 59.Lodge JK, Jackson-Machelski E, Toffaletti DL, Perfect JR, Gordon JI. Targeted gene replacement demonstrates that myristoyl-CoA:protein N-myristoyltransferase is essential for viability of Cryptococcus neoformans. Proc Natl Acad Sci U S A. 1994;91:12008–12012. doi: 10.1073/pnas.91.25.12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Price HP, Menon MR, Panethymitaki C, Goulding D, McKean PG, Smith DF. Myristoyl-CoA:protein N-myristoyltransferase, an essential enzyme and potential drug target in kinetoplastid parasites. J Biol Chem. 2003;278:7206–7214. doi: 10.1074/jbc.M211391200. [DOI] [PubMed] [Google Scholar]
- 61.Sikorski JA, Devadas B, Zupec ME, et al. Selective peptidic and peptidomimetic inhibitors of Candida albicans myristoylCoA:protein N-myristoyltransferase: a new approach to antifungal therapy. Biopolymers. 1997;43:43–71. doi: 10.1002/(SICI)1097-0282(1997)43:1<43::AID-BIP5>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 62.White TC, Marr KA, Bowden RA. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin Microbiol Rev. 1998;11:382–402. doi: 10.1128/cmr.11.2.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sogabe S, Masubuchi M, Sakata K, et al. Crystal structures of Candida albicans N-myristoyltransferase with two distinct inhibitors. Chem Biol. 2002;9:1119–1128. doi: 10.1016/s1074-5521(02)00240-5. [DOI] [PubMed] [Google Scholar]
- 64.Georgopapadakou NH. Antifungals targeted to protein modification: focus on protein N-myristoyltransferase. Expert Opin Investig Drugs. 2002;11:1117–1125. doi: 10.1517/13543784.11.8.1117. [DOI] [PubMed] [Google Scholar]
- 65.Rein A, McClure MR, Rice NR, Luftig RB, Schultz AM. Myristylation site in Pr65gag is essential for virus particle formation by Moloney murine leukemia virus. Proc Natl Acad Sci U S A. 1986;83:7246–7250. doi: 10.1073/pnas.83.19.7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Marc D, Drugeon G, Haenni AL, Girard M, van der Werf S. Role of myristoylation of poliovirus capsid protein VP4 as determined by site-directed mutagenesis of its N-terminal sequence. EMBO J. 1989;8:2661–2668. doi: 10.1002/j.1460-2075.1989.tb08406.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bryant M, Ratner L. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc Natl Acad Sci U S A. 1990;87:523–527. doi: 10.1073/pnas.87.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bohlen P, Stein S, Dairman W, Udenfriend S. Fluorometric assay of proteins in the nanogram range. Arch Biochem Biophys. 1973;155:213–220. doi: 10.1016/s0003-9861(73)80023-2. [DOI] [PubMed] [Google Scholar]