Abstract
As research progresses towards understanding the role of the amyloid-β (Aβ) in Alzheimer’s disease, certain aspects of the aggregation process for Aβ are still not clear. In particular, the accepted constitution of toxic aggregates in neurons has shifted towards small oligomers. However, the process of forming these oligomers in cells is still not fully clear. Even more interestingly, it has been implied that cell membranes, and, in particular, anionic lipids within those membranes, play a key role in the progression of Aβ aggregation, but the exact nature of the Aβ-membrane interaction in this process is still unknown. In this work, we use a thermodynamic cycle and umbrella sampling molecular dynamics to investigate dimerization of the 42-residue Aβ peptide on model zwitterionic dipalmitoylphosphatidylcholine (DPPC) or model anionic dioleoylphosphatidylserine (DOPS) bilayer surfaces. We determined that Aβ dimerization was strongly favored through interactions with the DOPS bilayer. Further, our calculations showed that the DOPS bilayer promoted strong protein-protein interactions within the Aβ dimer, while DPPC favored strong protein-lipid interactions. By promoting dimer formation and subsequent dimer release into the solvent, the DOPS bilayer acts as a catalyst in Aβ aggregation through converting Aβ monomers in solution into Aβ dimers in solution without substantial a free energy cost.
Keywords: Peptide-membrane interaction, Alzheimer’s disease, computer simulation, umbrella sampling, amyloid peptides, aggregation
Introduction
Aberrant protein aggregation and function are the hallmark of a variety of neurodegenerative disorders found in humans. In Alzheimer’s disease, the neural degeneration that characterizes this disease has been linked to the aggregation of the amyloid-β (Aβ) peptide, among other potential aggregate species in neurons1–6. Because of this direct link between properties of the Aβ peptide and progression of Alzheimer’s disease, the Aβ peptide has been at the center of extensive biological research over the last 30 years3–6. In particular, both experimental7–19 and computational20–43 biophysics approaches have focused on this peptide. Along with many other aspects of Aβ function and activity, the underlying processes connected to Aβ aggregation have been of substantial interest to researchers. A more thorough and clearer understanding of the aggregation pathway from Aβ monomer to full Aβ fibril is considered to be crucial to development of any targeted therapeutic against this aspect of Alzheimer’s disease.
As our understanding of the aggregation pathway of Aβ has progressed, our view of Aβ toxicity in Alzheimer’s disease has evolved7,8,44,45. Initially, it was believed that full Aβ fibrils or possibly protofibrils were the toxic species in Alzheimer’s disease. However, further investigation into this process shifted the focus from full fibrils to Aβ oligomers as the toxic species in neurons7,8,44. Research has shown that these oligomers were able to disrupt cell function and also disrupt homeostasis across the cell membrane16,45–48. Further, it has been postulated that these oligomers could form ion channels that would allow unregulated flow of ions such as calcium across the cell membrane46,47,49,50. Recent work has also shown that amyloid fibrils are not harmless but can act as reservoirs of oligomers that can be released if the fibrils are placed under stress51,52. Another interesting aspect of this system is the underlying structure of oligomers and fibrils. Aβ monomers have been shown to be mostly random coil in solution27,53 with some transient β-sheet or α-helical structure. The Aβ monomer structure can be altered by placing the protein in different environments, promoting either a α-helical or predominantly β-sheet structure54. However, for Aβ oligomers, the predicted structures of Aβ units are not as clear. The structures of Aβ oligomers have been shown to be highly variable48,55–58. Structures that are fibril-like have been observed57,58, as well as completely amorphous structures48,55–58, or cylindrical structures inserted in cell membranes41,50,59. Thus, it is expected that Aβ oligomer formation is highly heterogeneous and that ordered structure for Aβ is not locked until the protein begins to aggregate into a fibril. Even at the fibril level, there is substantial heterogeneity both on the scale of the fibril as a whole60,61, considering the size and shape of the fibril, and the predicted underlying structure of the Aβ units within a fibril62–65. Thus, a better understanding of the physical processes that dictate Aβ oligomerization and impart such a heterogeneous class of structures to the smallest oligomeric units is essential.
Aβ is a 38–43 amino acid cleavage product of the transmembrane Amyloid Precursor Protein3–5. Thus, the Aβ peptide contains significant portions of hydrophobic and hydrophilic residues and shows favorable interactions with cell membranes7,8,10–18,66. Further, the phenomena dictating the earliest stages of Aβ oligomerization are still not clear. While experimental work is able to replicate most aspects of in-vivo Aβ aggregation with in-vitro methods, these studies still require orders-of-magnitude higher Aβ concentrations to match the aggregation rate of Aβ in cells12,13. While many physical attributes of the system, such as cellular pH67, salt concentration68, and oxidation of methionine and other residues69, may play a role in promoting Aβ aggregation in vivo, interactions with cell membranes have also been postulated to assist in this process. Cell membranes can affect peptide aggregation through direct electrostatic, hydrophobic or hydrogen bonding interactions with residues of the peptide7,8,70–72, new partially folded free energy states at the bilayer surface7,8,70, faster aggregation rates over a two dimensional lipid surface7,8,70, higher transient peptide concentration as proteins are restricted to the surface, and lower surface pH due to the attraction of H+ ions in solution to anionic lipid headgroups11,71,72. All of these factors may play a role in Aβ aggregation on the bilayer surface. Previous experimental work7–9,11,13–17 has demonstrated that interactions between Aβ peptides and lipid vesicles strongly promote a conversion to an amyloid-like structure. Further research7,8,11,13–15,17,18 has shown that mixing anionic lipids with Aβ peptides leads to a substantial increase in aggregation rate. Finally, extensive interactions with cell membranes can lead to Aβ pore formation and disruption of ion balance across the membrane41,46,50,59. While these results demonstrate the importance of protein-lipid interactions in Aβ aggregation, one shortcoming of these methods is the lack of resolution that results from the low concentration of Aβ necessary to prevent substantial aggregation during the experiment. Thus, it is unclear what the aggregation state of Aβ is during these investigations using cell membranes. Further work with more detailed methods will be necessary to understand the particulars of atomic-level interactions during Aβ aggregation at the membrane surface.
Towards this end, we use molecular dynamics (MD) simulations to study the earliest step in Aβ aggregation: formation of a dimer. Computational studies have been used with great success to study Aβ structure at the monomer20,22–24,26–30,32,34–39,43,56, small oligomer21,25,28,29,33,39,40,43,56, and fibril level31,42,43, using either full-length Aβ20,21,24–27,33–38,40,43 or Aβ fragments22,23,28–32,39,42,43,56. These studies have used all-atom20–27,29–32,34–38,40,42,43,56 and coarse-grained28,33,39 techniques in various explicit22,23,26,27,29–38,40,56 or implicit20,21,24,25,28,39,42,43 solvents to understand this system. More recent works have used advanced computational techniques such as replica exchange for even more encompassing studies20–25,38,39,43. However, the extent of computational research34–38,41,59,73 on Aβ-lipid interactions is more restricted. Most research that has been previously performed on an Aβ-lipid system has involved use of Aβ monomers34–38,73. These works have also been heavily biased towards studying pre-inserted Aβ over interactions between Aβ and the bilayer surface. Our previous work with this system has investigated both the properties of Aβ binding to a model bilayer surface37 and the structure of Aβ on the bilayer surface38. Nevertheless, it is of upmost importance to begin to expand these studies to the small oligomer structure of Aβ. Work has been done towards predicting Aβ pore structure within the bilayer41,59, but there is a substantial dearth of investigations in the specific interactions between Aβ and the surface of cell membranes that may promote Aβ dimerization. Recent works48,55 have demonstrated the fundamental role that the Aβ dimer plays as a unit of Aβ aggregation. A more complete understanding of the physical processes that dictate Aβ dimer formation at the cell surface is necessary to fully appreciate Aβ aggregation. In this work, we use a thermodynamic cycle to calculate the free energy of dimerization on a model lipid bilayer for two specific Aβ dimers. The results of this work, including comprehensive information gleaned from each process of the thermodynamic cycle, provide molecular level details of Aβ aggregation that can be utilized towards a greater understanding of the earliest stages of aggregation and the role of cell membranes in this process.
System and Methods
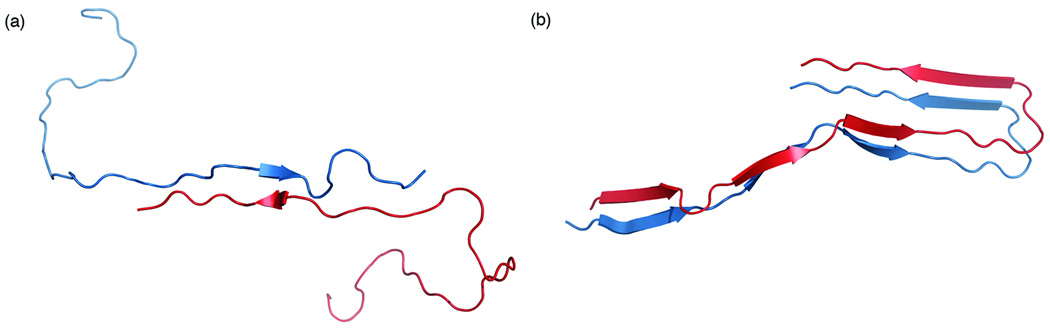
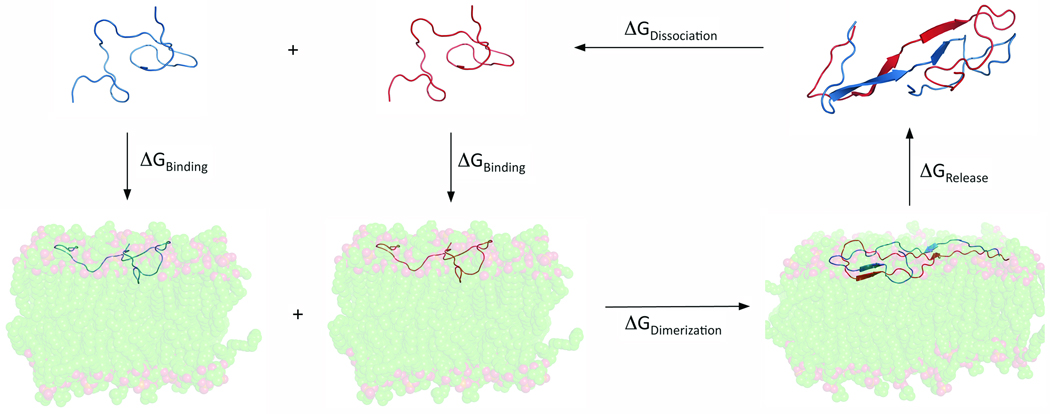
A full, detailed description of the molecular dynamics procedures used in this work is provided in the Supplementary Information. In short, two initial dimer structures, an extended structure (Figure 1a) based on electron microscopy data62 and a hairpin structure (Figure 1b) derived from NMR data65, were used to calculate a specific dimerization free energy of the 42 residue Aβ peptide on the surface of either a model zwitterionic dipalmitoylphosphatidylcholine (DPPC) or model anionic dioleoylphosphatidylserine (DOPS) bilayer. Further, due to the effect of anionic lipid headgroups on local pH 11, a neutral Aβ peptide was used in simulations with DOPS to represent the protonation of three histidine residues. For simulations with DPPC, the physiological charge of −3 was used for Aβ. In this work, we used a thermodynamic cycle:
| (1) |
to indirectly calculate ΔGDimerization through direct calculation of ΔGRelease + ΔGDissociation and use of ΔGBinding values calculated in a previous work37. A depiction of the steps of this cycle is provided in Figure 2. Using umbrella sampling techniques74,75, we were able to calculate ΔGRelease + ΔGDissociation and, thus, obtain ΔGDimerization with less systematic error than a comparable value achieved through direct calculation of ΔGDimerization using molecular dynamics techniques.
FIGURE 1.
Initial structures used for the (a) Extended and (b) Hairpin dimer simulations.
FIGURE 2.
Representation of the thermodynamic cycle used to approximate the free energy of dimerization for Aβ42 on a lipid bilayer.
Results
Equilibration Simulations
As described in the Methods section, two initial dimer structures (Figure 1) were used to calculate a specific dimerization free energy for each Aβ dimer on the bilayer surface. The first of these structures, the extended dimer (Figure 1a), represents shared C-termini between two Aβ monomers as predicted from electron microscopy data62. The other structure, the hairpin dimer (Figure 1b), represents two monomer units from within an Aβ fibril as determined by NMR65. Thus, intense equilibration is necessary in order to bring each structure to an equilibrated state on the bilayer surface from which our thermodynamic cycle calculations could begin. Initially, each structure was equilibrated in solution for a short time to remove any clashes due to the placement of each monomer in the dimer. Then, the dimer was placed on the surface of a model lipid bilayer and 150ns MD simulations were performed. Aside from just equilibration, these 150ns simulations provided an ideal opportunity to study the stability of the idealized dimer structures on the bilayer surface versus in solution. Along with the 150ns equilibration on the bilayer surface, we also continued the short equilibration in solution for an analogous 150ns. By comparing the evolution of the dimer secondary structure over these equilibration events, it is possible to study the stability of these specific, ideal dimer structures in either environment. Such a comparison is presented in the Supplementary Materials. A more thorough discussion and justification of our choice of these two dimer structures is presented in the Discussion section.
Thermodynamic Cycle Calculations
Once the dimers were appropriately equilibrated on the bilayer surface, the thermodynamic cycle calculations could be performed, as demonstrated in Figure 2. The thermodynamic cycle allowed us to calculate a quantity, ΔGDimerization, that would be very difficult to calculate directly due to bias created from the choice of reaction coordinate for the pulling procedure. Further, by calculating ΔGDimerization for two specific dimers instead of one generic dimer calculation, we can draw more specific conclusions regarding the effect of the bilayer on this process.
The results of these calculations are summarized in Table I. The dimerization free energy is calculated using the results as shown in Eq. 1. The free energies presented in Table I are calculated from the potentials of mean force as described in Supplementary Materials. All data for the monomer binding step is taken from a previous work37. For a thorough discussion of the monomer binding process, please see our previous work (note that Aβ charged is referred to a pH7 and Aβ neutral is referred to as pH5 in the previous work). To better understand the results in Table I, it is best to present each step individually and then investigate the calculated dimerization free energy.
TABLE I.
Free energies calculated for steps of the thermodynamic cycle shown in Figure 2. All values are given in kcal/mol.
| Dimer Structure | ΔGBinding | ΔGDimerization1 | ΔGRelease | ΔGDissociation | |
|---|---|---|---|---|---|
| Step 1 | Step 2 | 2*Step 1 + Step 2 | Step 3 | Step 4 | |
| Extended | |||||
| Charged Aβ + DPPC | −14.42 | −15.05 | −43.89 | 29.60 | 14.29 |
| Neutral Aβ + DOPS | −12.52 | 4.45 | −20.59 | 7.52 | 13.07 |
| Hairpin | |||||
| Charged Aβ + DPPC | −14.42 | −17.46 | −46.30 | 30.57 | 15.73 |
| Neutral Aβ + DOPS | −12.52 | −6.34 | −31.38 | 5.61 | 25.77 |
ΔGDimerization = −1 * (2*ΔGBinding + ΔGRelease + ΔGDissociation)
Dimer Release from the Bilayer Surface
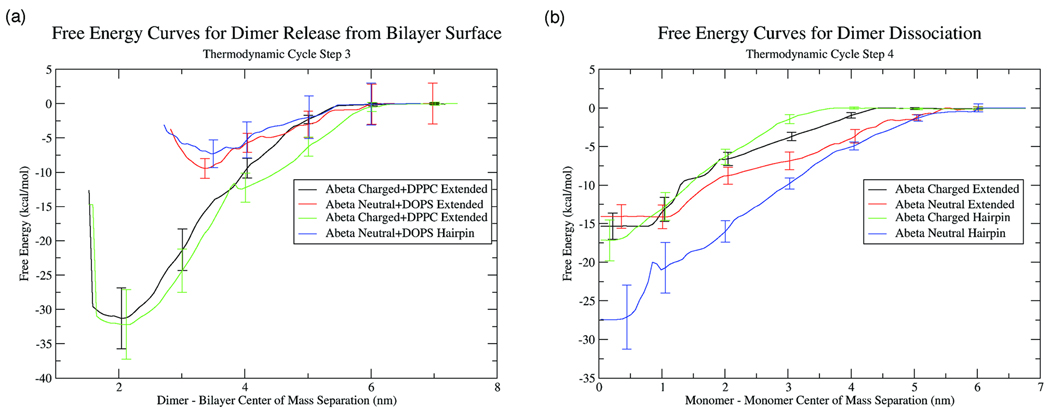
The release of the Aβ dimer from the bilayer surface is the third step in our cycle. The potentials of mean force calculated for this process are given in Figure 3a. From examining both the potential of mean force curves and the calculated free energies, it is clear that the largest source of distinction between systems is the charge on the lipid and, therefore, the charge on the peptide. For the charged peptide on DPPC, the structure of the dimer does not affect the dimer release free energy to any extent. The free energies for release are similar and the shapes of the potentials are also similar. Further, the release free energy of the dimer is quite large, implying a very strong attraction of the dimer to the bilayer surface. It is interesting that the magnitude of the release free energy is very close to twice the magnitude of the binding free energy of the monomer. Also, the potential curve for dimer release is remarkably similar to the monomer binding curves presented in our earlier work37. This implies that the Aβ dimer is still preferentially interacting with the interfacial region of the lipid bilayer and the protein-protein interaction within the dimer is not particularly strong. As the dimer is removed from the bilayer surface, the peptide-lipid interaction becomes weaker and the peptide-peptide interaction becomes dominant in order to retain the fidelity of the dimer.
FIGURE 3.
Potentials of mean force for (a) Aβ dimer release from the lipid surface into solution and (b) Aβ dimer dissolution in solution.
For the neutral Aβ dimer interacting with the DOPS bilayer, the structure of the dimer also does not effect the dimer release process. Both the extended and hairpin free energies of dimer release and the potentials of mean force curves are very similar. However, in contrast to the calculations on DPPC, the release free energy on DOPS is much smaller. Further, the potential of mean force curves are also more rugged near the free energy minimum. For the calculations on DPPC, there is a sharp drop in potential when approaching the minimum on the surface. Yet, on DOPS, there is no sharp drop in the potential and the dimer center of mass can explore approximately anywhere within a 2nm range of the free energy minimum without a substantial gain in free energy. All of these aspects of the potential of mean force curves imply a weaker interaction between the dimer and the bilayer surface. Also, the location of the free energy minimum provides evidence towards a weaker interaction. While the free energy minimum is sharply centered around a dimer-bilayer center of mass separation of 2nm on DPPC, the free energy minimum on DOPS is closer to a 3–3.5nm center of mass separation. In addition, visual inspection and analogies to our previous work37,38 show that, on DOPS, the dimer is not strongly interacting with the interfacial region of the bilayer, as in the case of the dimer on DPPC, but is only transiently interacting with the headgroup region of the bilayer. Further, the weak free energy of dimer release demonstrates that the DOPS bilayer biases the system towards strong peptide-peptide interactions due to weaker protein-lipid interactions. This is in direct contrast to the strong peptide-lipid and weak peptide-peptide interactions observed on the surface of the DPPC bilayer.
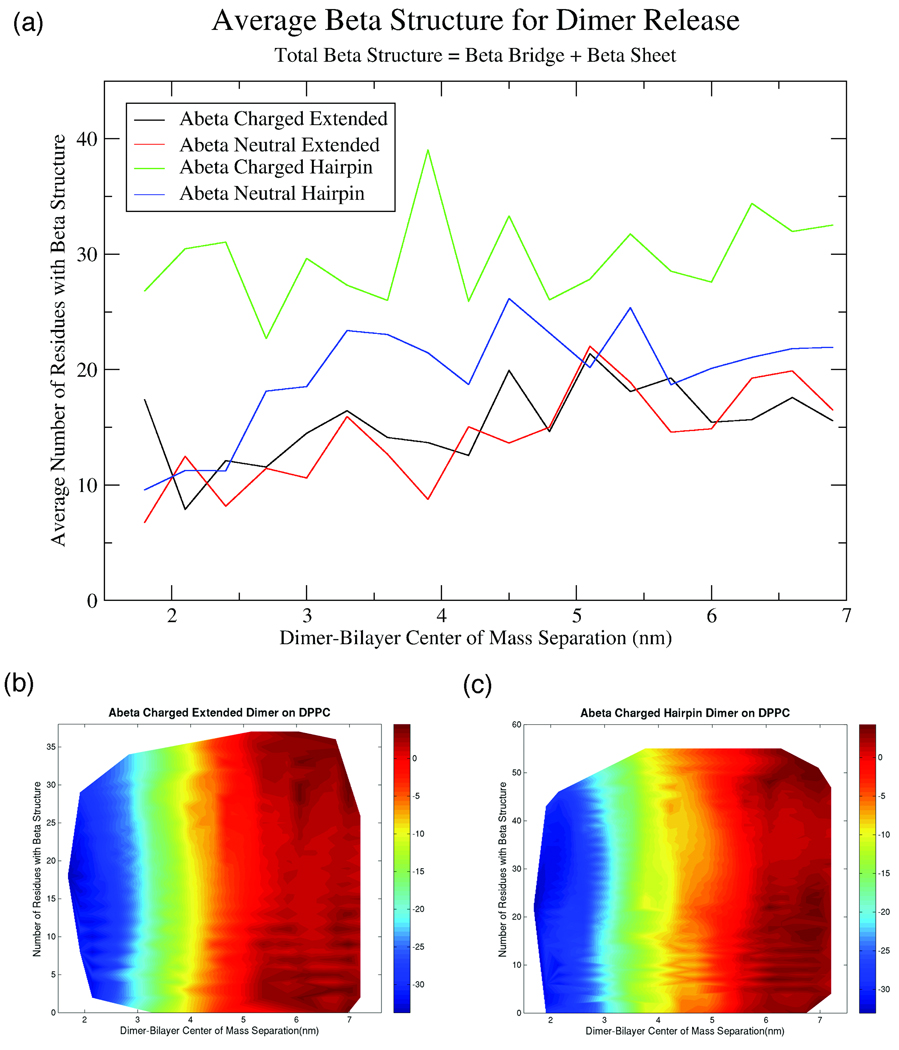
Along with the analysis of the free energies for dimer release and potential of mean force curves, we also investigated secondary structure change during the dimer release process. Use of umbrella sampling for free energy calculations provides 145ns simulations, after 5ns of equilibration, at a series of dimer-bilayer center of mass separations. By analyzing secondary structure content within each umbrella, we are able to obtain some insight into how secondary structure changes as the dimer is released from the bilayer surface. Figure 4a shows the average β-structure for each of the dimers studied as a function of center of mass separation. For these calculations, β-content is termed as any residue with β-bridge or β-sheet structure as determined by DSSP calculation. From this plot, it appears that the amount of β-content slowly increases as the dimer is removed from the bilayer surface. For the charged and neutral extended dimer and the neutral hairpin dimer, the average β-content is approximately the same, between 10–20 residues. For the charged hairpin dimer, the average β-content is somewhat higher than the same structure with a neutral charge. The average values for β-content do increase as the dimer is pulled from the surface, it is not clear if these averages are due to a population of a few structures centered on one average structure, or if the average is due to a large range of structures over a significant variation in β-content. To investigate this discrepancy, we calculated free energy surfaces as shown in Figures 4b and 4c. Figures 4b and 4c show the free energy surface for either the charged extended dimer (Figure 4b) or the charged hairpin dimer (Figure 4c) as a function of bilayer-dimer center of mass separation and the number of β-residues. It is obvious from these two plots that the free energy surfaces are very similar. In both cases, there are no substantial free energy wells at a given bilayer-center of mass separation. Actually, if the value of bilayer-center of mass separation is fixed, there appears to be no favored number of β-residues. This implies that the secondary structure content for either the extended dimer or the hairpin dimer is very fluid and there is not a “dominant” structure throughout this process. Also, this demonstrates that the apparent slight increase in average β-structure as the dimer is removed from the surface is not significant. It appears that the averages shown in Figure 4a represent numerical values with little physical significance, as the average is not representative of the expansive population of potential secondary structures available to the dimer at any dimer-bilayer center of mass separation. These results are consistent for the dimer release process of a neutral dimer from the DOPS bilayer as well. What can be concluded from these plots is that neither lipids on the bilayer surface or water molecules in solution greatly bias the dimer towards a specific secondary structure but allow for substantial structural flexibility. We also investigated other secondary structure elements, such as helix content, as it has been predicted that Aβ can adopt a helical shape when inserted near the bilayer interface 34. However, as the initial dimer structures were heavily biased towards β-sheets, the extent of helix structure was negligible and any helix structure that appeared was transient. For the dimer release process, it appears that the nature of the lipid headgroup significantly affects the energetics of the release process, but does not bias the system towards a favored secondary structure content during release.
FIGURE 4.
(a) Plot of average number of residues with β-structure, where β-structure contains β-Bridge and β-Sheet residues, as a function of dimer-bilayer center of mass separation for the dimer release step of the thermodynamic cycle. Also shown are 2D free energy surfaces of number of residues with β-structure and center of mass separation with free energy in units of kcal/mol for the (b) Extended charged dimer or (c) Hairpin charged dimer on DPPC.
Dimer Dissociation
The fourth step in the thermodynamic cycle involved calculation of the free energy required to dissolve the specific Aβ dimer structure in solution, leaving two non-interacting Aβ monomers. For this calculation, the final Aβ structure from the dimer release simulation at the largest bilayer-center of mass separation was used as the initial structure for the dimer dissociation calculation. The dimer was placed in a water box large enough so that periodic images would not interact at the largest monomer-monomer center of mass separation. Again, as in the dimer release calculation, umbrella sampling was used to calculate the potential of mean force curves (Figure 3b) and the ΔGDissociation was calculated.
From both the ΔGDissociation values and the potential of mean force curves, it is apparent that a significant amount of free energy is required to dissolve both dimer structures. In solution, the protein-protein interactions for both dimer structures are very strong, thus leading to the high free energy of dissociation. Further, the potentials of mean force for all four structures show a similar shape. The curves are smooth until close to the free energy minimum, where a sharper drop occurs until a broad free energy minimum is reached. It appears that the two monomers quickly form an ordered structure once the two monomers are close enough together.
For the charged extended, neutral extended and charged hairpin dimer structures, both the ΔGDissociation and the potential of mean forces curves are very similar. This is not surprising considering the interactions that are stabilizing the dimer. For both extended dimer structures, the monomers are overlapping from approximately residues 28 – 42 on the C-terminus. The majority of residues in this region of Aβ are hydrophobic and derive from the transmembrane portion of the APP peptide before secretase cleavage. As there are no histidines in this region of the peptide, there should be no difference within the overlapping regions of the monomer for the charged and neutral dimer. The difference in charge between these two species is at the N-terminus of the peptide and these regions of the dimer do not participate in any extensive interprotein interactions. This information also provides an important baseline for the strength of the hydrophobic interaction at the C-terminus of the peptide. With ΔGDissociation values between −15kcal/mol and −20kcal/mol, the hydrophobic interactions and potential hydrogen bonding interactions between the two C-termini of the monomers provide a substantial stabilizing force for this dimer structure.
Dimer dissociation for the neutral hairpin dimer is very different from the three other conditions. While the potential of mean force curve is similar in its shape, the ΔGDissociation for the neutral dimer is almost 50% larger than the ΔGDissociation of the charged peptide. Thus, extra interactions are occurring in the neutral dimer that are not available to charged dimer. As mentioned previously, the difference between the charged and neutral dimer are the protonation of three histidines (His 6, His 13 and His 14) on the N-terminal tail of Aβ. While, for the extended dimer, the geometry of the dimer prevents the two N-terminal tails from interacting, the hairpin dimer promotes N-terminal tail interaction due to the geometry. As mentioned in more detail in the Supplemental Materials, high temperatures are used in the equilibration of the hairpin dimer to randomize the structure of the N-terminal tails and prevent any bias due to the initial structure of the N-terminus. However, once the dimer is placed on the bilayer surface, it is possible for these tails to interact. Further, because of the strong interactions occurring between residues 28 – 42 of the two monomers, as demonstrated by the large ΔGDissociation of the extended dimers, the N-terminal tails are restricted in their motion. It is also a fallacy to state that these tails are fully charged or hydrophilic. While the tails do contain the majority of charged or hydrophilic residues in the peptide, a significant number of amino acids in the N-terminus are still hydrophobic. Thus, strong interactions could occur between the two N-terminus tails.
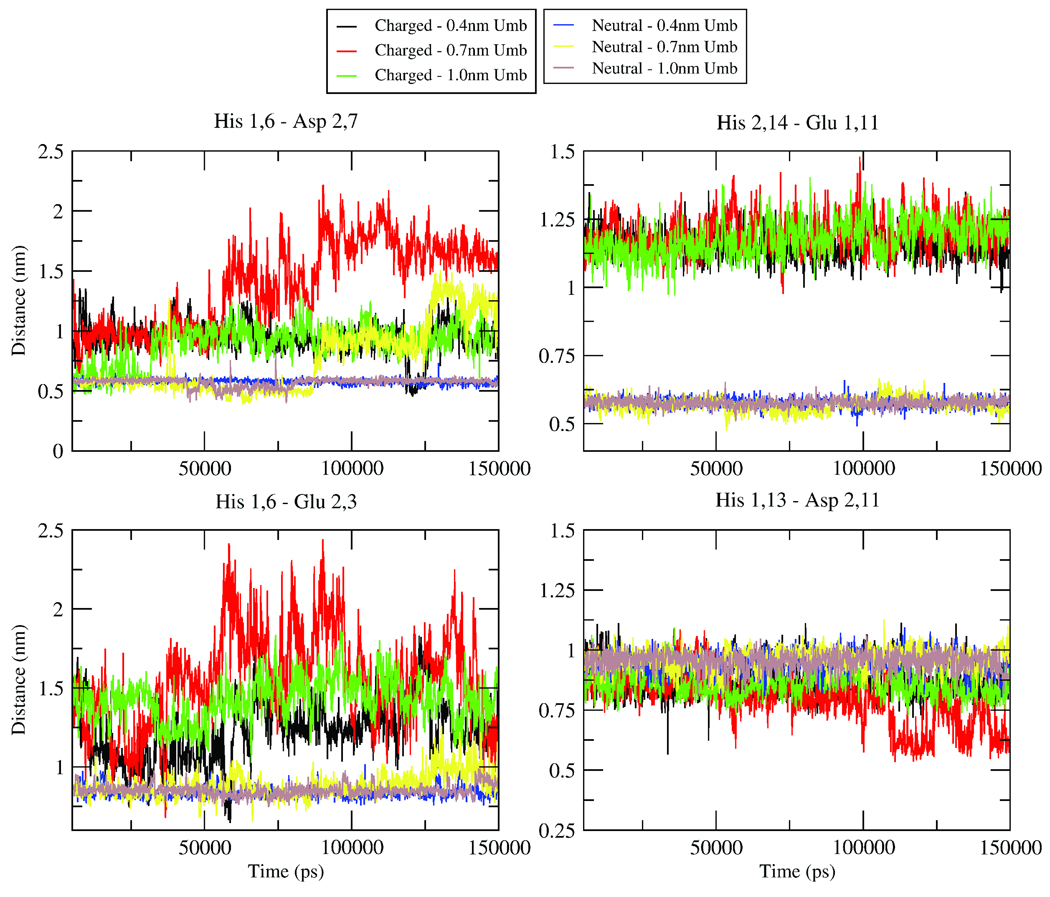
To investigate if the N-terminus residues are the cause of the significant difference between the ΔGDissociation of the charged and neutral hairpin dimers, we calculated center of mass distances between residues of the two monomers (Figure 5). The reasoning behind these calculations was to determine if specific interactions were occurring for the neutral peptide that were not occurring for the charged peptides due to the protonation of the three histidines. Figure 5 shows four representative plots for these distances. Also, for each pair of residues, these distances were calculated for three different windows. Each window corresponds approximately to the free energy minimum and the use of three different windows shows that these effects are not due the choice of a specific simulation. It can be observed for three of these plots that, for the neutral dimer, the two analyzed residues are much closer and the separation between them fluctuates much less, implying a more stable dimer structure. For His 1,6 and Asp 2,7 (where the first number identifies the monomer and the second number identifies the residue within that monomer) and His 1,6 and Glu 2,3, there is a much larger fluctuation for the charged tails. This demonstrates that the N-terminal tails of the charged dimer show some electrostatic repulsion between the two tails, which prevents them from coming close enough to form stable electrostatic or hydrogen bonding interactions. For His 2,14 and Glu 1,11, the fluctuations are less extreme than the first two examples for the charged dimer, but the extent of fluctuations is still larger than for the neutral dimer. The smaller fluctuations for these two residues for the charged dimer are likely due to the geometric restrictions of being closer to the “fixed” points of the hydrophobic region of the peptides. Further, the neutral dimer shows a smaller distance between the two residues, implying that a direct electrostatic interaction is formed. Once again, this illustrates that electrostatic interactions are occurring between residues of the N-terminal tails that act like a glue to stick the tails together, which helps to form hydrophobic or hydrogen-bonding interactions that are not seen between the two N-terminal tails for the charged dimer. The interaction between His 1,13 and Asp 2,11 is included to confirm that this pattern does not hold for all pairs of charged residues on the N-terminal tails. However, analysis of all pairs of nearby charged residues with His6, His13 and His14 in the N-terminal tails shows patterns closest to the first three plots shown in Figure 5 for the majority of pairs. Further, similar analysis was done with some hydrophobic pairs within the N-terminus. While the results were not as dramatic as the pair distance plots shown in Figure 5, the neutral dimer did show closer pair distances and less fluctuation then the charged dimer. These results imply that the much stronger ΔGDissociation for the neutral hairpin dimer is due to interactions between the N-terminus of the two monomers. They also emphasize the importance of the N-terminus and the role of pH and protein charge in the Aβ aggregation process.
FIGURE 5.
Distance measurements between the centers of mass of indicated residues for either charged or neutral Hairpin dimer in solution. Distances were calculated within the 0.4nm, 0.7nm and 1.0nm umbrellas to show that the explained phenomenona were not due to a specific structure or initial condition. Amino acids are listed with the monomer number followed by the residue number on that monomer. For example, Asp2,7 is actually Asp7 on the second monomer. The plots begin at 5000ps to indicate that the first 5ns were considered equilibration.
Dimerization on the Bilayer Surface
The value for ΔGDimerization is calculated using Eq. 1 from the free energy values calculated in this work and a previous work37. Further, in Table I, a value for ΔGDimerization + 2*ΔGBinding, which can be termed the total dimerization free energy, is also provided. This second value represents the free energy gain in binding two non-interacting Aβ monomers in solution into a specific dimer structure on the bilayer surface. The values for ΔGDimerization show that dimerization on the bilayer surface is favorable for most conditions. Even for the neutral extended dimer, the free energy gain upon monomer binding is able to drive a favorable dimerization process. It is important to consider this total dimerization free energy in comparison to the directly calculated ΔGDimerization as binding of the monomer to the bilayer surface may be one of the key aspects of membrane-assisted Aβ aggregation. Overall, these results of the calculations from the thermodynamic cycle presented in this work demonstrate the effect of the membrane on multiple aspects of Aβ dimerization process.
Discussion
Justification of Chosen Dimer Structures
The intention of our work was to calculate a ΔGDimerization for specific Aβ dimers on a membrane surface. We have strived to use two different structures to investigate protein-lipid interactions at the Aβ dimer level and to use the ΔGDimerization as a tool to differentiate the effects of lipid and peptide charge on this process. Therefore, one of the first, and most fundamental, points of discussion in this work is the choice of the two specific dimer structures. For this work, we are deriving no actual conclusions as to which of the structures is correct or which structure is most likely within a fibril or an oligomer. Previous work has demonstrated that the small oligomer structures of Aβ are very heterogeneous, including structures containing significant β-sheet, α-helix and unstructured sections. Due to the nature of the thermodynamic cycle chosen for this work, intermediate monomer structures, such as predominantly α-helical or β-sheet structures, would not effect the conclusions regarding dimerization free energies of the specific dimer structures that were chosen, as monomer structure is masked by the nature of the cycle. Because of the significant β-structure formed by Aβ mixed with anionic lipids11 and the proclivity of Aβ to form fibrils on the membrane surface9,66, we decided to use β-sheet models for the dimers studied in this work. However, extension of this work to other potential dimer structures, including dimers with significant α-helical character that has been observed in studies with Aβ monomers27,76, would be critical to gaining a better understanding of the structural diversity of Aβ when interacting with cell membranes.
Our choices for dimer structures were also motivated by the discrete structural differences between these dimers. The extended dimer is a more solvent-exposed dimer with the N-terminal sections of the peptide not able to interact. The hairpin dimer is more compact, has N-terminal sections that are able to interact and has a different potential β-structure as the two monomers are parallel versus antiparallel in the extended dimer. Direct comparison of these two dimers allowed us to study both the importance of N-terminus – N-terminus interactions in dimerization and the inherent stability of the interactions between the C-terminus regions of the two monomers.
The use of the thermodynamic cycle to calculate ΔGDimerization was another crucial step necessary to obtain the results presented in this work. Directly calculating a dimerization free energy on the membrane surface through a method such as umbrella sampling would suffer from substantial limitations associated with the reaction coordinate and with the sampling of the system. First, the reaction coordinate that is chosen for the pulling would highly bias the system to a particular dimer structure. With the current restrictions to sampling in all-atom molecular dynamics, pulling together two monomers on the bilayer surface will not give a global ΔGDimerization but give a path-dependent ΔGDimerization for a specific structure. However, within our thermodynamic cycle, the choice of reaction coordinate at each step is more obvious. The reaction coordinate for monomer binding is explained in a previous work37. Much like in the monomer binding study, the use of the dimer-bilayer center of mass separation is also a clear reaction coordinate for dimer release. For dimer dissolution, the reaction coordinate of monomer-monomer center of mass separation also appears to be fairly apparent. Because the end point of dimer dissolution is two non-interacting Aβ monomers, using monomer-monomer center of mass separation is an appropriate reaction coordinate. By using a thermodynamic cycle with more explicit reaction coordinates, we can calculate a ΔGDimerization for a specific dimer that is reaction coordinate independent.
As mentioned, another reason for utilizing the thermodynamic cycle was sampling issues. In a previous work, we observed that interactions with a cell membrane slow down structural transitions for monomeric Aβ38. This would imply that directly calculating ΔGDimerization through pulling two monomers together would suffer from this same sampling issue, where the restrictions of all-atom molecular dynamics would not allow enough conformational sampling in reasonable simulation times. For the other steps of the thermodynamic cycle, the conformational sampling issue is not as substantial. The error bars we calculate for our potentials of mean force are small and the simulations are of significant length, indicating that these are reasonable calculations for steps of the cycle. Additionally, for these systems, extensive equilibration is performed before the umbrella sampling to lessen bias due to initial conditions, but this bias will still exist to some extent. Within each step of the cycle, a full exploration of conformational space would require advanced MD techniques, such as a combination of umbrella sampling with replica exchange. However, while more sampling would be ideal for all steps of this calculation, the penalties for lacking of complete sampling for dimer release and dimer dissolution are much less significant than for a direct dimerization free energy calculation.
Dimer Release Process
Within the thermodynamic cycle, each step reveals interesting aspects of this process. A detailed analysis of monomer binding is provided in our previous work37. For the dimer release process, what appears to be the deciding factor dictating dimer release is the charge on the bilayer surface and not the specific dimer structure. On the DPPC surface, both the extended and hairpin dimer are tightly bound with a very large ΔGRelease. Similar to the results seen with monomeric Aβ, both the hairpin and extended dimer interact strongly with the interfacial region of the bilayer. Because of the zwitterionic nature of the phosphatidylcholine headgroup, lack of repulsion between the Aβ N-termini and the lipid headgroups allows the hydrophobic portions of the dimer to interact with the interfacial region of the lipids. Further, comparison of the ΔGBinding and ΔGRelease shows that, as ΔGRelease is very close to 2*ΔGBinding in magnitude, the monomers within the dimer are not undergoing strong protein-protein interactions on the membrane surface, but still act similar to the bound Aβ monomers. Thus, peptide-lipid interactions remain dominant on DPPC. However, on DOPS, the dimer is more weakly bound to the bilayer surface. From our simulations with neutral Aβ on DOPS, we found that, in comparison to the dimers on DPPC, ΔGRelease is much smaller then 2*ΔGBinding. Because the values for ΔGDissociation are large for the neutral dimer, we can assume that peptide-peptide interactions are favored on DOPS over much weaker peptide-lipid interactions. On DOPS, the dimer is not acting as two monomers but is acting as a dimeric unit that binds more weakly to the lipid surface.
Dimer Dissociation Process
In the dimer dissociation step of the thermodynamic cycle, it appears that the dimer structure begins to play a more decisive role. From both the ΔGDissociation and free energy profiles, it is clear that the neutral hairpin dimer has a drastically different dissociation free energy than either extended dimer. Further, the neutral hairpin dimer dissociation differs from the charged hairpin dimer; yet, the charged hairpin dimer dissociation is almost exactly dissociation in the same as the two extended dimer calculations. For the two extended dimer conditions, the N-termini are physically separated and are not able to interact. As the difference in charge between the neutral and charged Aβ dimers is localized to the N-termini, which are non-interacting for the extended dimers, there is no significant difference in the dissociation calculations for the two extended dimers. However, for the hairpin dimer, this is not the case. Due to the parallel β-sheet structure of the overlapping hairpins in the dimer, the N-terminal sections of both monomers are restricted to being in proximity. For the charged hairpin dimer, electrostatic repulsion between the tails prevents extensive interaction. Thus, the interactions, either electrostatic, hydrophobic or hydrogen-bond, that bind the two monomers together for the charged hairpin are isolated to the C-terminal halves of the monomers, which is the same region of interaction as the extended dimer. This is clearly seen in the similarities of the ΔGDissociation and free energy profiles of the charged hairpin and two extended dimers. However, for the neutral hairpin dimer, the charged histidines in the N-terminal tail of each monomer are now available for favorable electrostatic interactions with the anionic residues with the opposing monomer. By changing the N-terminal interaction from unfavorable to favorable and allowing for extensive electrostatic and hydrophobic interactions amongst the tails, the monomers are more tightly bound and the magnitude of ΔGDissociation increases significantly.
Our calculations show the importance of N-terminal tail charge state when studying Aβ peptide oligomerization and raise an important caveat to future experimental and computational studies of this system. While studies using Aβ fragments, especially C-terminal fragments, have been very important and necessary to extending our knowledge of this system, it is important to note that the N-terminal half of Aβ can play a significant role in this process. If studies are performed to investigate effects of peptide charge on aggregation, use of the full Aβ peptide will be crucial to obtaining a full understanding of this system.
Dimerization Free Energy
Using the thermodynamic cycle presented in this work, we have been able to estimate a dimerization free energy, ΔGDimerization, as presented in Table I. From this calculation, it appears that the formation of a dimer on the bilayer surface for three different conditions, the charged extended dimer and both hairpin dimers, is favorable while formation of the neutral extended dimer is not. However, we believe that this ΔGDimerization is a somewhat misleading number. While this number would provide insight for dimerization of nascent Aβ peptides after APP cleavage but before membrane release, it does not describe the effect of a cell membrane on soluble Aβ. To determine this value, it is best to look at the sum of ΔGDimerization + 2*ΔGBinding. Considering this value, dimerization is highly favorable for all potential dimer structures. Nevertheless, it is important to remember that our calculations are only for thermodynamic properties of the system and not kinetic properties, which may significantly bias the favored dimer structure for a given Aβ-membrane system. Thus, the most important result of this ΔGDimerization calculation is that the dimerization free energy for all dimer structures calculated here are favorable considering the full membrane binding and membrane-assisted dimerization process.
The limitations of this work are similar to previous computational studies. Simple one-dimensional reaction coordinates are not perfect for describing most biological systems and sampling will continue to be an issue for all-atom molecular dynamics calculations with the current computing environment. Because of these limitations, we were not able to directly calculate ΔGDimerization. However, it is our belief that use of the thermodynamic cycle helps to avoid these issues while still providing us with a substantial amount of data that is helpful towards understanding this system. In the future, use of coarse-grain, replica exchange or other techniques that expand the sampling capabilities of molecular dynamics would be ideal for studying this system and testing our results. The ideal test would be direct experimental validation of the patterns we observed in our data. Hopefully, experimental techniques will progress enough in the near future and resolution can be improved to the extent that direct Aβ protein-protein interactions on the cell surface can be studied. Our work demonstrates that the interaction between Aβ proteins and lipid membranes during the aggregation process is a delicate balance that encompasses a variety of interactions, from hydrophobic to hydrogen-bonding to electrostatic interactions, which dictate the structure and feasibility of Aβ aggregation on a cell membrane.
Conclusion
From the results presented in this work, it appears that the interactions between small Aβ aggregates and cell membranes play a significant role in Aβ aggregation. With the thermodynamic cycle calculations performed here, we demonstrate that Aβ dimers formed on DPPC lipids are not strong dimers, but are similar to two Aβ monomers with weak interpeptide interactions and strong peptide-lipid interactions. However, DOPS lipids favor strong protein-protein interactions and weak protein-lipid interactions during dimerization, implying that DOPS is more likely than DPPC to promote the formation of viable dimers for aggregation. In aggregation, we can speculate that Aβ peptides bind to DOPS lipids, where concentration effects increase local Aβ concentration, thus increasing the probability of two Aβ peptides coming into contact. Due to the anionic nature of the lipid headgroup, the local pH drops and Aβ can adopt a neutral charge. The DOPS lipids then promote strong protein-protein interactions between monomers while weakening protein-lipid interactions during dimerization. The nascent dimer is free to be released from the bilayer surface while maintaining very strong peptide-peptide interactions, acting as a seed for further aggregation in solution, or dimers could accumulate on the membrane surface, leading to fiber formation on the membrane. The complex interactions between Aβ and the membrane surface demonstrated in this work imply the importance of both lipid charge and properties of the Aβ peptide in aggregation.
Supplementary Material
Acknowledgements
This work was supported by the National Science Foundation under grant numbers MCB-0615469 and MCB-0950280 and by a National Institute of Health Ruth L. Kirschstein National Research Service Award to CHD. The authors would like to thank V. Williams and the UNC Research Computing Group for providing and maintaining the computing resources used in this work. The project described was supported by Award Number F31AG032819 from the National Institute On Aging. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute On Aging or the National Institutes of Health.
References
- 1.Thirumalai D, Klimov DK, Dima RI. Emerging ideas on the molecular basis of protein and peptide aggregation. Curr Opin Struc Biol. 2003;13(2):146–159. doi: 10.1016/s0959-440x(03)00032-0. [DOI] [PubMed] [Google Scholar]
- 2.Stefani M. Protein misfolding and aggregation: new examples in medicine and biology of the dark side of the protein world. Bba-Mol Basis Dis. 2004;1739(1):5–25. doi: 10.1016/j.bbadis.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 3.Kang J, Lemaire HG, Unterbeck A, Salbaum JM, Masters CL, Grzeschik KH, Multhaup G, Beyreuther K, Mullerhill B. The precursor of Alzheimer's-disease Amyloid-A4 protein resembles a cell-surface receptor. Nature. 1987;325(6106):733–736. doi: 10.1038/325733a0. [DOI] [PubMed] [Google Scholar]
- 4.Miller DL, Papayannopoulos IA, Styles J, Bobin SA, Lin YY, Biemann K, Iqbal K. Peptide compositions of the cerebrovascular and senile plaque core amyloid deposits of Alzheimer's-Disease. Arch Biochem Biophys. 1993;301(1):41–52. doi: 10.1006/abbi.1993.1112. [DOI] [PubMed] [Google Scholar]
- 5.Selkoe DJ. Alzheimer's disease: Genes, proteins, and therapy. Physiol Rev. 2001;81(2):741–766. doi: 10.1152/physrev.2001.81.2.741. [DOI] [PubMed] [Google Scholar]
- 6.Shankar GM, Li SM, Mehta TH, Garcia-Munoz A, Shepardson NE, Smith I, Brett FM, Farrell MA, Rowan MJ, Lemere CA, Regan CM, Walsh DM, Sabatini BL, Selkoe DJ. Amyloid-β protein dimers isolated directly from Alzheimer's brains impair synaptic plasticity and memory. Nat Med. 2008;14(8):837–842. doi: 10.1038/nm1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsuzaki K. Physicochemical interactions of amyloid β-peptide with lipid bilayers. Bba-Biomembranes. 2007;1768(8):1935–1942. doi: 10.1016/j.bbamem.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 8.Aisenbrey C, Borowik T, Bystrom R, Bokvist M, Lindstrom F, Misiak H, Sani MA, Grobner G. How is protein aggregation in amyloidogenic diseases modulated by biological membranes? Eur Biophys J Biophy. 2008;37(3):247–255. doi: 10.1007/s00249-007-0237-0. [DOI] [PubMed] [Google Scholar]
- 9.Yip CM, McLaurin J. Amyloid-β peptide assembly: A critical step in fibrillogenesis and membrane disruption. Biophys J. 2001;80(3):1359–1371. doi: 10.1016/S0006-3495(01)76109-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoda M, Miura T, Takeuchi H. Non-electrostatic binding and self-association of amyloid β-peptide on the surface of tightly packed phosphatidylcholine membranes. Biochem Bioph Res Co. 2008;376(1):56–59. doi: 10.1016/j.bbrc.2008.08.093. [DOI] [PubMed] [Google Scholar]
- 11.Terzi E, Holzemann G, Seelig J. Self-association of β-Amyloid peptide(1–40) in solution and binding to lipid-membranes. J Mol Biol. 1995;252(5):633–642. doi: 10.1006/jmbi.1995.0525. [DOI] [PubMed] [Google Scholar]
- 12.Terzi E, Holzemann G, Seelig J. Interaction of Alzheimer β-amyloid peptide(1–40) with lipid membranes. Biochemistry-Us. 1997;36(48):14845–14852. doi: 10.1021/bi971843e. [DOI] [PubMed] [Google Scholar]
- 13.McLaurin J, Chakrabartty A. Characterization of the interactions of Alzheimer β-amyloid peptides with phospholipid membranes. Eur J Biochem. 1997;245(2):355–363. doi: 10.1111/j.1432-1033.1997.t01-2-00355.x. [DOI] [PubMed] [Google Scholar]
- 14.Ege C, Majewski J, Wu GH, Kjaer K, Lee KYC. Templating effect of lipid membranes on Alzheimer's amyloid β peptide. Chemphyschem. 2005;6(2):226–229. doi: 10.1002/cphc.200400468. [DOI] [PubMed] [Google Scholar]
- 15.Bokvist M, Lindstrom F, Watts A, Grobner G. Two types of Alzheimer's β-amyloid (1–40) peptide membrane interactions: Aggregation preventing transmembrane anchoring Versus accelerated surface fibril formation. J Mol Biol. 2004;335(4):1039–1049. doi: 10.1016/j.jmb.2003.11.046. [DOI] [PubMed] [Google Scholar]
- 16.Ambroggio EE, Kim DH, Separovic F, Barrow CJ, Barrow CJ, Barnham KJ, Bagatolli LA, Fidelio GD. Surface behavior and lipid interaction of Alzheimer β-amyloid peptide 1–42: A membrane-disrupting peptide. Biophys J. 2005;88(4):2706–2713. doi: 10.1529/biophysj.104.055582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wong PT, Schauerte JA, Wisser KC, Ding H, Lee EL, Steel DG, Gafni A. Amyloid-β membrane binding and permeabilization are distinct processes influenced separately by membrane charge and fluidity. J Mol Biol. 2009;386(1):81–96. doi: 10.1016/j.jmb.2008.11.060. [DOI] [PubMed] [Google Scholar]
- 18.Dante S, Hauss T, Dencher NA. β-amyloid 25 to 35 is intercalated in anionic and zwitterionic lipid membranes to different extents. Biophys J. 2002;83(5):2610–2616. doi: 10.1016/S0006-3495(02)75271-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chauhan A, Ray I, Chauhan VPS. Interaction of amyloid β-protein with anionic phospholipids: Possible involvement of Lys(28) and C-terminus aliphatic amino acids. Neurochem Res. 2000;25(3):423–429. doi: 10.1023/a:1007509608440. [DOI] [PubMed] [Google Scholar]
- 20.Anand P, Nandel FS, Hansmann UHE. The Alzheimer's β amyloid (Aβ(1–39)) monomer in an implicit solvent. J Chem Phys. 2008;128(16):165102. doi: 10.1063/1.2907718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anand P, Nandel FS, Hansmann UHE. The Alzheimer β amyloid (Aβ(1–39)) dimer in an implicit solvent. J Chem Phys. 2008;129(19):195102. doi: 10.1063/1.3021062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu C, Murray MM, Bernstein SL, Condron MM, Bitan G, Shea JE, Bowers MT. The structure of Aβ42 C-terminal fragments probed by a combined experimental and theoretical study. J Mol Biol. 2009;387(2):492–501. doi: 10.1016/j.jmb.2009.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baumketner A, Shea JE. The structure of the Alzheimer amyloid β 10–35 peptide probed through replica-exchange molecular dynamics simulations in explicit solvent. J Mol Biol. 2007;366(1):275–285. doi: 10.1016/j.jmb.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 24.Yang MF, Teplow DB. Amyloid β-protein monomer folding: Free-energy surfaces reveal alloform-specific differences. J Mol Biol. 2008;384(2):450–464. doi: 10.1016/j.jmb.2008.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jang S, Shin S. Computational study on the structural diversity of amyloid beta peptide (Aβ(10–35)) oligomers. J Phys Chem B. 2008;112(11):3479–3484. doi: 10.1021/jp076450w. [DOI] [PubMed] [Google Scholar]
- 26.Sgourakis NG, Yan YL, McCallum SA, Wang CY, Garcia AE. The Alzheimer's peptides Aβ40 and 42 adopt distinct conformations in water: A combined MD/NMR study. J Mol Biol. 2007;368(5):1448–1457. doi: 10.1016/j.jmb.2007.02.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomaselli S, Esposito V, Vangone P, van Nuland NAJ, Bonvin AMJJ, Guerrini R, Tancredi T, Temussi PA, Picone D. The α-to-β conformational transition of Alzheimer's Aβ-(1–42) peptide in aqueous media is reversible: A step by step conformational analysis suggests the location of beta conformation seeding. Chembiochem. 2006;7(2):257–267. doi: 10.1002/cbic.200500223. [DOI] [PubMed] [Google Scholar]
- 28.Dong X, Chen W, Mousseau N, Derreumaux P. Energy landscapes of the monomer and dimer of the Alzheimer's peptide Aβ(1–28) J Chem Phys. 2008;128(12):125108. doi: 10.1063/1.2890033. [DOI] [PubMed] [Google Scholar]
- 29.Reddy G, Straub JE, Thirumalai D. Influence of preformed Asp23-Lys28 salt bridge on the conformational fluctuations of monomers and dimers of Aβ peptides with implications for rates of fibril formation. J Phys Chem B. 2009;113(4):1162–1172. doi: 10.1021/jp808914c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tarus B, Straub JE, Thirumalai D. Dynamics of Asp23-Lys28 salt-bridge formation in Aβ(10–35) monomers. J Am Chem Soc. 2006;128(50):16159–16168. doi: 10.1021/ja064872y. [DOI] [PubMed] [Google Scholar]
- 31.Buchete NV, Tycko R, Hummer G. Molecular dynamics simulations of Alzheimer's β-amyloid protofilaments. J Mol Biol. 2005;353(4):804–821. doi: 10.1016/j.jmb.2005.08.066. [DOI] [PubMed] [Google Scholar]
- 32.Cruz L, Urbanc B, Borreguero JM, Lazo ND, Teplow DB, Stanley HE. Solvent and mutation effects on the nucleation of amyloid β-protein folding. P Natl Acad Sci USA. 2005;102(51):18258–18263. doi: 10.1073/pnas.0509276102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Urbanc B, Cruz L, Ding F, Sammond D, Khare S, Buldyrev SV, Stanley HE, Dokholyan NV. Molecular dynamics simulation of amyloid β dimer formation. Biophys J. 2004;87(4):2310–2321. doi: 10.1529/biophysj.104.040980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu YC, Shen JJ, Luo XM, Zhu WL, Chen KX, Ma JP, Jiang HL. Conformational transition of amyloid β-peptide. P Natl Acad Sci USA. 2005;102(15):5403–5407. doi: 10.1073/pnas.0501218102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lemkul JA, Bevan DR. A comparative molecular dynamics analysis of the amyloid β-peptide in a lipid bilayer. Arch Biochem Biophys. 2008;470(1):54–63. doi: 10.1016/j.abb.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 36.Lemkul JA, Bevan DR. Perturbation of membranes by the amyloid beta-peptide - a molecular dynamics study. Febs J. 2009;276(11):3060–3075. doi: 10.1111/j.1742-4658.2009.07024.x. [DOI] [PubMed] [Google Scholar]
- 37.Davis CH, Berkowitz ML. Interaction between amyloid-β (1–42) peptide and phospholipid bilayers: A molecular dynamics study. Biophys J. 2009;96(3):785–797. doi: 10.1016/j.bpj.2008.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davis CH, Berkowitz ML. Structure of the Amyloid-β (1–42) Monomer Absorbed To Model Phospholipid Bilayers: A Molecular Dynamics Study. J Phys Chem B. 2009;113(43):14480–14486. doi: 10.1021/jp905889z. [DOI] [PubMed] [Google Scholar]
- 39.Chebaro Y, Mousseau N, Derreumaux P. Structures and Thermodynamics of Alzheimer's Amyloid-β Aβ(16–35) Monomer and Dimer by Replica Exchange Molecular Dynamics Simulations: Implication for Full-Length Aβ Fibrillation. J Phys Chem B. 2009;113(21):7668–7675. doi: 10.1021/jp900425e. [DOI] [PubMed] [Google Scholar]
- 40.Masman MF, Eisel ULM, Csizmadia IG, Penke B, Enriz RD, Marrink SJ, Luiten PGM. In Silico Study of Full-Length Amyloid β 1–42 Tri- and Penta-Oligomers in Solution. J Phys Chem B. 2009;113(34):11710–11719. doi: 10.1021/jp901057w. [DOI] [PubMed] [Google Scholar]
- 41.Jang HB, Zheng J, Lal R, Nussinov R. New structures help the modeling of toxic amyloidβ ion channels. Trends Biochem Sci. 2008;33(2):91–100. doi: 10.1016/j.tibs.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 42.O'Brien EP, Okamoto Y, Straub JE, Brooks BR, Thirumalai D. Thermodynamic Perspective on the Dock-Lock Growth Mechanism of Amyloid Fibrils. J Phys Chem B. 2009;113(43):14421–14430. doi: 10.1021/jp9050098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takeda T, Klimov DK. Interpeptide interactions induce helix to strand structural transition in Aβ peptides. Proteins. 2009;77(1):1–13. doi: 10.1002/prot.22406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klein WL, Stine WB, Teplow DB. Small assemblies of unmodified amyloid β-protein are the proximate neurotoxin in Alzheimer's disease. Neurobiol Aging. 2004;25(5):569–580. doi: 10.1016/j.neurobiolaging.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 45.Cappai R, Barnham KJ. Delineating the mechanism of Alzheimer's disease Aβ peptide neurotoxicity. Neurochem Res. 2008;33(3):526–532. doi: 10.1007/s11064-007-9469-8. [DOI] [PubMed] [Google Scholar]
- 46.Arispe N, Diaz JC, Simakova O. Aβ ion channels. Prospects for treating Alzheimer's disease with Aβ channel blockers. Bba-Biomembranes. 2007;1768(8):1952–1965. doi: 10.1016/j.bbamem.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 47.Simakova O, Arispe NJ. The cell-selective neurotoxicity of the Alzheimer's Aβ peptide is determined by surface phosphatidylserine and cytosolic ATP levels. Membrane binding is required for Aβ toxicity. J Neurosci. 2007;27(50):13719–13729. doi: 10.1523/JNEUROSCI.3006-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ono K, Condron MM, Teplow DB. Structure-neurotoxicity relationships of amyloid β-protein oligomers. P Natl Acad Sci USA. 2009;106(35):14745–14750. doi: 10.1073/pnas.0905127106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Valincius G, Heinrich F, Budvytyte R, Vanderah DJ, McGillivray DJ, Sokolov Y, Hall JE, Losche M. Soluble Amyloid β-Oligomers Affect Dielectric Membrane Properties by Bilayer Insertion and Domain Formation: Implications for Cell Toxicity. Biophys J. 2008;95(10):4845–4861. doi: 10.1529/biophysj.108.130997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Quist A, Doudevski L, Lin H, Azimova R, Ng D, Frangione B, Kagan B, Ghiso J, Lal R. Amyloid ion channels: A common structural link for protein-misfolding disease. P Natl Acad Sci USA. 2005;102(30):10427–10432. doi: 10.1073/pnas.0502066102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martins IC, Kuperstein I, Wilkinson H, Maes E, Vanbrabant M, Jonckheere W, Van Gelder P, Hartmann D, D'Hooge R, De Strooper B, Schymkowitz J, Rousseau F. Lipids revert inert Aβ amyloid fibrils to neurotoxic protofibrils that affect learning in mice. Embo J. 2008;27(1):224–233. doi: 10.1038/sj.emboj.7601953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xue WF, Hellewell AL, Gosal WS, Homans SW, Hewitt EW, Radford SE. Fibril Fragmentation Enhances Amyloid Cytotoxicity. J Biol Chem. 2009;284(49):34272–34282. doi: 10.1074/jbc.M109.049809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hou LM, Shao HY, Zhang YB, Li H, Menon NK, Neuhaus EB, Brewer JM, Byeon IJL, Ray DG, Vitek MP, Iwashita T, Makula RA, Przybyla AB, Zagorski MG. Solution NMR studies of the Aβ (1–40) and Aβ (1–42) peptides establish that the Met35 oxidation state affects the mechanism of amyloid formation. J Am Chem Soc. 2004;126(7):1992–2005. doi: 10.1021/ja036813f. [DOI] [PubMed] [Google Scholar]
- 54.Wahlstrom A, Hugonin L, Peralvarez-Marin A, Jarvet J, Graslund A. Secondary structure conversions of Alzheimer's Aβ(1–40) peptide induced by membrane-mimicking detergents. Febs J. 2008;275(20):5117–5128. doi: 10.1111/j.1742-4658.2008.06643.x. [DOI] [PubMed] [Google Scholar]
- 55.Bernstein SL, Dupuis NF, Lazo ND, Wyttenbach T, Condron MM, Bitan G, Teplow DB, Shea JE, Ruotolo BT, Robinson CV, Bowers MT. Amyloid-β protein oligomerization and the importance of tetramers and dodecamers in the aetiology of Alzheimer's disease. Nat Chem. 2009;1(4):326–331. doi: 10.1038/nchem.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miller Y, Ma BY, Nussinov R. Polymorphism of Alzheimer's Aβ(17–42) (p3) Oligomers: The Importance of the Turn Location and Its Conformation. Biophys J. 2009;97(4):1168–1177. doi: 10.1016/j.bpj.2009.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Picone P, Carrotta R, Montana G, Nobile MR, Biagio PLS, Di Carlo M. Aβ Oligomers and Fibrillar Aggregates Induce Different Apoptotic Pathways in LAN5 Neuroblastoma Cell Cultures. Biophys J. 2009;96(10):4200–4211. doi: 10.1016/j.bpj.2008.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Necula M, Kayed R, Milton S, Glabe CG. Small molecule inhibitors of aggregation indicate that amyloid β oligomerization and fibrillization pathways are independent and distinct. J Biol Chem. 2007;282(14):10311–10324. doi: 10.1074/jbc.M608207200. [DOI] [PubMed] [Google Scholar]
- 59.Jang H, Arce FT, Capone R, Ramachandran S, Lal R, Nussinov R. Misfolded Amyloid Ion Channels Present Mobile β-Sheet Subunits in Contrast to Conventional Ion Channels. Biophys J. 2009;97(11):3029–3037. doi: 10.1016/j.bpj.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Paravastu AK, Leapman RD, Yau WM, Tycko R. Molecular structural basis for polymorphism in Alzheimer's β-amyloid fibrils. P Natl Acad Sci USA. 2008;105(47):18349–18354. doi: 10.1073/pnas.0806270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meinhardt J, Sachse C, Hortschansky P, Grigorieff N, Fandrich M. Aβ(1–40) Fibril Polymorphism Implies Diverse Interaction Patterns in Amyloid Fibrils. J Mol Biol. 2009;386(3):869–877. doi: 10.1016/j.jmb.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sachse C, Fandrich M, Grigorieff N. Paired β-sheet structure of an Aβ(1–40) amyloid fibril revealed by electron microscopy. P Natl Acad Sci USA. 2008;105(21):7462–7466. doi: 10.1073/pnas.0712290105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Petkova AT, Ishii Y, Balbach JJ, Antzutkin ON, Leapman RD, Delaglio F, Tycko R. A structural model for Alzheimer's beta-amyloid fibrils based on experimental constraints from solid state NMR. P Natl Acad Sci USA. 2002;99(26):16742–16747. doi: 10.1073/pnas.262663499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Petkova AT, Yau WM, Tycko R. Experimental constraints on quaternary structure in Alzheimer's beta-amyloid fibrils. Biochemistry-Us. 2006;45(2):498–512. doi: 10.1021/bi051952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Luhrs T, Ritter C, Adrian M, Riek-Loher D, Bohrmann B, Doeli H, Schubert D, Riek R. 3D structure of Alzheimer's amyloid-β(1–42) fibrils. P Natl Acad Sci USA. 2005;102(48):17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yip CM, Darabie AA, McLaurin J. Aβ42-peptide assembly on lipid Bilayers. J Mol Biol. 2002;318(1):97–107. doi: 10.1016/S0022-2836(02)00028-1. [DOI] [PubMed] [Google Scholar]
- 67.Kirkitadze MD, Condron MM, Teplow DB. Identification and characterization of key kinetic intermediates in amyloid β-protein fibrillogenesis. J Mol Biol. 2001;312(5):1103–1119. doi: 10.1006/jmbi.2001.4970. [DOI] [PubMed] [Google Scholar]
- 68.Klement K, Wieligmann K, Meinhardt J, Hortschansky P, Richter W, Fandrich M. Effect of different salt ions on the propensity of aggregation and on the structure of Alzheimer's Aβ(1–40) amyloid fibrils. J Mol Biol. 2007;373(5):1321–1333. doi: 10.1016/j.jmb.2007.08.068. [DOI] [PubMed] [Google Scholar]
- 69.Smith DG, Cappai R, Barnham KJ. The redox chemistry of the Alzheimer's disease amyloid β peptide. Bba-Biomembranes. 2007;1768(8):1976–1990. doi: 10.1016/j.bbamem.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 70.Gorbenko GP, Kinnunen PKJ. The role of lipid-protein interactions in amyloid-type protein fibril formation. Chem Phys Lipids. 2006;141(1–2):72–82. doi: 10.1016/j.chemphyslip.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 71.Krishtalik LI, Cramer WA. On the physical basis for the cis-positive rule describing protein orientation in biological-membranes. Febs Lett. 1995;369(2–3):140–143. doi: 10.1016/0014-5793(95)00756-y. [DOI] [PubMed] [Google Scholar]
- 72.van Klompenburg W, de Kruijff B. The role of anionic lipids in protein insertion and translocation in bacterial membranes. J Membr Biol. 1998;162(1):1–7. doi: 10.1007/s002329900336. [DOI] [PubMed] [Google Scholar]
- 73.Mobley DL, Cox DL, Singh RR, Maddox MW, Longo ML. Modeling amyloid β-peptide insertion into lipid bilayers. Biophys J. 2004;86(6):3585–3597. doi: 10.1529/biophysj.103.032342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Patey GN, Valleau JP. Free-energy of spheres with dipoles - Monte-Carlo with multistage sampling. Chem Phys Lett. 1973;21(2):297–300. [Google Scholar]
- 75.Torrie GM, Valleau JP. Non-physical sampling distributions in Monte-Carlo free-energy estimation - umbrella sampling. J Comput Phys. 1977;23(2):187–199. [Google Scholar]
- 76.Coles M, Bicknell W, Watson AA, Fairlie DP, Craik DJ. Solution structure of amyloid β-peptide(1–40) in a water-micelle environment. Is the membrane-spanning domain where we think it is? Biochemistry-Us. 1998;37(31):11064–11077. doi: 10.1021/bi972979f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.