Abstract
All characterized major ampullate silks from orb-web weaving spiders are composites of primarily two different proteins: MaSp1 and MaSp2. The conserved association of MaSp1 and MaSp2 in these spider species, the highly conserved amino acid motifs, and variable ratios of MaSp1 to MaSp2 demonstrate the importance of both MaSp1 and MaSp2 to the strength and elasticity of the fiber. Computer simulated mechanical tests predicted differing roles for MaSp1 and MaSp2 in the mechanical properties of the fibers. Recombinant MaSp1 and MaSp2 proteins were blended and spun into fibers mimicking the computer-simulated conditions. Mechanical testing verified the differing roles of MaSp1 and MaSp2.
Keywords: spider silk, mechanical properties, MaSp1, MaSp2
Spider silks are composed almost entirely of protein (Peters 1955). Each protein posses a distinct substructure of highly conserved amino acid motifs providing the silk fiber with mechanical properties that correspond to its specific use (Vollrath 1992; Hayashi et al 1999). A conserved set of amino acid motifs which can be associated with specific mechanical properties provides a molecular architecture which can be utilized to understand, and potentially manipulate, the structure/function relationship that governs spider silk.
Major ampullate silk, which serves as the lifeline for the spider, has a stress capacity similar to Kevlar but with moderate elasticity (Gosline et al 1986). In all characterized orb-weaving species, a combination of primarily MaSp1 and MaSp2 are responsible for these properties (Xu and Lewis 1990; Hinman and Lewis 1992) although the precise ratio of the two proteins varies (Brooks et al 2005). These two silk proteins are composed of four amino acid motifs: a crystalline poly alanine or (GA)n motif, which forms β-sheets that correspond with strength (Gosline et al 1986; Hayashi et al 1998, 1999; Grubb and Ji 1999; Riekel et al 2000), a GPGXX motif, which likely forms stackable type II β-turns to confer elasticity, and GGX regions, that are believed to form glycine II helices that interact to stabilize the fiber (Dong et al 1991; Hayashi and Lewis 2000; Gosline et al 2002; Rising et al 2005). Combinations of these motifs produce a repetitive pattern that is responsible for the material properties of the biopolymer fiber.
Major ampullate silk fibers contain crystallites made from the poly-alanine sequences of MaSp1 and MaSp2 arranged in antiparallel β-sheets (Grubb and Jelinski 1997; Hayashi et al 1999; Rising et al 2005). This arrangement allows the proteins to “zipper” together on 2 sides of each peptide chain, with the side chain methyl groups occupying the void spaces near the alpha carbons of neighboring peptide chains thus combining hydrogen bonds and van der Waals interactions. This densely packed arrangement results in a structure that is impenetrable to water, a key feature of the resulting solid silk fiber.
Rigid crystallites embedded in a nano-structure cannot completely explain the material properties of the fiber. In fact, these crystallites are suspended in what is described as an “amorphous matrix” which is composed of the glycine rich regions of the constituent silk fibroin proteins. No definitive structure for this fraction of the silk material has been described, although the presence of beta turns has been detected (Ohgo et al 2006). The sequences of both major ampullate silk proteins show a pattern of alternating poly-alanine regions and glycine rich regions, suggesting that each molecule possesses a number of crystalline and amorphous regions. However, because of the extreme stiffness of the crystalline regions (Termonia 1994) and lack of any discernable structural changes with extension, the observed extension of silk fibers must arise from the amorphous regions.
Using amino acid and cDNA analyses (Anderson 1970; Tillinghast 1984; Tillinghast and Christenson 1984; Work and Young 1987; Lombardi and Kaplan 1990), the ratio of MaSp1 to MaSp2 can be determined and attempts have been made to correlate these ratios with the mechanical properties of major ampullate fibers from different species (Brooks et al 2005). Although the ratio of MaSp1 and MaSp2 as well as the pattern of amino acid motifs present in each protein is known for Nephila clavipes (Xu and Lewis 1990; Hinman and Lewis 1992) and correlations have been drawn as to the functions of the motifs (Brooks et al 2008; Dicko et al 2008; Liu et al 2008; Savage and Gosline 2008a, 2008b), the overall contribution of each protein to the mechanical function of the fiber remains elusive.
To unravel the inseparable proteins and determine if the mechanical properties of the major ampullate fiber are due to the occurrence of both proteins in the fiber or just the mere presence of the amino acid motifs, we used two approaches: molecular dynamics simulations and expressed protein fibers. Experiments were designed using three different ratios of recombinant MaSp1 to MaSp2: 100% MaSp1, 50%: 50%, and 100% MaSp2. In silico experiments, simulated silk fibrils were constructed by arranging 20 amino acid chains based on either MaSp1 or MaSp2 N. clavipes sequences, each comprising 120 amino acids with peptide chains arranged in anti-parallel fashion (supplementary material). The peptide arrangement was allowed to equilibrate for 20ns using the molecular dynamics simulation package, NAMD2 (Phillips et al 2005). Following these equilibration simulations, each construct was subjected to virtual mechanical testing using the steered molecular dynamics functionality of the NAMD2 package. One end of each peptide aggregate was fixed while the other was subjected to constant velocity pulling. The output from such a simulation then includes force and displacement data for the construct in addition to atomic coordinates (Figure 1).
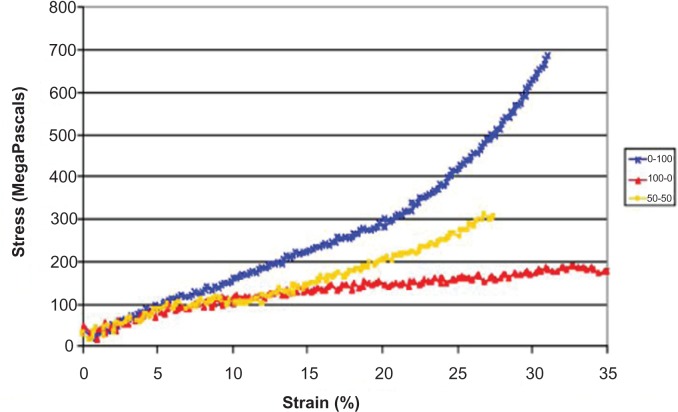
Figure 1.
Molecular dynamics simulations of stress/strain curves produced form either 100% MaSp2 (blue), 100% MaSp1 (red), 50% MaSp1 and 50% MaSp2 (yellow). This model shows discrete contributions of each protein to the overall mechanical functions.
The simulation clearly shows the two proteins have very different mechanical behavior as fibers suggesting that MaSp1 and MaSp2 have distinct contributions to the mechanical properties of a natural silk fiber (Figure 2). It also shows that the blend of the two proteins (n = 9) exhibits behavior intermediate between the two pure protein fibers. Contrary to expectations, the MaSp2 fibers (n = 2) exhibited much higher strength than the MaSp1 fibers (n = 1). Both types of fibers showed similar extension, which was also unexpected. These fibers all showed strains that are almost an order of magnitude lower than those observed for natural dragline silks (15%–30%). The initial modulus, or linear slope, for all three fiber types were similar; however, the shapes of the overall curves were different. The MaSp2 fiber showed an increasing modulus with extension, whereas the MaSp1 fiber showed a more typical two-phase slope where the modulus decreased after the initial extension. The pure MaSp1 constructs displayed an equal amount of extensibility with a much lower modulus. Interestingly, the blended fiber has a shape that looks like a combination of the two pure fibers with the initial extension to approximately 10% matching that of the MaSp1 fiber but then switching to the shape of the MaSp2 fiber. These experiments also confirmed the presence of polyalanine β-sheets embedded in an amorphous matrix. No synergistic properties were apparent upon mixing the two proteins. Notwithstanding disassociating secondary structures, it is important to note that the simulations were terminated after the designed peptides were fully extended but prior to stretching or breaking any covalent bonds. Consequently, comparison with natural major ampullate fibers showed lower stress and strain capacity (Figure 2).
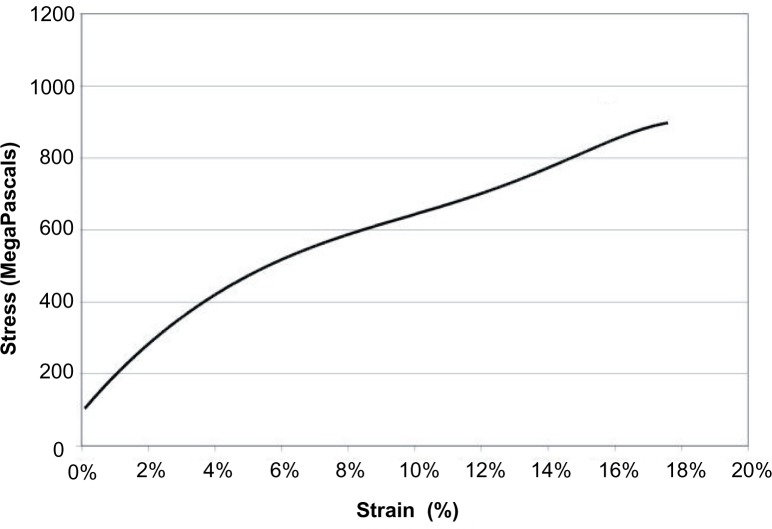
Figure 2.
Polynomial regression of natural Nephila clavipes major ampullate single fibers for comparison with simulations and synthetic fiber mechanical testing.
The recombinant protein blends (n = 9) were produced (Lewis et al 1996; Supplementary materials) based on N. clavipes MaSp1 and MaSp2 sequences (Hayashi and Lewis 1998; Supplementary materials) and were spun into fibers that were subsequently tested on the MTS Synergie 100. For each mechanical test an average stress/strain curve (Figure 3) was calculated using a polynomial regression. Each fiber was spun using a spinning apparatus from Nexia Biotechnologies (Montreal, QC, Canada) (Lazaris et al 2002), which was designed to regulate the force used to extrude the spin dope. Despite the mechanical spinning apparatus, fiber appearance as well as diameter remained highly variable, although in certain instances birefringence was noted. Birefringence is an indication of protein alignment as is seen in a natural N. clavipes major ampullate fiber (Um et al 2004).
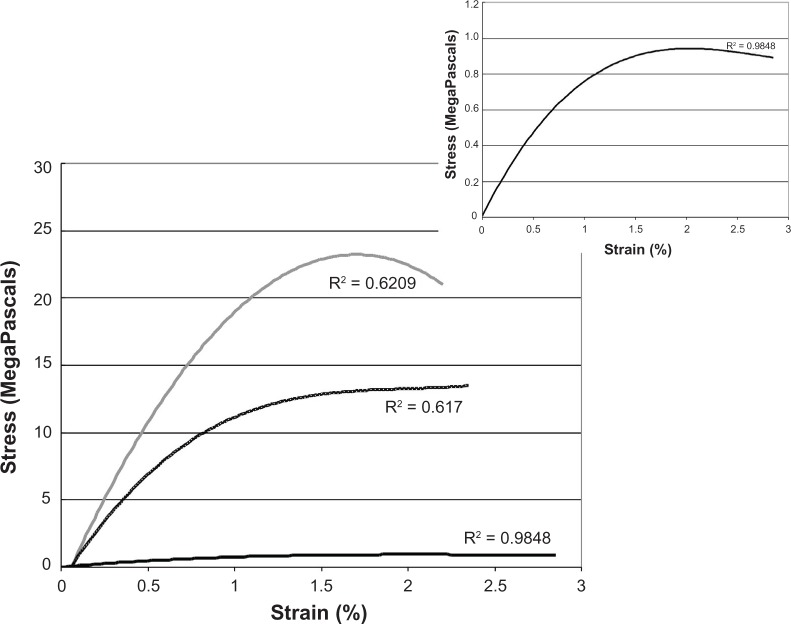
Figure 3.
Mechanical tests of MaSp1 and MaSp2 synthetic protein blends. We found that altering the ratio of the two proteins changed the response of the fiber to simple tensile testing; (light grey) represents Masp2 (2 fibers), (black) represents MaSp1 (1 fibers), and (dotted) shows a 50:50 MaSp1/MaSp2 blend (9 fibers). It is difficult to see the details of the MaSp1 (black) curve due to the extreme differences in stress; therefore, the inset represents a graph with only the MaSp1 fibers.
The recombinant fibers generated qualitatively similar curves but with distinct differences. The MaSp2 fiber again showed a higher stress to break with the MaSp1 fiber in this case much lower and the blended fiber having an intermediate curve. These fibers all showed much lower extension (2%–2.5%) than the computer simulations and the natural fibers (Supplementary Table 2). The shape of the curves also shows substantial differences from the computer simulation. The MaSp2 fiber, instead of an increasing modulus with strain, shows a decreasing modulus. The MaSp1 fiber has a shape similar to the MaSp2 curve with the modulus deceasing with strain (Figure 3 inset). The blended fiber curve again is a composite but shows behavior that is similar to MaSp2 in the initial extension and then like the MaSp1 fiber for further extension, the opposite of the simulation. As with the simulation no synergistic properties were observed with the blend of the two proteins.
| Stress (MPa) | Strain (%) | Young’s Modulus (MPa) | |
|---|---|---|---|
| MaSp1 | 0.83 | 2.9 | 0.7 |
| MaSp2 | 31.1 ± 8.8 | 1.8 ± 0.5 | 20 ± 10.5 |
| 50:50 Blend | 20.4 ± 22 | 1.4 ± 0.6 | 12.6 ± 4.5 |
| Natural Fiber | 806 ± 137 | 12.8 ± 4.5 | 8,520 ± 2,710 |
Discussion
The amino acid sequences of the silk proteins of orb weaving spiders have remained relatively unchanged for over 150 million years (Gatesy et al 2001; Garb et al 2006). Interestingly, the MaSp1 and MaSp2 proteins of major ampullate silk, which contain this conserved molecular architecture, are observed together in all studied major ampullate fibers. However, the varying mechanical properties of single major ampullate silk fibers between different species (Brooks et al 2005; Swanson et al 2006), coupled with species-dependent alterations in the ratio of MaSp1 to MaSp2 supports a hypothesized structure/function relationship. Considering the different motif content of the MaSp1 and MaSp2 proteins allows major ampullate silk to be conceptualized as a nanostructured composite fiber in which each protein makes a distinct contribution to the mechanical attributes of the fiber. Confirming and understanding the discrete contributions necessitates the use of both molecular dynamics simulations as well as mechanical testing of synthetic silk-like fibers.
Both molecular dynamics simulations and laboratory experiments demonstrate that the different the amino motifs that compose MaSp1 and MaSp2 do indeed contribute distinct aspects to the mechanical properties of a silk fiber. The MaSp1:MaSp2 blend fiber has properties that are a combination of pure MaSp1 and pure MaSp2. Thus, it is evident from the results presented here that the mechanical properties (ie, the balance of strength and elasticity) observed in natural major ampullate fibers are in large part due to the proper ratio of MaSp1 and MaSp2. The results from this study substantiate the plausibility of using silk proteins as templates for creating designer materials.
The data from the simulation studies indicates that computer modeling can reveal the basic features of the fibers. The contradiction to expectations for both the MaSp1 and MaSp2 fibers is likely rooted in the unknown interactions between the proteins in the fiber. Additional studies are necessary to consider these interactions, as well as other solvent effects, that likely contribute to the mechanical properties of synthetic fibers. The secondary structure of the poly-Ala motif is clearly a ß-sheet with most of these sheets oriented parallel to the fiber axis, which forms a crystalline block and defines one set of interactions between protein molecules. As is observed for the natural fibers during X-ray diffraction studies, the simulations show that the crystalline regions are maintained during the extension. However, it is not clear what the relative preference for interactions is: intra- and intermolecular either between the same protein or with the other protein in the fiber. The computer simulation was not sufficient to identify these factors. Importantly, the simulation was arranged such that the individual proteins were well mixed initially. If the proteins were not well mixed initially in the lumen of the gland (Vollrath 1992) then such a design would lead to a disparity with the natural fiber.
The GGX motif is most likely in a Gly II helix conformation (Tatham and Shewry 2000), but its orientation relative to the fiber axis is unknown. It is also unknown what its intra- or intermolecular interactions are. It has been suggested that this motif would form anti-parallel supercoils as is seen in GGX synthetic peptides (Tamburro et al 1991). These particular interactions were not obvious in the simulation and if present clearly would influence the mechanical properties of both the MaSp1 and blend fibers. In the case of MaSp2, this would involve the GPGXX sequence, which is highly likely to be in a ß-spiral conformation as is indicated for elastin. However the differing amino acid sequences of the MaSp2 and elastin motifs suggest that the MaSp2 motif structure could involve intermolecular interactions as well as intramolecular ones. As with the GGX motif these are not clearly seen in the simulation.
The data from the recombinant protein fibers was consistent in many ways with the simulation. However, the synthetic fibers themselves vastly under performed the natural fibers in all regards. It is surprising that pure MaSp2 fibers show a qualitatively greater stress to break (or load to break) as well as a higher Young’s modulus (slope of the initial linear region). Both observations seem to indicate a more crystalline fiber than both MaSp1 fibers and blended fibers. MaSp2 contains the polyalanine regions, which form β-sheets and if fiber formation did not allow the correct structure for the GPGXX motifs to form then they would not be available to provide the natural mechanical properties. It has been observed that post-spin draw greatly enhances the properties of many fibers and that process might also lead to removal of possible entanglement of the protein chains, which could occur during the spinning process. Both of these factors could lead to a fiber showing a high crystalline fraction and a limited extension.
The novel finding that synthetic MaSp1 protein fibers, which are composed of the GGX and polyalanine motifs associated with rigid crystalline (Hayashi et al 1999), show a lower Young’s modulus and stress to break appears initially contradictory. This is especially true since the strain to break (indication of the extension of the fiber prior to breaking and a rudimentary measure of elasticity) is approximately the same regardless of the fiber composition (Figure 3). One plausible explanation for this apparent contradiction is the protein folding back on itself, creating a stack of adjacent β-sheet crystals. These β-sheet stacks do not provide additional strength to the fiber; thus, as tension on the fiber is increased, the protein and particularly the β-sheet stacks unfold according to a hierarchy of structures. The lack of orientation of the fibers parallel to the fiber axis, which seems likely based on the very low birefringence observed for these fibers, would also contribute to lesser mechanical properties for all three fibers. A post-spin draw process might alleviate these difficulties.
Although the necessity for two separate proteins to contribute the constituent amino acid motifs to a fiber is not clear, the conservation of both the sequences of MaSp1 and MaSp2 and their combination to form dragline silk for over 100 MY clearly delineates their importance to the mechanical properties of that fiber. This study provides data on the reasons behind this conservation, but clearly there are key factors still to be identified. Regardless, these new insight into the molecular basis for the outstanding material properties of major ampullate silk can now be incorporated into the design of novel biomaterials.
Materials
Recombinant proteins were produced (Tamburro et al 1991; Supplementary materials) based on N. clavipes MaSp1 and MaSp2 sequences (Grubb and Jelinski 1997; Supplementary materials). After expression and purification (Supplementary materials), a 12%–15% (w/v) spin dope was made from each individual protein using lyophilized protein and HFIP (1,3,3-Hexaflouroisopropanol; TCI America, Portland, OR, USA). For protein mixtures lyophilized protein was mixed in the appropriate molar ratio prior to dissolving in HFIP. Each protein powder took about 4 days to completely dissolve in the HFIP (supplementary material) and reach the appropriate viscosity for spinning.
Each fiber was spun using a spinning apparatus from Nexia Biotechnologies (Lazaris et al 2002), which was designed to regulate the force used to extrude the spin dope from a 1 mL Hamilton syringe through 0.005 inch Peek tubing (VWR, West Chester, PA, USA) into an isopropanol coagulation bath. Following fiber formation, the strand was removed from the coagulation bath and mounted on a card with a minimum length opening of 2.5 cm by taping the ends and reinforcing with superglue, as described previously (Brooks et al 2005). Each fiber was viewed with a microscope to determine the cross-sectional area and the length of the fiber was measured. The MTS Synergie 100 (MTS corporation, Eden Prairie, MN, USA) with a custom-built 10-gram load cell (Transducer Techniques, Temecula, CA, USA) was used to test the fibers at 2 mm/min extension.
Supplementary Material
Methods and Materials
All chemicals used were obtained from Sigma Aldrich (St. Louis, MO, USA) unless otherwise stated.
Protein expression
Each protein was produced from approximately 12L of E. coli bacterial culture. The culture was permitted to reach an OD600 of approximately 1 upon which IPTG (isopropyl-beta-D-thiogalactopyranoside) (Biosynth AG, Switzerland) was added to a 1 mM final concentration. After induction, the culture was allowed to produce the synthetic silk protein for 2.5 hours at which time the cells were harvested by centrifugation at 5500 rpm for 20 minutes. Cell pellets were resuspended in a three to one weight to volume ratio of binding buffer (5 mM imidazole, 0.5 M NaCl, 20 mM Tris (Fisher Chemicals, Fair Lawn, NJ, USA) pH 7.9) and frozen at −80 °C until purification.
Purification
Purification was accomplished using the N-terminal histidine tags provided by the pET vector system. To purify the silk-like protein of interest, nickel affinity chromatography (Ni-NTA resin; Novagen, Gibbstown, NJ, USA) was utilized. The manufacturer’s protocol was followed with the following modifications for the specific protein. The cells were lysed chemically with 0.5% lysozyme, 0.01% DNase, and 2% deoxycholate acid sodium salt monohydrate (MP Biomedicals, Aurora, OH, USA). Additionally, to ensure complete lysis, the resuspended cells were also sonicated (Misonix Sonicator 3000) for approximately five minutes. The soluble and insoluble fractions were separated using centrifugation (9500 rpm for approximately 10 minutes). Protein in the soluble fraction was allowed to bind to the resin for approximately 30 minutes on an orbital shaker (S500, VWR, West Chester, PA, USA) and then purified. To compete with nonspecific interactions a 30 mM imidazole wash (30 mM imidazole [Fisher Chemicals, Fair Lawn, NJ, USA], 0.5 M NaCl, 20 mM Tris pH 7.9) followed by a 60 mM imidazole (60 mM imidazole, 0.5 M NaCl, 20 mM Tris pH 7.9) were used. The strip fraction (100 mM EDTA, 0.2 M Tris, 0.5 M NaCl) relied on the chelating affects of EDTA (Fisher Chemicals, Fair Lawn, NJ, USA) to elute the protein. This fraction was dialyzed against water using a stirred cell with a 10 K membrane (Millipore Amicon, Billerica, MA, USA) and subsequently lyophilized.
Results
Amino acid sequence for the MaSp1 and MaSp2 repeat units from Nephila clavipes used for both the synthetic constructs as well as the molecular dynamics simulations. All MaSp1 fibers were spun from a construct with 8 repeat units to produce a protein of approximately 55 kD; whereas all MaSp2 fibers were spun from a construct with 16 repeat units to produce a protein of approximately 67 kD. Accession numbers U20329, M92913.
| MaSp1 | GGAGQGGYGGLGSQGAGRGGLGGQGAGAAAAAA |
| MaSp2 | GPGQQGPGGYGPGQQGPGGYGPGQQGPSGPGSAAAAAAAA |
Comparison of the values for several mechanical properties for natural versus synthetic silk fibers. Natural fibers outperform synthetic versions in every property considered by at least one order of magnitude.
Acknowledgments
The NIH, the NSF, and the Department of Defense supported this work. Amanda E Brooks and Shane R Nelson contributed equally to this work. The authors report no conflicts of interest in this work.
References
- Anderson SO. Amino acid composition of spider silks. Comp Biochem Physiol. 1970;35:705. [Google Scholar]
- Brooks AE, Creager MS, Lewis RV. Altering the mechanics of spider silk through methanol post-spin drawing. Biomed Sci Instrum. 2005;41:1–6. [PubMed] [Google Scholar]
- Brooks AE, Steinkraus HB, Nelson SR, et al. An investigation of the divergence of major ampullate silk fibers from Nephila clavipes and Argiope aurantia. Biomacromolecules. 2005;6:3095–9. doi: 10.1021/bm050421e. [DOI] [PubMed] [Google Scholar]
- Brooks AE, Stricker SM, Joshi SB, et al. Properties of synthetic spider silk fibers based on Argiope aurantia MaSp2. Biomacromolecules. 2008;9:1506–10. doi: 10.1021/bm701124p. [DOI] [PubMed] [Google Scholar]
- Dicko C, Porter D, Bond J, et al. Structural disorder in silk proteins reveals the emergence of elastomericity. Biomacromolecules. 2008;9:216–21. doi: 10.1021/bm701069y. [DOI] [PubMed] [Google Scholar]
- Dong Z, Lewis RV, Middaugh CR. Molecular mechanism of spider silk elasticity. Arch Biochem Biophys. 1991;284:53–7. doi: 10.1016/0003-9861(91)90262-h. [DOI] [PubMed] [Google Scholar]
- Garb JE, Dimauro T, Vo V, et al. Silk genes support the single origin of orb webs. Science. 2006;312:1762. doi: 10.1126/science.1127946. [DOI] [PubMed] [Google Scholar]
- Gatesy J, Hayashi C, Motriuk D, et al. Extreme diversity, conservation, and convergence of spider silk fibroin sequences. Science. 2001;291:2603–5. doi: 10.1126/science.1057561. [DOI] [PubMed] [Google Scholar]
- Gosline J, Lillie M, Carrington E, et al. Elastic proteins: biological roles and mechanical properties. Philos Trans R Soc Lond B Biol Sci. 2002;357:121–32. doi: 10.1098/rstb.2001.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosline JM, DeMont ME, Denny MW. The structure and properties of spider silk. Endevour. 1986;10:37–43. [Google Scholar]
- Grubb DT, Jelinski L. Fiber morphology of spider silk: the effects of tensile deformation. Macromolecules. 1997;30:2860–7. [Google Scholar]
- Grubb DT, Ji G. Molecular chain orientation in supercontracted and re-extended spider silk. Int J Biol Macromol. 1999;24(2–3):203–10. doi: 10.1016/s0141-8130(98)00086-5. [DOI] [PubMed] [Google Scholar]
- Hayashi CY, Lewis RV. Evidence from flagelliform silk cDNA for the structural basis of elasticity and modular nature of spider silks. J Mol Biol. 1998;275:773–84. doi: 10.1006/jmbi.1997.1478. [DOI] [PubMed] [Google Scholar]
- Hayashi CY, Lewis RV. Molecular architecture and evolution of a modular spider silk protein gene. Science. 2000;287:1477–9. doi: 10.1126/science.287.5457.1477. [DOI] [PubMed] [Google Scholar]
- Hayashi CY, Shipley NH, Lewis RV. Hypotheses that correlate the sequence, structure, and mechanical properties of spider silk proteins. Int J Biol Macromol. 1999;24:271–5. doi: 10.1016/s0141-8130(98)00089-0. [DOI] [PubMed] [Google Scholar]
- Hinman MB, Lewis RV. Isolation of a clone encoding a second dragline silk fibroin. Nephila clavipes dragline silk is a two-protein fiber. J Biol Chem. 1992;267:19320–4. [PubMed] [Google Scholar]
- Lazaris A, Arcidiacono S, Huang Y, et al. Spider silk fibers spun from soluble recombinant silk produced in mammalian cells. Science. 2002;295:472–6. doi: 10.1126/science.1065780. [DOI] [PubMed] [Google Scholar]
- Lewis RV, Hinman M, Kothakota S, et al. Expression and purification of a spider silk protein: a new strategy for producing repetitive proteins. Protein Expr Purif. 1996;7:400–6. doi: 10.1006/prep.1996.0060. [DOI] [PubMed] [Google Scholar]
- Liu Y, Shao Z, Vollrath F. Elasticity of spider silks. Biomacromolecules. 2008;9:1782–6. doi: 10.1021/bm7014174. [DOI] [PubMed] [Google Scholar]
- Lombardi SJ, Kaplan DL. The amino acid composition of major ampullate gland silk (Dragline) of Nephila clavipes (Araneae, Tetragnathidae) J Arachnol. 1990;18:297–306. [Google Scholar]
- Ohgo K, Kawase T, Ashida J, et al. Solid-state NMR analysis of a peptide (Gly-Pro-Gly-Gly-Ala)6-Gly derived from a flagelliform silk sequence of Nephila clavipes. Biomacromolecules. 2006;7:1210–14. doi: 10.1021/bm0600522. [DOI] [PubMed] [Google Scholar]
- Peters HM. Über den spinnapparat von Nephila madagascariensis. Z Naturforsch. 1955;10:395–404. [Google Scholar]
- Phillips JC, Braun R, Wang W, et al. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riekel C, Madsen B, Knight D, et al. X-ray diffraction on spider silk during controlled extrusion under a synchrotron radiation X-ray beam. Biomacromolecules. 2000;1:622–6. doi: 10.1021/bm000047c. [DOI] [PubMed] [Google Scholar]
- Rising A, Nimmervoll H, Grip S, et al. Spider silk proteins-mechanical property and gene sequence. Zoolog Sci. 2005;22:273–81. doi: 10.2108/zsj.22.273. [DOI] [PubMed] [Google Scholar]
- Savage KN, Gosline JM. The effect of proline on the network structure of major ampullate silks as inferred from their mechanical and optical properties. J Exp Biol. 2008a;211(Pt 12):1937–47. doi: 10.1242/jeb.014217. [DOI] [PubMed] [Google Scholar]
- Savage KN, Gosline JM. The role of proline in the elastic mechanism of hydrated spider silks. J Exp Biol. 2008b;211(Pt 12):1948–57. doi: 10.1242/jeb.014225. [DOI] [PubMed] [Google Scholar]
- Swanson BO, Blackledge TA, Beltan J, et al. Variation in the material properties of spider dragline silk across species. Appl Phys A. 2006;82:213–8. [Google Scholar]
- Tamburro AM, Guantieri V, Scopa A, et al. Polypeptide models of elastin: CD and NMR studies on synthetic poly(X-Gly-Gly) Chirality. 1991;3:318–23. doi: 10.1002/chir.530030417. [DOI] [PubMed] [Google Scholar]
- Tatham AS, Shewry PR. Elastomeric proteins: biological roles, structures and mechanisms. Trends Biochem Sci. 2000;25:567–71. doi: 10.1016/s0968-0004(00)01670-4. [DOI] [PubMed] [Google Scholar]
- Termonia Y. Molecular modeling of spider silk elasticity. Macromolecules. 1994;27:7378–81. [Google Scholar]
- Tillinghast EK, Christenson T. Observations on the chemical composition of the web of Nephila clavipes (Araneae, Araneidae) J Arachnol. 1984;12:69–74. [Google Scholar]
- Tillinghast EK. The chemical fractionation of the orb web of Argiope spiders. Insect Biochem. 1984;14:115–20. [Google Scholar]
- Um IC, Ki CS, Kweon H, et al. Wet spinning of silk polymer. II. Effect of drawing on the structural characteristics and properties of filament. Int J Biol Macromol. 2004;34:107–19. doi: 10.1016/j.ijbiomac.2004.03.011. [DOI] [PubMed] [Google Scholar]
- Vollrath F. Spider webs and silks. Scientific American. 1992;70 [Google Scholar]
- Work RW, Young CT. The amino acid compositions of major and minor ampullate silks of certain orb-web-building spiders (Araneae, Araneidae) J Arachnology. 1987;15:65–80. [Google Scholar]
- Xu M, Lewis RV. Structure of a protein superfiber: spider dragline silk. Proc Natl Acad Sci U S A. 1990;87:7120–4. doi: 10.1073/pnas.87.18.7120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Amino acid sequence for the MaSp1 and MaSp2 repeat units from Nephila clavipes used for both the synthetic constructs as well as the molecular dynamics simulations. All MaSp1 fibers were spun from a construct with 8 repeat units to produce a protein of approximately 55 kD; whereas all MaSp2 fibers were spun from a construct with 16 repeat units to produce a protein of approximately 67 kD. Accession numbers U20329, M92913.
| MaSp1 | GGAGQGGYGGLGSQGAGRGGLGGQGAGAAAAAA |
| MaSp2 | GPGQQGPGGYGPGQQGPGGYGPGQQGPSGPGSAAAAAAAA |
Comparison of the values for several mechanical properties for natural versus synthetic silk fibers. Natural fibers outperform synthetic versions in every property considered by at least one order of magnitude.