Abstract
Autophagy is the process by which organelles and portions of the cytoplasm are degraded in lysosomes. Several different forms of autophagy are known that are distinguishable chiefly by the mode in which cargo is delivered to the lysosome for degradation. Ubiquilin was recently reported to regulate macroautophagy, the form of autophagy in which cytosolic cargo is packaged in a double-membrane structure or autophagosome that fuses with lysosomes for degradation. We confirm here using different morphological and biochemical procedures that ubiquilin is present in autophagosomes in HeLa cells and in brain and liver tissue of mouse. Coimmunoprecipitation studies indicated that ubiquilin binds the autophagosome marker LC3 in a complex and that reduction of ubiquilin expression reduces autophagosome formation, which correlates with a reduction in maturation of LC3-I to the LC3-II form of the protein. We found that ubiquilin is degraded during both macroautophagy and during chaperone-mediated autophagy (CMA), the latter of which involves the active transport of proteins into lysosomes. We discuss the implication of this degradation in mediating cross-talk between macroautophagy and CMA. Finally, we demonstrate that ubiquilin protects cells against starvation-induced cell death propagated by overexpression of mutant Alzheimer's disease PS2N141I protein and green fluorescent protein (GFP)-huntingtin exon-1 fusion protein containing 74 polyglutamines.
INTRODUCTION
Ubiquilin is present in all eukaryotes and appears to function in protein degradation pathways (1–3). Humans contain four ubiquilin genes, each encoding a separate protein. The proteins are approximately 600 amino acids in length and share extensive homology with each other over their entire sequence. They are characterized by containing an N-terminal sequence that is very similar to ubiquitin, called the ubiquitin-like domain (UBL), followed by a longer more variable central domain containing several heat shock chaperonin-binding motifs (STI1 repeats), and terminate with a conserved 50 amino acid sequence called a ubiquitin-associated domain (UBA). This structural organization is characteristic of proteins that function to deliver ubiquitinated proteins to the proteasome for degradation (4,5).
Indeed, studies of ubiquilin protein have shown that its UBL domain binds subunits of the proteasome and its UBA domain binds polyubiquitin chains (6–8). Moreover, recent studies have shown that ubiquilin regulates ER-associated protein degradation (ERAD), the pathway by which misfolded proteins are dislocated from the ER to the cytoplasm for degradation by proteasomes (9,10). Apart from this function, ubiquilin has also been linked to macroautophagy, an alternative degradation pathway by which cellular cargos are sequestered in double membrane structures called autophagosomes, which subsequently fuse with lysosomes that harbor the acid hydrolases involved in protein degradation (11–13). Early suspicions that ubiquilin might be involved in macroautophagy was the discovery that ubiquilin binds mTOR (mammalian target of rapamycin) kinase (14), a key negative regulator of macroautophagy (15). However, the authors of that study did not find any evidence that mTOR activity was altered by ubiquilin overexpression. Recently, more convincing evidence linking ubiquilin to autophagy was obtained by the demonstration that ubiquilin colocalizes with the autophagosome marker LC3 and that knockdown of ubiquilin-1 reduces, and overexpression of ubiquilin enhances, starvation-induced macroautophagy and its protective effect against cell death (16). In this work, we have confirmed the involvement of ubiquilin in macroautophagy by showing that the protein localizes and copurifies with autophagosomes in HeLa cells and mouse tissue samples, respectively. We further demonstrate that ubiquilin binds the autophagosomal protein component LC3 in a complex and show that silencing of ubiquilin expression leads to a reduction in autophagosome formation that correlates with a defect in maturation of LC3-I to LC3-II. Interestingly, we found that ubiquilin is degraded during both macroautophagy and by chaperone-mediated autophagy (CMA), which is a different form of autophagy (17). Finally, we demonstrate that ubiquilin functions in cytoprotection against mutant presenilin and huntingtin proteins.
RESULTS
Evidence that ubiquilin binds LC3 in a complex
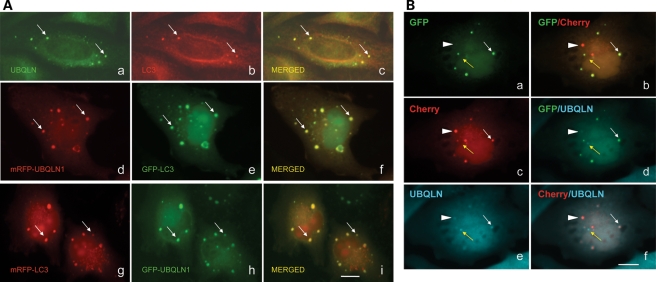
Our immunofluorescence localization studies revealed that endogenous ubiquilin protein is present in both the cytoplasm and nucleus of cells (2). A striking feature of the localization was the enrichment of the protein in puncta of variable size and number (2,14). We used double immunofluorescence staining to identify the nature of the ubiquilin-containing puncta. Of the different antibodies specific for proteins in different organelles tested (autophagosomes, endosomes, perixosomes, mitochondria), only antibodies against LC3, a well-established marker of autophagosomes, decorated the ubiquilin-containing puncta (Fig. 1Aa–c and data not shown). To ensure that the colocalization was not an artifact of the staining procedure, we cotransfected cells with cDNAs encoding ubiquilin-1 or LC3 proteins that had been tagged with either GFP or monomeric red fluorescent protein (mRFP) and examined the cells to see whether the two fluorescent proteins were colocalized. As shown in Figure 1Ad–i, cells cotransfected with the two constructs contained puncta that exhibited both red and green fluorescence, even after the tags were switched. These puncta were not seen in cells coexpressing unfused GFP and mRFP proteins, as expected (data not shown).
Figure 1.
Ubiquilin colocalizes with LC3 in autophagosomes. (A) Immunofluorescence microscopy of HeLa cells stained for endogenous ubiquilin (a) and LC3 proteins (b) and the result of merging of the two images (c). Arrows in this and subsequent panels show examples of colocalization of ubiquilin and LC3 in characteristic autophagosome-like structures. Fluorescent images of a HeLa cell transfected with mRFP- and GFP-tagged ubiquilin-1 (d) and LC3 (e) proteins and the resulting merged image (f). Fluorescent images of HeLa cells transfected with mRFP- and GFP-tagged LC3 (g) and ubiquilin-1 (h) proteins and the resulting merged image (i). (B) Immunofluorescence microscopy of HeLa cells transfected with double-tagged mCherry-GFP-LC3 reporter and stained for ubiquilin. The left-hand panels show the images of the primary fluorescence of GFP (a), mCherry (c) and ubiquilin staining (e) and the right-hand images are the result produced after merging two of the three primary images. The white arrow shows the colocalization of ubiquilin with GFP and mCherry fluorescence. The arrowhead shows colocalization of ubiquilin with predominantly mCherry and faint GFP fluorescence. The yellow arrow indicates rare ubiquilin-positive puncta in which GPF or mCherry fluorescence was not detected. Bar, 5 µm.
We next examined whether ubiquilin is present during maturation of autophagosomes to autolysosomes during macroautophagy. To do so, we utilized the tandem-tagged pH sensitive mCherry-GFP-LC3 autophagosome reporter construct to follow maturation of autophagosomes to autolysosomes (18). As shown previously, fluorescence of both the mCherry and GFP moieties in the tandem tagged LC3 protein is visible in autophagosomes, whereas the acidic-rich environment of autolysosomes destroys that of GFP fluorescence, but does not affect mCherry fluorescence, which remains stable (18). As shown in Figure 1B, autophagosomes were clearly distinguishable with this reporter by colocalization of the mCherry and GFP fluorescence in a subset of the puncta. Autolysosomes were also clearly distinguishable as puncta that contained mCherry, but not GFP fluorescence. Counterstaining of the transfected cells with an anti-ubiquilin-specific antibody revealed the presence of endogenous ubiquilin protein in both autophagosomes and autolysosomes, indicating that ubiquilin is present in both early and late organelles involved in macroautophagy. Interestingly, we detected occasional puncta of similar size to autophagosomes that stained for ubiquilin but which neither exhibited any GFP or mCherry fluorescence despite being clearly transfected with the GFP-mCherry-reporter construct, the identity of which is not known (Fig. 1B, yellow arrow).
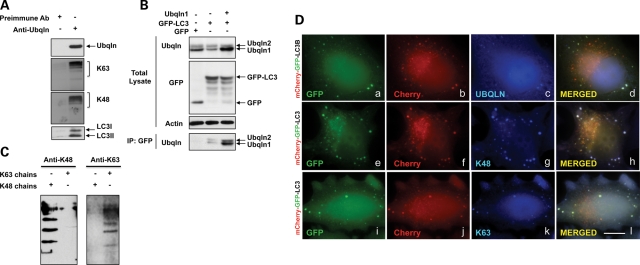
We next used coimmunoprecipitation assays to determine whether ubiquilin and LC3 are bound together in a complex in cells. As shown in Figure 2A, immunoprecipitates prepared from HeLa cells with a ubiquilin-specific antibody contained LC3-I and -II isoforms. The specificity of this immunoprecipitation is evident from the failure to detect ubiquilin and LC3 proteins in parallel immunoprecipitates made with the pre-immune serum. However, for unknown reasons, we were unable to detect coimmunoprecipitation of ubiquilin by the reverse immunoprecipitation. We postulated that this might have arisen from obstruction of the epitope recognized by our LC3 antibody by ubiquilin binding. Accordingly, we transfected HeLa cells with either GFP-LC3 or GFP expression constructs and examined GFP immunoprecipitates from the cells for ubiquilin presence. As shown in Figure 2B, detectable amounts of ubiquilin were coimmunoprecipitated from cells transfected with GFP-LC3, but not from the cells transfected with GFP alone. This interaction was substantiated by overexpression of ubiquilin, which led to increased coimmunoprecipitation of ubiquilin in cells cotransfected with GFP-LC3. These results demonstrate ubiquilin binds both the unmodified and lipid-conjugated forms of LC3.
Figure 2.
Ubiquilin, LC3 and polyubiquitinated proteins bind together in a complex. (A) Equal portions of a HeLa cell lysates were used to immunoprecipitate proteins with either an anti-ubiquilin antibody or its pre-immune serum and the resulting precipitates were probed with antibodies to detect different proteins, as indicated. (B) Cultures of HeLa cells were transfected with cDNAs encoding GFP alone, or GFP-LC3, or GFP-LC3 and ubiquilin-1 (+ indicates lanes in which the cDNAs were transfected). GFP proteins were immunoprecipitated from lysates of the cultures and the resulting precipitates were examined for coimmunoprecipitation of ubiquilin by immunoblotting (bottom panel). Also shown are blots of the total lysates probed with different antibodies, as indicated. (C) Duplicate samples of K48 and K63 in vitro assembled ubiquitin chains (− and + indicate lanes in which the chains were omitted or added, respectively) were immunoblotted with antibodies that specifically recognize K48 or K63 chains, illustrating their specificity. (D) HeLa cells grown on coverslips were transfected with cDNA encoding mCherry-GFP-LC3 and after 24 h the cells were fixed and stained with antibodies to specifically detect either ubiquilin (a–d), or K48-ubiquitin chains (e–h), or K63-ubiquitin chains (i–j). GFP (a, e and i), mCherry (b, f and j) and Cy5.5 (c, g and k) fluorescence images were captured and used to examine whether ubiquilin, K48 or K63 chains colocalized with the mCherry-GFP-LC3 reporter. The right-hand panels are the images produced after merging the three images shown to the left of each of them. Bar, 5 µm.
Ubiquilin binds proteins containing K48- and K63-linked ubiquitin chains that are enriched in autophagosomes
Because ubiquilin possesses a UBA domain that has been shown to bind different ubiquitin moieties in vitro (7,8,19) we asked whether ubiquilin protein in cells binds proteins containing K48- and/or K63-linked ubiquitin chains and whether proteins with these different ubiquitin linkages are contained in autophagosomes. Using antibodies that specifically recognize either K48 or K63 ubiquitin-linked chains (20), we found that endogenous ubiquilin protein that was immunoprecipitated from HeLa cells contained an abundance of proteins tagged with either K48- or K63-linked chains (Fig. 2A and C). To determine whether these chains are present in autophagosomes, we stained HeLa cells transfected with the tandem-tagged GFP-mCherry-LC3 reporter to see whether the chains were localized in autophagosomes. As shown in Figure 2D, autophagosomes identified using the LC3 reporter were found to stain for both K48- and K63-linked ubiquitin chains, suggesting that proteins containing either or both types of linkages are present and degraded by macroautophagy.
Knockdown of ubiquilin expression leads to a reduction in autophagosome number from a presumed failure of maturation of LC3-I to LC3-II
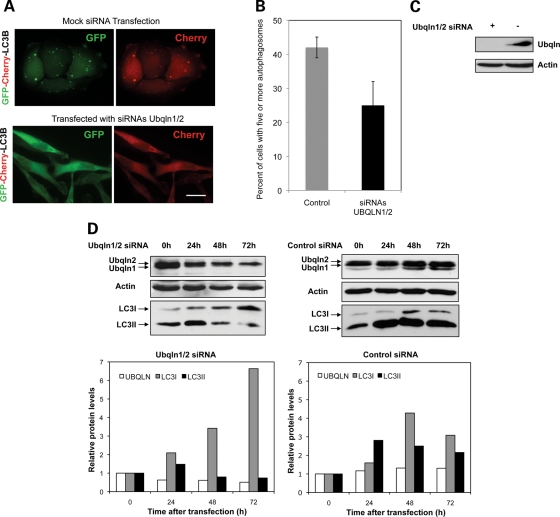
Detection of ubiquilin in different organelles of the autophagic pathway could simply be due to the normal clearance of this long-lived protein (half life of >20 h (2)) by macroautophagy or because ubiquilin actively participates in macroautophagy. To distinguish between these and other possibilities, we asked whether the reduction of ubiquilin levels in cells affects autophagosome formation. Accordingly, we counted the number of cells containing five or more autophagosomes (revealed by dual fluorescence of GFP and mCherry from the mCherry-GFP-LC3 reporter) in cells that had been transfected with siRNAs to knockdown ubiquilin 1 and 2 proteins and in cells in which the proteins had not been targeted for silencing (Fig. 3A). The quantification revealed a dramatic 60% reduction in cells containing five or more autophagosomes after ubiquilin knockdown (Fig. 3B). Immunoblots confirmed that the siRNAs directed against ubiquilin 1 and 2 proteins were indeed effective in reducing ubiquilin protein levels by over 80% (Fig. 3C). To examine whether the reduction of ubiquilin proteins affects LC3 levels, we immunoblotted protein lysates from cells for changes in LC3 proteins over a 72 h period of ubiquilin knockdown. Remarkably, ubiquilin knockdown led to a time-dependent decrease in LC3-II levels, which was especially evident at the 48 and 72 h time points, and this was accompanied by a concomitant increase in the immature form of LC3 (Fig. 3D). The results suggest that ubiquilin knockdown might directly prevent processing and/or conjugation of LC3-I to phosphatidyl ethanolamine to generate LC3-II.
Figure 3.
Knockdown of ubiquilin inhibits autophagosome formation and maturation of LC3 protein. (A–C) HeLa cells were either mock transfected or transfected with siRNAs to specifically knockdown ubiquilin-1 and -2 proteins and 24 h later they were both re-transfected with cDNA encoding mCherry-GFP-LC3. Twenty hours later, the cells were fixed and the percent of GFP transfected cells that displayed five or more GFP puncta was quantified. (A) Representative images of cells showing knockdown of ubiquilin reduces autophagosome formation as measured using the mCherry-GFP-LC3 reporter. Bar, 10 µm. (B) Graphs of the quantification of cells with autophagosomes described in (A) obtained from three independent experiments. The graphs in this and subsequent figures show the mean and the error bars show the standard deviation. (C) Immunoblots showing effective knockdown of ubiquilin proteins in the cultures after 44 h of knockdown (+ and − indicate the cells that were transfected with siRNAs to knockdown ubiquilin-1 and -2 proteins, or which were mock-transfected, respectively). (D) Cultures of HeLa cells were transfected with siRNAs to knockdown ubiquilin-1 and -2 proteins or with negative control siRNAs and lysates were collected 0, 24, 48 and 72 h later. The lysates containing equal amounts of protein were immunoblotted with antibodies to detect ubiquilin, actin and LC3 proteins. Also shown are graphs of the quantification of the ubiquilin and LC3-I and -II bands. Similar results were seen in two separate experiments.
Ubiquilin protein is degraded during macroautophagy
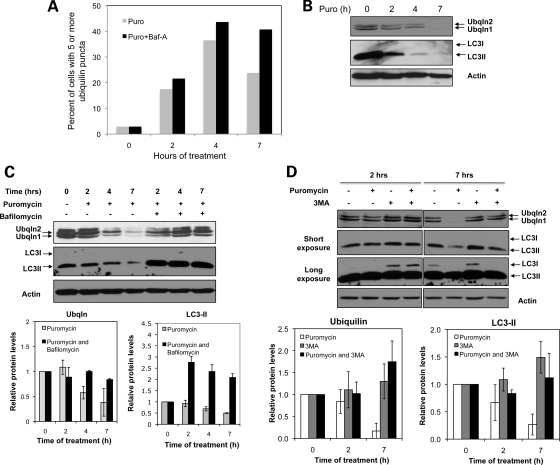
The above studies indicated that ubiquilin is involved in regulating macroautophagy, which is in accord with the conclusions reported by N'Diaye et al. (16). We next examined whether the ubiquilin protein is consumed during macroautophagy, as this would provide clues about the fate of the protein during the process. Because ubiquilin is normally a very stable protein, we stimulated autophagy by treating cells for different periods of time with puromycin, which causes protein misfolding due to premature chain termination of translation (18). As expected, puromycin treatment increased macroautophagy as evident by a progressive time-dependent increase in the percentage of cells containing five or more ubiquilin-positive autophagosomes (Fig. 4A). Peak induction was observed at 4 h and by 7 h the numbers began to decline, presumably because autophagosome clearance exceeded the rate of their formation. Interestingly, immunoblots of protein lysates prepared from the cultures revealed that ubiquilin and LC3-II protein levels both declined progressively during the same 7 h period (Fig. 4B). The immediate decline in LC3 and ubiquilin levels after puromycin treatment suggests that clearance of the proteins is rapid and precedes autophagosome formation. To evaluate whether the loss of LC3-II and ubiquilin proteins arises from consumption of the proteins in macroautophagy, we repeated the experiment, but this time added bafilomycin A1, which acts to block macroautophagy by inhibiting acidification of autolysosomes (21–23). As shown in Figure 4C, bafilomycin A1 treatment prevented the time-dependent decline in ubiquilin and LC3-II levels for the 7 h of puromycin treatment. Similar stabilization of ubiquilin and LC3-II proteins was found by inhibiting macroautophagy with 3MA, which inhibits class-III phosphoinositide 3-kinase (PI3K) Vps34 by a totally different molecular mechanism to bafilomycin A1 (24,25). This is most clearly evident by examining the proteins levels in cells treated with and without 3MA (Fig. 4D). The above experiments strongly suggest ubiquilin is not only required, but is actively degraded during macroautophagy.
Figure 4.
Ubiquilin is degraded during macroautophagy. (A) HeLa cell cultures were treated with puromycin or puromycin and bafilomycin A1 for the time periods as indicated. At the intervals shown, the cells were fixed and stained for ubiquilin. The percent of cells displaying five or more ubiquilin puncta was quantified after capturing images under the microscope with a ×40 objective lens. (B) Immunoblots of equal amounts of protein lysate made from HeLa cell cultures treated for 0–7 h with puromycin and probed for ubiquilin, LC3 or actin. (C) Same as in (B) except that additional lysates from cultures that were treated for 2, 4, and 7 h with both puromycin and bafilomycin A1 were analyzed. Also shown are graphs of the quantification of the ubiquilin [lower two bands corresponding to ubiquilin-1 and -2 proteins (from three separate experiments), and LC3-II bands (from two separate experiments)]. (D) Immunoblots of equal amounts of protein lysate prepared from HeLa cell cultures treated for 2 or 7 h with different combinations of puromycin and 3MA. Actin was used to monitor protein loading. Also shown are graphs of the quantification of the ubiquilin and LC3-II bands obtained from three separate experiments.
Ubiquilin is a substrate of CMA
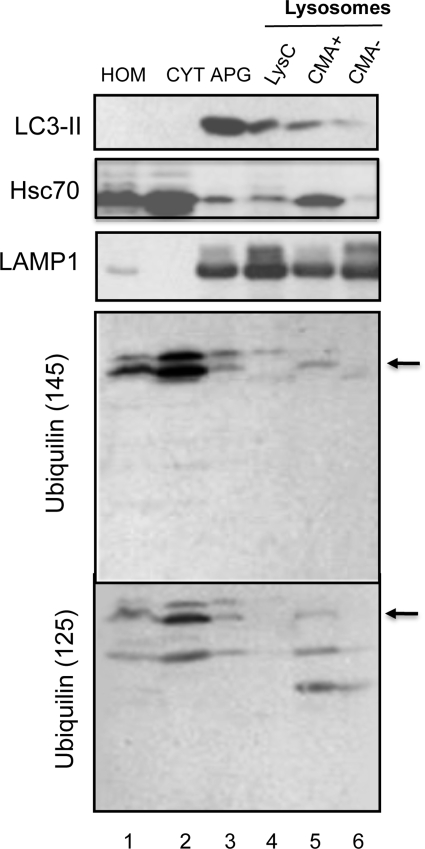
Besides its clear localization with autophagosome markers, we were intrigued by the presence of ubiquilin in puncta that lacked mCherry or GFP fluorescence using the tandem-tagged mCherry-GFP-LC3 autophagosome/autolysosomal reporter (Fig. 1B, yellow arrows), which we theorized could be secondary lysosomes. To confirm the lysosomal nature of this compartment, we isolated autophagosomes (AV) and different classes of lysosomes from mouse liver and examined them for ubiquilin presence by immunoblotting (Fig. 5). Three different lysosome preparations were examined that differed according to their autophagic activities (26,27). The differences between these lysosomes were described previously (27) (see also Fig. 5 legend). The immunoblots showed that our autophagosome and lysosome preparations were heavily enriched for LC3 and LAMP1, respectively, which is consistent with the known enrichment of the proteins in the compartments (Fig. 5). Consistent with its localization seen by fluorescence microscopy, we detected ubiquilin immunoreactive bands in our autophagosome preparation using two different anti-ubiquilin-1 antibodies (UM145 and UM125 made to different regions of ubiquilin-1 protein), strongly indicating ubiquilin is indeed involved in macroautophagy in vivo. Multiple ubiquilin-immunoreactive bands were seen in the blots, the upper two of which we presume correspond to the full length (indicated with an arrow in Fig. 5) and a modified form of ubiquilin, whereas the lower band seen mostly with the UM125 antibody is probably a breakdown product. We next examined whether ubiquilin is also present in the lysosomal preparations from mouse liver possessing different autophagic activities. In agreement with our immunofluorescence data, a fraction of ubiquilin was detectable in the two groups of lysosomes that are chiefly involved in macroautophagy (Lys C and CMA−, Fig. 5) (27). However, and interestingly, levels of ubiquilin were higher in the third group of lysosomes involved in CMA+ (Fig. 5) (27). In contrast to micro- and macroautophagy, both of which involve sequestration and degradation of bulk proteins and organelles, CMA involves the selective transport of individual proteins into lysosomes for degradation. CMA is distinguishable by its strict dependence on hsc70 and LAMP-2A proteins for substrate protein targeting to lysosomes and binding/transport across the lysosomal membrane, respectively (28). Analysis of the amino acid sequence of human ubiquilin-1 revealed that it contains two pentapeptide sequences (ILKDQ and EVRFQ) both of which conform to the sequence requirements for selective recognition and degradation of cytosolic proteins by CMA (17).
Figure 5.
Ubiquilin is enriched in purified autophagosomes and lysosomes active for CMA. Subcellular fractions from mouse liver were isolated as described under material and methods and 100 µg protein of each fraction were subjected to immunoblot analysis for the indicated proteins. Hom, homogenate; Cyt, cytosol; AV, autophagosomes; Lys C, lysosomes; CMA−, lysosomes with low activity for CMA; CMA+, lysosomes active for CMA.
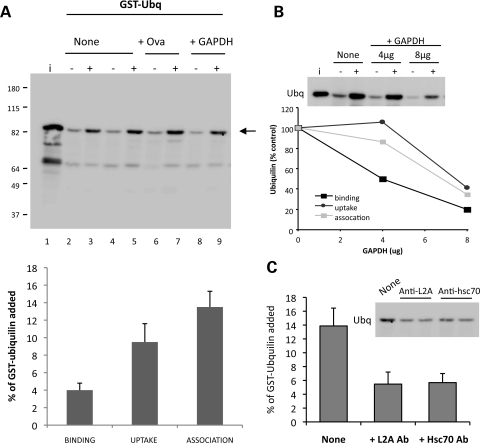
To directly test whether ubiquilin is a substrate of CMA and to determine whether it is transported into lysosomes independently of macroautophagy, we used an in vitro assay to determine whether the recombinant GST-ubiquilin-1 purified protein is bound to and degraded by isolated lysosomes by CMA (29). In this assay, candidate substrates are incubated with lysosomes in the presence or absence of protease inhibitors in an isosomotic media optimized for CMA. After the assay, lysosomes are recovered and examined for binding and transport of the substrate. In the absence of protease inhibitors, any substrate protein that is recovered in the lysosome pellet usually corresponds to the protein that is bound to the lysosomal membrane, whereas the portion that is transported into the lysosomal lumen is typically degraded. In contrast, in the presence of protease inhibitors, both the transported and membrane-bound substrate is protected from degradation. Uptake is thus calculated by subtracting the substrate remaining in lysosomes in the absence of the inhibitors from that seen after the addition of the protease inhibitors (29). As shown in Figure 6A, ubiquilin binds to the lysosomal membrane and ∼10% of the protein added reaches the lysosomal lumen. Internalization of ubiquilin can be competed in a dose-dependent manner with glyceraldehydes-3-phosphate dehydrogenase (Fig. 6B), a well-characterized CMA substrate, but not with ovalbumin, a protein that does not contain a CMA targeting motif (30,31). Moreover, association of ubiquilin with lysosomes was inhibited with antibodies to hsc70 and LAMP2A, two essential components of CMA (Fig. 6C) (28), further supporting that ubiquilin is translocated into lysosomes via CMA. These results confirm that ubiquilin is a bonafide CMA substrate and that part of the protein reaches the lysosomal compartment in vivo where it can be degraded.
Figure 6.
Ubiquilin is a substrate of CMA. (A) GST-ubiquilin-1 (2 µg) was incubated with freshly isolated intact lysosomes (30 µg) treated (+) or not (−) with protease inhibitors. Where indicated, a non-CMA substrate Ovalbumin (Ova) (lanes 6–7) or a CMA substrate GAPDH (lanes 8–9) were added to the incubation media. i: input (0.5 µg). (Bottom) Quantification of binding, uptake and association of GST-ubiquilin-1 to lysosomes. Values are expressed as percentage of the protein added into the reaction and are mean + SE of the densitometric analysis of the four separate experiments. (B) Competition of CMA uptake of GST-ubiquilin-1 by increasing concentrations of GAPDH. GST-ubiquilin-1 (2 µg) was incubated with lysosomes either alone or in the presence of increasing concentrations (as indicated) of GAPDH in an experiment similar to that shown in (A). The graph shows the quantification of binding and uptake of GST-Ubiquilin by lysosomes in the presence of GAPDH. Values were obtained from the densitometric analysis of three different experiments. Inset shows a representative immunoblot. (C) Blockage of ubiquilin CMA with antibodies against LAMP-2A and hsc70. GST-ubiquilin-1 (2 µg) was incubated with lysosomes as in (A) in the absence or presence of neutralizing antibodies against LAMP-2A or hsc70. Quantification of association of GST-ubiquilin-1 to lysosomes is shown. Values are mean + SE and were obtained from the densitometric analysis of three different experiments. Inset shows a representative immunoblot.
Overexpression of ubiquilin-1 reduces cell death induced by AD-mutant presenilin-2 protein
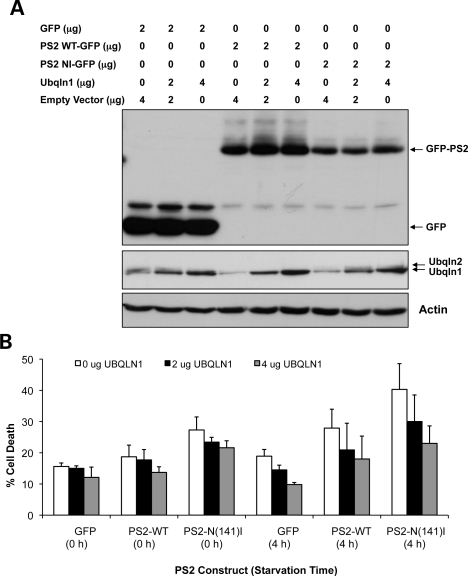
Because ubiquilin binds presenilin proteins (2), we next investigated whether modulation of ubiquilin levels affects cell death caused by overexpression of presenilin-2 proteins. In an elegant study, N'Diaye et al. (16) showed that starvation of HeLa cells induces an increase in cell death due to activation of autophagy and that cell death can be prevented by overexpression of ubiquilin-1. We surmised that ubiquilin function in autophagy might likewise affect presenilin-induced cell death. Previously, we found that overexpression of PS2 harboring the Alzheimer's disease linked N141I mutation in HeLa cells propagates increased cell death compared with the wild-type protein (32). We therefore examined whether PS2 induction of cell death is affected by starvation and whether PS2-induced cell death is suppressed by overexpression of ubiquilin. In accord with our previous findings, the PS2N(141)I mutant induced an increase in cell death compared with the wild-type protein, and this difference was also evident after 4 h of starvation when cell death was clearly amplified (Fig. 7A and B). Importantly, overexpression of ubiquilin-1 suppressed induction of cell death caused by overexpression of PS2 in a dose-dependent manner, which was especially evident at 4 h after starvation (Fig. 7A and B). Unfortunately, we could not reliably quantify the consequence of ubiquilin silencing because of massive cell loss of ubiquilin-knockdown cells following transfection with the PS2 cDNAs. Instead, we examined how survival of cells stably expressing mutant huntingtin (htt) protein is affected by knockdown of ubiquilin, as described below.
Figure 7.
Ubiquilin overexpression reduces starvation-induced cell death caused by overexpression of presenilin-2 proteins. (A and B) Cultures of HeLa cells were transfected with a total of 6 µg of DNA, 2 µg of which corresponded to an expression plasmid encoding either GFP alone, or wild-type PS2-GFP, or PS2N141I-GFP protein and the remainder of which contained either 0, 2 or 4 µg of cDNA encoding ubiquilin-1 protein or with empty vector DNA. Twenty hours after transfection, the cultures were washed and incubated in starvation medium for either 0 or 4 h after which time cell death of GFP expressing cells was quantified. (A) Immunoblots of lysates from the transfected cultures blotted for expression of the GFP-tagged proteins, ubiquilin proteins and actin. (B) Quantification of cell death of GFP expressing cells in the cultures obtained in three independent experiments.
Loss of ubiquilin expression increases cell death in cells expressing GFP-htt-exon-1 protein containing 74 polyglutamines
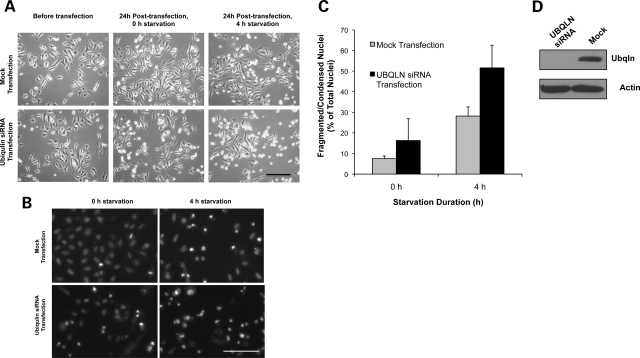
Loss of ubiquilin expression was studied using a HeLa cell line that stably expresses GFP-htt-exon-1 fragment containing 74 polyglutamine repeats (GFPHtt74Q). In a previous report, we showed that overexpression of ubiquilin-1 reduces oxidative stress-induced cell death, whereas knockdown of the protein enhances cell death (33). However, the role of ubiquilin in autophagic regulation of starvation-induced cell death was not tested. Accordingly, we measured starvation-induced cell death in the Htt74Q cell line after knockdown of ubiquilin-1 and -2 proteins. As shown in Figure 8A–D, transfection of cells with siRNAs against ubiquilin-1 and -2 almost doubled starvation-induced cell death compared with mock-transfected cells. Immunoblots confirmed that ubiquilin-1 protein was knocked down in the experiments (Fig. 8D).
Figure 8.
Knockdown of ubiquilin expression enhances starvation-induced cell death in HeLa cells stably expressing GFP-Htt74Q protein. (A–C) Cultures of HeLa cells stably expressing GFP-Htt74Q protein were either mock transfected or transfected with siRNAs to specifically knockdown ubiquilin-1 and -2 proteins. Twenty-four hours after transfection, the medium was replaced with starvation medium and cell death assays were performed at 0 and 4 h after starvation. (A) Phase-contrast microscopy images of cells just before transfection and 24 h post-transfection after culturing for 0 and 4 h in starvation medium. Bar, 100 µm. (B) Hoechst staining of cells at 0 and 4 h of starvation. Bar, 100 µm. (C) Quantification of cell death seen in the cultures after 24 h post-transfection. (D) Immunoblots showing knockdown of ubiquilin in lysates from the cultures.
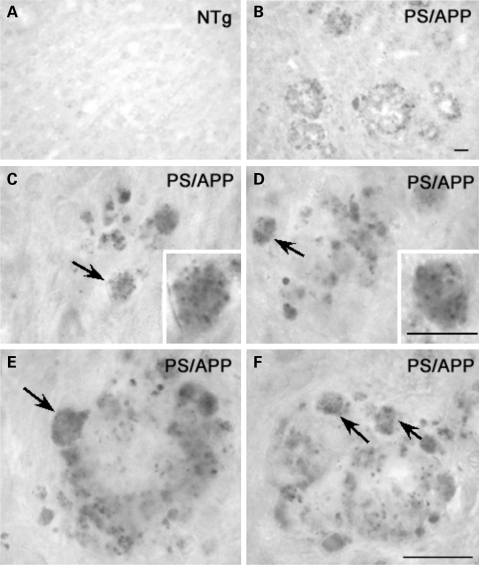
Ubiquilin is enriched in autophagosomes that accumulate in dystrophic neurites of brain of presenilin/amyloid precursor protein double transgenic mice
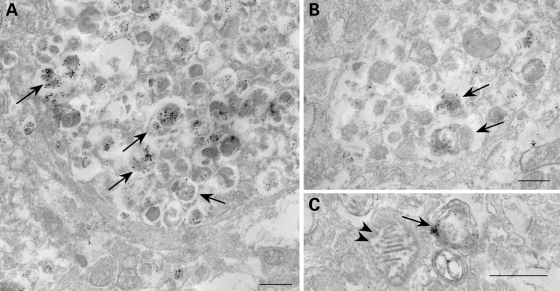
To obtain further evidence in support of ubiquilin involvement in autophagy, we stained mouse brains of presenilin-1/amyloid precursor protein (APP) double transgenic mice for ubiquilin. The PS1/APP mice exhibit abnormal autophagosome and plaque buildup (34,35) providing a powerful system to determine whether the endogenous ubiquilin protein in mouse brain is associated with neuropathology. As shown in Figure 9A and B, we observed strong staining of ubiquilin in brains of 12-month-old PS/APP mice, but not in age-matched non-transgenic mice. The staining was particularly strong around plaques in the PS/APP mice, and examination at higher magnification revealed clear evidence of punctate staining in neurites (Fig. 9C–F). Further immunogold localization by electron microscopy indicated that the ubiquilin staining was mainly present in autophagosomes, but not in other organelles, such as mitochondria (Fig. 10A–C). The results provide convincing evidence that ubiquilin is enriched in autophagosomes in a mouse model of Alzheimer's disease.
Figure 9.
Ubiquilin immunoreactivity is associated with plaques in the brain of PS1/APP mice. (A–F) Mouse brain sections from a 12-month-old PS1/APP mouse (B–F) and an age-matched non-transgenic control (A) stained with antibodies against ubiquilin by immunohostochemistry. Examinations of plaques at higher magnifications reveal neurites demonstrating a clear, punctate labeling pattern (C–F, arrows and C and D, inset). Bars A–F: 20 µm; C inset and D inset: 10 µm.
Figure 10.
Immunogold localization of ubiquilin in dystrophic neurites of brain from a 12-month-old PS/APP mouse using a pre-embedding silver enhancement technique. Ubiquilin immunoreactivity is highly specific to AVs within dystrophic neurites (A–C, arrows) and is absent in other organelles (C, arrowheads). Bar: 500 nm.
DISCUSSION
Macroautophagy is a highly regulated pathway involving multiple proteins that function to sequester cellular proteins and organelles in a double membrane organelle for degradation by lysosomes. Ubiquilin-1 protein was recently reported to regulate macroautophagy (16). Our results described here, conducted in HeLa cells and mice, support this conclusion and provide additional information about ubiquilin's function in macroautophagy. We have demonstrated for the first time that ubiquilin and LC3 bind together in a complex and that the loss of ubiquilin appears to inhibit conversion of LC3-I to LC3-II, the latter of which is important for macroautophagy. We found that ubiquilin is a substrate of CMA and that ubiquilin is degraded during autophagy. Consistent with a role in macroautophagy, we found that ubiquilin is enriched upon purification of autophagosomes from mouse liver and that the protein is localized in autophagosomes of dystrophic neurites in neuropathological lesions of a PS/APP mouse model of AD.
Compelling evidence that ubiquilin is found in autophagosomes is based on multiple criteria, including colocalization of the endogenous and transfected proteins with the autophagosome marker LC3, strong enrichment of ubiquilin during purification of autophagosomes from mouse liver and immunogold localization of ubiquilin in autophagosomes in mouse brain tissue. Furthermore, our results suggest that ubiquilin that is packaged in autophagosomes is eventually degraded in lysosomes, based on its detection during all stages of maturation of autophagosome to lysosome as revealed using the mCherry-GFP-LC3 autophagosome–lysosome reporter, stabilization of ubiquilin degradation by inhibition of autophagosome maturation with bafilomycin A1 or inhibition of autophagosome formation by inhibition of PI3K with 3MA.
Interestingly, we have also shown here that a portion of intracellular ubiquilin undergoes degradation via a different autophagic pathway, CMA. The presence of two sequences in ubiquilin that match the pentapeptide motif implicated in CMA—also present in ubiquilin-2, 3 and 4 proteins—and the preferential association of ubiquilin in lysosomes active for CMA is in accord with ubiquilin being a substrate of CMA. Furthermore, utilizing an in vitro assay, we have shown that ubiquilin is actively transported and degraded by lysosomes by CMA. The dual degradation of ubiquilin by both macroautophagy and CMA might have important implications in the regulation of these two autophagic processes. For example, we previously showed that blockage of CMA in cultured cells leads to upregulation of macroautophagy suggesting cross-talk between these two forms of autophagy (36). However, the molecular mediators of this cross-talk remain unknown. We speculate that degradation of ubiquilin by CMA could potentially reduce the total intracellular levels of ubiquilin that is necessary for macroautophagy, and vice versa. Likewise inhibition of ubiquilin degradation by CMA could lead to increased accumulation of ubiquilin to levels sufficient to support macroautophgy. If correct, it will be interesting to discover how ubiquilin is selected for CMA and macroautophgy.
The destruction of ubiquilin during macroautophagy suggests that the protein cannot be recycled after carrying out its function in autophagy. We speculate that this occurs because ubiquilin is either transported or packaged into the lumen of the lysosome where it cannot be retrieved. The consumption of ubiquilin during macroautophagy suggests that strong upregulation of macroautophagy would exhaust ubiquilin supply unless new protein were made. Interestingly, ubiquilin is induced by ER stress (10), which is closely linked to induction of macroautophagy (37,38).
Several questions arise as to what role ubiquilin plays in autophagy. A clear indication that ubiquilin is important for autophagy is that knockdown of the protein in cells reduces autophagosome formation. The knockdown studies revealed that the inhibition might occur early on during autophagosome biogenesis by inhibiting maturation of LC3 to LC3-II. The conversion of LC3 to LC3-II involves at least two important steps. The first step is the cleavage of LC3 protein by Atg4 and the second step is the covalent conjugation of the lipid phosphatidylethanolamine (PE) to the C-terminus of the cleaved protein by Atg7 and Atg3 proteins. Our studies do not distinguish whether ubiquilin is required for any of these steps. Unfortunately, this issue was not clarified by any selective binding of the LC3 isoforms with ubiquilin because both LC3-I and -II were found to coimmunoprecipitate with ubiquilin. Coimmunoprecipitation of both isoforms would support the idea that ubiquilin might bind both isoforms equally well and independent of the presence of the PE modification. We, therefore, tested whether purified bacterially expressed ubiquilin and LC3 proteins bind directly to one another via GST-pulldown assays, but could not detect any strong binding between the proteins (data not shown). The results suggest that ubiquilin might bind LC3 indirectly.
Another possible function of ubiquilin might be to bind and transport ubiquitinated proteins to autophagosomes. Ubiquilin contains a UBA domain that is known to bind different ubiquitin moieties with high specificity (7,8,19). By coimmunoprecipitation studies, we found that proteins containing K48- and K63-linked ubiquitin chains bind ubiquilin and that both ubiquilin and the chains are present together in autophagosomes. Based on these observations, we speculate that ubiquilin binds and targets misfolded proteins tagged with K48 and K63 ubiquitin chains for autophagic degradation. This function appears analogous to p62, a protein that contains a UBA that has been shown to bind ubiquitin moieties and target them to autophagosomes (39–41). Interestingly, p62, like ubiquilin, has been implicated not only in autophagy, but also in proteasome degradation. The dual role played by proteins in these processes is likely related to the common domains shared by the proteins: they both contain a UBA domain at their C-terminus and a UBL at their N-terminus (a classical UBL domain in ubiquilin and a UBL-related PB1 domain in p62), which has been implicated in binding subunits of the proteasome (3,6,8,40,42). Recent reports have shown that ubiquilin functions in ERAD, the pathway by which misfolded proteins are exported from the ER to the cytoplasm for degradation by proteasomes. To our knowledge, we are not aware of any study showing p62 is involved in ERAD. Nevertheless, if the proteins share similar functions then two questions arise. One, how do the proteins choose between autophagy and the proteasome pathways? Is there a signal that directs the proteins to utilize one pathway in preference to the other, or is the choice random? Two, are the two proteins truly redundant in function, or do they carry out independent functions? We do not know the answers to these questions; however, we speculate that p62 and ubiquilin might execute independent functions in autophagy because silencing of ubiquilin expression by itself was sufficient to inhibit autophagosome formation.
Based on its involvement in both ERAD and autophagy it is not surprising that ubiquilin has been found to be cytoprotective against toxic proteins linked to different diseases. In previous studies, we showed that overexpression of ubiquilin-1 prevents oxidative stress-induced cell death caused by overexpression of proteins containing expanded polyalanine or polyglutamine tracts, whereas the loss of its expression exacerbated cell death (33,43). Our new findings presented here demonstrate that ubiquilin is cytoprotective against overexpression of mutant PS2141I protein linked to AD. The dual capacity of ubiquilin to function in either autophagy/lysosome or proteasome degradation pathways makes it very difficult to pinpoint whether the protective effect exerted by ubiquilin against the toxic proteins is due to its specific function in one or both pathways. An indication that ubiquilin might be protective in autophagy is that overexpression of the protein reduced PS2-induced cell death under conditions of starvation, which is known to activate the autophagy pathway. Likewise, knockdown of ubiquilin expression increased starvation induced cell death in cells expressing GFP-Htt74Q protein.
The strong detection of endogenous ubiquilin protein in autophagosome-like structures in dystrophic neurites in brain tissue of APP/PS1 transgenic mice, but not in non-transgenic mice, provides valuable new information regarding the involvement of ubiquilin and autophagy in neurodegeneration. The increased staining would support the idea that autophagosome accumulation is abnormally increased during neurodegeneration in APP/PS1 mice (34). Similar abnormalities in autophagosome accumulation have been detected in the brains of humans afflicted with AD (44,45). This might explain why we observed increased staining of ubiquilin in AD brain (2). Thus anti-ubiquilin staining might serve as a useful diagnostic tool for detecting autophagosomes in AD, similar to mitochondrial DNA and lipoic acid that were found to accumulate within autophagosomes in AD (46,47).
MATERIALS AND METHODS
Cell culture and immunofluorescence microscopy
HeLa cells were grown in DMEM medium supplemented with 10% fetal bovine serum. The cultures were treated with puromycin (5 µg/ml final conc., Sigma-Aldrich, St Louis, MO, USA), bafilomycin A1 (200 nm final conc., Sigma-Aldrich) or 3-methyladenine (10 mm final conc, Sigma-Aldrich) in the combinations indicated in the figures for different time periods. For immunofluorescence microscopy, cells were plated on glass coverslips and then transfected with plasmid DNA by the calcium phosphate coprecipitation method (33). Approximately 20 h after transfection, the cells were then fixed with 4% paraformaldehyde in 1× PBS and were then either first stained with appropriate primary and secondary antibodies or directly mounted on slides and imaged by microscopy as described previously (10). Cell death assays of the GFP-HttQ74-3 expressing cell line were conducted using the procedure described previously (33), except that the cells were starved by incubation in PBS for different lengths of time prior to conducting the cell death assays. Silencing of ubiquilin expression was achieved by transfection of siRNAs directed against ubiquilin-1 and ubiquilin-2 following the procedure described previously (33). Briefly, cells were transfected with 20 nM of ON-TARGETplus SMART pool siRNAs against ubiquilin-1 and -2 (L-041012-01 and L-042279-01) (Dharmacon, Inc., Chicago, IL, USA) using DharmaFECT 1 following the method provided by the manufacturer. Control transfections were performed using ON-TARGETplus non-targeting pool siRNAs (D-001810-10) that were designed to have no known target in HeLa cells. Our immunoblots of cell lysates suggest that two of the four ubiquilin genes found in humans are expressed in HeLa cells, with ubiquilin-1 protein, the smaller and faster migrating, form being more prominent than the slightly larger ubiquilin-2 protein. Ubiquilin-3 is a testis-specific isoform that is not expressed in HeLa cells. Ubiquilin-4 is probably expressed at low levels, if at all, because silencing of ubiquilin-1 and -2 is sufficient to eliminate ubiquilin immunoreactivity (Fig. 3C). In some of our gels, three bands are sometimes seen, the uppermost of which we believe could be a modified form of ubiquilin-1 or -2 protein. Importantly, in our knockdown experiments, we have been able to achieve approximately 60–90% knockdown of ubiquilin proteins after 72 h transfection with siRNAs directed against ubiquilin-1 and -2 (43). Cell death assays of PS2 overexpressing cells were performed by transfecting HeLa cells with a constant amount of GFP-tagged wild-type and N(141)I mutant PS2 expression constructs (32) and with varying amounts of plasmid DNA encoding ubiquilin-1 or empty vector as shown in the figures, ensuring that the total amount of DNA transfected in each culture was the same. One day after transfection, the cells were starved by replacing the culture medium with a 1 X PBS solution (137 mm NaCl, 3.5 mm KCl, 0.9 mm MgCl2, 0.9 mm CaCl2, 20 mm NaPO4, pH 7.5) and at appropriate time intervals, Hoechst 33342 (1 µg/ml) was added and the cultures incubated for another 15 min prior to acquisition of DNA and GFP fluorescent images by microscopy (33). Cell death was quantified by counting the percent of GFP-expressing cells that displayed bright compacted DNA staining indicative of dying cells.
SDS–PAGE and immunoblotting
The standard protocol for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS–PAGE) and immunoblotting was followed (10). Antibodies used were: rabbit anti-ubiquilin 145 and 125 [made against the central and C-terminal regions of ubiquilin, respectively, by the M.J. Monteiro laboratory (2)], rabbit anti-GFP [made against GST-GFP, by the M.J. Monteiro laboratory (33)], rabbit anti-LC3 (Cell Signaling Technology, Inc. Danvers, MA, USA), mouse anti-ubiquilin (Invitrogen, Carlsbad, CA, USA), rabbit anti-ubiquitin Lys63-specific Clone Apu3 and Lys48-specific Clone Apu2 (both from Millpore, Temecula, CA, USA), goat anti-actin (Santa Cruz Biotechnology, Santa Cruz, CA, USA), rabbit anti-LAMP-2A [made against the C-terminal tail of LAMP-2A (48)], mouse anti-LAMP-1 (Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA, USA) and mouse anti-hsc70 (Maine Biotechnology Services, Inc., Portland, ME, USA).
In vitro assay of CMA
Transport of purified proteins into isolated lysosomes was analyzed using a previously described in vitro system (27). Briefly, GST-ubiquilin-1 was incubated with freshly isolated rat liver lysosomes in MOPS buffer (10 mm 3-(N-morpholino) propanesulfonic acid (MOPS) pH 7.3, 0.3 M sucrose) for 20 min at 37°C. Where indicated, lysosomes were pre-incubated with a cocktail of proteinase inhibitors for 10 min at 0°C. At the end of the incubation, lysosomes were collected by centrifugation, washed with MOPS buffer and subjected to SDS–PAGE and immunoblotted with an antibody specific for ubiquilin. Transport was calculated by subtracting the protein associated with lysosomes in the absence of inhibitors of lysosomal proteases from the amount found in the presence of the inhibitors (29). Where indicated, blockage of CMA components was attained by pre-incubation of lysosomes with antibodies against LAMP-2A or hsc70 for 15 min at room temperature. The lysosomes decorated with the bound antibodies were recovered by centrifugation and added to the transport assay reaction as described above.
Isolation of autophagosomes and lysosomes from mouse
Autophagic vacuoles were isolated from male C57BL/6 mouse livers by centrifugation in discontinuous density gradients, following an isolation protocol modified from Marzella et al. (26) as previously described (49). Briefly, a fraction enriched in autophagic vacuoles and lysosomes was isolated by differential centrifugation of liver homogenates (2000 g for 5 min and 17 000 g for 12 min). Autophagic vacuoles in the 17 000 g pellet were separated from the other components in the fraction by centrifugation at 247 000 g for 3 h in a discontinuous metrizamide gradient (26, 24, 20 and 15% metrizamide). Autophagic vacuoles were collected from the 15 to 20% and 20 to 24% interfaces and lysosomes from the 24 to 26% interface. A fraction enriched in endoplasmic reticulum resealed vesicles (microsomes) was prepared as a pellet by centrifugation of the supernatant of the 17 000 g centrifugation at 100 000 g for 1 h. Fractions were washed in 0.25 m sucrose and collected by centrifugation at 24 000 g for 10 min. Two other lysosomal fractions with different autophagic activity were isolated from mouse liver from a light mitochondrial-lysosomal fraction in a discontinuous metrizamide density gradient (50). Lysosomal fractions with different activities for CMA were separated as described (27). In all cases, preparations with more than 10% broken lysosomes, measured as β-hexosaminidase latency, were discarded.
Immunoelectron microscopy
Mice were perfused intra-cardially with a fixative containing 0.1% glutaraldehyde and 4% paraformaldehyde in 0.1 m sodium cacodylate at pH 7.4. Brains were removed and post-fixed at room temperature overnight. Brains were sectioned at 50 µm on a vibratome (Leica, Germany). Sections were washed in PBS and treated with 0.1% sodium borohydride, permeabilized in 0.05% triton and blocked in BSA. Primary antibody was applied overnight at 4°C. Sections were washed and further incubated overnight with secondary antibody conjugates to 6 nm gold. Immunolabeled sections were fixed in 2% glutaraldehyde and silver enhanced for 60 min, osmicated in 1% Osmium tetroxide solution for 10 min, dehydrated and flat embedded in Epon. Ultrathin sections were stained with uranyl acetate and lead citrate. Specimens were visualized on a FEI CM10 transmission electon microscope equipped with an AMT CCD camera.
Immunohistochemistry of mouse brain
Twelve month-old PS1/APP mice (51) and their littermate controls were intra-cardially perfused with 4% paraformaldehyde. Following 48 h post-fix, 40 um sections were cut on a vibratome. Sections were incubated in 0.3% hydrogen peroxide diluted in methanol, subsequently in 20% normal goat serum to block non-specific binding. Sections were incubated for 3 days at 4°C with anti-ubiquilin (1:500), diluted in TBS/0.9% NaCl/ 2% BSA/1% normal goat serum and 0.4% Triton X-100. Sections were incubated in biotinylated anti-mouse for 30 min, in ABC reagent for 1 h (Vector Labs, Burlingame, CA, USA) and visualized with DAB (Vector Labs). In parallel, sections were processed in the absence of primary antibody serum to test specificity of antibody binding. Microscopic analyses were performed using a Zeiss Axioskop II equipped with an AxioCam HRm digital camera (Carl Zeiss, Thornwood, NY, USA).
FUNDING
This work was supported by the National Institutes of Health grants GM066287 (M.J.M.), AG016839 (M.J.M.), GM06696 (S.F.) and AG02190 (A.M.C.). S.K. is supported by a NIA T32 grant AG023475.
ACKNOWLEDGEMENTS
We thank Dr Terje Johansen for kindly providing the mCherry-GFP-LC3 reporter and Dr Howard Doong for conducting the GFP immunoprecipitation.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Wu A.L., Wang J., Zheleznyak A., Brown E.J. Ubiquitin-related proteins regulate interaction of vimentin intermediate filaments with the plasma membrane. Mol. Cell. 1999;4:619–625. doi: 10.1016/s1097-2765(00)80212-9. doi:10.1016/S1097-2765(00)80212-9. [DOI] [PubMed] [Google Scholar]
- 2.Mah A.L., Perry G., Smith M.A., Monteiro M.J. Identification of ubiquilin, a novel presenilin interactor that increases presenilin protein accumulation. J. Cell Biol. 2000;151:847–862. doi: 10.1083/jcb.151.4.847. doi:10.1083/jcb.151.4.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kleijnen M.F., Shih A.H., Zhou P., Kumar S., Soccio R.E., Kedersha N.L., Gill G., Howley P.M. The hPLIC proteins may provide a link between the ubiquitination machinery and the proteasome. Mol. Cell. 2000;6:409–419. doi: 10.1016/s1097-2765(00)00040-x. doi:10.1016/S1097-2765(00)00040-X. [DOI] [PubMed] [Google Scholar]
- 4.Miller J., Gordon C. The regulation of proteasome degradation by multi-ubiquitin chain binding proteins. FEBS Lett. 2005;579:3224–3230. doi: 10.1016/j.febslet.2005.03.042. doi:10.1016/j.febslet.2005.03.042. [DOI] [PubMed] [Google Scholar]
- 5.Elsasser S., Finley D. Delivery of ubiquitinated substrates to protein-unfolding machines. Nat. Cell Biol. 2005;7:742–749. doi: 10.1038/ncb0805-742. doi:10.1038/ncb0805-742. [DOI] [PubMed] [Google Scholar]
- 6.Kleijnen M.F., Alarcon R.M., Howley P.M. The ubiquitin-associated domain of hPLIC-2 interacts with the proteasome. Mol. Biol. Cell. 2003;14:3868–3875. doi: 10.1091/mbc.E02-11-0766. doi:10.1091/mbc.E02-11-0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Massey L.K., Mah A.L., Ford D.L., Miller J., Liang J., Doong H., Monteiro M.J. Overexpression of ubiquilin decreases ubiquitination and degradation of presenilin proteins. J. Alzheimers Dis. 2004;6:79–92. doi: 10.3233/jad-2004-6109. [DOI] [PubMed] [Google Scholar]
- 8.Ko H.S., Uehara T., Tsuruma K., Nomura Y. Ubiquilin interacts with ubiquitylated proteins and proteasome through its ubiquitin-associated and ubiquitin-like domains. FEBS Lett. 2004;566:110–114. doi: 10.1016/j.febslet.2004.04.031. [DOI] [PubMed] [Google Scholar]
- 9.Kim T.Y., Kim E., Yoon S.K., Yoon J.B. Herp enhances ER-associated protein degradation by recruiting ubiquilins. Biochem. Biophys. Res. Commun. 2008;369:741–746. doi: 10.1016/j.bbrc.2008.02.086. doi:10.1016/j.bbrc.2008.02.086. [DOI] [PubMed] [Google Scholar]
- 10.Lim P.J., Danner R., Liang J., Doong H., Harman C., Srinivasan D., Rothenberg C., Wang H., Ye Y., Fang S., et al. Ubiquilin and p97/VCP bind erasin, forming a complex involved in ERAD. J. Cell Biol. 2009;187:201–217. doi: 10.1083/jcb.200903024. doi:10.1083/jcb.200903024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie Z., Klionsky D.J. Autophagosome formation: core machinery and adaptations. Nat. Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. doi:10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 12.Levine B., Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. doi:10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ravikumar B., Futter M., Jahreiss L., Korolchuk V.I., Lichtenberg M., Luo S., Massey D.C., Menzies F.M., Narayanan U., Renna M., et al. Mammalian macroautophagy at a glance. J. Cell Sci. 2009;122:1707–1711. doi: 10.1242/jcs.031773. doi:10.1242/jcs.031773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu S., Mikhailov A., Kallo-Hosein H., Hara K., Yonezawa K., Avruch J. Characterization of ubiquilin 1, an mTOR-interacting protein. Biochim. Biophys. Acta. 2002;1542:41–56. doi: 10.1016/s0167-4889(01)00164-1. [DOI] [PubMed] [Google Scholar]
- 15.Jung C.H., Ro S.H., Cao J., Otto N.M., Kim D.H. mTOR regulation of autophagy. FEBS Lett. 584:1287–1295. doi: 10.1016/j.febslet.2010.01.017. doi:10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.N'Diaye E.N., Kajihara K.K., Hsieh I., Morisaki H., Debnath J., Brown E.J. PLIC proteins or ubiquilins regulate autophagy-dependent cell survival during nutrient starvation. EMBO Rep. 2009;10:173–179. doi: 10.1038/embor.2008.238. doi:10.1038/embor.2008.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cuervo A.M. Chaperone-mediated autophagy: selectivity pays off. Trends Endocrinol. Metab. 2009;21:142–150. doi: 10.1016/j.tem.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pankiv S., Clausen T.H., Lamark T., Brech A., Bruun J.A., Outzen H., Overvatn A., Bjorkoy G., Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. doi:10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 19.Raasi S., Varadan R., Fushman D., Pickart C.M. Diverse polyubiquitin interaction properties of ubiquitin-associated domains. Nat. Struct. Mol. Biol. 2005;12:708–714. doi: 10.1038/nsmb962. doi:10.1038/nsmb962. [DOI] [PubMed] [Google Scholar]
- 20.Newton K., Matsumoto M.L., Wertz I.E., Kirkpatrick D.S., Lill J.R., Tan J., Dugger D., Gordon N., Sidhu S.S., Fellouse F.A., et al. Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell. 2008;134:668–678. doi: 10.1016/j.cell.2008.07.039. doi:10.1016/j.cell.2008.07.039. [DOI] [PubMed] [Google Scholar]
- 21.Yoshimori T., Yamamoto A., Moriyama Y., Futai M., Tashiro Y. Bafilomycin A1, a specific inhibitor of vacuolar-type H(+)-ATPase, inhibits acidification and protein degradation in lysosomes of cultured cells. J. Biol. Chem. 1991;266:17707–17712. [PubMed] [Google Scholar]
- 22.Yamamoto A., Tagawa Y., Yoshimori T., Moriyama Y., Masaki R., Tashiro Y. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct. Funct. 1998;23:33–42. doi: 10.1247/csf.23.33. doi:10.1247/csf.23.33. [DOI] [PubMed] [Google Scholar]
- 23.Klionsky D.J., Elazar Z., Seglen P.O., Rubinsztein D.C. Does bafilomycin A1 block the fusion of autophagosomes with lysosomes? Autophagy. 2008;4:849–950. doi: 10.4161/auto.6845. [DOI] [PubMed] [Google Scholar]
- 24.Seglen P.O., Bohley P. Autophagy and other vacuolar protein degradation mechanisms. Experientia. 1992;48:158–172. doi: 10.1007/BF01923509. doi:10.1007/BF01923509. [DOI] [PubMed] [Google Scholar]
- 25.Blommaart E.F., Luiken J.J., Meijer A.J. Autophagic proteolysis: control and specificity. Histochem. J. 1997;29:365–385. doi: 10.1023/a:1026486801018. doi:10.1023/A:1026486801018. [DOI] [PubMed] [Google Scholar]
- 26.Marzella L., Ahlberg J., Glaumann H. Isolation of autophagic vacuoles from rat liver: morphological and biochemical characterization. J. Cell Biol. 1982;93:144–154. doi: 10.1083/jcb.93.1.144. doi:10.1083/jcb.93.1.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cuervo A.M., Dice J.F., Knecht E. A population of rat liver lysosomes responsible for the selective uptake and degradation of cytosolic proteins. J. Biol. Chem. 1997;272:5606–5615. doi: 10.1074/jbc.272.9.5606. [DOI] [PubMed] [Google Scholar]
- 28.Kon M., Cuervo A.M. Chaperone-mediated autophagy in health and disease. FEBS Lett. 2009;584:1399–1404. doi: 10.1016/j.febslet.2009.12.025. doi:10.1016/j.febslet.2009.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaushik S., Cuervo A.M. Methods to monitor chaperone-mediated autophagy. Methods Enzymol. 2009;452:297–324. doi: 10.1016/S0076-6879(08)03619-7. doi:10.1016/S0076-6879(08)03619-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aniento F., Roche E., Cuervo A.M., Knecht E. Uptake and degradation of glyceraldehyde-3-phosphate dehydrogenase by rat liver lysosomes. J. Biol. Chem. 1993;268:10463–10470. [PubMed] [Google Scholar]
- 31.Cuervo A.M., Terlecky S.R., Dice J.F., Knecht E. Selective binding and uptake of ribonuclease A and glyceraldehyde-3-phosphate dehydrogenase by isolated rat liver lysosomes. J. Biol. Chem. 1994;269:26374–26380. [PubMed] [Google Scholar]
- 32.Janicki S., Monteiro M.J. Increased apoptosis arising from increased expression of the Alzheimer's disease-associated presenilin-2 mutation (N141I) J. Cell Biol. 1997;139:485–495. doi: 10.1083/jcb.139.2.485. doi:10.1083/jcb.139.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang H., Lim P.J., Yin C., Rieckher M., Vogel B.E., Monteiro M.J. Suppression of polyglutamine-induced toxicity in cell and animal models of Huntington's disease by ubiquilin. Hum. Mol. Genet. 2006;15:1025–1041. doi: 10.1093/hmg/ddl017. doi:10.1093/hmg/ddl017. [DOI] [PubMed] [Google Scholar]
- 34.Yu W.H., Cuervo A.M., Kumar A., Peterhoff C.M., Schmidt S.D., Lee J.H., Mohan P.S., Mercken M., Farmery M.R., Tjernberg L.O., et al. Macroautophagy—a novel Beta-amyloid peptide-generating pathway activated in Alzheimer's disease. J. Cell Biol. 2005;171:87–98. doi: 10.1083/jcb.200505082. doi:10.1083/jcb.200505082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boland B., Kumar A., Lee S., Platt F.M., Wegiel J., Yu W.H., Nixon R.A. Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer's disease. J. Neurosci. 2008;28:6926–6937. doi: 10.1523/JNEUROSCI.0800-08.2008. doi:10.1523/JNEUROSCI.0800-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaushik S., Massey A.C., Mizushima N., Cuervo A.M. Constitutive activation of chaperone-mediated autophagy in cells with impaired macroautophagy. Mol. Biol. Cell. 2008;19:2179–2192. doi: 10.1091/mbc.E07-11-1155. doi:10.1091/mbc.E07-11-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yorimitsu T., Nair U., Yang Z., Klionsky D.J. Endoplasmic reticulum stress triggers autophagy. J. Biol. Chem. 2006;281:30299–30304. doi: 10.1074/jbc.M607007200. doi:10.1074/jbc.M607007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bernales S., McDonald K.L., Walter P. Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol. 2006;4:e423. doi: 10.1371/journal.pbio.0040423. doi:10.1371/journal.pbio.0040423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Donaldson K.M., Li W., Ching K.A., Batalov S., Tsai C.C., Joazeiro C.A. Ubiquitin-mediated sequestration of normal cellular proteins into polyglutamine aggregates. Proc. Natl Acad. Sci. USA. 2003;100:8892–8897. doi: 10.1073/pnas.1530212100. doi:10.1073/pnas.1530212100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seibenhener M.L., Babu J.R., Geetha T., Wong H.C., Krishna N.R., Wooten M.W. Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol. Cell Biol. 2004;24:8055–8068. doi: 10.1128/MCB.24.18.8055-8068.2004. doi:10.1128/MCB.24.18.8055-8068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim P.K., Hailey D.W., Mullen R.T., Lippincott-Schwartz J. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc. Natl Acad. Sci. USA. 2008;105:20567–20574. doi: 10.1073/pnas.0810611105. doi:10.1073/pnas.0810611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Geetha T., Seibenhener M.L., Chen L., Madura K., Wooten M.W. p62 serves as a shuttling factor for TrkA interaction with the proteasome. Biochem. Biophys. Res. Commun. 2008;374:33–37. doi: 10.1016/j.bbrc.2008.06.082. doi:10.1016/j.bbrc.2008.06.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang H., Monteiro M.J. Ubiquilin overexpression reduces GFP-polyalanine-induced protein aggregates and toxicity. Exp. Cell Res. 2007;313:2810–2820. doi: 10.1016/j.yexcr.2007.04.006. doi:10.1016/j.yexcr.2007.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nixon R.A., Wegiel J., Kumar A., Yu W.H., Peterhoff C., Cataldo A., Cuervo A.M. Extensive involvement of autophagy in Alzheimer disease: an immuno-electron microscopy study. J. Neuropathol. Exp. Neurol. 2005;64:113–122. doi: 10.1093/jnen/64.2.113. [DOI] [PubMed] [Google Scholar]
- 45.Nixon R.A. Autophagy, amyloidogenesis and Alzheimer disease. J. Cell Sci. 2007;120:4081–4091. doi: 10.1242/jcs.019265. doi:10.1242/jcs.019265. [DOI] [PubMed] [Google Scholar]
- 46.Hirai K., Aliev G., Nunomura A., Fujioka H., Russell R.L., Atwood C.S., Johnson A.B., Kress Y., Vinters H.V., Tabaton M., et al. Mitochondrial abnormalities in Alzheimer's disease. J. Neurosci. 2001;21:3017–3023. doi: 10.1523/JNEUROSCI.21-09-03017.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moreira P.I., Siedlak S.L., Wang X., Santos M.S., Oliveira C.R., Tabaton M., Nunomura A., Szweda L.I., Aliev G., Smith M.A., et al. Autophagocytosis of mitochondria is prominent in Alzheimer disease. J. Neuropathol. Exp. Neurol. 2007;66:525–532. doi: 10.1097/01.jnen.0000240476.73532.b0. doi:10.1097/01.jnen.0000240476.73532.b0. [DOI] [PubMed] [Google Scholar]
- 48.Cuervo A.M., Dice J.F. A receptor for the selective uptake and degradation of proteins by lysosomes. Science. 1996;273:501–503. doi: 10.1126/science.273.5274.501. doi:10.1126/science.273.5274.501. [DOI] [PubMed] [Google Scholar]
- 49.Singh R., Kaushik S., Wang Y., Xiang Y., Novak I., Komatsu M., Tanaka K., Cuervo A.M., Czaja M.J. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. doi:10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wattiaux R., Wattiaux-De Coninck S., Ronveaux-dupal M.F., Dubois F. Isolation of rat liver lysosomes by isopycnic centrifugation in a metrizamide gradient. J. Cell Biol. 1978;78:349–368. doi: 10.1083/jcb.78.2.349. doi:10.1083/jcb.78.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cataldo A.M., Peterhoff C.M., Schmidt S.D., Terio N.B., Duff K., Beard M., Mathews P.M., Nixon R.A. Presenilin mutations in familial Alzheimer disease and transgenic mouse models accelerate neuronal lysosomal pathology. J. Neuropathol. Exp. Neurol. 2004;63:821–830. doi: 10.1093/jnen/63.8.821. [DOI] [PubMed] [Google Scholar]