Introduction
The peptidoglycan is the polymer that surrounds, contains, and protects the cytoplasm of bacteria. The structure of this polymer consists of glycan strands, cross-linked to each other via oligopeptide stems attached to alternate glycans of the strands. As the peptidoglycan is a unique structure of bacteria, and thus is without a eukaryotic counterpart, small molecule inhibition of the enzymatic events of peptidoglycan biosynthesis coincides with antibacterial activity.1–3 For this reason, our laboratory has focused on the understanding of key events that involve biological recognition of the peptidoglycan. Examples of these recognition events include the peptidoglycan (and its biosynthetic precursors) acting as a host for small molecule binding, and the peptidoglycan acting as a guest wherein it is recognized by protein macromolecules. Peptidoglycan recognition as a guest occurs during its biosynthesis, during the activation of bacterial resistance pathways, and in the innate immune response of eukaryotes to the presence of bacterial infection. In this Perspective we emphasize the multi-disciplinary approaches that are now being used toward the molecular-level understanding of the host-guest chemistry of the peptidoglycan, and with emphasis on the central place of this structure in the continuing pursuit of improved antibacterial chemotherapy.
The Bacterial Peptidoglycan
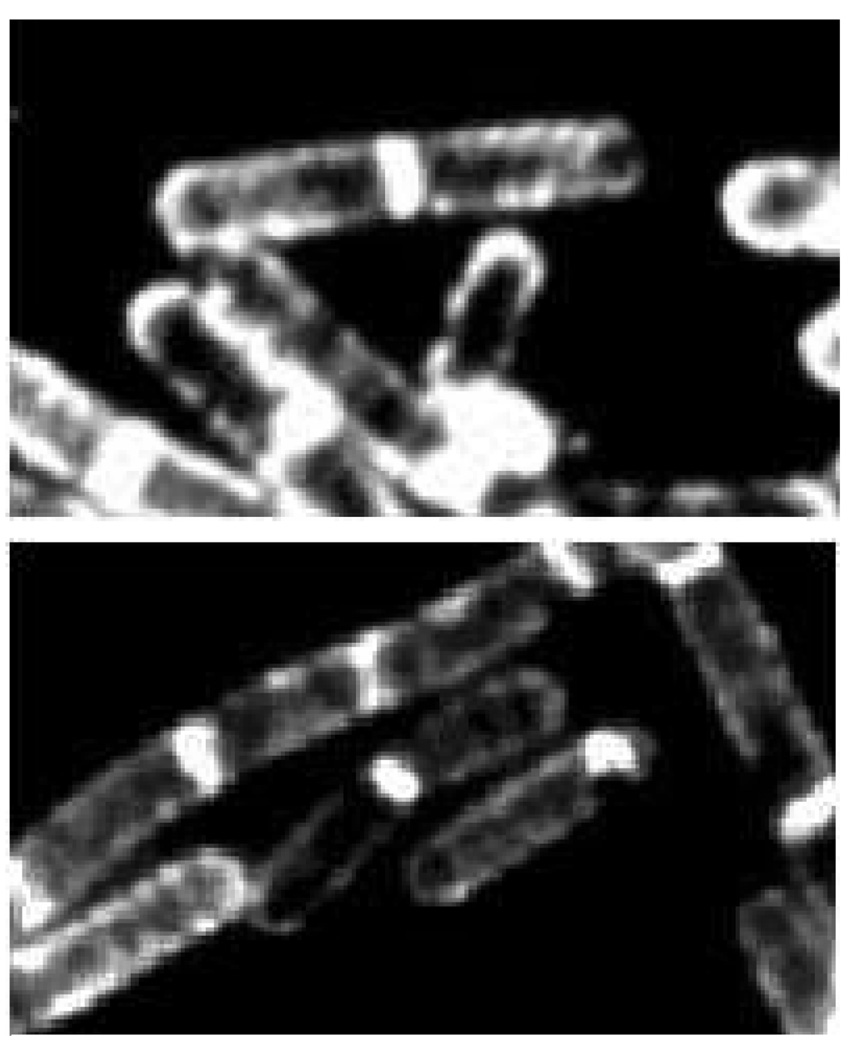
The peptidoglycan is the exterior surface of the Gram-positive bacterium. In these bacteria, it overlays the cell membrane while leaving a space (termed the “periplasmic” space) to accommodate inter alia the macromolecule proteins of small molecule influx and efflux, and of cell envelope biosynthesis. Microscopic images (Figure 1) of the rod-shaped Gram-positive bacterium Bacillus subtilis, taken in the presence of sub-lethal concentrations of a fluorescent derivative of the peptidoglycan-binding antibiotic ramoplanin, show the location of active peptidoglycan synthesis in this bacterium.4,5 In contrast to the Gram-positive bacterium, the exterior surface of the Gram-negative bacteria is a membrane, underneath which are found sequentially the peptidoglycan, the periplasmic space, and a second (or cellular) membrane. Together, the peptidoglycan and membrane(s) comprise the cell envelope that surrounds the cytoplasm of these bacteria. Although the evolutionary deletion of peptidoglycan biosynthesis has occurred in most plants, following incorporation of the genome of an endosymbiotic cyanobacterium into that of the plant, expression of peptidoglycan structure still occurs in the plastids of certain mosses.6 Conversely, while creation of the peptidoglycan polymer appears not to occur in several pathogenic bacterial species that have adapted to living within the cells of host species (the “cell wall-less” bacteria), portions of the peptidoglycan biosynthetic pathway are essential to their viability.7,8
Figure 1.
Visualization of the external peptidoglycan surface of Gram-positive bacteria by the use of fluorescently-labeled peptidoglycan-binding antibiotics. These fluorescent microscopy images, reproduced from the study by Tiyanont et al. Proc. Natl Acad. Sci. U. S. A. 2006, 103, 11033–11038 © 2006 National Academy of Sciences, show rod-shaped B. subtilis (the bacterium is approximately 2 µm in length) bacteria stained with fluorescein-labeled ramoplanin. The peptidoglycan is intensely stained at the newly forming division septum, and to a lesser extent on the sidewalls and the old pole. These locations coincide with the sub-cellular locations of Lipid II, the key biosynthetic precursor of the peptidoglycan (see Scheme 1). The sidewall staining is suggestive of a helical pattern for peptidoglycan growth during sidewall elongation.
The primary role of the peptidoglycan in Gram-positive and Gram-negative bacteria is believed to be structural, and a synonymous term for the peptidoglycan is the bacterial cell wall. The peptidoglycan contains the turgor pressure of the bacterium, and provides a scaffold for the many proteins, enzymes, and structural complexes that locate within the cell envelope for communication by the bacterium with its environment.9 Yet at the same time as the peptidoglycan fulfills this structural role, it undergoes dynamic transformation, structurally accommodating the fundamental processes of bacterial cell growth and division. During cell growth, all of the structural components of the cell envelope must grow in unison. During bacterial cell division, all must separate in unison, without compromise of the integrity of either cell. For these reasons, it is hardly surprising that circumstances leading to the loss of the integrity of the peptidoglycan are potentially lethal circumstances for the bacterium. Indeed, many of the most useful antibiotics used to control bacterial infections—such as the penicillin and cephalosporin β-lactam antibiotics, and the vancomycin and teicoplanin glycopeptide antibiotics—use molecular mechanisms that disrupt specific processes involving the peptidoglycan. The uniqueness of the peptidoglycan as a characteristic structure of bacteria explains its value as an antibacterial target. The processes of peptidoglycan biosynthetic growth as a polymer, and eventual separation during bacterial division, demand the orchestrated interplay of specific and structurally complementary enzymes. Accordingly, the integrity of the peptidoglycan structure is monitored by the bacterium, by pathways that remain very poorly understood, as a means of activating the defense systems it possesses to resist these antibiotics. Likewise, as the peptidoglycan is a structure unique to bacteria, its degradation provides several small molecule structures that are used by eukaryotes for immune surveillance against bacterial infection. All of these events raise fundamental questions concerning the chemistry of the peptidoglycan. How are the events of its growth, surveillance, and separation accommodated within the structural details of its molecular structure? And how can these events be exploited to the advantage of human and animal health, by a molecular-level understanding of its structure as a host for antibacterial recognition, and as a guest for recognition by macromolecules?
The Structure of the Peptidoglycan
The basic bonding arrangement of the bacterial cell wall polymer has been known for some time, and is reflected by the term peptidoglycan.10 The peptidoglycan is a polymer consisting of glycan strands, synthesized by the transglycosylase activity of bifunctional enzymes. The glycan strands are subsequently cross-linked by the conjoining of oligopeptide stems attached to alternate glycans of the strand. The process achieving this cross-linking is a transpeptidation reaction, catalyzed by a separate domain of these same bifunctional peptidoglycan-biosynthesizing enzymes.11–14 The glycan structure is substantially identical for both Gram-negative and Gram-positive bacteria, and consists of repeating units of a disaccharide composed of N-acetyl-2-amino-2-deoxyglucosamine (abbreviated NAG) in β-1,4-glycosidic linkage to the saccharide N-acetylmuramic acid (abbreviated NAM). NAM, a derivative of NAG, has an additional ether-linked O-3 lactyl substituent. The carboxylate of the O-3 lactyl group is further substituted by an oligopeptide. The terminus of this oligopeptide is a repeat of the d-amino acid, d-alanine. This -d-Ala-d-Ala stem terminus is key to the cross-linking chemistry of the transpeptidation. During transpeptidation, the penultimate d-Ala terminus of the one glycan strand acylates the amine terminus of a stem peptide provided by an adjacent glycan strand, with departure of the ultimate -d-Ala as a leaving group. With the important exception of some new antibiotic-resistant Gram-positive bacteria,15 the use of a -d-Ala-d-Ala stem terminus for the transpeptidase-catalyzed cross-linking of glycan strands in cell wall biosynthesis is common to both Gram-positive and Gram-negative bacteria. Although there are strong similarities in the structures of the rest of the oligopeptide stems of the Gram-negative and Gram-positive bacteria, significant variations in the composition, length, and stereochemistry of the oligopeptide structure are encountered.16 These variations reflect both innate differences in biosynthesis, as well as resistance adaptations.17–20 Indeed, there is no better evidence as to how much more there is to understand concerning the peptidoglycan than the recent discovery that d-amino acids other than d-Ala are important modulators of its structure.21,22
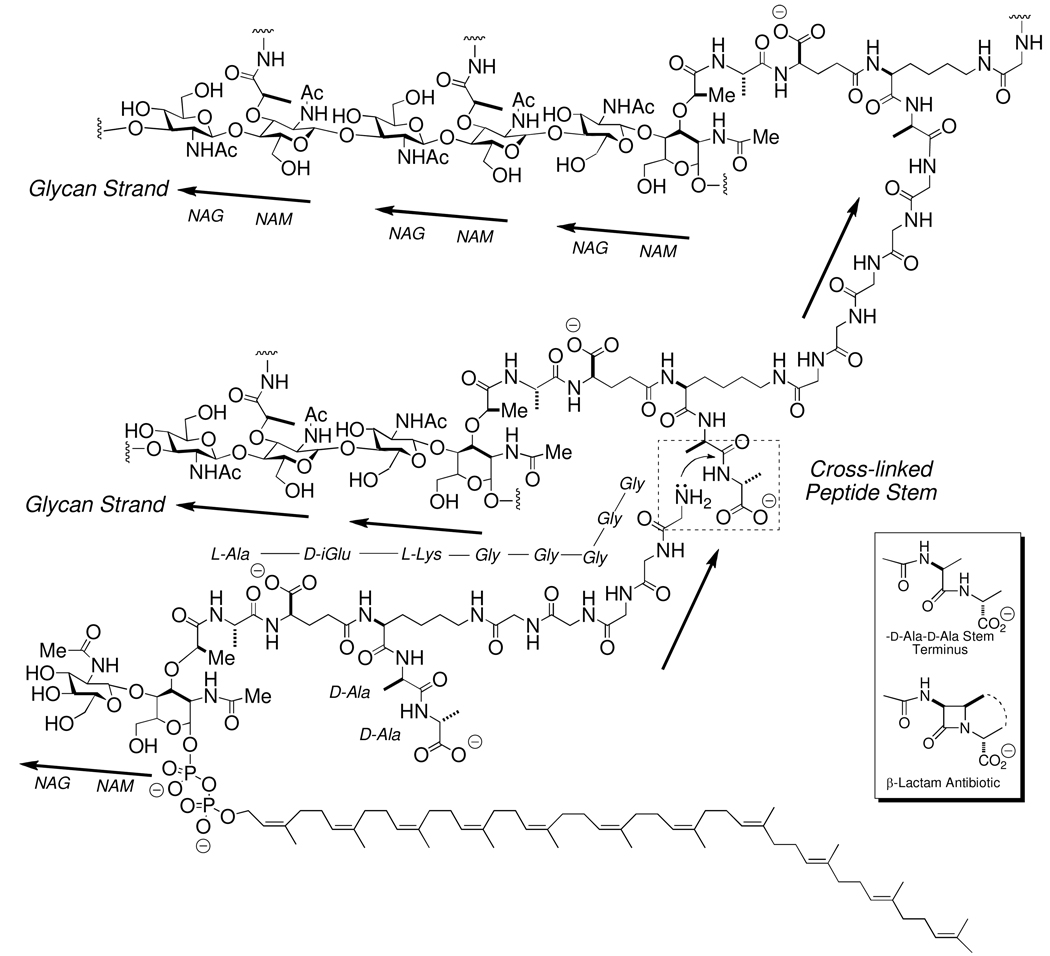
A sense of the biosynthetic complexity of these events is shown in Scheme 1, which shows the key structures involved in the biosynthesis and cross-linking of three glycan strands of the cell wall of the Gram-positive bacterium Staphyloccus aureus. The structure in the lower left of this chart is the key biosynthetic precursor for cell wall biosynthesis, Lipid II. In the Lipid II of S. aureus, the peptide stem contains additionally a -Gly5 “bridge” pentapeptide.23–26 The amine terminus of this pentaglycine bridge is the nucleophile acceptor in cross-linking to the d-alanyl acyl donor. Lipid II itself is assembled in the cytoplasm and inner leaflet of the bacterial cell membrane by a series of enzymatic reactions.27–29 In the final steps of its biosynthesis, the hydrophobic undecaprenol segment of Lipid II embeds into the inner leaflet of the cell membrane, exposing the hydrophilic NAG-NAM disaccharide to the cytoplasm. Sequential assembly of the oligopeptide stem and bridge structures on the NAM saccharide completes the Lipid II structure. Lipid II is then translocated to the outer leaflet of the cell membrane, by a poorly characterized lipid translocase.30–35 Following this translocation, lengthening of the glycan strand occurs by transglycosylase catalysis36–39 wherein one molecule of Lipid II acts as a NAG-NAM donor, releasing the undecaprenylpyrophosphate for return to the cytoplasm and re-use in Lipid II (and in Lipid A, an intermediate used in the biosynthesis of the lipopolysaccharides of the exterior membrane of Gram-negative bacteria) synthesis.40–43 The result of transglycosylase catalysis is pairwise NAG-NAM lengthening of the glycan strands.38,44 As previously noted, the transglycosylase and transpeptidase enzymatic activities of peptidoglycan biosynthesis are catalyzed by separate, and by spatially distanced, domains of a single enzyme structure.13,45 These enzymes are called the high-molecular weight penicillin-binding proteins (HMW PBPs), wherein their name reflects their preeminence as the molecular target of the β-lactam (which include the penicillin and cephalosporin) antibiotics. The β-lactams engage in the functionally irreversible acylation of the active site serine of the PBP transpeptidase domain. As a result, further catalysis of the transpeptidation event of peptidoglycan synthesis is blocked.46–48 The inability of the bacterium to complete this final event of cell wall biosynthesis either arrests bacterial growth, or is bactericidal to the bacterium, depending on the structure of the β-lactam and the particular PBP enzymes (for example, the Gram-negative bacterium Neisseria gonorrhoeae has four, while the Gram-positive bacterium Bacillus subtilis has at least sixteen) which are inactivated.49 The key mimicry used by the β-lactam in this deception is with the -d-Ala-d-Ala peptidoglycan stem terminus, as first suggested by Tipper and Strominger.50–53 During normal peptidoglycan biosynthesis, the active site serine of the PBP transpeptidase domain is acylated by the -d-Ala-d-Ala stem, with departure of the terminal -d-Ala as a leaving group. Interception of the -d-alaninyl acyl-enzyme by the amino terminus of an amino acid from an adjoining peptidoglycan strand cross-links the two strands. The Gram-positive bacterium S. aureus uses the amine of the N-terminal glycine of a pentaglycine bridge extension added to the ε-amine of the l-lysine of the oligopeptide stem of the NAM saccahride. In other bacteria (such as the Gram-negative bacterium E. coli) a bridge extension is not used, and meso-diaminopimelic acid is used instead of l-lysine. The β-lactams, acting as -d-Ala-d-Ala mimetics (Scheme 1), efficiently acylate the same active site serine used during catalytic transpeptidation, but as the β-lactam introduces a steric impediment not found in -d-Ala-d-Ala, acyl-transfer to the amine provided by a neighboring glycan strand is not possible.54–57 The catalytic activity of the transpeptidase domain of the PBP is thus inactivated.
Scheme 1.
The cross-linking event in peptidoglycan biosynthesis by the Gram-positive bacterium S. aureus. The structure of the immediate biosynthetic precursor for the peptidoglycan of S. aureus, Lipid II-Gly5 is shown as the bottom structure. The cross-linking event is highlighted within the hashed-edged box shown in the center right. This stem cross-linking event, wherein the amine terminus of the pentaaglycine displaces the d-alanine terminus on the stem of an adjacent glycan strand, is catalyzed by the transpeptidase domain of a high molecular mass PBP enzyme. This reaction proceeds through an acyl-enzyme intermediate (not depicted). Lipid II-Gly5 participation in transpeptidase cross-linking is shown only for the purpose of illustration. Peptide stem cross-linking almost certainly occurs subsequent to Lipid II-Gly5-dependent, transglycosylase-catalyzed elongation of the glycan strand. The inset box (lower right) shows the β-lactam antibiotic structure as a mimetic of a conformation of the -d-Ala-d-Ala stem used in the cross-linking.
Scheme 1 leaves absent any sense of a higher order structure to the peptidoglycan polymer. The three-dimensional structure of the peptidoglycan is not known. As the last major biopolymer with an unknown three-dimensional structure, it is hardly surprising that a full breadth of experimental methodology has been applied toward this objective. The key unanswered questions regarding the three-dimensional structure of the peptidoglycan include the length of the glycan strands, the mechanism by which glycan length is controlled,58 the orientations of the glycan strands as they straddle the transpeptidase domain of the PBP for cross-linking, the spatial orientation of the interconnected stems to the glycan strands,26 and the spatial orientation of the entire peptidoglycan with respect to the cell membrane.59–61 This structural problem may be further divided to encompass possible structural differences between the “static” regions of the peptidoglycan, where the cell wall growth does not occur or is completed, and the “dynamic” regions where the peptidoglycan is growing for eventual septation into a daughter cell. Moreover, it is now known that the biosynthesis of the cell envelope components is coordinated by cytoskeletal assemblies within the cytoplasm of the bacterium.12,62–69 With respect to the peptidoglycan as a component of the cell envelope, the remarkable structural similarities within the PBP family of enzymes45 suggests commonality (of at least some structural features) among all of the bacteria. Nonetheless, bacteria exemplify such diversity of shape—including spherical, rod, curved and spiral morphologies70–72—that variation in the peptidoglycan structure may be needed to accommodate these shapes.73 The merits of several limiting possibilities to the three-dimensional peptidoglycan have been extensively discussed,45,59,60 and have stimulated the creation of sophisticated peptidoglycan models that valiantly attempt to reconcile current knowledge concerning peptidoglycan strand interconnection, constituent stoichiometry, extent of cross-linking, shape, growth, and division.26,61,74–76
A quantitative sense of the dimensions of the peptidoglycan polymer is available from microscopy. The peptidoglycan of the Gram-negative rod-shaped bacterium E. coli forms a remarkably consistent (in terms of depth) shell around the entire bacterium, as seen by cryo-electron tomography.77 The depth of the E. coli peptidoglycan is approximately 6.4 nm, while the depth of the peptidoglycan of a second rod-shaped Gram-negative bacterium, Pseudomonas aeuruginosa, is less (2.4 nm) by cryo-electron microscopy.78,79 In these images the peptidoglycan is visualized as closely apposing the outer membrane of these two bacteria, and above a periplasmic space. Microscopy studies of other Gram-negative bacteria suggest a more central location of the peptidoglycan within the periplasmic space is also possible.80–82 In contrast to the relatively thin peptidoglycan dimensions seen for Gram-negative bacteria, the depth of the mature peptidoglycan exoskeleton of the Gram-positive S. aureus cocci is greater (19 nm), and the peptidodglycan depth is even greater at the cross-wall formed during S. aureus cell division.83,84 Atomic force microscopy of the S. aureus peptidoglycan85 and of germinating Gram-positive spores86 show the surface of the Gram-positive peptidoglycan exoskeleton as a porous network of peptidoglycan strands.
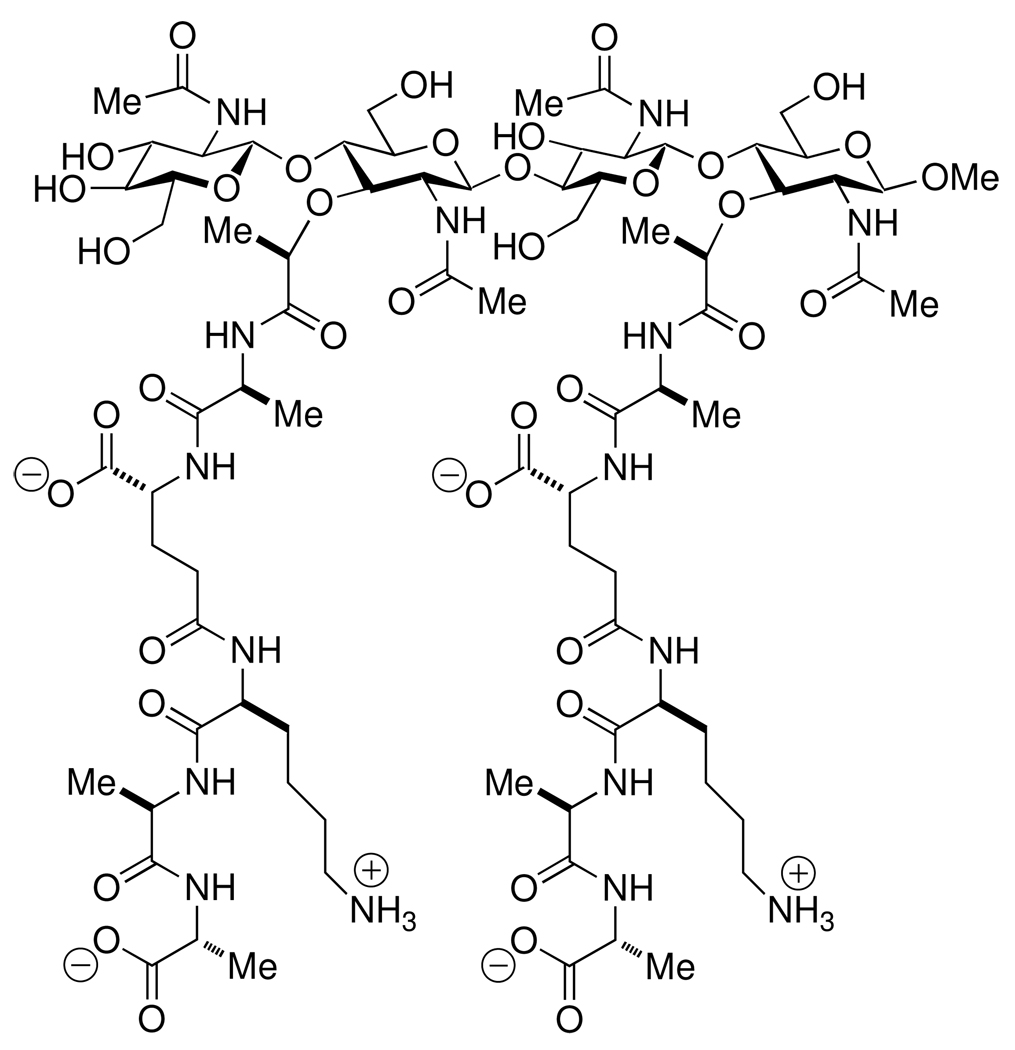
Molecular-level experimental evidence concerning the three-dimensional structure of the peptidoglycan is being sought. A notable contribution from our own laboratory was the NMR determination of the solution structure of a single [NAG-NAM]2 peptidoglycan strand (Scheme 2).87,88 This tetrasaccharide was sufficiently small (Mr = 1,930) to be within the reach of synthesis, but yet sufficiently large to demonstrate the innate conformational preference of an isolated peptidoglycan strand. The observed conformation for this strand was a right-handed helical glycan, exhibiting three-fold rotational periodicity for its peptide stem. It is most curious that several proteins that interact directly with the peptidoglycan via binding domains (such as the Braun lipoprotein of Escherichia coli,89 the Omp porins of Gram-negative bacteria,90–92 and the TolC channel protein of efflux assemblies93–95) oligomerize with three-fold symmetry. The relevance of the three-fold rotational periodicity seen for the peptidoglycan stems for the structure of Scheme 2, with respect to biological recognition of the un-crosslinked peptidodglycan structure, is attested to by a breadth of experimental studies involving both bacterial and eukaryotic proteins, as discussed below.
Scheme 2.
The synthetic [NAG-NAM]2 mimetic of the non-crosslinked peptidoglycan of the cell wall, used for the NMR determination of the solution conformation of a peptidoglycan strand.
The solid-state NMR spectrum of the isolated peptidoglycan polymer of E. coli—a single molecule having an approximate molecular mass of 3 × 109 Da—shows remarkable simplicity.96 Spin diffusion NMR spectra of the intact 13C-glycine-labeled S. aureus peptidoglycan is most consistent with close spatial proximity between the pentaglycyl bridge and the glycan strand, interpreted as requiring that all of the peptide stems lie parallel a plane perpendicular to the glycan.97 Given the ability of the peptidoglycan to accommodate extraordinary variation within the peptide stem,18,25 as evidenced by the facile adaptation by S. aureus to the use of both short and long peptide bridges,98,99 expectations for a single consensus three-dimensional peptidoglycan structure may be unreasonable. Perhaps the most promising future efforts toward this objective are those examining the direct interaction of the peptidoglycan with bacterial hyperstructure. For example, substantial recent progress has been made toward the molecular interaction of the peptidoglycan with the stator protein of the flagellum hyperstructure, a protein that possesses a well defined peptidoglycan-binding domain100–103 in order to anchor the flagellum hyperstructure to the cell wall.104–106
The Peptidoglycan as Host
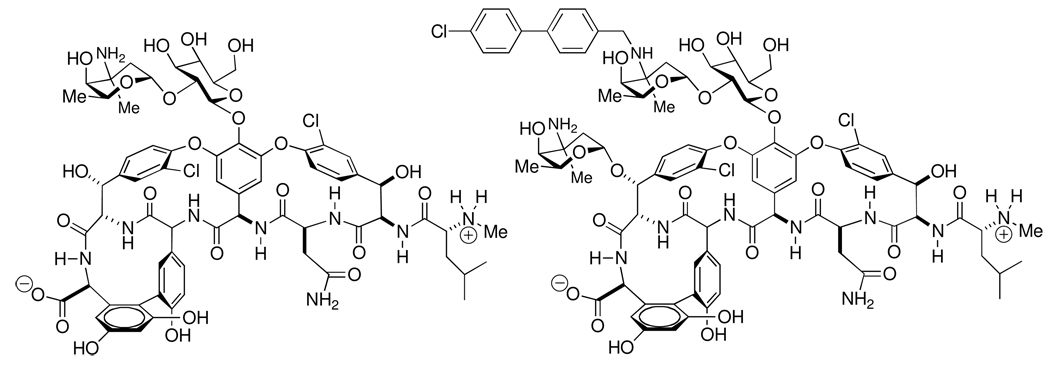
Given the necessity of peptidglycan integrity to bacterial viability, it is not surprising that the peptidoglycan is a target for antibiotics. Indeed, the clinical development of new antibiotics having the peptidoglycan as their molecular target remains no less compelling an objective today, than it was at the dawn of the 20th Century antibiotic era. Antibiotic recognition of the peptidoglycan occurs at the early stages of peptidoglycan biosynthesis, as well as by the polymeric peptidoglycan. A particularly important point for antibiotic recognition of the peptidoglycan occurs at the PBP-dependent events of transglycosylation and transpeptidation.40,107 As briefly discussed above, the chemotherapeutic value of the β-lactam antibiotics rests on their ability to function as mimetics of the unique –d-Ala-d-Ala motif used during this transpeptidation reaction (additional aspects of the relationship between the β-lactams and the peptidoglycan are covered in the section on the peptidoglycan as guest). For clarity of focus, our discussion on the peptidoglycan as a host structure is restricted to the single example of vancomycin, as a chemotherapeutically important example of non-covalent peptidoglycan recognition. Numerous examples of antibiotics other than the glycopeptides (as exemplified by vancomycin and teicoplanin) exploiting non-covalent peptidoglycan host-guest recognition are known, and include the closely-related (but mechanistically separate) lipoglycopeptides (exemplified by oritavancin and telavancin),108 the lipoglycodepsipeptides (exemplified by ramoplanin),109–111 the mannopeptimycins,112,113 and the peptide-derived lantibiotics (exemplified by nisin and clausin).114–118 Of these antibiotic classes, the molecular recognition mechanism of the vancomycin class of antibiotics (Scheme 3) against Gram-positive pathogens, is the best understood.119–122 Vancomycin recognizes primarily the acyl-d-Ala-d-Ala motif at the peptide stem terminus of the NAM residue of Lipid II, the key biosynthetic intermediate in peptidoglycan biosynthesis. Consequently, this aspect of the molecular mechanism of vancomycin is exerted largely through steric interference by the bound antibiotic at the transglycosylase stage of peptidoglycan biosynthesis.123 The complex formed between the -d-Ala-d-Ala motif and vancomycin is non-covalent, and is characterized by strong electrostatic (including both ion pair and hydrogen-bond) interactions.124–127 The thermodynamics of vancomycin binding in solution with the [NAG-NAM]2 cell wall mimetic shown in Scheme 2 correspond to a 1.0 µM dissociation constant (ΔG° = −34.5 kJ mol−1) that is dominated by favorable enthalpy (ΔH° = −41.2 kJ mol−1) at a relatively small entropic cost (TΔS° = −6.7 kJ mol−1).128 Although this in vitro dissociation constant is notably greater than the nanomolar benchmark that customarily defines strong biological interaction, the effective concentration of the -d-Ala-d-Ala motif encountered by vancomycin, as it engages the peptidoglycan surface of Gram-positive bacteria, is high. The supramolecular organization of the vancomycin–N-acetyl-d-Ala-d-Ala dipeptide complex formed in vitro shows back-to-back and side-to-side intermolecular contact between the vancomycin molecules of the complex.129–131 These solid-state structures were precedented by the observation that vancomycin poorly self-associates to form a dimer in solution, in the absence of peptidoglycan structure.119,132–135 Covalent vancomycin dimers with strong antibiotic activity are known.136 Exploitation of the vancomycin-peptidoglycan complex for the purpose of delivering a second antibacterial agent, representing the concept of hybrid antibacterial design,137 also has been demonstrated by a covalently linked vancomycin-cephalosporin hybrid with potent Gram-positive activity.138,139
Scheme 3.
Structures of vancomycin (left) and oritavancin (right).152
NMR analyses of vancomycin (and its derivatives) bound to the intact peptidoglycan of S. aureus indicate that the pentaglycine bridge that is unique to S. aureus further contributes to the binding interaction.140,141 A recent—and surprising—structure-activity development within the vancomycin class is the discovery that the interactions of vancomycin and of oritavancin, a semi-synthetic lipoglycopeptide derivative of vancomycin (Scheme 3),142–147 are fundamentally different. Oritavancin exhibits much faster bactericidal activity against methicillin-resistant S. aureus (MRSA) and vancomycin-resistant enterococci (VRE) than does vancomycin.143 Solid-state NMR evaluation of oritavancin derivatives148,149 indicates that oritavancin has two separate binding interactions, one using the -d-Ala-d-Ala and the other the pentaglycine bridge motifs. Whereas both vancomycin and oritavancin have comparable inhibition of the transglycosylase step of peptidoglycan biosynthesis, as a result of the pentaglycine interaction oritavancin is the better transpeptidase inhibitor.149 Using 19F REDOR NMR of a fluorinated oritavancin derivative, Kim et al. observe oritavancin to preferentially bind to the mature, as distinct from the nascent, peptidoglycan of S. aureus protoplast membranes.150 Direct NMR comparison of vancomycin and oritavancin binding to whole cells of a second Gram-positive pathogen (Enterococcus faecium) that does not use a pentaglycine bridge further suggests the value of a secondary binding site interaction with respect to antibacterial potency.123 Glycopeptide binding to the E. faecium peptidoglycan occurs only to nascent peptidoglycan, and further correlates to preferential transpeptidase inhibition, in contrast with preferential transglycosylase inhibition by vancomycin.123 Independent evidence supporting different binding interactions for oritavancin compared to vancomycin, using the peptidoglycan structures of both methicillin-resistant S. aureus and vancomycin-resistant Entercoccus faecalis, is also provided by transmission electron microscopy.151 With both the MRSA and VRE strains, oritavancin showed enhanced affinity for binding to the peptidoglycan at the septum, the eventual point of cell lysis.151 The importance of further understanding of the structure-activity basis for peptidoglycan-glycopeptide host-guest chemistry, toward improvement in the breadth and potency of these (and other) antibacterials, cannot be overstated.
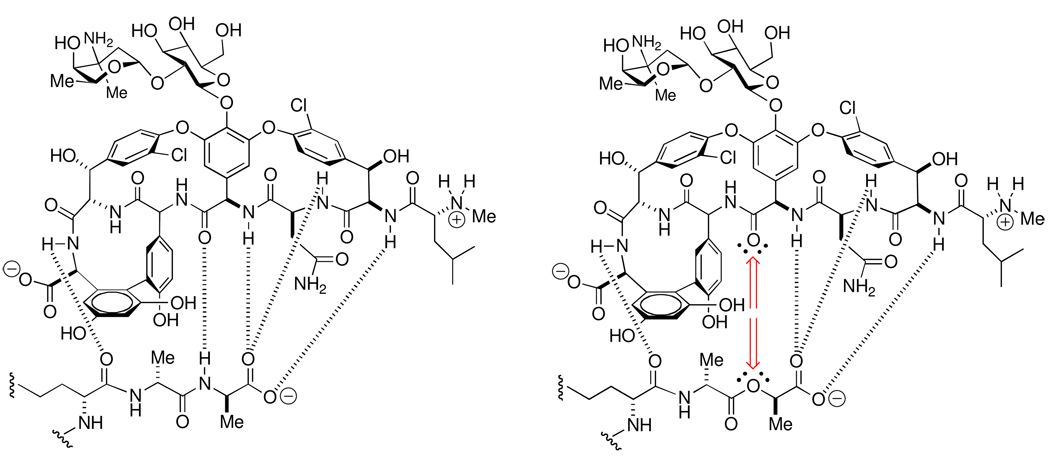
A direct relevance of peptidoglycan-vancomycin host-guest chemistry to the understanding of the resistance mechanisms against the vancomycin antibacterial class is already proven. Several of the resistance mechanisms against vancomycin correlate directly to lowering the effectiveness of its complex with the peptidoglycan.153–155 One mechanism is the thickening of the cell wall, resulting in greater propensity for non-productive complex formation on the bacterial surface, while simultaneously limiting the access of vancomycin to the site of cell wall biosynthesis at the base of the cell wall.156–159 Two other mechanisms involve modification of the peptide stem. Replacement of the acyl-d-Ala-d-Ala stem terminus with a depsipeptide (ester-containing) acyl– d-Ala-d-Lac terminus abolishes the central hydrogen bond between the vancomycin amide carbonyl and the acyl-d-Ala-d-Ala amide NH,160 while concomitantly introducing unfavorable non-bonding interactions (Scheme 4).161–163 As a result the affinity of vancomycin for the peptidoglycan decreases by three orders of magnitude, matching the decrease in antibiotic efficacy.164 This vancomycin resistance mechanism is now common in the enterococci, and has been recently observed in S. aureus. The biosynthetic enzymatic activities that are necessary to accomplish this change in peptide stem structure may have originated by adaptation of the self-resistance mechanism used by the microorganisms that biosynthesize these glycopeptides.165 A third resistance mechanism, used by the enterococci, involves peptidoglycan crosslinking to the acyl-enzyme formed from the stem acyl-l-lysine-d-Ala amide bond, rather than the acyl-d-Ala-d-Ala amide bond. This cross-linking is catalyzed by an l,d-transpeptidase enzyme,166 rather than the d,d-transpeptidase activity of the PBPs. This change in acyl donor evades the consequence of the vancomycin complexation, and results in high-level resistance of the enterococci to both the β-lactam and the glycopeptide classes of antibiotics.167
Scheme 4.
Hydrogen-bonding interactions in the -d-Ala-d-Ala peptidoglycan-vancomycin host-guest complex (left) compared to the -d-Ala-d-Lac peptidoglycan-vancomycin host-guest complex (right). The loss of a key hydrogen bond, and its replacement by unfavorable non-bonding repulsive interactions, is indicated by the red-colored arrows in the -d-Ala-d-Lac depsipeptide complex.161
As important as the events of molecular recognition are to antibiotic activity, it is now generally recognized that the antibiotic recognition event—whether the antibiotic is a β-lactam (peptidoglycan targeted), a glycopeptide (peptidoglycan targeted), an aminoglycoside (ribosome targeted) or other antibiotic/target pairing—serves primarily as initiating events. Antibiotic activity results from a much further dowstream disruption of homeostasis within the metabolic pathways of the bacterium. These disruptions compromise the ability of the bacterium to grow (antibiotic is bacteriostatic) or to survive (bactericidal). At the molecular level, however, we presently have almost no understanding as to how antibiotic recognition event generates a signal, how this signal is propagated, and how this propagation ultimately results in the loss of homeostasis for a particular pathway(s). Moreover, we further suffer in this ignorance since it is likely that the molecular events of signal propagation leading to cell disruption may also coincide with the molecular events leading to expression of mutations allowing resistance, or to the expression of a resistance response. An exciting recent development is recognition that the bacterial SOS response, and in particular the generation of reactive oxygen species, are central events to the mechanism of action of many antibiotics, including that of vancomycin.168–173
A final example of the application of peptidoglycan host-guest chemistry is the visualization of the peptidoglycan surface. Peptidoglycan biosynthesis is coordinated with the the biosynthesis of the other components of the cell envelope.12,65,174–182 The ability of the peptidoglycan to recognize fluorescent conjugates of vancomycin and of ramoplanin (see Figure 1) has allowed visualization of the pattern of peptidoglycan biosynthesis5,12,21,174,176,183 and correlation of antibiotic location with relative resistance.159 Complementary fluorescence microscopy images suggest a helical pattern for cell envelope assembly in rod-shaped Gram-positive (such as B. subtilis) and Gram-negative (such as E. coli) bacteria, as also visualized from the perspective of the membrane lipids184,185 and of the chemoreceptor proteins.186
The Peptidoglycan as Guest
A preponderance of research effort on the peptidoglycan has focused on the structural relationship between the β-lactam antibiotics and the acyl-d-Ala-d-Ala stem structure46,47,187 in relation to β-lactam inactivation of the transpeptidation/carboxypeptidase activities of the PBPs.13 While this focus is by no means misplaced, a richer perspective on the guest chemistry of the peptidoglycan is now evident. Peptidoglycan recognition embraces not just the PBPs of cell wall biosynthesis, but also a sweeping breadth of bacterial enzymes that monitor peptidoglycan integrity (so as to detect the presence of antibiotics that would compromise peptidoglycan integrity) and for activating resistance mechanisms, and for controlling peptidoglycan degradation (during septation, and for some bacteria the recycling of old peptidoglycan into new peptidoglycan). All of these events are now understood to be highly regulated and interdependent, in ways that we are only now beginning to understand. Moreover, the detection of several of the structural components of the peptidoglycan by protein recognition is a key mechanism for eukaryotic activation of innate immunity in response to bacterial infection. An evaluation of the full interplay of these processes is beyond the scope of this perspective. Our emphasis here is the growing appreciation of the value of chemical synthesis of discrete peptidoglycan structures (“muropeptides”) as a prerequisite to the understanding of the specific roles played by the key proteins and enzymes of these processes.
The value of peptidoglycan of defined structure to elucidation of the extraordinary breadth of its host-guest chemistry is proven in terms of understanding its biosynthesis,37,188–192 its recognition by enzymes,193 and its immune recognition.194–202 These examples include peptidoglycan structures obtained by both chemoenzymatic and total synthesis. Our laboratory has used the total synthesis of peptidoglycan structures to address a diversity of important problems concerning peptidoglycan host-guest chemistry. These problems include the substrate specificity of the ampD amidase of peptidoglycan recycling, the structural basis for peptidoglycan binding by the peptidoglycan recognition proteins of innate immunity, the structural basis for peptidoglycan recognition by a phage endolysin, and peptidoglycan recognition by a PBP.
An emerging area is the signaling processes by which antibiotics induce defensive and virulence responses by bacteria. The integrity of the peptidoglycan structure is monitored continuously by the bacterium, and we know little as to how this monitoring is done. In many Gram-negative bacterial pathogens, β-lactamase induction as a resistance mechanism occurs through a muropeptide-signaling pathway.203–205 The β-lactamases are defensive enzymes that recognize β-lactam antibiotics as substrates, and catalyze the hydrolytic opening of the β-lactam ring. This destruction of the pivotal structural feature of the β-lactams, used for PBP inactivation, abolishes their antibiotic activity. An antagonist of this signaling could have great value by preserving β-lactam activity against these bacteria, by suppression of β-lactamase expression. Although key events in the pathway are known, how these events interconnect remains largely unknown. Clearly, the pivotal event is initial detection of the β-lactam antibiotic. In resistant Gram-positive bacteria β-lactam recognition occurs by acylation of a serine residue located in a surface-exposed transmembrane signaling protein.206–213 The triggering event in β-lactam recognition by Gram-negative bacteria now appears also to involve serine acylation, but here of specific members of the PBP family. The PBP enzyme family is divided between the high-molecular mass enzymes (having both transglycosylase and transpeptidase activity) and the low-molecular mass enzymes (having primarily carboxypeptidase activity, catalyzed by domains with very strong structural homology to the transpeptidase domain of the high-Mr PBPs). At least one functional high-Mr PBP enzyme is essential for bacterial viability while the low-Mr PBPs, although having ancillary roles relating to shape214–216 and septation,217 are regarded as non-critical, notwithstanding their evolutionary conservation.218 In the Gram-negative bacterium Pseudomonas aeruginosa, β-lactam acylation of a specific low-Mr PBP senses the presence of β-lactam antibiotics.219 Structural analysis of the cognate Haemophilus influenzae PBP suggests a conformational translocation involving a specific phenylalanine—a residue that is not found in other low-Mr PBPs—in signaling.220 The next step in signal transduction is unknown. Loss of the activity of this PBP is, however, ultimately coincides with a change in the steady-state muropeptide catabolite levels governed by a muropeptide recycling pathway, and interpreted to result in derepression of the β-lactamase gene.
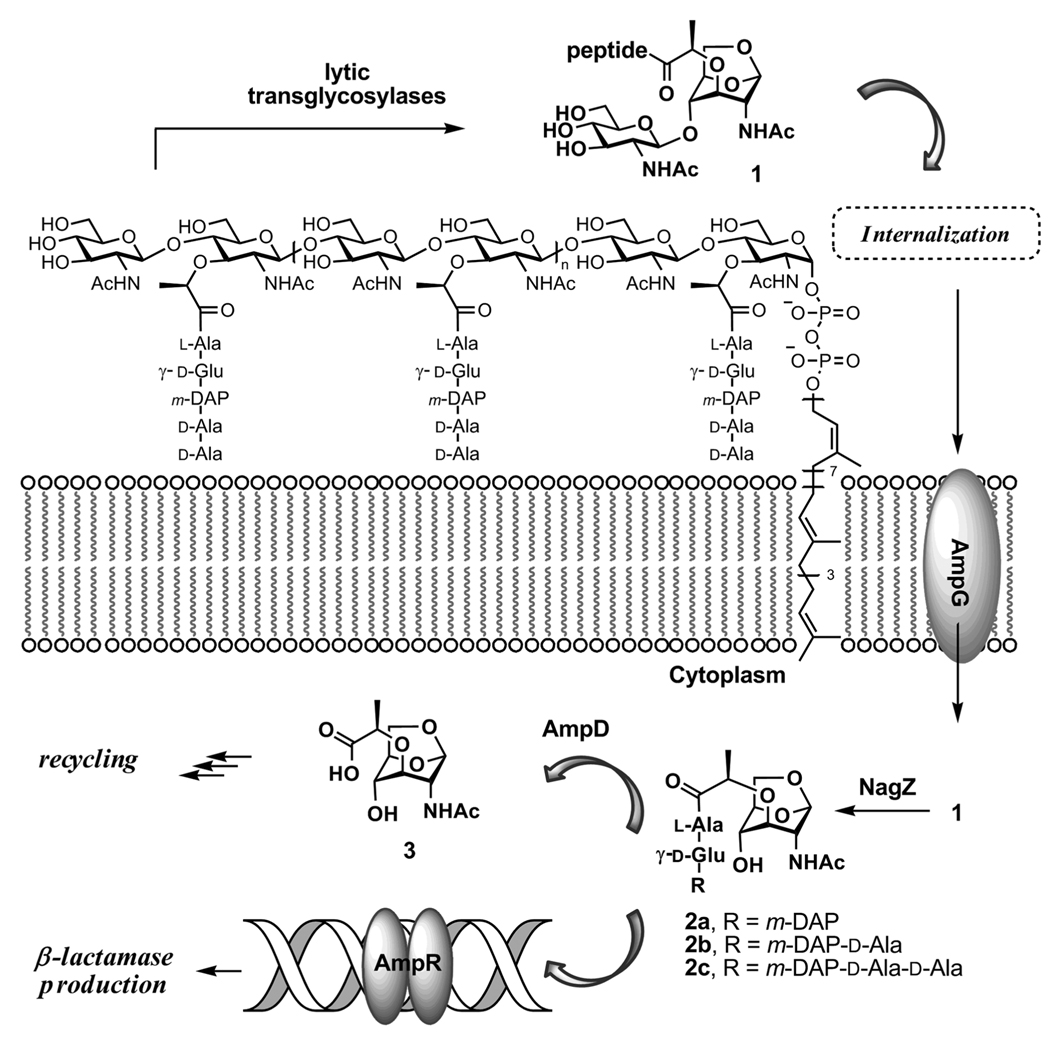
As the Gram-negative peptidoglycan-recycling pathway that regulates β-lactamase expression is now understood, recycling of the glycan strands of the peptidoglycan gives a steady-state muropeptide pool.221,222 The initial step in muropeptide recycling is catalyzed by a family of enzymes found in the periplasm, the lytic transglycosylases, that cleave the β-(1,4) NAG-NAM disaccharide bond to give muropeptides having as a terminus the unusual bicyclic 1,6-anhydromuramamide saccharide. The mechanism for formation of the 1,6-anhydromuramamide by these enzymes is intramolecular interception by the C-6 alcohol of an incipient oxocarbenium cation species.223,224 While the presence of anhydromuramic terminii in endogenous Gram-negative muropeptides is long recognized, the circumstances governing their appearance are unknown. The 1,6-anhydromuramamide muropeptides enter the cytoplasm from the periplasmic space, by passage through the proton-motive driven AmpG transporter.225,226 Within the cytoplasm, the terminal 1,6-anhydromuramamide is separated from the glycan by the NagZ glycosylase.227,228 A 1,6-anhydromuramamide structure is believed to bind to the AmpR transcription factor so as to derepress β-lactamase expression.229 The final step in this portion of the pathway for peptidoglycan recycling is hydrolytic cleavage of the lactyl amide of 1,6-anhydromuramamide, catalyzed by AmpD amidase, and giving the 1,6-anhydromuramic monosaccharide and the free peptide stem as products. A schematic overview of these events is shown in Scheme 5.
Scheme 5.
Peptidoglycan fragmentation leads to NAG-NAM segments, which are transported across plasma membrane to initiate the recycling of these segments and gene induction for production of the β-lactamase enzyme.
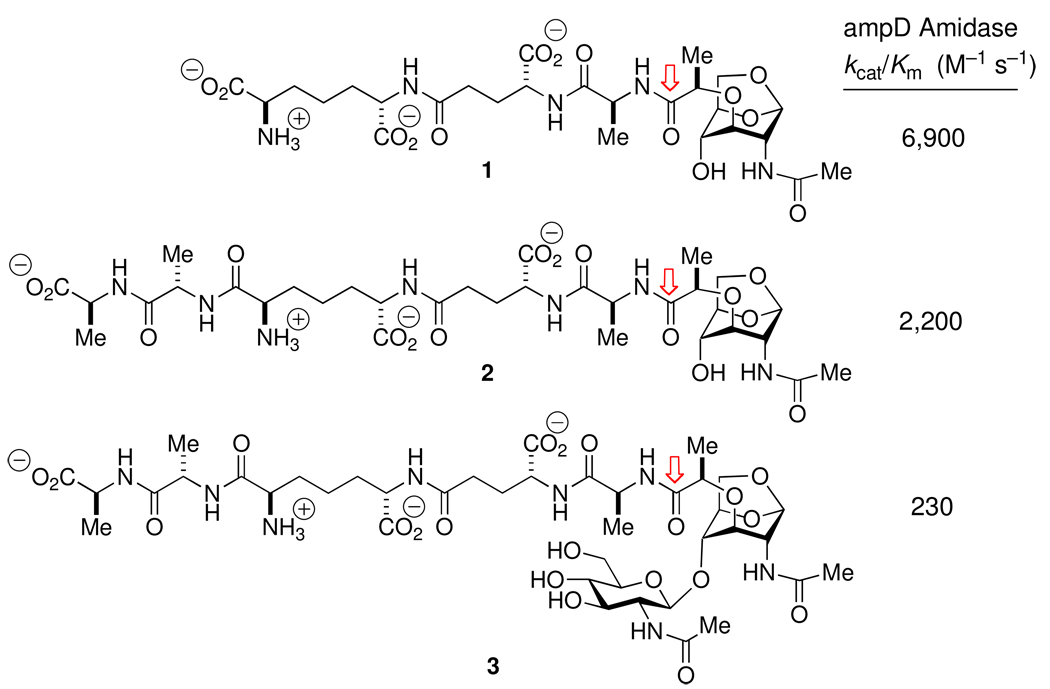
Seminal studies on this pathway used the 1,6-anhydroMurNAc-l-Ala-d-iGln-mA2pm muropeptide (a variant of structure 1 of Scheme 6 with d-iGln in place of d-iGlu), isolated from bacterium following deletion of the ampD gene.230 The value of a more complete set of 1,6-anhydroMurNAc muropeptides, in order to clarify key structural questions within this pathway, was evident to us. Two examples of such questions include the confirmation of the endo- and exo-glycosylase preferences within the lytic transglycosylase family of enzymes,231 and correlation of the substrate specificity of AmpD with the structure recognized by the AmpR transcription factor. Accordingly, adaptation of outstanding synthetic precedent232 provided the set of muropeptides shown in Scheme 6.233 Evaluation of this set with the AmpD enzyme led to two conclusions.234 The first conclusion (comparing 1 and 2 to 3) strongly suggests that AmpD functions in the recycling pathway after NagZ, as previously believed.235 The second conclusion, comparing the very similar kcat/Km values for 1 and 2, is that the substrate specificity of this enzyme correlates more closely to the presence of a (1,6-anhydromuramyl)alaninyl peptide, than to the peptide structure beyond the alanine residue. Since muramyl peptide 2 accumulates following exposure of the bacterium to β-lactams, and is only slightly less capable as a substrate for AmpD, the structure of the AmpR ligand used in derepressing the β-lactamase gene remains open. Clearly, a detailed understanding of the interplay within the NagZ-AmpD-AmpR nexus will be critical to identifying opportunities for understanding the role of muropeptides in intracellular communication between prokaryotes236–238 and in developing new means for antibiotic management and discovery against emerging Gram-negative pathogens.239,240 A sense of these latter opportunities is evidenced by the observation of antibacterial synergy between NagZ inhibitors and the β-lactams.241–243
Scheme 6.
Substrate specificity, measured by kcat/Km, for Citrobacter freundii AmpD amidase-catalyzed amide bond hydrolysis (cleavage point indicated by the arrows) of three anhydromuramyl muropeptide structures.
The peptidoglycan structure is not only used in bacteria-to-bacteria communication, but by eukaryotes engaging in bacterial symbiosis244,245 and by eukaryotes in the immunological detection of, and response to, bacterial infection. Mammals possess a complex and interdependent array of peptidoglycan-binding proteins for innate immunity, including proteins involved in immune activation246–249 and proteins (some are enzymes) which are also intrinsically bactericidal (peptidoglycan recognition proteins, PGRPs) by a vancomycin-like mechanism of peptidoglycan adherence.250–257 An understanding of the structural specificity of this large family of proteins is relevant to the mechanisms by which bacterial pathogens are recognized196,258,259 and how some evade immune surveillance,260–263 and for the purpose of identifying adjuvants for immune stimulation.199,202,264 In insects, muropeptide recognition by the insect PGRPs appears to be the exclusive mechanism by which bacterial infection is detected.265 Some insect PGRPs have enzymatic activity, while others are protein receptors for the muropeptide. Some are specific for lysine-containing muropeptides (lysine is found in the peptide stem of many Gram-positive bacteria), while others are specific for diaminopimelate-containing muropeptides (as are found in Gram-negative, and rod-shaped Gram-positive, bacteria).266 Muropeptide binding to the insect PGRP results in PGRP heterodimerization, culminating in an innate immune response synthesis of antimicrobial defensive peptides.
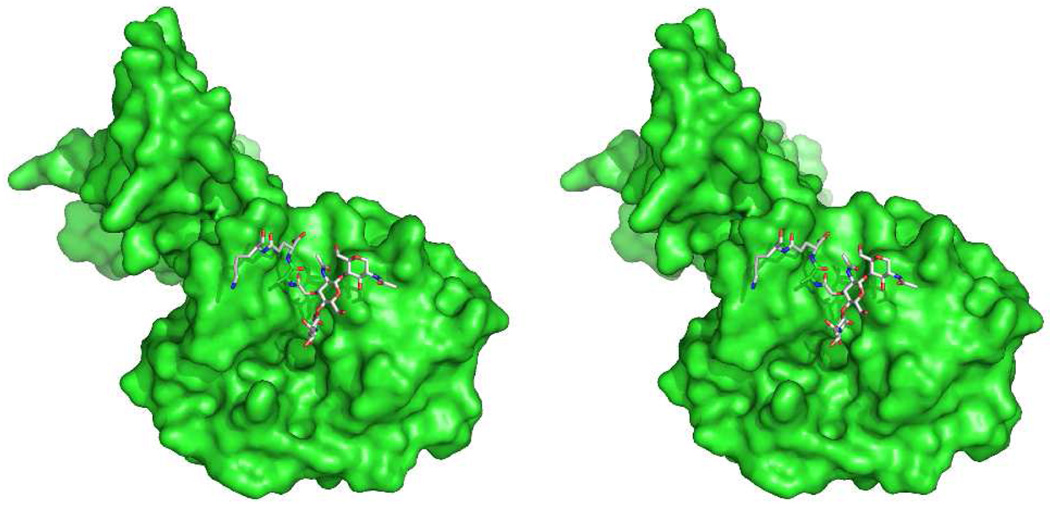
The mechanism of PGRP signaling in humans is different from that of insects. Human muropeptide recognition by PGRPs is one component of a much more complex innate immune response to bacteria (other bacteria-derived structure also contributes to human recognition of bacteria, including the lipopolysaccharides, the cell-wall protein derived lipopeptides, mannan saccharides, and proteins of the flagellum). Human PGRP interaction with bacteria is directly bactericidal253 in a zinc-dependent process.267 The structural basis for peptidoglycan recognition by the PGRPs has been studied using synthetic peptidoglycan fragments195 and by the crystal structure determination of uncomplexed PGRPs,198,268,269 and peptidoglycan-complexed insect PGRPs270,271 and human PGRPs.272 These structures indicate that these PGRP use both the glycan and peptide stem sub-structures of the muropeptides in recognition. In collaboration with the Mariuzza group at the University of Maryland, we determined the structure of human PGRP-1β complexed with a NAG-NAM-l-Ala-γ-d-Glu-l-Lys-d-Ala-d-Ala muropeptide.273 The bound glycan conformation of this complex is virtually identical to the conformation of the [NAG-NAM]2 peptidoglycan (structure of Scheme 2) seen in solution.88 Indeed, the occurrence of this same glycan conformation in peptidoglycan complexes with a phage endolysin and with a PBP (discussed below), and by others with yet other peptidoglycan-recognizing enzymes,44,274 suggest that this conformation of the peptidoglycan may be the rule, and not the exception. A computational model using the [NAG-NAM]2 structure bound to this PGRP, based on crystallographic data of a partial peptiodglycan stucture seen in the electron density, is shown in Figure 2. In contrast to the glycan conformation, a different peptide stem sub-structure is seen as a result of a steric imposition of a different conformation on the lactyl amide of the NAM. These observations are consistent with the PGRP itself acting as a steric impediment for completion of cell wall synthesis while simultaneously stabilizing a peptide stem conformation that is unproductive for transpeptidase-dependent cross-linking.
Figure 2.
Computational stereo structure of the [NAG-NAM]2 structure (see Scheme 2) in complex with the human PGRP-1® peptidodglycan recognition protein, based on an X-ray structure of a smaller synthetic fragment bound to the protein (PDB Code 2EAX).273 The approximate diameter of the protein is 4 nm. The perspective shown for the protein orients the peptidoglycan cleft from top to bottom.
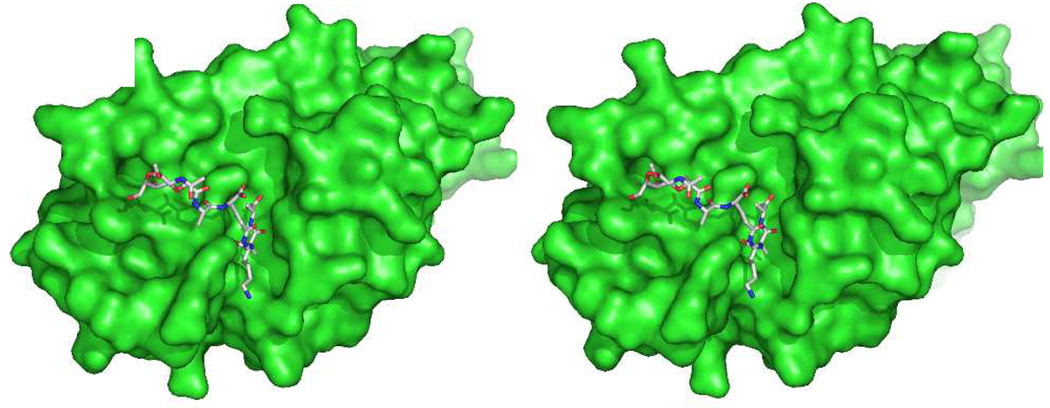
A complementary perspective for macromolecule recognition of the peptidoglycan structure is provided by the structure of this same [NAG-NAM]2 peptidoglycan strand bound to a catalytically inactive (E94Q) mutant of the pneumococcal phage Cpl-1 lysozyme, solved in collaboration with the Hermoso laboratory at the Istituto de Chimica-Fisica Rocasolano, CSIC in Madrid. The Cpl-1 lysozyme is an endolysin that catalyzes hydrolytic cleavage of the peptidoglycan at the NAG-NAM glycosidic bond, functioning in the final cell lysis step of the phage reproductive cycle.275 Cpl-1 has a two-domain structure, with one domain anchoring the enzyme to the cell wall and the second domain containing the active site. Five saccharides of the peptidoglycan flank the active site during catalysis. The [NAG-NAM]2 tetrasaccharide is bound by the inactive E94Q mutant (Figure 3) in positions +1 to +3 (NAG-NAM-NAG, with the trailing NAM disordered) of the active site.276 Again, this bound conformation is identical to the conformation encountered for the glycan strand in solution. The existence of a common peptidoglycan recognition pattern, encompassing peptidoglycan recognition by the PBP glycosyltransferase domains (which are related to the phage λ-lysozyme), the PGRPs, and the Cpl-1 endolysin, is implied.
Figure 3.
The catalytic module of the Cpl-1 endolysin in complex with peptidoglycan (PDB Code 2J8G).276 A comparison of this protein structure with those of Figs. 2 and 4 emphasizes the evolutionary convergence of different protein motifs for the purpose of peptidoglycan binding.
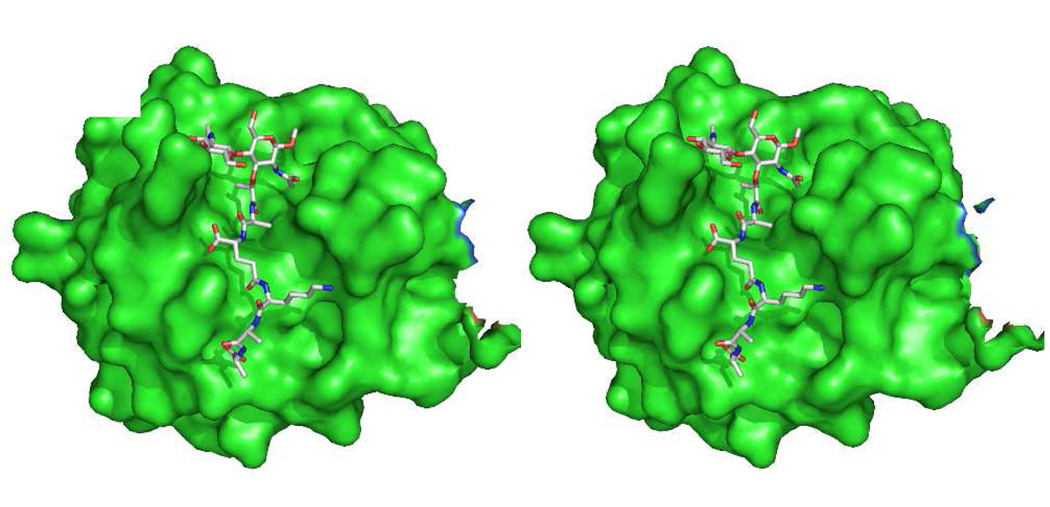
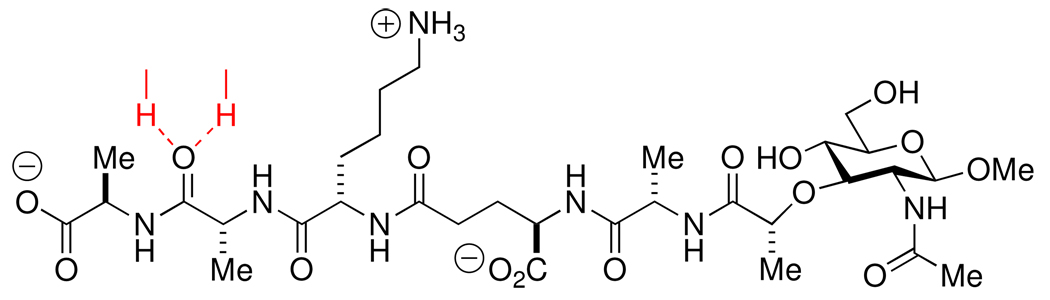
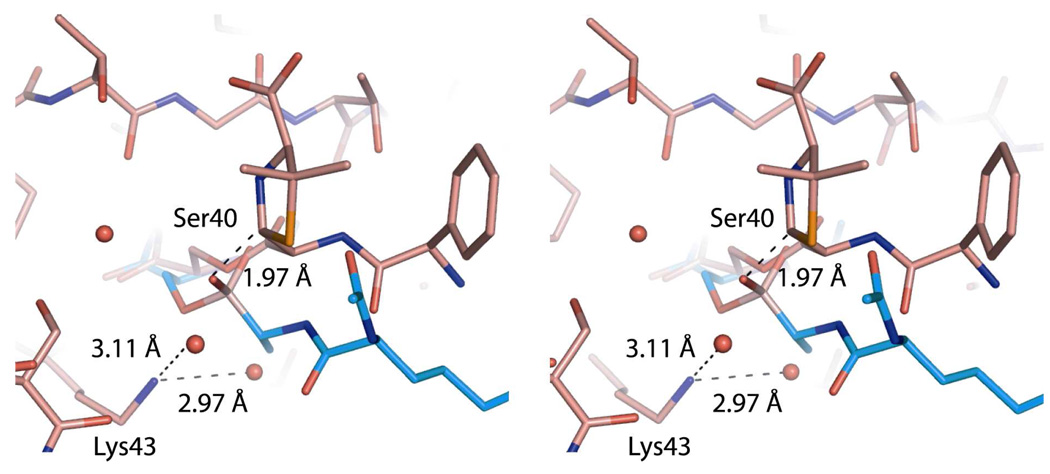
Recent experiments suggest inclusion of the transpeptidase domain of the PBPs in this list. In collaboration with the Shoichet laboratory at the University of California, San Francisco a peptidoglycan segment (Scheme 7) occupied the active site of the E. coli PBP6 low-Mr PBP when diffused into the crystalline enzyme at a catalytically sub-optimal pH (Figure 4).57 This peptidoglycan segment has the -d-Ala-d-Ala motif recognized by this PBP for its catalysis. In the resulting PBP6 complex, the carbonyl of the scissile d-Ala fully engages the catalytic residues of the active site. This complex strongly resembles our expectations for the Michaelis complex (pre-acylation complex) of a peptidoglycan with this enzyme. The inability of the crystalline PBP6 to process this peptidoglycan as a substrate likely reflects a combination of factors, including the sub-optimal pH used for crystallization relative to catalysis and the intrinsic limitation of this peptidoglycan as a substrate of PBP6 (kcat/Km = 20 M−1 s−1). A comparison of the structure of Figure 4 with the crystal structure of a penicillin-inactivated PBP5 carboxypeptidase was especially rewarding (Figure 5). In the PBP5 acyl-enzyme, the steric clash between the bridging carbon atom (between the N and S atoms of the thiazolidine ring of the ring-opened penicillin) and the position otherwise required for the catalytic water molecule—a clash anticipated previously by Silvaggi et al.47—adequately explains the molecular mechanism for β-lactam inactivation of the PBPs. The anticipation that other peptidoglycan-binding proteins recognize the peptidoglycan in the same conformation as is found in solution for a single peptidoglycan strand is supported by yet other recent experimental44,277 and computational studies.278
Scheme 7.
Structure of the synthetic peptidoglycan82,273 used in the determination of the crystal structure of a Michaelis complex-like structure with E. coli PBP6. The red-colored hydrogen bonds drawn to the (scissile) amide carbonyl of the penultimate d-Ala residue indicates its occupancy of the oxyanion hole of the active site.
Figure 4.
Complex of E. coli PBP6 (catalytic domain of monomer B, PDB Code 3ITB) with the peptidoglycan structure that is shown in Scheme 7.57
Figure 5.
Stereo view of the PBP6 acyl-enzyme complex compared to that of the deacylation transition state. A PBP5 transition-state analog structure (PDB Code 1Z6F) is superimposed onto the PBP6 structure with ampicillin (PDB Code 3ITA), showing active site residues 39, 40, 43, 108 and 210. The transition-state analog and the catalytic serine of PBP5 are shown in blue, whereas ampicillin and PBP6 are shown in pink. Water molecules are represented as red spheres. Key distances are displayed as dashed lines. The amine group of Lys43 engages in hydrogen bonds with two water molecules in the PBP6 acyl-enzyme complex. The distance between the β-lactam-thiazolidine bridging carbon atom of ampicillin, and the boronic acid oxygen of the superimposed transition state analog, is only 1.97 Å. This carbon presents a steric impediment to hydrolytic deacylation.47,57
Host-Guest Chemistry of the Peptidoglycan
The definition of the bacterial peptidoglycan as an amorphous polymer, tasked with the simple (albeit essential) role of a structural scaffold and barrier, is inadequate. The solitary glycan strand in solution has an ordered conformation that is directly relevant to a host of biological roles, and it is now reasonable to believe that this conformation will continue to have relevance to the host-guest chemistry of the peptidoglycan. While the conformations of the peptidoglycan polymer is likely different in ways we cannot yet anticipate, experiments where the peptidoglycan serves as host for antibiotics (as exemplified by vancomycin) support the relevance of in vitro study of the peptidoglycan structure. Likewise, the chemical synthesis of peptidoglycan fragments having defined structure has opened a vast realm to more precisely explore peptidoglycan recognition by macromolecules. The peptidoglycan is one of the last remaining biological polymers of unknown three-dimensional structure. An extraordinary diversity of opportunities, seen in terms of the recognition chemistry of the peptidoglycan structure, are now coming within experimental reach.
Acknowledgement
We gratefully acknowledge continuing support of this research by the National Institutes of Health.
Abbreviations used
- NAG
N-Acetyl-2-amino-2-deoxyglucosamine
- NAM
N-acetylmuramic acid
- PBP
penicillin binding protein
- MRSA
methicillin-resistant Staphylococus aureus
- VRE
vancomycin-resistant enterococci
- PGRP
peptidoglycan recognition protein
Biographies
Jed F. Fisher has engaged almost the full breadth of biological chemistry (including research on antibiotics, lipids, peptides, saccharides, drug candidates, coenzymes and enzymes) in the course of a career embracing bioorganic mechanism and medicinal chemistry. The enigma of the peptidoglycan engages this entire breadth.
Shahriar Mobashery received his undergraduate and graduate degrees from the University of Southern California and the University of Chicago, respectively. Following a short postdoctoral stay at the Rockefeller University, he served on the faculty at Wayne State University before relocating to the University of Notre Dame in 2003. He thrives on multidisciplinary research encompassing organic chemistry, biochemistry, molecular biology, microbiology and computation. He has had the good fortune of working with outstanding researchers in these disciplines, within and without his own research group.
References
- 1.Koch AL. Bacterial wall as target for attack: past, present, and future research. Clin. Microbiol. Rev. 2003;16:673–687. doi: 10.1128/CMR.16.4.673-687.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coyette J, van der Ende A. Peptidoglycan: the bacterial Achilles heel. FEMS Microbiol. Rev. 2008;32:147–148. doi: 10.1111/j.1574-6976.2008.00108.x. [DOI] [PubMed] [Google Scholar]
- 3.Schneider T, Sahl H-G. An oldie but a goodie - cell wall biosynthesis as antibiotic target pathway. Int. J. Med. Microbiol. 2010;300:161–169. doi: 10.1016/j.ijmm.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 4.Daniel RA, Errington J. Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell. 2003;113:767–776. doi: 10.1016/s0092-8674(03)00421-5. [DOI] [PubMed] [Google Scholar]
- 5.Tiyanont K, Doan T, Lazarus MB, Fang X, Rudner DZ, Walker S. Imaging peptidoglycan biosynthesis in Bacillus subtilis with fluorescent antibiotics. Proc. Natl. Acad. Sci. U.S.A. 2006;103:11033–11038. doi: 10.1073/pnas.0600829103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takano H, Takechi K. Plastid peptidoglycan. Biochim. Biophys. Acta. 2010;1800 doi: 10.1016/j.bbagen.2009.07.020. 1444-1151. [DOI] [PubMed] [Google Scholar]
- 7.Casadesus J. Bacterial L-forms require peptidoglycan synthesis for cell division. Bioessays. 2007;29:1189–1191. doi: 10.1002/bies.20680. [DOI] [PubMed] [Google Scholar]
- 8.Henrichfreise B, Schiefer A, Schneider T, Nzukou E, Poellinger C, Hoffmann TJ, Johnston KL, Moelleken K, Wiedemann I, Pfarr K, Hoerauf A, Sahl HG. Functional conservation of the lipid II biosynthesis pathway in the cell wall-less bacteria Chlamydia and Wolbachia: why is lipid II needed? Mol. Microbiol. 2009;73:913–923. doi: 10.1111/j.1365-2958.2009.06815.x. [DOI] [PubMed] [Google Scholar]
- 9.Koch AL. The exocytoskeleton. J Mol. Microbiol. Biotechnol. 2006;11:115–125. doi: 10.1159/000094048. [DOI] [PubMed] [Google Scholar]
- 10.Walsh CT. Antibiotics: Actions, Origins, Resistance (ISBN-10: 1-55581-254-6) Washington, DC: ASM Press; 2003. p. 336. [Google Scholar]
- 11.Holtje JV. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol. Mol. Biol. Rev. 1998;62:181–203. doi: 10.1128/mmbr.62.1.181-203.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scheffers DJ, Pinho MG. Bacterial cell wall synthesis: new insights from localization studies. Microbiol. Mol. Biol. Rev. 2005;69:585–607. doi: 10.1128/MMBR.69.4.585-607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Macheboeuf P, Contreras-Martel C, Job V, Dideberg O, Dessen A. Penicillin binding proteins: key players in bacterial cell cycle and drug resistance processes. FEMS Microbiol. Rev. 2006;30:673–691. doi: 10.1111/j.1574-6976.2006.00024.x. [DOI] [PubMed] [Google Scholar]
- 14.Vollmer W. Structure and biosynthesis of the murein (peptidoglycan) sacculus. In: Ehrmann M, editor. The Periplasm. Washington, DC: ASM Press; 2007. pp. 198–213. [Google Scholar]
- 15.Sacco E, Hugonnet JE, Josseaume N, Cremniter J, Dubost L, Marie A, Patin D, Blanot D, Rice LB, Mainardi JL, Arthur M. Activation of the l,d-transpeptidation peptidoglycan cross-linking pathway by a metallo-d,d-carboxypeptidase in Enterococcus faecium. Mol. Microbiol. 2010;75:874–885. doi: 10.1111/j.1365-2958.2009.07014.x. [DOI] [PubMed] [Google Scholar]
- 16.Vollmer W. Structural variation in the glycan strands of bacterial peptidoglycan. FEMS Microbiol. Rev. 2008;32:287–306. doi: 10.1111/j.1574-6976.2007.00088.x. [DOI] [PubMed] [Google Scholar]
- 17.Goffin C, Ghuysen JM. Biochemistry and comparative genomics of SxxK superfamily acyltransferases offer a clue to the mycobacterial paradox: presence of penicillin-susceptible target proteins versus lack of efficiency of penicillin as therapeutic agent. Microbiol. Mol. Biol. Rev. 2002;66:702–738. doi: 10.1128/MMBR.66.4.702-738.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arbeloa A, Hugonnet JE, Sentilhes AC, Josseaume N, Dubost L, Monsempes C, Blanot D, Brouard JP, Arthur M. Synthesis of mosaic peptidoglycan cross-bridges by hybrid peptidoglycan assembly pathways in Gram-positive bacteria. J. Biol. Chem. 2004;279:41546–41556. doi: 10.1074/jbc.M407149200. [DOI] [PubMed] [Google Scholar]
- 19.Lloyd AJ, Gilbey AM, Blewett AM, De Pascale G, El Zoeiby A, Levesque RC, Catherwood AC, Tomasz A, Bugg TD, Roper DI, Dowson CG. Characterization of tRNA-dependent peptide bond formation by MurM in the synthesis of Streptococcus pneumoniae peptidoglycan. J. Biol. Chem. 2008;283:6402–6417. doi: 10.1074/jbc.M708105200. [DOI] [PubMed] [Google Scholar]
- 20.van Dam V, Olrichs N, Breukink E. Specific labeling of peptidoglycan precursors as a tool for bacterial cell wall studies. ChemBioChem. 2009;10:617–624. doi: 10.1002/cbic.200800678. [DOI] [PubMed] [Google Scholar]
- 21.Lam H, Oh DC, Cava F, Takacs CN, Clardy J, de Pedro MA, Waldor MK. d-Amino acids govern stationary phase cell wall remodeling in bacteria. Science. 2009;325:1552–1555. doi: 10.1126/science.1178123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blanke SR. Cell biology. Expanding functionality within the looking-glass universe. Science. 2009;325:1505–1506. doi: 10.1126/science.1180332. [DOI] [PubMed] [Google Scholar]
- 23.Rohrer S, Berger-Bachi B. FemABX peptidyl transferases: a link between branched-chain cell wall peptide formation and β-lactam resistance in Gram-positive cocci. Antimicrob. Agents Chemother. 2003;47:837–846. doi: 10.1128/AAC.47.3.837-846.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schneider T, Senn MM, Berger-Bachi B, Tossi A, Sahl HG, Wiedemann I. In vitro assembly of a complete, pentaglycine interpeptide bridge containing cell wall precursor (Lipid II-Gly5) of Staphylococcus aureus. Mol. Microbiol. 2004;53:675–685. doi: 10.1111/j.1365-2958.2004.04149.x. [DOI] [PubMed] [Google Scholar]
- 25.Mainardi JL, Villet R, Bugg TD, Mayer C, Arthur M. Evolution of peptidoglycan biosynthesis under the selective pressure of antibiotics in Gram-positive bacteria. FEMS Microbiol. Rev. 2008;386–408:32. doi: 10.1111/j.1574-6976.2007.00097.x. [DOI] [PubMed] [Google Scholar]
- 26.Sharif S, Kim SJ, Labischinski H, Schaefer J. Characterization of peptidoglycan in fem-deletion mutants of methicillin-resistant Staphylococcus aureus by solid-state NMR. Biochemistry. 2009;48:3100–3108. doi: 10.1021/bi801750u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Heijenoort J. Lipid intermediates in the biosynthesis of bacterial peptidoglycan. Microbiol. Mol. Biol. Rev. 2007;71:620–635. doi: 10.1128/MMBR.00016-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barreteau H, Kovac A, Boniface A, Sova M, Gobec S, Blanot D. Cytoplasmic steps of peptidoglycan biosynthesis. FEMS Microbiol. Rev. 2008;32:168–172. doi: 10.1111/j.1574-6976.2008.00104.x. [DOI] [PubMed] [Google Scholar]
- 29.Bouhss A, Trunkfield AE, Bugg TD, Mengin-Lecreulx D. The biosynthesis of peptidoglycan lipid-linked intermediates. FEMS Microbiol. Rev. 2008;32:208–233. doi: 10.1111/j.1574-6976.2007.00089.x. [DOI] [PubMed] [Google Scholar]
- 30.Doerrler WT. Lipid trafficking to the outer membrane of Gram-negative bacteria. Mol. Microbiol. 2006;60:542–552. doi: 10.1111/j.1365-2958.2006.05130.x. [DOI] [PubMed] [Google Scholar]
- 31.Daleke DL. Phospholipid flippases. J. Biol. Chem. 2007;282:821–825. doi: 10.1074/jbc.R600035200. [DOI] [PubMed] [Google Scholar]
- 32.Poulsen LR, Lopez-Marques RL, Palmgren MG. Flippases: still more questions than answers. Cell. Mol. Life Sci. 2008;65:3119–3125. doi: 10.1007/s00018-008-8341-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruiz N. Bioinformatics identification of MurJ (MviN) as the peptidoglycan lipid II flippase in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 2008;105:15553–15557. doi: 10.1073/pnas.0808352105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruiz N. Streptococcus pyogenes YtgP (Spy_0390) complements Escherichia coli strains depleted of the putative peptidoglycan flippase MurJ. Antimicrob. Agents Chemother. 2009;53:3604–3605. doi: 10.1128/AAC.00578-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fay A, Dworkin J. Bacillus subtilis homologs of MviN (MurJ), the putative Escherichia coli lipid II flippase, are not essential for growth. J. Bacteriol. 2009;191:6020–6028. doi: 10.1128/JB.00605-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spencelayh MJ, Cheng Y, Bushby RJ, Bugg TD, Li JJ, Henderson PJ, O'Reilly J, Evans SD. Antibiotic action and peptidoglycan formation on tethered lipid bilayer membranes. Angew. Chem. Int. Ed. 2006;45:2111–2116. doi: 10.1002/anie.200504035. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y, Fechter EJ, Wang TS, Barrett D, Walker S, Kahne DE. Synthesis of heptaprenyl-Lipid IV to analyze peptidoglycan glycosyltransferases. J. Am. Chem. Soc. 2007;129:3080–3081. doi: 10.1021/ja069060g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuan Y, Barrett D, Zhang Y, Kahne D, Sliz P, Walker S. Crystal structure of a peptidoglycan glycosyltransferase suggests a model for processive glycan chain synthesis. Proc. Natl. Acad. Sci. U. S. A. 2007;104:5348–5353. doi: 10.1073/pnas.0701160104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lovering AL, de Castro LH, Lim D, Strynadka NC. Structural insight into the transglycosylation step of bacterial cell-wall biosynthesis. Science. 2007;315:1402–1405. doi: 10.1126/science.1136611. [DOI] [PubMed] [Google Scholar]
- 40.Ostash B, Walker S. Bacterial transglycosylase inhibitors. Curr. Opin. Chem. Biol. 2005;9:459–466. doi: 10.1016/j.cbpa.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 41.Lehrer J, Vigeant KA, Tatar LD, Valvano MA. Functional characterization and membrane topology of Escherichia coli WecA, a sugar-phosphate transferase initiating the biosynthesis of enterobacterial common antigen and O-antigen lipopolysaccharide. J. Bacteriol. 2007;189:2618–2628. doi: 10.1128/JB.01905-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Valvano MA. Undecaprenyl phosphate recycling comes out of age. Mol. Microbiol. 2008;67:232–235. doi: 10.1111/j.1365-2958.2007.06052.x. [DOI] [PubMed] [Google Scholar]
- 43.Touze T, Tran AX, Hankins JV, Mengin-Lecreulx D, Trent MS. Periplasmic phosphorylation of lipid A is linked to the synthesis of undecaprenyl phosphate. Mol. Microbiol. 2008;67:264–277. doi: 10.1111/j.1365-2958.2007.06044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sung MT, Lai YT, Huang CY, Chou LY, Shih HW, Cheng WC, Wong CH, Ma C. Crystal structure of the membrane-bound bifunctional transglycosylase PBP1b from Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2009;106:8824–8829. doi: 10.1073/pnas.0904030106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol. Rev. 2008;32:234–258. doi: 10.1111/j.1574-6976.2008.00105.x. [DOI] [PubMed] [Google Scholar]
- 46.Waxman DJ, Yocum RR, Strominger JL. Penicillins and cephalosporins are active site-directed acylating agents: evidence in support of the substrate analogue hypothesis. Phil. Trans. R. Soc. Lond. B. 1980;289:257–271. doi: 10.1098/rstb.1980.0044. [DOI] [PubMed] [Google Scholar]
- 47.Silvaggi NR, Josephine HR, Kuzin AP, Nagarajan R, Pratt RF, Kelly JA. Crystal structures of complexes between the R61 dd-peptidase and peptidoglycan-mimetic β-lactams: a non-covalent complex with a "perfect penicillin". J. Mol. Biol. 2005;345:521–533. doi: 10.1016/j.jmb.2004.10.076. [DOI] [PubMed] [Google Scholar]
- 48.Zapun A, Contreras-Martel C, Vernet T. Penicillin-binding proteins and β-lactam resistance. FEMS Microbiol. Rev. 2008;32:361–385. doi: 10.1111/j.1574-6976.2007.00095.x. [DOI] [PubMed] [Google Scholar]
- 49.Chung HS, Yao Z, Goehring NW, Kishony R, Beckwith J, Kahne D. Rapid β-lactam-induced lysis requires successful assembly of the cell division machinery. Proc. Natl. Acad. Sci. U.S.A. 2009;106:21872–21877. doi: 10.1073/pnas.0911674106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tipper DJ, Strominger JL. Mechanism of action of penicillins: a proposal based on their structural similarity to acyl-d-alanyl-d-alanine. Proc. Natl. Acad. Sci. U.S.A. 1965;54:1133–1141. doi: 10.1073/pnas.54.4.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee B. Conformation of penicillin as a transition-state analog of the substrate of peptidoglycan transpeptidase. J. Mol. Biol. 1971;61:463–469. doi: 10.1016/0022-2836(71)90393-7. [DOI] [PubMed] [Google Scholar]
- 52.Sweet RM. Chemical and biological activity: Inferences from X-ray crystal structures. In: Flynn EH, editor. Cephalosporins and Penicillins: Chemistry and Biology. New York: Academic Press; 1972. pp. 281–309. [Google Scholar]
- 53.Pratt RF. Functional evolution of the serine β-lactamase active site. J. Chem. Soc., Perkin Trans. 2. 2002:851–861. [Google Scholar]
- 54.Silvaggi NR, Kaur K, Adediran SA, Pratt RF, Kelly JA. Toward better antibiotics: crystallographic studies of a novel class of dd-peptidase/β-lactamase inhibitors. Biochemistry. 2004;43:7046–7053. doi: 10.1021/bi049612c. [DOI] [PubMed] [Google Scholar]
- 55.Sauvage E, Powell AJ, Heilemann J, Josephine HR, Charlier P, Davies C, Pratt RF. Crystal structures of complexes of bacterial dd-peptidases with peptidoglycan-mimetic ligands: the substrate specificity puzzle. J. Mol. Biol. 2008;381:383–393. doi: 10.1016/j.jmb.2008.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pratt RF. Substrate specificity of bacterial dd-peptidases (penicillin-binding proteins) Cell. Mol. Life Sci. 2008;65:2138–2155. doi: 10.1007/s00018-008-7591-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen Y, Zhang W, Shi Q, Hesek D, Lee M, Mobashery S, Shoichet BK. Crystal structures of penicillin-binding protein 6 from Escherichia coli. J. Am. Chem. Soc. 2009;131:14345–14354. doi: 10.1021/ja903773f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang TS, Manning SA, Walker S, Kahne D. Isolated peptidoglycan glycosyltransferases from different organisms produce different glycan chain lengths. J. Am. Chem. Soc. 2008;130:14068–14069. doi: 10.1021/ja806016y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vollmer W, Bertsche U. Murein (peptidoglycan) structure, architecture and biosynthesis in Escherichia coli. Biochim. Biophys. Acta. 2008;1778:1714–1734. doi: 10.1016/j.bbamem.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 60.Dmitriev B, Toukach F, Ehlers S. Towards a comprehensive view of the bacterial cell wall. Trends Microbiol. 2005;13:569–574. doi: 10.1016/j.tim.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 61.Vollmer W, Seligman SJ. Architecture of peptidoglycan: more data and more models. Trends Microbiol. 2010;18:59–66. doi: 10.1016/j.tim.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 62.Varma A, Young KD. FtsZ collaborates with penicillin binding proteins to generate bacterial cell shape in Escherichia coli. J. Bacteriol. 2004;186:6768–6774. doi: 10.1128/JB.186.20.6768-6774.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Scheffers DJ. Cell wall growth during elongation and division: one ring to bind them? Mol. Microbiol. 2007;64:877–880. doi: 10.1111/j.1365-2958.2007.05731.x. [DOI] [PubMed] [Google Scholar]
- 64.Norris V, den Blaauwen T, Cabin-Flaman A, Doi RH, Harshey R, Janniere L, Jimenez-Sanchez A, Jin DJ, Levin PA, Mileykovskaya E, Minsky A, Saier MJ, Skarstad K. Functional taxonomy of bacterial hyperstructures. Microbiol. Mol. Biol. Rev. 2007;71:230–253. doi: 10.1128/MMBR.00035-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.den Blaauwen T, de Pedro MA, Nguyen-Distèche M, Ayala JA. Morphogenesis of rod-shaped sacculi. FEMS Microbiol. Rev. 2008;32:321–344. doi: 10.1111/j.1574-6976.2007.00090.x. [DOI] [PubMed] [Google Scholar]
- 66.Kawai Y, Daniel RA, Errington J. Regulation of cell wall morphogenesis in Bacillus subtilis by recruitment of PBP1 to the MreB helix. Mol. Microbiol. 2009;71:1131–1144. doi: 10.1111/j.1365-2958.2009.06601.x. [DOI] [PubMed] [Google Scholar]
- 67.Adams DW, Errington J. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat. Rev. Microbiol. 2009;7:642–653. doi: 10.1038/nrmicro2198. [DOI] [PubMed] [Google Scholar]
- 68.Gamba P, Veening JW, Saunders NJ, Hamoen LW, Daniel RA. Two-step assembly dynamics of the Bacillus subtilis divisome. J. Bacteriol. 2009;191:4186–4194. doi: 10.1128/JB.01758-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cabeen MT, Charbon G, Vollmer W, Born P, Ausmees N, Weibel DB, Jacobs-Wagner C. Bacterial cell curvature through mechanical control of cell growth. EMBO J. 2009;28:1208–1219. doi: 10.1038/emboj.2009.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cabeen MT, Jacobs-Wagner C. Bacterial cell shape. Nat. Rev. Microbiol. 2005;3:601–610. doi: 10.1038/nrmicro1205. [DOI] [PubMed] [Google Scholar]
- 71.Young KD. The selective value of bacterial shape. Microbiol. Mol. Biol. Rev. 2006;70:660–703. doi: 10.1128/MMBR.00001-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Young KD. Bacterial morphology: why have different shapes? Curr. Opin. Microbiol. 2007;10:596–600. doi: 10.1016/j.mib.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Young KD. Too many strictures on structure. Trends Microbiol. 2006;14:155–156. doi: 10.1016/j.tim.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 74.Hayhurst EJ, Kailas L, Hobbs JK, Foster SJ. Cell wall peptidoglycan architecture in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 2008;105:14603–14608. doi: 10.1073/pnas.0804138105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gan L, Chen S, Jensen GJ. Molecular organization of Gram-negative peptidoglycan. Proc. Natl. Acad. Sci. U. S. A. 2008;105:18953–18957. doi: 10.1073/pnas.0808035105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huang KC, Mukhopadhyay R, Wen B, Gitai Z, Wingreen NS. Cell shape and cell-wall organization in Gram-negative bacteria. Proc. Natl. Acad. Sci. U. S. A. 2008;105:19282–19287. doi: 10.1073/pnas.0805309105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ting CS, Hsieh C, Sundararaman S, Mannella C, Marko M. Cryo-electron tomography reveals the comparative three-dimensional architecture of Prochlorococcus, a globally important marine cyanobacterium. J. Bacteriol. 2007;189:4485–4493. doi: 10.1128/JB.01948-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Beveridge TJ. Visualizing bacterial cell walls and biofilms. Microbe. 2006;1:279–284. [Google Scholar]
- 79.Matias VR, Al-Amoudi A, Dubochet J, Beveridge TJ. Cryo-transmission electron microscopy of frozen-hydrated sections of Escherichia coli and Pseudomonas aeruginosa. J. Bacteriol. 2003;185:6112–6118. doi: 10.1128/JB.185.20.6112-6118.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Borgnia MJ, Subramaniam S, Milne JL. Three-dimensional imaging of the highly bent architecture of Bdellovibrio bacteriovorus by using cryo-electron tomography. J. Bacteriol. 2008;190:2588–2596. doi: 10.1128/JB.01538-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Milne JL, Subramaniam S. Cryo-electron tomography of bacteria: progress, challenges and future prospects. Nat. Rev. Microbiol. 2009;7:666–675. doi: 10.1038/nrmicro2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Izard J, Renken C, Hsieh CE, Desrosiers DC, Dunham-Ems S, La Vake C, Gebhardt LL, Limberger RJ, Cox DL, Marko M, Radolf JD. Cryo-electron tomography elucidates the molecular architecture of Treponema pallidum, the syphilis spirochete. J. Bacteriol. 2009;191:7566–7580. doi: 10.1128/JB.01031-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Matias VRF, Beveridge TJ. Native cell wall organization shown by cryo-electron microscopy confirms the existence of a periplasmic space in Staphylococcus aureus. J. Bacteriol. 2006;188:1011–1021. doi: 10.1128/JB.188.3.1011-1021.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Matias VR, Beveridge TJ. Cryo-electron microscopy of cell division in Staphylococcus aureus reveals a mid-zone between nascent cross walls. Mol. Microbiol. 2007;64:195–206. doi: 10.1111/j.1365-2958.2007.05634.x. [DOI] [PubMed] [Google Scholar]
- 85.Touhami A, Jericho MH, Beveridge TJ. Atomic force microscopy of cell growth and division in Staphylococcus aureus. J. Bacteriol. 2004;186:3286–3295. doi: 10.1128/JB.186.11.3286-3295.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Plomp M, Leighton TJ, Wheeler KE, Hill HD, Malkin AJ. In vitro high-resolution structural dynamics of single germinating bacterial spores. Proc. Natl. Acad. Sci. U. S. A. 2007;104:9644–9649. doi: 10.1073/pnas.0610626104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hesek D, Lee M, Morio K-i, Mobashery S. Synthesis of a fragment of bacterial cell wall. J. Org. Chem. 2004;69:2137–2146. doi: 10.1021/jo035583k. [DOI] [PubMed] [Google Scholar]
- 88.Meroueh SO, Bencze KZ, Hesek D, Lee M, Fisher JF, Stemmler TL, Mobashery S. Three-dimensional structure of the bacterial cell wall peptidoglycan. Proc. Natl. Acad. Sci. U. S. A. 2006;103:4404–4409. doi: 10.1073/pnas.0510182103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shu W, Liu J, Ji H, Lu M. Core structure of the outer membrane lipoprotein from Escherichia coli at 1.9 Å resolution. J. Mol. Biol. 2000;299:1101–1112. doi: 10.1006/jmbi.2000.3776. [DOI] [PubMed] [Google Scholar]
- 90.Lambert O, Benabdelhak H, Chami M, Jouan L, Nouaille E, Ducruix A, Brisson A. Trimeric structure of OprN and OprM efflux proteins from Pseudomonas aeruginosa, by 2D electron crystallography. J. Struct. Biol. 2005;150:50–57. doi: 10.1016/j.jsb.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 91.Smith SG, Mahon V, Lambert MA, Fagan RP. A molecular Swiss army knife: OmpA structure, function and expression. FEMS Microbiol. Lett. 2007;273:1–11. doi: 10.1111/j.1574-6968.2007.00778.x. [DOI] [PubMed] [Google Scholar]
- 92.Jaroslawski S, Duquesne K, Sturgis JN, Scheuring S. High-resolution architecture of the outer membrane of the Gram-negative bacteria Roseobacter denitrificans. Mol. Microbiol. 2009;74:1211–1222. doi: 10.1111/j.1365-2958.2009.06926.x. [DOI] [PubMed] [Google Scholar]
- 93.Koronakis V, Eswaran J, Hughes C. Structure and function of TolC: the bacterial exit duct for proteins and drugs. Annu. Rev. Biochem. 2004;73:467–489. doi: 10.1146/annurev.biochem.73.011303.074104. [DOI] [PubMed] [Google Scholar]
- 94.Bavro VN, Pietras Z, Furnham N, Perez-Cano L, Fernandez-Recio J, Pei XY, Misra R, Luisi B. Assembly and channel opening in a bacterial drug efflux machine. Mol. Cell. 2008;30:114–121. doi: 10.1016/j.molcel.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Masi M, Duret G, Delcour AH, Misra R. Folding and trimerization of signal sequence-less mature TolC in the cytoplasm of Escherichia coli. Microbiology. 2009;155:1847–1857. doi: 10.1099/mic.0.027219-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kern T, Hediger S, Muller P, Giustini C, Joris B, Bougault C, Vollmer W, Simorre JP. Toward the characterization of peptidoglycan structure and protein-peptidoglycan interactions by solid-state NMR. J. Am. Chem. Soc. 2008;130:5618–5619. doi: 10.1021/ja7108135. [DOI] [PubMed] [Google Scholar]
- 97.Sharif S, Singh M, Kim SJ, Schaefer J. Staphylococcus aureus peptidoglycan tertiary structure from C-13 spin diffusion. J. Am. Chem. Soc. 2009;131:7023–7030. doi: 10.1021/ja808971c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Stranden AM, Ehlert K, Labischinski H, Berger-Bachi B. Cell wall monoglycine cross-bridges and methicillin hypersusceptibility in a femAB null mutant of methicillin-resistant Staphylococcus aureus. J. Bacteriol. 1997;179:9–16. doi: 10.1128/jb.179.1.9-16.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hubscher J, Jansen A, Kotte O, Schafer J, Majcherczyk PA, Harris LG, Bierbaum G, Heinemann M, Berger-Bachi B. Living with an imperfect cell wall: compensation of femAB inactivation in Staphylococcus aureus. BMC Genomics. 2007;8:307. doi: 10.1186/1471-2164-8-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Minamino T, Imada K, Namba K. Molecular motors of the bacterial flagella. Curr. Opin. Struct. Biol. 2008;18:693–701. doi: 10.1016/j.sbi.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 101.Roujeinikova A. Crystal structure of the cell wall anchor domain of MotB, a stator component of the bacterial flagellar motor: implications for peptidoglycan recognition. Proc. Natl. Acad. Sci. U.S.A. 2008;105:10348–10353. doi: 10.1073/pnas.0803039105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kojima S, Shinohara A, Terashima H, Yakushi T, Sakuma M, Homma M, Namba K, Imada K. Insights into the stator assembly of the Vibrio flagellar motor from the crystal structure of MotY. Proc. Natl. Acad. Sci. U.S.A. 2008;105:7696–7701. doi: 10.1073/pnas.0800308105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kojima S, Imada K, Sakuma M, Sudo Y, Kojima C, Minamino T, Homma M, Namba K. Stator assembly and activation mechanism of the flagellar motor by the periplasmic region of MotB. Mol. Microbiol. 2009;73:710–718. doi: 10.1111/j.1365-2958.2009.06802.x. [DOI] [PubMed] [Google Scholar]
- 104.Manson MD. How 34 pegs fit into 26 + 8 holes in the flagellar motor. J. Bacteriol. 2007;189:291–293. doi: 10.1128/JB.01556-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Manson MD, Harlow ML. A grand view of the flagellar motor. J. Bacteriol. 2009;191:5023–5025. doi: 10.1128/JB.00695-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Liu J, Lin T, Botkin DJ, McCrum E, Winkler H, Norris SJ. Intact flagellar motor of Borrelia burgdorferi revealed by cryo-electron tomography: evidence for stator ring curvature and rotor/C-ring assembly flexion. J. Bacteriol. 2009;191:5026–5036. doi: 10.1128/JB.00340-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Breukink E, de Kruijff B. Lipid II as a target for antibiotics. Nat. Rev. Drug Discov. 2006;5:321–323. doi: 10.1038/nrd2004. [DOI] [PubMed] [Google Scholar]
- 108.Lunde CS, Hartouni SR, Janc JW, Mammen M, Humphrey PP, Benton BM. Telavancin disrupts the functional integrity of the bacterial membrane through targeted interaction with the cell wall precursor lipid II. Antimicrob. Agents Chemother. 2009;53:3375–3383. doi: 10.1128/AAC.01710-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fang X, Tiyanont K, Zhang Y, Wanner J, Boger D, Walker S. The mechanism of action of ramoplanin and enduracidin. Mol. BioSys. 2006;2:69–76. doi: 10.1039/b515328j. [DOI] [PubMed] [Google Scholar]
- 110.Fang X, Nam J, Shin D, Rew Y, Boger DL, Walker S. Functional and biochemical analysis of a key series of ramoplanin analogues. Bioorg. Med. Chem. Lett. 2009;19:6189–6191. doi: 10.1016/j.bmcl.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hamburger JB, Hoertz AJ, Lee A, Senturia RJ, McCafferty DG, Loll PJ. A crystal structure of a dimer of the antibiotic ramoplanin illustrates membrane positioning and a potential Lipid II docking interface. Proc. Natl. Acad. Sci. U.S.A. 2009;106:13759–13764. doi: 10.1073/pnas.0904686106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ruzin A, Singh G, Severin A, Yang Y, Dushin RG, Sutherland AG, Minnick A, Greenstein M, May MK, Shlaes DM, Bradford PA. Mechanism of action of the mannopeptimycins, a novel class of glycopeptide antibiotics active against vancomycin-resistant gram-positive bacteria. Antimicrob. Agents Chemother. 2004;48:728–738. doi: 10.1128/AAC.48.3.728-738.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Koehn FE. New strategies and methods in the discovery of natural product anti-infective agents: the mannopeptimycins. J. Med. Chem. 2008;51:2613–2617. doi: 10.1021/jm070432l. [DOI] [PubMed] [Google Scholar]
- 114.Breukink E. A lesson in efficient killing from two-component lantibiotics. Mol. Microbiol. 2006;61:271–273. doi: 10.1111/j.1365-2958.2006.05239.x. [DOI] [PubMed] [Google Scholar]
- 115.Martin NI, Breukink E. Expanding role of lipid II as a target for lantibiotics. Future Microbiol. 2007;2:513–525. doi: 10.2217/17460913.2.5.513. [DOI] [PubMed] [Google Scholar]
- 116.Arnusch CJ, Bonvin AM, Verel AM, Jansen WT, Liskamp RM, de Kruijff B, Pieters RJ, Breukink E. The vancomycin-nisin(1–12) hybrid restores activity against vancomycin resistant Enterococci. Biochemistry. 2008;47:12661–12663. doi: 10.1021/bi801597b. [DOI] [PubMed] [Google Scholar]
- 117.Bouhss A, Al-Dabbagh B, Vincent M, Odaert B, Aumont-Nicaise M, Bressolier P, Desmadril M, Mengin-Lecreulx D, Urdaci MC, Gallay J. Specific interactions of clausin, a new lantibiotic, with lipid precursors of the bacterial cell wall. Biophys. J. 2009;97:1390–1397. doi: 10.1016/j.bpj.2009.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bierbaum G, Sahl HG. Lantibiotics: mode of action, biosynthesis and bioengineering. Curr. Pharm. Biotechnol. 2009;10:2–18. doi: 10.2174/138920109787048616. [DOI] [PubMed] [Google Scholar]
- 119.Williams DH, Bardsley B. The vancomycin group of antibiotics and the fight against resistant bacteria. Angew. Chem. Int. Ed. 1999;38:1172–1193. doi: 10.1002/(SICI)1521-3773(19990503)38:9<1172::AID-ANIE1172>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 120.Loll PJ, Axelsen PH. The structural biology of molecular recognition by vancomycin. Annu. Rev. Biophys. Biomol. Struct. 2000;29:265–289. doi: 10.1146/annurev.biophys.29.1.265. [DOI] [PubMed] [Google Scholar]
- 121.Kahne D, Leimkuhler C, Lu W, Walsh CT. Glycopeptide and lipoglycopeptide antibiotics. Chem. Rev. 2005;105:425–448. doi: 10.1021/cr030103a. [DOI] [PubMed] [Google Scholar]
- 122.Levine DP. Vancomycin: a history. Clin. Infect. Dis. 2006;42 Suppl. 1:S5–S12. doi: 10.1086/491709. [DOI] [PubMed] [Google Scholar]
- 123.Patti GJ, Kim SJ, Yu TY, Dietrich E, Tanaka KS, Parr TRJ, Far AR, Schaefer J. Vancomycin and oritavancin have different modes of action in Enterococcus faecium. J. Mol. Biol. 2009;392:1178–1191. doi: 10.1016/j.jmb.2009.06.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Cooper MA, Williams DH. Binding of glycopeptide antibiotics to a model of a vancomycin-resistant bacterium. Chem. Biol. 1999;6:891–899. doi: 10.1016/s1074-5521(00)80008-3. [DOI] [PubMed] [Google Scholar]
- 125.Yang Z, Vorpagel ER, Laskin J. Experimental and theoretical studies of the structures and interactions of vancomycin antibiotics with cell wall analogues. J. Am. Chem. Soc. 2008;130:13013–13022. doi: 10.1021/ja802643g. [DOI] [PubMed] [Google Scholar]
- 126.Yang Z, Vorpagel ER, Laskin J. Influence of the charge state on the structures and interactions of vancomycin antibiotics with cell-wall analogue peptides: experimental and theoretical studies. Chem. Eur. J. 2009;15:2081–2090. doi: 10.1002/chem.200802010. [DOI] [PubMed] [Google Scholar]
- 127.Leung SSF, Tirado-Rives J, Jorgensen WL. Vancomycin analogs: Seeking improved binding of d-Ala-d-Ala and d-Ala-d-Lac peptides by side-chain and backbone modifications. Bioorg. Med. Chem. 2009;17:5874–5886. doi: 10.1016/j.bmc.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rekharsky M, Hesek D, Lee M, Meroueh SO, Inoue Y, Mobashery S. Thermodynamics of interactions of vancomycin and synthetic surrogates of bacterial cell wall. J. Am. Chem. Soc. 2006;128:7736–7737. doi: 10.1021/ja061828+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Nahoum V, Spector S, Loll PJ. Structure of ristocetin A in complex with a bacterial cell-wall mimetic. Acta Crystallogr. D Biol. Crystallogr. 2009;65:832–838. doi: 10.1107/S0907444909018344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Loll PJ, Derhovanessian A, Shapovalov MV, Kaplan J, Yang L, Axelsen PH. Vancomycin forms ligand-mediated supramolecular complexes. J. Mol. Biol. 2009;385:200–211. doi: 10.1016/j.jmb.2008.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Nitanai Y, Kikuchi T, Kakoi K, Hanamaki S, Fujisawa I, Aoki K. Crystal structures of the complexes between vancomycin and cell-wall precursor analogs. J. Mol. Biol. 2009;385:1422–1432. doi: 10.1016/j.jmb.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 132.Williams DH, Searle MS, Mackay JP, Gerhard U, Maplestone RA. Toward an estimation of binding constants in aqueous solution: studies of associations of vancomycin group antibiotics. Proc. Natl. Acad. Sci. U.S.A. 1993;90:1172–1178. doi: 10.1073/pnas.90.4.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Rao J, Yan L, Lahiri J, Whitesides GM, Weis RM, Warren HS. Binding of a dimeric derivative of vancomycin to l-Lys-d-Ala-d-lactate in solution and at a surface. Chem. Biol. 1999;6:353–359. doi: 10.1016/S1074-5521(99)80047-7. [DOI] [PubMed] [Google Scholar]
- 134.Jusuf S, Loll PJ, Axelsen PH. Configurational entropy and cooperativity between ligand binding and dimerization in glycopeptide antibiotics. J. Am. Chem. Soc. 2003;125:3988–3994. doi: 10.1021/ja027780r. [DOI] [PubMed] [Google Scholar]
- 135.Ghetta A, Prosperi D, Mantegazza F, Panza L, Riva S, Bellini T. Light scattered by model phantom bacteria reveals molecular interactions at their surface. Proc. Natl. Acad. Sci. U.S.A. 2005;102:15866–15870. doi: 10.1073/pnas.0505877102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lu J, Yoshida O, Hayashi S, Arimoto H. Synthesis of rigidly-linked vancomycin dimers and their in vivo efficacy against resistant bacteria. Chem. Commun. 2007:251–253. doi: 10.1039/b613319c. [DOI] [PubMed] [Google Scholar]
- 137.Barbachyn MR. Recent advances in the discovery of hybrid antibacterial agents. Annu. Rep. Med. Chem. 2008;43:281–290. [Google Scholar]
- 138.Long DD, Aggen JB, Christensen BG, Judice JK, Hegde SS, Kaniga K, Krause KM, Linsell MS, Moran EJ, Pace JL. A multivalent approach to drug discovery for novel antibiotics. J. Antibiot. 2008;61:595–602. doi: 10.1038/ja.2008.79. [DOI] [PubMed] [Google Scholar]
- 139.Long DD, Aggen JB, Chinn J, Choi SK, Christensen BG, Fatheree PR, Green D, Hegde SS, Judice JK, Kaniga K, Krause KM, Leadbetter M, Linsell MS, Marquess DG, Moran EJ, Nodwell MB, Pace JL, Trapp SG, Turner SD. Exploring the positional attachment of glycopeptide/β-lactam heterodimers. J. Antibiot. 2008;61:603–614. doi: 10.1038/ja.2008.80. [DOI] [PubMed] [Google Scholar]
- 140.Kim SJ, Cegelski L, Preobrazhenskaya M, Schaefer J. Structures of Staphylococcus aureus cell-wall complexes with vancomycin, eremomycin, and chloroeremomycin derivatives by 13C,19F and 15N,19F rotational-echo double resonance. Biochemistry. 2006;45:5235–5250. doi: 10.1021/bi052660s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kim SJ, Matsuoka S, Patti GJ, Schaefer J. Vancomycin derivative with damaged d-Ala-d-Ala binding cleft binds to cross-linked peptidoglycan in the cell wall of Staphylococcus aureus. Biochemistry. 2008;47:3822–3831. doi: 10.1021/bi702232a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Poulakou G, Giamarellou H. Oritavancin: a new promising agent in the treatment of infections due to Gram-positive pathogens. Expert Opin. Investig. Drugs. 2008;17:225–243. doi: 10.1517/13543784.17.2.225. [DOI] [PubMed] [Google Scholar]
- 143.Belley A, Neesham-Grenon E, McKay G, Arhin FF, Harris R, Beveridge T, Parr TRJ, Moeck G. Oritavancin kills stationary-phase and biofilm Staphylococcus aureus cells in vitro. Antimicrob. Agents Chemother. 2009;53:918–925. doi: 10.1128/AAC.00766-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Arhin FF, Sarmiento I, Parr TRJ, Moeck G. Comparative in vitro activity of oritavancin against Staphylococcus aureus strains that are resistant, intermediate or heteroresistant to vancomycin. J. Antimicrob. Chemother. 2009;64(4):868–870. doi: 10.1093/jac/dkp286. [DOI] [PubMed] [Google Scholar]
- 145.McKay GA, Beaulieu S, Arhin FF, Belley A, Sarmiento I, Parr TJ, Moeck G. Time-kill kinetics of oritavancin and comparator agents against Staphylococcus aureus Enterococcus faecalis and Enterococcus faecium. J. Antimicrob. Chemother. 2009;63:1191–1199. doi: 10.1093/jac/dkp126. [DOI] [PubMed] [Google Scholar]
- 146.Arhin FF, Draghi DC, Pillar CM, Parr TRJ, Moeck G, Sahm DF. Comparative in vitro activity profile of oritavancin against recent Gram-positive clinical isolates. Antimicrob. Agents Chemother. 2009;53:4762–4771. doi: 10.1128/AAC.00952-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Cornaglia G, Rossolini GM. Forthcoming therapeutic perspectives for infections due to multidrug-resistant Gram-positive pathogens. Clin. Microbiol. Infect. 2009;15:218–223. doi: 10.1111/j.1469-0691.2009.02740.x. [DOI] [PubMed] [Google Scholar]
- 148.Toke O, Cegelski L, Schaefer J. Peptide antibiotics in action: Investigation of polypeptide chains in insoluble environments by rotational-echo double resonance. Biochim. Biophys. Acta. 2006;1758:1314–1329. doi: 10.1016/j.bbamem.2006.02.031. [DOI] [PubMed] [Google Scholar]
- 149.Kim SJ, Cegelski L, Stueber D, Singh M, Dietrich E, Tanaka KS, Parr TRJ, Far AR, Schaefer J. Oritavancin exhibits dual mode of action to inhibit cell-wall biosynthesis in Staphylococcus aureus. J. Mol. Biol. 2008;377:281–293. doi: 10.1016/j.jmb.2008.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kim SJ, Singh M, Schaefer J. Oritavancin binds to isolated protoplast membranes but not intact protoplasts of Staphylococcus aureus. J. Mol. Biol. 2009;391:414–425. doi: 10.1016/j.jmb.2009.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Belley A, Harris R, Beveridge T, Parr TJ, Moeck G. Ultrastructural effects of oritavancin on methicillin-resistant Staphylococcus aureus and vancomycin-resistant Enterococcus. Antimicrob. Agents Chemother. 2009;53:800–804. doi: 10.1128/AAC.00603-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Allen NE, Nicas TI. Mechanism of action of oritavancin and related glycopeptide antibiotics. FEMS Microbiol. Rev. 2003;26:511–532. doi: 10.1111/j.1574-6976.2003.tb00628.x. [DOI] [PubMed] [Google Scholar]
- 153.Woodford N. Biological counterstrike: antibiotic resistance mechanisms of Gram-positive cocci. Clin. Microbiol. Infect. 2005;11 Suppl. 3:2–21. doi: 10.1111/j.1469-0691.2005.01140.x. [DOI] [PubMed] [Google Scholar]
- 154.Courvalin P. Vancomycin resistance in Gram-positive cocci. Clin. Infect. Dis. 2006;42 Suppl. 1:S25–S34. doi: 10.1086/491711. [DOI] [PubMed] [Google Scholar]
- 155.de Lencastre H, Oliveira D, Tomasz A. Antibiotic resistant Staphylococcus aureus: a paradigm of adaptive power. Curr. Opin. Microbiol. 2007;10:428–435. doi: 10.1016/j.mib.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Sieradzki K, Tomasz A. Alterations of cell wall structure and metabolism accompany reduced susceptibility to vancomycin in an isogenic series of clinical isolates of Staphylococcus aureus. J. Bacteriol. 2003;185:7103–7110. doi: 10.1128/JB.185.24.7103-7110.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Cui L, Iwamoto A, Lian JQ, Neoh HM, Maruyama T, Horikawa Y, Hiramatsu K. Novel mechanism of antibiotic resistance Originating in vancomycin-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 2006;50:428–438. doi: 10.1128/AAC.50.2.428-438.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Cui L, Tominaga E, Neoh HM, Hiramatsu K. Correlation between reduced daptomycin susceptibility and vancomycin resistance in vancomycin-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 2006;50:1079–1082. doi: 10.1128/AAC.50.3.1079-1082.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Pereira PM, Filipe SR, Tomasz A, Pinho MG. Fluorescence ratio imaging microscopy shows decreased access of vancomycin to cell wall synthetic sites in vancomycin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2007;51:3627–3633. doi: 10.1128/AAC.00431-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Bugg TD, Wright GD, Dutka-Malen S, Arthur M, Courvalin P, Walsh CT. Molecular basis for vancomycin resistance in Enterococcus faecium BM4147: biosynthesis of a depsipeptide peptidoglycan precursor by vancomycin resistance proteins VanH and VanA. Biochemistry. 1991;30:10408–10415. doi: 10.1021/bi00107a007. [DOI] [PubMed] [Google Scholar]
- 161.McComas CC, Crowley BM, Boger DL. Partitioning the loss in vancomycin binding affinity for d-Ala-d-Lac into lost H-bond and repulsive lone pair contributions. J. Am. Chem. Soc. 2003;125:9314–9315. doi: 10.1021/ja035901x. [DOI] [PubMed] [Google Scholar]
- 162.Crowley BM, Boger DL. Total synthesis and evaluation of [Ψ[CH2NH]Tpg4] vancomycin aglycon: Reengineering vancomycin for dual d-Ala-d-Ala and d-Ala-d-Lac binding. J. Am. Chem. Soc. 2006;128:2885–2892. doi: 10.1021/ja0572912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Crane CM, Boger DL. Synthesis and evaluation of vancomycin aglycon analogues that bear modifications in the N-Terminal d-leucyl amino acid. J. Med. Chem. 2009;52:1471–1476. doi: 10.1021/jm801549b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Bugg TD, Walsh CT. Intracellular steps of bacterial cell wall peptidoglycan biosynthesis: enzymology, antibiotics, and antibiotic resistance. Nat. Prod. Rep. 1992;9:199–215. doi: 10.1039/np9920900199. [DOI] [PubMed] [Google Scholar]
- 165.Beltrametti F, Consolandi A, Carrano L, Bagatin F, Rossi R, Leoni L, Zennaro E, Selva E, Marinelli F. Resistance to glycopeptide antibiotics in the teicoplanin producer is mediated by van gene homologue expression directing the synthesis of a modified cell wall peptidoglycan. Antimicrob. Agents Chemother. 2007;51:1135–1141. doi: 10.1128/AAC.01071-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Biarrotte-Sorin S, Hugonnet JE, Delfosse V, Mainardi JL, Gutmann L, Arthur M, Mayer C. Crystal structure of a novel β-lactam-insensitive peptidoglycan transpeptidase. J. Mol. Biol. 2006;359:533–538. doi: 10.1016/j.jmb.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 167.Cremniter J, Mainardi JL, Josseaume N, Quincampoix JC, Dubost L, Hugonnet JE, Marie A, Gutmann L, Rice LB, Arthur M. Novel mechanism of resistance to glycopeptide antibiotics in Enterococcus faecium. J. Biol. Chem. 2006;281:32254–32262. doi: 10.1074/jbc.M606920200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kohanski MA, Dwyer DJ, Wierzbowski J, Cottarel G, Collins JJ. Mistranslation of membrane proteins and two-component system activation trigger antibiotic-mediated cell death. Cell. 2008;135:679–690. doi: 10.1016/j.cell.2008.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Dwyer DJ, Kohanski MA, Collins JJ. Role of reactive oxygen species in antibiotic action and resistance. Curr. Opin. Microbiol. 2009;12:482–489. doi: 10.1016/j.mib.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Davies BW, Kohanski MA, Simmons LA, Winkler JA, Collins JJ, Walker GC. Hydroxyurea induces hydroxyl radical-mediated cell death in Escherichia coli. Mol. Cell. 2009;36:845–860. doi: 10.1016/j.molcel.2009.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kohanski MA, Depristo MA, Collins JJ. Sublethal antibiotic treatment leads to multidrug resistance via radical-induced mutagenesis. Mol. Cell. 2010;37:311–320. doi: 10.1016/j.molcel.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Kaufmann BB, Hung DT. The fast track to multidrug resistance. Mol. Cell. 2010;37:297–298. doi: 10.1016/j.molcel.2010.01.027. [DOI] [PubMed] [Google Scholar]
- 173.Bizzini A, Zhao C, Auffray Y, Hartke A. The Enterococcus faecalis superoxide dismutase is essential for its tolerance to vancomycin and penicillin. J. Antimicrob. Chemother. 2009;64:1196–1202. doi: 10.1093/jac/dkp369. [DOI] [PubMed] [Google Scholar]
- 174.Pinho MG, Errington J. Dispersed mode of Staphylococcus aureus cell wall synthesis in the absence of the division machinery. Mol. Microbiol. 2003;50:871–881. doi: 10.1046/j.1365-2958.2003.03719.x. [DOI] [PubMed] [Google Scholar]
- 175.Carballido-Lopez R. The bacterial actin-like cytoskeleton. Microbiol. Mol. Biol. Rev. 2006;70:888–909. doi: 10.1128/MMBR.00014-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Bhavsar AP, Brown ED. Cell wall assembly in Bacillus subtilis: how spirals and spaces challenge paradigms. Mol. Microbiol. 2006;60:1077–1090. doi: 10.1111/j.1365-2958.2006.05169.x. [DOI] [PubMed] [Google Scholar]
- 177.Michie KA, Lowe J. Dynamic filaments of the bacterial cytoskeleton. Annu. Rev. Biochem. 2006;75:467–492. doi: 10.1146/annurev.biochem.75.103004.142452. [DOI] [PubMed] [Google Scholar]
- 178.Varma A, de Pedro MA, Young KD. FtsZ directs a second mode of peptidoglycan synthesis in Escherichia coli. J. Bacteriol. 2007;189:5692–5704. doi: 10.1128/JB.00455-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Graumann PL. Cytoskeletal elements in bacteria. Annu. Rev. Microbiol. 2007;61:589–618. doi: 10.1146/annurev.micro.61.080706.093236. [DOI] [PubMed] [Google Scholar]
- 180.Gitai Z. Diversification and specialization of the bacterial cytoskeleton. Curr. Opin. Cell Biol. 2007;19:5–12. doi: 10.1016/j.ceb.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 181.Dye NA, Shapiro L. The push and pull of the bacterial cytoskeleton. Trends Cell Biol. 2007;17:239–245. doi: 10.1016/j.tcb.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 182.Pogliano J. The bacterial cytoskeleton. Curr. Opin. Cell Biol. 2008;20:19–27. doi: 10.1016/j.ceb.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 183.van der Donk WA. Lighting up the nascent cell wall. ACS Chem. Biol. 2006;1:425–428. doi: 10.1021/cb600308w. [DOI] [PubMed] [Google Scholar]
- 184.Ghosh AS, Young KD. Helical disposition of proteins and lipopolysaccharide in the outer membrane of Escherichia coli. J. Bacteriol. 2005;187:1913–1922. doi: 10.1128/JB.187.6.1913-1922.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Barak I, Muchova K, Wilkinson AJ, O'Toole PJ, Pavlendova N. Lipid spirals in Bacillus subtilis and their role in cell division. Mol. Microbiol. 2008;68:1315–1327. doi: 10.1111/j.1365-2958.2008.06236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Shiomi D, Yoshimoto M, Homma M, Kawagishi I. Helical distribution of the bacterial chemoreceptor via colocalization with the Sec protein translocation machinery. Mol. Microbiol. 2006;60:894–906. doi: 10.1111/j.1365-2958.2006.05145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Kumar I, Josephine HR, Pratt RF. Reactions of peptidoglycan-mimetic β-lactams with penicillin-binding proteins in vivo and in membranes. ACS Chem. Biol. 2007;2:620–624. doi: 10.1021/cb7001347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Narayan RS, VanNieuwenhze MS. Synthesis of substrates and biochemical probes for study of the peptidoglycan biosynthetic pathway. Eur. J. Org. Chem. 2007:1399–1414. doi: 10.1002/ejoc.200600750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Perlstein DL, Zhang Y, Wang TS, Kahne DE, Walker S. The direction of glycan chain elongation by peptidoglycan glycosyltransferases. J. Am. Chem. Soc. 2007;129:12674–12675. doi: 10.1021/ja075965y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Schouten JA, Bagga S, Lloyd AJ, de Pascale G, Dowson CG, Roper DI, Bugg TD. Fluorescent reagents for in vitro studies of lipid-linked steps of bacterial peptidoglycan biosynthesis: derivatives of UDPMurNAc-pentapeptide containing d-cysteine at position 4 or 5. Mol. Biosyst. 2006;2:484–491. doi: 10.1039/b607908c. [DOI] [PubMed] [Google Scholar]
- 191.De Pascale G, Lloyd AJ, Schouten JA, Gilbey AM, Roper DI, Dowson CG, Bugg TD. Kinetic characterization of lipid II-Ala:alanyl-tRNA ligase (MurN) from Streptococcus pneumoniae using semisynthetic aminoacyl-lipid II substrates. J. Biol. Chem. 2008;283:34571–34579. doi: 10.1074/jbc.M805807200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Vinatier V, Blakey CB, Braddick D, Johnson BR, Evans SD, Bugg TD. In vitro biosynthesis of bacterial peptidoglycan using d-Cys-containing precursors: fluorescent detection of transglycosylation and transpeptidation. Chem. Commun. 2009:4037–4039. doi: 10.1039/b819869a. [DOI] [PubMed] [Google Scholar]
- 193.Lupoli TJ, Taniguchi T, Wang TS, Perlstein DL, Walker S, Kahne DE. Studying a cell division amidase using defined peptidoglycan substrates. J. Am. Chem. Soc. 2009;131:18230–18231. doi: 10.1021/ja908916z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Roychowdhury A, Wolfert MA, Boons GJ. Synthesis and proinflammatory properties of muramyl tripeptides containing lysine and diaminopimelic acid moieties. ChemBioChem. 2005;6:2088–2097. doi: 10.1002/cbic.200500181. [DOI] [PubMed] [Google Scholar]
- 195.Kumar S, Roychowdhury A, Ember B, Wang Q, Guan R, Mariuzza RA, Boons GJ. Selective recognition of synthetic lysine and meso-diaminopimelic acid-type peptidoglycan fragments by human peptidoglycan recognition proteins Iα and S. J. Biol. Chem. 2005;280:37005–37012. doi: 10.1074/jbc.M506385200. [DOI] [PubMed] [Google Scholar]
- 196.Swaminathan CP, Brown PH, Roychowdhury A, Wang Q, Guan R, Silverman N, Goldman WE, Boons GJ, Mariuzza RA. Dual strategies for peptidoglycan discrimination by peptidoglycan recognition proteins (PGRPs) Proc. Natl. Acad. Sci. U.S.A. 2006;103:684–689. doi: 10.1073/pnas.0507656103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Asong J, Wolfert MA, Maiti KK, Miller D, Boons GJ. Binding and cellular activation studies reveal that toll-like receptor 2 can differentially recognize peptidoglycan from Gram-positive and Gram-negative bacteria. J. Biol. Chem. 2009;284:8643–8653. doi: 10.1074/jbc.M806633200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Park JW, Je BR, Piao S, Inamura S, Fujimoto Y, Fukase K, Kusumoto S, Ha NC, Soderhall K, Ha NC, Lee BL. A synthetic peptidoglycan fragment as a competitive inhibitor of the melanization cascade. J. Biol. Chem. 2006;281:7747–7755. doi: 10.1074/jbc.M510058200. [DOI] [PubMed] [Google Scholar]
- 199.Fujimoto Y, Inamura S, Kawasaki A, Shiokawa Z, Shimoyama A, Hashimoto T, Kusumoto S, Fukase K. Chemical synthesis of peptidoglycan fragments for elucidation of the immunostimulating mechanism. J. Endotoxin Res. 2007;13:189–196. doi: 10.1177/0968051907080739. [DOI] [PubMed] [Google Scholar]
- 200.Hasegawa M, Kawasaki A, Yang K, Fujimoto Y, Masumoto J, Breukink E, Nunez G, Fukase K, Inohara N. A role of lipophilic peptidoglycan-related molecules in induction of Nod1-mediated immune responses. J. Biol. Chem. 2007;282:11757–11764. doi: 10.1074/jbc.M700846200. [DOI] [PubMed] [Google Scholar]
- 201.Kawasaki A, Karasudani Y, Otsuka Y, Hasegawa M, Inohara N, Fujimoto Y, Fukase K. Synthesis of diaminopimelic acid containing peptidoglycan fragments and tracheal cytotoxin (TCT) and investigation of their biological functions. Chem. Eur. J. 2008;14:10318–10330. doi: 10.1002/chem.200801121. [DOI] [PubMed] [Google Scholar]
- 202.Fujimoto Y, Konishi Y, Kubo O, Hasegawa M, Inohara N, Fukase K. Synthesis of crosslinked peptidoglycan fragments for investigation of their immunobiological functions. Tetrahedron Lett. 2009;50:3631–3634. [Google Scholar]
- 203.Uehara T, Suefuji K, Valbuena N, Meehan B, Donegan M, Park JT. Recycling of the anhydro-N-acetylmuramic acid derived from cell wall murein involves a two-step conversion to N-acetylglucosamine-phosphate. J. Bacteriol. 2005;187:3643–3649. doi: 10.1128/JB.187.11.3643-3649.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Uehara T, Park JT. Growth of Escherichia coli: significance of peptidoglycan degradation during elongation and septation. J. Bacteriol. 2008;190:3914–3922. doi: 10.1128/JB.00207-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Lister PD, Wolter DJ, Hanson ND. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin. Microbiol. Rev. 2009;22:582–610. doi: 10.1128/CMR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Golemi-Kotra D, Cha JY, Meroueh SO, Vakulenko SB, Mobashery S. Resistance to β-lactam antibiotics and its mediation by the sensor domain of the transmembrane BlaR signaling pathway in Staphylococcus aureus. J. Biol. Chem. 2003;278:18419–18425. doi: 10.1074/jbc.M300611200. [DOI] [PubMed] [Google Scholar]
- 207.Kerff F, Charlier P, Colombo ML, Sauvage E, Brans A, Frère JM, Joris B, Fonze E. Crystal structure of the sensor domain of the BlaR penicillin receptor from Bacillus licheniformis. Biochemistry. 2003;42:12835–12843. doi: 10.1021/bi034976a. [DOI] [PubMed] [Google Scholar]
- 208.Birck C, Cha JY, Cross J, Schulze-Briese C, Meroueh SO, Schlegel HB, Mobashery S, Samama JP. X-ray crystal structure of the acylated β-lactam sensor domain of BlaR1 from Staphylococcus aureus and the mechanism of receptor activation for signal transduction. J. Am. Chem. Soc. 2004;126:13945–13947. doi: 10.1021/ja044742u. [DOI] [PubMed] [Google Scholar]
- 209.Wilke MS, Hills TL, Zhang HZ, Chambers HF, Strynadka NC. Crystal structures of the apo and penicillin-acylated forms of the BlaR1 β-lactam sensor of Staphylococcus aureus. J. Biol. Chem. 2004;279:47278–47287. doi: 10.1074/jbc.M407054200. [DOI] [PubMed] [Google Scholar]
- 210.Fuda CC, Fisher JF, Mobashery S. β-Lactam resistance in Staphylococcus aureus: the adaptive resistance of a plastic genome. Cell Mol. Life Sci. 2005;62:2617–2633. doi: 10.1007/s00018-005-5148-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Thumanu K, Cha J, Fisher JF, Perrins R, Mobashery S, Wharton C. Discrete steps in sensing of β-lactam antibiotics by the BlaR1 protein of the methicillin-resistant Staphylococcus aureus bacterium. Proc. Natl. Acad. Sci. U.S.A. 2006;103:10630–10635. doi: 10.1073/pnas.0601971103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Cha JY, Mobashery S. Lysine Nε-decarboxylation in the BlaR1 protein from Staphylococcus aureus at the root of its function as an antibiotic sensor. J. Am. Chem. Soc. 2007;129:3834–3835. doi: 10.1021/ja070472e. [DOI] [PubMed] [Google Scholar]
- 213.McCallum N, Berger-Bachi B, Senn MM. Regulation of antibiotic resistance in Staphylococcus aureus. Int. J. Med. Microbiol. 2010;300:118–129. doi: 10.1016/j.ijmm.2009.08.015. [DOI] [PubMed] [Google Scholar]
- 214.Nelson DE, Young KD. Penicillin binding protein 5 affects cell diameter, contour, and morphology of Escherichia coli. J. Bacteriol. 2000;182:1714–1721. doi: 10.1128/jb.182.6.1714-1721.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Ghosh AS, Young KD. Sequences near the active site in chimeric penicillin binding proteins 5 and 6 affect uniform morphology of Escherichia coli. J. Bacteriol. 2003;185:2178–2186. doi: 10.1128/JB.185.7.2178-2186.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Popham DL, Young KD. Role of penicillin-binding proteins in bacterial cell morphogenesis. Curr. Opin. Microbiol. 2003;6:594–599. doi: 10.1016/j.mib.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 217.Priyadarshini R, Popham DL, Young KD. Daughter cell separation by penicillin-binding proteins and peptidoglycan amidases in Escherichia coli. J. Bacteriol. 2006;188:5345–5355. doi: 10.1128/JB.00476-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Ghosh AS, Chowdhury C, Nelson DE. Physiological functions of d-alanine carboxypeptidases in Escherichia coli. Trends Microbiol. 2008;16:309–317. doi: 10.1016/j.tim.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 219.Moya B, Dotsch A, Juan C, Blazquez J, Zamorano L, Haussler S, Oliver A. β-Lactam resistance response triggered by inactivation of a nonessential penicillin-binding protein. PLoS Pathog. 2009;5:e1000353. doi: 10.1371/journal.ppat.1000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Kawai F, Clarke TB, Roper DI, Han GJ, Hwang KY, Unzai S, Obayashi E, Park SY, Tame JR. Crystal structures of penicillin-binding proteins 4 and 5 from Haemophilus influenzae. J. Mol. Biol. 2010;396:634–645. doi: 10.1016/j.jmb.2009.11.055. [DOI] [PubMed] [Google Scholar]
- 221.Jacobs C, Joris B, Jamin M, Klarsov K, Van Beeumen J, Mengin-Lecreulx D, van Heijenoort J, Park JT, Normark S, Frère JM. AmpD, essential for both β-lactamase regulation and cell wall recycling, is a novel cytosolic N-acetylmuramyl-l-alanine amidase. Mol. Microbiol. 1995;15:553–559. doi: 10.1111/j.1365-2958.1995.tb02268.x. [DOI] [PubMed] [Google Scholar]
- 222.Jacobs C, Frère JM, Normark S. Cytosolic intermediates for cell wall biosynthesis and degradation control inducible β-lactam resistance in Gram-negative bacteria. Cell. 1997;88:823–832. doi: 10.1016/s0092-8674(00)81928-5. [DOI] [PubMed] [Google Scholar]
- 223.Scheurwater E, Reid CW, Clarke AJ. Lytic transglycosylases: Bacterial space-making autolysins. Int. J. Biochem. Cell Biol. 2008;40:586–591. doi: 10.1016/j.biocel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 224.Scheurwater EM, Clarke AJ. The C-terminal domain of Escherichia coli YfhD functions as a lytic transglycosylase. J. Biol. Chem. 2008;283:8363–8373. doi: 10.1074/jbc.M710135200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Cheng Q, Park JT. Substrate specificity of the AmpG permease required for recycling of cell wall anhydro-muropeptides. J. Bacteriol. 2002;184:6434–6436. doi: 10.1128/JB.184.23.6434-6436.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Chahboune A, Decaffmeyer M, Brasseur R, Joris B. Membrane topology of the Escherichia coli AmpG permease required for recycling of cell wall anhydromuropeptides and AmpC β-lactamase induction. Antimicrob Agents Chemother. 2005;49:1145–1149. doi: 10.1128/AAC.49.3.1145-1149.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Cheng Q, Li H, Merdek K, Park JT. Molecular characterization of the β-N-acetylglucosaminidase of Escherichia coli and its role in cell wall recycling. J. Bacteriol. 2000;182:4836–4840. doi: 10.1128/jb.182.17.4836-4840.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Votsch W, Templin MF. Characterization of a β-N-acetylglucosaminidase of Escherichia coli and elucidation of its role in muropeptide recycling and β-lactamase induction. J. Biol. Chem. 2000;275:39032–39038. doi: 10.1074/jbc.M004797200. [DOI] [PubMed] [Google Scholar]
- 229.Kong KF, Jayawardena SR, Indulkar SD, Del Puerto A, Koh CL, Hoiby N, Mathee K. Pseudomonas aeruginosa AmpR is a global transcriptional factor that regulates expression of AmpC and PoxB β-lactamases, proteases, quorum sensing, and other virulence factors. Antimicrob. Agents Chemother. 2005;49:4567–4575. doi: 10.1128/AAC.49.11.4567-4575.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Jacobs C, Huang LJ, Bartowsky E, Normark S, Park JT. Bacterial cell wall recycling provides cytosolic muropeptides as effectors for β-lactamase induction. EMBO J. 1994;13:4684–4694. doi: 10.1002/j.1460-2075.1994.tb06792.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Suvorov M, Lee M, Hesek D, Boggess B, Mobashery S. Lytic transglycosylase MltB of Escherichia coli and its role in recycling of peptidoglycan strands of bacterial cell wall. J. Am. Chem. Soc. 2008;130:11878–11879. doi: 10.1021/ja805482b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Paulsen H, Himpkamp P, Peters T. Building units of oligosaccharides. LXIX. Synthesis of 1,6-anhydromuramyl peptides. Liebigs Ann. Chem. 1986:664–674. [Google Scholar]
- 233.Hesek D, Lee M, Zhang W, Noll BC, Mobashery S. Total synthesis of N-acetylglucosamine-1,6-anhydro-N-acetylmuramylpentapeptide and evaluation of its turnover by AmpD from Escherichia coli. J. Am. Chem. Soc. 2009;131:5187–5193. doi: 10.1021/ja808498m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Lee M, Zhang W, Hesek D, Noll BC, Boggess B, Mobashery S. Bacterial AmpD at the crossroads of peptidoglycan recycling and manifestation of antibiotic resistance. J. Am. Chem. Soc. 2009;131:8742–8743. doi: 10.1021/ja9025566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Park JT, Uehara T. How bacteria consume their own exoskeletons (turnover and recycling of cell wall peptidoglycan) Microbiol. Mol. Biol. Rev. 2008;72:211–227. doi: 10.1128/MMBR.00027-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Cloud-Hansen KA, Peterson SB, Stabb EV, Goldman WE, McFall-Ngai MJ, Handelsman J. Breaching the great wall: peptidoglycan and microbial interactions. Nat. Rev. Microbiol. 2006;4:710–716. doi: 10.1038/nrmicro1486. [DOI] [PubMed] [Google Scholar]
- 237.Garcia DL, Dillard JP. Mutations in ampG or ampD affect peptidoglycan fragment release from Neisseria gonorrhoeae. J. Bacteriol. 2008;190:3799–3807. doi: 10.1128/JB.01194-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Cloud-Hansen KA, Hackett KT, Garcia DL, Dillard J. P, Neisseria gonorrhoeae uses two lytic transglycosylases to produce cytotoxic peptidoglycan monomers. J. Bacteriol. 2008;190:5989–5994. doi: 10.1128/JB.00506-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2009;48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 240.Fischbach MA, Walsh CT. Antibiotics for emerging pathogens. Science. 2009;325:1089–1093. doi: 10.1126/science.1176667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Stubbs KA, Balcewich M, Mark BL, Vocadlo DJ. Small molecule inhibitors of a glycoside hydrolase attenuate inducible AmpC-mediated β-lactam resistance. J. Biol. Chem. 2007;282:21382–21391. doi: 10.1074/jbc.M700084200. [DOI] [PubMed] [Google Scholar]
- 242.Balcewich MD, Stubbs KA, He Y, James TW, Davies GJ, Vocadlo DJ, Mark BL. Insight into a strategy for attenuating AmpC-mediated β-lactam resistance: structural basis for selective inhibition of the glycoside hydrolase NagZ. Protein Sci. 2009;18:1541–1551. doi: 10.1002/pro.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Asgarali A, Stubbs KA, Oliver A, Vocadlo DJ, Mark BL. Inactivation of the glycoside hydrolase NagZ attenuates anti-Pseudomonal β-lactam resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2009;53:2274–2282. doi: 10.1128/AAC.01617-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Adin DM, Engle JT, Goldman WE, McFall-Ngai MJ, Stabb EV. Mutations in ampG and lytic transglycosylase genes affect the net release of peptidoglycan monomers from Vibrio fischeri. J. Bacteriol. 2009;191:2012–2022. doi: 10.1128/JB.01547-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Troll JV, Bent EH, Pacquette N, Wier AM, Goldman WE, Silverman N, McFall-Ngai MJ. Taming the symbiont for coexistence: a host PGRP neutralizes a bacterial symbiont toxin. Environ. Microbiol. 2010 doi: 10.1111/j.1462-2920.2009.02121.x. in press (Doi: 10.1111/j.1462-2920.2009.02121.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Delbridge LM, O'Riordan MX. Innate recognition of intracellular bacteria. Curr. Opin. Immunol. 2007;19:10–16. doi: 10.1016/j.coi.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 247.Rietdijk ST, Burwell T, Bertin J, Coyle AJ. Sensing intracellular pathogens-NOD-like receptors. Curr. Opin. Pharmacol. 2008;8:261–266. doi: 10.1016/j.coph.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 248.Franchi L, Warner N, Viani K, Nunez G. Function of Nod-like receptors in microbial recognition and host defense. Immunol. Rev. 2009;227:106–128. doi: 10.1111/j.1600-065X.2008.00734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Rosenstiel P, Schreiber S. NOD-like receptors-pivotal guardians of the immunological integrity of barrier organs. Adv. Exp. Med. Biol. 2009;653:35–47. doi: 10.1007/978-1-4419-0901-5_3. [DOI] [PubMed] [Google Scholar]
- 250.Dziarski R. Peptidoglycan recognition proteins (PGRPs) Mol. Immunol. 2004;40:877–886. doi: 10.1016/j.molimm.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 251.Dziarski R, Gupta D. The peptidoglycan recognition proteins (PGRPs) Genome Biol. 2006;7:232. doi: 10.1186/gb-2006-7-8-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Dziarski R, Gupta D. Mammalian PGRPs: novel antibacterial proteins. Cell Microbiol. 2006;8:1059–1069. doi: 10.1111/j.1462-5822.2006.00726.x. [DOI] [PubMed] [Google Scholar]
- 253.Lu X, Wang M, Qi J, Wang H, Li X, Gupta D, Dziarski R. Peptidoglycan recognition proteins are a new class of human bactericidal proteins. J. Biol. Chem. 2006;281:5895–5907. doi: 10.1074/jbc.M511631200. [DOI] [PubMed] [Google Scholar]
- 254.Guan R, Mariuzza RA. Peptidoglycan recognition proteins of the innate immune system. Trends Microbiol. 2007;15:127–134. doi: 10.1016/j.tim.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 255.Gupta D. Peptidoglycan recognition proteins-maintaining immune homeostasis and normal development. Cell Host Microbe. 2008;3:273–274. doi: 10.1016/j.chom.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 256.Boneca IG. Mammalian PGRPs in the spotlight. Cell Host Microbe. 2009;5:109–111. doi: 10.1016/j.chom.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 257.Troll JV, Adin DM, Wier AM, Paquette N, Silverman N, Goldman WE, Stadermann FJ, Stabb EV, McFall-Ngai MJ. Peptidoglycan induces loss of a nuclear peptidoglycan recognition protein during host tissue development in a beneficial animal-bacterial symbiosis. Cell. Microbiol. 2009;11:1114–1127. doi: 10.1111/j.1462-5822.2009.01315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Cooney R, Baker J, Brain O, Danis B, Pichulik T, Allan P, Ferguson DJ, Campbell BJ, Jewell D, Simmons A. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat. Med. 2010;16:90–97. doi: 10.1038/nm.2069. [DOI] [PubMed] [Google Scholar]
- 259.Travassos LH, Carneiro LA, Ramjeet M, Hussey S, Kim YG, Magalhaes JG, Yuan L, Soares F, Chea E, Le Bourhis L, Boneca IG, Allaoui A, Jones NL, Nunez G, Girardin SE, Philpott DJ. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat. Immunol. 2010;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- 260.Finlay BB, McFadden G. Anti-immunology: evasion of the host immune system by bacterial and viral pathogens. Cell. 2006;124:767–782. doi: 10.1016/j.cell.2006.01.034. [DOI] [PubMed] [Google Scholar]
- 261.Boneca IG, Dussurget O, Cabanes D, Nahori MA, Sousa S, Lecuit M, Psylinakis E, Bouriotis V, Hugot JP, Giovannini M, Coyle A, Bertin J, Namane A, Rousselle JC, Cayet N, Prevost MC, Balloy V, Chignard M, Philpott DJ, Cossart P, Girardin SE. A critical role for peptidoglycan N-deacetylation in Listeria evasion from the host innate immune system. Proc. Natl. Acad. Sci. U.S.A. 2007;104:997–1002. doi: 10.1073/pnas.0609672104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Wolfert MA, Roychowdhury A, Boons GJ. Modification of the structure of peptidoglycan is a strategy to avoid detection by nucleotide-binding oligomerization domain protein 1. Infect. Immun. 2007;75:706–713. doi: 10.1128/IAI.01597-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Cigana C, Curcuru L, Leone MR, Ierano T, Lore NI, Bianconi I, Silipo A, Cozzolino F, Lanzetta R, Molinaro A, Bernardini ML, Bragonzi A. Pseudomonas aeruginosa exploits lipid A and muropeptides modification as a strategy to lower innate immunity during cystic fibrosis lung infection. PLoS One. 2009;4:e8439. doi: 10.1371/journal.pone.0008439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Geddes K, Magalhaes JG, Girardin SE. Unleashing the therapeutic potential of NOD-like receptors. Nat. Rev. Drug Discov. 2009;8:465–479. doi: 10.1038/nrd2783. [DOI] [PubMed] [Google Scholar]
- 265.Chang CI, Deisenhofer J. The peptidoglycan recognition proteins LCa and LCx. Cell Mol. Life Sci. 2007;64:1395–1402. doi: 10.1007/s00018-007-6567-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Filipe SR, Tomasz A, Ligoxygakis P. Requirements of peptidoglycan structure that allow detection by the Drosophila Toll pathway. EMBO Rep. 2005;6:327–333. doi: 10.1038/sj.embor.7400371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Wang M, Liu LH, Wang S, Li X, Lu X, Gupta D, Dziarski R. Human peptidoglycan recognition proteins require zinc to kill both gram-positive and gram-negative bacteria and are synergistic with antibacterial peptides. J. Immunol. 2007;178:3116–3125. doi: 10.4049/jimmunol.178.5.3116. [DOI] [PubMed] [Google Scholar]
- 268.Reiser JB, Teyton L, Wilson IA. Crystal structure of the Drosophila peptidoglycan recognition protein (PGRP)-SA at 1.56 Å resolution. J. Mol. Biol. 2004;340:909–917. doi: 10.1016/j.jmb.2004.04.077. [DOI] [PubMed] [Google Scholar]
- 269.Sharma P, Singh N, Sinha M, Sharma S, Perbandt M, Betzel C, Kaur P, Srinivasan A, Singh TP. Crystal structure of the peptidoglycan recognition protein at 1.8 Å resolution reveals dual strategy to combat infection through two independent functional homodimers. J. Mol. Biol. 2008;378:921–930. doi: 10.1016/j.jmb.2008.03.018. [DOI] [PubMed] [Google Scholar]
- 270.Lim JH, Kim MS, Kim HE, Yano T, Oshima Y, Aggarwal K, Goldman WE, Silverman N, Kurata S, Oh BH. Structural basis for preferential recognition of diaminopimelic acid-type peptidoglycan by a subset of peptidoglycan recognition proteins. J. Biol. Chem. 2006;281:8286–8295. doi: 10.1074/jbc.M513030200. [DOI] [PubMed] [Google Scholar]
- 271.Chang CI, Chelliah Y, Borek D, Mengin-Lecreulx D, Deisenhofer J. Structure of tracheal cytotoxin in complex with a heterodimeric pattern-recognition receptor. Science. 2006;311:1761–1764. doi: 10.1126/science.1123056. [DOI] [PubMed] [Google Scholar]
- 272.Guan R, Roychowdhury A, Ember B, Kumar S, Boons GJ, Mariuzza RA. Structural basis for peptidoglycan binding by peptidoglycan recognition proteins. Proc. Natl. Acad. Sci. U.S.A. 2004;101:17168–17173. doi: 10.1073/pnas.0407856101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Cho S, Wang Q, Swaminathan CP, Hesek D, Lee M, Boons GJ, Mobashery S, Mariuzza RA. Structural insights into the bactericidal mechanism of human peptidoglycan recognition proteins. Proc. Natl. Acad. Sci. U.S.A. 2007;104:8761–8766. doi: 10.1073/pnas.0701453104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Kerff F, Petrella S, Mercier F, Sauvage E, Herman R, Pennartz A, Zervosen A, Luxen A, Frere JM, Joris B, Charlier P. Specific structural features of the N-acetylmuramoyl-L-alanine amidase AmiD from Escherichia coli and mechanistic implications for enzymes of this family. J. Mol. Biol. 2010;397:249–259. doi: 10.1016/j.jmb.2009.12.038. [DOI] [PubMed] [Google Scholar]
- 275.Hermoso JA, Garcia JL, Garcia P. Taking aim on bacterial pathogens: from phage therapy to enzybiotics. Curr. Opin. Microbiol. 2007;10:461–472. doi: 10.1016/j.mib.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 276.Perez-Dorado I, Campillo NE, Monterroso B, Hesek D, Lee M, Paez JA, Garcia P, Martinez-Ripoll M, Garcia JL, Mobashery S, Menendez M, Hermoso JA. Elucidation of the molecular recognition of bacterial cell wall by modular pneumococcal phage endolysin CPL-1. J. Biol. Chem. 2007;282:24990–24999. doi: 10.1074/jbc.M704317200. [DOI] [PubMed] [Google Scholar]
- 277.Perez-Dorado I, Gonzalez A, Morales M, Sanles R, Striker W, Vollmer W, Mobashery S, Garcia JL, Martinez-Ripoll M, Garcia P, Hermoso JA. Insights into pneumococcal fratricide from the crystal structures of the modular killing factor LytC. Nat. Struct. Mol. Biol. 2010 doi: 10.1038/nsmb.1817. in press (Doi: 10.1038/nsmb.1817) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Shi Q, Meroueh SO, Fisher JF, Mobashery S. Transpeptidase reaction of penicillin-binding protein 1b of Streptococcus pneumoniae and cross-linking of the cell wall. 2010 unpublished data. [Google Scholar]