Abstract
Cognitive impairment is highly frequent in the elderly. The high estimates of conversion to dementia have spurred the interest in identification of genetic risk factors associated with development of cognitive impairment and or its progression. However, despite notable achievements in human genetics over the years, in particular technological advances in gene mapping and in statistical methods that relate genetic variants to disease, to date only a small proportion of the genetic contribution to late-life cognitive impairment can be explained. A likely explanation for the difficulty in gene identification is that it is a multifactorial disorder with both genetic and environmental components, in which several genes with small effects each are likely to contribute to the quantitative traits associated with the disease.
The motivation for identifying the underlying genetic risk factors elderly is clear. Not only could it shed light on disease pathogenesis, but it may also provide potential targets for effective treatment, screening, and prevention. In this article we review the current knowledge on underlying genetic variants and the usefulness of genetic variation as diagnostic tools and biomarkers. In addition, we discuss the potentials and difficulties researchers face in designing appropriate studies for gene discovery.
INTRODUCTION
Late-onset Alzheimer’s disease (LOAD) is among the most common diseases in aging societies. It is estimated that approximately five million people in the United States and 17 million people worldwide suffer from the disease. By age 85 years and older 15–30% are affected, and the incidence rate increases from about 1% among people aged 65–70 years to approximately 6–8% for people aged 85 years and older.[1, 2] It is expected that these numbers will quadruple by the year 2040, by which 1 out of 45 Americans will be affected, leading to a considerable public health burden.[3]
To date, there are no definitive diagnostic tests or biological markers of the disease. The diagnosis of LOAD during life is based on clinical examination using the criteria of the National Institute of Neurological and Communicative Disorders and Stroke (NINCDS) and the Alzheimer’s Disease and Related Disorders Association (ADRDA) Work Group.[4] Although these criteria have good reliability[5–7] and validity, [8, 9] any measure that would allow detection at an early stage and would increase diagnostic sensitivity and specificity, would help improve early therapeutic intervention.
Mild cognitive impairment (MCI) is a clinical diagnostic entity that may represent this early stage. It refers to individuals who have cognitive deficits but who do not fulfill a diagnosis of dementia.[10–12] Studies using the criteria by Petersen et al. for diagnosing MCI in clinical and epidemiological settings, [11, 13] report an incidence rate of 9.9/1,000 person-years for MCI among nondemented elderly, [14] and an annual conversion rate of 10% to 12% to AD in subjects with MCI, particularly amnestic MCI, in contrast to a conversion rate of 1% to 2% in the normal elderly population.[11] The high estimates of conversion rate of MCI to dementia has spurred the interest in establishing preclinical prognostic markers for MCI and the progression from MCI to dementia.[14, 15]
Twin studies suggest that 37% to as much as 78% of the variance in the age-at-onset of cognitive impairment can be attributed to additive genetic effects.[16] As a consequence, genes involved in MCI or LOAD could be highly valuable diagnostic tools. The usefulness of genetic variation as biomarkers for cognitive impairment is further supported by the fact that genetic variation is stable across the life span and the disease process, and is not influenced by confounding factors. Despite available improved analytic techniques, the continued pursuit of genetic variants associated with cognitive impairment has, however, been limited. To date, only two genes have been implicated in the cause: the Apolipoprotein (APOE)-ε4 allele and the Sortilin-related receptor (SORL1) gene. Together these reported genes explain only a small proportion of the genetic contribution to cognitive impairment in late-life leaving several genetic risk factors to be identified.
In this article we review the genetic risk factors that have been implicated in MCI, LOAD and progression of MCI to dementia, and review their usefulness as diagnostic tools and biomarkers. In addition, we discuss the difficulties researchers face when performing studies for gene discovery in common complex diseases such as late-life cognitive impairment.
GENETICS OF LOAD AND MILD COGNITIVE IMPAIRMENT
Most of the studies assessing the role of genetic variation in cognitive impairment have used the diagnostic criteria of LOAD or endophenotypes of cognition such as age-at-onset of dementia or cognitive test performance. The motivation for use of endophenotypes is that quantitative traits provide more accurate phenotypes than simply considering affection status as a dichotomized variable, and thus provide more statistical power to detect small polygenic effects. Few studies have used MCI as the phenotype or have assessed the influence of genetic variation on progression of MCI to dementia.
1. Apolipoprotein E (APOE)
APOE, which maps to chromosome 19 in a cluster with Apolipoprotein C1 and Apolipoprotein C2, is a lipid-binding protein and is expressed in humans as three common isoforms coded for by three alleles, APOEε2, ε3, and ε4. Early studies linked the APOE genotype with LOAD and found a significant increase in the frequency of the APOEε4 allele in patients with the disease compared to healthy controls. The large body of epidemiologic data that subsequently accumulated clarified this effect by demonstrating that APOEε4 decreases the age-at-onset of LOAD in a gene dosage-dependent manner, [17–26] that APOEε4 is associated with lower cognitive performance, in particular the memory domain, that it is associated with MCI, the prodromal stage of LOAD, [27–30] and that it is associated with progression from MCI to dementia.[27–37] It is thought that APOE may account for as much as 20–50% of LOAD risk.[38, 39]
In vitro studies have indicated that the APOE-ε4 isoform binds Aβ peptides with a higher avidity compared to APOE-ε3.[40] Furthermore, there is a strong correlation between the presence of an APOE-ε4 allele and a higher Aβ burden in the brains of AD patients, [41, 42] suggesting that APOE interacts with Aβ in enhancing its deposition in plaques. This is supported by the observation that homozygous APOE knockout (APOE −/−) mice develop fewer and more diffuse, non-fibrillar Aβ deposits.[43–45] Some but not all studies assessing the effect of different APOE isoforms on Aβ fibrillization showed that the ε4 isoform leads to increased Aβ aggregation in vitro.[46, 47] Similarly, in vivo studies in APOE −/− mice indicated that Aβ fibrillization and plaques formation was increased in mice expressing human APOE-ε4 (APPV717F+/−, apo E−/−) compared to mice not expressing human APOE.[48, 49] Still, it is possible that APOE exerts its effects through different mechanisms, e.g. APOE is a major cholesterol transporter and high cholesterol levels have been associated with an increased Aβ load in animal models[50, 51] and changes in APP processing.[52, 53] Thus, APOE isoform-specific changes in cholesterol binding and transport in brain might also affect plaque formation in AD brains.
LOAD as the phenotype
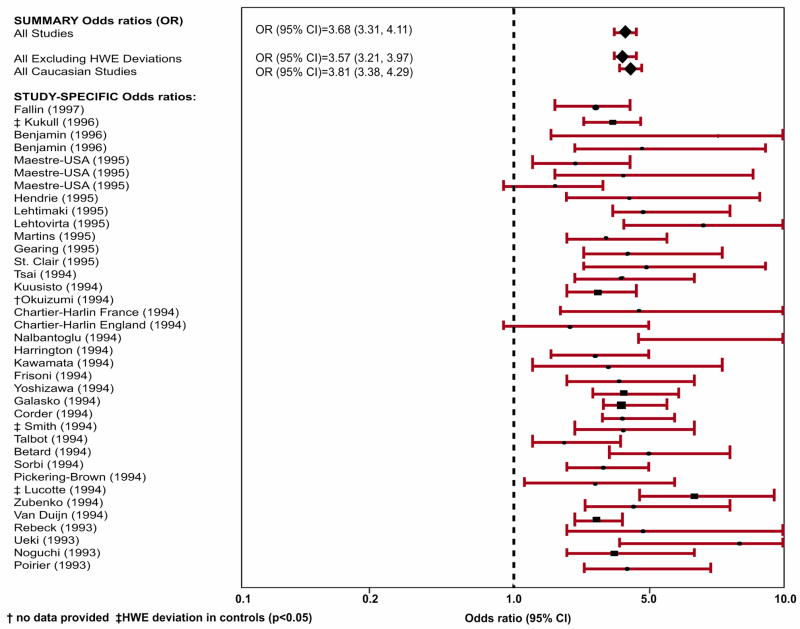
A large amount of studies assessed the relation between APOE genotypes and LOAD in population-based settings. In a meta analysis[54] that included data from 40 research teams on 5930 patients with LOAD and 8607 controls without dementia who were recruited from clinical, community, and brain bank sources, the risk of LOAD was significantly increased for Caucasians with genotypes ε2/ε4 (OR=2.6, 95% CI=1.6–4.0), ε3/ε4 (OR=3.2, 95% CI=2.8–3.8), and ε4/ε4 (OR=14.9, 95% CI= 10.8–20.6), whereas the ORs were decreased for people with genotypes ε2/ε2 (OR=0.6, 95% CI=0.2–2.0) and ε2/ε3 (OR=0.6, 95% CI=0.5–0.8). The association between the APOE-ε4 allele and LOAD was weaker among African Americans and Hispanics, but there was significant heterogeneity in ORs among studies of African Americans (p<0.03). In Japanese subjects, the association was stronger than in Caucasian subjects (ε3/ε4: OR=5.6, 95% CI=3.9–8.0; ε4/ε4: OR=33.1, 95% CI=13.6–80.5). The ε2/ε3 genotype appeared equally protective across ethnic groups. Figure 1 shows the pooled odds ratios (95% CI) of the 40 studies included in the paper. Taken together, it seems that one ε4 allele is associated with a 2-3-fold increased risk, while having two copies is associated with a 5-10-fold increase.
Figure 1.
Pooled odds ratios (95% CI) of the 40 studies included in the meta-analysis by Farrer et al.[54] relating APOE genotype with LOAD (ε4 allele vs. ε3 allele). †No data provided; ‡HWE deviation in controls (p≤0.05)
Age-at-onset as the phenotype
In the vast majority of studies, both clinical and epidemiological, age-at-onset of LOAD was strongly related to the presence of the APOE-ε4 allele (table 1).[17–26] Taken together, these studies which include both clinical and epidemiological studies, suggest that APOEε4 may decrease the age-at-onset by as much as 7 to 9 years per allele. They further suggest that this effect is present across the life span including children and adolescents[22, 23, 55–60] and across various ethnic groups although it may be stronger in Caucasians and Hispanics than African Americans.[26] Mak et al., [61] studied the APOE allele frequencies in Hong Kong elderly Chinese (65 LOAD patients and 82 age- and sex-matched controls). Both the mean and the median age-at-onset tended to be lower in subjects with one or two copies of ε4 compared to persons without ε4 allele (mean age-of-onset (SD) no ε4 vs. one ε4, one ε4 vs. two ε4s: 73.3 (8.5) vs. 72.0 (6.4) vs. 71.2 (5.0)). There was in addition a tendency for the mean and median ages at onset to be higher in subjects with ε2/ε2 or ε2/ε3 than in subjects with ε3/ε3. Although these differences only approached statistical significance (p = 0.078, Z = 1.419) these findings suggest that APOE also exerts its effect in Chinese populations. This notion is supported by the fact that in the same study the APOE-ε4 allele frequency was significantly higher in the AD group than in the control group (0.169 versus 0.067, p < 0.01), and the fact that in Chinese the ε4 frequency is low which decreases the power to obtain statistical significant results.[62]
Table 1.
Summary of studies relating APOE genotype with LOAD, LOAD endophenotypes, MCI and progression from MCI to Dementia
| Author | Subjects | Age in years, mean (range) | Endophenotype | Finding |
|---|---|---|---|---|
| AGE-AT-ONSET | ||||
| Lehtovirta et al.[147],1995 | 202 Finnish LOAD patients and 55 age-and sex-matched controls | Disease onset: ε4: −/− 76 ± 10, −/+: 77 ± 8, 2, +/+ 71 ± 7 | Age-at-onset | age-at-onset decreased from 76 to 69 as the number of ε4 alleles increased from 0 to 2 |
| Gomez-Isla et al., [148]1996 | 359 patients LOAD, age and sex matched 129 controls | LOAD group: mean age of 77.8 years; control group: mean age of 77.8 years | Age-at-onset | Age of onset declined significantly as number of ε4 alleles increased (p < 0.0001 for linear contrast ε3/ε3 to ε3ε4 to ε4/ε4 |
| Holmes et al., [20] 1996 | 164 patients | 60 years and older | Age-at-onset | trend for decreasing age-at-onset of 3 to 4 years in carriers of the APOEε4 allele (mean age (SD): no ε4− vs ε4: 78.7 (7.9) vs. 75.5 (5.9), p=0.004)) |
| Murman et al., [23] 1996 | 107 normal, elderly control subjects and 123 LOAD patients | 45 years and older | Age-at-onset | increased APOEε4 frequencies associated with onset ages of 55 and 75 years, but not at the extremes of onset ages (i.e. onset between 45 and 54 years of age and after age 75) |
| Breitner et al., [17] 1999 | 5,677 elderly residents of Cache County, Utah | 65 years and older | Prevalence and Age-at-onset | age-specific prevalence of LOAD reached in APOEε4 heterozygotes the maximum at age 87, in homozygotes at age 73 and in non-carriers at age 95 |
| Tang et al., [26] 1996 | 305 LOAD patients, 485 nondemented controls | LOAD cases: 76.4 ± 9.1 years, controls: 72.9 ± 6.7 years | Relative risk of LOAD, Age-at-onset | RR for LOAD associated with APOEε4 homozygosity increased in all ethnic groups (African American relative risk [RR]=3.0; 95% confidence interval [CI]=1.5–5.9; Caucasian RR=7.3, 95% CI=2.5–21.6; and Hispanic RR=2.5, 95% CI=1.1–5.7), compared with those with APOE-epsilon 3/epsilon 3 genotypes. The risk was also increased for APOE-epsilon 4 heterozygous Caucasians (RR=2.9, 95% CI=1.7–5.1) and Hispanics (RR=1.6, 95% CI=1.1–2.3), but not for African Americans (RR=0.6, 95% CI=0.4–0.9). The age distribution of the proportion of Caucasians and Hispanics without LOAD was consistently lower for ε4 homozygous and heterozygous individuals than for those with other APOE genotypes |
| Kurz et al., [22] 1996 | 91 patients, 69 healthy age-matched controls | 44 to 95 years | Age-at-onset | inheritance of at least one ε4 allele associated with significant reduction of age-at-onset by 7.7 years among patients 83 years or older, and a weaker relationship among patients aged 44–63 year |
| Poirier et al., [24] 1993 | 91 patients with LOAD and 74 controls | mean age (SD): 75.1 (10.3) | Prevalence of LOAD, Age-at-onset | significant association between ε4 and sporadic LOAD (ε4 frequency 0.380 in LOAD and 0.122 in controls, p < 0.01). Age-at-onset in ε4 carriers earlier than in ε2 or ε4 carriers |
| Mak et al., [61] 1996 | 65 LOAD patients and 82 controls | mean age of 76.5 years | Age-at-onset | Tendency towards lower age-at-onset in subjects with one or two copies of ε4 (mean age-of-onset (SD) −/− vs. 4/− vs. 4/4: 73.3 (8.5) vs. 72.0 (6.4) vs. 71.2 (5.0)), and higher in subjects with ε2/ε2 or ε2/ε3 than in subjects with ε3/ε3 but differences not statistically significant (p = 0.078, Z = 1.419) |
| do Couto et al.[63], 1998 | 68 patients with LOAD | mean age (SD):68.8 (7.9) | Age-at-onset | Age-at-onset significantly higher in patients bearing the APOEε4 allele (ε3/ε4 and ε4/ε4, 65.7 (7.1), n=40) compared with patients without ε4 allele (ε3/ε3, 61.6 (7.6), n=28, p<0.05) |
| Dal Forno et al., [64]1996 | 101 LOAD subjects | mean age: 69.6 years | Age-at-onset | Age-at-onset highest for ε4 heterozygous subjects and least for ε4 negative subjects. Heterozygous subjects declined more rapidly on MMSE and the Category Fluency Test than subjects without ε4 or ε4 homozygosity |
| COGNITIVE PERFORMANCE | ||||
| Welsh-Bomer et l., [65] 2008 | 507 participants of the Cache County Study of Memory in Aging (CCMS) | 70–110 years | Cognitive performance | No association |
| Salo et al., [66] 2001 | 46 nondemented persons | >85 years | Memory performance | No association |
| Murphy et al., [67] 1997 | 86 subjects with LOAD | Mean age of onset (SD): based on caregiver report: 65.3 (7.4); based on age when MMSE < 23: 68.8 (7.0) | Rate of decline on MMSE | No association |
| Cosentino et al., [68] 2008 | one incident (n=199) and two prevalent samples (n=215, n=156) of LOAD patients | age 65 years and older | Memory performance | presence of an APOE ε4 allele associated with a more rapid decline in memory perfomance over a 7-year follow-up period |
| Wehling et al., [60] 2007 | 70 LOAD patients | 50–75 years | Cognitive performance | APOEε4 carriers had slightly poorer performance than non-carriers on the MMSE (27.5 vs. 28.4, p=0.03) and learning trials of the California Verbal Learning Test (CVLT, (F (1,68) = 5.46, p = 0.022) |
| Hirono et al., [69] 2003 | 64 LOAD patients | 60 years or older | Memory performance | presence of the APOEε4 allele in dose-response fashion associated with accelerated memory decline on Word Recall subtest of ADAS-Cog (mean score −/− vs. 4/− vs 4/4: −0.2 vs. 0.4 vs 1.0, p=0.008) |
| Mayeux et al., [70] 2001 | 563 healthy elderly without LOAD or questionable dementia | 65 years and older | Memory performance over 7-year follow-up | APOEε4 allele associated with a more rapid decline in memory performance |
| Wilson et al., [71] 2002 | 669 participants from the Religious Order Study | 65 years and older | summary measures of episodic memory, semantic memory, working memory, perceptual speed, and visuospatial ability | average annual increase of 0.016 units in the ε2 subgroup and annual decreases of 0.022 units in those with ε3/3 and of 0.073 units in the ε4 subgroup |
| Lehman et al., [72] 2006 | 2181 elderly of the Hordaland Health Study | 70–74 years | episodic memory | APOEε4 effect on episodic memory: OR of cognitive impairment in women 1.8 (95% CI: 1.1–2.8) for heterozygotes and 1.1 (0.3–3.7) for homozygotes; OR in men 1.1 (95% CI 0.6–2.1) for heterozygotes and 10.7 (95% CI 4.7–24) for homozygotes |
| Liu et al., [58] 2008 | 2208 related individuals | 50 years and older | Cognitive performance | APOEε4 significantly associated with reduced test scores for Adult Verbal Learning Test, particularly on the memory and learning sub domains |
| Bondi et al., [73] 1995 | 52 elderly non-demented | 59–83 years | Performance on California Verbal Learning Test (CVLT | APOEε4 associated with poorer performance on CVLT. Six of the 14 APOEε4 subjects developed either LOAD or questionable LOAD, whereas none of the 26 non APOEε4 subjects demonstrated any cognitive decline |
| Dik et al., [149] 2000 | 1,243 subjects with a MMSE score between 21 and 30 | 62–85 years | Memory decline | APOEε4 allele associated with memory decline in cognitively impaired subjects (decline on immediate recall: OR:3.8 (1.,10.0); decline on delayed recall; OR:2.9 (1.2,7.0); decline on retention: OR 3.3 (1.1,10. 1), but not in cognitively normal subjects (MMSE score, 27 to 30) |
| Dik et al., [74] 2001 | 1,168 subjects from the population-based Longitudinal Aging Study Amsterdam | 62 to 85 years | Performance on MMSE, immediate recall and delayed recall, and the Alphabet Coding Task-15 | APOEε4 carriers had a greater rate of cognitive decline shown by MMSE scores and slower information processing speeds after 6 years. The effects of both memory complaints and APOEε4 allele carriage were additive: subjects with both factors had a two times higher cognitive decline than did subjects without both factors |
| Caselli et al., [56] 1999 | 100 nondemented individuals | mean age 56 years | Immediate and delayed recall | tests sensitive to immediate and delayed recall showed significant negative correlation with age in the APOEε4 homozygote group relative to the noncarrier group |
| Flory et al., [57] 2000 | 220 non-demented non-Hispanic Caucasian men and women | aged 24–60 (average age = 46) | verbal learning and memory (e. g., learning a list of words and recalling them 30 min later), visual memory (e.g., reproducing a previously copied figure from memory), and attention span memory | performance on learning and memory tasks was significantly poorer in adults having any APOEε4 allele, relative to adults with APOEε2 or APOEε3 genotypes (p <.01) |
| Reynolds et al., [75] 2006 | 478 non-demented twins from the Swedish Adoption/Twin Study of Aging (SATSA) | 50 years and older | memory performance over 13 years | APOEε4 associated with working and recall memory ability levels and working memory rate of change, with ε4 homozygotes exhibiting the worst performance at all ages over 13 year follow up |
| Schultz et al., [59] 2008 | 626 male twins randomly drawn from the Vietnam Era Twin (VET) Registry | In their 50s | memory performance | ε4-carriers: lower performance on immediate and delayed recall than non-carriers (mean (SD) comparing ε4+ vs. ε4−: immediate recall 22.19 (5.37) vs. 23.8 (6.2); delayed recall: 19.5 (5.9) vs. 20.12 (6.6)) |
| Small et al., [150] 2000 | 413 older adults from the Charlotte County Healthy Aging Study | mean age: 72.9 | cognitive functioning, including episodic memory, implicit memory, psychomotor speed, and attention | no association between APOEε4 allele and cognitive functioning |
| Farlow et al., [151] 2004 | 494 MCI subjects | 55–85 years, mean age: 70.8 | cognitive functioning | APOEε4 carriers had lower MMSE (p = 0.01), higher ADAS-cog (p < 0.0001) scores, greater deficits on cued SRT and ADCS-ADL scale (p < 0.001), and smaller hippocampal volumes (p = 0.002) than non-carriers |
| Kleiman et al., [152] 2006 | 366 AD patients | ε4 non-carriers: 73.4±9.6, ε4-heterozygotes 74.4±7.3, ε4 homozygotes: 71.6±7.0: | Progression of cognitive/function al decline in AD patients | No association between APOEε4 and cognitive or functional decline |
| Caselli et al., [153] 2007 | 43 e4 homozygotes, 59 e4 heterozygotes, and 112 noncarriers without cognitive imapirment | 50–69 years | Progression of cognitive/function al decline | Cognitive domain decline occurred in 4 of 10 APOEε4 homozygotes 60 years and older at entry (40.0%) compared with 5 of 66 APOEε4 heterozygotes and noncarriers (7.6%) (P = .02) |
| MCI/PROGRESSION FROM MCI TO DEMENTIA | ||||
| Petersen et al., [28] 1995 | 66 patients with MCI from Mayo Clinic | mean age: 79.8 years | Conversion from MCI to dementia | APOEε4 strong predictor for conversion to dementia |
| Tierney et al., 1996[37] | 107 patients with memory impairment but no dementia | Patients developing AD:74.4±7.1, subjects developing MCI: 71.5±7.8 | Conversion from memory impairment to dementia | APOEε4 allele predicts development of AD in memory-impaired individuals |
| Jack et al.[35], 1999 | 80 consecutive patients with MCI | Mean age: 77.7 ± 6.8 | Conversion from MCI to dementia | APOE genotype was reliable prognostic indicator of development of AD |
| Hsiung et al., [34] 2004 | 1469 cases with cognitive impairment, 582 controls | control group: mean age 75.6, group with CIND: mean age 77.8, group with AD: mean age 82.7 | progression from normal cognition to CIND and from CIND to AD or VaD, age-at-onset of LOAD | possession of an APOEε4 allele associated with increased risk of LOAD developing from CIND (OR 2.6, 95% CI 1.48–4.92), and associated with decrease in the age-at-onset of LOAD |
| Devanand et al., [32] 2005 | 136 patients with MCI and 57 age-and sex-matched healthy controls | mean age 66 years | Conversion from MCI to dementia | APOEε4 carrier status associated with conversion to AD in older patients after controlling for confounders (RR:2.77; 95% CI: 1.1–7.3; P = 0.03), but not by itself |
| Aggarwal et al., [154] 2005 | 181 patients with MCI from the Religious Order Study | mean age:78.7± 6.9 | Conversion from MCI to dementia | APOEε4 allele associated with a 93% increase in the risk of developing AD (95% CI; 1.02, 2.63) |
| Tschanz et al., [155] 2006 | 120 participants with cognitive impairment from the Cache County Study | ≥65 years at baseline | Conversion from MCI to dementia | Among individuals with ≥1 APOEε4 allele, those with prodromal AD or other cognitive syndromes exhibited a 22-to 25-fold higher risk of dementia than cognitively unimpaired individuals (vs 5-to 10-fold higher risk in those without epsilon4) |
| Tyas et al., [30] 2007 | 470 nondemented participants from the Nun Study | >75 years | Risk of MCI and progression to dementia | APOEε4 allele significantly associated with MCI but not with progression to dementia. |
| Barabash et al., [27] 2007 | 89 patients with amnestic MCI | mean age: 75±7.1 | Risk of MCI and progression to dementia | APOEε4 allele associated with an increased risk of MCI (OR: 6.04, 95% CI: 2.76–3.23; p<0.001) but not with progression to AD |
| Fleisher et al., [156] 2007 | 539 participants with amnestic MCI | Mean age: Progressors: 74.9±6.6, non-progressors: 71.5±7.4 | Progression from amnestic MCI to AD | Progression from amnestic MCI to AD was best predicted by combining APOE status and the Symbol Digit Modalities Test, Delayed 10-Word List Recall, NYU Delayed Paragraph Recall Test, and the ADAS-cog total score (estimated predictive accuracy:81% (95% CI: 0.79 to 0.83)) |
| Ramakers et al., [36] 2008 | 180 subjects with MCI | Mean age: ε4 non-carriers: 55.2±9.2, ε4-carriers: 58.9±9.9 | cognitive functioning | APOEε4 allele strongly related to subjective organization in middle-aged subjects (p = 0.011) and strongly related to delayed recall performance in elderly subjects (p = 0.02) |
| van der Flier et al., [157] 2008 | 749 memory clinic patients and 2,233 controls | mean age:66.0±11.0 | subjective complaints, MCI, AD, other types of dementia | Compared with controls (15%) the prevalence of APOEε4 increased among patients with subjective complaints (22%), MCI (36%), AD (42%) and other types of dementia (25%) |
| Sasaki et al., [29] 2009 | 1433 Japanese subjects | ≥65 years | MCI | frequency of APOEε4 allele higher in persons with amnestic MCI compared to persons with non-amnestic MCI or controls |
| Blom et al., [31] 2009 | 47 AD patients, 58 patients with MCI and 35 healthy control subjects | Mean age: controls: 57.0±8.1, MCI: 62.9±8.2, AD:71.7±8.1 | Conversion from MCI to dementia | MCI subjects with high CSF T-tau or P-tau and APOEε4 homozygosity progressed faster from MCI to AD |
In contrast to these studies, two studies found a higher age-at-onset for patients bearing the APOEε4 allele. In a study by do Couto et al.[63] among 68 patients with LOAD, the age-at-onset of disease was significantly higher in the patients with the ε4 allele (mean onset (SD) of ε3/ε4 and ε4/ε4, 65.7 (7.1), n=40) compared with patients without the ε4 allele (mean onset (SD) ε3/ε3, 61.6 (7.6), n=28, p<0.05, two tailed Student’s t test). Among 101 LOAD patients[64] age-at-onset was highest for the ε4-heterozygous subjects and lowest for the ε4-negative subjects. The heterozygous subjects declined more rapidly on the Mini-Mental State Examination and the Category Fluency Test than the subjects without the ε4 allele or with ε4 homozygosity. The homozygous subjects declined only faster on the Physical Capacity subscale of the Psychogeriatric Dependency Rating Scale. It is important to note that these two studies included relatively younger patients. It remains possible that the presence of the ε4 allele represents a particularly high risk in the older patients. The bulk of data on age-at-onset is consistent with the large body of studies showing an association between the APOEε4 allele and risk of LOAD, and suggests that the ε4 allele decreases age-at-onset of LOAD in a dose-dependent manner.
Cognitive performance as the phenotype
Few studies, including the Cache County Study of Memory in Aging (CCMS), [65] a study among 46 nondemented persons aged 85 years or over from a randomly selected group of 128 subjects in Vantaa, Finland, [66] and the study by Murphy et al., [67] observed no effect of the APOE locus on the rate of cognitive decline. It is important to note that these studies either had unspecific assessment of memory, [67] small sample sizes[66, 67] or consisted of samples prone to survival bias[65] which may limit their ability to detect harmful associations. However, most studies exploring the association of APOE with cognitive performance were consistent with the studies reporting an association of the APOE genotype with LOAD or age-at-onset of LOAD, and showed a harmful effect of the APOEε4 variant with a dose-response-relationship of the effect (table 1). In general, these studies can be divided into studies including and excluding subjects with cognitive impairment or dementia. Studies that explore the effect of APOE on cognitive performance in non-demented subjects provide the ability to draw conclusions about the effect of genetic risk factors on cognition in cognitively normal persons or the preclinical stage of the disease.
Studies including subjects with cognitive impairment or dementia
Cosentino et al.[68] examined the impact of the APOEε4 variant on the rate of cognitive change in one incident (n=199) and two prevalent samples (n=215, n=156) of LOAD patients 65 years and older. The presence of at least one ε4 allele was associated with faster cognitive decline in the incident LOAD group (p = 0.01). Similar results were observed for the two prevalent dementia samples when adjusting for disease severity or excluding the most impaired participants from the analyses, indicating that the APOEε4 may influence the rate of cognitive decline in both the early and late stages of LOAD. In a study by Wehling et al., [60] which comprised 70 consecutively referred patients aged 50–75 years, APOEε4 carriers showed a slightly poorer performance than non-carriers on the MMSE (27.5 vs. 28.4, p=0.03) and learning trials of the California Verbal Learning Test (CVLT; F (1,68) = 5.46, p = 0.022). Hirono et al., [69] who explored the effect of APOE on cognition in 64 LOAD patients using the Alzheimer Disease Assessment Scale-Cognitive subscale (ADAS-Cog), observed that the presence of the ε4 allele was in a dose-response fashion associated with accelerated memory decline (mean ADAS-Cog score −/− vs. 4/− vs 4/4: −0.2 vs. 0.4 vs 1.0, p=0.008).
Studies excluding subjects with cognitive impairment or dementia
Most studies exploring these associations among non-demented subjects yielded consistent results, indicating that APOE also exerts its effect in cognitively normal subjects or preclinical stages of the disease. In a study by Mayeux et al.[70] presence of an APOEε4 allele was in 563 non-demented elderly associated with a more rapid decline in a composite score of memory performance over a 7-year follow-up period. Among 669 participants of the Religious Order Study, [71] possession of one or more ε4 alleles was over a 8-year follow-up associated with faster decline in episodic memory compared to the ε3/3 genotype, while possession of one or more APOEε2 alleles was associated with reduced decline. The rate of change in episodic memory were an average annual increase of 0.016 units in the ε2 subgroup and annual decreases of 0.022 units in those with ε3/3 and of 0.073 units in the ε4 subgroup. In 2,181 elderly participants (aged 70–74 years) from the Hordaland Health Study the APOEε4 allele was in a dose-dependent fashion also associated with lower episodic memory performance. The strongest effect was seen in homozygous men (OR 10.7; 95% CI 4.7–24.0).[72] In a Dutch sample of 2,208 related individuals, the ε4 variant was associated with reduced test scores for the Adult Verbal Learning Test, and within this test strongest for the memory and learning sub domains.[58] Bondi et al.[73] explored the effect of APOE on cognition in 52 non-demented elderly using the California Verbal Learning Test (CVLT). Consistent with the studies described above, APOEε4 carriers demonstrated significantly poorer mean performances than non-carriers. Six of the 14 APOEε4 carriers who completed annual follow-up evaluations developed either LOAD or questionable LOAD, whereas none of the 26 non-carriers demonstrated any cognitive decline.
The longitudinal population-based Longitudinal Aging Study Amsterdam[74] explored to what extent subjective memory complaints and APOEε4 allele carriage interact in their prediction of future cognitive decline. In this study of 1,168 elderly subjects, APOEε4 carriers had after a six year follow-up a greater rate of cognitive decline measured by MMSE scores and slower information processing speeds. This effect appeared to be additive with the effect of memory complaints: subjects with both factors showed a two times higher cognitive decline than did subjects without memory complaints and ε4 allele.
In the Canadian Study of Health and Aging[34] and a consecutive sample of 66 patients from the Mayo Clinic Alzheimer’s Disease Center/Alzheimer’s Disease Patient Registry who met criteria for a diagnosis of a mild cognitive impairment (MCI) and who had at least one clinical reevaluation, [28] possession of an APOEε4allele increased the risk of conversion from cognitive impairment no dementia (CIND) or MCI to LOAD. In the Canadian Study of Health and Aging the presence of the APOEε4 allele was also associated with a decrease in the age-at-onset of LOAD.[34]
In two cross-sectional studies in younger subjects (average ages 46 and 56)[56, 57] the APOEε4 allele was relative to the noncarrier group associated with significantly poorer performance on learning and memory tasks and immediate and delayed recall, suggesting that age-related memory decline occurs earlier in cognitively healthy APOEε4 carriers than in noncarriers, and precedes clinically detectable LOAD.
Finally, these findings could also be replicated by twin studies. In a longitudinal study over 13 years[75] among 478 twins from the Swedish Adoption/Twin Study of Aging (SATSA), the APOEε4 variant was in a dose-dependent fashion at all ages associated with worse working and recall memory, and rate of change in working memory. In a second longitudinal twin study among 626 twins in their 50s[59] ε4-carriers showed significantly lower performance on immediate and delayed recall than non-carriers (mean (SD) comparing ε4+ vs. ε4−: immediate recall 22.19 (5.37) vs. 23.8 (6.2); delayed recall: 19.5 (5.9) vs. 20.12 (6.6)), supporting the genetic contribution of APOE to LOAD.
MCI and progression of MCI to dementia as the phenotype
Fewer studies assessed the relation between APOE genotypes and MCI or progression of MCI to dementia in population-based settings. The vast majority of these studies observed an increase in MCI risk or progression from MCI to dementia in ε4-carriers.[27–37] Two studies[27, 30] observed increased risks of MCI in ε4-carriers compared to non-carriers but no association between the APOE genotype and progression to dementia. Potential reasons for these negative findings are the limited number of patients included in the studies, the short time of follow-up, the insufficient control of potentially confounding factors and the lack of consensus criteria for MCI leading to considerable heterogeneity. It has been well established that dementia risk varies with the definition of MCI used.
Sensitivity and Specificity of APOE
Studies assessing the usefulness of the APOE genotype (ie. APOE genetic testing) in the diagnosis of Alzheimer’s disease among persons with dementia, reported specificities of the e4 allele between 81 and 100%[76–79] when used in combination with clinical or autopsy criteria, but lower specificities when used alone.[77] Sensitivity estimates, were lower and ranged between 19–75%.[76, 77, 79, 80] These estimates and the relatively low frequency of the ε4-allele in persons with AD and the general population limit the utility of APOE genetic testing. As described above the ε4-allele increases the risk of developing AD but is neither necessary nor sufficient, meaning that not all persons with APOE*E4 alleles will develop Alzheimer’s disease. If considered, genetic testing should only be undertaken after carefully discussing the benefits, ethical issues and risks (ie. potential harm such as anxiety through revealing the test results) with a physician or genetic counselor.
2. Sortilin-related receptor (SORL1)
Identification of APP, presenilin 1 (PSEN1), presenilin 2 (PSEN2) as susceptibility genes for early-onset AD (EOAD) has led to the initiation of the “amyloid cascade”, the basic biochemical formula for production of Aβ, the putative culprit of AD. The amyloid pathway involves two enzymatic steps: In the first β-cleavage step, BACE cleaves APP near the N terminus of the Aβ peptide; in the second γ-cleavage step, the membrane-bound C-terminal APP fragment is cleaved by γ-secretase, a complex composed of transmembrane proteins presenilin 1 and 2, nicastrin, APH1, TMP21, and PEN2.[81]
It is notable that APP and the secretases are all integral transmembrane proteins. Further, they are dynamically sorted through the plasma membrane and the membranes of intracellular organelles, and the liberation of Aβ involves a transmembrane secretase enzyme acting on a transmembrane APP CTF substrate. Thus, from a cell biology perspective, sorting mechanisms that cause APP and the secretases to colocalize in the same membranous compartment are expected to play important roles in the regulation of Aβ production. Over 30% of all proteins are transmembrane proteins, [82] and most are typically sorted via the secretory and endocytic pathways.[83, 84] During the last two decades, the trans-Golgi network and the endosome were identified as the key organelles organizing the complex movement of the transmembrane proteins via secretory and endocytic pathways. Important coat complexes initiating the transport of APP and BACE through this sorting itinerary are the clathrin coat and the retromer.[85–87] Clathrin coats are involved in the endocytic pathway connecting the cell surface to the endosome, and the pathway connecting the trans-Golgi network to the endosome.[88] The retromer is involved in the trafficking from the endosome to the trans-Golgi network.
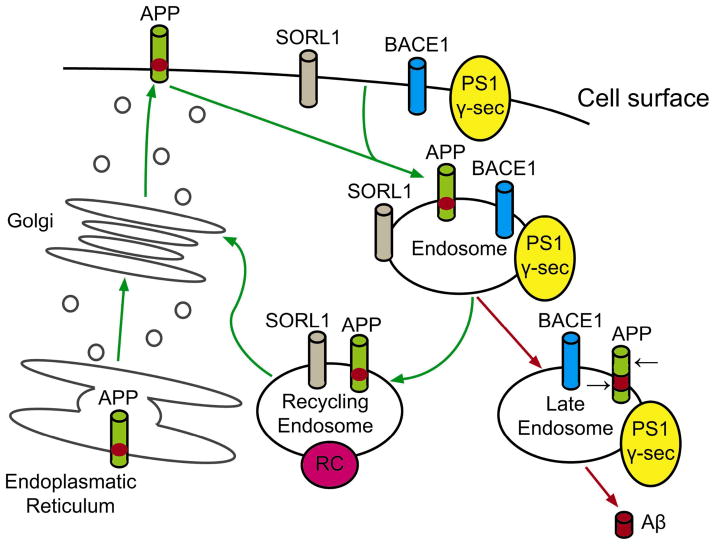
Recent studies showed that SORL1 is involved in trafficking of APP from the cell surface to the golgi-endoplasmic reticulum complex (Figure 2). SORL1 belongs to the VPS10 vacuolar protein sorting receptor family, [87] which in turn belongs to a group of protein trafficking molecules that are in the endocytic and retromer pathways, and are highly expressed in the central nervous system.[87] It is clear that the subcellular domains of these trafficking molecules are important sites for the generation of the amyloid β-peptide (Aβ), the main putative culprit in the pathogenesis of AD. The sub-cellular domain of SORL1 interacts with the amyloid precursor protein (APP) and directs its trafficking into recycling pathways. As a consequence, APP is sorted into Aβ-generating compartments when SORL1 is under-expressed, leading to an increased risk of AD[89–91] Accordingly, knockout of SORL1 in mice is associated with increased brain Aβ levels.[89] It is also possible that SORL1 contributes to the risk of AD through the nerve growth factor (NGF)-neurotrophin system. NGF promotes cell survival via binding to the tyrosine kinase receptor A (TrkA). Sortilin receptors bind, as a trimolecular complex, with p75NTR to its precursor (proNGF) initiating apoptosis.[92, 93] Whether proNGF can also act neurotrophically following binding to TrkA is still unclear.
Figure 2.
Role of SORL1 in transmembrane sorting of APP. The green arrows track re-entry of APP from the cell surface when SORL1 is present. The red arrows show that, when SORL1 is absent, more APP moves into domains such as the late endosome/lysosome, where the black arrows show how it is subsequently cut by beta-secretase (BACE1) and gamma-secretase (PS1 γ-sec), generating the neurotoxic amyloid beta-peptide (Aβ). [Illustration adapted from Rogaeva et al.[89]]
Most studies exploring the effect of SORL1 on cognitive impairment or dementia used LOAD as a dichotomized trait in the analyses. Rogaeva and colleagues[89] first reported the allelic and haplotypic associations between LOAD and variants in SORL1 (table 2). Subsequently several studies supported the initial finding by showing that genetic variants in SORL1 contribute toward LOAD.[94–100] The original study included four different ethnic groups, ranging from North American and European Caucasians, Caribbean Hispanics, African-Americans, and Israeli-Arabs. With this investigation on over 6,000 subjects, two different sets of haplotypes were identified: (1) SNPs in the 5′ end of the gene (SNP 8–10; 120873131 bp-120886175 bp) among Caribbean Hispanics (family study), Caucasians (case-control study), and Israeli-Arabs (case-control study); and (2) SNPs in the 3′ end of the gene (SNP 22–25; 120962172 bp-120988611 bp) among multiple Caucasian samples (family and case-control studies) and African-Americans (family study). Haplotype analysis strengthened the statistical support further. However, as observed in many common diseases, these candidate SNPs confer a modestly elevated risk of LOAD, ranging from an odds ratio of 1.4 to 2.2, and the allelic association was not uniform across datasets or ethnic groups. The authors strengthened their allelic association findings by functional cell biology findings which showed that suppression of SORL1 led to elevation of amyloid β levels.[89] Two subsequent studies by the same group broadly supported one or both haplotypes or some variations of the two: Haplotype C-G-C at SNPs 8–10, or haplotype T-T-C at SNPs 23–25, or both. Lee and colleagues[96] showed that the same set of SNPs at SNPs 23–25 were associated with LOAD in Caucasians residing in northern Manhattan. They then confirmed the allelic and haplotypic associations in autopsy confirmed cases of Caucasian ethnicity for haplotype at SNPs 8–10 and haplotype at SNPs 23–25.[95]
Table 2.
Summary of studies relating SORL1 with LOAD. No study specifically assessed the association between genetic variation in SORL1 and MCI or Progression from MCI to Dementia.
| Author (Year) | Age | Haplotype 1 | Haplotype 2 | Other Significant SNPs | ||||
|---|---|---|---|---|---|---|---|---|
| rs668387 SNP 8 (1,2) | rs689021 SNP 9 | rs641120 SNP 10 | rs3824968 SNP 23 | rs2282649 SNP 24 | rs1010159 SNP 25 | |||
| SIGNIFICANT ASSOCIATION | ||||||||
| Rogaeva et al. (2007) | Mean AAO: 70±9–77±8 | |||||||
| Caucasians (family dataset) | T | T | C | |||||
| Caribbean Hispanics | C | G | C | |||||
| Caucasians (case-control datasets) | C | G | C | T | T | C | ||
| Israeli Arabs | C | G | C | |||||
| African-Americans | ||||||||
| Lee et al. (2007) - Northern Manhattan | Mean AAO: 79.1±5.1 – 84.4±8.0 | |||||||
| Caucasians | C | A | T | T | T | C | rs3824966 (SNP 20) | |
| Hispanics | rs12285364 (SNP 12) | |||||||
| African Americans | C | G | T | C | C | rs12285364, rs1784933 (SNP 26) | ||
| Meng et al. (2007) (2) | Not released | |||||||
| Caucasians | + | + | + | |||||
| Lee et al. (2007) - Autopsy | Mean AAO: cases: 80.5, controls: 79.9 | |||||||
| Caucasians | C | G | C | A | T | C | ||
| Tan et al. (2007) | Mean AAO: 71.2 ± 8.9 | |||||||
| Han Chinese | A | T | ||||||
| Seshadri et al. (2007) (3) | Mean age: 62 +9 | |||||||
| Caucasians | + | rs1131497 (SNP29) | ||||||
| Bettens et al. (2008) | Mean AAO: 79.0 ± 5.2 | |||||||
| Caucasians | C | G | C | rs560573 (SNP 6), rs1614735 (SNP 27) | ||||
| Lee et al.(2008), reanalyzing data by Shibata et al. (2008) | ||||||||
| Japanese | C | T | ||||||
| Koelsch et al. (2009) | ||||||||
| Caucasians | Mean age: cases: 71.91 ± 8.2 years; controls: 71.56 years | SORL1-18ex26 (SNP21), haplotype: rs2070045 (SNP19)/SORL1-18ex26 (SNP21)/rs3824968 (SNP23):T/G/A | ||||||
| WEAK ASSOCIATION | ||||||||
| Webster et al. (2007) (2) | Age ≥ 65 years | |||||||
| Caucasians | + | + | ||||||
| Li Y, Grupe, et al. (2008) (2) | Mean age: 77.4 ± 7.5 (WU), 76.4 ± 6.1 (UK1), 76.5 ± 5.6 (UK2) | |||||||
| Caucasians | T | rs2070045 (SNP 19) | ||||||
| NO ASSOCIATION | ||||||||
| Li H, Roses et al (2008) (4) | Mean AAO ≥60 | |||||||
| Caucasians | ||||||||
| Houlihan et al.(2008) | Mean age: 70 | |||||||
| Caucasians | ||||||||
| Minster et al. (2008) | Mean AAO: cases: 72.8 ± 6.2 years; controls: 74.1 ± 6.2 years | |||||||
| Caucasians | ||||||||
SNP numbers from Rogaeva et al (2007) are presented. Alleles are presented only when significant Alleles in bold were statistically significant in either allelic, genotypic, or haplotypic analysis.
Used the nearest SNPs (indicated with a “+” sign) when different SNPs were used.
Endophenotype was studied
No specific marker information for SORL1 was available from the paper. AAO=age of onset
Subsequently various other groups examined the relation between SORL1 and LOAD or LOAD endophenotypes in different populations (table 2).[94, 98–105] Four replication studies supported the initial findings, while the remaining showed either negative or weak results. Three clearly positive studies included one by Bettens et al, [94] Tan et al, [99] Seshadri et al, [98] and Koelsch et al.[100] Bettens and colleagues[94] directly replicated SNPs 8 through 10 and showed support for SNPs 25–27 in 550 Belgians with LOAD and 637 unaffected individuals. Tan et al. examined 223 cases and 263 controls from a Han Chinese population to show that haplotype G-C-A at SNP 19-22-23 were associated with LOAD (OR=1.35; 1.04–1.74), but none of the haplotypes in SNP 8 to SNP 10 were associated. In the study by Koelsch et al. which included 349 AD patients and 483 controls recruited from a multicenter study of the German Competence Network Dementias, [100] the SNP21G-allele and a SORL1 haplotype consisting of the SNP19 T-allele, SNP21 G-allele and SNP23 A-allele (T/G/A) were associated with an increased risk of LOAD and an earlier age at onset (SNP21: p=0.002; T/G/A haplotype: p=0.007). This effect was most pronounced in carriers of an additional APOE4 allele (SNP21: p=0.003; T/G/A haplotype: p=0.005). Webster et al.[103] and Li. et al.[102] reported weak associations.
Li et al, [101] Houlihan et al, [104] Minster et al, [105] and Shibata et al.[106] reported no associations between SORl1 and LOAD. However, in the latter study the negative results were based on genotypic association analyses only. When Lee et al.[107] re-analyzed the data of this study using allelic association tests, SNPs 8 and 24 were significantly associated with LOAD supporting the association in both the 3′ and 5′ regions of SORL1.
Cognitive performance as the phenotype
Using the Framingham community based family samples, Seshadri et al.[98] extended the existing studies using cognitive performance as an endophenotype. The authors reported that SORL1 was significantly associated with abstract reasoning ability as measured by the Similarity test (p=3.2×10−6). However, they did not observe an association with memory. A possible explanation for this discrepancy may be that this sample consisted of 705 related persons, which can lead to limited power to uncover associations as compared to larger samples that include unrelated subjects.
MCI and progression of MCI to dementia as the phenotype
No study specifically explored the association between variation in SORL1 and MCI, or SORL1 and progression from MCI to dementia. However, in a study by Sager et al.[108]who explored the association between SORL1 expression in brain tissue and MCI in participants from the Religious Order Study, SORL1 expression was heterogeneous, forming low- and high-level SORL1 subgroups. MCI subjects with low SORL1 were significantly more cognitively impaired than the high SORL1subjects, suggesting that that reduced SORL1 levels reflect disease severity and may predict progression to AD in a subgroup of individuals with MCI.
Overall, these genetic and functional genomic studies provide compelling evidence for a role of SORL1 in LOAD. Putative variants and their sensitivity and specificity for LOAD diagnosis, however, remain to be identified as the reported variants do not affect coding sequence or splicing. In any case, the results of the above summarized studies imply that there are several different LOAD–associated allelic variants in distinct regions of the SORL1 gene in different populations, that these variants are likely to be in intronic regulatory sequences that might govern cell type–specific or tissue-specific expression of SORL, and that these variants affect this risk by altering the physiological role of SORL1 in the processing of APP holoprotein.
3. Other genes
In addition to APOE and SORL1, several genes and putative loci have been reported, but independent replication remains inconsistent. There is little concordance between case-control and family-based studies[109–113] suggesting that both clinical and genetic heterogeneity influence the outcome of these analyses. The P86L polymorphism in the calcium homeostasis modulator 1 (CALHM1), which encodes an essential component of a previously uncharacterized cerebral Ca2+ channel that may control Aβ levels, has been putatively associated with LOAD.[114] The GRB2-associated binding protein 2 (GAB2) may modify the risk of LOAD in APOEε4 carriers and has been associated with hyperphosporylation of tau protein.[115] The butyrylcholinesterase K variant (BCHE K) allele has been shown to act in synergy with the APOEε4 allele to promote risk for AD.[116] BCHE blocks aggregation of Aβ less aggressive long fibrils[117] and possession of the K variant allele is known to result in a 30% reductionin serum cholinesterase activity.[117] The low density lipoprotein receptor-related protein 6 (LRP6), a coreceptor for Wnt signaling, has been associated with LOAD and confirmed in a case-control analysis.[118] Additional loci that have been reported but remain to be confirmed include the toll-like receptor 4 (TLR4), the cholinergic receptor (nicotinic beta 2, CHRNB2), alpha-2-macroglobulin (A2M), catenin (CTNNA3), glutathione S-transferase omega 1 and 2 (GSTO1, GSTO2) and the glyceraldehyde-3-phosphate dehydrogenase (GAPD)[119–122] and loci at at 6p, 9q, 10q and 12p and 19q.[123–133]
Genes that have been reported to be associated with MCI or risk of progression from MCI to dementia include alpha1-antichymotripsin (ACT), [134] cholinergic receptor (nicotinic alpha 7, CHRNA7)[135], peptidylprolyl cis-trans isomerase (PIN-1)[27], transforming growth factor-beta 1 (TGF-beta), [136] vascular endothelial growth factor (VEGF), [137] a member of the cytochrome P450 superfamily (Cyp46A1)[138], and nitric oxide synthase 3 (NOS3)[139]. In particular genes mapping to chromosome 10q21–25, have been reported to influence amyloid β levels in cognitive impairment. In a study by Ertekin-Taner et al., [140] amyloid β42 levels were related to a missense C/T polymorphism in exon 6 of the in the urokinas18 and 19 of the revised manuscript.e-type plasminogen activator (PLAU) gene at chromosome 10q24. In a second study by the same group genetic variants in a haplotype block spanning the insulin degrading enzyme (IDE) mapping to 10q23–25 were significantly associated with plasma amyloid β42 levels.[141] The latter finding is consistent with a study by Farris et al.[142] demonstrating that partial loss-of-function mutations in IDE, that induce diabetes, also impair degradation of amyloid β protein. PLAU [140] and IDE[141] were also associated with an increased risk of LOAD and cognitive impairment, supporting the usefulness of amyloid β levels as an endophenotype in cognitive impairment.
DISCUSSION
One of the most important challenges in research on cognitive impairment in the elderly is to identify genes that predispose to MCI and could be used to predict which subjects will progress to dementia. Individuals genetically predisposed to evolve to dementia could benefit of therapeutic intervention in the early stages of the disease in which the neurodegeneration has not progressed. Early intervention could significantly prevent or delay the onset, which in turn would improve quality of life of the patient and their relatives and would significantly reduce the public health burden.
However, in contrast to EOAD, which is caused by mutations in APP, PSEN1 and PSEN2 that have almost complete penetrance (>85%), and a clear cut autosomal dominant pattern of inheritance, several issues in research on late-onset cognitive impairment lead to significant difficulties in gene identification. First, current knowledge suggests that a variety of mechanisms underlie the various pathological and clinical changes, and that these have different genetic and environmental components. Thus, it is likely that late-onset cognitive impairment is a complex genetic disorder characterized by an interaction of multiple genes and the environment leading to genotypes with incomplete penetrance and a low magnitude associated risk. Consistent with this notion is the fact that to date only two genes (APOE and SORL1)with modest effect sizes each have been firmly identified as genetic risk factors although segregation analyses conducted in families of patients with LOAD support the presence of at least 4 to 6 major genes.[143, 144] With a population attributable risk that is estimated at 20–50%, [38, 39] the APOEε4 allele increases risk of cognitive impairment, LOAD, and age-of onset of cognitive impairment in a dose-dependent fashion: one ε4 allele is associated with a 2–3 fold increased risk, having two copies is associated with a 5–10 fold increase. Similar effect sizes have been observed for progression of cognitive impairment to dementia. The two haplotypes in the 3′ and 5′ regions of SORL1 that repeatedly were found to be associated with LOAD have effect sizes ranging from odds ratios of 1.4 to 2.2; their associations with MCI and progression to dementia remain yet to be determined. The facts that both APOE and SORL1 have only moderate diagnostic sensitivity and specificity, increase risk of cognitive impairment in a non-Mendelian fashion, are not fully penetrant, and that they are neither necessary nor sufficient by themselves to cause impairment further support the notion of a complex genetic mechanism. The same is likely to be true for the remaining, yet to be identified, genetic factors associated with cognitive decline. Additional genes and genetic loci that have been reported but remain to be confirmed include TLR4, CHRNB2, A2M, CTNNA3, GSTO1, GSTO2, GAPD, ACT, [134] CHRNA7, PIN-1, TGF-beta, VEGF, Cyp46A1, and NOS3, PLAU, IDE and loci at 6p, 9q, 10q and 12p and 19q.
Additional factors hampering genetic research on late–life cognitive impairment are pleiotropic effects, locus or allelic heterogeneity, small sample sizes leading to insufficient power to detect the expected small-moderate effect sizes, uncontrolled population stratification, and the failure to develop better quantitative endophenotypes. Endophenotypes are closer to the action of the gene than affection status, exhibit higher genetic signal-to-noise ratios, [145] and thus provide greater power to localize and identify the various disease-related quantitative trait loci (QTLs) associated with the disease such as memory performance, amyloid/tau pathology or hippocampal atrophy than does affection status alone.[146] It is possible that the endophenotypes that are commonly used in research on late-life cognitive impairment are too heterogeneous to be informative.
Four additional phenomena particularly complicating genetic research on rate of progression are the beginning of the observations in the middle of a developing pathologic process, survival bias, uncertainty in the timing of disease diagnosis, and nonlinear disease progression trajectories. MCI presumably is diagnosed in the middle of an ongoing, accumulating, pathologic process, which introduces problems if unmeasured factors influence both MCI onset and rate of disease progression. For example, among newly diagnosed MCI cases, APOEε4 status may be associated with unmeasured causes of MCI, even though it is independent from these causes in the population. As a consequence, in a study in which the sample is selected conditional on MCI diagnosis, the unmeasured factors will confound analyses of the association of disease progression and APOEε4 status (“selection bias”). Survival bias is often induced by selecting primarily elderly participants, as persons who did not die but survived into the study are more likely to have a lower frequency of risk factors associated with cognitive decline. Uncertainty in the timing of disease diagnosis is caused by the fact that cognitive functioning in patients with incipient MCI frequently fluctuates, for example due to intermittent periods of depression or medication changes. Thus, the threshold for MCI diagnosis is not clearly demarcated, and as a result, individuals may be prematurely diagnosed with MCI during brief periods of impaired functioning that subsequently remit, or may remain undiagnosed because they were assessed on a particularly lucid day. The consequence of premature diagnosis in turn would be that the apparent decline trajectory post diagnosis appears flatter than the true decline trajectory and that the time to dementia appears longer. In contrast, the consequence of delayed diagnosis would be that the apparent decline trajectory post diagnosis appears steeper than the true decline trajectory and that the time to dementia appears shorter. Nonlinear progression of cognitive impairment can occur if compensatory or resilience processes buffer functional consequences of neurologic damage in early disease. Neurologic damage may accumulate until the brain loses resilience to further damage and decline. The decline trajectory will be relatively flat in early stages and then suddenly collapse. Alternatively, the trajectory may flatten at the end stages of disease, when there is little function remaining to lose. When the decline trajectory is nonlinear, variables associated with where in the trajectory an individual is first observed will tend to predict subsequent rate of change. Modeling transition to dementia as a function of prior cognition without attempting to measure rate of decline directly can help circumvent this problem.
Although -due to the low specificity and sensitivity- SORL1 and APOE are probably not suitable as diagnostic markers, they may be targets for prevention and treatment. However, several issues must be resolved before development of a drug based on these genes can be considered. First, for both genes, it is necessary to clarify the exact mechanisms through which they increase risk of cognitive impairment. Second, it is necessary to further characterize the molecular pathways in which they are involved or with which they interact. Clarification of the biological functions, risk-factor activities and pathways of SORL1 and APOE will help to understand their role in cognitive impairment and dementia and can provide targets for effective intervention. Third, for SORL1, the precise putative genetic variants have to be identified. The reported variants are nonfunctional and do not affect coding sequence or splicing. Fourth, the additional risk factor genes need to be known. The accurate risks associated with each gene involved can only be estimated when all putative and protective genetic variants are known. Finally, it has to be determined whether SORL1 and APOE are unique to cognitive impairment in LOAD, or are shared by other diseases such as Dementia with Lewy Bodies, Parkinson’s disease or depression. Lewy body inclusions and Lewy neurites, the key pathological hallmarks of dementia with Lewy Bodies and Parkinson’s disease, are a frequent coexistent pathologic change observed in autopsy-confirmed LOAD.
The issues posed above raise considerable challenges for investigators aiming to clarify the genetic complexity of cognitive impairment. Only when these issues are better understood, development of preventive and treatment strategies based on genetic risk factors, including SORL1 and APOE, can be considered. Nevertheless, the major advantage of genetic studies is the ability to overcome limitations of classic epidemiological techniques, in particular residual confounding and reverse causation. Among the various genetic epidemiologic approaches, candidate gene studies with subsequent confirmation in independent datasets and functional analyses, is probably the method with the highest statistical power. If correctly conducted and carefully interpreted, the merge of modern functional genomics with large-scale studies of genetically at-risk samples and sophisticated statistical algorithms can be a powerful tool for identification of genes, and therefore biomarkers, associated with common complex diseases such as cognitive impairment.
Acknowledgments
This work was supported by federal grants from the National Institute on Aging of the National Institutes of Health (P01AG07232, R37AG15473, P50 AG08702) and by grants from the Alzheimer Association, the Blanchette Hooker Rockefeller Fund, the Robertson Gift from the Banbury Fund and the Merrill Lynch Foundation. Dr. Reitz was further supported by a Paul B. Beeson Career Development Award (K23AG034550).
References
- 1.Ferri CP, Prince M, Brayne C, Brodaty H, Fratiglioni L, Ganguli M, Hall K, Hasegawa K, Hendrie H, Huang Y, Jorm A, Mathers C, Menezes PR, Rimmer E, Scazufca M. Global prevalence of dementia: a Delphi consensus study. Lancet. 2005;366:2112–2117. doi: 10.1016/S0140-6736(05)67889-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fratiglioni L, De Ronchi D, Aguero-Torres H. Worldwide prevalence and incidence of dementia. Drugs Aging. 1999;15:365–375. doi: 10.2165/00002512-199915050-00004. [DOI] [PubMed] [Google Scholar]
- 3.Brookmeyer R, Gray S, Kawas C. Projections of Alzheimer’s disease in the United States and the public health impact of delaying disease onset. Am J Public Health. 1998;88:1337–1342. doi: 10.2105/ajph.88.9.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer’s disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 5.Lopez OL, Swihart AA, Becker JT, Reinmuth OM, Reynolds CF, 3rd, Rezek DL, Daly FL., 3rd Reliability of NINCDS-ADRDA clinical criteria for the diagnosis of Alzheimer’s disease. Neurology. 1990;40:1517–1522. doi: 10.1212/wnl.40.10.1517. [DOI] [PubMed] [Google Scholar]
- 6.Kukull WA, Larson EB, Reifler BV, Lampe TH, Yerby M, Hughes J. Interrater reliability of Alzheimer’s disease diagnosis. Neurology. 1990;40:257–260. doi: 10.1212/wnl.40.2.257. [DOI] [PubMed] [Google Scholar]
- 7.Schofield PW, Tang M, Marder K, Bell K, Dooneief G, Lantigua R, Wilder D, Gurland B, Stern Y, Mayeux R. Consistency of clinical diagnosis in a community-based longitudinal study of dementia and Alzheimer’s disease. Neurology. 1995;45:2159–2164. doi: 10.1212/wnl.45.12.2159. [DOI] [PubMed] [Google Scholar]
- 8.Morris JC, McKeel DW, Jr, Fulling K, Torack RM, Berg L. Validation of clinical diagnostic criteria for Alzheimer’s disease. Ann Neurol. 1988;24:17–22. doi: 10.1002/ana.410240105. [DOI] [PubMed] [Google Scholar]
- 9.Burns A, Luthert P, Levy R, Jacoby R, Lantos P. Accuracy of clinical diagnosis of Alzheimer’s disease. Bmj. 1990;301:1026. doi: 10.1136/bmj.301.6759.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petersen RC. Mild cognitive impairment as a diagnostic entity. J Intern Med. 2004;256:183–194. doi: 10.1111/j.1365-2796.2004.01388.x. [DOI] [PubMed] [Google Scholar]
- 11.Petersen RC, Smith GE, Waring SC, Ivnik RJ, Tangalos EG, Kokmen E. Mild cognitive impairment: clinical characterization and outcome. Arch Neurol. 1999;56:303–308. doi: 10.1001/archneur.56.3.303. [DOI] [PubMed] [Google Scholar]
- 12.Petersen RC, Stevens JC, Ganguli M, Tangalos EG, Cummings JL, DeKosky ST. Practice parameter: early detection of dementia: mild cognitive impairment (an evidence-based review). Report of the Quality Standards Subcommittee of the American Academy of Neurology. Neurology. 2001;56:1133–1142. doi: 10.1212/wnl.56.9.1133. [DOI] [PubMed] [Google Scholar]
- 13.Petersen RC, Doody R, Kurz A, Mohs RC, Morris JC, Rabins PV, Ritchie K, Rossor M, Thal L, Winblad B. Current concepts in mild cognitive impairment. Arch Neurol. 2001;58:1985–1992. doi: 10.1001/archneur.58.12.1985. [DOI] [PubMed] [Google Scholar]
- 14.Larrieu S, Letenneur L, Orgogozo JM, Fabrigoule C, Amieva H, Le Carret N, Barberger-Gateau P, Dartigues JF. Incidence and outcome of mild cognitive impairment in a population-based prospective cohort. Neurology. 2002;59:1594–1599. doi: 10.1212/01.wnl.0000034176.07159.f8. [DOI] [PubMed] [Google Scholar]
- 15.Palmer K, Wang HX, Backman L, Winblad B, Fratiglioni L. Differential evolution of cognitive impairment in nondemented older persons: results from the Kungsholmen Project. Am J Psychiatry. 2002;159:436–442. doi: 10.1176/appi.ajp.159.3.436. [DOI] [PubMed] [Google Scholar]
- 16.Meyer JM, Breitner JC. Multiple threshold model for the onset of Alzheimer’s disease in the NAS-NRC twin panel. Am J Med Genet. 1998;81:92–97. doi: 10.1002/(sici)1096-8628(19980207)81:1<92::aid-ajmg16>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 17.Breitner JC, Wyse BW, Anthony JC, Welsh-Bohmer KA, Steffens DC, Norton MC, Tschanz JT, Plassman BL, Meyer MR, Skoog I, Khachaturian A. APOE-epsilon4 count predicts age when prevalence of AD increases, then declines: the Cache County Study. Neurology. 1999;53:321–331. doi: 10.1212/wnl.53.2.321. [DOI] [PubMed] [Google Scholar]
- 18.Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 19.Gomez-Isla T, West HL, Rebeck GW, Harr SD, Growdon JH, Locascio JJ, Perls TT, Lipsitz LA, Hyman BT. Clinical and pathological correlates of apolipoprotein E epsilon 4 in Alzheimer’s disease. Ann Neurol. 1996;39:62–70. doi: 10.1002/ana.410390110. [DOI] [PubMed] [Google Scholar]
- 20.Holmes C, Levy R, McLoughlin DM, Powell JF, Lovestone S. Apolipoprotein E: non-cognitive symptoms and cognitive decline in late onset Alzheimer’s disease. J Neurol Neurosurg Psychiatry. 1996;61:580–583. doi: 10.1136/jnnp.61.6.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hyman BT, Gomez-Isla T, Rebeck GW, Briggs M, Chung H, West HL, Greenberg S, Mui S, Nichols S, Wallace R, Growdon JH. Epidemiological, clinical, and neuropathological study of apolipoprotein E genotype in Alzheimer’s disease. Ann N Y Acad Sci. 1996;802:1–5. doi: 10.1111/j.1749-6632.1996.tb32592.x. [DOI] [PubMed] [Google Scholar]
- 22.Kurz A, Altland K, Lautenschlager N, Zimmer R, Busch R, Gerundt I, Lauter H, Muller U. Apolipoprotein E type 4 allele and Alzheimer’s disease: effect on age at onset and relative risk in different age groups. J Neurol. 1996;243:452–456. doi: 10.1007/BF00900498. [DOI] [PubMed] [Google Scholar]
- 23.Murman DL, Foster NL, Kilgore SP, McDonagh CA, Fink JK. Apolipoprotein E and Alzheimer’s disease: strength of association is related to age at onset. Dementia. 1996;7:251–255. doi: 10.1159/000106888. [DOI] [PubMed] [Google Scholar]
- 24.Poirier J, Davignon J, Bouthillier D, Kogan S, Bertrand P, Gauthier S. Apolipoprotein E polymorphism and Alzheimer’s disease. Lancet. 1993;342:697–699. doi: 10.1016/0140-6736(93)91705-q. [DOI] [PubMed] [Google Scholar]
- 25.Roses AD. Alzheimer’s disease: the genetics of risk. Hosp Pract (Minneap) 1997;32:51–55. 58–63, 67–59. doi: 10.1080/21548331.1997.11443525. [DOI] [PubMed] [Google Scholar]
- 26.Tang MX, Maestre G, Tsai WY, Liu XH, Feng L, Chung WY, Chun M, Schofield P, Stern Y, Tycko B, Mayeux R. Relative risk of Alzheimer disease and age-at-onset distributions, based on APOE genotypes among elderly African Americans, Caucasians, and Hispanics in New York City. Am J Hum Genet. 1996;58:574–584. [PMC free article] [PubMed] [Google Scholar]
- 27.Barabash A, Marcos A, Ancin I, Vazquez-Alvarez B, de Ugarte C, Gil P, Fernandez C, Encinas M, Lopez-Ibor JJ, Cabranes JA. APOE, ACT and CHRNA7 genes in the conversion from amnestic mild cognitive impairment to Alzheimer’s disease. Neurobiol Aging. 2007 doi: 10.1016/j.neurobiolaging.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 28.Petersen RC, Smith GE, Ivnik RJ, Tangalos EG, Schaid DJ, Thibodeau SN, Kokmen E, Waring SC, Kurland LT. Apolipoprotein E status as a predictor of the development of Alzheimer’s disease in memory-impaired individuals. JAMA. 1995;273:1274–1278. [PubMed] [Google Scholar]
- 29.Sasaki M, Kodama C, Hidaka S, Yamashita F, Kinoshita T, Nemoto K, Ikejima C, Asada T. Prevalence of four subtypes of mild cognitive impairment and APOE in a Japanese community. Int J Geriatr Psychiatry. 2009 doi: 10.1002/gps.2234. [DOI] [PubMed] [Google Scholar]
- 30.Tyas SL, Salazar JC, Snowdon DA, Desrosiers MF, Riley KP, Mendiondo MS, Kryscio RJ. Transitions to mild cognitive impairments, dementia, and death: findings from the Nun Study. Am J Epidemiol. 2007;165:1231–1238. doi: 10.1093/aje/kwm085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blom ES, Giedraitis V, Zetterberg H, Fukumoto H, Blennow K, Hyman BT, Irizarry MC, Wahlund LO, Lannfelt L, Ingelsson M. Rapid progression from mild cognitive impairment to Alzheimer’s disease in subjects with elevated levels of tau in cerebrospinal fluid and the APOE epsilon4/epsilon4 genotype. Dement Geriatr Cogn Disord. 2009;27:458–464. doi: 10.1159/000216841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Devanand DP, Pelton GH, Zamora D, Liu X, Tabert MH, Goodkind M, Scarmeas N, Braun I, Stern Y, Mayeux R. Predictive utility of apolipoprotein E genotype for Alzheimer disease in outpatients with mild cognitive impairment. Arch Neurol. 2005;62:975–980. doi: 10.1001/archneur.62.6.975. [DOI] [PubMed] [Google Scholar]
- 33.Hamalainen A, Grau-Olivares M, Tervo S, Niskanen E, Pennanen C, Huuskonen J, Kivipelto M, Hanninen T, Tapiola M, Vanhanen M, Hallikainen M, Helkala EL, Nissinen A, Vanninen RL, Soininen H. Apolipoprotein E epsilon 4 allele is associated with increased atrophy in progressive mild cognitive impairment: a voxel-based morphometric study. Neurodegener Dis. 2008;5:186–189. doi: 10.1159/000113698. [DOI] [PubMed] [Google Scholar]
- 34.Hsiung GY, Sadovnick AD, Feldman H. Apolipoprotein E epsilon4 genotype as a risk factor for cognitive decline and dementia: data from the Canadian Study of Health and Aging. CMAJ. 2004;171:863–867. doi: 10.1503/cmaj.1031789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jack CR, Jr, Petersen RC, Xu YC, O’Brien PC, Smith GE, Ivnik RJ, Boeve BF, Waring SC, Tangalos EG, Kokmen E. Prediction of AD with MRI-based hippocampal volume in mild cognitive impairment. Neurology. 1999;52:1397–1403. doi: 10.1212/wnl.52.7.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramakers IH, Visser PJ, Aalten P, Bekers O, Sleegers K, van Broeckhoven CL, Jolles J, Verhey FR. The association between APOE genotype and memory dysfunction in subjects with mild cognitive impairment is related to age and Alzheimer pathology. Dement Geriatr Cogn Disord. 2008;26:101–108. doi: 10.1159/000144072. [DOI] [PubMed] [Google Scholar]
- 37.Tierney MC, Szalai JP, Snow WG, Fisher RH, Tsuda T, Chi H, McLachlan DR, St George-Hyslop PH. A prospective study of the clinical utility of ApoE genotype in the prediction of outcome in patients with memory impairment. Neurology. 1996;46:149–154. doi: 10.1212/wnl.46.1.149. [DOI] [PubMed] [Google Scholar]
- 38.Slooter AJ, Cruts M, Kalmijn S, Hofman A, Breteler MM, Van Broeckhoven C, van Duijn CM. Risk estimates of dementia by apolipoprotein E genotypes from a population-based incidence study: the Rotterdam Study. Arch Neurol. 1998;55:964–968. doi: 10.1001/archneur.55.7.964. [DOI] [PubMed] [Google Scholar]
- 39.Ashford JW, Mortimer JA. Non-familial Alzheimer’s disease is mainly due to genetic factors. J Alzheimers Dis. 2002;4:169–177. doi: 10.3233/jad-2002-4307. [DOI] [PubMed] [Google Scholar]
- 40.Strittmatter WJ, Weisgraber KH, Huang DY, Dong LM, Salvesen GS, Pericak-Vance M, Schmechel D, Saunders AM, Goldgaber D, Roses AD. Binding of human apolipoprotein E to synthetic amyloid beta peptide: isoform-specific effects and implications for late-onset Alzheimer disease. Proc Natl Acad Sci U S A. 1993;90:8098–8102. doi: 10.1073/pnas.90.17.8098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmechel DE, Saunders AM, Strittmatter WJ, Crain BJ, Hulette CM, Joo SH, Pericak-Vance MA, Goldgaber D, Roses AD. Increased amyloid beta-peptide deposition in cerebral cortex as a consequence of apolipoprotein E genotype in late-onset Alzheimer disease. Proc Natl Acad Sci U S A. 1993;90:9649–9653. doi: 10.1073/pnas.90.20.9649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rebeck GW, Reiter JS, Strickland DK, Hyman BT. Apolipoprotein E in sporadic Alzheimer’s disease: allelic variation and receptor interactions. Neuron. 1993;11:575–580. doi: 10.1016/0896-6273(93)90070-8. [DOI] [PubMed] [Google Scholar]
- 43.Bales KR, Verina T, Cummins DJ, Du Y, Dodel RC, Saura J, Fishman CE, DeLong CA, Piccardo P, Petegnief V, Ghetti B, Paul SM. Apolipoprotein E is essential for amyloid deposition in the APP(V717F) transgenic mouse model of Alzheimer’s disease. Proc Natl Acad Sci U S A. 1999;96:15233–15238. doi: 10.1073/pnas.96.26.15233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bales KR, Verina T, Dodel RC, Du Y, Altstiel L, Bender M, Hyslop P, Johnstone EM, Little SP, Cummins DJ, Piccardo P, Ghetti B, Paul SM. Lack of apolipoprotein E dramatically reduces amyloid beta-peptide deposition. Nat Genet. 1997;17:263–264. doi: 10.1038/ng1197-263. [DOI] [PubMed] [Google Scholar]
- 45.Kindy MS, Rader DJ. Reduction in amyloid A amyloid formation in apolipoprotein-E-deficient mice. Am J Pathol. 1998;152:1387–1395. [PMC free article] [PubMed] [Google Scholar]
- 46.Ma J, Yee A, Brewer HB, Jr, Das S, Potter H. Amyloid-associated proteins alpha 1-antichymotrypsin and apolipoprotein E promote assembly of Alzheimer beta-protein into filaments. Nature. 1994;372:92–94. doi: 10.1038/372092a0. [DOI] [PubMed] [Google Scholar]
- 47.Sanan DA, Weisgraber KH, Russell SJ, Mahley RW, Huang D, Saunders A, Schmechel D, Wisniewski T, Frangione B, Roses AD, et al. Apolipoprotein E associates with beta amyloid peptide of Alzheimer’s disease to form novel monofibrils. Isoform apoE4 associates more efficiently than apoE3. J Clin Invest. 1994;94:860–869. doi: 10.1172/JCI117407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Holtzman DM, Bales KR, Tenkova T, Fagan AM, Parsadanian M, Sartorius LJ, Mackey B, Olney J, McKeel D, Wozniak D, Paul SM. Apolipoprotein E isoform-dependent amyloid deposition and neuritic degeneration in a mouse model of Alzheimer’s disease. Proc Natl Acad Sci U S A. 2000;97:2892–2897. doi: 10.1073/pnas.050004797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holtzman DM, Bales KR, Wu S, Bhat P, Parsadanian M, Fagan AM, Chang LK, Sun Y, Paul SM. Expression of human apolipoprotein E reduces amyloid-beta deposition in a mouse model of Alzheimer’s disease. J Clin Invest. 1999;103:R15–R21. doi: 10.1172/JCI6179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sparks DL, Scheff SW, Hunsaker JC, 3rd, Liu H, Landers T, Gross DR. Induction of Alzheimer-like beta-amyloid immunoreactivity in the brains of rabbits with dietary cholesterol. Exp Neurol. 1994;126:88–94. doi: 10.1006/exnr.1994.1044. [DOI] [PubMed] [Google Scholar]
- 51.Refolo LM, Malester B, LaFrancois J, Bryant-Thomas T, Wang R, Tint GS, Sambamurti K, Duff K, Pappolla MA. Hypercholesterolemia accelerates the Alzheimer’s amyloid pathology in a transgenic mouse model. Neurobiol Dis. 2000;7:321–331. doi: 10.1006/nbdi.2000.0304. [DOI] [PubMed] [Google Scholar]
- 52.Bodovitz S, Klein WL. Cholesterol modulates alpha-secretase cleavage of amyloid precursor protein. J Biol Chem. 1996;271:4436–4440. doi: 10.1074/jbc.271.8.4436. [DOI] [PubMed] [Google Scholar]
- 53.Howland DS, Trusko SP, Savage MJ, Reaume AG, Lang DM, Hirsch JD, Maeda N, Siman R, Greenberg BD, Scott RW, Flood DG. Modulation of secreted beta-amyloid precursor protein and amyloid beta-peptide in brain by cholesterol. J Biol Chem. 1998;273:16576–16582. doi: 10.1074/jbc.273.26.16576. [DOI] [PubMed] [Google Scholar]
- 54.Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, Myers RH, Pericak-Vance MA, Risch N, van Duijn CM. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. Jama. 1997;278:1349–1356. [PubMed] [Google Scholar]
- 55.Gozal D, Capdevila OS, Kheirandish-Gozal L, Crabtree VM. APOE epsilon 4 allele, cognitive dysfunction, and obstructive sleep apnea in children. Neurology. 2007;69:243–249. doi: 10.1212/01.wnl.0000265818.88703.83. [DOI] [PubMed] [Google Scholar]
- 56.Caselli RJ, Graff-Radford NR, Reiman EM, Weaver A, Osborne D, Lucas J, Uecker A, Thibodeau SN. Preclinical memory decline in cognitively normal apolipoprotein E-epsilon4 homozygotes. Neurology. 1999;53:201–207. doi: 10.1212/wnl.53.1.201. [DOI] [PubMed] [Google Scholar]
- 57.Flory JD, Manuck SB, Ferrell RE, Ryan CM, Muldoon MF. Memory performance and the apolipoprotein E polymorphism in a community sample of middle-aged adults. Am J Med Genet. 2000;96:707–711. doi: 10.1002/1096-8628(20001204)96:6<707::aid-ajmg1>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 58.Liu F, Pardo LM, Schuur M, Sanchez-Juan P, Isaacs A, Sleegers K, de Koning I, Zorkoltseva IV, Axenovich TI, Witteman JC, Janssens AC, van Swieten JC, Aulchenko YS, Oostra BA, van Duijn CM. The apolipoprotein E geneand its age-specific effects on cognitive function. Neurobiol Aging. 2008 doi: 10.1016/j.neurobiolaging.2008.09.015. [DOI] [PubMed] [Google Scholar]
- 59.Schultz MR, Lyons MJ, Franz CE, Grant MD, Boake C, Jacobson KC, Xian H, Schellenberg GD, Eisen SA, Kremen WS. Apolipoprotein E genotype and memory in the sixth decade of life. Neurology. 2008;70:1771–1777. doi: 10.1212/01.wnl.0000286941.74372.cc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wehling E, Lundervold AJ, Standnes B, Gjerstad L, Reinvang I. APOE status and its association to learning and memory performance in middle aged and older Norwegians seeking assessment for memory deficits. Behav Brain Funct. 2007;3:57. doi: 10.1186/1744-9081-3-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mak YT, Chiu H, Woo J, Kay R, Chan YS, Hui E, Sze KH, Lum C, Kwok T, Pang CP. Apolipoprotein E genotype and Alzheimer’s disease in Hong Kong elderly Chinese. Neurology. 1996;46(1):146–149. doi: 10.1212/wnl.46.1.146. [DOI] [PubMed] [Google Scholar]
- 62.Hallman DM, Boerwinkle E, Saha N, Sandholzer C, Menzel HJ, Csazar A, Utermann G. The apolipoprotein E polymorphism: a comparison of allele frequencies and effects in nine populations. Am J Hum Genet. 1991;49:338–349. [PMC free article] [PubMed] [Google Scholar]
- 63.do Couto FS, de Mendonca A, Garcia C, Rocha L, Lechner MC. Age of onset in patients with Alzheimer’s disease with different apoE genotypes. J Neurol Neurosurg Psychiatry. 1998;64:817. doi: 10.1136/jnnp.64.6.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dal Forno G, Rasmusson DX, Brandt J, Carson KA, Brookmeyer R, Troncoso J, Kawas CH. Apolipoprotein E genotype and rate of decline in probable Alzheimer’s disease. Arch Neurol. 1996;53:345–350. doi: 10.1001/archneur.1996.00550040085017. [DOI] [PubMed] [Google Scholar]
- 65.Welsh-Bohmer KA, Ostbye T, Sanders L, Pieper CF, Hayden KM, Tschanz JT, Norton MCFTCCSG. Neuropsychological performance in advanced age: Influences of Demographic factors and Apolipoprotein E: Findings from the Cache County Memory Study. Clin Neuropsychol. 2008:1–23. doi: 10.1080/13854040801894730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Salo A, Ylikoski R, Verkkoniemi A, Polvikoski T, Juva K, Rastas S, Kontula K, Kainulainen K, Niinisto L, Notkola IL, Sulkava R. Does apolipoprotein E influence learning and memory in the nondemented oldest old? Int Psychogeriatr. 2001;13:451–459. doi: 10.1017/s1041610201007864. [DOI] [PubMed] [Google Scholar]
- 67.Murphy GM, Jr, Taylor J, Kraemer HC, Yesavage J, Tinklenberg JR. No association between apolipoprotein E epsilon 4 allele and rate of decline in Alzheimer’s disease. Am J Psychiatry. 1997;154:603–608. doi: 10.1176/ajp.154.5.603. [DOI] [PubMed] [Google Scholar]
- 68.Cosentino S, Scarmeas N, Helzner E, Glymour MM, Brandt J, Albert M, Blacker D, Stern Y. APOE epsilon 4 allele predicts faster cognitive decline in mild Alzheimer disease. Neurology. 2008;70:1842–1849. doi: 10.1212/01.wnl.0000304038.37421.cc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hirono N, Hashimoto M, Yasuda M, Kazui H, Mori E. Accelerated memory decline in Alzheimer’s disease with apolipoprotein epsilon4 allele. J Neuropsychiatry Clin Neurosci. 2003;15:354–358. doi: 10.1176/jnp.15.3.354. [DOI] [PubMed] [Google Scholar]
- 70.Mayeux R, Small SA, Tang M, Tycko B, Stern Y. Memory performance in healthy elderly without Alzheimer’s disease: effects of time and apolipoprotein-E. Neurobiol Aging. 2001;22:683–689. doi: 10.1016/s0197-4580(01)00223-8. [DOI] [PubMed] [Google Scholar]
- 71.Wilson RS, Bienias JL, Berry-Kravis E, Evans DA, Bennett DA. The apolipoprotein E epsilon 2 allele and decline in episodic memory. J Neurol Neurosurg Psychiatry. 2002;73:672–677. doi: 10.1136/jnnp.73.6.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lehmann DJ, Refsum H, Nurk E, Warden DR, Tell GS, Vollset SE, Engedal K, Nygaard HA, Smith AD. Apolipoprotein E epsilon4 and impaired episodic memory in community-dwelling elderly people: a marked sex difference. The Hordaland Health Study. J Neurol Neurosurg Psychiatry. 2006;77:902–908. doi: 10.1136/jnnp.2005.077818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bondi MW, Salmon DP, Monsch AU, Galasko D, Butters N, Klauber MR, Thal LJ, Saitoh T. Episodic memory changes are associated with the APOE-epsilon 4 allele in nondemented older adults. Neurology. 1995;45:2203–2206. doi: 10.1212/wnl.45.12.2203. [DOI] [PubMed] [Google Scholar]
- 74.Dik MG, Jonker C, Comijs HC, Bouter LM, Twisk JW, van Kamp GJ, Deeg DJ. Memory complaints and APOE-epsilon4 accelerate cognitive decline in cognitively normal elderly. Neurology. 2001;57:2217–2222. doi: 10.1212/wnl.57.12.2217. [DOI] [PubMed] [Google Scholar]
- 75.Reynolds CA, Prince JA, Feuk L, Brookes AJ, Gatz M, Pedersen NL. Longitudinal memory performance during normal aging: twin association models of APOE and other Alzheimer candidate genes. Behav Genet. 2006;36:185–194. doi: 10.1007/s10519-005-9027-6. [DOI] [PubMed] [Google Scholar]
- 76.Saunders AM, Hulette O, Welsh-Bohmer KA, Schmechel DE, Crain B, Burke JR, Alberts MJ, Strittmatter WJ, Breitner JC, Rosenberg C. Specificity, sensitivity, and predictive value of apolipoprotein-E genotyping for sporadic Alzheimer’s disease. Lancet. 1996;348:90–93. doi: 10.1016/s0140-6736(96)01251-2. [DOI] [PubMed] [Google Scholar]
- 77.Mayeux R, Saunders AM, Shea S, Mirra S, Evans D, Roses AD, Hyman BT, Crain B, Tang MX, Phelps CH. Utility of the apolipoprotein E genotype in the diagnosis of Alzheimer’s disease. Alzheimer’s Disease Centers Consortium on Apolipoprotein E and Alzheimer’s Disease. N Engl J Med. 1998;338:506–511. doi: 10.1056/NEJM199802193380804. [DOI] [PubMed] [Google Scholar]
- 78.Roses AD. Apolipoprotein E genotyping in the differential diagnosis, not prediction, of Alzheimer’s disease. Ann Neurol. 1995;38:6–14. doi: 10.1002/ana.410380105. [DOI] [PubMed] [Google Scholar]
- 79.Slooter AJ, Breteler MB, Ott A, Van Broeckhoven C, van Duijn CM. APOE genotyping in differential diagnosis of Alzheimer’s disease. Lancet. 1996;348:334. doi: 10.1016/s0140-6736(05)64501-1. [DOI] [PubMed] [Google Scholar]
- 80.Kakulas BA, Wilton SD, Fabian VA, Jones TM. Apolipoprotein-E genotyping in diagnosis of Alzheimer’s disease. Lancet. 1996;348:483. doi: 10.1016/s0140-6736(05)64588-6. [DOI] [PubMed] [Google Scholar]
- 81.Edbauer D, Winkler E, Regula JT, Pesold B, Steiner H, Haass C. Reconstitution of gamma-secretase activity. Nat Cell Biol. 2003;5:486–488. doi: 10.1038/ncb960. [DOI] [PubMed] [Google Scholar]
- 82.Cobbold C, Monaco AP, Sivaprasadarao A, Ponnambalam S. Aberrant trafficking of transmembrane proteins in human disease. Trends Cell Biol. 2003;13:639–647. doi: 10.1016/j.tcb.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 83.Harter C, Reinhard C. The secretory pathway from history to the state of the art. Subcell Biochem. 2000;34:1–38. doi: 10.1007/0-306-46824-7_1. [DOI] [PubMed] [Google Scholar]
- 84.Le Borgne R, Hoflack B. Protein transport from the secretory to the endocytic pathway in mammalian cells. Biochim Biophys Acta. 1998;1404:195–209. doi: 10.1016/s0167-4889(98)00057-3. [DOI] [PubMed] [Google Scholar]
- 85.Chen WJ, Goldstein JL, Brown MS. NPXY, a sequence often found in cytoplasmic tails, is required for coated pit-mediated internalization of the low density lipoprotein receptor. J Biol Chem. 1990;265:3116–3123. [PubMed] [Google Scholar]
- 86.He X, Li F, Chang WP, Tang J. GGA proteins mediate the recycling pathway of memapsin 2 (BACE) J Biol Chem. 2005;280:11696–11703. doi: 10.1074/jbc.M411296200. [DOI] [PubMed] [Google Scholar]
- 87.Small SA, Gandy S. Sorting through the cell biology of Alzheimer’s disease: intracellular pathways to pathogenesis. Neuron. 2006;52:15–31. doi: 10.1016/j.neuron.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Traub LM. Common principles in clathrin-mediated sorting at the Golgi and the plasma membrane. Biochim Biophys Acta. 2005;1744:415–437. doi: 10.1016/j.bbamcr.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 89.Rogaeva E, Meng Y, Lee JH, Gu Y, Kawarai T, Zou F, Katayama T, Baldwin CT, Cheng R, Hasegawa H, Chen F, Shibata N, Lunetta KL, Pardossi-Piquard R, Bohm C, Wakutani Y, Cupples LA, Cuenco KT, Green RC, Pinessi L, Rainero I, Sorbi S, Bruni A, Duara R, Friedland RP, Inzelberg R, Hampe W, Bujo H, Song YQ, Andersen OM, Willnow TE, Graff-Radford N, Petersen RC, Dickson D, Der SD, Fraser PE, Schmitt-Ulms G, Younkin S, Mayeux R, Farrer LA, St George-Hyslop P. The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat Genet. 2007;39:168–177. doi: 10.1038/ng1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Andersen OM, Reiche J, Schmidt V, Gotthardt M, Spoelgen R, Behlke J, von Arnim CA, Breiderhoff T, Jansen P, Wu X, Bales KR, Cappai R, Masters CL, Gliemann J, Mufson EJ, Hyman BT, Paul SM, Nykjaer A, Willnow TE. Neuronal sorting protein-related receptor sorLA/LR11 regulates processing of the amyloid precursor protein. Proc Natl Acad Sci U S A. 2005;102:13461–13466. doi: 10.1073/pnas.0503689102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Andersen OM, Schmidt V, Spoelgen R, Gliemann J, Behlke J, Galatis D, McKinstry WJ, Parker MW, Masters CL, Hyman BT, Cappai R, Willnow TE. Molecular dissection of the interaction between amyloid precursor protein and its neuronal trafficking receptor SorLA/LR11. Biochemistry. 2006;45:2618–2628. doi: 10.1021/bi052120v. [DOI] [PubMed] [Google Scholar]
- 92.Clewes O, Fahey MS, Tyler SJ, Watson JJ, Seok H, Catania C, Cho K, Dawbarn D, Allen SJ. Human ProNGF: biological effects and binding profiles at TrkA, P75NTR and sortilin. J Neurochem. 2008;107:1124–1135. doi: 10.1111/j.1471-4159.2008.05698.x. [DOI] [PubMed] [Google Scholar]
- 93.Nykjaer A, Lee R, Teng KK, Jansen P, Madsen P, Nielsen MS, Jacobsen C, Kliemannel M, Schwarz E, Willnow TE, Hempstead BL, Petersen CM. Sortilin is essential for proNGF-induced neuronal cell death. Nature. 2004;427:843–848. doi: 10.1038/nature02319. [DOI] [PubMed] [Google Scholar]
- 94.Bettens K, Brouwers N, Engelborghs S, De Deyn PP, Van Broeckhoven C, Sleegers K. SORL1 is genetically associated with increased risk for late-onset Alzheimer disease in the Belgian population. Hum Mutat. 2008;29:769–770. doi: 10.1002/humu.20725. [DOI] [PubMed] [Google Scholar]
- 95.Lee JH, Cheng R, Honig LS, Vonsattel JP, Clark L, Mayeux R. Association between genetic variants in SORL1 and autopsy-confirmed Alzheimer disease. Neurology. 2008;70:887–889. doi: 10.1212/01.wnl.0000280581.39755.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lee JH, Cheng R, Schupf N, Manly J, Lantigua R, Stern Y, Rogaeva E, Wakutani Y, Farrer L, St George-Hyslop P, Mayeux R. The association between genetic variants in SORL1 and Alzheimer disease in an urban, multiethnic, community-based cohort. Arch Neurol. 2007;64:501–506. doi: 10.1001/archneur.64.4.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Meng Y, Lee JH, Cheng R, St George-Hyslop P, Mayeux R, Farrer LA. Association between SORL1 and Alzheimer’s disease in a genome-wide study. Neuroreport. 2007;18:1761–1764. doi: 10.1097/WNR.0b013e3282f13e7a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Seshadri S, DeStefano AL, Au R, Massaro JM, Beiser AS, Kelly-Hayes M, Kase CS, D’Agostino RB, Sr, Decarli C, Atwood LD, Wolf PA. Genetic correlates of brain aging on MRI and cognitive test measures: a genome-wide association and linkage analysis in the Framingham Study. BMC Med Genet. 2007;8(Suppl 1):S15. doi: 10.1186/1471-2350-8-S1-S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tan EK, Lee J, Chen CP, Teo YY, Zhao Y, Lee WL. SORL1 haplotypes modulate risk of Alzheimer’s disease in Chinese. Neurobiol Aging. 2007 doi: 10.1016/j.neurobiolaging.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 100.Kolsch H, Jessen F, Wiltfang J, Lewczuk P, Dichgans M, Teipel SJ, Kornhuber J, Frolich L, Heuser I, Peters O, Wiese B, Kaduszkiewicz H, van den Bussche H, Hull M, Kurz A, Ruther E, Henn FA, Maier W. Association of SORL1 gene variants with Alzheimer’s disease. Brain Res. 2009 doi: 10.1016/j.brainres.2009.01.044. [DOI] [PubMed] [Google Scholar]
- 101.Li H, Wetten S, Li L, St Jean PL, Upmanyu R, Surh L, Hosford D, Barnes MR, Briley JD, Borrie M, Coletta N, Delisle R, Dhalla D, Ehm MG, Feldman HH, Fornazzari L, Gauthier S, Goodgame N, Guzman D, Hammond S, Hollingworth P, Hsiung GY, Johnson J, Kelly DD, Keren R, Kertesz A, King KS, Lovestone S, Loy-English I, Matthews PM, Owen MJ, Plumpton M, Pryse-Phillips W, Prinjha RK, Richardson JC, Saunders A, Slater AJ, St George-Hyslop PH, Stinnett SW, Swartz JE, Taylor RL, Wherrett J, Williams J, Yarnall DP, Gibson RA, Irizarry MC, Middleton LT, Roses AD. Candidate single-nucleotide polymorphisms from a genomewide association study of Alzheimer disease. Arch Neurol. 2008;65:45–53. doi: 10.1001/archneurol.2007.3. [DOI] [PubMed] [Google Scholar]
- 102.Li Y, Rowland C, Catanese J, Morris J, Lovestone S, O’Donovan MC, Goate A, Owen M, Williams J, Grupe A. SORL1 variants and risk of late-onset Alzheimer’s disease. Neurobiol Dis. 2008;29:293–296. doi: 10.1016/j.nbd.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Webster JA, Myers AJ, Pearson JV, Craig DW, Hu-Lince D, Coon KD, Zismann VL, Beach T, Leung D, Bryden L, Halperin RF, Marlowe L, Kaleem M, Huentelman MJ, Joshipura K, Walker D, Heward CB, Ravid R, Rogers J, Papassotiropoulos A, Hardy J, Reiman EM, Stephan DA. Sorl1 as an Alzheimer’s disease predisposition gene? Neurodegener Dis. 2008;5:60–64. doi: 10.1159/000110789. [DOI] [PubMed] [Google Scholar]
- 104.Houlihan LM, Harris SE, Luciano M, Gow AJ, Starr JM, Visscher PM, Deary IJ. Replication study of candidate genes for cognitive abilities: the Lothian Birth Cohort 1936. Genes Brain Behav. 2009;8:238–247. doi: 10.1111/j.1601-183X.2008.00470.x. [DOI] [PubMed] [Google Scholar]
- 105.Minster RL, DeKosky ST, Kamboh MI. No association of SORL1 SNPs with Alzheimer’s disease. Neurosci Lett. 2008;440:190–192. doi: 10.1016/j.neulet.2008.05.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shibata N, Ohnuma T, Baba H, Higashi S, Nishioka K, Arai H. Genetic association between SORL1 polymorphisms and Alzheimer’s disease in a Japanese population. Dement Geriatr Cogn Disord. 2008;26:161–164. doi: 10.1159/000149821. [DOI] [PubMed] [Google Scholar]
- 107.Lee JH, Shibata N, Cheng R, Mayeux R. Possible Association between SORL1 and Alzheimer Disease?. Reanalysing the Data of Shibata et al. Dement Geriatr Cogn Disord. 2008;26:482. doi: 10.1159/000167792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sager KL, Wuu J, Leurgans SE, Rees HD, Gearing M, Mufson EJ, Levey AI, Lah JJ. Neuronal LR11/sorLA expression is reduced in mild cognitive impairment. Ann Neurol. 2007;62:640–647. doi: 10.1002/ana.21190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet. 2007;39:17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- 110.Grupe A, Abraham R, Li Y, Rowland C, Hollingworth P, Morgan A, Jehu L, Segurado R, Stone D, Schadt E, Karnoub M, Nowotny P, Tacey K, Catanese J, Sninsky J, Brayne C, Rubinsztein D, Gill M, Lawlor B, Lovestone S, Holmans P, O’Donovan M, Morris JC, Thal L, Goate A, Owen MJ, Williams J. Evidence for novel susceptibility genes for late-onset Alzheimer’s disease from a genome-wide association study of putative functional variants. Hum Mol Genet. 2007;16:865–873. doi: 10.1093/hmg/ddm031. [DOI] [PubMed] [Google Scholar]
- 111.Holmans P, Hamshere M, Hollingworth P, Rice F, Tunstall N, Jones S, Moore P, Wavrant DeVrieze F, Myers A, Crook R, Compton D, Marshall H, Meyer D, Shears S, Booth J, Ramic D, Williams N, Norton N, Abraham R, Kehoe P, Williams H, Rudrasingham V, O’Donovan M, Jones L, Hardy J, Goate A, Lovestone S, Owen M, Williams J. Genome screen for loci influencing age at onset and rate of decline in late onset Alzheimer’s disease. Am J Med Genet B Neuropsychiatr Genet. 2005;135:24–32. doi: 10.1002/ajmg.b.30114. [DOI] [PubMed] [Google Scholar]
- 112.Liu F, Arias-Vasquez A, Sleegers K, Aulchenko YS, Kayser M, Sanchez-Juan P, Feng BJ, Bertoli-Avella AM, van Swieten J, Axenovich TI, Heutink P, van Broeckhoven C, Oostra BA, van Duijn CM. A genomewide screen for late-onset Alzheimer disease in a genetically isolated dutch population. Am J Hum Genet. 2007;81:17–31. doi: 10.1086/518720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Myers A, Wavrant De-Vrieze F, Holmans P, Hamshere M, Crook R, Compton D, Marshall H, Meyer D, Shears S, Booth J, Ramic D, Knowles H, Morris JC, Williams N, Norton N, Abraham R, Kehoe P, Williams H, Rudrasingham V, Rice F, Giles P, Tunstall N, Jones L, Lovestone S, Williams J, Owen MJ, Hardy J, Goate A. Full genome screen for Alzheimer disease: stage II analysis. AmJ Med Genet. 2002;114:235–244. doi: 10.1002/ajmg.10183. [DOI] [PubMed] [Google Scholar]
- 114.Dreses-Werringloer U, Lambert JC, Vingtdeux V, Zhao H, Vais H, Siebert A, Jain A, Koppel J, Rovelet-Lecrux A, Hannequin D, Pasquier F, Galimberti D, Scarpini E, Mann D, Lendon C, Campion D, Amouyel P, Davies P, Foskett JK, Campagne F, Marambaud P. A polymorphism in CALHM1 influences Ca2+ homeostasis, Abeta levels, and Alzheimer’s disease risk. Cell. 2008;133:1149–1161. doi: 10.1016/j.cell.2008.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Reiman EM, Webster JA, Myers AJ, Hardy J, Dunckley T, Zismann VL, Joshipura KD, Pearson JV, Hu-Lince D, Huentelman MJ, Craig DW, Coon KD, Liang WS, Herbert RH, Beach T, Rohrer KC, Zhao AS, Leung D, Bryden L, Marlowe L, Kaleem M, Mastroeni D, Grover A, Heward CB, Ravid R, Rogers J, Hutton ML, Melquist S, Petersen RC, Alexander GE, Caselli RJ, Kukull W, Papassotiropoulos A, Stephan DA. GAB2 alleles modify Alzheimer’s risk in APOE epsilon4 carriers. Neuron. 2007;54:713–720. doi: 10.1016/j.neuron.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.McIlroy SP, Crawford VL, Dynan KB, McGleenon BM, Vahidassr MD, Lawson JT, Passmore AP. Butyrylcholinesterase K variant is genetically associated with late onset Alzheimer’s disease in Northern Ireland. J Med Genet. 2000;37:182–185. doi: 10.1136/jmg.37.3.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bartels CF, Jensen FS, Lockridge O, van der Spek AF, Rubinstein HM, Lubrano T, La Du BN. DNA mutation associated with the human butyrylcholinesterase K-variant and its linkage to the atypical variant mutation and other polymorphic sites. Am J Hum Genet. 1992;50:1086–1103. [PMC free article] [PubMed] [Google Scholar]
- 118.De Ferrari GV, Papassotiropoulos A, Biechele T, Wavrant De-Vrieze F, Avila ME, Major MB, Myers A, Saez K, Henriquez JP, Zhao A, Wollmer MA, Nitsch RM, Hock C, Morris CM, Hardy J, Moon RT. Common genetic variation within the Low-Density Lipoprotein Receptor-Related Protein 6 and late-onset Alzheimer’s disease. Proc Natl Acad Sci U S A. 2007;104:9434–9439. doi: 10.1073/pnas.0603523104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Blacker D, Wilcox MA, Laird NM, Rodes L, Horvath SM, Go RC, Perry R, Watson B, Jr, Bassett SS, McInnis MG, Albert MS, Hyman BT, Tanzi RE. Alpha-2 macroglobulin is genetically associated with Alzheimer disease. Nat Genet. 1998;19:357–360. doi: 10.1038/1243. [DOI] [PubMed] [Google Scholar]
- 120.Giedraitis V, Hedlund M, Skoglund L, Blom E, Ingvast S, Brundin R, Lannfelt L, Glaser A. New Alzheimer’s disease locus on chromosome 8. J Med Genet. 2006;43:931–935. doi: 10.1136/jmg.2006.043000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Li Y, Nowotny P, Holmans P, Smemo S, Kauwe JS, Hinrichs AL, Tacey K, Doil L, van Luchene R, Garcia V, Rowland C, Schrodi S, Leong D, Gogic G, Chan J, Cravchik A, Ross D, Lau K, Kwok S, Chang SY, Catanese J, Sninsky J, White TJ, Hardy J, Powell J, Lovestone S, Morris JC, Thal L, Owen M, Williams J, Goate A, Grupe A. Association of late-onset Alzheimer’s disease with genetic variation in multiple members of the GAPD gene family. Proc Natl Acad Sci U S A. 2004;101:15688–15693. doi: 10.1073/pnas.0403535101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ozturk A, Desai PP, Minster RL, Dekosky ST, Kamboh MI. Three SNPs in the GSTO1, GSTO2 and PRSS11 genes on chromosome 10 are notassociated with age-at-onset of Alzheimer’s disease. Neurobiol Aging. 2005;26:1161–1165. doi: 10.1016/j.neurobiolaging.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 123.Bertram L, Hiltunen M, Parkinson M, Ingelsson M, Lange C, Ramasamy K, Mullin K, Menon R, Sampson AJ, Hsiao MY, Elliott KJ, Velicelebi G, Moscarillo T, Hyman BT, Wagner SL, Becker KD, Blacker D, Tanzi RE. Family-based association between Alzheimer’s disease and variants in UBQLN1. N Engl J Med. 2005;352:884–894. doi: 10.1056/NEJMoa042765. [DOI] [PubMed] [Google Scholar]
- 124.Blacker D, Bertram L, Saunders AJ, Moscarillo TJ, Albert MS, Wiener H, Perry RT, Collins JS, Harrell LE, Go RC, Mahoney A, Beaty T, Fallin MD, Avramopoulos D, Chase GA, Folstein MF, McInnis MG, Bassett SS, Doheny KJ, Pugh EW, Tanzi RE. Results of a high-resolution genome screen of 437 Alzheimer’s disease families. Hum Mol Genet. 2003;12:23–32. doi: 10.1093/hmg/ddg007. [DOI] [PubMed] [Google Scholar]
- 125.Farrer LA, Bowirrat A, Friedland RP, Waraska K, Korczyn AD, Baldwin CT. Identification of multiple loci for Alzheimer disease in a consanguineous Israeli-Arab community. Hum Mol Genet. 2003;12:415–422. doi: 10.1093/hmg/ddg037. [DOI] [PubMed] [Google Scholar]
- 126.Hahs DW, McCauley JL, Crunk AE, McFarland LL, Gaskell PC, Jiang L, Slifer SH, Vance JM, Scott WK, Welsh-Bohmer KA, Johnson SR, Jackson CE, Pericak-Vance MA, Haines JL. A genome-wide linkage analysis of dementia in the Amish. Am J Med Genet B Neuropsychiatr Genet. 2006;141:160–166. doi: 10.1002/ajmg.b.30257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lee JH, Cheng R, Santana V, Williamson J, Lantigua R, Medrano M, Arriaga A, Stern Y, Tycko B, Rogaeva E, Wakutani Y, Kawarai T, St George-Hyslop P, Mayeux R. Expanded genomewide scan implicates a novel locus at 3q28 among Caribbean hispanics with familial Alzheimer disease. Arch Neurol. 2006;63:1591–1598. doi: 10.1001/archneur.63.11.1591. [DOI] [PubMed] [Google Scholar]
- 128.Pericak-Vance MA, Grubber J, Bailey LR, Hedges D, West S, Santoro L, Kemmerer B, Hall JL, Saunders AM, Roses AD, Small GW, Scott WK, Conneally PM, Vance JM, Haines JL. Identification of novel genes in late-onset Alzheimer’s disease. Exp Gerontol. 2000;35:1343–1352. doi: 10.1016/s0531-5565(00)00196-0. [DOI] [PubMed] [Google Scholar]
- 129.Rademakers R, Cruts M, Sleegers K, Dermaut B, Theuns J, Aulchenko Y, Weckx S, De Pooter T, Van den Broeck M, Corsmit E, De Rijk P, Del-Favero J, van Swieten J, van Duijn CM, Van Broeckhoven C. Linkage and association studies identify a novel locus for Alzheimer disease at 7q36 in a Dutch population-based sample. Am J Hum Genet. 2005;77:643–652. doi: 10.1086/491749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Scott WK, Hauser ER, Schmechel DE, Welsh-Bohmer KA, Small GW, Roses AD, Saunders AM, Gilbert JR, Vance JM, Haines JL, Pericak-Vance MA. Ordered-subsets linkage analysis detects novel Alzheimer disease loci on chromosomes 2q34 and 15q22. Am J Hum Genet. 2003;73:1041–1051. doi: 10.1086/379083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Li M, Atmaca-Sonmez P, Othman M, Branham KE, Khanna R, Wade MS, Li Y, Liang L, Zareparsi S, Swaroop A, Abecasis GR. CFH haplotypes without the Y402H coding variant show strong association with susceptibility to age-related macular degeneration. Nat Genet. 2006;38:1049–1054. doi: 10.1038/ng1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sweet RA, Nimgaonkar VL, Devlin B, Jeste DV. Psychotic symptoms in Alzheimer disease: evidence for a distinct phenotype. Mol Psychiatry. 2003;8:383–392. doi: 10.1038/sj.mp.4001262. [DOI] [PubMed] [Google Scholar]
- 133.Wijsman EM, Daw EW, Yu CE, Payami H, Steinbart EJ, Nochlin D, Conlon EM, Bird TD, Schellenberg GD. Evidence for a novel late-onset Alzheimer disease locus on chromosome 19p13.2. Am J Hum Genet. 2004;75:398–409. doi: 10.1086/423393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Arosio B, Segat L, Milanese M, Galimberti L, Calabresi C, Zanetti M, Trabattoni D, Annoni G, Crovella S, Vergani C. PIN-1 promoter polymorphisms in mild cognitive impairment and susceptibility to Alzheimer’s disease: a preliminary report. Aging Clin Exp Res. 2007;19:406–409. doi: 10.1007/BF03324722. [DOI] [PubMed] [Google Scholar]
- 135.Barabash A, Marcos A, Ancin I, Vazquez-Alvarez B, de Ugarte C, Gil P, Fernandez C, Encinas M, Lopez-Ibor JJ, Cabranes JA. APOE, ACT and CHRNA7 genes in the conversion from amnestic mild cognitive impairment to Alzheimer’s disease. Neurobiol Aging. 2009;30:1254–1264. doi: 10.1016/j.neurobiolaging.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 136.Arosio B, Bergamaschini L, Galimberti L, La Porta C, Zanetti M, Calabresi C, Scarpini E, Annoni G, Vergani C. +10 T/C polymorphisms in the gene of transforming growth factor-beta1 are associated with neurodegeneration and its clinical evolution. Mech Ageing Dev. 2007;128:553–557. doi: 10.1016/j.mad.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 137.Chiappelli M, Borroni B, Archetti S, Calabrese E, Corsi MM, Franceschi M, Padovani A, Licastro F. VEGF gene and phenotype relation with Alzheimer’s disease and mild cognitive impairment. Rejuvenation Res. 2006;9:485–493. doi: 10.1089/rej.2006.9.485. [DOI] [PubMed] [Google Scholar]
- 138.Fernandez Del Pozo V, Alvarez Alvarez M, Fernandez Martinez M, Galdos Alcelay L, Gomez Busto F, Pena JA, Alfonso-Sanchez MA, Zarranz Imirizaldu JJ, de Pancorbo MM. Polymorphism in the cholesterol 24S-hydroxylase gene (CYP46A1) associated with the APOEpsilon3 allele increases the risk of Alzheimer’s disease and of mild cognitive impairment progressing to Alzheimer’s disease. Dement Geriatr Cogn Disord. 2006;21:81–87. doi: 10.1159/000090215. [DOI] [PubMed] [Google Scholar]
- 139.Sole-Padulles C, Bartres-Faz D, Junque C, Via M, Matarin M, Gonzalez-Perez E, Moral P, Moya A, Clemente IC. Poorer cognitive performance in humans with mild cognitive impairment carrying the T variant of the Glu/Asp NOS3 polymorphism. Neurosci Lett. 2004;358:5–8. doi: 10.1016/j.neulet.2003.12.044. [DOI] [PubMed] [Google Scholar]
- 140.Ertekin-Taner N, Ronald J, Feuk L, Prince J, Tucker M, Younkin L, Hella M, Jain S, Hackett A, Scanlin L, Kelly J, Kihiko-Ehman M, Neltner M, Hersh L, Kindy M, Markesbery W, Hutton M, de Andrade M, Petersen RC, Graff-Radford N, Estus S, Brookes AJ, Younkin SG. Elevated amyloid beta protein (Abeta42) and late onset Alzheimer’s disease are associated with single nucleotide polymorphisms in the urokinase-type plasminogen activator gene. Hum Mol Genet. 2005;14:447–460. doi: 10.1093/hmg/ddi041. [DOI] [PubMed] [Google Scholar]
- 141.Ertekin-Taner N, Allen M, Fadale D, Scanlin L, Younkin L, Petersen RC, Graff-Radford N, Younkin SG. Genetic variants in a haplotype block spanning IDE are significantly associated with plasma Abeta42 levels and risk for Alzheimer disease. Hum Mutat. 2004;23:334–342. doi: 10.1002/humu.20016. [DOI] [PubMed] [Google Scholar]
- 142.Farris W, Mansourian S, Leissring MA, Eckman EA, Bertram L, Eckman CB, Tanzi RE, Selkoe DJ. Partial loss-of-function mutations in insulin-degrading enzyme that induce diabetes also impair degradation of amyloid beta-protein. Am J Pathol. 2004;164:1425–1434. doi: 10.1016/s0002-9440(10)63229-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Daw EW, Heath SC, Wijsman EM. Multipoint oligogenic analysis of age-at-onset data with applications to Alzheimer disease pedigrees. Am J Hum Genet. 1999;64:839–851. doi: 10.1086/302276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Daw EW, Payami H, Nemens EJ, Nochlin D, Bird TD, Schellenberg GD, Wijsman EM. The number of trait loci in late-onset Alzheimer disease. Am J Hum Genet. 2000;66:196–204. doi: 10.1086/302710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Gottesman II, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- 146.Blangero J, Williams JT, Almasy L. Novel family-based approaches to genetic risk in thrombosis. J Thromb Haemost. 2003;1:1391–1397. doi: 10.1046/j.1538-7836.2003.00310.x. [DOI] [PubMed] [Google Scholar]
- 147.Lehtovirta M, Helisalmi S, Mannermaa A, Soininen H, Koivisto K, Ryynanen M, Riekkinen P., Sr Apolipoprotein E polymorphism and Alzheimer’s disease in eastern Finland. Neurosci Lett. 1995;185:13–15. doi: 10.1016/0304-3940(94)11213-3. [DOI] [PubMed] [Google Scholar]
- 148.Geschwind DH, Miller BL, DeCarli C, Carmelli D. Heritability of lobar brain volumes in twins supports genetic models of cerebral laterality and handedness. Proc Natl Acad Sci U S A. 2002;99:3176–3181. doi: 10.1073/pnas.052494999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Dik MG, Jonker C, Bouter LM, Geerlings MI, van Kamp GJ, Deeg DJ. APOE-epsilon4 is associated with memory decline in cognitively impaired elderly. Neurology. 2000;54:1492–1497. doi: 10.1212/wnl.54.7.1492. [DOI] [PubMed] [Google Scholar]
- 150.Small BJ, Graves AB, McEvoy CL, Crawford FC, Mullan M, Mortimer JA. Is APOE--epsilon4 a risk factor for cognitive impairment in normal aging? Neurology. 2000;54:2082–2088. doi: 10.1212/wnl.54.11.2082. [DOI] [PubMed] [Google Scholar]
- 151.Farlow MR, He Y, Tekin S, Xu J, Lane R, Charles HC. Impact of APOE in mild cognitive impairment. Neurology. 2004;63:1898–1901. doi: 10.1212/01.wnl.0000144279.21502.b7. [DOI] [PubMed] [Google Scholar]
- 152.Kleiman T, Zdanys K, Black B, Rightmer T, Grey M, Garman K, Macavoy M, Gelernter J, van Dyck C. Apolipoprotein E epsilon4 allele is unrelated to cognitive or functional decline in Alzheimer’s disease: retrospective and prospective analysis. Dement Geriatr Cogn Disord. 2006;22:73–82. doi: 10.1159/000093316. [DOI] [PubMed] [Google Scholar]
- 153.Caselli RJ, Reiman EM, Locke DE, Hutton ML, Hentz JG, Hoffman-Snyder C, Woodruff BK, Alexander GE, Osborne D. Cognitive domain decline in healthy apolipoprotein E epsilon4 homozygotes before the diagnosis of mild cognitive impairment. Arch Neurol. 2007;64:1306–1311. doi: 10.1001/archneur.64.9.1306. [DOI] [PubMed] [Google Scholar]
- 154.Aggarwal NT, Wilson RS, Beck TL, Bienias JL, Berry-Kravis E, Bennett DA. The apolipoprotein E epsilon4 allele and incident Alzheimer’s disease in persons with mild cognitive impairment. Neurocase. 2005;11:3–7. doi: 10.1080/13554790490903038. [DOI] [PubMed] [Google Scholar]
- 155.Tschanz JT, Welsh-Bohmer KA, Lyketsos CG, Corcoran C, Green RC, Hayden K, Norton MC, Zandi PP, Toone L, West NA, Breitner JC. Conversion to dementia from mild cognitive disorder: the Cache County Study. Neurology. 2006;67:229–234. doi: 10.1212/01.wnl.0000224748.48011.84. [DOI] [PubMed] [Google Scholar]
- 156.Fleisher AS, Sowell BB, Taylor C, Gamst AC, Petersen RC, Thal LJ. Clinical predictors of progression to Alzheimer disease in amnestic mild cognitive impairment. Neurology. 2007;68:1588–1595. doi: 10.1212/01.wnl.0000258542.58725.4c. [DOI] [PubMed] [Google Scholar]
- 157.van der Flier WM, Pijnenburg YA, Schoonenboom SN, Dik MG, Blankenstein MA, Scheltens P. Distribution of APOE genotypes in a memory clinic cohort. Dement Geriatr Cogn Disord. 2008;25:433–438. doi: 10.1159/000124750. [DOI] [PubMed] [Google Scholar]