Abstract
Background
Capping protein (CP), a heterodimer of α and β subunits, is found in all eukaryotes. CP binds to the barbed ends of actin filaments in vitro and controls actin assembly and cell motility in vivo. Vertebrates have three isoforms of CPβ produced by alternatively splicing from one gene; lower organisms have one gene and one isoform.
Results
We isolated genomic clones corresponding to the β subunit of mouse CP and identified its chromosomal location by interspecies backcross mapping.
Conclusions
The CPβ gene (Cappb1) mapped to Chromosome 4 between Cdc42 and D4Mit312. Three mouse mutations, snubnose, curly tail, and cribriform degeneration, map in the vicinity of the β gene.
Background
Capping protein (CP) is a ubiquitous actin binding protein that regulates actin assembly and cell motility. CP is a heterodimer composed of α and β subunits, each approximately 30 kD. Lower organisms, including Saccharomyces cerevisiae, Caenorhabditis elegans and Drosophila melanogaster, have one gene and one isoform for each of the CPα and β subunits [1,2,3]. Vertebrates contain three α subunit isoforms encoded by three different genes and three β subunit isoforms (β1, β2, β3) produced from one gene by alternative splicing [4,5,6].
Results and Discussion
To confirm the genomic complexity of the β subunit of mouse CP, Southern blots of mouse genomic DNA were cleaved with several restriction enzymes and independently probed with mouse β1 and β2 cDNAs. The β1 and β2 probes each hybridized to one identical band. To obtain genomic clones that correspond to this pattern, a 129SV genomic library in the lambda FIXII vector (Stratagene) was screened using the complete β1 cDNA as probe. Positive plaques were purified according to standard procedures [7] and the phage DNA isolated using DEAE-cellulose [8]. Comparison of the hybridization pattern for the genomic clones with the mouse genomic DNA revealed that the patterns were identical, confirming the presence of a single β gene in the mouse genome.
In chicken, the β1 and β2 isoforms have been described as the differentially spliced products of a single gene, with the β1 cDNA containing a 113 bp exon that is absent in the β2 cDNA [4]. To determine if the murine β2 isoform is spliced in a manner identical to that of chicken, the sequence of the β2 cDNA was compared to the corresponding genomic region. Sequence comparison revealed that alternative splicing of the β2 gene occurs in an identical manner in mice. The mouse intron is also 113 bp in length with the exon sequence and surrounding intron sequence containing the canonical splice donor (AGA) and acceptor (AGG) sites at either end of the intron. The human gene for CPβ has a similar structure, based on the sequence of clone HS657E11, located at Chr1 p35.1-p36.23, from the Sanger Centre (http://www.sanger.ac.uk/HGP/Chr1/).
To determine the chromosomal location of marine Cappb1, we identified interspecies variations of genomic sequences and mapped their chromosomal locations using an interspecies backcross panel from Jackson Laboratory [9]. The mapping panel is anchored to maps from crosses by various known genes, retroviral loci, and the D#Mit loci. The strain distribution pattern of each polymorphism in the interspecific backcross was determined and used to position the Cappb1 gene on the map (Figure 1).
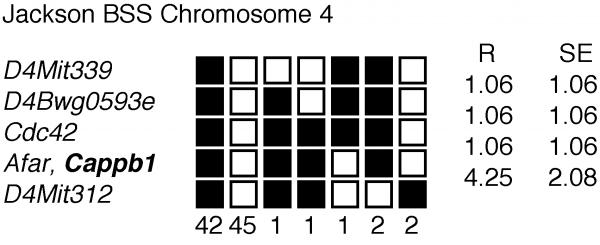
Figure 1.
Haplotype data for the mapping of the mouse CPβ gene showing a portion of Chromosome 4 with loci linked to Cappb1. Loci are listed in order with the most proximal at the top. The black boxes represent the C57BL6/J allele and the white boxes the SPRET/Ei allele. The number of animals with each haplotype is given at the bottom of each column of boxes. The percent recombination (R) between adjacent loci is listed to the right, with the standard error (SE) for each R. Missing typings were inferred from surrounding data where assignment was unambiguous.
A region corresponding to the 3' UTR of the Cappb1 gene was PCR-amplified using parental genomic DNA and gene-specific primers (5'TTTTCCCTCTTCCTTTCC3' and 5'ACTCCAAGCAACTCCCACAC3'). Direct sequencing of the PCR product identified two polymorphisms: 1) Nucleotides 1220-1224 (referring to Genbank Acc. No. U10406), C57BL/6J: CCCCC; M. spretus: CCCC, 2) Nucleotides 1336 1349: C57BL/6J: GGTGTGTGA-GAGAA and M. spretus: GGTGTGTGAAAGAGAA. Genomic DNA from the panel of 94 backcross animals was PCR-amplified using the aforementioned primers and then hybridized via dotblot analysis with four oligonucleotides: 5'AAGGAAGGGGGACAGG3', 5'AAGGAAGGGGACAGG3', 5'CTCTCACACA3', 5'CCACACACTTTCTCTT3', which specifically bound to each of the four polymorphic sequences, respectively. The resulting allele patterns were compared to those of other loci previously mapped in this cross to detect linkage.
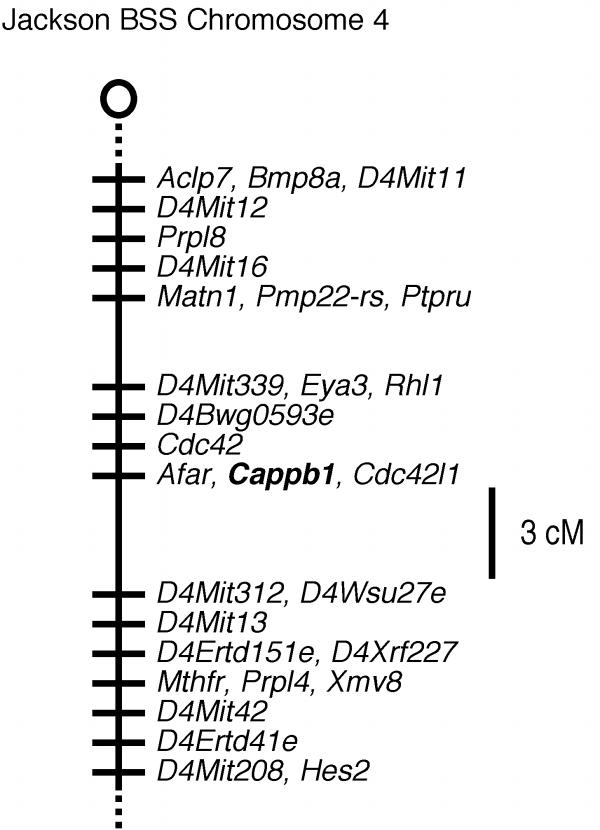
The CPβ gene (Cappb1) mapped to Chromosome 4 between D4Mitl6 and D4Mit13. Additional linkage information is available at http://www.jax.org./resources/documents/cmdata. Mouse Genome Database accession numbers for the mapping data are J:55295 (Cappb1). The order of loci (with recombination distances in centimorgans, cM and standard error) and intergenic distances for the CPβ gene (Figure 1 and 2) is as follows: Cappb1: Nearby genes include Cdc42 and Afar. D4Mit339- (1.06 ± 1.06 cM) -D4Bwg0593e- (1.06 ± 1.06 cM) -Cdc42- (1.06 ± 1.06 cM) -Afar, Cappb1- (4.25 ± 2.08 cM)- D4Mit312.
Figure 2.
Map from the Jackson BSS backcross showing part of Chromosome 4. The centromere is toward the top. Loci mapping to the same position are listed in alphabetical order. Missing typings were inferred from surrounding data where assignment was unambiguous. Raw data from Jackson Laboratory were obtained from http://www.jax.org./resources/documents/cmdata.
To determine if the Cappb1 gene was equivalent to a locus that had previously been mapped thereby identifying a possible association with an existing mutant, the Jackson Laboratory Mouse Genome Database (MGD) and the Mouse Chromosome Committee Report (1999) were scanned for loci that lie within 10 cM of the map positions of Cappb1. snubnose (sn), curly tail (ct), and cribriform degeneration (cri) are candidate mutants for Cappb1.
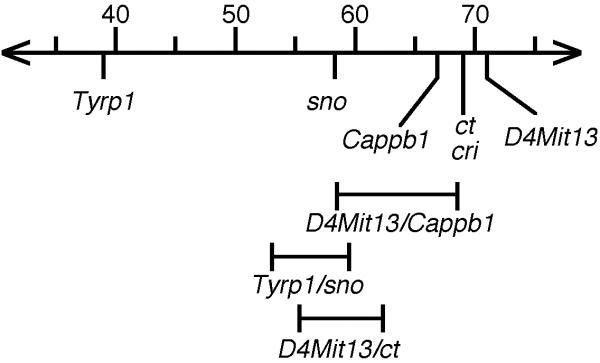
The likelihood that Cappb1 and sno, ct and cri map to the same position was evaluated by determining the confidence intervals associated with their map positions (Figure 3). Cappb1 (66.8 cM, MGD) was mapped relative to the well-established anchoring locus D4Mitl3 (71 cM, MGD), with 5 recombinants in 94 backcross samples. At 95% confidence, the upper and lower limits are 2.5 and 12.5, respectively [10]. sno (58.3 cM, MGD) was mapped relative to tyrosinase related protein, Tyrp1, (39 cM, MGD) with 92 recombinants in 535 backcross animals providing confidence limits of 14 and 20.5 [11]. cri (69 cM, MGD) was mapped relative to Tyrp1, but raw mapping data were not reported, which precludes determination of the confidence limits [12]. ct (69 cM) was mapped relative to D4Mitl3 with 26 recombinants in 272 backcross animals providing confidence limits of 6.6 and 13.7. The confidence intervals of ct and Cappb1 overlap, indicating that equivalency of these loci are not ruled out by the mapping data. Because the confidence interval of the mapping of cri could not be determined, the equivalency of cri and Cappb1 cannot be ruled out. The confidence intervals of sno and Cappb1 do not overlap, indicating that these loci are not equivalent.
Figure 3.
The confidence limits of the mapping of Cappb1 and its candidate genes: snubnose (sno), curly tail (ct), and cribriform degeneration (cri). The chromosomal region where Cappb1 and the candidate genes reside is depicted. The MGD map positions are indicated above. 95% confidence intervals based on specific mapping experiments, discussed in the text, are indicated below.
Analysis of the mutant phenotype for cri and ct is a way to evaluate their potential roles. Mutations in cri have a severe neurological defect that can be traced to vacuolar degeneration in white and gray matter of the spinal cord and brainstem [13]. Mutations in ct have some degree of spina bifida which results in a curly tail and occasionally, exencephaly [14]. Mutations in sno are recognized by their short noses and have spina bifida occulta in their lumbar, posterior thoracic and sacral vertebrae [15]. Additional studies will be necessary to determine whether these loci encode or map to Cappb1.
Comparative gene mapping between mouse and human has revealed numerous regions of homologous genome organization [16]. The mapping of Cappb1 supports the evolutionary conservation between the two species. The chromosomal assignment of the mouse CPβ gene to Chromosome 4 is consistent with the localization of the human CPβ gene to chromosome 1 position p36.1. These regions of the mouse and human chromosomes have conserved synteny.
Conclusions
The CPβ gene (Cappb1) mapped to Chromosome 4 between Cdc42 and D4Mit312. Three mouse mutations, snubnose, curly tail, and cribriform degeneration, map in the vicinity of the β gene.
Acknowledgments
Acknowledgments
We thank Lucy Rowe and Mary Barter of the Jackson Laboratory for their advice and assistance with the backcross panels and the interpretation of the genetic mapping data. This work was supported by a grant from NIH (GM38542). M.H. was supported by an American Heart Association Post-Doctoral Fellowship and was a member of the Lucille P. Markey Pathway for Human Pathobiology. J.A.C. was an Established Investigator of the American Heart Association.
Contributor Information
Marilyn C Hart, Email: mhart@VAX2.WINONA.MSUS.EDU.
Yulia O Korshunova, Email: yuliak@sequencer.wustl.edu.
John A Cooper, Email: jcooper@cellbio.wustl.edu.
References
- Waddle JA, Cooper JA, Waterston RH. The α and β subunits of nematode actin capping protein function in yeast. Molecular Biology of the Cell. 1993;4:907–917. doi: 10.1091/mbc.4.9.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopmann R, Cooper JA, Miller KG. Actin organization, bristle morphology, and viability are affected by actin capping protein mutations in Drosophila. Journal of Cell Biology. 1996;133:1293–1305. doi: 10.1083/jcb.133.6.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amatruda JF, Cannon JF, Tatchell K, Hug C, Cooper JA. Disruption of the actin cytoskeleton in yeast capping protein mutants. Nature. 1990;344:352–354. doi: 10.1038/344352a0. [DOI] [PubMed] [Google Scholar]
- Schafer DA, Korshunova YO, Schroer TA, Cooper JA. Differential localization and sequence analysis of capping protein β-subunit isoforms of vertebrates. Journal of Cell Biology. 1994;127:453–465. doi: 10.1083/jcb.127.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart MC, Korshunova YO, Cooper JA. Vertebrates have conserved capping protein alpha isoforms with specific expression patterns. Cell Motil Cytoskeleton. 1997;38:120–132. doi: 10.1002/(SICI)1097-0169(1997)38:2<120::AID-CM2>3.3.CO;2-L. [DOI] [PubMed] [Google Scholar]
- von Bulow M, Rackwitz HR, Zimbelmann R, Franke WW. CPβ-3, a Novel Isoform of an Actin- Binding Protein, is a Component of the CytoskeletalCalyx of the Mammalian Sperm Head. Experimental Cell Research. 1997;233:216–224. doi: 10.1006/excr.1997.3564. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning A Laboratory Manual, Second edn Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989.
- Helms C, Dutchik JE, Olson MV. A lambda DNA protocol based on purification of phage on DEAE-cellulose. Methods Enzymol. 1987;153:69–82. doi: 10.1016/0076-6879(87)53048-8. [DOI] [PubMed] [Google Scholar]
- Rowe LB, Nadeau JH, Turner R, Frankel WN, Letts VA, Eppig JT, Ko MSH, Thurston SJ, Birkenmeier EH. Maps from two interspecific backcross DNA panels available as a community genetic mapping resource. Mammalian Genome. 1994;5:253–274. doi: 10.1007/BF00389540. [DOI] [PubMed] [Google Scholar]
- Silver LM. Mouse genetics: concepts and applications New York: Oxford University Press; 1995.
- Hollander WF. Research notes: testcross linkage data for snubnose and brown. Mouse News Letter. 1966;34 [Google Scholar]
- Green MN. Cribriform degeneration (cri). Mouse News Letter. 1972;47:37. [Google Scholar]
- Green MC, Sidman RL, Pivetta OH. Cribriform degeneration (cri): a new recessive neurological mutation in the mouse. Science. 1972;176:800–803. doi: 10.1126/science.176.4036.800. [DOI] [PubMed] [Google Scholar]
- Gruneberg H. Genetical studies on the skeleton of the mouse. VIII. Curlytail. J Genet. 1954;52:52–67. [Google Scholar]
- Hollander WF. Genetic spina bifida occulta in the mouse. Am J Anat. 1976;146:173–179. doi: 10.1002/aja.1001460205. [DOI] [PubMed] [Google Scholar]
- O'Brien SJ, Womack JE, Lyons LA, Moore KJ, Jenkins NA, Copeland NG. Anchored reference loci for comparative mapping in mammals. Nature Genetics. 1993;3:103–112. doi: 10.1038/ng0293-103. [DOI] [PubMed] [Google Scholar]