Abstract
In this paper, we have examined two cysteine modifications resulting from sample preparation for protein characterization by MS: (1) a previously observed conversion of cysteine to dehydroalanine, now found in the case of disulfide mapping and (2) a novel modification corresponding to conversion of cysteine to alanine. Using model peptides, the conversion of cysteine to dehydroalanine via β-elimination of a disulfide bond was seen to result from the conditions of typical tryptic digestion (37 °C, pH 7.0– 9.0) without disulfide reduction and alkylation.. Furthermore, the surprising conversion of cysteine to alanine was shown to occur by heating cysteine containing peptides in the presence of a phosphine (TCEP). The formation of alanine from cysteine, investigated by performing experiments in H2O or D2O, suggested a radical-based desulfurization mechanism unrelated to β-elimination. Importantly, an understanding of the mechanism and conditions favorable for cysteine desulfurization provides insight for the establishment of improved sample preparation procedures of protein analysis.
INTRODUCTION
Cysteine residues have highly reactive thiol groups and are thus prone to modifications, including oxidation1, nitrosylation2, or glutathionylation.3 For example, disulfide bond formation, due to the oxidation of pairs of cysteines, is critical for protein folding and maintaining three-dimensional structures.4–7 Additionally, disulfide heterogeneity, including formation of non-native disulfide bonds from disulfide scrambling, as well as the occurrence of lanthionines8 or trisulfide bridges9, 10 is of concern in the production and shelf life of protein reagents and pharmaceuticals.11 Therefore, the identification and prevention of cysteine modifications is an important part of comprehensive protein characterization.
Mass spectrometry-based methods have been widely used in the analysis disulfide bridges.12–19 Conventional methods for disulfide mapping include enzymatic digestion of non-reduced proteins by pepsin at pH 2 to 4.13 However, pepsin generates non-specific cleavages, which increase the complexity of the digests. Alternatively, enzymes such as trypsin or Asp-N can provide specific cleavages; however, these enzymes prefer basic pH, which may lead to disulfide bond scrambling.20, 21 It was found that blocking free thiol groups by reagents such as N-ethylmaleimide (NEM) effectively prevents disulfide bond scrambling.22 To locate the exact linkage positions of disulfides, ESI-MS/MS in the negative mode has been suggested.23 Recently, electron capture dissociation (ECD)24 and electron transfer dissociation (ETD)17, 25 have been reported to facilitate the identification of disulfide linkages.
Besides mapping of disulfide linkages, additional modifications of cysteines have often been observed.1 In a recent paper, we applied a novel algorithm, termed Search for Modified Peptides (SeMoP), to discover unexpected modifications of peptides in MS data.26 Two unusual modifications, resulting in the loss of sulfur from a cysteine residue, were identified. The first desulfurization, corresponding to cysteine to dehydroalanine conversion, was associated with the known heat and alkaline induced β-elimination of disulfide bridges.12, 27–29 The other desulfurization detected by the SeMoP algorithm corresponded to the unexpected conversion of cysteine to alanine, which could be erroneously assumed to be a mutation resulting from a single nucleotide polymorphism (SNP). However, such a mutation is highly unlikely because two nucleotides would have to be changed to convert the codon for cysteine to alanine.30, 31
In this paper, we have examined the above two desulfurization reactions of cysteine in the context of sample handling for MS protein characterization in order to provide insight into proper sample preparation protocols. Atriopeptin, and briefly, lysozyme and somatostatin, were selected as model compounds to generate disulfide linked peptides. Tryptic digests under nonreduced conditions of these model compounds were characterized, including scrambled disulfides, and then subjected to heating at various temperatures with or without reducing reagents to study the formation of the modifications. In addition, we examined a thiyl radical-based mechanism for the conversion of cysteine to alanine.
EXPERIMENTAL
Materials and Methods
Atriopeptin and somatostatin were purchased from Anaspec (San Jose, CA). Egg white lysozyme, Tris buffer, tris (2-carboxyethyl) phosphine hydrochloride (TCEP), ethylenediaminetetraacetic acid (EDTA), iodoacetamide (IAM), ammonium bicarbonate, α-cyano-4-hydroxycinnamic acid (CHCA) and heavy water (D2O 99.9%), were obtained from Sigma-Aldrich (St Louis, MO). Trypsin Gold, mass spectrometry grade, was purchased from Promega (Madison, WI). Ellman’s reagent (DTNB, 5, 5 -dithiobis-(2-nitrobenzoic acid)], HPLC grade acetonitrile (ACN), trifluoroacetic acid (TFA) and formic acid (FA) were from Thermo Fisher Scientific (Fair Lawn, NJ).
Preparation of Model Compounds
Separate stock solutions of atriopeptin, lysozyme and somatostatin (1 μg/μL) were prepared in deionized water and stored at − 20 °C. Stock solutions of reducing reagent, 50 mM DTT and TCEP, were freshly made by dissolving DTT or TCEP in 100 mM ammonium bicarbonate. Atriopeptin (0.1 μg/μL) was subjected to trypsin digestion for one hour with a 50:1 (w/w) substrate to enzyme ratio at 37 °C in 100 mM ammonium bicarbonate buffer, pH 8.0. Lysozyme was digested with a 30:1 (w/w) substrate to enzyme ratio at 37 °C for 4 hours. Digestion was stopped by addition of 1% FA.
Cysteine Desulfurization
Forty μL of a tryptic digest of atriopeptin and separately lysozyme was heated at 60 °C or 90 °C for up to 2 hours, either without reduction or in the presence of reducing reagent (5 mM of TCEP or DTT). The atriopeptin digest was also prepared in D2O using the following procedure. Atriopeptin digest (40 μL) was dried to remove H2O and then resuspended in 100 mM ammonium bicarbonate in D2O (pH 8). Peptides in D2O were dried to remove residual H2O and again resuspended in 40 μL of 100 mM ammonium bicarbonate prepared in D2O. Four μL of TCEP in 100 mM ammonium bicarbonate in D2O was added to 40 μL of atriopeptin solution. The atrioepeptin solution was heated in the presence of TCEP at 90 °C for 20 minutes, and then the peptides were extracted with a C18 ZipTip (Millipore, Billerica, MA) to remove TCEP and resuspended in 50% ACN : 0.1% TFA in H2O (50 : 50). Finally, the samples were dried and resuspended in 0.1% FA (in H2O) before MALDI MS analysis. The somatostatin peptide without digestion was heated at 90 °C for up to 2 hours alone or in the presence of 5 mM of TCEP. All reactions were performed in 100 mM ammonium bicarbonate at pH 8.0 unless otherwise stated.
Analysis of Free Sulfhydryl Groups
Ellman’s reagent was used to determine the free sulfhydryl groups in the atriopeptin digest, before and after reduction, following the manufacturer’s protocol. The absorbance of the sample was measured at 412 nm in a Nanodrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE). The concentration of sulfhydryl groups in the sample was calculated using the molar extinction coefficient of 2-nitro-5-thiobenzoic acid (14,150 M−1cm−1).
Mass Spectrometry
Samples were desalted by ZipTip μ–C18 pipette tips (Millipore) and spotted on the MALDI plate followed by addition of MALDI matrix consisting of 10 mg/mL of CHCA in 50% ACN: 0.1% TFA. Peptide MS and MS/MS spectra were obtained using a MALDI-TOF/TOF 4700 mass spectrometer (Applied Biosystems, Framingham, MA). The instrument was externally calibrated with Glu-fibrinopeptide B (m/z=1570. 67).
For ESI -MS analysis, 2 pmol/μL of sample was infused into a LTQ-FT MS (Thermo Fisher Scientific, San Jose, CA). Data were acquired in the data-dependent mode with a survey scan using the FT MS, and then the ten most intense ions were fragmented for MS/MSanalysis using the LTQ (28% normalized collision energy).
RESULTS AND DISCUSSION
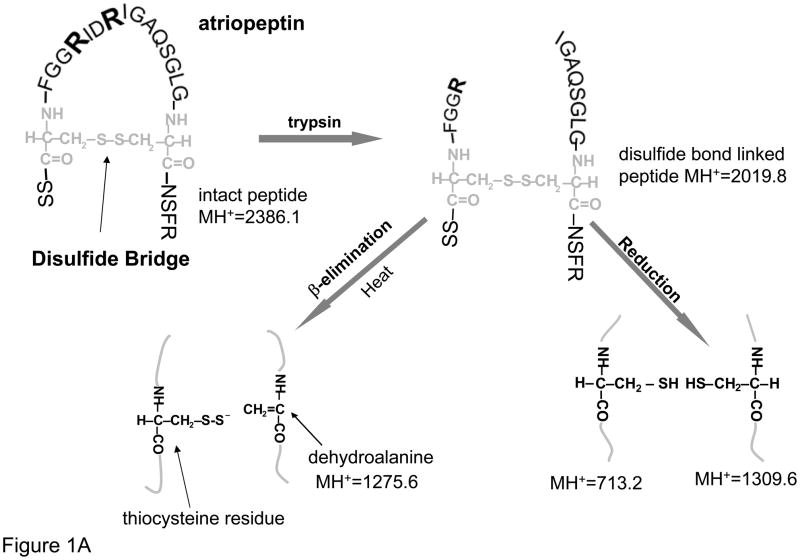
The goal of this paper was to examine cysteine modifications induced during the sample preparation for protein characterization, specifically the conversion of cysteine to dehydroalanine for example during disulfide mapping; and cysteine to alanine26 when heating with TCEP. To study these modifications, atriopeptin, a small 23 amino acid peptide was first selected as a model compound. Atriopeptin contains only a single disulfide bond, and thus, after tryptic digestion without reduction, two peptides linked by a disulfide bond are formed, with minimal potential interference of the non-cysteine peptides, see Figure 1A. Initially, the tryptic digest of atriopeptin was analyzed by MS to establish a baseline for comparison with the same sample processed under various conditions suitable for cysteine desulfurization.
Figure 1.
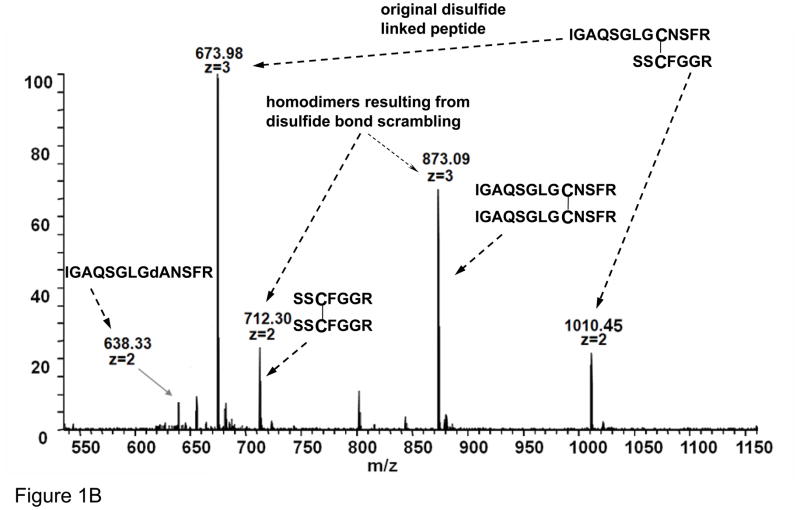
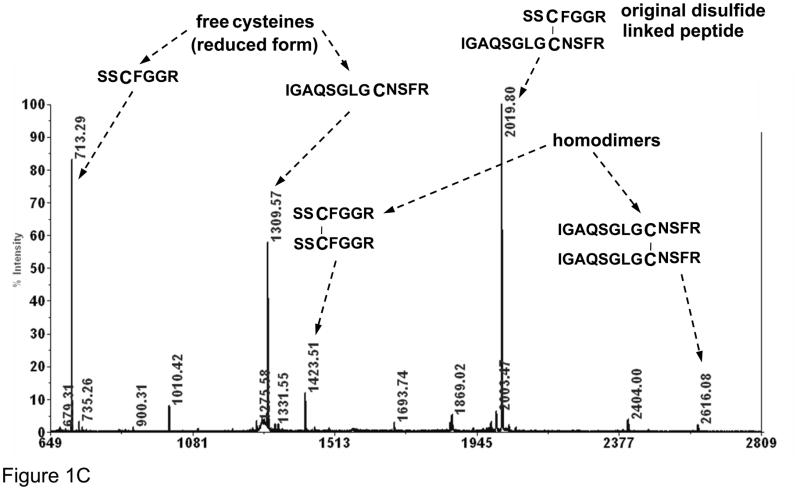
(A) Schematic of atriopeptin digestion by trypsin without reduction that is further subjected to heat induced β-elimination and reduction; (B) ESI spectrum of non-reduced atriopeptin digest, peak at m/z=712.3 (2+) and 873.1 (3+) are homodimers and (C) MALDI MS of atriopeptin digest. (dA represents dehydroalanine).
MS Analysis of the Tryptic Digest of Non-Reduced Atriopeptin
Atriopeptin was first subjected to tryptic digestion for 1 hour at pH 8.0 without reduction, and Figure 1B shows the ESI-MS spectrum of this directly infused digest. Peaks at m/z=674.0 (3+) and 1010.5 (2+) correspond to the fully digested product. Two peaks at m/z=712.3 (2+) and 873.1 (3+) were assigned to disulfide connected homodimers of atriopeptin tryptic peptides. Since larger amounts of the homodimers were found with longer digestion times (data not shown), disulfide scrambling is likely a consequence of the digestion conditions (37 °C, pH 8.0).13 It should be noted that since atriopeptide contains only a single pair of cysteines, the homodimer formation should, in theory, not occur before the digestion. Since it has been reported that the presence of an unpaired cysteine can facilitate disulfide bond scrambling,20 Ellman’s reagent was used to determine the amount of the free sulfhydryl groups in the atriopeptin tryptic digest. It was estimated that there were roughly 2% of free cysteines in the non-reduced digest solution, and the sulfhydryl group likely triggered the disulfide scrambling during digestion, resulting in the formation of the homodimers. Alkylation of atriopeptin with IAA before digestion (without reduction)was found to effectively eliminate the homodimers as expected (data not shown).22
The same non-reduced atriopeptin digest sample was next analyzed by MALDI-MS. Figure 1C shows the tryptic digestion product of atriopeptin peak at m/z=2019.8 (1+), and the miscleaved digestion peak at m/z=2404.0 (1+). The two homodimers with m/z =1423.5 (1+) and 2616.1 (1+) were observed as well. (There was no alkylation of the peptides before digestion.) Besides the above ions found in the ESI-MS spectrum, two peaks with relatively high intensities at m/z=713.2 (1+) and 1309.6 (1+) were seen. These two ions were found to be single cysteine containing peptides that were likely formed as the result of in-source fragmentation of the disulfide bond of atriopeptin.32 Importantly, these free cysteine-containing peptides were not observed in the ESI-MS spectrum (Figure 1B), likely due to the lower energy in the ionization process for ESI compared to MALDI. In summary, this simple example illustrates that a number of artifacts can occur when analyzing digests of non-reduced proteins, including scrambled disulfides and, in the case of MALDI MS analysis, the presence of ions generated by in-source fragmentation.
Conversion of Cysteine to Dehydroalanine in Non-Reduced Tryptic Digests
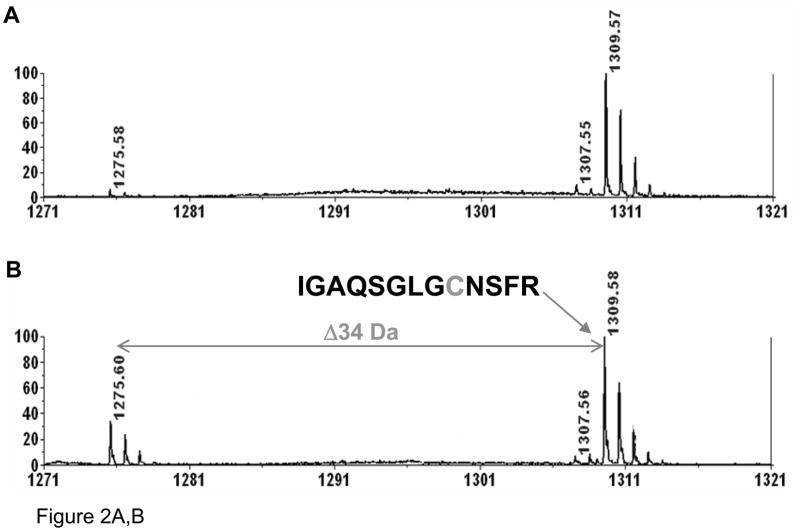
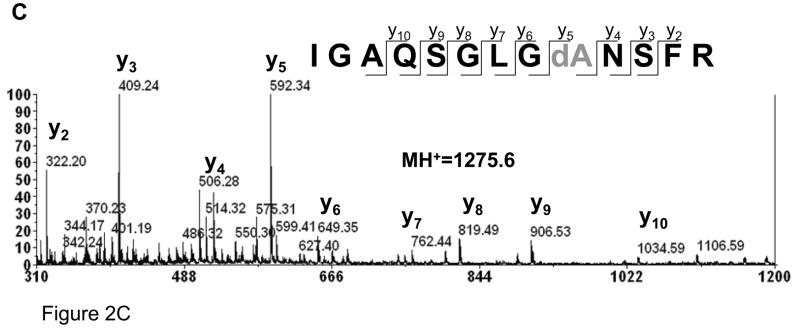
Of significance, a small peak at m/z=638.3 (2+) in the ESI (Fig. 1B) and 1275.6 (1+) in the MALDI spectrum (Fig. 1C and Fig. 2A) was found in the digest. Presumably, this ion corresponds to the loss of 34 Da from one of atriopeptin peptides, i.e., conversion of cysteine to dehydroalanine that was generated during the tryptic digestion by heat induced β-elimination.12 In order to confirm this, aliquots of the sample were collected during the digestion every 10 minutes for 1 hour and analyzed by MALDI MS. The peak at 1275.6 (1+) was not observed at the beginning of digestion, but was detected after 10 minutes, and the intensity of this peak increased with time. This result suggests that the dehydroalanine product was indeed generated during the digestion process. In order to accelerate this conversion to produce a sufficient amount of modified peptide that could be confidently identified by MALDI MS/MS analysis, atriopeptin digest was next heated at 60 °C in ammonium bicarbonate buffer (pH 8.0). As shown in Figure 2B, after heating the atriopeptin digest for 1 hour, the expected dehydroalanine product at m/z=1275.6 (1+), (loss of 34 Da) was observed. The MS/MS spectrum shown in Figure 2C confirmed the location of the dehydroalanine residue, i.e. IGAQSGLVdANSFR. Since thermal decomposition of ammonium bicarbonate buffer during heating could alter pH of the digestion solution, and thus the rate of β-elimination. The experiments were thus performed in thermally stable Tris-buffer with pH adjusted to 7, 8 and 9, and it was found that there was no change in the extent of cysteine to dehydroalanine conversion. It should be noted that the 7–9 pH range was selected because it is generally preferred for trypsin digestion; however, it is expected that higher pH could lead to an increased rate of β-elimination and disulfide scrambling.
Figure 2.
MALDI spectra of (A) non-reduced atriopeptin digested at 37 °C for 1 hour; (B) non-reduced atriopeptin digest heated at 60 °C for 1 hour; (C) MALDI MS/MS spectrum of a peak at m/z=1275.6 (1+), corresponding to the dehydroalanine containing peptide.
The dehydroalanine containing peptide at m/z=1275.6 (1+) could be generated via β-elimination from two disulfide linked tryptic peptides, (a) the original disulfide linked peptide with m/z=2019.8 (1+) (Figure 1C) and (b) the homodimer resulting from disulfide scrambling for the species with m/z=2616.1 (1+) (Figure 1C). However, during heating at pH 8.0, atriopeptin homodimers are generated by disulfide scrambling and at the same time, these homodimers could then undergo decomposition to dehydroalanine or thiocysteine.33 Thus, the change in the abundance of disulfide linked peptides will be the result of multiple processes, i.e. generation of disulfide linked peptides and their decomposition, and is thus difficult to quantitate. Nevertheless, it was estimated that less than 5% of cysteines were converted to dehydroalanines during 1-hour heating at 37 °C. However, the conversion can be as high as 30% for heating for 1 hour at 60 °C, see Figure 2. Prolonged digestion times, for example, overnight tryptic digestion at 37 °C of non-reduced proteins was found to lead to accumulation of detectable amounts of dehydroalanine products resulting from β-elimination (data not shown). In addition, it should be noted that besides widely used enzymatic digestion conditions, other commonly employed techniques such as denaturation of proteins by heating20 and long time storage8 have been shown to lead to formation of dehydroalanine. Interestingly, dehydroalanine was detected in serum albumin isolated from human plasma probably reflecting the long life time of albuminin the plasma29 As a result, the presence of dehydroalanine should always be considered when analyzing non-reduced samples. Reduction of disulfides followed by alkylation will prevent β-elimination of disulfides;34, 35 however, in the case of mapping of disulfide bonds, this is not possible.
Cysteine Desulfurization in the Presence of Reducing Reagents
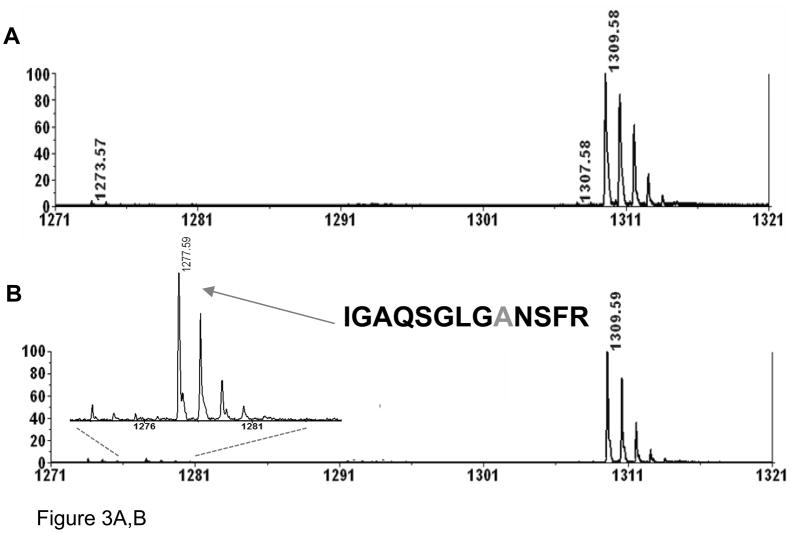
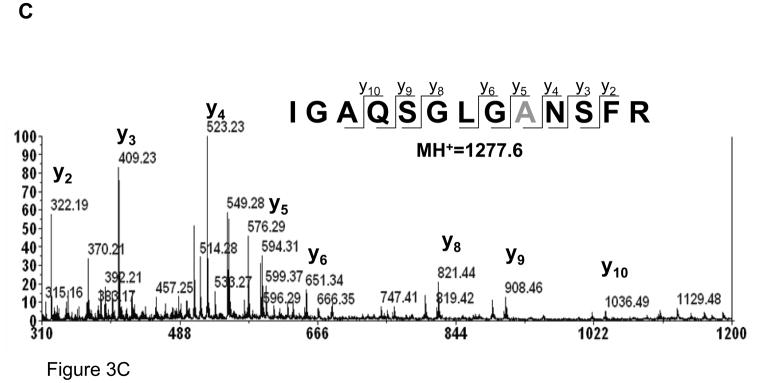
Since the β-elimination reaction requires disulfide bridges, reduction of disulfides should prevent cysteine desulfurization.34 As can be seen from Figure 3A, no formation of dehydroalanine was detected under commonly applied experimental conditions for protein reduction, i.e. DTT at 60 °C for 30 minutes. TCEP, a water-soluble phosphine, is often used as an alternative reducing reagent for reduction of disulfide bonds; but unlike DTT, the reduction by TCEP should be effective at room temperature. Nevertheless, TCEP has been mistakenly used at elevated temperatures. In order to evaluate the effect of TCEP on the sample at elevated temperature, a tryptic digest of non-reduced atriopeptin (pH 8.0) was heated at 60 °C for 1 hour in the presence of TCEP. In contrast to DTT, heating the peptide with TCEP led to the formation of a new modification corresponding to the loss of 32 Da (Figure 3B), suggesting conversion of cysteine to alanine. To confirm this modification, the peak at m/z=1277.6, i.e. loss of 32 Da from the peptide IGAQSGLVCNSFR (m/z=1309.6), was subjected to MALDI MS/MS analysis, Figure 3C. It can be seen that, starting from the y5 fragment ion, corresponding to the position of the cysteine residue, the masses of fragments are shifted by 2 Da with respect to the dehydroalanine product, which occurs in the absence of TCEP (Figure 2C), supporting the conversion of the cysteine to alanine.
Figure 3.
MALDI spectra of atriopeptin digest heated at 60°C for 1 hour in the presence of reducing reagent (A) 5 mM DTT; (B) 5 mM TCEP; and (C) MS/MS spectrum of the product of desulfurization at m/z=1277.6 (1+), corresponding to the conversion of cysteine to alanine.
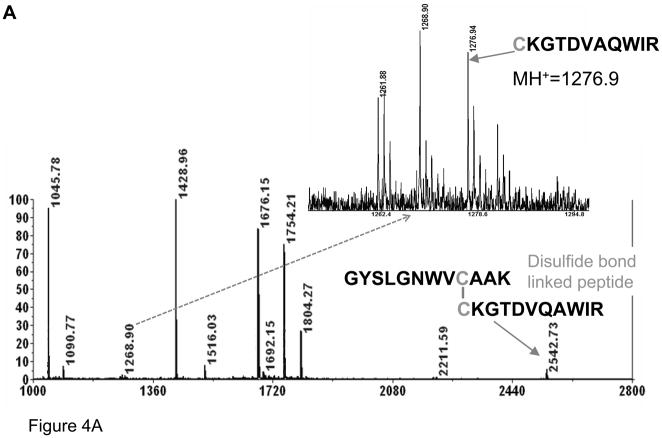
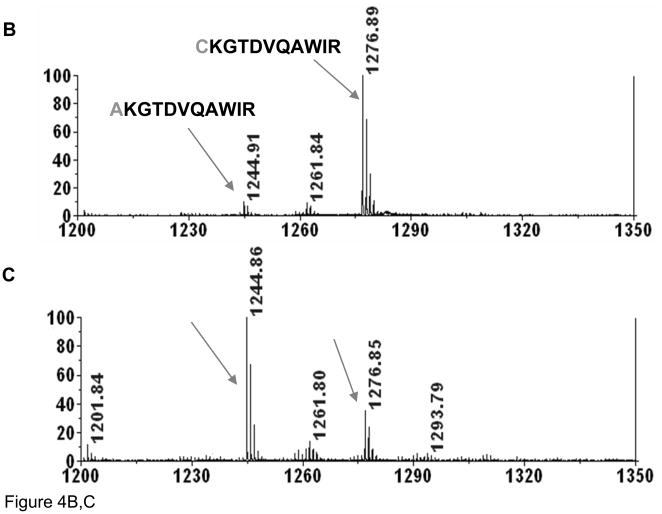
To further explore this unusual conversion, a similar experiment was next performed using a tryptic digest of lysozyme. MALDI-MS spectra were collected at different digestion times to assess the rate of cysteine desulfurization, see Figure 4. A peak, corresponding to two lysozyme tryptic peptides connected by a disulfide bridge, m/z=2542.7 (1+), could be clearly observed when digesting at 37 °C, pH 8.0, for 4 hours while only small peaks corresponding to reduced peptides (Figure 4A). On the other hand, after 10 minutes of heating at 90 °C in the presence of TCEP (Figure 4B), a strong ion at m/z=1276.9 (1+), corresponding to the reduced form of the 2542.5 (1+) ion, appeared along with a peak of relatively low intensity at m/z=1244.9 (1+), representing an alanine containing peptide (confirmed by MS/MS). It should be stressed that heating at 90 °C was utilized to generate sufficient amount of modified peptide for the MS/MS analysis using the AB4700 instrument. Nevertheless, brief heating to 90 °C is used in various protocols. The peak corresponding to the disulfide bond- linked peptide ion, m/z=2542.5 (1+), was not detected, indicating that this peptide was completely reduced. The intensity of the ion corresponding to the desulfurized peptide, m/z=1244.9 (1+), increased with additional heating, as shown in Figure 4C. Importantly, the peak corresponding to the alanine containing peptide increased after the disulfide bond was completely reduced, suggesting that the disulfide bond is not required for this conversion, i.e., that it could even occur for free cysteines. It should be noted that alkylation of free cysteines should prevent formation of this modification. Based on this result, the mechanism of conversion of cysteine to alanine is clearly different from that of β–elimination.
Figure 4.
MALDI spectra of (A) lysozyme digested by trypsin at 37 °C for 4 hours; (B) lysozyme digest heated in the presence of TCEP at 90°C for 10 minutes; and (C) for 120 minutes.
Mechanism of Cysteine to Alanine Conversion
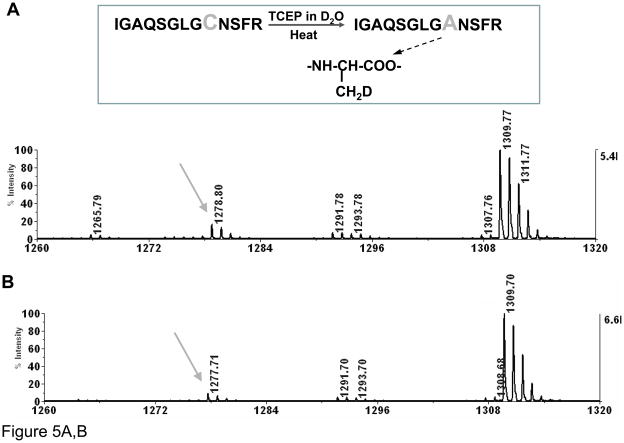
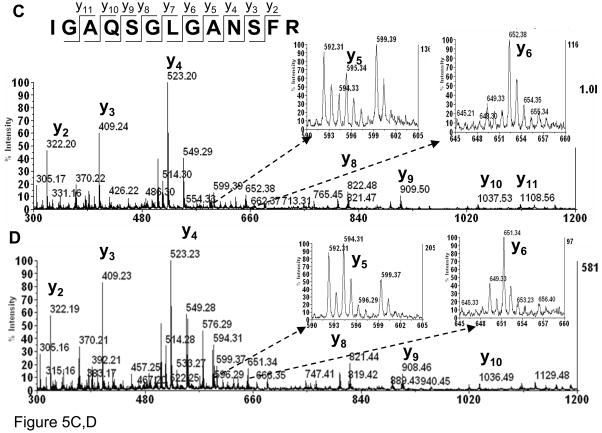
Since we found that the conversion of cysteine to alanine can be induced by heating in the presence of TCEP, it was of interest to investigate the mechanism of this conversion. A desulfurization of cysteine to alanine has been reported in glycan-peptide synthesis36, 37. Based on that work, we hypothesized that the cysteine to alanine modification in the example with atriopeptin follows a similar radical mechanism. In order to determine if this is the case, the reaction was conducted in D2O. As shown in Figure 5A (insert), the direct conversion in D2O without formation of dehydroalanine intermediate should lead to incorporation of a single non-exchangeable deuterium atom, increasing the mass of the newly formed alanine peptide by a 1 Da compared to the alanine product obtained in H2O. On the other hand, the mechanism that would involve formation of dehydroalanine would lead to incorporation of two non-exchangeable deuterium atoms, thus increasing the mass by 2 Da. This mass difference can be distinguished by MS. The mass spectra for reactions conducted in D2O and H2O are shown in Figures 5A and B, respectively. It can be seen that the mass of the original cysteine-containing peptide (m/z=1309.7) did not change when D2O was the solvent, suggesting that the deuterium atoms on amino acids that are not involved in the cysteine to alanine conversion can be back exchanged to hydrogen after the peptide is resuspended in H2O. However, a 1 Da shift was observed for the alanine containing peptide generated in D2O, m/z=1278.6 (1+) in Figure 5A vs. m/z=1277.6 (1+) in Figure 5B, indicating incorporation of a single deuterium atom. Furthermore, MS/MS spectra of the alanine products generated in both D2O and H2O were obtained, Figures 5C and 5D, respectively. Comparing Figures 5C and 5D, there is only a 1 Da shift for y5, whereas masses of y2, y3 and y4 remain unchanged. In summary, these data support that the modification is the direct conversion of cysteine to alanine without formation of dehydroalanine.
Figure 5.
MALDI MS of atriopeptin digest (A) heated at 90°C with TCEP in ammonium bicarbonate in D2O for 20 minutes; (B) heating at 90°C with TCEP in ammonium bicarbonate in H2O for 20 minutes; (C) MS/MS of precursor ion at m/z=1278.7 from (A); and (D) MS/MS of precursor ion at m/z=1277.7 from (B).
Based on the above result, we propose that the conversion follows the thiyl radical mechanism36, 37 as shown in Figure 6. Generally, the cysteine thiyl radical can be generated by interaction with oxidant such as oxygen or UV light. Next, the thiyl radical can react with alkylphosphine (TCEP) to form a phosphoranyl species and subsequent abstraction of a hydrogen atom, for example from a cysteinyl thiol via hemolytic cleavage.38 This reaction leads to the formation of the desulfurized product, alanine. Importantly, as we have shown, this mechanism does not require a disulfide bridge, and thus any free cysteine residue should be accessible for this reaction.
Figure 6.
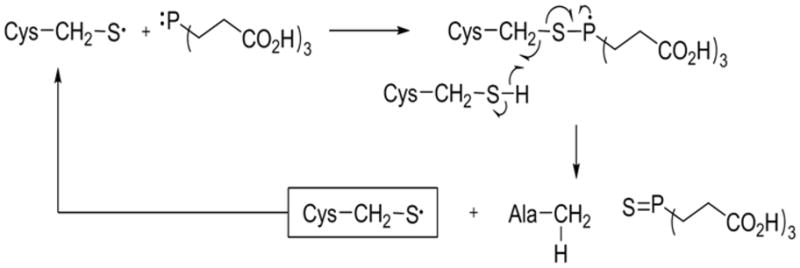
Proposed radical mechanism for the conversion of cysteine to alanine.
In summary, these results indicate that while heating disulfide cysteine containing peptides in the presence of DTT efficiently results in reduction without desulfurization. On the other hand, heating in the presence of TCEP should be avoided because it could lead to the conversion of cysteine to alanine. Importantly, even in the case when the TCEP reduction is performed at room temperature, failure to remove TCEP after the reduction could still lead to desulfurization of non-alkylated cysteines during subsequent heating steps. These results are important because experimental protocols that utilize TCEP do not emphasize that samples should not be heated. Finally, the rates of desulfurization of cysteine are likely dependent on the peptide sequences and conformation. Hence, other peptides may be more prone to this modification at lower temperature.
Cleavage of the Peptide Bond Induced by TCEP and Heat
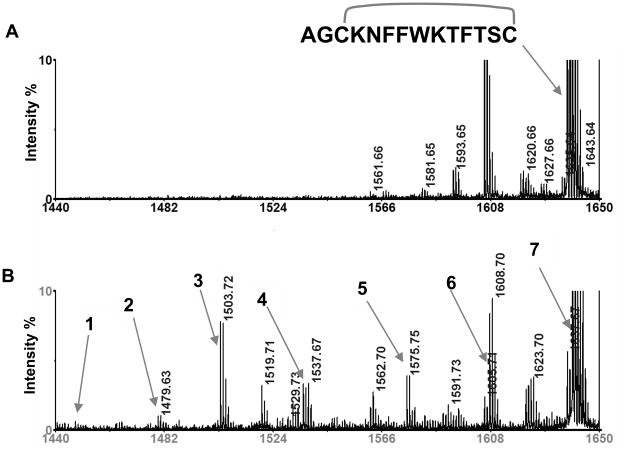
We observed that, besides conversion of cysteine to alanine, heating cysteine containing peptides in the presence of TCEP resulted in the formation of additional peaks, indicating other modifications. It has been reported that heating of disulfide containing proteins or peptides can lead to peptide bond cleavage with the formation of an amide at the C-terminus and a pyruvoyl residue at the N-terminus of the cleaved bond.8, 11 To examine these possibilities, somatostatin, a peptide with an intra-disulfide bond, was heated with and without TCEP. Figure 7 shows the MALDI MS spectra after 30 minutes of heating, indicating the formation of a number of peaks (Figure 7B), while a relatively clean spectrum is observed without TCEP (Figure 7A). In order to determine the identities of these newly formed species, individual peaks in Figure 7B were subjected to MS/MS analysis. It was determined that peaks labeled 1–4 corresponded to cleavages of peptide bonds, as described previously.8 Interestingly, the rate of formation of these cleavage products was significantly enhanced in the presence of TCEP. After 30 minutes of heating with TCEP, the hydrolytic cleavage products can be clearly observed, while in the sample without TCEP, the products were detected only after extending the heating time to one hour (data not shown). In summary, for TCEP containing samples, all sample preparation steps should be performed at room temperature to minimize these side reactions.
Figure 7.
MALDI mass spectra of (A) intact somatostatin heated at 90 °C, pH 8.0 for 30 minutes as control (intact peptide at m/z 1637.7); (B) somatostatin heated at 90 °C for 30 minutes in the presence of TCEP and shows additional peaks 1–7 identified as follows: 1. CH3COCO-KNFFWKTFTSA, m/z=1447.7; 2. CH3COCO-KNFFWKTFTSC, m/z=1479.7; 3. AGAKNFFWKTFTS-NH2, m/z=1503.7; 4. AGCKNFFWKTFTS-NH2, m/z=1535.7; 5. AGAKNFFWKTFTSA, m/z=1575.7; 6. AGCKNFFWKTFTSA and AGAKNFFWKTFTSC, m/z=1607.7; and 7. AGCKNFFWKTFTSC (reduced somatostatin), m/z =1639.7.
CONCLUSIONS
In this paper, we have studied cysteine modifications induced in the sample preparation step for MS protein characterization. Specifically, we examined two modifications, i.e. conversion of cysteine to dehydroalanine and cysteine to alanine, caused by sample preparation. Our results indicated that since the disulfide bond containing peptides are prone to β-elimination and thus the formation of dehydroalanine, special care should be taken to avoid exposure of disulfide bond containing samples to heat at basic pH. Though the extent of β-elimination at room temperature is small, overnight protein digestion with trypsin at pH=8.0 and temperature 37 °C without the use of reducing reagent such as DTT may lead to formation of peptide with dehydroalanine residue detectable by mass spectrometry. In addition, it is expected that heat-induced protein denaturation performed without reduction of disulfide bond (e.g. 90 °C for 10 minutes) may lead to relatively high levels of desulfurization and thus should be avoided. Formation of dehydroalanine involves breakage of disulfide bonds, and thus heating of a sample in basic media (e.g. during trypsin digestion) can contribute to disulfide scrambling, even if free cysteines are not blocked.
Next, similarly to reports by others,34 it was found that addition of reducing reagents, specifically DTT, is an effective way to eliminate cysteine desulfurization since the reduction of disulfide bonds will prevent β-elimination with heating. On the other hand, it was found that though TCEP is an effective reducing reagent at room temperature, an artifact corresponding to the conversion of cysteine to alanine can occur after heating cysteine containing peptides in the presence of TCEP. Therefore, special care should be taken to avoid heating peptides with free cysteines in the presence of TCEP, which is not emphasized in the sample preparation protocols. In addition, it was found that hydrolytic cleavage of peptide bonds observed previously8 is significantly enhanced by heating in the presence of TCEP. These results demonstrate that sample preparation steps for MS can lead to erroneous conclusions concerning protein structure and that detailed characterization of each step is necessary to assure that the results reflect the true protein structure.
Acknowledgments
This research was support in part by the Barnett Fund for Innovative Research (TR) and by NIH GM15847 (BLK) and 1R01AI05814 (ZSZ). Contribution No. 954 from the Barnett Institute.
References
- 1.Leichert LI, Jakob U. Protein thiol modifications visualized in vivo. Plos Biology. 2004;2(11):1723–1737. doi: 10.1371/journal.pbio.0020333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stamler JS. Redox singnaling-nitrosylation and related target interactions of nitric-oxide. Cell. 1994;78(6):931–936. doi: 10.1016/0092-8674(94)90269-0. [DOI] [PubMed] [Google Scholar]
- 3.Klatt P, Lamas S. Regulation of protein function by S-glutathiolation in response to oxidative and nitrosative stress. European Journal of Biochemistry. 2000;267(16):4928–4944. doi: 10.1046/j.1432-1327.2000.01601.x. [DOI] [PubMed] [Google Scholar]
- 4.Arolas JL, Aviles FX, Chang JY, Ventura S. Folding of small disulfide-rich proteins: clarifying the puzzle. Trends in Biochemical Sciences. 2006;31(5):292–301. doi: 10.1016/j.tibs.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 5.Bardwell JCA. The dance of disulfide formation. Nature Structural & Molecular Biology. 2004;11(7):582–583. doi: 10.1038/nsmb0704-582. [DOI] [PubMed] [Google Scholar]
- 6.Daggett V, Fersht AR. Is there a unifying mechanism for protein folding? Trends in Biochemical Sciences. 2003;28(1):18–25. doi: 10.1016/s0968-0004(02)00012-9. [DOI] [PubMed] [Google Scholar]
- 7.Giles NM, Watts AB, Giles GI, Fry FH, Littlechild JA, Jacob C. Metal and redox modulation of cysteine protein function. Chemistry & Biology. 2003;10(8):677–693. doi: 10.1016/s1074-5521(03)00174-1. [DOI] [PubMed] [Google Scholar]
- 8.Cohen SL, Price C, Vlasak J. beta-elimination and peptide bond hydrolysis: Two distinct mechanisms of human IgG1 hinge fragmentation upon storage. Journal of the American Chemical Society. 2007;129(22):6976. doi: 10.1021/ja0705994. [DOI] [PubMed] [Google Scholar]
- 9.Pristatsky P, Cohen S, Krantz D, Acevedo J, Ionescu R, Vlasak J. Evidence for Trisulfide Bonds in a Recombinant Variant of a Human IgG2 Monoclonal Antibody. Analytical Chemistry. 2009 July 10; doi: 10.1021/ac9006254. [DOI] [PubMed] [Google Scholar]
- 10.CanovaDavis E, Baldonado IP, Chloupek RC, Ling VT, Gehant R, Olson K, GilleceCastro BL. Confirmation by mass spectrometry of a trisulfide variant in methionyl human growth hormone biosynthesized in Escherichia coli. Analytical Chemistry. 1996;68(22):4044–4051. doi: 10.1021/ac9605915. [DOI] [PubMed] [Google Scholar]
- 11.Wang W, Singh S, Zeng DL, King K, Nema S. Antibody structure, instability, and formulation. Journal of Pharmaceutical Sciences. 2007;96(1):1–26. doi: 10.1002/jps.20727. [DOI] [PubMed] [Google Scholar]
- 12.Kim JS, Kim HJ. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometric observation of a peptide triplet induced by thermal cleavage of cystine. Rapid Commun Mass Spectrom. 2001;15(23):2296–300. doi: 10.1002/rcm.509. [DOI] [PubMed] [Google Scholar]
- 13.Gorman JJ, Wallis TP, Pitt JJ. Protein disulfide bond determination by mass spectrometry. Mass Spectrometry Reviews. 2002;21(3):183–216. doi: 10.1002/mas.10025. [DOI] [PubMed] [Google Scholar]
- 14.Loo JA. Eeffect of reduceing disulfide-containing proteins on elcetrospray ionizaton mass-spectra. Anal Chem. 1990;(62):693–698. doi: 10.1021/ac00206a009. [DOI] [PubMed] [Google Scholar]
- 15.Yu HQ, Murata K, Hedrick JL, Almaraz RT, Xiang F, Franz AH. The disulfide bond pattern of salmon egg lectin 24 K from the Chinook salmon Oncorhynchus tshawytscha (vol 463, pg 1, 2007) Archives of Biochemistry and Biophysics. 2008;479(2):179–179. doi: 10.1016/j.abb.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 16.Yan BX, Valliere-Douglass J, Brady L, Steen S, Han M, Pace D, Elliott S, Yates Z, Han YH, Balland A, Wang WC, Pettit D. Analysis of post-translational modifications in recombinant monoclonal antibody IgG1 by reversed-phase liquid chromatography/mass spectrometry. Journal of Chromatography A. 2007;1164(1–2):153–161. doi: 10.1016/j.chroma.2007.06.063. [DOI] [PubMed] [Google Scholar]
- 17.Wu SL, Jiang HT, Lu QZ, Dai SJ, Hancock WS, Karger BL. Mass Spectrometric Determination of Disulfide Linkages in Recombinant Therapeutic Proteins Using Online LC-MS with Electron-Transfer Dissociation. Analytical Chemistry. 2009;81(1):112–122. doi: 10.1021/ac801560k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones MD, Patterson SD, Lu HS. Determination of disulfide bonds in highly bridged disulfide-linked peptides by matrix-assisted laser desorption/ionization mass spectrometry with postsource decay. Analytical Chemistry. 1998;70(1):136–143. doi: 10.1021/ac9707693. [DOI] [PubMed] [Google Scholar]
- 19.Kim HI, Beauchamp JL. Mapping Disulfide Bonds in Insulin with the Route 66 Method: Selective Cleavage of S-C Bonds Using Alkali and Alkaline Earth Metal Enolate Complexes. Journal of the American Society for Mass Spectrometry. 2009;20(1):157–166. doi: 10.1016/j.jasms.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 20.Liu HC, Gaza-Bulseco G, Chumsae C, Newby-Kew A. Characterization of lower molecular weight artifact bands of recombinant monoclonal IgG1 antibodies on non-reducing SDS-PAGE. Biotechnology Letters. 2007;29(11):1611–1622. doi: 10.1007/s10529-007-9449-8. [DOI] [PubMed] [Google Scholar]
- 21.Pompach P, Man P, Kavan D, Hofbauerová K, Kumar V, Bezouška K, Havlíček V, Novák P. Modified electrophoretic and digestion conditions allow a simplified mass spectrometric evaluation of disulfide bonds. Journal of the American Society for Mass Spectrometry. 2009 doi: 10.1002/jms.1609. [DOI] [PubMed] [Google Scholar]
- 22.Taylor FR, Prentice HL, Garber EA, Fajardo HA, Vasilyeva E, Pepinsky RB. Suppression of sodium dodecyl sulfate-polyacrylamide gel electrophoresis sample preparation artifacts for analysis of IgG4 half-antibody. Analytical Biochemistry. 2006;353(2):204–208. doi: 10.1016/j.ab.2006.02.022. [DOI] [PubMed] [Google Scholar]
- 23.Chelius D, Wimer MEH, Bondareriko PV. Reversed-phase liquid chromatography in-line with negative ionization electrospray mass spectrometry for the characterization of the disulfide-linkages of an immunoglobulin gamma antibody. Journal of the American Society for Mass Spectrometry. 2006;17(11):1590–1598. doi: 10.1016/j.jasms.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 24.Zubarev RA, Kruger NA, Fridriksson EK, Lewis MA, Horn DM, Carpenter BK, McLafferty FW. Electron capture dissociation of gaseous multiply-charged proteins is favored at disulfide bonds and other sites of high hydrogen atom affinity. Journal of the American Chemical Society. 1999;121(12):2857–2862. [Google Scholar]
- 25.Chrisman PA, Pitteri SJ, Hogan JM, McLuckey SA. SO2-* electron transfer ion/ion reactions with disulfide linked polypeptide ions. J Am Soc Mass Spectrom. 2005;16(7):1020–30. doi: 10.1016/j.jasms.2005.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baumgartner C, Rejtar T, Kullolli M, Akella LM, Karger BL. SeMoP: a new computational strategy for the unrestricted search for modified peptides using LC-MS/MS data. J Proteome Res. 2008;7(9):4199–208. doi: 10.1021/pr800277y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Friedman M. Chemistry, biochemistry, nutrition, and microbiology of lysinoalanine, lanthionine, and histidinoalanine in food and other proteins. Journal of Agricultural and Food Chemistry. 1999;47(4):1295–1319. doi: 10.1021/jf981000+. [DOI] [PubMed] [Google Scholar]
- 28.Nakanishi T, Sato T, Sakoda S, Yoshioka M, Shimizu A. Modification of cysteine residue in transthyretin and a synthetic peptide: analyses by electrospray ionization mass spectrometry. Biochimica Et Biophysica Acta-Proteins and Proteomics. 2004;1698(1):45–53. doi: 10.1016/j.bbapap.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 29.Bar-Or R, Rael LT, Bar-Or D. Dehydroalanine derived from cysteine is a common post-translational modification inhuman serum albumin. Rapid Communications in Mass Spectrometry. 2008;22(5):711–716. doi: 10.1002/rcm.3421. [DOI] [PubMed] [Google Scholar]
- 30.Gloss LM, Spencer DE, Kirsch JF. Cysteine-191 in aspartate aminotransferases appears to be conserved due to the lack of a neutral mutation pathway to the functional equivalent, alanine-191. Proteins-Structure Function and Genetics. 1996;24(2):195–208. doi: 10.1002/(SICI)1097-0134(199602)24:2<195::AID-PROT6>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 31.Ohno S. Ancient linkage groups and frozen accidents. Nature. 1973:244–259. doi: 10.1038/244259a0. [DOI] [PubMed] [Google Scholar]
- 32.Patterson SD, Katta V. Prompt fragmentation of disulfide-linked peptides during matrix-assisted laser desorption ionization mass spectrometry. Analytical Chemistry. 1994;66(21):3727–32. doi: 10.1021/ac00093a030. [DOI] [PubMed] [Google Scholar]
- 33.Parker AJKN. The scission of the sulfur-sulfur bond. Chemical Reviews. 1969:59. [Google Scholar]
- 34.Herbert B, Hopwood F, Oxley D, McCarthy J, Laver M, Grinyer J, Goodall A, Williams K, Castagna A, Righetti PG. Beta-elimination: an unexpected artefact in proteome analysis. Proteomics. 2003;3(6):826–31. doi: 10.1002/pmic.200300414. [DOI] [PubMed] [Google Scholar]
- 35.Herbert B, Galvani M, Hamdan M, Olivieri E, MacCarthy J, Pedersen S, Righetti PG. Reduction and alkylation of proteins in preparation of two-dimensional map analysis: why, when, and how? Electrophoresis. 2001;22(10):2046–57. doi: 10.1002/1522-2683(200106)22:10<2046::AID-ELPS2046>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 36.Wan Q, Danishefsky SJ. Free-radical-based, specific desulfurization of cysteine: A powerful advance in the synthesis of polypeptides and glycopolypeptides. Angewandte Chemie-International Edition. 2007;46(48):9248–9252. doi: 10.1002/anie.200704195. [DOI] [PubMed] [Google Scholar]
- 37.Alferiev IS, Connolly JM, Levy RJ. A novel mercapto-bisphosphonate as an efficient anticalcification agent for bioprosthetic tissues. Journal of Organometallic Chemistry. 2005;690:2543–2547. [Google Scholar]
- 38.Schoneich C. Mechanisms of protein damage induced by cysteine thiyl radical formation. Chemical Research in Toxicology. 2008;21(6):1175–1179. doi: 10.1021/tx800005u. [DOI] [PubMed] [Google Scholar]