Abstract
The three homologous members of the p160 SRC family (SRC-1, SRC-2 and SRC-3) mediate the transcriptional functions of nuclear receptors and other transcription factors, and are the most studied of all transcriptional coactivators. Recent work has indicated that the SRC genes are subject to amplification and overexpression in various human cancers. Some of the molecular mechanisms responsible for SRC overexpression along with the mechanisms by which SRCs promote breast and prostate cancer cell proliferation and survival have been identified, as have the specific contributions of individual SRC family members in spontaneous breast and prostate carcinogenesis in genetically manipulated mouse models. These studies have identified new challenges for cancer research and therapy.
Introduction
Nuclear hormone receptors (NRs) are ligand-dependent, DNA-binding transcription factors that regulate gene expression and various physiological functions. Understanding how NRs control gene transcription has been a long and difficult journey. Initially, it was thought that NRs enabled general transcription factors (GTFs) and RNA polymerase II to assemble at the promoter to turn on mRNA synthesis. However, work began in the 1970s to search for nuclear non histone helper proteins that were thought to aid the binding to DNA and the transcriptional function of NRs 1, 2. The finding that activation of one overexpressed NR could indirectly inhibit the transcriptional activity of another NR3, 4 and that in vitro transcription systems consisting of purified NRs and GTFs were inefficient further suggested that additional transcription activators were required for efficient hormone-induced transcriptional activation 5. In 1995, the steroid receptor coactivator-1 (SRC-1) was cloned as the first authentic NR coactivator. SRC-1 interacted with steroid receptors in a hormone-dependent manner and robustly increased the transcriptional activities of steroid receptors 6. Soon after, two other homologous proteins, SRC-2 (TIF2 or GRIP1) 7, 8 and SRC-3 (p/CIP, RAC3, AIB1, ACTR or TRAM-1) 9–13, were characterized as NR coactivators. These three homologous proteins comprise the p160 SRC family. In this review, we summarize our current knowledge regarding the molecular features, functional mechanisms, posttranslational modifications and physiological functions of the SRC family members. Our major emphasis will be on the contributions and mechanisms of the SRC family in cancer.
Structural and functional characteristics of the SRC family
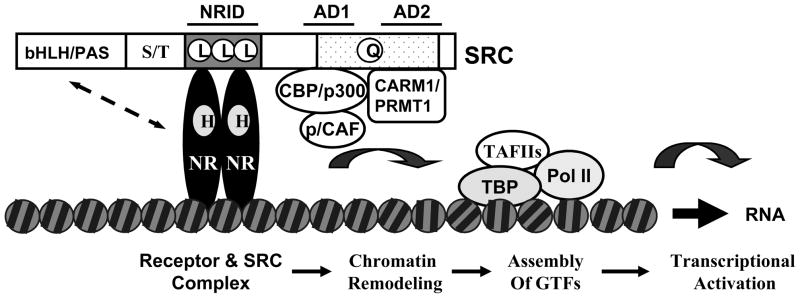
SRCs contain three structural domains. The N-terminal basic helix-loop-helix-Per/ARNT/Sim (bHLH-PAS) domain is the most conserved region and is required for protein-protein interactions 14–16. The bHLH-PAS domain can interact with several transcription factors such as myogenin, MEF-2C and TEF to potentiate transcription 17, 18. The central region contains three LXXLL (L for leucine and X for any amino acid) motifs, which form amphipathic α-helices and are responsible for interacting with NRs 19–21. The C-terminus contains two transcriptional activation domains (AD1 and AD2). AD1 binds CBP and p300 histone acetyltransferase (HAT), and the recruitment of CBP or p300 by SRCs to the chromatin context is essential for SRC-mediated transcriptional activation. AD2 interacts with coactivator-associated arginine methyltransferase 1 (CARM1) and protein arginine methyltransferases (PRMT1), which are histone methyltransferases 22–29. The C-termini of SRC-1 and SRC-3 contain HAT activity domains, although their cellular substrates are incompletely identified 12, 30. These molecular features provide SRCs with a suitable structural base for recruiting additional coregulators and general transcription factors, which in turn results in chromatin remodeling, assembly of general transcription factors and recruitment of RNA Polymerase II for transcriptional activation 31, 32. The basic SRC structural domains and the simplified functional mechanisms for SRCs in NR-dependent transcriptional activation are sketched in Fig. 1.
Fig. 1.
Molecular structure of SRCs and their functional mechanisms in steroid hormone-induced gene expression. The locations of basic structural and functional domains of SRCs are indicated. Upon hormone (H) binding, the hormone nuclear receptors (NR) expose their coactivator-binding motifs in their ligand-binding domains and allow SRCs to be recruited to the enhancer region of the NR target genes. SRCs further interact with CBP, p300, p/CAF, CARM1 and PRMT1 and recruit these common coactivators to the chromatin to build up a steroid receptor-directed transcriptional activation complex. This protein complex uses its protein acetyltransferase and methyltransferase activities to remodel the chromatin structure and to facilitate the assembly of general transcription factors and RNA polymerase II on the promoter for transcriptional activation. Of note, in addition to interactions between NR and the NRID domain of SRCs, interactions between NR and the bHLH/PAS domain of SRCs have been documented and may be important for function (see dotted line with arrowheads). Abbreviations: NRID, NR interaction domain; AD1 and AD2, activation domains 1 and 2; bHLH/PAS, the basis helix-loop-helix/Per-Ah receptor nuclear translocator-Sim domain; S/T, the serine and threonine-rich domain; L, L and L, the three LXXLL motifs responsible for interaction with nuclear receptors; Q, the glutamine-rich region; HAT, the histone acetyltransferase domain; CBP, the CREB (cAMP response element-binding protein) binding protein; p300, the 300 kDa protein homologous to CBP; p/CAF, the p300 and CBP-associated factor; CARM1, the coactivator-associated arginine methyltransferase 1; PRMT1, the protein arginine methyltransferase 1; TBP, the TATA binding protein; TAFIIs, TBP-associated general transcription factors (GTFs); Pol II, RNA polymerase II.
In addition to serving as coactivators for NRs, SRCs also serve as coactivators for other transcription factors, including NF-κB, Smads, E2F1, STATs, HIF1, p53 and RB (Table 1). SRCs also can promote gene transcription by interacting with kinases, phosphatases, ubiquitin/sumo ligases, histone acetyltransferases and histone methyltransferases (Table 1). By modulating gene expression controlled by a broad range of NRs and non-NR transcription factors, SRCs regulate many diverse physiological functions.
Table 1.
List of proteins that interact with SRCs
| Categories | Functional consequence | Method of detection | References |
|---|---|---|---|
| Transcription factors | |||
| Nuclear receptors | activation | Co-IP, in vitro | 150, 151 |
| AhR/ARNT | activation | Co-IP, in vitro | 152–154 |
| AP-1 | activation | Yeast 2-hybrid, in vitro | 155 |
| Brn-3 | activation | Co-IP | 156 |
| β-catenin | activation | Mammalian 2-hybrid, in vitro | 157 |
| c-Ets | activation | Co-IP | 158 |
| E2F1 | activation | Co-IP, in vitro | 135 |
| HNF4 | activation | Yeast 2-hybrid | 159, 160 |
| IRF3 | activation | Yeast 2-hybrid | 161 |
| MEF-2C | activation | Mammalian 2-hybrid, Co-IP, in vitro | 18 |
| NF-κB | activation | Co-IP | 162 |
| Smad3 | activation | Co-IP, mammalian 1-hybrid | 163 |
| Tat | activation | Co-IP, in vitro | 164 |
| TEF-4 | activation | Yeast 2-hybrid, in vitro | 17 |
| TTF1 | activation | in vitro | 165 |
| Oncogene/Tumor suppressor | |||
| HPV E7 | suppression | Co-IP, in vitro | 166 |
| p53 | activation | in vitro | 167 |
| Rb | activation | Co-IP, in vitro | 168 |
| Kinase/Phosphatase | |||
| aPKC | phosphorylation, stabilization | Co-IP | 56 |
| c-Abl | phosphoryaltion, activation | Co-IP | 50 |
| GSK3 | phosphorylation, activation, degradation | enzyme-substrate interaction | 54 |
| IKK | phosphorylation, activation | Co-IP | 61 |
| MAPK | phosphorylation, activation | enzyme-substrate interaction | 42, 69 |
| PDXP | dephosphorylation, suppression | enzyme-substrate interaction | 55 |
| PKA | phosphorylation, activation | enzyme-substrate interaction | 43, 169 |
| PP1 | dephosphorylation, stabilization | enzyme-substrate interaction | 55 |
| PP2A | dephosphorylation, suppression | enzyme-substrate interaction | 55 |
| Ub/Sumo enzymes | |||
| ARIP3/PIASxα | sumoylation, activation | in vitro | 74 |
| E6AP | ubiquitination, degradation | Co-IP, in vitro | 170 |
| Fbw7 | ubiquitination, activation, degradation | Co-IP | 54 |
| UBCH7 | activation | Co-IP, in vitro | 171 |
| Coregulators | |||
| ANCOS | suppression | Yeast 2-hybrid, Co-IP, in vitro | 172 |
| ASC | activation | Yeast 2-hybrid, in vitro | 173 |
| CARM1 | activation, methylation, suppression | Yeast 2-hybrid, Co-IP | 174 |
| CoCoA | activation | Co-IP, in vitro | 15 |
| GAC63 | activation | Co-IP, in vitro | 14 |
| GCN5 | activation | Co-IP, yeast 2-hybrid | 23 |
| MMS19 | activation | Yeast 2-hybrid, in vitro | 175 |
| p300/CBP | activation, acetylation, suppression | Co-IP, in vitro | 35 |
| PGC1 | activation | in vitro | 142 |
| Others | |||
| Cyclin T1 | activation | in vitro | 164 |
| FLASH | activation, suppression | Yeast 2-hybrid, in vitro | 176, 177 |
| GAS | activation | in vitro | 178 |
| JAB1 | activation | Yeast 2-hybrid, in vitro | 179 |
| Pin1 | activation, degradation | in vitro | 180 |
| REGγ | degradation | Co-IP | 77 |
| SRA | activation | IP | 181 |
| TIA, TIAR | translational inhibition | Co-IP, in vitro | 63 |
This is an incomplete list of SRC interactive proteins representing distinct categories. Co-IP, coimmunoprecipitation; IP, immunoprecipitation; in vitro, in vitro protein-protein interaction assay; AhR/ARNT, aryl hydrocarbon receptor/aryl hydrocarbon receptor nuclear translocator; AP-1, activator protein 1; Brn-3, POU domain, class 4, transcription factor 1; c-Ets, mammalian protooncogene homologue of the avian v-ets; E2F1, E2F transcription factor 1; HNF4, hepatic nuclear factor 4; IRF3, interferon regulatory factor 3; MEF-2C, myelin basic protein expression factor 2; NF-κB, nuclear factor of kappa light polypeptide gene enhancer in B-cells 1; Smad3, MAD homolog 3; Tat, Human immunodeficiency virus transactivator protein; TEF-4, transcriptional enhancer factor family of transcription factors; TTF1, thyroid transcription factor 1; p53, transformation related protein 53; Rb, retinoblastoma 1; APKC, atypical protein kinase C; c-Abl, c-abl oncogene 1, receptor tyrosine kinase; GSK3, glycogen synthase kinase 3; IKK, inhibitor of kappaB kinase; MAPK, mitogen-activated protein kinase; PDXP, pyridoxal (pyridoxine, vitamin B6) phosphatase; PKA, protein kinase A; PP1, protein phosphatase 1; PP2A, protein phosphatase 2; ARIP3/PIASxα, protein inhibitor of activated STAT 2; E6AP, ubiquitin protein ligase E3A; Fbw7, F-box and WD-40 domain protein 7; UBCH7, ubiquitin-conjugating enzyme E2L 3; ANCOS, ankyrin repeats containing cofactors; ASC, Activating signal cointegrator 1; CARM1, coactivator-associated arginine methyltransferase 1; CoCoA, calcium binding and coiled coil domain 1; GAC63, solute carrier family 30 (zinc transporter), member 9; GCN5, lysine acetyltransferase 2A; MMS19, MET18 S. cerevisiae; p300/CBP, E1A binding protein p300/CREB binding protein; PGC1, peroxisome proliferative activated receptor, gamma, coactivator 1; FLASH, caspase 8 associated protein 2; GAS, glutamate-rich coactivator interacting with SRC1; JAB1, JUN activation binding protein 1; Pin1, protein (peptidyl-prolyl cis/trans isomerase) NIMA-interacting 1; REGγ, proteaseome (prosome, macropain) 28 subunit, 3; SRA, steroid receptor RNA activator; TIA, TIAR, T-cell intracellular antigen 1 and related protein.
The molecular targets of SRCs are numerous (Box 1). Genetic ablation of SRC-1 altered gene expression patterns involved in cell cycle and energy metabolism pathways such as glycolysis, glycogen synthesis and fatty acid synthesis 33. Genetic ablation of SRC-2 increased gene expression for energy expenditure but decreased gene expression for energy storage 33. SRC-3 is required for the expression of several genes for cell cycle and apoptosis in breast cancer cells 34.
Box 1. SRC knockout mice.
Although SRC-1 null (Src1−/−) mice display no gross defects in development and growth, careful analyses revealed that SRC-1 plays important in vivo roles in organ physiology. In reproductive organs, SRC-1 deficiency reduces oestrogen-induced uterine growth, oestrogen- and progesterone-dependent uterine decidual response, mammary gland ductal side branching and alveolar formation, and testosterone-stimulated prostate growth 139. In addition to partial resistance to sex steroids, partial resistance to thyroid hormone (RTH) also exists in Src1−/− mice140,141. In the liver, SRC-1 is an essential coactivator for CEBPα, which together with PPARγ regulates the pyruvate carboxylase gene, the limiting enzyme for gluconeogenesis. In brown fat, activation of PPARγ triggers the recruitment of a coactivator complex containing PGC-1, SRC-1 and CBP/p300 142 and inactivation of SRC-1 impairs the thermogenic activity of PGC-1, decreasing energy expenditure and resulting in obesity with a high fat diet 143.
Assessment of Src2−/− (Tif2−/−) mice demonstrated an important function of SRC-2 in reproductive organs and in the regulation of metabolism. Src2−/− Sertoli cells are unable to support normal spermatogenesis144 and SRC-2 is the dominant member of the SRC family that mediates androgen receptor function in the testis 145. The hypofertility of female Src2−/− mice is partially due to a placental hypoplasia 144 and specific ablation of SRC-2 in the PR-positive cell linage inhibits the progesterone-induced decidual response and causes a block in embryo implantation. SRC-2 ablation also results in reduced ductal side branching and alveologenesis in the mammary gland 137. In white adipose tissue, SRC-2 deficiency increases leptin expression and decreases the expression of genes responsible for anti-lipolysis and fatty acid uptake and trapping. In brown adipose tissue, SRC-2 deficiency increases levels of UCP-1, PGC-1 and acetyl CoA oxidase, promoting energy expenditure. As a result, Src2−/− mice exhibit higher body temperature under cold conditions, less fat accumulation, lower levels of fasting glycemia and triglycerides, and higher insulin sensitivity 143. In the liver, SRC-2 enhances RORα-mediated G6Pase expression to regulate fasting hepatic glucose release from the liver. SRC-2 ablation recapitulates the human syndrome of Von Gierke’s disease (glycogen storage disease-1a) 146.
Src3−/− mice show growth retardation, delayed puberty, reduced female reproductive function and blunted mammary gland development 147, 148. In agreement with the growth retardation, circulating IGF-I levels in Src3−/− mice are significantly reduced. SRC-3 is required for vitamin D receptor-mediated expression of IGFBP-3, a binding protein maintaining IGF-I stability in the circulation 149. In the mammary gland, SRC-3 deficiency decreases oestrogen and progesterone-induced mammary ductal growth and alveologenesis, revealing SRC-3’s role in ER and PR function 147. SRC-3 is not required for castration-induced regression and testosterone-stimulated regeneration of the prostate in mice, suggesting that SRC-3 is not essential for androgen and androgen receptor-dependent prostate morphogenesis 118.
Four SRC-1 splicing isoforms have been reported 35. In comparison with SRC-1a, SRC-1b lacks a N-terminal region, while SRC-1c, SRC-1d and SRC-1e differ from SRC-1a and from each other at their unique C-terminal sequences. It has been shown that SRC-1a and SRC-1b have different abilities to enhance ERα activity in cultured cells 36. A SRC-3 isoform, AIB1-Δ3, also has been reported 37. AIB1-Δ3 lacks the N-terminal PAS/bHLH domain and it may be a more active coactivator for ERα and PR compared with the full-length SRC-3. However, the in vivo expression profiles and physiological significances of these SRC-1 and SRC-3 isoforms are currently unclear.
Posttranslational modifications of SRCs
The limiting concentrations of cellular SRCs suggest that changes in SRC levels and/or activities are efficient means for the cell to regulate gene expression. A number of studies have demonstrated that signaling pathways activated by extracellular stimuli such as hormones, growth factors and cytokines induce multiple posttranslational modifications of SRCs, including phosphorylation, ubiquitination, sumoylation, acetylation and methylation (Fig. 2). These dynamic and often reversible post translational modifications have crucial roles in determining the protein stability, the transcription factor-interactive specificity and the transcriptional activity of SRCs, and they link SRC functions to cellular responses to environmental cues. Deregulated post translational modifcation of SRC molecules have significant implications in human diseases such as cancer.
Fig. 2. Posttranslational modifications of SRCs.
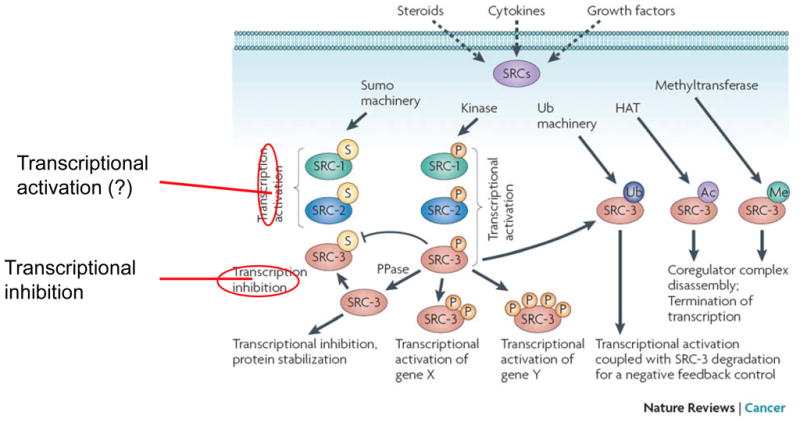
In general, phosphorylation results in activation of SRCs. In the case of SRC-3, phosphorylation determines the selectivity of SRC-3 for different transcription factors, promotes sequential ubiquitination of SRC-3 from mono-ubiquitination (activation) to poly-ubiquitination (degradation), and controls the duration of transcriptional activation by SRC-3. Conversely, de-phosphorylation by phosphatase (PPase) promotes SRC-3 sumoylation, stabilizes SRC-3 protein, and inhibits SRC-3 activity. Depending on specific kinases and phosphorylation sites, phosphorylation could either increase or decrease SRC-3 stability and cellular levels. Sumoylation enhances SRC-1 and SRC-2 activities (the “?” indicates that the conclusion is based on a limited amount of data), but it inhibits SRC-3 activity. SRC-3 acetylation and methylation cause a disassembly of the transcription complex and promote transcriptional termination.
Phosphorylation
Phosphorylation of SRCs changes their affinity for select NRs and modulates NR-dependent gene expression 38–41. Epidermal growth factor (EGF), interleukin 6 (IL-6) and cAMP treatments stimulate proline-directed kinase phosphorylation of SRC-1 on Thr1179 and Ser1185 and phosphorylated SRC-1 has higher coactivator function in both ligand-dependent and ligand-independent NR pathways 39, 42. Interestingly, EGF induced phosphorylation of SRC-1 enhances progesterone receptor (PR)-dependent transcription 39; IL6 mediated SRC-1 phosphorylation promotes androgen receptor (AR)-dependent transcription in a ligand-independent manner 42; and cAMP induced phosphorylation of SRC-1 enables recruitment of p300 and CBP and PR-dependent transcription in a ligand-independent manner 43. Furthermore, MAPK-mediated phosphorylation of SRC-1 on Thr1179 and Ser1185 increase its affinity for AR in prostate cancer cells, perhaps contributing to prostate cancer recurrence 42, 44. In endometrial cancer cells, Src-mediated SRC-1 phosphorylation significantly increases the agonist activity of tamoxifen, indicating that SRC-1 has a role in tamoxifen-induced endometrial proliferation and increase the risk of endometrial cancer associated with use of tamoxifen 45, 46.
For SRC-2, Ser736 is phosphorylated by MAPKs, including oestrogen-induced p38 and EGF-induced ERKs. Phosphorylation of Ser736 enhances SRC-2 and p300/CBP interaction and increases SRC-2 coactivation function for oestrogen receptor α (ERα), PR and AR 38, 44, 47. Protein kinase A (PKA) also phosphorylates SRC-2, resulting in a rapid increase in SRC-2 coactivator activity followed by accelerated SRC-2 degradation 48, 49.
In SRC-3, seven Ser/Thr (Thr24, Ser505, Ser543, Ser601, Ser857, Ser860 and Ser867) and one Tyr (Tyr1357) phosphorylation sites are functionally important 40, 41, 50. The kinases that phosphorylate and activate SRC-3 include MAPK, IKK, Akt (by inhibiting GSK3) and CK1δ, suggesting a role for SRC-3 in accepting signals from multiple pathways. SRC-3 is also a target of the c-Abl tyrosine kinase that can be activated by oestrogen and growth factors. Phosphorylation of Tyr1357 by c-Abl facilitates SRC-3 action in mediating ERα, PR and NF-κB-dependent transcription by enhancing SRC-3 binding to p300 and transcription factors 50. Importantly, Tyr1357 phosphorylation is elevated in ERBB2-induced mouse breast tumors, suggesting that phosphor-Tyr1357 plays a role in oncogenesis. Moreover, levels of SRC-3 Tyr1357 phosphorylation might serve as a marker for evaluating the efficacy of tyrosine kinase inhibitors.
Clinical studies have shown that SRC-3 expression levels are increased in a subset of breast tumors, and high SRC-3 levels are commonly associated with ERBB2 expression, tamoxifen resistance, disease recurrence and poor prognosis 51–53. This unfavorable outcome can be explained by a series of linked molecular events. ERBB2 expression activates MAPK and Akt and causes phosphorylation of SRC-3 and ER, resulting in transcriptional activation and cell proliferation. Furthermore, Akt can stabilize SRC-3 by inhibiting GSK3 54, suggesting a positive feedback mechanism between SRC-3 levels and Akt activity. Akt over activation is frequently observed in human cancers.
Interestingly, SRC-3 function and cellular concentrations are counter-regulated by kinases and phosphatases. Phosphatases PDXP and PP2A dephosphorylate SRC-3, inhibit SRC-3 interaction with ER and reduce SRC-3 coactivator activity 55. Phosphatase PP1 de-phosphorylates phospho-Serines 101 and 102, which decreases SRC-3 transcriptional activity and increases SRC-3 stability 55 (Fig. 2). In contrast, atypical PKC (aPKC)-mediated SRC-3 phosphorylation stabilizes SRC-3 by targeting an acidic region of amino acid residues 1031–1097 and preventing SRC-3 from interacting with the C8 subunit of the 20S core proteasome 56. Since both aPKC and SRC-3 are frequently overexpressed in cancers, they may synergistically promote carcinogenesis.
The transcriptional activities of SRCs also are regulated by subcellular localization and intracellular trafficking. SRCs contain nuclear import and export signals and locate in both cytoplasm and nucleus 57–59. Post translational modifications can alter the availability of SRCs in a subcellular compartment by regulating their nucleo-cytoplasmic trafficking. For example, TNFα and EGF-induced phopshorylation of SRC-3 causes its redistribution from cytoplasm to nucleus 60, 61.In the cytoplasm, oestrogen-induced SRC-3 phosphorylation leads to SRC-3 and ERα interaction, suggesting that activation of SRC-3 may play a role in oestrogen-induced non-genomic effects 62. SRC-3 also is associated with and phosphorylated by IKK and c-Abl in the cytoplasm in response to TNFα or growth factor stimulation, indicating a role for SRC-3 in these kinase pathways 50, 61. Furthermore, SRC-3 can interact with TIA-1 and TIAR, two translational repressors associated with AU-rich regions in the 3’-UTR of cytokine mRNAs in the cytoplasm, thereby inhibiting translation of IL-1, IL-6 and TNF-α mRNAs 63.
Ubiquitination
26S proteasome-mediated degradation regulates the function of NRs and the turnover of activated NRs and SRCs 64–68. Recently, two SCF-dependent mono-ubiquitination sites (Lys723 and Lys786) within the NR interaction domain of SRC-3 were identified 54. Phosphorylation of Ser505 by GSK3 and Ser860 by p38 MAPK not only enhances SRC-3 interaction with ER and AR but also regulates its ubiquitination and protein stability 54, 69. Sequential phosphorylation-dependent mono- to poly-ubiquitination events couple transcriptional activation with SRC-3 degradation, ensuring a proper termination of transcription (Fig. 2). In contrast to GSK3-mediated phosphorylation that causes SRC-3 degradation, aPKC-mediated SRC-3 phosphorylation protects SRC-3 from proteasomal degradation in an ERα-dependent manner, leading to an increased oestrogen-induced breast cancer cell growth 56.
Although the structure of the small ubiqitin-related modifier (SUMO) is similar to ubiquitin, the fate of a sumoylated protein is usually distinct from that of a ubiquitinated protein. Sumoylation can either antagonize SRC ubiquitination by targeting a common lysine substrate to prevent degradation and enhance its concentration, or change SRC into an inactive confirmation 70–76 (Fig. 2). In addition to ubiquitin-dependent turnover, REGγ, a proteasome activator that stimulates the trypsin-like activity of the 20S proteasome, binds and promotes SRC-3 degradation in a ubiquitin- and ATP-independent manner 77.
Acetylation and methylation
The function of SRCs can be modulated by acetylation. After transcriptional initiation, SRC-3 is acetylated by p300/CBP, resulting in disassembly of the NR and SRC-3 complex and termination of transcription 78 (Fig. 2). After initial stimulation, oestrogen treatment enhances CARM-1-mediated SRC-3 methylation on Arg1171 and terminates transcription by disassembling the SRC-3 coactivator complexes 79 and increasing SRC-3 degradation 80 (Fig. 2).
Combined posttranslational modifications
Different combinations of posttranslational modifications determine SRC coactivator potency and selectivity and allow SRCs to integrate multiple upstream signals into finely tuned regulation of gene expression. For example, retinoic acid and p38 MAPK-induced SRC-3 phosphorylation initially enhances SRC-3 interaction with retinoic acid receptor α (RARα) to activate transcription. Subsequently, the phosphorylated SRC-3 is subjected to ubiquitination and degradation, thereby leading to transcriptional termination. Another study demonstrated that phosphorylation of SRC-3 on Ser509 primed it for phosphorylation on Ser505. This then triggered SRC-3 mono- and subsequent poly-ubiquitination, which are responsible for activation and degradation of SRC-3, respectively 54 (Fig. 2). Oestrogen-induced MAPK-dependent SRC-3 phosphorylation inhibited SRC-3 sumoylation conversely, de-phosphorylation increased SRC-3 sumoylation and decreased SRC-3 activity 76. As ubiquitination and sumoylation can occur on the same lysine residue, different phosphorylation codes may determine whether SRC-3 is ubiquitinated or sumoylated, methylated or acetylated. As postranslational modifications largely influence both the cellular concentrations and the coactivator activities of SRCs, interventions that modulate SRC post translational modification have potential for controlling the detrimental roles of overexpressed SRCs in cancer.
In vivo functions of SRC Family Members
Our knowledge of the diverse in vivo functions of the SRC family is primarily gleaned from the characterization of knockout mouse models (Box 1). These studies indicate that each SRC family member has specific physiological functions. However, the additive severity of the phenotypes observed in double knockout mice indicates certain cooperative physiological functions among SRC family members. Most Src1−/−;Src2−/− mice do not survive and male and female Src1+/−;Src2−/− mice are completely infertile81. SRC-1 and SRC-3 cooperatively regulate viability, metabolism and energy balance. Most Src1−/−;Src3−/− mice die before birth, and mice that do survive exhibit compromised regulation of selective PPARγ target genes involved in adipogenesis and mitochondrial uncoupling, a higher leptin level, a developmental arrest in interscapular brown fat and a defect in adaptive thermogenesis. As a result, these mice eat more, but are lean and resistant to high-fat-diet-induced obesity due to a high basal metabolic rate and increased physical activity 82.
SRC genes in cancer
Each SRC has been found to be overexpressed in many types of human cancer, and in steroid hormone-promoted breast and prostate cancers in particular. Amplification of SRC1 or SRC2 is rare in cancers — by contrast, SRC3 on human chromosome 20q21 is more frequently amplified in cancer (Table 2, a–c). The precise mechanisms that underlie over expression of SRCs in human cancer are still unclear. Many studies have investigated the mechanisms through which SRCs promote carcinogenesis and indicate that SRCs have important and distinct roles in promoting cancer initiation, progression and metastasis through alterations of multiple signaling pathways (Fig. 3).
Table 2a.
Gene Amplification and Expression of SRC1 in Human Tumors
| Measurements | Methods | Amp. or Exp. Changes (n) | Molecular Association | Pathology Association | Refs |
|---|---|---|---|---|---|
| Breast cancer | |||||
| mRNA E | RT-PCR | lower in tumor (21) | ND | better tamoxifen response with high SRC-1 | 182 |
| Protein O | IHC | high in 28% tumors (25) | ERβ+, SRC-2+ | ND | 86 |
| Protein E | IHC | high in 29% tumors (52) | ND | LN+, recurrence tamoxifen resistance | 83 |
| Protein E | IHC | high in 25.7% tumors (70) | HER2+, PEA3+ | recurrence, poor DFS | 84 |
| Protein E | IHC | high in 19% tumors (150) | ERβ-HER2+ | recurrence, poor DFS | 85 |
| Endometrial Carcinoma | |||||
| Protein E | IHC | decrease in tumor (58) | ND | ND | 121 |
| mRNA E | qRT-PCR | increase in tumor (30) | ND | ND | 120 |
| Meningioma | |||||
| Protein E | WB, IHC | high in 81% tumors (21) | PR+ | ND | 183 |
| Ovarian cancer | |||||
| mRNA E | RT-PCR | no change in tumors (17) | ND | ND | 184 |
| Prostate cancer | |||||
| Protein E | IHC, WB | high in recurrent PCa (8) | ND | high tumor grade | 110 |
| mRNA E | RT-PCR | high in high grade cancer (20) | ND | high tumor grade | 111 |
| Protein E | IHC | high in high grade tumors (500) | ND | high Gleason score | 109 |
| Mutation | sequencing | mutated in 1% tumors (81) | Val156Ile | ND | 108 |
| Gene A | FISH | in 2.8% tumors (70) | ND | ND | 108 |
| Protein E | IHC | high in 48% tumors (254) | more nuclear SRC-1 | androgen resistance | 108 |
| mRNA E | qRT-PCR | ND (148) | ND | no significant correlation | 112 |
Amp. or A, amplification; O, overexpression; Exp. Or E, expression; DFS, disease-free survival; n, number of samples; refs, references; LN+, lymph node positive metastasis; ISH, in situ hybridization; NB, Northern blot; SB, Southern blot; IHC, immunohistochemistry; WB, Western blot; FISH, fluorescence in situ hybridization; CGH, comparative genomic hybridization; ND, not determined.
Table 2c.
Gene Amplification and Expression of SRC3 in Human Tumors
| Measurements | Methods | Amp. or Exp. Changes (n) | Molecular Association | Pathology Association | Refs |
|---|---|---|---|---|---|
| Breast cancer | |||||
| Gene A | FISH | in 9.5% tumors (105) | ND | ND | 11 |
| mRNA O | ISH | high in 64% tumors (75) | ND | big tumor size | 11 |
| Gene A | SB | in 4.8% tumors (1157) | ER+, PR+, MDM2 & FGFR1 A | big tumor size | 97 |
| mRNA O | ISH | high in 31% tumors (83) | ER−, PR−, p53+, HER2+ | high tumor grade | 51 |
| Gene A | PCR | no change (127) | ND | ND | 99 |
| mRNA O | RT-PCR | high in 13% tumors (23) | ND | ND | 99 |
| Protein O | IHC | high in 9.8% tumors (41) | ND | ND | 87 |
| Protein E | WB | high in 25% tumors (316 mix) | HER2+, PR+ | high proliferation | |
| high in 25% (119 no treat) | ND | better DFS | |||
| high in 25% (187 Tam treat) | HER2+ | worse DFS, Tam resistance | 52 | ||
| Protein O | IHC | high in 20% tumors (25) | CBP/p300 | high tumor grade | 86 |
| mRNA | qRT-PCR | high in 31.6% tumors (38) | ND | poor DFS | 98 |
| mRNA E | qRT-PCR | ND (99 ER+) | HER2+ | ND | 96 |
| Protein E | IHC | high in 73.8% tumors (560) | MMP2/9+, PEA3+ | ND | 88 |
| Colorectal Cancer | |||||
| Gene A | FISH | in 10% samples (85) | ND | ND | 119 |
| Protein O | IHC | high in 35% samples (85) | p53+, DNA aneuploidy | higher tumor grade | 119 |
| Gene A | CGH | in 32% samples (22) | ND | ND | 185 |
| Endometrial Carcinoma | |||||
| Gene A | PCR | no change (30) | ND | ND | 99 |
| mRNA O | RT-PCR | high in 17% samples (30) | ND | ND | 99 |
| mRNA E | qRT-PCR | increase in carcinomas (30) | ND | ND | 120 |
| Protein E | IHC | increase in 97% samples (84) | nuclear ER+, PRB+ | poor prognosis | 122 |
| Esophageal squamous cell carcinoma | |||||
| Gene A | SB, FISH | in 4.9% carcinomas (41) | 20q A | metastasis, poor prognosis | 186 |
| Gene A | FISH | in 13% carcinomas (221) | ND | ND | 123 |
| Protein O | IHC | high in 46% carcinomas (221) | ND | high proliferation | 123 |
| Gastric cancer | |||||
| Gene A | SB, FISH | in 7% cancer (72) | ND | ND | 124 |
| mRNA O | NB, RT-PCR | high in 40% cancer (72) | ND | metastasis, poor prognosis | 124 |
| Hepatocellular carcinoma (HCC) | |||||
| Gene A | FISH | in 23% primary HCC (35) | ND | ND | |
| in 41% metastatic HCC (33) | ND | ND | |||
| in 60% recurrent HCC (17) | ND | poor prognosis | 187 | ||
| Meningioma | |||||
| Protein E | WB, IHC | high in 76% samples (21) | ND | ND | 183 |
| Oral squamous cell carcinoma | |||||
| Gene A | CGH | ND (13) | ND | LN+ | 188 |
| Ovarian cancer | |||||
| Gene A | SB | in 7.4% samples (122) | ND | ND | 97 |
| Gene A | CGH, FISH | in 25% samples (24) | ER+ | poor prognosis | 189 |
| mRNA E | RT-PCR | no change (17) | ND | ND | 184 |
| Protein O | IHC | in 64% high grade cancer (83) | ND | high tumor grade | 125 |
| Pancreatic cancer | |||||
| Gene A | FISH | in 37% samples (46) | ND | ND | 126 |
| mRNA, Protein O | ISH, IHC | high in 65% samples (78) | ND | cancer progression | 126 |
| Prostate cancer | |||||
| mRNA, protein O | ISH, IHC | high in 38% samples (37) | ND | high tumor grade, poor DFS | 115 |
| Protein O | IHC | high in advanced cancer (480) | PSA recurrence, high pAKT | high proliferation | 116 |
Amp. or A, amplification; O, overexpression; Exp. Or E, expression; DFS, disease-free survival; n, number of samples; refs, references; LN+, lymph node positive metastasis; ISH, in situ hybridization; NB, Northern blot; SB, Southern blot; IHC, immunohistochemistry; WB, Western blot; FISH, fluorescence in situ hybridization; CGH, comparative genomic hybridization; ND, not determined.
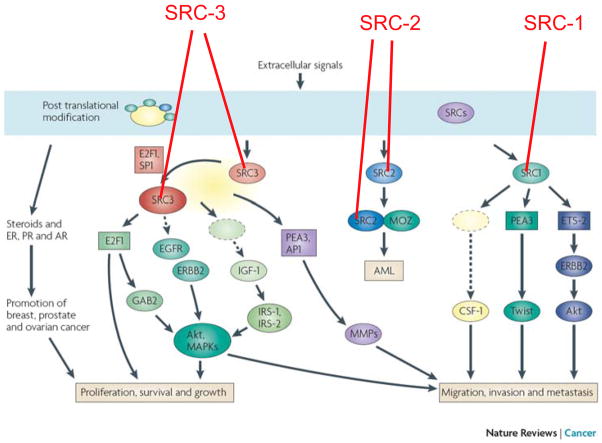
Fig. 3. SRCs promote carcinogenesis through multiple pathways.
Extracellular signals and their signaling pathways cause posttranslational modifications of SRCs, which regulate the cellular concentrations, activities and specificities of SRCs. In general, SRCs enhance steroid receptor functions and facilitate hormonal promotion of breast, prostate and ovarian cancers. Specifically, SRC-1 enhances Ets-2-mediated HER2 expression and PEA3-mediated Twist expression and upregulates CSF-1 expression to promote breast tumor cell migration, invasion and metastasis. MOZ and SRC-2 fusion gene causes AML (acute myeloid leukemia). SRC-3 upregulates its own expression through serving as a coactivator for E2F1 and SP1. The overexpressed SRC-3 enhances PEA3 and AP-1 mediated MMP expression to promote breast and prostate tumor cell metastasis. SRC-3 also enhances E2F1-mediated cell cycle progression and Gab2 expression that activates Akt. In addition, SRC-3 upregulates IGF-I, IRS-1 and IRS-2 to promote the IGF-I signaling pathway and to activate EGFR and ERBB2 to enhance Akt and MAPK activities, resulting in hyperactivation of Akt and MAPK which contribute to cancer cell proliferation, growth, survival, migration, invasion and metastasis.
SRC expression and function in breast cancer
In normal human breast, the levels of the three SRC proteins in epithelial cells are variable, but they are usually low or undetectable as assayed by immunohistochemistry (IHC) 83–88. A number of studies examined the expression profiles of SRC mRNA and protein levels in human breast tumors and these data are considered below alongside data from mouse models and cell lines.
SRC1
Several studies demonstrated that SRC-1 protein is significantly elevated in 19% to 29% of breast tumors 83–86. More importantly, SRC-1 elevation positively correlates with ERBB2 expression, lymph node metastasis, disease recurrence and poor disease-free survival (DFS) (Table 2a) 83–85. Interestingly, a recent clinical study revealed elevated SRC-1 to be an excellent independent predictor of breast cancer recurrence following therapy 89.
In vitro, SRC-1 plays a role in cancer cell proliferation and invasion through multiple pathways. In MCF-7 breast cancer cells, SRC-1 overexpression potentiates cell growth stimulated by oestrogen in accordance with an increase in expression of oestrogen-responsive genes, indicating that SRC-1 plays an important role in oestrogen receptor-mediated cell growth90. In contrast, reduction of SRC-1 in MCF-7 cells decreases oestrogen-dependent DNA synthesis and the oestrogen-responsive pS2 gene expression 91. Interestingly, MUC1, an ERα interactive protein overexpressed in breast carcinomas, can stimulate ERα-mediated transcription by aiding SRC-1 recruitment to oestrogen-responsive promoters, suggesting that SRC-1 acts in MUC1-stimulated cell growth 92. Furthermore, MCF-7 cells lacking SRC-1 do not show increases in oestrogen-induced SDF-1α expression and cell proliferation and invasion, suggesting that SRC-1 may regulate cell proliferation and invasion through autocrine/paracrine activity of the SDF-1α–CXCL12 signaling pathway 93.
In MMTV-polyoma middle T (PyMT) mammary tumor-prone mice, SRC-1 levels are increased during tumorigenesis. Knockout of Src1 does not affect PyMT-induced mammary tumor initiation and growth, but it helps to maintain epithelial differentiation and polarity, and importantly, prevents tumor cell metastasis to the lung. Further analyses revealed that SRC-1 deficiency reduced ERBB2 expression and Akt activation and also inhibited colony stimulating factor 1 (CSF-1) expression and macrophage recruitment to the tumor site 94. In addition, a recent study demonstrated that SRC-1 promotes epithelial-mesenchymal transition (EMT), migration, invasion and metastasis of mammary tumor cells by coactivating PEA3-mediated Twist expression 95.
SRC2
One study reported no significant change in the levels of SRC-2 protein in breast tumors, while another study reported the correlation of SRC-2 with cyclin D1 expression in ERα+ breast tumors 86, 96 (Table 2b). In MCF-7 breast cancer cells, knockdown of SRC-2 reduces oestrogen-induced cell proliferation and target gene expression 91 and like SRC-1, SRC-2 overexpression may also promote cell proliferation and invasion through inducing SDF-1α expression 93.
Table 2b.
Gene Amplification and Expression of SRC2 in Human Tumors
| Measurements | Methods | Amp. or Exp. Changes (n) | Molecular Association | Pathology Association | Refs |
|---|---|---|---|---|---|
| Breast cancer | |||||
| Protein E | IHC | no change (25) | SRC-1+ | LN+ | 86 |
| mRNA E | qRT-PCR | ND (99, ER+) | PR+, high cyclin D1 | ND | 96 |
| Endometrial Carcinoma | |||||
| mRNA E | qRT-PCR | increase in carcinomas (30) | ND | ND | 120 |
| Meningioma | |||||
| Protein E | WB, IHC | high in76% samples (21) | PR+ | ND | 183 |
| Ovarian cancer | |||||
| mRNA E | RT-PCR | no change (17) | ND | ND | 184 |
| Prostate cancer | |||||
| Protein E | IHC, WB | high in high grade tumors (16) | ND | high tumor grade | 110 |
| Protein E | IHC | variable (518) | ND | recurrence & proliferation | 114 |
| mRNA E | qRT-PCR | ND (148) | ND | no significant correlation | 112 |
Amp. or A, amplification; O, overexpression; Exp. Or E, expression; DFS, disease-free survival; n, number of samples; refs, references; LN+, lymph node positive metastasis; ISH, in situ hybridization; NB, Northern blot; SB, Southern blot; IHC, immunohistochemistry; WB, Western blot; FISH, fluorescence in situ hybridization; CGH, comparative genomic hybridization; ND, not determined.
SRC3
In separate studies of breast tumors, the frequencies of SRC3 amplification were 9.5% as measured by FISH 11 and 4.8% as measured by Southern blot 97. The levels of SRC-3 mRNA and protein in breast cancer have been extensively analyzed. SRC-3 mRNA overexpression was found in 64% 11, 31.6% 98, 31% 51 and 13% 99 of independent breast tumor cohorts. SRC-3 protein overproduction was identified in 9.8% 87 and 20% 86 of two different breast tumor cohorts. Interestingly, SRC-3 overexpression in breast cancer usually correlates with the expression of ERBB2, matrix metalloproteinase 2 (MMP2), MMP9 and PEA3 and with larger tumor size, higher tumor grade, and/or poor DFS 11, 51, 52, 86, 88, 96–98 (Table 2c). In tamoxifen-treated patients, high levels of SRC-3 expression are associated with tamoxifen resistance and a poorer DFS. Importantly, patients with high levels of both SRC-3 and ERBB2 exhibit early and severe resistance to selective oestrogen receptor modulators (SERM) therapy 52. As ERBB2 activates MAPK, which in turn phosphorylates ER and activates SRC-3 100, the overexpression of both ERBB2 and SRC-3 significantly enhances the agonist activity of tamoxifen and therefore, reduces its antitumor activity in patients with breast cancer 101.
In breast cancer cells, SRC-3 can be recruited to the oestrogen responsive CCND1 promoter to enhance cyclin D1 expression 102 – depletion of SRC-3 in MCF-7 cells significantly reduces oestrogen-mediated cell proliferation and survival. Down-regulation of SRC-3 in MCF-7 cells also reduces oestrogen-dependent colony formation in soft agar and tumor growth in nude mice 103.
The in vivo role of SRC-3 in breast cancer initiation and progression has been investigated in multiple mouse models. In mice harboring the MMTV–v-ras (ras) transgene, breast tumor incidence was reduced dramatically in Src3−/−;ras virgin mice and inhibited completely in ovariectomized Src3−/−;ras mice 104. Breast tumor latency and growth were delayed significantly in Src3−/−;ras virgin mice with natural oestrous cycles, multiparous mice with cyclically elevated reproductive hormones and virgin mice bearing pituitary isografts with persistently elevated hormones. Interestingly, SRC-3 deficiency did not alter the expression of oestrogen and progesterone-responsive genes in the mammary gland and tumors, but it caused partial resistance to insulin-like growth factor I (IGF1) because of a significant reduction in insulin receptor substrate-1 (IRS-1) and IRS-2. The impaired IGF-I signaling pathway in Src3−/−;ras mammary epithelium and tumor cells may be responsible in part for the suppression of mammary tumorigenesis and metastasis.
In a second model, the role of SRC-3 in susceptibility of the mammary gland to chemical carcinogens was characterized 105. This study demonstrated that mammary ductal outgrowths emanating from the Src3−/− mammary epithelial transplants in WT mice were attenuated, indicating that the role of SRC-3 in mammary ductal growth is mammary epithelial cell autonomous. In mice treated with the chemical carcinogen 7,12-dimethylbenz[a]anthracene (DMBA), SRC-3 deficiency protected the mammary gland, but not the skin, from tumorigenesis. In this model, SRC-3 deficiency also suppressed the up-regulation of both IRS-1 and IRS-2 and thereby inhibited the activation of Akt, the expression of cyclin D1 and cell proliferation.
In a third model, the role of SRC-3 in breast tumorigenesis was assessed in MMTV-Erbb2-induced mammary tumorigenesis in WT and Src3−/− mice 106. This study showed that ERBB2-induced mammary tumor development was significantly delayed in Src3+/− mice and completely suppressed in Src3−/− mice. In comparison with WT;Erbb2 tumors, Src3+/−;Erbb2 tumors exhibited a decrease in phosphorylated ERBB2, cyclin D1 and cyclin E, reduced activity of Akt and JNK, and decreased cell proliferation. These findings suggest that SRC-3 is required for ERBB2 oncogenic activity and SRC-3 reduction in the mammary epithelium should potentiate therapies aimed at inhibiting ERBB2 signaling in breast cancer 106.
In a fourth model, the role of SRC-3 in breast cancer metastasis was investigated by using MMTV-PyMT mice 88. This study demonstrated that genetic ablation of Src3 in MMTV-PyMT mice significantly reduced lung metastasis and the effect was cell autonomous. Cellular and molecular analyses revealed that SRC-3 interacted with PEA3 and directly enhanced the activity of the MMP2 and MMP9 promoters and increased the expression of MMP2 and MMP9 in WT;PyMT tumor cells. Therefore, SRC-3 can aid breast cancer metastasis through MMP2- and MMP9-mediated EMT and cell invasiveness. This correlates with data from human breast tumors where SRC-3 expression is associated with high PEA3, MMP2 and MMP9 expression levels 88.
Finally and importantly, the oncogenic role of SRC-3 overexpression in the mammary gland was directly demonstrated by generating and characterizing MMTV-Src3 transgenic mice. This study demonstrated that SRC-3 overexpression caused mammary hypertrophy, hyperplasia, abnormal post-weaning involution, and the spontaneous development of malignant mammary tumors. Tumor incidence was increased in other organs, including the pituitary and uterus 107. In agreement with the down-regulation of the IGF-I signaling pathway in Src3−/−;ras mammary tumors 104, SRC-3 overexpression-induced mammary tumors have a hyperactive IGF-I signaling pathway 107.
In summary, consensus results indicate that both SRC-1 and SRC-3 are overexpressed in a subpopulation of breast cancers without clear relevance to ER and PR expression. Overproduction of SRC-1 and SRC-3 are generally detrimental to patients. SRC-2 expression profiles in breast cancer are less conclusive due to lack of sufficient data. Data from mouse models and cell lines indicate that SRC-1 and SRC-3 have important functions in breast cancer, including tumour initiation and progression. Data from SRC-3 mouse models highlight the oncogenic nature of this protein when its expression is deregulated.
Prostate cancer
The expression profiles of SRC family members have been investigated in prostate tumors (Table 2, a–c). Amplification of SRC-1 is rare in human tumors and one study reported that SRC1 was amplified in only 2 out of 70 specimens that included prostate cancer cell lines, xenografts and primary tumors108. However, several studies have found that the expression levels of both SRC-1 mRNA and protein are positively correlated with prostate tumor grades 109–111. In addition, one study found more nuclear localized SRC-1 protein in androgen-independent prostate tumors 108, while another study using quantitative RT-PCR failed to detect significant correlations between SRC-1 mRNA and prostate tumor progression 112. Overall, SRC-1 is thought to be elevated in certain prostate tumors, but its overexpression frequency has not been accurately determined.
In prostate cancer cells, SRC-1 is able to enhance AR-mediated cell proliferation in culture. Src1 knockdown inhibits proliferation of the androgen-dependent LNCaP cells in culture. More interestingly, SRC-1 knockdown inhibits the growth of C4-2 prostate cancer cells that depend on AR for growth in androgen-depleted medium 109. However, reduction of SRC-1 has no effect on the growth of the AR-negative PC-3 and DU145 prostate cancer cells 109. These results suggest that SRC-1 promotes prostate cancer growth through enhancing AR function in an androgen-dependent and -independent manner.
The in vivo role of SRC-1 in prostate cancer has been investigated in transgenic adenocarcinoma of mouse prostate (TRAMP) mice harboring the SV40 large and small T antigen transgene driven by a probasin promoter113. During prostate carcinogenesis, SRC-1 protein levels remained constant in prostate tumors of TRAMP mice. TRAMP;Src1−/− mice exhibited similar prostate tumor initiation, progression and metastasis when compared to that observed in WT mice. Interestingly, in both WT and Src1−/− TRAMP mice, prostate carcinogenesis induced SRC-3 overexpression. Thus, the role of SRC-1 in murine prostate carcinogenesis may be nonessential due to possible compensations from SRC-3 or overexpression of other coactivators.
SRC-2 expression in prostate tumors was found to be elevated but the frequency of SRC-2 overexpression has not been characterized well. It was reported that SRC-2 expression in a subgroup of prostate cancer was positively associated with high tumor cell proliferation, high tumor grade and/or disease recurrence 110, 114. In AR-positive prostate cancer cells, high levels of androgen repress SRC-2 expression. Depletion of SRC-2 reduces AR target gene expression, and inhibits proliferation of AR-dependent and AR-independent prostate cancer cells, suggesting that SRC-2 also enhances prostate cancer cell growth through both AR-dependent and AR-independent pathways 114.
SRC-3 expression in prostate tumors has been examined in two studies. One study with a small number of samples found that SRC-3 protein was overproduced in 38% of tumor samples 115. The other study with a large number of tumor samples found that SRC-3 levels were positively correlated with increasing PSA levels indicating tumour recurrence, Akt activation and tumor cell proliferation 116. In prostate cancer cells, SRC-3 activates the Akt–mTOR signaling pathway and stimulates cell growth by increasing cell size. SRC-3 knockdown decreases cancer cell proliferation and xenograft tumor growth in nude mice and increases apoptosis 116, 117. The role of SRC-3 in spontaneous prostate cancer was investigated by comparing tumor initiation and progression in TRAMP and TRAMP;Src3−/− mice 118. SRC-3 was nonessential for normal androgen–AR-dependent prostate growth and high levels of SRC-3 expression were found in stromal and basal cells, but not in luminal epithelial cells. Of note, high levels of SRC-3 expression were detected in prostate tumor cells during progression toward more malignant stages. In agreement with this finding, development of prostate epithelial hyperplasia and the initiation of well-differentiated tumors were not affected by loss of SRC3, but it efficiently arrested prostate tumor growth and progression at the well-differentiated stage and significantly extended the animal life 118. These results indicate that de novo induction of SRC-3 expression in partially transformed epithelial cells is essential for progression of prostate tumorigenesis into poorly differentiated carcinoma. Inhibition of SRC-3 expression or function in the prostate epithelium may be a potential strategy to suppress prostate cancer progression. Taken together, these findings suggest that overexpressed SRC family members have detrimental roles in promotion of prostate cancer initiation and progression.
Other cancers
The expression profiles of SRC family members in other types of cancers are summarized in Table 2, a–c). Briefly, in colorectal cancer, high levels of SRC-3 protein were detected in 35% tumors and were positively associated with tumor grades 119. In endometrial carcinoma, one study reported an increase in SRC-1 and SRC-2 mRNAs 120, while another study reported a decrease in SRC-1 protein 121. SRC-3 mRNA and protein levels were found to be increased in endometrial carcinomas, and various overexpression frequencies were reported in different studies 99, 120, 122. SRC-3 overexpression was detected in 46% of esophageal squamous cell carcinomas and 40% of gastric cancers, and SRC-3 expression levels were positively associated with higher cell proliferation, metastasis and poor prognosis 123, 124. Up-regulated SRC-3 protein was found in 64% of high-grade ovarian cancer specimens and 65% of pancreatic cancer specimens; SRC-3 levels in these cancers were positively correlated with disease degrees125, 126.
An abnormal chromosomal rearrangement of inv(8)(p11p13) has been identified in a subset of patients with acute myeloid leukemia (AML) 127, 128. This rearrangement results in a fusion between the 5’ portion of MOZ mRNA and the 3’ portion of SRC2 mRNA; the fusion encodes a transcriptional activator domain that binds nucleosomes through MOZ and recruits CBP through a SRC-2 domain 127–129. MOZ–SRC-2-transduced myeloid progenitors can be continuously passaged in culture and induce acute myeloid leukemia in vivo 130. These findings reinforce the concept that deregulation of a transcriptional coactivator can lead to malignant disease in humans.
Intriguingly, one study reported that SRC-3 could inhibit IκB kinase to stabilize IκB and suppress NF-κB activation in the lymphoid lineage 131. Genetic ablation of SRC-3 resulted in the release of IκB kinase inhibition and constitutive NF-κB activation. Consequently, a subset of old Src3−/− mice developed lymphocyte proliferation and spontaneous β-cell lymphoma 131. Another study demonstrated that SRC-3 deficiency in macrophages did not affect NF-κB-mediated gene transcription, but inhibited the production of proinflammatory cytokines such as IL-1, IL-6 and TNF-α at the translational level 63. It is currently unknown if the increased inflammatory cytokines in Src3−/− mice are responsible in part for the development of β-cell lymphoma. Taken together, these findings suggest that SRC-3 regulates cell proliferation in a cellular and signaling context-dependent manner, ranging from proliferative and tumorigenenic effects in most endocrine target organ epithelial cells to paradoxical antiproliferative effects in lymphoid cells.
Overall, one can conclude that miss-expression of SRC family members occurs not only in steroid hormone-promoted breast and prostate cancers but also in steroid-independent cancers, suggesting SRC family members promote carcinogenesis through both steroid dependent and independent pathways.
Summary and perspectives
Since SRC-1 was identified as a NR coactivator in 1995 6, the NR coregulator field has experienced an explosion in research aimed at understanding the mechanism of NR coactivator function, much of which has been applied to studies of hormone-related cancers. Emanating from these studies, many previously unknown proteins such as SRCs, MTA1 and PGC-1 have been found to be ‘Master Genetic Regulators’ 2. In this review, we have summarized our present knowledge about the molecular features, post translational modifications (Fig. 2), interactive proteins (Table 1), molecular targets and physiological functions of SRCs. By interacting with and coactivating most NRs and certain other transcription factors, SRCs coordinate expression of many genes directed to accomplishing larger physiological goals. Importantly, overexpression of SRC-3 in mammary epithelial cells has been shown to be sufficient to induce spontaneous mammary tumors in mice, indicating that overexpression of SRC-3 is oncogenic 107. Conversely, knockout of SRC-3 in mice suppresses oncogene- and carcinogen-induced breast cancer initiation, progression and metastasis 88, 104–106. However, knockout of SRC-3 only inhibits prostate cancer progression and metastasis and does not affect the initiation of prostate tumorigenesis in TRAMP mice 118. Interestingly, knockout of SRC-1 only suppresses mammary tumor metastasis without affecting primary tumor formation, and does not affect prostate cancer initiation and progression 94, 95, suggesting differing contributions from SRC-1 and SRC-3.
Many studies have investigated the mechanisms responsible for SRCs to promote carcinogenesis. These studies revealed that SRC-1 can enhance the expression of ERBB2, colony stimulating factor 1 (CSF-1) and TWIST expression to promote breast cancer metastasis 94, 95. SRC-3 overexpression can enhance v-ras-mediated cell transformation and activate ERα, EGFR and cyclin D1 expression, as well as the Akt pathway, MMPs and FAK to promote tumor initiation, growth and metastasis 88, 102, 104–106, 117, 132, 133.
Although progress has been made in understanding the function of SRCs in various cancers and the molecular mechanisms by which SRCs influence carcinogenesis, important questions remain to be addressed. First, the molecular mechanisms responsible for SRC-1 and SRC-3 gene amplification and mRNA overexpression in cancers are still not fully understood. A limited number of studies suggested that SRC-3 elevation in cancers might be attributed to both transcriptional activation and posttranslational stabilization. On one hand, SRC-3 serves as a coactivator for E2F1, which drives cell proliferation 134, 135. Both E2F1 and SRC-3 can be tethered to the proximal region of the SRC-3 gene promoter through interacting with SP1, which enhances SRC-3 mRNA transcription. In this manner, E2F1 and SRC-3 form a positive regulatory loop that can constitutively enhance SRC-3 overexpression in cancer cells (Fig. 3) 136. On the other hand, regulation of SRC-3 protein stability by posttranslational modifications represents another important mechanism for increasing SRC-3 protein levels in cancer (reviewed in preceding sections). Further investigations on the molecular mechanisms responsible for the miss-expression of SRC-3 and other SRC family members may provide approaches to reprogram their expression levels for cancer therapy.
Second, although some genes important for carcinogenesis and metastasis have been identified as SRC-regulated genes, a complete catalog of the direct targets of SRCs in cancer cells is still unavailable. Characterization of all SRC target genes and a proteomic profile of the transcription factors that work with SRCs will provide important insights into the gene networks and pathways used by SRCs to promote carcinogenesis. Third, most in vivo experiments to date have been carried out in knockout mice where certain systemic effects, such as changes in IGF-1 activity, exist. It will be important to further assess and validate the roles of SRCs in carcinogenesis by greater use of conditional knockout mice 137, 138 with cell type-specific Src ablations. In addition, development of stage-specific inactivation (or overexpression) of SRCs in breast and prostate tumors in mice will allow us to determine if SRCs are stage-dependent therapeutic targets. Finally, the most challenging of all tasks will be to translate the knowledge obtained from basic research to clinical applications and to develop clinically deliverable reagents that can counteract the excessive SRC activities in SRC-promoted cancers.
Online 'at-a-glance' summary.
SRC-1 was the first cloned steroid receptor coactivator that interacts with steroid hormone receptors to promote transcriptional activation in a hormone-dependent manner.
The p160 SRC family contains three homologous members, SRC-1, SRC-2 and SRC-3. These SRCs interact with nuclear receptors and certain other transcription factors and recruit chromatin-remodeling and other transcriptional enzymes to facilitate the assembly of general transcription factors for transcriptional activation.
SRCs are targets of multiple signaling pathways for posttranslational modifications. These posttranslational modifications determine or modulate SRC protein stability, subcellular localization, functional specificity, coactivator activity and/or coactivator complex assembly or disassembly.
The phenotypes of knockout mice for individual and combinatorial SRC family genes revealed that SRCs are involved in many physiological processes and each SRC has both specific and redundant physiological functions in development and organ function.
SRC-1 expression is elevated in a subset of breast cancers and is positively correlated with ERBB2 positivity and poor disease-free survival rate. Knockdown of SRC-1 in breast cancer cells inhibits cell proliferation.
Knockout of SRC-1 in MMTV-PyMT mice suppresses metastasis without affecting primary tumor formation. SRC-1 promotes breast cancer metastasis through upregulating ERBB2, CSF-1 and Twist expression.
Both gene amplification and overexpression of SRC-3 are present in a subset of breast cancers. SRC-3 overexpression usually correlates with the expression of ERBB2, MMP2, MMP9 and PEA3 and with larger tumor size, higher tumor grade, and/or poor disease-free survival.
SRC-3 plays an important role to promote breast tumor cell proliferation, migration, invasion and metastasis through many mechanisms such as enhancing ERα and E2F1 functions, IGF-I signaling pathway, EGF receptor and ERBB2 activation, as well as the expression of MMPs.
Knockout of SRC-3 in mice suppresses mammary tumor initiation, growth and metastasis, while overexpression of SRC-3 in mouse mammary epithelial cells is sufficient to induce spontaneous mammary tumorigenesis.
SRC-3 expression is elevated during prostate tumorigenesis in mice. Knockout of SRC-3 efficiently arrests prostate tumor progression at a well-differentiated stage.
Acknowledgments
This work was funded by National Institutes of Health grants (R01DK058242, R01CA112403 and R01CA119689 to J.X.; P01DK059820, R01HD07857 and R01HD08818 to B.W.O.), NIDDK-NURSA, an American Cancer Society Research Scholar Award (ACS #RSG-05-082-01 to J.X.), and a Susan Komen for the Cure Award (BCTR0707225 to R.W.).
Glossary terms
- pS2 gene
The human pS2 gene in estrogen receptor α (ERα)-positive breast cancer cells such as MCF-7 cells is a direct target gene of ERα. Upon estrogen treatment, pS2 mRNA expression can be significantly induced within 15 minutes
- Pituitary isograft
A pituitary gland isolated from a syngenic donor mouse is implanted into the kidney capsule of the recipient mouse. Upon the stimulation of the implanted pituitary isograft, the recipient mouse shows significantly increased levels of prolactin, progesterone and estradiol
Biographies
Jianming Xu: Jianming Xu got his Ph.D. from Clarkson University and W. Alton John’s Cell Science Center, NY, USA and completed postdoctoral training in Baylor College of Medicine, TX, USA. He is currently an associate professor in Baylor College of Medicine and a guest professor in Luzhou Medical College. His research interests include the roles of transcriptional coregulators in development, organ function and breast and prostate cancers, as well as transcriptional control of epithelial-mesenchymal transition and cancer metastasis.
Ray-Chang Wu: Ray-Chang Wu got his Ph.D. from University of Southern California, CA, USA and completed his postdoctoral training in Baylor College of Medicine, TX, USA. He is currently an assistant professor in Baylor College of Medicine. His research interests include the signaling and function of nuclear hormone receptors and coactivators in tumorigenesis.
Bert W. O’Malley: Bert W. O’Malley is the Tom C. Thompson Professor and Chair of the Department of Molecular and Cellular Biology at Baylor College of Medicine, TX, USA. He is a pioneer contributor to the field of hormone action, molecular endocrinology and endocrine cancers. His current research interests include the mechanistic, physiologic and pathologic roles of transcriptional coregulators.
Footnotes
Further Information
J. Xu’s homepage: http://www.bcm.edu/mcb/?PMID=7714
B.W. O’Malley’s homepage: http://www.bcm.edu/mcb/?PMID=7694
Contributor Information
Jianming Xu, Email: jxu@bcm.tmc.edu.
Ray-Chang Wu, Email: rwu@bcm.tmc.edu.
Bert W. O’Malley, Email: berto@bcm.tmc.edu.
References
- 1.Spelsberg TC, Steggles AW, O'Malley BW. Progesterone-binding components of chick oviduct. 3. Chromatin acceptor sites. J Biol Chem. 1971;246:4188–97. [PubMed] [Google Scholar]
- 2.O'Malley BW. Coregulators: from whence came these “master genes”. Mol Endocrinol. 2007;21:1009–13. doi: 10.1210/me.2007-0012. [DOI] [PubMed] [Google Scholar]
- 3.Meyer ME, et al. Steroid hormone receptors compete for factors that mediate their enhancer function. Cell. 1989;57:433–42. doi: 10.1016/0092-8674(89)90918-5. [DOI] [PubMed] [Google Scholar]
- 4.Halachmi S, et al. Estrogen receptor-associated proteins: possible mediators of hormone-induced transcription. Science. 1994;264:1455–8. doi: 10.1126/science.8197458. [DOI] [PubMed] [Google Scholar]
- 5.Klein-Hitpass L, et al. The progesterone receptor stimulates cell-free transcription by enhancing the formation of a stable preinitiation complex. Cell. 1990;60:247–57. doi: 10.1016/0092-8674(90)90740-6. [DOI] [PubMed] [Google Scholar]
- 6.Onate SA, Tsai SY, Tsai MJ, O'Malley BW. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–7. doi: 10.1126/science.270.5240.1354. This article identified SRC-1 as the first steroid receptor coactivator. [DOI] [PubMed] [Google Scholar]
- 7.Voegel JJ, Heine MJ, Zechel C, Chambon P, Gronemeyer H. TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. Embo J. 1996;15:3667–75. [PMC free article] [PubMed] [Google Scholar]
- 8.Hong H, Kohli K, Garabedian MJ, Stallcup MR. GRIP1, a transcriptional coactivator for the AF-2 transactivation domain of steroid, thyroid, retinoid, and vitamin D receptors. Mol Cell Biol. 1997;17:2735–44. doi: 10.1128/mcb.17.5.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Torchia J, et al. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–84. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 10.Li H, Gomes PJ, Chen JD. RAC3, a steroid/nuclear receptor-associated coactivator that is related to SRC-1 and TIF2. Proc Natl Acad Sci U S A. 1997;94:8479–84. doi: 10.1073/pnas.94.16.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anzick SL, et al. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277:965–8. doi: 10.1126/science.277.5328.965. This article first reported that SRC-3/AIB1 was amplified and overexpressed in breast cancer. [DOI] [PubMed] [Google Scholar]
- 12.Chen H, et al. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–80. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 13.Takeshita A, Cardona GR, Koibuchi N, Suen CS, Chin WW. TRAM-1, A novel 160-kDa thyroid hormone receptor activator molecule, exhibits distinct properties from steroid receptor coactivator-1. J Biol Chem. 1997;272:27629–34. doi: 10.1074/jbc.272.44.27629. [DOI] [PubMed] [Google Scholar]
- 14.Chen YH, Kim JH, Stallcup MR. GAC63, a GRIP1-dependent nuclear receptor coactivator. Mol Cell Biol. 2005;25:5965–72. doi: 10.1128/MCB.25.14.5965-5972.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim JH, Li H, Stallcup MR. CoCoA, a nuclear receptor coactivator which acts through an N-terminal activation domain of p160 coactivators. Mol Cell. 2003;12:1537–49. doi: 10.1016/s1097-2765(03)00450-7. [DOI] [PubMed] [Google Scholar]
- 16.Lee YH, Campbell HD, Stallcup MR. Developmentally essential protein flightless I is a nuclear receptor coactivator with actin binding activity. Mol Cell Biol. 2004;24:2103–17. doi: 10.1128/MCB.24.5.2103-2117.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Belandia B, Parker MG. Functional interaction between the p160 coactivator proteins and the transcriptional enhancer factor family of transcription factors. J Biol Chem. 2000;275:30801–5. doi: 10.1074/jbc.C000484200. [DOI] [PubMed] [Google Scholar]
- 18.Chen SL, Dowhan DH, Hosking BM, Muscat GE. The steroid receptor coactivator, GRIP-1, is necessary for MEF-2C-dependent gene expression and skeletal muscle differentiation. Genes Dev. 2000;14:1209–28. [PMC free article] [PubMed] [Google Scholar]
- 19.Darimont BD, et al. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 1998;12:3343–56. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heery DM, Kalkhoven E, Hoare S, Parker MG. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–6. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 21.Voegel JJ, et al. The coactivator TIF2 contains three nuclear receptor-binding motifs and mediates transactivation through CBP binding-dependent and -independent pathways. Embo J. 1998;17:507–19. doi: 10.1093/emboj/17.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anafi M, et al. GCN5 and ADA adaptor proteins regulate triiodothyronine/GRIP1 and SRC-1 coactivator-dependent gene activation by the human thyroid hormone receptor. Mol Endocrinol. 2000;14:718–32. doi: 10.1210/mend.14.5.0457. [DOI] [PubMed] [Google Scholar]
- 23.Brown K, Chen Y, Underhill TM, Mymryk JS, Torchia J. The coactivator p/CIP/SRC-3 facilitates retinoic acid receptor signaling via recruitment of GCN5. J Biol Chem. 2003;278:39402–12. doi: 10.1074/jbc.M307832200. [DOI] [PubMed] [Google Scholar]
- 24.Huang SM, Stallcup MR. Mouse Zac1, a transcriptional coactivator and repressor for nuclear receptors. Mol Cell Biol. 2000;20:1855–67. doi: 10.1128/mcb.20.5.1855-1867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koh SS, Chen D, Lee YH, Stallcup MR. Synergistic enhancement of nuclear receptor function by p160 coactivators and two coactivators with protein methyltransferase activities. J Biol Chem. 2001;276:1089–98. doi: 10.1074/jbc.M004228200. [DOI] [PubMed] [Google Scholar]
- 26.Liu PY, Hsieh TY, Chou WY, Huang SM. Modulation of glucocorticoid receptor-interacting protein 1 (GRIP1) transactivation and co-activation activities through its C-terminal repression and self-association domains. Febs J. 2006;273:2172–83. doi: 10.1111/j.1742-4658.2006.05231.x. [DOI] [PubMed] [Google Scholar]
- 27.Ma H, et al. Multiple signal input and output domains of the 160-kilodalton nuclear receptor coactivator proteins. Mol Cell Biol. 1999;19:6164–73. doi: 10.1128/mcb.19.9.6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Surapureddi S, et al. Identification of a transcriptionally active peroxisome proliferator-activated receptor alpha -interacting cofactor complex in rat liver and characterization of PRIC285 as a coactivator. Proc Natl Acad Sci U S A. 2002;99:11836–41. doi: 10.1073/pnas.182426699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yao TP, Ku G, Zhou N, Scully R, Livingston DM. The nuclear hormone receptor coactivator SRC-1 is a specific target of p300. Proc Natl Acad Sci U S A. 1996;93:10626–31. doi: 10.1073/pnas.93.20.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spencer TE, et al. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–8. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 31.Xu J, Li Q. Review of the in vivo functions of the p160 steroid receptor coactivator family. Mol Endocrinol. 2003;17:1681–92. doi: 10.1210/me.2003-0116. [DOI] [PubMed] [Google Scholar]
- 32.Zhang H, et al. Differential gene regulation by the SRC family of coactivators. Genes Dev. 2004;18:1753–65. doi: 10.1101/gad.1194704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jeong JW, et al. The genomic analysis of the impact of steroid receptor coactivators ablation on hepatic metabolism. Mol Endocrinol. 2006;20:1138–52. doi: 10.1210/me.2005-0407. [DOI] [PubMed] [Google Scholar]
- 34.Oh A, et al. The nuclear receptor coactivator AIB1 mediates insulin-like growth factor I-induced phenotypic changes in human breast cancer cells. Cancer Res. 2004;64:8299–308. doi: 10.1158/0008-5472.CAN-04-0354. [DOI] [PubMed] [Google Scholar]
- 35.Kamei Y, et al. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–14. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 36.Kalkhoven E, Valentine JE, Heery DM, Parker MG. Isoforms of steroid receptor co-activator 1 differ in their ability to potentiate transcription by the oestrogen receptor. Embo J. 1998;17:232–43. doi: 10.1093/emboj/17.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reiter R, Wellstein A, Riegel AT. An isoform of the coactivator AIB1 that increases hormone and growth factor sensitivity is overexpressed in breast cancer. J Biol Chem. 2001;276:39736–41. doi: 10.1074/jbc.M104744200. [DOI] [PubMed] [Google Scholar]
- 38.Lopez GN, Turck CW, Schaufele F, Stallcup MR, Kushner PJ. Growth factors signal to steroid receptors through mitogen-activated protein kinase regulation of p160 coactivator activity. J Biol Chem. 2001;276:22177–82. doi: 10.1074/jbc.M010718200. [DOI] [PubMed] [Google Scholar]
- 39.Rowan BG, Weigel NL, O'Malley BW. Phosphorylation of steroid receptor coactivator-1. Identification of the phosphorylation sites and phosphorylation through the mitogen-activated protein kinase pathway. J Biol Chem. 2000;275:4475–83. doi: 10.1074/jbc.275.6.4475. [DOI] [PubMed] [Google Scholar]
- 40.Wu RC, et al. Selective phosphorylations of the SRC-3/AIB1 coactivator integrate genomic reponses to multiple cellular signaling pathways. Mol Cell. 2004;15:937–49. doi: 10.1016/j.molcel.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 41.Giamas G, et al. CK1delta modulates the transcriptional activity of ERalpha via AIB1 in an estrogen-dependent manner and regulates ERalpha-AIB1 interactions. Nucleic Acids Res. 2009;37:3110–23. doi: 10.1093/nar/gkp136. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Ueda T, Mawji NR, Bruchovsky N, Sadar MD. Ligand-independent activation of the androgen receptor by interleukin-6 and the role of steroid receptor coactivator-1 in prostate cancer cells. J Biol Chem. 2002;277:38087–94. doi: 10.1074/jbc.M203313200. [DOI] [PubMed] [Google Scholar]
- 43.Rowan BG, Garrison N, Weigel NL, O'Malley BW. 8-Bromo-cyclic AMP induces phosphorylation of two sites in SRC-1 that facilitate ligand-independent activation of the chicken progesterone receptor and are critical for functional cooperation between SRC-1 and CREB binding protein. Mol Cell Biol. 2000;20:8720–30. doi: 10.1128/mcb.20.23.8720-8730.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gregory CW, et al. Epidermal growth factor increases coactivation of the androgen receptor in recurrent prostate cancer. J Biol Chem. 2004;279:7119–30. doi: 10.1074/jbc.M307649200. [DOI] [PubMed] [Google Scholar]
- 45.Shang Y, Brown M. Molecular determinants for the tissue specificity of SERMs. Science. 2002;295:2465–8. doi: 10.1126/science.1068537. [DOI] [PubMed] [Google Scholar]
- 46.Shah YM, Rowan BG. The Src kinase pathway promotes tamoxifen agonist action in Ishikawa endometrial cells through phosphorylation-dependent stabilization of estrogen receptor (alpha) promoter interaction and elevated steroid receptor coactivator 1 activity. Mol Endocrinol. 2005;19:732–48. doi: 10.1210/me.2004-0298. [DOI] [PubMed] [Google Scholar]
- 47.Frigo DE, et al. p38 mitogen-activated protein kinase stimulates estrogen-mediated transcription and proliferation through the phosphorylation and potentiation of the p160 coactivator glucocorticoid receptor-interacting protein 1. Mol Endocrinol. 2006;20:971–83. doi: 10.1210/me.2004-0075. [DOI] [PubMed] [Google Scholar]
- 48.Borud B, et al. The nuclear receptor coactivators p300/CBP/cointegrator-associated protein (p/CIP) and transcription intermediary factor 2 (TIF2) differentially regulate PKA-stimulated transcriptional activity of steroidogenic factor 1. Mol Endocrinol. 2002;16:757–73. doi: 10.1210/mend.16.4.0799. [DOI] [PubMed] [Google Scholar]
- 49.Hoang T, et al. cAMP-dependent protein kinase regulates ubiquitin-proteasome-mediated degradation and subcellular localization of the nuclear receptor coactivator GRIP1. J Biol Chem. 2004;279:49120–30. doi: 10.1074/jbc.M409746200. [DOI] [PubMed] [Google Scholar]
- 50.Oh AS, et al. Tyrosine phosphorylation of the nuclear receptor coactivator AIB1/SRC-3 is enhanced by Abl kinase and is required for its activity in cancer cells. Mol Cell Biol. 2008;28:6580–93. doi: 10.1128/MCB.00118-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bouras T, Southey MC, Venter DJ. Overexpression of the steroid receptor coactivator AIB1 in breast cancer correlates with the absence of estrogen and progesterone receptors and positivity for p53 and HER2/neu. Cancer Res. 2001;61:903–7. [PubMed] [Google Scholar]
- 52.Osborne CK, et al. Role of the estrogen receptor coactivator AIB1 (SRC-3) and HER-2/neu in tamoxifen resistance in breast cancer. J Natl Cancer Inst. 2003;95:353–61. doi: 10.1093/jnci/95.5.353. [DOI] [PubMed] [Google Scholar]
- 53.Schiff R, Massarweh S, Shou J, Osborne CK. Breast cancer endocrine resistance: how growth factor signaling and estrogen receptor coregulators modulate response. Clin Cancer Res. 2003;9:447S–54S. [PubMed] [Google Scholar]
- 54.Wu RC, Feng Q, Lonard DM, O'Malley BW. SRC-3 coactivator functional lifetime is regulated by a phospho-dependent ubiquitin time clock. Cell. 2007;129:1125–40. doi: 10.1016/j.cell.2007.04.039. [DOI] [PubMed] [Google Scholar]
- 55.Li C, et al. Essential phosphatases and a phospho-degron are critical for regulation of SRC-3/AIB1 coactivator function and turnover. Mol Cell. 2008;31:835–49. doi: 10.1016/j.molcel.2008.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yi P, et al. Atypical protein kinase C regulates dual pathways for degradation of the oncogenic coactivator SRC-3/AIB1. Mol Cell. 2008;29:465–76. doi: 10.1016/j.molcel.2007.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Amazit L, et al. Subcellular localization and mechanisms of nucleocytoplasmic trafficking of steroid receptor coactivator-1. J Biol Chem. 2003;278:32195–203. doi: 10.1074/jbc.M300730200. [DOI] [PubMed] [Google Scholar]
- 58.Hermanson O, Glass CK, Rosenfeld MG. Nuclear receptor coregulators: multiple modes of modification. Trends Endocrinol Metab. 2002;13:55–60. doi: 10.1016/s1043-2760(01)00527-6. [DOI] [PubMed] [Google Scholar]
- 59.Qutob MS, Bhattacharjee RN, Pollari E, Yee SP, Torchia J. Microtubule-dependent subcellular redistribution of the transcriptional coactivator p/CIP. Mol Cell Biol. 2002;22:6611–26. doi: 10.1128/MCB.22.18.6611-6626.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Amazit L, et al. Regulation of SRC-3 intercompartmental dynamics by estrogen receptor and phosphorylation. Mol Cell Biol. 2007;27:6913–32. doi: 10.1128/MCB.01695-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu RC, et al. Regulation of SRC-3 (pCIP/ACTR/AIB-1/RAC-3/TRAM-1) Coactivator activity by I kappa B kinase. Mol Cell Biol. 2002;22:3549–61. doi: 10.1128/MCB.22.10.3549-3561.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zheng FF, Wu RC, Smith CL, O'Malley BW. Rapid estrogen-induced phosphorylation of the SRC-3 coactivator occurs in an extranuclear complex containing estrogen receptor. Mol Cell Biol. 2005;25:8273–84. doi: 10.1128/MCB.25.18.8273-8284.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yu C, et al. An essential function of the SRC-3 coactivator in suppression of cytokine mRNA translation and inflammatory response. Mol Cell. 2007;25:765–78. doi: 10.1016/j.molcel.2007.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baumann CT, et al. The glucocorticoid receptor interacting protein 1 (GRIP1) localizes in discrete nuclear foci that associate with ND10 bodies and are enriched in components of the 26S proteasome. Mol Endocrinol. 2001;15:485–500. doi: 10.1210/mend.15.4.0618. [DOI] [PubMed] [Google Scholar]
- 65.Shao W, Keeton EK, McDonnell DP, Brown M. Coactivator AIB1 links estrogen receptor transcriptional activity and stability. Proc Natl Acad Sci U S A. 2004;101:11599–604. doi: 10.1073/pnas.0402997101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yan F, Gao X, Lonard DM, Nawaz Z. Specific ubiquitin-conjugating enzymes promote degradation of specific nuclear receptor coactivators. Mol Endocrinol. 2003;17:1315–31. doi: 10.1210/me.2002-0209. [DOI] [PubMed] [Google Scholar]
- 67.Imhof MO, McDonnell DP. Yeast RSP5 and its human homolog hRPF1 potentiate hormone-dependent activation of transcription by human progesterone and glucocorticoid receptors. Mol Cell Biol. 1996;16:2594–605. doi: 10.1128/mcb.16.6.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lonard DM, Nawaz Z, Smith CL, O'Malley BW. The 26S proteasome is required for estrogen receptor-alpha and coactivator turnover and for efficient estrogen receptor-alpha transactivation. Mol Cell. 2000;5:939–48. doi: 10.1016/s1097-2765(00)80259-2. [DOI] [PubMed] [Google Scholar]
- 69.Gianni M, et al. P38MAPK-dependent phosphorylation and degradation of SRC-3/AIB1 and RARalpha-mediated transcription. Embo J. 2006;25:739–51. doi: 10.1038/sj.emboj.7600981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Desterro JM, Rodriguez MS, Hay RT. SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol Cell. 1998;2:233–9. doi: 10.1016/s1097-2765(00)80133-1. [DOI] [PubMed] [Google Scholar]
- 71.Kirsh O, et al. The SUMO E3 ligase RanBP2 promotes modification of the HDAC4 deacetylase. Embo J. 2002;21:2682–91. doi: 10.1093/emboj/21.11.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chauchereau A, Amazit L, Quesne M, Guiochon-Mantel A, Milgrom E. Sumoylation of the progesterone receptor and of the steroid receptor coactivator SRC-1. J Biol Chem. 2003;278:12335–43. doi: 10.1074/jbc.M207148200. [DOI] [PubMed] [Google Scholar]
- 73.Jimenez-Lara AM, Heine MJ, Gronemeyer H. PIAS3 (protein inhibitor of activated STAT-3) modulates the transcriptional activation mediated by the nuclear receptor coactivator TIF2. FEBS Lett. 2002;526:142–6. doi: 10.1016/s0014-5793(02)03154-x. [DOI] [PubMed] [Google Scholar]
- 74.Kotaja N, Vihinen M, Palvimo JJ, Janne OA. Androgen receptor-interacting protein 3 and other PIAS proteins cooperate with glucocorticoid receptor-interacting protein 1 in steroid receptor-dependent signaling. J Biol Chem. 2002;277:17781–8. doi: 10.1074/jbc.M106354200. [DOI] [PubMed] [Google Scholar]
- 75.Kotaja N, Karvonen U, Janne OA, Palvimo JJ. The nuclear receptor interaction domain of GRIP1 is modulated by covalent attachment of SUMO-1. J Biol Chem. 2002;277:30283–8. doi: 10.1074/jbc.M204768200. [DOI] [PubMed] [Google Scholar]
- 76.Wu H, et al. Coordinated regulation of AIB1 transcriptional activity by sumoylation and phosphorylation. J Biol Chem. 2006;281:21848–56. doi: 10.1074/jbc.M603772200. [DOI] [PubMed] [Google Scholar]
- 77.Li X, et al. The SRC-3/AIB1 coactivator is degraded in a ubiquitin- and ATP-independent manner by the REGgamma proteasome. Cell. 2006;124:381–92. doi: 10.1016/j.cell.2005.11.037. [DOI] [PubMed] [Google Scholar]
- 78.Chen H, Lin RJ, Xie W, Wilpitz D, Evans RM. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell. 1999;98:675–86. doi: 10.1016/s0092-8674(00)80054-9. [DOI] [PubMed] [Google Scholar]
- 79.Feng Q, Yi P, Wong J, O'Malley BW. Signaling within a coactivator complex: methylation of SRC-3/AIB1 is a molecular switch for complex disassembly. Mol Cell Biol. 2006;26:7846–57. doi: 10.1128/MCB.00568-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Naeem H, et al. The activity and stability of the transcriptional coactivator p/CIP/SRC-3 are regulated by CARM1-dependent methylation. Mol Cell Biol. 2007;27:120–34. doi: 10.1128/MCB.00815-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mark M, et al. Partially redundant functions of SRC-1 and TIF2 in postnatal survival and male reproduction. Proc Natl Acad Sci U S A. 2004;101:4453–8. doi: 10.1073/pnas.0400234101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang Z, et al. Critical roles of the p160 transcriptional coactivators p/CIP and SRC-1 in energy balance. Cell Metab. 2006;3:111–22. doi: 10.1016/j.cmet.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 83.Fleming FJ, Hill AD, McDermott EW, O'Higgins NJ, Young LS. Differential recruitment of coregulator proteins steroid receptor coactivator-1 and silencing mediator for retinoid and thyroid receptors to the estrogen receptor-estrogen response element by beta-estradiol and 4-hydroxytamoxifen in human breast cancer. J Clin Endocrinol Metab. 2004;89:375–83. doi: 10.1210/jc.2003-031048. [DOI] [PubMed] [Google Scholar]
- 84.Fleming FJ, et al. Expression of SRC-1, AIB1, and PEA3 in HER2 mediated endocrine resistant breast cancer; a predictive role for SRC-1. J Clin Pathol. 2004;57:1069–74. doi: 10.1136/jcp.2004.016733. This article reported that SRC-1 expression in breast cancer was correlated with HER2 positivity and worse DFS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Myers E, et al. Inverse relationship between ER-beta and SRC-1 predicts outcome in endocrine-resistant breast cancer. Br J Cancer. 2004;91:1687–93. doi: 10.1038/sj.bjc.6602156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hudelist G, et al. Expression of sex steroid receptors and their co-factors in normal and malignant breast tissue: AIB1 is a carcinoma-specific co-activator. Breast Cancer Res Treat. 2003;78:193–204. doi: 10.1023/a:1022930710850. [DOI] [PubMed] [Google Scholar]
- 87.List HJ, Reiter R, Singh B, Wellstein A, Riegel AT. Expression of the nuclear coactivator AIB1 in normal and malignant breast tissue. Breast Cancer Res Treat. 2001;68:21–8. doi: 10.1023/a:1017910924390. [DOI] [PubMed] [Google Scholar]
- 88.Qin L, et al. The AIB1 oncogene promotes breast cancer metastasis by activation of PEA3-mediated matrix metalloproteinase 2 (MMP2) and MMP9 expression. Mol Cell Biol. 2008;28:5937–50. doi: 10.1128/MCB.00579-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Redmond AM, et al. Coassociation of Estrogen Receptor and p160 Proteins Predicts Resistance to Endocrine Treatment; SRC-1 is an Independent Predictor of Breast Cancer Recurrence. Clin Cancer Res. 2009;15:2098–2106. doi: 10.1158/1078-0432.CCR-08-1649. [DOI] [PubMed] [Google Scholar]
- 90.Tai H, Kubota N, Kato S. Involvement of nuclear receptor coactivator SRC-1 in estrogen-dependent cell growth of MCF-7 cells. Biochem Biophys Res Commun. 2000;267:311–6. doi: 10.1006/bbrc.1999.1954. [DOI] [PubMed] [Google Scholar]
- 91.Cavarretta IT, et al. Reduction of coactivator expression by antisense oligodeoxynucleotides inhibits ERalpha transcriptional activity and MCF-7 proliferation. Mol Endocrinol. 2002;16:253–70. doi: 10.1210/mend.16.2.0770. [DOI] [PubMed] [Google Scholar]
- 92.Wei X, Xu H, Kufe D. MUC1 oncoprotein stabilizes and activates estrogen receptor alpha. Mol Cell. 2006;21:295–305. doi: 10.1016/j.molcel.2005.11.030. [DOI] [PubMed] [Google Scholar]
- 93.Kishimoto H, et al. The p160 family coactivators regulate breast cancer cell proliferation and invasion through autocrine/paracrine activity of SDF-1alpha/CXCL12. Carcinogenesis. 2005;26:1706–15. doi: 10.1093/carcin/bgi137. [DOI] [PubMed] [Google Scholar]
- 94.Wang S, et al. Disruption of the SRC-1 gene in mice suppresses breast cancer metastasis without affecting primary tumor formation. Proc Natl Acad Sci U S A. 2009;106:151–6. doi: 10.1073/pnas.0808703105. This article first reported that SRC-1 deficiency in mice strongly suppressed breast cancer metastasis in MMTV-PyMT mice by inhibiting HER2 and CSF-1 expression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Qin L, Liu Z, Chen H, Xu J. The steroid receptor coactivator-1 (SRC-1) regulates Twist expression and promotes breast cancer metastasis. Cancer Res. 2009;69:3819–3827. doi: 10.1158/0008-5472.CAN-08-4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Girault I, et al. Expression analysis of estrogen receptor alpha coregulators in breast carcinoma: evidence that NCOR1 expression is predictive of the response to tamoxifen. Clin Cancer Res. 2003;9:1259–66. [PubMed] [Google Scholar]
- 97.Bautista S, et al. In breast cancer, amplification of the steroid receptor coactivator gene AIB1 is correlated with estrogen and progesterone receptor positivity. Clin Cancer Res. 1998;4:2925–9. [PubMed] [Google Scholar]
- 98.Zhao C, et al. Elevated expression levels of NCOA3, TOP1, and TFAP2C in breast tumors as predictors of poor prognosis. Cancer. 2003;98:18–23. doi: 10.1002/cncr.11482. [DOI] [PubMed] [Google Scholar]
- 99.Glaeser M, Floetotto T, Hanstein B, Beckmann MW, Niederacher D. Gene amplification and expression of the steroid receptor coactivator SRC3 (AIB1) in sporadic breast and endometrial carcinomas. Horm Metab Res. 2001;33:121–6. doi: 10.1055/s-2001-14938. [DOI] [PubMed] [Google Scholar]
- 100.Font de Mora J, Brown M. AIB1 is a conduit for kinase-mediated growth factor signaling to the estrogen receptor. Mol Cell Biol. 2000;20:5041–7. doi: 10.1128/mcb.20.14.5041-5047.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Smith CL, Nawaz Z, O'Malley BW. Coactivator and corepressor regulation of the agonist/antagonist activity of the mixed antiestrogen, 4-hydroxytamoxifen. Mol Endocrinol. 1997;11:657–66. doi: 10.1210/mend.11.6.0009. [DOI] [PubMed] [Google Scholar]
- 102.Planas-Silva MD, Shang Y, Donaher JL, Brown M, Weinberg RA. AIB1 enhances estrogen-dependent induction of cyclin D1 expression. Cancer Res. 2001;61:3858–62. [PubMed] [Google Scholar]
- 103.List HJ, et al. Ribozyme targeting demonstrates that the nuclear receptor coactivator AIB1 is a rate-limiting factor for estrogen-dependent growth of human MCF-7 breast cancer cells. J Biol Chem. 2001;276:23763–8. doi: 10.1074/jbc.M102397200. [DOI] [PubMed] [Google Scholar]
- 104.Kuang SQ, et al. AIB1/SRC-3 deficiency affects insulin-like growth factor I signaling pathway and suppresses v-Ha-ras-induced breast cancer initiation and progression in mice. Cancer Res. 2004;64:1875–85. doi: 10.1158/0008-5472.can-03-3745. This article first demonstrated that SRC-3/AIB1 deficiency suppressed oncogene-induced mammary tumor initiation, growth and metastasis and inhibited the IGF-I signaling pathways by downregulating IGF-1, IRS-1 and IRS-2. [DOI] [PubMed] [Google Scholar]
- 105.Kuang SQ, et al. Mice lacking the amplified in breast cancer 1/steroid receptor coactivator-3 are resistant to chemical carcinogen-induced mammary tumorigenesis. Cancer Res. 2005;65:7993–8002. doi: 10.1158/0008-5472.CAN-05-1179. This article demonstrated that SRC-3/AIB1 deficiency specifically protected mouse mammary gland from chemical carcinogen-induced tumorigenesis. [DOI] [PubMed] [Google Scholar]
- 106.Fereshteh MP, et al. The nuclear receptor coactivator amplified in breast cancer-1 is required for Neu (ErbB2/HER2) activation, signaling, and mammary tumorigenesis in mice. Cancer Res. 2008;68:3697–706. doi: 10.1158/0008-5472.CAN-07-6702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Torres-Arzayus MI, et al. High tumor incidence and activation of the PI3K/AKT pathway in transgenic mice define AIB1 as an oncogene. Cancer Cell. 2004;6:263–74. doi: 10.1016/j.ccr.2004.06.027. This article demonstrated that overexpression of SRC-3/AIB1 in mouse mammary epithelial cells caused spontaneous mammary tumors, suggesting overexpressed SRC-3 is oncogenic. [DOI] [PubMed] [Google Scholar]
- 108.Maki HE, et al. Screening of genetic and expression alterations of SRC1 gene in prostate cancer. Prostate. 2006;66:1391–8. doi: 10.1002/pros.20427. [DOI] [PubMed] [Google Scholar]
- 109.Agoulnik IU, et al. Role of SRC-1 in the promotion of prostate cancer cell growth and tumor progression. Cancer Res. 2005;65:7959–67. doi: 10.1158/0008-5472.CAN-04-3541. [DOI] [PubMed] [Google Scholar]
- 110.Gregory CW, et al. A mechanism for androgen receptor-mediated prostate cancer recurrence after androgen deprivation therapy. Cancer Res. 2001;61:4315–9. [PubMed] [Google Scholar]
- 111.Fujimoto N, Mizokami A, Harada S, Matsumoto T. Different expression of androgen receptor coactivators in human prostate. Urology. 2001;58:289–94. doi: 10.1016/s0090-4295(01)01117-7. [DOI] [PubMed] [Google Scholar]
- 112.Mori R, et al. Prognostic value of the androgen receptor and its coactivators in patients with D1 prostate cancer. Anticancer Res. 2008;28:425–30. [PubMed] [Google Scholar]
- 113.Tien JCY, Zhou S, Xu J. The role of SRC-1 in murine prostate carcinogenesis is nonessential due to a possible compensation of SRC-3/AIB1 overexpression. Int J Biol Sci. 2009;5:256–264. doi: 10.7150/ijbs.5.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Agoulnik IU, et al. Androgens modulate expression of transcription intermediary factor 2, an androgen receptor coactivator whose expression level correlates with early biochemical recurrence in prostate cancer. Cancer Res. 2006;66:10594–602. doi: 10.1158/0008-5472.CAN-06-1023. [DOI] [PubMed] [Google Scholar]
- 115.Gnanapragasam VJ, Leung HY, Pulimood AS, Neal DE, Robson CN. Expression of RAC 3, a steroid hormone receptor co-activator in prostate cancer. Br J Cancer. 2001;85:1928–36. doi: 10.1054/bjoc.2001.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhou HJ, et al. SRC-3 is required for prostate cancer cell proliferation and survival. Cancer Res. 2005;65:7976–83. doi: 10.1158/0008-5472.CAN-04-4076. [DOI] [PubMed] [Google Scholar]
- 117.Zhou G, Hashimoto Y, Kwak I, Tsai SY, Tsai MJ. Role of the steroid receptor coactivator SRC-3 in cell growth. Mol Cell Biol. 2003;23:7742–55. doi: 10.1128/MCB.23.21.7742-7755.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chung AC, et al. Genetic ablation of the amplified-in-breast cancer 1 inhibits spontaneous prostate cancer progression in mice. Cancer Res. 2007;67:5965–75. doi: 10.1158/0008-5472.CAN-06-3168. This article first reported that SRC-3 deficiency in mice arrested spontaneous prostate cancer progression at a well-differentiated stage. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Xie D, et al. Correlation of AIB1 overexpression with advanced clinical stage of human colorectal carcinoma. Hum Pathol. 2005;36:777–83. doi: 10.1016/j.humpath.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 120.Kershah SM, Desouki MM, Koterba KL, Rowan BG. Expression of estrogen receptor coregulators in normal and malignant human endometrium. Gynecol Oncol. 2004;92:304–13. doi: 10.1016/j.ygyno.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 121.Uchikawa J, et al. Expression of steroid receptor coactivators and corepressors in human endometrial hyperplasia and carcinoma with relevance to steroid receptors and Ki-67 expression. Cancer. 2003;98:2207–13. doi: 10.1002/cncr.11760. [DOI] [PubMed] [Google Scholar]
- 122.Balmer NN, et al. Steroid receptor coactivator AIB1 in endometrial carcinoma, hyperplasia and normal endometrium: Correlation with clinicopathologic parameters and biomarkers. Mod Pathol. 2006;19:1593–605. doi: 10.1038/modpathol.3800696. [DOI] [PubMed] [Google Scholar]
- 123.Xu FP, et al. SRC-3/AIB1 protein and gene amplification levels in human esophageal squamous cell carcinomas. Cancer Lett. 2007;245:69–74. doi: 10.1016/j.canlet.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 124.Sakakura C, et al. Amplification and over-expression of the AIB1 nuclear receptor co-activator gene in primary gastric cancers. Int J Cancer. 2000;89:217–23. [PubMed] [Google Scholar]
- 125.Yoshida H, et al. Steroid receptor coactivator-3, a homolog of Taiman that controls cell migration in the Drosophila ovary, regulates migration of human ovarian cancer cells. Mol Cell Endocrinol. 2005;245:77–85. doi: 10.1016/j.mce.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 126.Henke RT, et al. Overexpression of the nuclear receptor coactivator AIB1 (SRC-3) during progression of pancreatic adenocarcinoma. Clin Cancer Res. 2004;10:6134–42. doi: 10.1158/1078-0432.CCR-04-0561. [DOI] [PubMed] [Google Scholar]
- 127.Carapeti M, Aguiar RC, Goldman JM, Cross NC. A novel fusion between MOZ and the nuclear receptor coactivator TIF2 in acute myeloid leukemia. Blood. 1998;91:3127–33. [PubMed] [Google Scholar]
- 128.Liang J, Prouty L, Williams BJ, Dayton MA, Blanchard KL. Acute mixed lineage leukemia with an inv(8)(p11q13) resulting in fusion of the genes for MOZ and TIF2. Blood. 1998;92:2118–22. [PubMed] [Google Scholar]
- 129.Deguchi K, et al. MOZ-TIF2-induced acute myeloid leukemia requires the MOZ nucleosome binding motif and TIF2-mediated recruitment of CBP. Cancer Cell. 2003;3:259–71. doi: 10.1016/s1535-6108(03)00051-5. [DOI] [PubMed] [Google Scholar]
- 130.Huntly BJ, et al. MOZ-TIF2, but not BCR-ABL, confers properties of leukemic stem cells to committed murine hematopoietic progenitors. Cancer Cell. 2004;6:587–96. doi: 10.1016/j.ccr.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 131.Coste A, et al. Absence of the steroid receptor coactivator-3 induces B-cell lymphoma. Embo J. 2006;25:2453–64. doi: 10.1038/sj.emboj.7601106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lahusen T, Fereshteh M, Oh A, Wellstein A, Riegel AT. Epidermal growth factor receptor tyrosine phosphorylation and signaling controlled by a nuclear receptor coactivator, amplified in breast cancer 1. Cancer Res. 2007;67:7256–65. doi: 10.1158/0008-5472.CAN-07-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yan J, et al. Steroid receptor coactivator-3/AIB1 promotes cell migration and invasiveness through focal adhesion turnover and matrix metalloproteinase expression. Cancer Res. 2008;68:5460–8. doi: 10.1158/0008-5472.CAN-08-0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Louie MC, Revenko AS, Zou JX, Yao J, Chen HW. Direct control of cell cycle gene expression by proto-oncogene product ACTR, and its autoregulation underlies its transforming activity. Mol Cell Biol. 2006;26:3810–23. doi: 10.1128/MCB.26.10.3810-3823.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Louie MC, Zou JX, Rabinovich A, Chen HW. ACTR/AIB1 functions as an E2F1 coactivator to promote breast cancer cell proliferation and antiestrogen resistance. Mol Cell Biol. 2004;24:5157–71. doi: 10.1128/MCB.24.12.5157-5171.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Mussi P, Yu C, O'Malley BW, Xu J. Stimulation of steroid receptor coactivator-3 (SRC-3) gene overexpression by a positive regulatory loop of E2F1 and SRC-3. Mol Endocrinol. 2006;20:3105–19. doi: 10.1210/me.2005-0522. [DOI] [PubMed] [Google Scholar]
- 137.Mukherjee A, et al. Steroid receptor coactivator 2 is critical for progesterone-dependent uterine function and mammary morphogenesis in the mouse. Mol Cell Biol. 2006;26:6571–83. doi: 10.1128/MCB.00654-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Liu Z, Liao L, Zhou S, Xu J. Generation and validation of a mouse line with a floxed SRC-3/AIB1 allele for conditional knockout. Int J Biol Sci. 2008;4:202–7. doi: 10.7150/ijbs.4.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Xu J, et al. Partial hormone resistance in mice with disruption of the steroid receptor coactivator-1 (SRC-1) gene. Science. 1998;279:1922–5. doi: 10.1126/science.279.5358.1922. This article first reported the phenotype of SRC-1 knockout mice and demonstrated an important physiologic role of the coactivator in vivo. [DOI] [PubMed] [Google Scholar]
- 140.Weiss RE, et al. Mice deficient in the steroid receptor co-activator 1 (SRC-1) are resistant to thyroid hormone. Embo J. 1999;18:1900–4. doi: 10.1093/emboj/18.7.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kamiya Y, et al. Modulation by steroid receptor coactivator-1 of target-tissue responsiveness in resistance to thyroid hormone. Endocrinology. 2003;144:4144–53. doi: 10.1210/en.2003-0239. [DOI] [PubMed] [Google Scholar]
- 142.Puigserver P, et al. Activation of PPARgamma coactivator-1 through transcription factor docking. Science. 1999;286:1368–71. doi: 10.1126/science.286.5443.1368. [DOI] [PubMed] [Google Scholar]
- 143.Picard F, et al. SRC-1 and TIF2 control energy balance between white and brown adipose tissues. Cell. 2002;111:931–41. doi: 10.1016/s0092-8674(02)01169-8. [DOI] [PubMed] [Google Scholar]
- 144.Gehin M, et al. The function of TIF2/GRIP1 in mouse reproduction is distinct from those of SRC-1 and p/CIP. Mol Cell Biol. 2002;22:5923–37. doi: 10.1128/MCB.22.16.5923-5937.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ye X, et al. Roles of steroid receptor coactivator (SRC)-1 and transcriptional intermediary factor (TIF) 2 in androgen receptor activity in mice. Proc Natl Acad Sci U S A. 2005;102:9487–92. doi: 10.1073/pnas.0503577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Chopra AR, et al. Absence of the SRC-2 coactivator results in a glycogenopathy resembling Von Gierke's disease. Science. 2008;322:1395–9. doi: 10.1126/science.1164847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Xu J, et al. The steroid receptor coactivator SRC-3 (p/CIP/RAC3/AIB1/ACTR/TRAM-1) is required for normal growth, puberty, female reproductive function, and mammary gland development. Proc Natl Acad Sci U S A. 2000;97:6379–84. doi: 10.1073/pnas.120166297. This article first reported the phenotype of SRC-3 knockout mice and demonstrated that SRC-3 played an important role in growth and mammary gland development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wang Z, et al. Regulation of somatic growth by the p160 coactivator p/CIP. Proc Natl Acad Sci U S A. 2000;97:13549–54. doi: 10.1073/pnas.260463097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Liao L, Chen X, Wang S, Parlow AF, Xu J. Steroid receptor coactivator 3 maintains circulating insulin-like growth factor I (IGF-I) by controlling IGF-binding protein 3 expression. Mol Cell Biol. 2008;28:2460–9. doi: 10.1128/MCB.01163-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Brzozowski AM, et al. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature. 1997;389:753–8. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- 151.Shiau AK, et al. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell. 1998;95:927–37. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 152.Beischlag TV, et al. Recruitment of the NCoA/SRC-1/p160 family of transcriptional coactivators by the aryl hydrocarbon receptor/aryl hydrocarbon receptor nuclear translocator complex. Mol Cell Biol. 2002;22:4319–33. doi: 10.1128/MCB.22.12.4319-4333.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Carlson DB, Perdew GH. A dynamic role for the Ah receptor in cell signaling? Insights from a diverse group of Ah receptor interacting proteins. J Biochem Mol Toxicol. 2002;16:317–25. doi: 10.1002/jbt.10051. [DOI] [PubMed] [Google Scholar]
- 154.Kumar MB, Perdew GH. Nuclear receptor coactivator SRC-1 interacts with the Q-rich subdomain of the AhR and modulates its transactivation potential. Gene Expr. 1999;8:273–86. [PMC free article] [PubMed] [Google Scholar]
- 155.Lee SK, et al. Steroid receptor coactivator-1 coactivates activating protein-1-mediated transactivations through interaction with the c-Jun and c-Fos subunits. J Biol Chem. 1998;273:16651–4. doi: 10.1074/jbc.273.27.16651. [DOI] [PubMed] [Google Scholar]
- 156.Dennis JH, Budhram-Mahadeo V, Latchman DS. Functional interaction between Brn-3a and Src-1 co-activates Brn-3a-mediated transactivation. Biochem Biophys Res Commun. 2002;294:487–95. doi: 10.1016/S0006-291X(02)00500-4. [DOI] [PubMed] [Google Scholar]
- 157.Song LN, Gelmann EP. Interaction of beta-catenin and TIF2/GRIP1 in transcriptional activation by the androgen receptor. J Biol Chem. 2005;280:37853–67. doi: 10.1074/jbc.M503850200. [DOI] [PubMed] [Google Scholar]
- 158.Goel A, Janknecht R. Concerted activation of ETS protein ER81 by p160 coactivators, the acetyltransferase p300 and the receptor tyrosine kinase HER2/Neu. J Biol Chem. 2004;279:14909–16. doi: 10.1074/jbc.M400036200. [DOI] [PubMed] [Google Scholar]
- 159.Martinez-Jimenez CP, Castell JV, Gomez-Lechon MJ, Jover R. Transcriptional activation of CYP2C9, CYP1A1, and CYP1A2 by hepatocyte nuclear factor 4alpha requires coactivators peroxisomal proliferator activated receptor-gamma coactivator 1alpha and steroid receptor coactivator 1. Mol Pharmacol. 2006;70:1681–92. doi: 10.1124/mol.106.025403. [DOI] [PubMed] [Google Scholar]
- 160.Wang JC, Stafford JM, Granner DK. SRC-1 and GRIP1 coactivate transcription with hepatocyte nuclear factor 4. J Biol Chem. 1998;273:30847–50. doi: 10.1074/jbc.273.47.30847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Reily MM, Pantoja C, Hu X, Chinenov Y, Rogatsky I. The GRIP1:IRF3 interaction as a target for glucocorticoid receptor-mediated immunosuppression. Embo J. 2006;25:108–17. doi: 10.1038/sj.emboj.7600919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Gao Z, et al. Coactivators and corepressors of NF-kappaB in IkappaB alpha gene promoter. J Biol Chem. 2005;280:21091–8. doi: 10.1074/jbc.M500754200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Li G, Heaton JH, Gelehrter TD. Role of steroid receptor coactivators in glucocorticoid and transforming growth factor beta regulation of plasminogen activator inhibitor gene expression. Mol Endocrinol. 2006;20:1025–34. doi: 10.1210/me.2005-0145. [DOI] [PubMed] [Google Scholar]
- 164.Kino T, Slobodskaya O, Pavlakis GN, Chrousos GP. Nuclear receptor coactivator p160 proteins enhance the HIV-1 long terminal repeat promoter by bridging promoter-bound factors and the Tat-P-TEFb complex. J Biol Chem. 2002;277:2396–405. doi: 10.1074/jbc.M106312200. [DOI] [PubMed] [Google Scholar]
- 165.Yi M, Tong GX, Murry B, Mendelson CR. Role of CBP/p300 and SRC-1 in transcriptional regulation of the pulmonary surfactant protein-A (SP-A) gene by thyroid transcription factor-1 (TTF-1) J Biol Chem. 2002;277:2997–3005. doi: 10.1074/jbc.M109793200. [DOI] [PubMed] [Google Scholar]
- 166.Baldwin A, Huh KW, Munger K. Human papillomavirus E7 oncoprotein dysregulates steroid receptor coactivator 1 localization and function. J Virol. 2006;80:6669–77. doi: 10.1128/JVI.02497-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Lee SK, Kim HJ, Kim JW, Lee JW. Steroid receptor coactivator-1 and its family members differentially regulate transactivation by the tumor suppressor protein p53. Mol Endocrinol. 1999;13:1924–33. doi: 10.1210/mend.13.11.0365. [DOI] [PubMed] [Google Scholar]
- 168.Batsche E, Desroches J, Bilodeau S, Gauthier Y, Drouin J. Rb enhances p160/SRC coactivator-dependent activity of nuclear receptors and hormone responsiveness. J Biol Chem. 2005;280:19746–56. doi: 10.1074/jbc.M413428200. [DOI] [PubMed] [Google Scholar]
- 169.Brosens JJ, Hayashi N, White JO. Progesterone receptor regulates decidual prolactin expression in differentiating human endometrial stromal cells. Endocrinology. 1999;140:4809–20. doi: 10.1210/endo.140.10.7070. [DOI] [PubMed] [Google Scholar]
- 170.Mani A, et al. E6AP mediates regulated proteasomal degradation of the nuclear receptor coactivator amplified in breast cancer 1 in immortalized cells. Cancer Res. 2006;66:8680–6. doi: 10.1158/0008-5472.CAN-06-0557. [DOI] [PubMed] [Google Scholar]
- 171.Verma S, et al. The ubiquitin-conjugating enzyme UBCH7 acts as a coactivator for steroid hormone receptors. Mol Cell Biol. 2004;24:8716–26. doi: 10.1128/MCB.24.19.8716-8726.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Zhang A, et al. Identification of a novel family of ankyrin repeats containing cofactors for p160 nuclear receptor coactivators. J Biol Chem. 2004;279:33799–805. doi: 10.1074/jbc.M403997200. [DOI] [PubMed] [Google Scholar]
- 173.Lee SK, et al. A nuclear factor, ASC-2, as a cancer-amplified transcriptional coactivator essential for ligand-dependent transactivation by nuclear receptors in vivo. J Biol Chem. 1999;274:34283–93. doi: 10.1074/jbc.274.48.34283. [DOI] [PubMed] [Google Scholar]
- 174.Chen D, et al. Regulation of transcription by a protein methyltransferase. Science. 1999;284:2174–7. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- 175.Wu X, Li H, Chen JD. The human homologue of the yeast DNA repair and TFIIH regulator MMS19 is an AF-1-specific coactivator of estrogen receptor. J Biol Chem. 2001;276:23962–8. doi: 10.1074/jbc.M101041200. [DOI] [PubMed] [Google Scholar]
- 176.Kino T, Chrousos GP. Tumor necrosis factor alpha receptor- and Fas-associated FLASH inhibit transcriptional activity of the glucocorticoid receptor by binding to and interfering with its interaction with p160 type nuclear receptor coactivators. J Biol Chem. 2003;278:3023–9. doi: 10.1074/jbc.M209234200. [DOI] [PubMed] [Google Scholar]
- 177.Kino T, Ichijo T, Chrousos GP. FLASH interacts with p160 coactivator subtypes and differentially suppresses transcriptional activity of steroid hormone receptors. J Steroid Biochem Mol Biol. 2004;92:357–63. doi: 10.1016/j.jsbmb.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 178.Liang J, Zhang H, Zhang Y, Zhang Y, Shang Y. GAS, a new glutamate-rich protein, interacts differentially with SRCs and is involved in oestrogen receptor function. EMBO Rep. 2009;10:51–7. doi: 10.1038/embor.2008.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Chauchereau A, Georgiakaki M, Perrin-Wolff M, Milgrom E, Loosfelt H. JAB1 interacts with both the progesterone receptor and SRC-1. J Biol Chem. 2000;275:8540–8. doi: 10.1074/jbc.275.12.8540. [DOI] [PubMed] [Google Scholar]
- 180.Yi P, et al. Peptidyl-prolyl isomerase 1 (Pin1) serves as a coactivator of steroid receptor by regulating the activity of phosphorylated steroid receptor coactivator 3 (SRC-3/AIB1) Mol Cell Biol. 2005;25:9687–99. doi: 10.1128/MCB.25.21.9687-9699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Lanz RB, et al. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell. 1999;97:17–27. doi: 10.1016/s0092-8674(00)80711-4. [DOI] [PubMed] [Google Scholar]
- 182.Berns EM, van Staveren IL, Klijn JG, Foekens JA. Predictive value of SRC-1 for tamoxifen response of recurrent breast cancer. Breast Cancer Res Treat. 1998;48:87–92. doi: 10.1023/a:1005903226483. [DOI] [PubMed] [Google Scholar]
- 183.Carroll RS, et al. Expression of a subset of steroid receptor cofactors is associated with progesterone receptor expression in meningiomas. Clin Cancer Res. 2000;6:3570–5. [PubMed] [Google Scholar]
- 184.Hussein-Fikret S, Fuller PJ. Expression of nuclear receptor coregulators in ovarian stromal and epithelial tumours. Mol Cell Endocrinol. 2005;229:149–60. doi: 10.1016/j.mce.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 185.Lassmann S, et al. Array CGH identifies distinct DNA copy number profiles of oncogenes and tumor suppressor genes in chromosomal- and microsatellite-unstable sporadic colorectal carcinomas. J Mol Med. 2007;85:293–304. doi: 10.1007/s00109-006-0126-5. [DOI] [PubMed] [Google Scholar]
- 186.Fujita Y, et al. Chromosome arm 20q gains and other genomic alterations in esophageal squamous cell carcinoma, as analyzed by comparative genomic hybridization and fluorescence in situ hybridization. Hepatogastroenterology. 2003;50:1857–63. [PubMed] [Google Scholar]
- 187.Wang Y, et al. Prognostic significance of c-myc and AIB1 amplification in hepatocellular carcinoma. A broad survey using high-throughput tissue microarray. Cancer. 2002;95:2346–52. doi: 10.1002/cncr.10963. [DOI] [PubMed] [Google Scholar]
- 188.Chen YJ, et al. Genome-wide profiling of oral squamous cell carcinoma. J Pathol. 2004;204:326–32. doi: 10.1002/path.1640. [DOI] [PubMed] [Google Scholar]
- 189.Tanner MM, et al. Frequent amplification of chromosomal region 20q12–q13 in ovarian cancer. Clin Cancer Res. 2000;6:1833–9. [PubMed] [Google Scholar]