Abstract
Mutations in the LMNA gene that cause Hutchinson-Gilford progeria syndrome (HGPS) lead to expression of a protein called progerin with 50 amino acids deleted from the tail of prelamin A. In cells from patients with HGPS, both the amount and distribution of heterochromatin are altered. We designed in vitro assays to ask whether such alterations might reflect changes in chromatin, DNA and/or histone binding properties of progerin compared to wild-type lamin C-terminal tails. We show that progerin tail has a reduced DNA/chromatin binding capacity and modified trimethylated H3K27 binding pattern, offering a molecular mechanism for heterochromatin alterations related to HGPS.
Keywords: chromatin, histone methylation, lamins, premature senescence, protein interaction
1. Introduction
A-type and B-type lamins are nuclear intermediate filaments that polymerize to form a meshwork between chromatin and the inner nuclear membrane. A-type lamins are encoded by the LMNA gene which generates lamins A and C by alternative RNA splicing in most terminally differentiated cells [1], whereas B-type lamins are encoded by two different genes (LMNB1 and LMNB2), which are expressed in all or most somatic cells [2,3]. While few disorders are associated with alterations in the LMNB1 and LMNB2 genes [4,5], a wide variety of diseases including myopathies, lipodystrophies and premature aging syndromes are caused by mutations throughout the LMNA gene [6]. In Hutchinson-Gilford progeria syndrome (HGPS), cells express progerin, a prelamin A variant from which 50 amino acids (aa) are deleted from the C-terminal tail (del aa 607–656) [7–8]. We have previously shown that progerin disturbs the segregation between A-type and B-type lamin homopolymers [9].
While the lamina is normally associated with chromatin in cells, many studies have shown that wild-type lamins bind chromatin in vitro [10–12]. However, in the pathological context of progeria, the abnormal structure of the lamina has been correlated with a loss of heterochromatin and perturbations in histones H3 and H4 epigenetic marks [13–15].
Here we asked whether the alterations of chromatin in cells from patients with HGPS reflect alterations in the chromatin binding properties of progerin. While the nuclear localization signal (NLS) region of lamins may represent a “basic binding motif” for chromatin and histones [10–12], another chromatin binding site has been suggested to reside in the C-terminal region of Xenopus lamin A tail [16]. The corresponding region in human lamin A contains the 50 amino acids deleted from progerin. To investigate the role of the 50 amino acids deleted in progerin in chromatin binding, we performed in vitro assays with recombinant C-terminal domains (wild-type or progerin) and chromatin, or isolated DNA and histones. We focused on the intrinsic properties of the C-terminal lamin sequences, as no attempt to farnesylate recombinant prelamin A or progerin tails was performed. We show that DNA/chromatin binding properties of the progerin tail are distinct from those of wild-type Atype and B-type lamin tails.
2. Materials and Methods
Detailed procedures are given in the supplementary material.
2.1. Preparations of histone complexes
Histone octamers used for dinucleosomes preparations were purified from duck erythrocytes as described [17]. Instead, histones H3, H4, H2A, H2B from calf thymus (Roche) were used in GST pull-down experiments, assembled in histone octamers following a protocol adapted from two published methods [18,19]. Histone octamers were stored at −80 °C.
2.2. DNA and dinucleosomes preparations
The 357 base pairs (bp) and 146 bp DNA fragments were purified, dephosphorylated and 5′-end labeled with 32P-ATP as described previously [20]. A dinucleosome preparation was obtained by mixing 32P-DNA fragments of 357 bp and histone octamers with a histone/DNA weight ratio of 1.5 [20].
2.3. Recombinant GST fusion proteins and peptides
cDNAs encoding the C-terminal tail of prelamin A, lamin A, lamin C, lamin B1 and progerin were cloned into pGEX-4T or pGEX-2T vectors that encode GST. Escherichia coli strain BL21 was transformed with the diverse plasmids. GST-HP1α construction was described previously [21]. Expression and purification of GST fusion proteins were performed using Glutathione Sepharose 4B according to manufacturer’s instructions (GE Healthcare, Bio-Sciences AB, Sweden). Purified proteins were analyzed by SDS-PAGE and their concentrations estimated after Coomassie blue staining. Protein aliquots were stored at −80 °C.
Peptides corresponding to aa 1–20 and aa 17–31 of the N-terminal tail of H3, and containing unmodified (H3K9; H3K27) or trimethylated lysines 9 and 27 (H3K9me3; H3K27me3), respectively, (Peptide Specialty Laboratories, Heidelberg, Germany) were solubilized at a concentration of 50 mg/ml in 50 mM Tris-HCl, pH8, 1 mM EDTA, 1 mM DTT, 1 mM 4-(2-aminoethyl) benzenesulfonyl fluoride (AEBSF), and 200 mM NaCl; peptide aliquots were stored at −80 °C.
2.4. Protein-DNA interactions and electrophoretic mobility shift assays (EMSA)
Recombinant GST-lamin tails were diluted in a Tris-NaCl buffer [10 mM Tris-HCl, pH 8, 50 mM NaCl, 1 mM EDTA, 0.1% Triton X-100 and 1 mM AEBSF], to different concentrations (21, 42, 84 and 168 nM for dinucleosome binding experiments and 26, 52, 104 and 208 nM for DNA binding experiments). They were incubated at room temperature for 3 hours with 32P-labeled DNA fragments or reconstituted dinucleosomes (26 nM and 10.5 nM, respectively).
For EMSA, protein-DNA complexes were analyzed on native polyacrylamide gels and DNA retardation was detected as described previously [20]. Measurements of the radioactive DNA signals were performed with a STORM 860 scanner (Amersham) using the ImageQuant software (Molecular Dynamics, Inc.).
2.5. GST pull-down assays
Recombinant GST-lamin tails, GST-HP1α or GST alone were incubated with histone octamers in equal stoechiometry (1.44 μM) in binding buffer [50 mM Tris-HCl, pH 8, 0.5% Triton X-100, 1 mM AEBSF and 1 mM DTT] containing 200 mM NaCl for 3 hours at 4 °C. The complexes were mixed with Glutathione Sepharose beads for one hour at 4 °C and centrifuged at 800g for 5 min at 4 °C. After 6 washes in binding buffer containing 300 mM NaCl, the proteins bound to the beads were analyzed by 12.5% SDS-PAGE. After proteins staining with SYPRO Ruby (Biorad), gels were scanned on a Typhoon Trio scanner (GE Health Care) and signal intensities were quantified using the Image J software.
2.6. Surface plasmon resonance experiment
Surface plasmon resonance (SPR) experiments were carried out on a BIAcore 3000 (BIAcore, Uppsala, Sweden) using a hydrophobic HPA sensor chip. Layouts of histone peptides diluted at 3 μM were performed at 25 °C in running buffer [HEPES 10 mM, pH 7.4, 150 mM NaCl]. Saturation coverage was obtained at 1200 resonance units (RU). GST-C-ter lamins were injected at 5 nM to obtain complete saturation. Kinetics analysis were performed by linearization using “fit kinetics Langmuir binding type” of BIAevaluation software, characteristic of a simple bimolecular interaction. The association rate of each protein was expressed as ng.mm−2 since their irreversible adsorption on histone peptides was not compatible with KD values determination as discussed in [22].
3. Results
3.1. Progerin tail has a binding affinity for chromatin close to that of A-type lamin tails, and much higher than that of lamin B1 tail
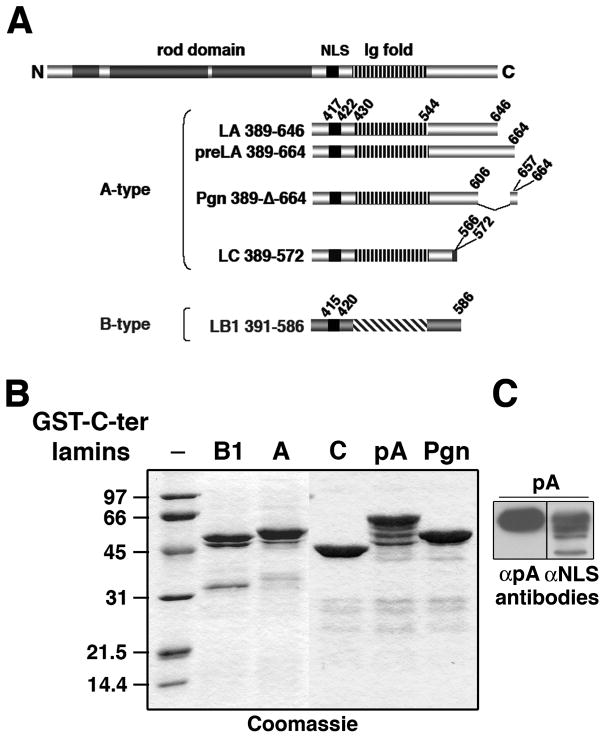
We first compared the affinities of A-type and B-type lamin tails for chromatin. To do this, we prepared pure recombinant GST fusion proteins containing the C-terminal tail of lamin A (aa 389–646), prelamin A (aa 389–664), progerin (aa 389-Δ-lamin C (aa 389–572), or lamin B1 (aa 391–586) (Fig. 1A-B). The presence of the region (aa 646–664) specific of prelamin A was confirmed by immunoblotting using anti-prelamin A antibodies (Fig. 1C).
Fig. 1.
Purification of proteins containing the C-terminal domains of A-type and B-type lamins fused to GST.
(A) Schematic representation of human lamin A secondary structure. Downstream of the rod domain (dark gray boxes), the C-terminal end contains the NLS (black boxes) and an Ig-like domain (Ig fold, streaked boxes). Peptides of the C-terminal tails of A-type and B-type lamins used here, lamin A (LA, aa 389–646), prelamin A (preLA, aa 389–664), progerin (Pgn, aa 389-Δ-664), lamin C (LC, aa 389–572) and lamin B1 (LB1, aa 391–586) are indicated. Positions of specific C-terminus of lamin C (aa 566–572) and the 50 amino acids deletion in the progerin mutant (Δ607–656) are indicated.
(B) Fusion proteins. GST-C-ter lamin B1 (B1), lamin A (A), lamin C (C), prelamin A (pA), and progerin (Pgn) were resolved by SDS-PAGE and stained with Coomassie blue. (C) Immunoblotting analysis of the GST-C-ter prelamin A (pA). Samples containing 100 ng of GST-C-ter pA (lanes 7 and 8) were analyzed by 12.5% SDS-PAGE followed by immunoblotting using either rabbit polyclonal antibodies directed against the NLS of lamin A/C (αNLS, lane 8) or goat antibodies raised against the last amino acids of WT prelamin A (αC-ter pA, lane 7).
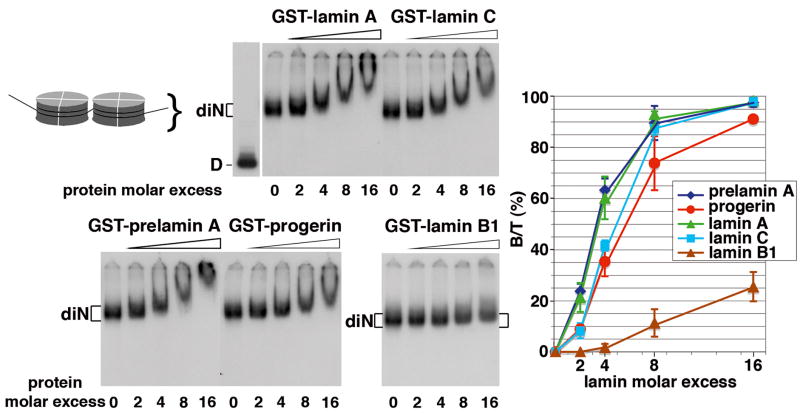
Dinucleosomes were used as chromatin sample. They were incubated with increasing amounts of GST-lamin tails. Figure 2 shows formation of complexes with dinucleosomes that occurred with a 4-fold molar excess of GST-lamin A, GST-lamin C, GST-prelamin A and GST-progerin tails whereas it required a 16-fold molar excess of GST-lamin B1 tail. No complex was detected with GST alone (Fig. S1A). The binding efficiency of lamin tails to dinucleosomes was in the order: lamin A and prelamin A > lamin C > progerin ≫ lamin B1 (Fig.2, graph). The KD values (Table 1) confirmed that prelamin A and lamin A tails affinities for dinucleosomes (KD = 21 and 25 nM, respectively) were 2 to 3-fold higher than that of lamin C and progerin tails (KD = 53 and 73 nM, respectively), and 100-fold higher than that of the lamin B1 tail (KD = 2 μM). While the low affinity of the lamin B1 tail for dinucleosomes clearly differentiates it from A-type lamins, the progerin tail appears distinct with an intermediate affinity for dinucleosomes.
Fig. 2.
The C-terminal domains of lamin isoforms bind dinucleosomes (diN) with different efficiencies.
The 32P-radiolabeled dinucleosomes were incubated in the absence (lanes 0) or presence (lanes 2, 4, 8 and 16) of increasing molar excess of GST-fusion proteins containing the C-terminal tails of lamin A, lamin C, lamin B1, prelamin A, and progerin. Complexes were resolved on a 5% non denaturing polyacrylamide gel and signals revealed by autoradiography. D refers to naked DNA. The graph represents the percentage of labeled diN bound to the GST-C-ter lamins (B/T) plotted as a function of the molar excess of the GST-C-ter lamins. Vertical bars indicated the standard deviation.
Table 1.
Binding affinities (KD) of the C-terminal tails of lamins for DNA and dinucleosomes.
Free and bound 32P DNA signals were quantified for 2 molar excess (DNA-lamin curves, Fig. 3) and 4 molar excess (diN-lamin curves, Fig. 2) of lamin tails.
| C-ter lamin |
KD |
||||
|---|---|---|---|---|---|
| dinucleosomes | DNA | ||||
| prelamin A | 20.9 nM ± 4.1 |
|
|
12.7 nM ± 3.7 |
|
| progerin | 73.1 nM ± 17.7 | 67.0 nM ± 8.8 | |||
| lamin A | 24.9 nM ± 8.3 |
|
8.3 nM ± 3.5 |
|
|
| lamin C | 53.0 nM ± 4.7 | 11.3 nM ± 3.8 | |||
| lamin B1 | 2.2 μM ± 0.8 | 80.0 nM ± 17 | |||
The dissociation constant KD was calculated using the equation , where [D] is the total DNA concentration, [P] the total protein concentration, and [DP] the protein-DNA complex concentration. The mean values obtained from three or four independent experiments are presented with standard deviations. (* and **) correspond to significant differences p < 0.01 and p < 0.001, respectively (student t-test).
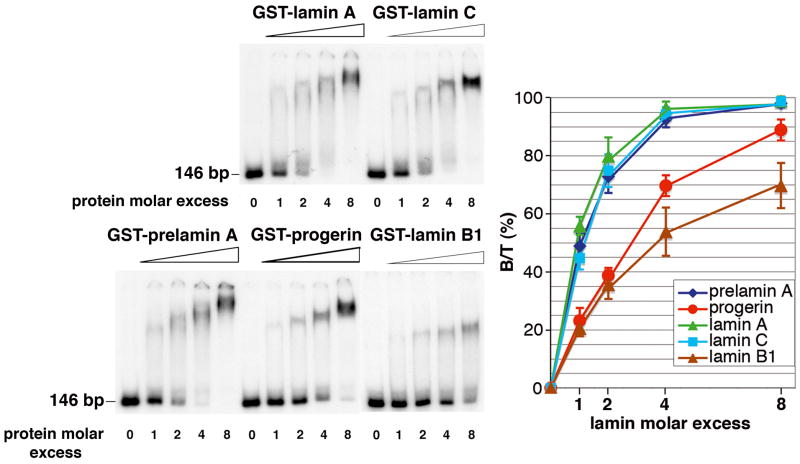
3.2. Progerin tail affinity for DNA is close to that of B-type lamin tails, and lower than that of A-type lamins
Increasing amounts of GST-lamin tails were incubated with a 146 bp DNA fragment. Figure 3 shows that 4-fold molar excess of GST-lamin A, GST-lamin C and GST-prelamin A tails were sufficient to complex all DNA, whereas 8-fold molar excess of GST-progerin tail was required to complex 90% of DNA. GST-lamin B1 failed to saturate DNA. No complex was detected with GST alone (Fig. S1B). The binding efficiencies of lamin tails to DNA (Fig.3, graph) were in the following order: lamin A, prelamin A and lamin C > progerin > lamin B1. KD values (Table 1) confirmed that prelamin A, lamin A and lamin C tails affinities for DNA (KD = 13, 8 and 11 nM, respectively) were 5 to 10-fold higher than those determined for progerin and lamin B1 tails (KD = 67 and 80 nM, respectively). Thus, affinity of the progerin tail for DNA appears close to that of the lamin B1 tail.
Fig. 3.
The C-terminal domains of lamin isoforms bind DNA with different efficiencies. A 146 bp 32P-radiolabeled DNA fragment was incubated in the absence (lanes 0) or presence (lanes 1, 2, 4, and 8) of increasing molar excess of GST-C-ter lamin A, GST-C-ter lamin C, GST-C-ter lamin B1, GST-C-ter prelamin A, and GST-C-ter progerin. The complexes were resolved on a 4% non denaturing polyacrylamide gel and the signals revealed by autoradiography. The percentage of labeled DNA bound to the GST-C-ter lamins (B/T) was plotted as a function of the molar excess of the GST-C-ter lamins. Vertical bars indicated the standard deviation.
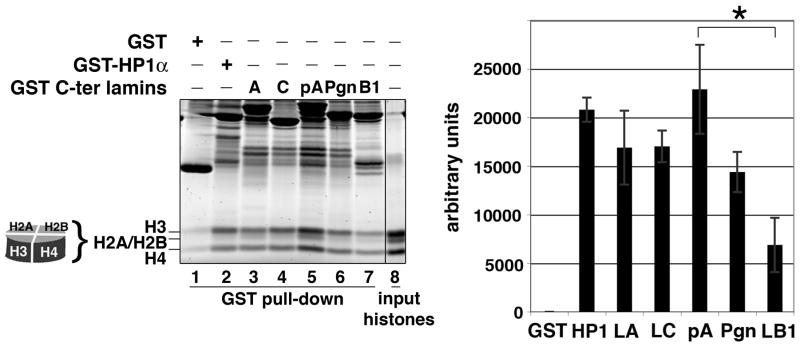
3.3. Progerin tail interaction with histone octamers is similar to that of A-type lamins, and higher to that of B-type lamins
Figure 4 shows that, as the control HP1α (lane 2), A-type lamin tails including progerin were more efficient than the lamin B1 tail to bind histones octamers (Fig. 4, lanes 3 to 7 and graph). Thus, the histone octamer binding capacity of the progerin tail appeared closer to that of A-type lamins. These results show that when compared to A-type lamins, the low binding of lamin B1 tail to chromatin (dinucleosomes) relies on a weaker interaction with both DNA and histone octamers, whereas the intermediate binding of progerin tail to chromatin relies mainly on a weaker interaction with DNA. Since the epigenetic marks of the histones used here are unknown, our chromatin binding assay cannot address the effects of epigenetic modification of chromatin on lamin binding.
Fig. 4.
The C-terminal domains of lamin isoforms bind the histone octamers.
GST-fusion proteins containing the C-terminal tails of lamin A (A), lamin C (C), prelamin A (pA), progerin (Pgn), and lamin B1 (B1), HP1α or GST protein alone (GST), were incubated with histone octamers. The pulled down complexes were separated by SDS-PAGE. Sample loaded in lane 8 represent 20% of the input. Data presented in the graph are the mean values of signal intensities of pulled down octamers expressed as arbitrary units and correspond to three independent experiments. Background values corresponding to octamers pulled down by GST were subtracted. Vertical bars indicate the standard deviation. (*) corresponds to significant difference p < 0.01 (student t-test).
3.4 Role of epigenetic marks on histone H3 in the binding of progerin tail to histones
A global loss of peripheral heterochromatin correlated with loss of trimethylated H3K9 and H3K27 (markers of constitutive and facultative heterochromatin, respectively) has been reported in cells from patients with progeria [13–15]. As lamins bind to histones, we reasoned that distribution of these modified histones may be influenced by their local association with specific lamin isoforms. We asked whether progerin that accumulates abnormally in progeria, would have a reduced capacity to recognize these epigenetic marks and thus contribute to their instability and progressive loss.
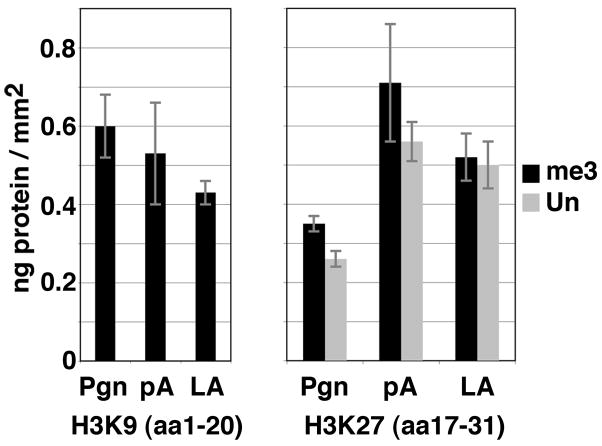
Interactions of GST-C-ter lamin tails to peptides mimicking the trimethylation of H3K9 and H3K27 versus the unmodified H3K27 peptide were investigated using surface plasmon resonance experiments (SPR) and reported as adsorption densities of the lamin tails expressed in ng/mm2. Figure 5 shows that the H3 peptide (aa 1–20) trimethylated on K9 bound slightly more efficiently GST-C-ter progerin than GST-C-ter lamin A. However, the binding capacities of that peptide for GST-C-ter progerin and GST-C-ter prelamin A were similar. The H3 peptide (aa 17–31) trimethylated on K27 bound less efficiently GST-C-ter progerin than GST-C-ter prelamin A and GST-C-ter lamin A. Interestingly, the unmodified H3 peptide (aa 17–31) presented the same lamin binding capacities as the K27me3 corresponding peptide. We conclude that, due to its deleted C-terminal region, progerin has an abnormally reduced capacity to bind the H3 peptide (aa 17–31), which occurs independently of the H3 trimethylation on K27, whereas it keeps a high capacity to bind the H3 peptide (aa 1–20) that bears trimethylation on K9.
Fig. 5.
Adsorption density of GST-C-ter progerin, prelamin A, and lamin A on peptides encoding histone N-terminal tails bearing specific epigenetic modifications. The adsorption density of the GST-C-ter progerin (Pgn), prelamin A (pA) and lamin A (LA) tails on the trimethylated (me3) H3K9 or H3K27 and unmodified (Un) H3K27 peptides was expressed as ng/mm2. Vertical bars indicate standard errors.
4. Discussion
We have demonstrated differences between A-type and B-type lamin tails in their DNA, histone and chromatin binding properties in vitro. In vivo, one cannot exclude that these differences may be modulated by the contribution of the N-terminal and/or the rod domains of lamins in binding to DNA or histones. Nevertheless, these results can provide a mechanistic explanation for the non-redundancy of the roles of A-type and B-type lamins in chromatin organization in cells. Compared to A-type lamins, progerin has two distinct features. First, it remains farnesylated in cells and second, it lacks 50 amino acids within the tail. These two features likely contribute to the HGPS phenotype both by impeding the normal assembly of the nuclear envelope associated lamina [9] and by disturbing the chromatin organization [13–15]. It has been shown that the toxicity of the progerin is not restricted to its farnesylation status [23] suggesting that the deletion of 50 amino acids might have an impact on interactions with specific chromatin partners.
We have shown that the DNA/chromatin binding properties of non-farnesylated progerin tail in vitro are reduced in comparison to wild-type A-type lamin tails. We conclude that the amino acid stretch (aa 607–646) of lamin A constitutes a binding site for DNA and chromatin. However, progerin still retains some capacity to bind DNA, histones and chromatin due to the presence of a basal binding site located in the basic NLS region (aa 389–422) as previously shown [10–12]. We propose that the amino acid sequence (aa 607–646) enhance the interaction of lamin A and prelamin A with DNA/chromatin. A weaker DNA binding affinity was found for the minimal Ig-fold lamin domain (aa 411–553) in our previous study [20] which may be related to the absence of the amino acid sequence (aa 607–646) as well as to a partial deletion (del aa 389–411) of the basal binding site mentioned above. In addition, we could not exclude that the GST-C-ter lamin tails dimerized through their GST moiety in reduced conditions may favor DNA binding compared to Ig-fold lamin dimers formed in the absence of DTT by formation of a disulfide bond through cysteine 522 [20].
Our experiments with histone H3 peptides with or without epigenetic modifications highlighted that, in comparison to wild-type immature and mature lamin A, progerin bound less efficiently the H3 peptide sequence (aa 17–30), either unmodified or trimethylated on K27. This suggest that the H3 sequence (aa 17–30) would contain a binding element for wild-type lamin A and that the lamin region (aa 607–656) that is deleted in progerin, largely contributes to the interaction with the H3 region (aa 17–30). In HGPS cells, it was reported that the decrease in H3K27me3 is an initial event that precedes the reduced amount of heterochromatin and its loss of interaction with the lamina [14]. Extrapolating our in vitro data to the situation reported in cells, we propose that the mechanism by which the amount of H3K27me3 decreases could result from the inability of progerin to bind and stabilize this mark efficiently. In contrast, the remodeling of heterochromatin in HGPS cells would not rely on a loss of interaction of H3K9me3 mark with progerin. In conclusion, our data illustrate the extent to which the non-farnesylated progerin tail has in vitro chromatin binding properties distinct from both A-type and B-type lamin tails, thus offering a molecular explanation for the heterochromatin alterations induced by progerin in HGPS cells.
Our in vitro studies were limited to non-farnesylated progerin, whereas in cells of human subjects with HGPS progerin is farnesylated. In this regard, it is important to note that non-farnesylated progerin elicits disease in a mouse model of HGPS, although the phenotype appears milder than if fanesylated progerin is expressed [23]. Furthermore, while treatment with protein farnesyltransferase inhibitors of mice with a genetically engineered HGPS mutation improves the progeroid phenotype, it does not completely reverse it [24]. Our experiments on non-farnesylated progerin therefore have relevance to the pathogenesis of HGPS and further suggest that blocking farnesylation of progerin may not be a “cure” for HGPS as the non-farnesylated protein has abnormal DNA/chromatin binding properties.
Supplementary Material
Acknowledgments
Authors are particularly indebted to Dr J.-C. Courvalin for enthusiastic discussions. We thank S. Zinn-Justin for helpful discussions and P. Vicart for encouragements. This work was supported by the “Centre National de la Recherche Scientifique” (C.N.R.S.), the “Institut National de la Santé et de la Recherche Médicale” (I.N.S.E.R.M.) and by grants from the “Association Française de Lutte contre les Myopathies” (A.F.M. to I. D.-G.). H.J.W. and C.Ö. were supported by a grant from the National Institutes of Health (AG025240). The support of the “Plateforme d’ingénierie des protéines (IFR83), service d’interactions des biomolécules, Institut de biologie intégrative, Université Pierre et Marie Curie, Paris 6” is acknowledged.
Abbreviations
- AEBSF
4-(2-aminoethyl)benzenesulfonyl fluoride
- aa
amino acids
- del
deletion
- ECL
enhanced chemiluminescence
- EMSA
electrophoretic mobility shift assay
- GST
glutathione S-transferase
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- HGPS
Hutchinson-Gilford progeria syndrome
- HP1
heterochromatin protein 1
- HPA
hydrophobic biosensor;
- LMNA
lamin A/C gene
- LMNB1
lamin B1 gene
- LMNB2
lamin B2 gene
- NLS
nuclear localization signal
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lin F, Worman HJ. Structural organization of the human gene encoding nuclear lamin A and nuclear lamin C. J Biol Chem. 1993;268:16321–16326. [PubMed] [Google Scholar]
- 2.Lin F, Worman HJ. Structural organization of the human gene (LMNB1) encoding nuclear lamin B1. Genomics. 1995;27:230–236. doi: 10.1006/geno.1995.1036. [DOI] [PubMed] [Google Scholar]
- 3.Höger TH, Zatloukal K, Waizenegger I, Krohne G. Characterization of a second highly conserved B-type lamin present in cells previously thought to contain only a single B-type lamin. Chromosoma. 1990;99:379–390. doi: 10.1007/BF01726689. [DOI] [PubMed] [Google Scholar]
- 4.Hegele RA, Cao H, Liu DM, Costain GA, Charlton-Menys V, Rodger NW, Durrington PN. Sequencing of the reannotated LMNB2 gene reveals novel mutations in patients with acquired partial lipodystrophy. Am J Hum Genet. 2006;79:383–389. doi: 10.1086/505885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Padiath QS, Saigoh K, Schiffmann R, Asahara H, Yamada T, Koeppen A, Hogan K, Ptácek LJ, Fu YH. Lamin B1 duplications cause autosomal dominant leukodystrophy. Nat Genet. 2006;38:1114–1123. doi: 10.1038/ng1872. [DOI] [PubMed] [Google Scholar]
- 6.Worman HJ, Fong LG, Muchir A, Young SG. Laminopathies and the long strange trip from basic cell biology to therapy. J Clin Invest. 2009;119:1825–36. doi: 10.1172/JCI37679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Sandre-Giovannoli A, Bernard R, Cau P, Navarro C, Amiel J, Boccaccio I, Lyonnet S, Stewart CL, Munnich A, Le Merrer M, Levy N. Lamin A truncation in Hutchinson-Gilford progeria. Science. 2003;300:2055. doi: 10.1126/science.1084125. [DOI] [PubMed] [Google Scholar]
- 8.Eriksson M, Brown WT, Gordon LB, Glynn MW, Singer J, Scott L, Erdos MR, Robbins CL, Moses TY, et al. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 2003;423:293–298. doi: 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delbarre E, Tramier M, Coppey-Moisan M, Gaillard C, Courvalin JC, Buendia B. The truncated prelamin A in Hutchinson-Gilford progeria syndrome alters segregation of A-type and B-type lamin homopolymers. Hum Mol Genet. 2006;15:1113–1122. doi: 10.1093/hmg/ddl026. [DOI] [PubMed] [Google Scholar]
- 10.Taniura H, Glass C, Gerace L. A chromatin binding site in the tail domain of nuclear lamins that interacts with core histones. J Cell Biol. 1995;131:33–44. doi: 10.1083/jcb.131.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldberg M, Harel A, Brandeis M, Rechsteiner T, Richmond TJ, Weiss AM, Gruenbaum Y. The tail domain of lamin Dm0 binds histones H2A and H2B. Proc Natl Acad Sci USA. 1999;96:2852–2857. doi: 10.1073/pnas.96.6.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mattout A, Goldberg M, Tzur Y, Margalit A, Gruenbaum Y. Specific and conserved sequences in D. melanogaster and C. elegans lamins and histone H2A mediate the attachment of lamins to chromosomes. J Cell Sci. 2006;120:77–85. doi: 10.1242/jcs.03325. [DOI] [PubMed] [Google Scholar]
- 13.Columbaro M, Capanni C, Mattioli E, Novelli G, Parnaik VK, Squarzoni S, Maraldi NM, Lattanzi G. Rescue of heterochromatin organization in Hutchinson-Gilford progeria by drug treatment. Cell Mol Life Sci. 2005;62:2669–2678. doi: 10.1007/s00018-005-5318-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shumaker DK, Dechat T, Kohlmaier A, Adam SA, Bozovsky MR, Erdos MR, Eriksson M, Goldman AE, Khuon S, Collins FS, et al. Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc Natl Acad Sci USA. 2006;103:8703–8708. doi: 10.1073/pnas.0602569103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scaffidi P, Misteli T. Reversal of the cellular phenotype in the premature aging disease Hutchinson-Gilford progeria syndrome. Nat Med. 2005;11:440–445. doi: 10.1038/nm1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Höger TH, Krohne G, Kleinschmidt JA. Interaction of Xenopus lamin A and LII with chromatin in vitro mediated by a sequence element in the carboxyterminal domain. Exp Cell Res. 1991;197:280–289. doi: 10.1016/0014-4827(91)90434-v. [DOI] [PubMed] [Google Scholar]
- 17.Zivanovic Y, Duband-Goulet I, Schultz P, Stofer E, Oudet P, Prunell A. Histone H5 dependence of DNA supercoiling in the nucleosome. J Mol Biol. 1990;214:479–495. doi: 10.1016/0022-2836(90)90195-R. [DOI] [PubMed] [Google Scholar]
- 18.Ausio J, Moore SC. Reconstitution of chromatin complexes from high performance liquid chromatography-purified histones. Methods. 1998;15:333–342. doi: 10.1006/meth.1998.0637. [DOI] [PubMed] [Google Scholar]
- 19.Tanaka Y, Tawaramoto-Sasanuma M, Kawaguchi S, Ohta T, Yoda K, Kurumizaka H, Yokoyama S. Expression and purification of recombinant human histones. Methods. 2004;33:3–11. doi: 10.1016/j.ymeth.2003.10.024. [DOI] [PubMed] [Google Scholar]
- 20.Stierlé V, Couprie J, Östlund C, Krimm I, Zinn-Justin S, Hossenlopp P, Worman HJ, Courvalin JC, Duband-Goulet I. The carboxyl-terminal region common to lamins A and C contains a DNA binding domain. Biochemistry. 2003;42:4819–4828. doi: 10.1021/bi020704g. [DOI] [PubMed] [Google Scholar]
- 21.Minc E, Allory Y, Worman HJ, Courvalin JC, Buendia B. Localization and phosphorylation of HP1 proteins during the cell cycle in mammalian cells. Chromosoma. 1999;108:220–234. doi: 10.1007/s004120050372. [DOI] [PubMed] [Google Scholar]
- 22.Noinville S, Bruston F, El Amri C, Baron D, Nicolas P. Conformation, Orientation, and Adsorption Kinetics of Dermaseptin B2 onto Synthetic Supports at Aqueous/Solid Interface. Biophys J. 2003;85:1196–1206. doi: 10.1016/S0006-3495(03)74555-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang SH, Andres DA, Spielmann HP, Young SG, Fong LG. Progerin elicits disease phenotypes of progeria in mice whether or not it is farnesylated. J Clin Invest. 2008;118:3291–3300. doi: 10.1172/JCI35876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang SH, Meta M, Qiao X, Frost D, Bauch J, Coffinier C, Majumdar S, Bergo MO, Young SG, Fong LG. A farnesyltransferase inhibitor improves disease phenotypes in mice with a Hutchinson-Gilford progeria syndrome mutation. J Clin Invest. 2006;116:2115–2121. doi: 10.1172/JCI28968. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.