Abstract
Background
At least five bHLH genes regulate cell fate determination and differentiation of sensory neurons, hair cells and supporting cells in the mammalian inner ear. Cross-regulation of Atoh1 and Neurog1 results in hair cell changes in Neurog1 null mice although the nature and mechanism of the cross-regulation has not yet been determined. Neurod1, regulated by both Neurog1 and Atoh1, could be the mediator of this cross-regulation.
Methodology/Principal Findings
We used Tg(Pax2-Cre) to conditionally delete Neurod1 in the inner ear. Our data demonstrate for the first time that the absence of Neurod1 results in formation of hair cells within the inner ear sensory ganglia. Three cell types, neural crest derived Schwann cells and mesenchyme derived fibroblasts (neither expresses Neurod1) and inner ear derived neurons (which express Neurod1) constitute inner ear ganglia. The most parsimonious explanation is that Neurod1 suppresses the alternative fate of sensory neurons to develop as hair cells. In the absence of Neurod1, Atoh1 is expressed and differentiates cells within the ganglion into hair cells. We followed up on this effect in ganglia by demonstrating that Neurod1 also regulates differentiation of subtypes of hair cells in the organ of Corti. We show that in Neurod1 conditional null mice there is a premature expression of several genes in the apex of the developing cochlea and outer hair cells are transformed into inner hair cells.
Conclusions/Significance
Our data suggest that the long noted cross-regulation of Atoh1 expression by Neurog1 might actually be mediated in large part by Neurod1. We suggest that Neurod1 is regulated by both Neurog1 and Atoh1 and provides a negative feedback for either gene. Through this and other feedback, Neurod1 suppresses alternate fates of neurons to differentiate as hair cells and regulates hair cell subtypes.
Introduction
Neuronal and hair cell development of the inner ear critically depends on the basic Helix-Loop-Helix (bHLH) genes Neurog1 and Atoh1, respectively [1], [2]. However, while several bHLH genes are known in the otocyst, their interplay in prosensory cells to determine neuronal and hair cell differntiation in interaction with other factors remains unclear [3], [4], [5]. Previous work has identified several genes that are co-expressed in both neurosensory primordia in the ear as well as delaminating and differentiating sensory neurons outside the ear, indicating that common upstream regulatory elements may exist for these topologically distinct cells that apparently differentiate into unique adult cells [6], [7], [8], [9]. For example, neurotrophins delineate future sensory areas and are transiently expressed in delaminating neurons that exit the ear adjacent to or overlapping with prosensory regions [10]. Based on this circumstantial evidence it was suggested that these delaminating neurons may have some lineal relationship with the prosensory areas in the ear [6]. If true, ES or iPS cells could be made to develop into both neurons and hair cells, and could regenerate all neurosensory cells lost in deaf patients [11], [12], [13].
This idea of some relationship of neurosensory precursors was further substantiated by studies of two inner ear bHLH genes (Neurog1, Neurod1). Loss of either gene affects both sensory neuron and hair cells to a variable degree across all epithelia [14], [15], [16]. Neurog1 is the earlier expressed of the two genes and its absence substantially reduces hair cells in all sensory epithelia [2], [14]. In addition, non-sensory cells such as cells in the cruciate eminence [14], greater epithelial ridge and ductus reuniens [17] are converted into hair cells. In contrast Neurod1, which is regulated by Neurog1 expression in neurons, shows a less profound effect on sensory epithelia [15], [16]. Neurog1 or Neurod1 may have distinct cell-autonomous effect and thus not all cells that are positive for Neurog1 will be positive for Neurod1 which results in some disparity between these two mutations. The simplest explanation for this combined effect on sensory neurons and hair cells by these two bHLH genes is a possible lineage or even clonal relationships of some sensory neuron and hair cell precursors [18], [19]. The observed reduction in hair cells in the respective null mutants could be a consequence of loss of neurosensory precursors [14] or their conversion into hair cells [17]. This idea of lineage relationship is supported by lineage tracing for some neurons and hair cells in mice [20], [21] and the clonal relationship of a small set of neurons and hair cells has been established in chicken [22]. However, technical limitations have thus far precluded establishing unequivocally the degree of this lineage/clonal relationship between all sensory neurons and hair cells.
While these data establish some molecular and in certain cases, lineage and clonal relationship of neurons and hair cells, the molecular basis for the distinct differentiation of either cell type has not been investigated beyond the transcriptional regulation of Neurog1 and Atoh1 [20] or short range interactions mediated by delta-notch [23]. This could either happen through de novo differentiation of distinct, unspecified otic cells or through successive refinement of cell fate within a given lineage of potentially ambivalent precursor cells. Using a conditional deletion approach we provide here evidence that Neurod1, a gene regulated by Neurog1 in neurons [2] and by Atoh1 in hair cells [17], suppresses an alternate Atoh1-mediated hair cell fate in cells within the ganglia and aids in differentiation of specific hair cell types in the cochlea. Neurod1 is regulating multiple transcription factors in the neurosensory precursors, which are prematurely expressed in a different pattern in the absence of Neurod1. These data provide for the first time a detailed molecular mechanism for cell fate switching in neurosensory precursors of the mammalian inner ear and show that it hinges on suppression of alternate fates in neurosensory precursors by Neurod1.
Materials and Methods
Ethics Statement
All animal procedures were approved by the University of Iowa Animal Care and Use Committee (IACUC) and conducted according to their guidelines (ACURF #0804066).
Mice and genotyping for generation of conditional Neurod1 knockout mice (CKO)
Previously, lethality of newborn Neurod1 systemic null mice due to severe diabetes arrested the analysis in postnatal mice. To overcome this problem, we extended our analysis in the inner ear using Neurod1 conditional knockout mice [Neurod1f/f,Tg(Pax2-cre)]. By generating the Neurod1 conditional knockout (CKO) mice we could successfully circumvent the effect of Neurod1 in pancreatic β-cell development and could rescue mice to adulthood in Mendelian ratio.
To generate the Neurod1 conditional knockout mice we crossed the Pax2-cre line [24] with the floxed Neurod1 line [25]. For this study we used crosses between homozygotic floxed Neurod1 mice (Neurod1f/f) with heterozygous Neurod1f/+,Tg(Pax2-cre) mice. The resulting Neurod1f/f,Tg(Pax2-cre) mice are conditional knockout (CKO) mutant and the Neurod1f/+,Tg(Pax2-cre) heterozygous siblings serve as controls, here referred to as wild-types. To show the endogenous Neurod1 expression by the lacZ reporter, we have used Neurod1f/z,Tg(Pax2-cre) mice as mutant and Neurod1+/z mice as control.
We also analyzed the Neurod1 CKO using Tg(Atoh1-cre). We generated the mice by breeding the homozygous floxed Neurod1 (Neurod1f/f) [25] and Tg(Atoh1-cre) with a ROSA26 reporter [17] as previously described [26].
Offspring were genotyped by PCR analysis of tail DNA using Cre-specific primers which produce a 280 bp product, and Neurod1-specific primers which produce a 400 bp product from Neurod1 coding region and a 600 bp product from the floxed allele. Embryos were collected from timed pregnant females at embryonic day 10.5 (E10.5), E11.5, E12.5, E14.5, E16.5 and E18.5 counting noon of the day the vaginal plug was found as E0.5. We have also analyzed post-natal day 0 (P0), P7, P14, P16 and P30 mice. Pregnant mothers or juvenile mice were anesthetized with a lethal dose of Avertin (1.25% of 2.2.2-tribromoethanol at a dose of 0.025 ml/g of body weight). Embryos were dissected from the uterus and perfused with 4% paraformaldehyde (PFA) in 0.1 M phosphate buffer (pH 7.4) using a peristaltic pump. Heads were isolated and fixed in 4% PFA for further analysis.
X-gal staining
After perfusion with 4% PFA, mice were hemisected and ears were dissected in 0.4% PFA. After brief washes with phosphate buffer, the samples were stained in a solution containing 0.1 M phosphate buffer, 0.01% deoxycholic acid, 0.02% NP40, 2 mM magnesium chloride, 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide and 0.1 mg/ml X-gal (5-bromo-4-chloro-3-indolyl- β -D-galactoside) for up to 24 hours at room temperature [27].
In situ hybridization
In situ hybridization was performed using the RNA probe labeled with digoxigenin. The plasmids containing the cDNAs were used to generate the RNA probe by in vitro transcription. The following probes were graciously provided: Atoh1; Dr. Zoghbi; Neurog1, Dr. Ma; Fgf8, Dr. Pirvola; Pou4f3, Dr. Xiang; Nhlh1 and Nhlh2, Dr. Braun; Sox2, Dr. Cheah. The dissected ears were fixed in 0.4% paraformaldehyde, dehydrated in 100% methanol and rehydrated and then digested briefly with 20 µg/ml of Proteinase K (Ambion, Austin, TX, USA) for 15–20 minutes. Then the samples were hybridized overnight at 60°C to the riboprobe in hybridization solution containing 50% (v/v) formamide, 50% (v/v) 2X saline sodium citrate (Roche) and 6% (w/v) dextran sulphate. After washing off the unbound probe, the samples were incubated overnight with an anti-digoxigenin antibody (Roche Diagnostics GmbH, Mannheim, Germany) conjugated with alkaline phosphatase. After a series of washes, the samples were reacted with nitroblue phosphate/5-bromo, 4-chloro, 3-indolil phosphate (BM purple substrate, Roche Diagnostics, Germany) which is enzymatically converted to a purple colored product. The ears were mounted flat in glycerol and viewed in a Nikon Eclipse 800 microscope using differential interference contrast microscopy and images were captured with Image-Pro software.
Immunofluorescence
For immunofluorescence staining, the ears were dehydrated in graded ethanol overnight and rehydrated in graded ethanol and PBS. Samples were then blocked with 0.25% normal goat serum in PBS containing 0.01% Triton-X-100 for 1 hour. Then the primary antibodies for Myo VIIa (Myosin VIIa, Proteus Biosciences), Tubulin (Sigma), Caspase 3 (Cell Signaling Technology) and espin (a gift from Dr. J. Bartles JR) were used in dilutions of 1∶200, 1∶800, 1∶100 and 1∶5 respectively and incubated for 24–48 hours at 4°C. After several washes with PBS, corresponding secondary antibodies (1∶500) (Alexa fluor molecular probe 647 or 532 or 488; Invitrogen) were added and incubated overnight at 4°C. The ears were washed with PBS and mounted in glycerol and images were taken with a Leica TCS SP5 confocal microscope.
Plastic embedding and Stevenel's Blue staining
The end organs of ears were fixed in 2.5% glutaraldehyde overnight followed by several washes with 0.1 M phosphate buffer and then fixed with 1% osmium tetroxide for up to 1 hour. Samples were then washed with deionized water and dehydrated in graded ethanol followed by propylene oxide, embedded with Epon 812 in beam capsules and baked at 60°C for 48 hours. 2 µm sections were cut using a Reichert Ultratome and stained with Stevenel's Blue [28] made of 2% potassium permanganate and 1.3% methylene blue.
For higher resolution and co-localization of probes and proteins, we performed in situ hybridization for Fgf8, followed by Myo VIIa immunocytochemistry on same ears. Some of these ears were embedded in resin, sectioned and imaged with epifluorescent and transmitted light on a Nikon E800. Some of these sections were counterstained with Stevenel's blue for more detailed histology.
SEM
P30 mice were lethally anesthetized and perfused with 4% PFA. Ears were dissected, decalcified in EDTA and osmicated [1% OsO4 in 0.1 M phosphate buffer (pH 7.4)]. Osmicated ears were washed several times in distilled water to remove all ions, dehydrated in a graded ethanol, critically point dried, mounted on stubs and coated with gold/palladium. Stubs were viewed with a Hitachi S-3400N Scanning Electron Microscope with 2MeV acceleration.
Results
Ectopic hair cells form in sensory ganglia of Neurod1 conditional null mice
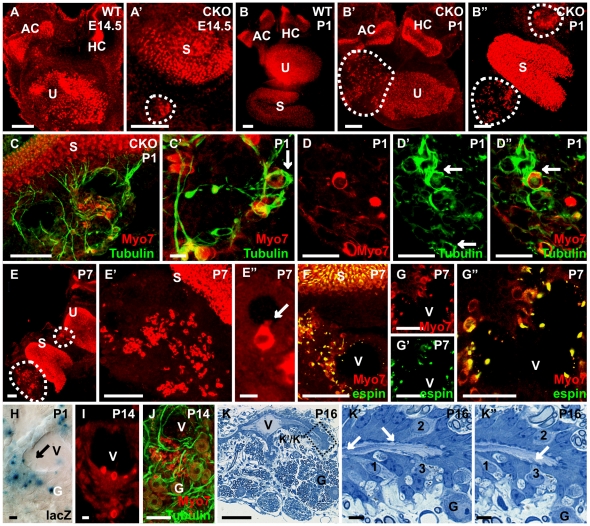
Previous work has shown that most cochlear and many vestibular sensory neurons are lost in Neurod1 null mice [15] with the surviving neurons projecting aberrantly to the sensory epithelia of the ear and into the brain [29]. When we studied the detailed distribution of sensory epithelia in whole mounted ears using Myo VIIa as a marker for hair cells to quantify the effects of loss of Neurod1 on hair cell development, we found numerous Myo VIIa positive cells scattered in the two remaining neuronal aggregations of vestibular and cochlear sensory neurons near the saccule and utricle (Fig. 1A, B’–K’). We also observed the appearance of these Myo VIIa positive cells in ganglia at different stages of Neurod1 CKO mice as early as E14.5 to near adult (E14.5, P0, P7, P14 and P30; Fig. 1). Many of these cells were grouped around multiple vesicles that were present within these ganglia (Fig. 1H,I). Immunofluorescence labeling with anti-β-tubulin antibody revealed that these cells were densely innervated and occasionally showed formation of a calyx (Fig. 1C’,D’, D”). Some of them had hair like bundles of apical specializations projecting from their apex into these vesicles (Fig. 1E”, I). To confirm that these apical processes were stereocilia, not microvilli, we next performed anti-espin immunofluorescence staining, a marker for stereocilia [30]. We detected espin immunostaining in stereocilia of these cells which, combined with the Myo VIIa marker identified them as hair cells (Fig. 1F–G”). The hair cells around the vesicles could be detected at least until P30 (P16 shown in Fig. 1K–K”) and the vesicles inside the ganglia persisted for about 9 month after birth (data not shown). These cells were surrounded by other cells with luminal contact, had ciliary protrusions into the vesicle and their base showed what appeared to be enlarged synaptic boutons (Fig. 1K’,K”). The cells outlining these vesicles seemed to be in contact with perineurial fibrocytes (Fig. 1K–K”).
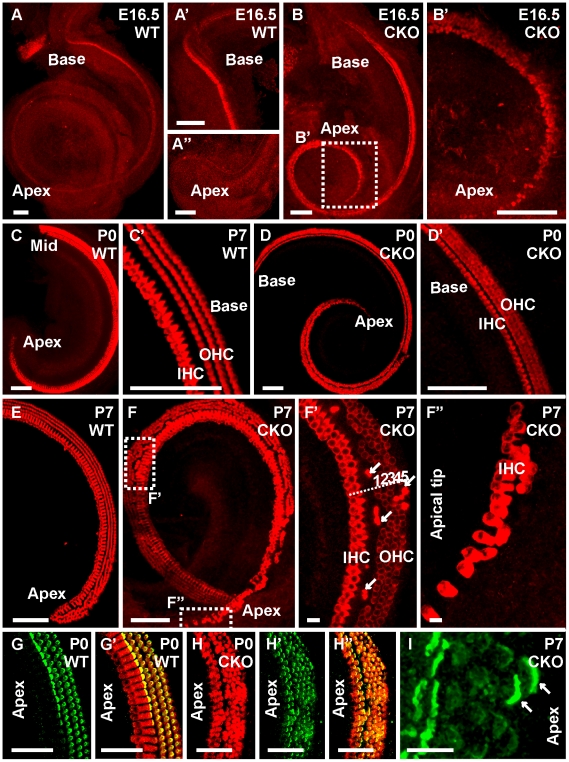
Figure 1. Absence of Neurod1 results in formation of MyoVIIa positive ectopic hair cells in inner ear ganglia.
Whole mount Immunofluorescence labeling with Myo VIIa labels the hair cells in the vestibular epithelia of wild-type mice exclusively inside the sensory epithelia (A,B). In contrast, in Neurod1 CKO mice, Myo VIIa positive cells are also found outside the sensory epithelia interspersed among the remaining vestibular ganglia near the utricle and the saccule from E14.5 until adulthood (A’, B’, B”, E,E’, I). These Myo VIIa positive cells are often grouped around intraganglionic vesicles (C, E”, F, G, G’, G”, I, J). Immunofluorescence labeling with anti-β-tubulin shows innervation of these Myo VIIa positive cells, some of them apparently form a calyx (arrows in C’, D’, D”). Closer examination shows that some of these cells assume a hair cell like shape with the formation of apical specialization protruding into vesicles (arrows in E”, H). We confirm with anti-espin antibody that these protrusions are stereocilia of these hair cell-like cells (F–G”). Formation of vesicles is also apparent in Neurod1-lacZ histochemically reacted ears (H) where β-galactosidase positive cells also protrude stereocilia into the vesicular lumen (arrow in H). These vesicles persist within the vestibular ganglia near the saccule (J). The vesicles are surrounded by cells forming an epithelial layer with hair cells displaying apical specializations protruding into the lumen shown in thin plastic sections (K, K’, K”). I,2,3 in (K’, K”) indicates three ‘intraganglionic hair cells’ in two consecutive sections showing apical protrusions into the vesicular lumen as well as enlarged contacts. AC, anterior canal crista; HC, horizontal canal crista S, saccule; U, utricle; G, ganglion cell; V, intraganglionic vesicle, Bar indicates 100 µm (except 50 µm in G” and10 µm in E”, C’, H and K’, K”).
In conclusion, our data suggest that Neurod1 expression in differentiating sensory neurons suppresses the development of Myo VIIa positive hair cells within inner ear ganglia. Alternatively, neural crest derived Schwann cells or mesenchyme derived fibroblasts, cell types that never express Neurod1, could be transformed into hair cells through the lack of interaction with Neurod1 containing neurons. We next tested more definitive markers for hair cell fate acquisition to obtain more insights into this transformation process.
Gene expression suggests that lack of Neurod1 transforms some surviving neurons into hair cells
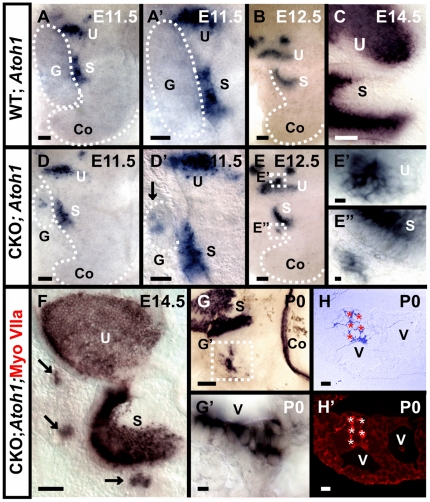
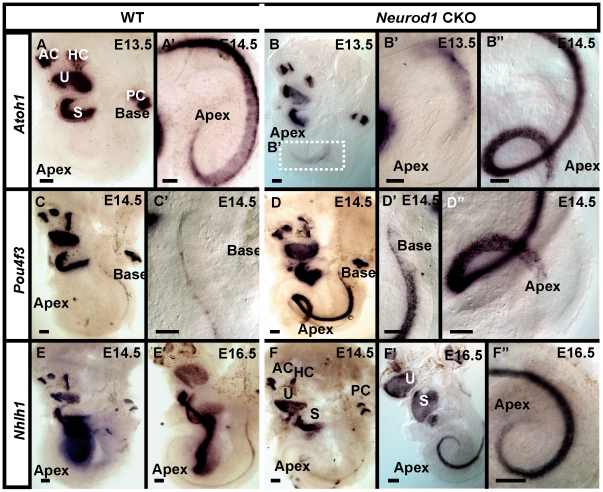
Atoh1 is a hair cell differentiation marker with a well established role in the ear only in the differentiation of hair cells [1], [31], [32]. Only limited expression of Atoh1 has been reported with sophisticated techniques in sensory neurons [17] and none in the neural crest derived Schwann cells or mesenchyme derived perneurial fibrocytes. Using in situ hybridization we observed only a transient Atoh1 expression in the vestibular ganglia of wild-type mice at E11.5 (Fig. 2A, A’). However, in the Neurod1 CKO mice, Atoh1 expression continued in these ganglia past the transient expression found in control animals (Fig. 2D, D’). Atoh1 expression was more profound in later stages and was found in a cluster of cells in the remaining ganglia next to the utricle and saccule, mostly around the ectopic vesicles (Fig. 2E–G’). Immunofluorescence labeling with Myo VIIa antibody revealed that each of the Atoh1 positive cells was also immunopositive for Myo VIIa (Fig. 2 H,H’).
Figure 2. Persistent Atoh1 expression in remaining ganglia relates to transformation of ganglionic cells into hair cells in Neurod1 mutant.
In situ hybridization of Atoh1 shows a faint and transient expression at E11.5 in some vestibular ganglion cells in wild-type mice (A, A’). This expression is more profound and continues in the ganglia of Neurod1 CKO mice (D, D’). In later stages, Atoh1 in situ signal appears in a cluster of cells in CKO mutants near the utricle and saccule (E–E’, F, G–G’). Some of the Atoh1 positive cells are aligned along the vesicular lumen similar to the Myo VIIa positive cells shown in Fig. 1 (G). To investigate co-localization, we labeled Atoh1 in situ reacted ears with anti-Myo VIIa antibody, embedded in plastic and sectioned. The sections reveal co-localization of Myo VIIa with Atoh1 in these cells (H,H’) thereby providing strong evidence that these cells are hair cells. S, saccule; U, utricle; G, ganglia; V, intraganglionic vesicle. Bar indicates 100 µm except E, E’ and 10 µm in E, E’.
While these data would normally be considered as proof of the hair cell nature of these cells [12], we worked with additional specific hair cells markers to establish that these cells were indeed hair cells and not transformed neurons with abnormal properties. Two POU domain factors are uniquely expressed in hair cells and neurons [33], [34], Pou4f3 and Pou4f1 (formerly Brn3c and Brn3a, respectively). We demonstrated that Pou4f3, an exclusive marker for hair cells in the ear with limited expression outside the ear [33], [35], was expressed in hair cell-like cells in the remaining ganglia of the Neurod1 CKO ear (Fig. 3H) and colocalized with Myo VIIa (data not shown) identical to Atoh1 (Fig. 2H, H’). The expression of these three markers (Atoh1, Myo VIIa and Pou4f3) is uniquely associated with hair cells in wild-type inner ears (Fig. 2C, 3). Therefore, their expression in cells within the remaining ganglia in Neurod1 CKO mice provides evidence that these cells are hair cells. Differentiation of these hair cells started around the same time those markers were upregulated in the nearby vestibular sensory epithelia and, transiently, in the delaminating sensory neurons.
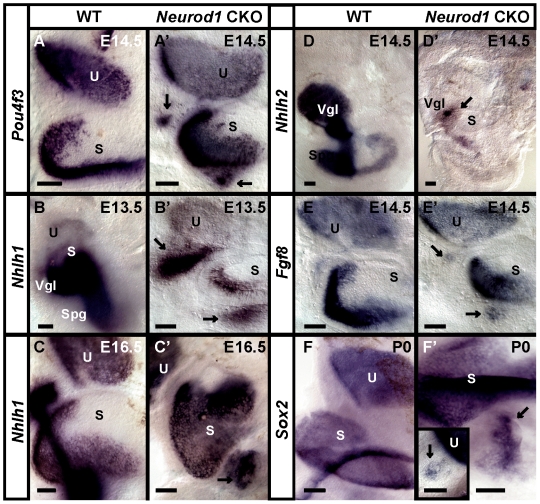
Figure 3. Neurod1 suppresses hair cell specific genes in ganglia.
While Myo VIIa and Atoh1 are good indicators for the possible hair cell formation inside the ganglia of Neurod1 CKO mice, we investigate the expression of other hair cell specific genes. Pou4f3 and Nhlh1 are expressed in hair cells (A–C) and responsible for their differentiation and are found in the ganglia in CKO mutants (A’–C’). In addition to hair cell marker, we also examine the neuronal marker Nhlh2 (D,D’) and neurosensory marker Fgf8 (E,E’) and Sox2 (F,F’). We found only few Nhlh2 positive neurons in the same topology near the intraganglionic vesicles in mutants suggesting a mix of hair cells and neurons near these vesicles (D’) as is clearly the case in histological sections (Fig. 1 K,K’, K”). Fgf8 (E’) and Sox2 (F’, insert in F’), which participate in neurosensory development, are also found near intraganglionic vesicles near the utricle and saccule. However, Sox2 expression may also indicate formation of some supporting cells in those vesicles. (F’, insert in F’). S, saccule; U, utricle; Vgl, vestibular ganglia; Spg, spiral ganglia. Arrows indicate labeled cells inside the ganglia of Neurod1 CKO mice. S, saccule; U, utricle; Vgl, vestibular ganglia; Spg, spiral ganglia. Bar indicates 100 µm.
To further expand the notion that these cells were genuine hair cells, we next studied another set of bHLH genes Nhlh1 and Nhlh2 which are associated with hair cells and neurons, respectively [36]. Nhlh1 and Nhlh2 are primarily expressed in sensory neurons in early embryos with an additional later expression of Nhlh1 in inner ear sensory epithelia [36]. We observed Nhlh1 mRNA expression in the vestibular ganglia and in delaminating cells in early embryos with progressive upregulation in the sensory epithelia in later stages (Fig. 3B,C). In the absence of Neurod1 the expression of Nhlh1 was massively reduced in neurons with some residual expression in cells below the saccule and utricle (Fig. 3B’,C’). In contrast to Nhlh1, Nhlh2 expression was exclusively in sensory neurons (Fig. 3D). In Neurod1 CKO mice, Nhlh2 expression was retained only in a small set of cells near the utricle (Fig. 3D’). Loss of expression of both genes in neurons was likely associated with early onset of apoptosis in Neurod1 mutant [29].
We also investigated Sox2, a protein required for hair cell differentiation [37] and later is highly expressed in supporting cells in the ear [38]. Expression of this gene was also found within the ganglia next to the utricle and saccule (Fig. 3F’). This could indicate that some cells in these ganglia were possibly supporting cells.
Another factor exclusively expressed in some hair cells and only transiently in sensory neurons is Fgf8 [39]. Consistent with data on several transcription factors, we found transient expression of Fgf8 in delaminating sensory neurons of both wild-type and Neurod1 CKO mice (Fig. 4H–J’). At later stages, when other markers for hair cells are expressed, we found Fgf8 expression in the ganglia in a pattern reminiscent of the hair cells identified by other markers (Fig. 3 E’).
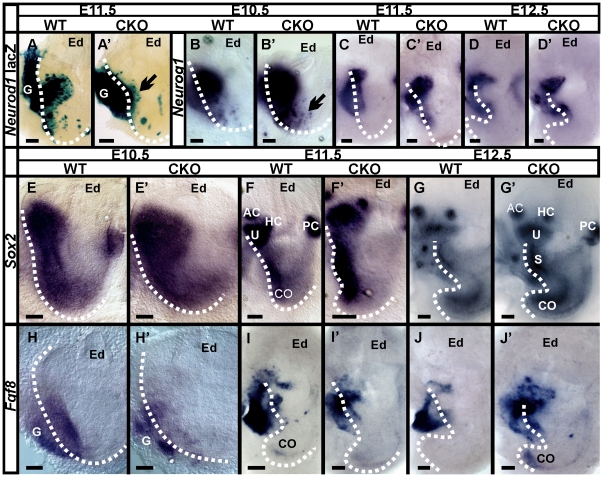
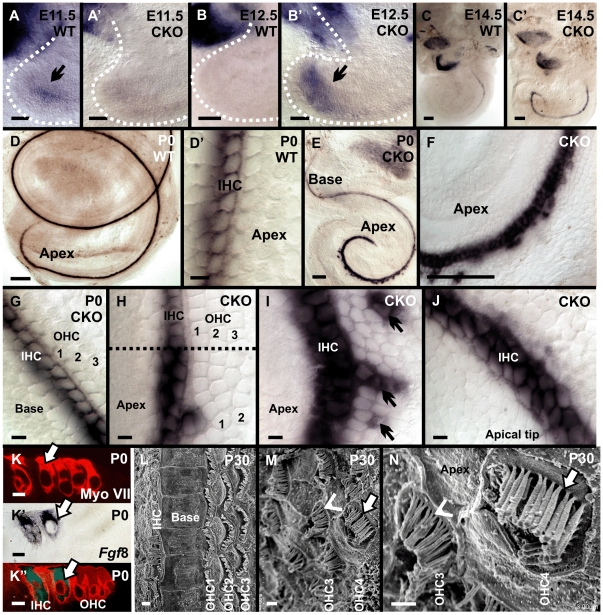
Figure 4. Absence of Neurod1 results in aberrant expression of genes responsible for prosensory specification.
At E11.5 embryo, β-galactosidase histochemistry show Neurod1-lacZ expression in the delaminating neuroblast (A, A’) and in the vestibular ganglion (G) which is moderately larger in the Neurod1 CKO mice (A’). In contrast, expression in the otic vesicle is reduced in Neurod1 CKO mice (arrow in A’). Neurog1, an upstream regulator of Neurod1, show a larger expression in the absence of Neurod1 (B’, C’, D’). In particular the prosensory domain is remarkably enlarged inside the otic vesicle with aberrant migration of the prosensory precursors (arrow in B’) in Neurod1 CKO mice. After the specification of the sensory epithelia, Neurog1 expression is progressively downregulated in wild-type mice (C, D) but some expression remains in the Neurod1 CKO mutant (C’, D’). In the mutant, Sox2 expression is expanded in the otocyst (compare E, E’) and shows more profound expression in the neurosensory precursor domain (F’,G’). We also investigate Fgf8 expression which is strongly positive in the delaminating neuroblasts both in wild-type and in Neurod1 CKO mice (H–J’). In the wild-type, Fgf8 disappears in the otic vesicle after E11.5 but remains transiently restricted to the ganglia (I, J). In contrast, this Fgf8 expression remains in the absence of Neurod1 inside the otic vesicle in areas identified to be composed of sensory precursor cells (Fig. H’, I’, J’). G, vestibular ganglion; AC, anterior canal crista; HC, horizontal canal crista; PC, posterior canal crista; S, saccule; U, utricle;Co, cochlea; Ed, endolymphatic duct. Boundary of otic vesicle is marked with dotted lines. Bar indicates 100 µm.
In summary, the presence of MyoVIIa, Atoh1, Pou4f3, Sox2, Nhlh1, Nhlh2 and Fgf8 expression in cells near intraganglionic vesicles inside the remaining ganglia implied a substantial modification of the cellular identity of these cells in Neurod1 CKO mice. We suggest that at least some surviving neurons are converted into hair cells which organize the surrounding tissue into vesicles inside ganglia and possibly regulate supporting cell differentiation of nearby neurons, fibroblasts or Schwann cells to form epithelia-like structures. Such organizing capacity of Atoh1 expressing cells has already been demonstrated in vitro [40] and is known for Fgf8 in vivo [41]. Since none of these markers ever appear in neural crest derived Schwann cells or mesenchyme derived fibroblasts, it seems unlikely that they are de novo expressed in these cells in the absence of Neurod1 expression in differentiating neurons. In contrast, several of these factors are known to be expressed in sensory neurons [17]. We therefore suggested that among the three cell types found in wild-type ear ganglia, it is the sensory neurons that are converted into hair cells. While highly suggestive of a neuronal origin, these data cannot fully exclude the alternative but more complex scenario of a Schwann cell or fibroblast transformation into hair cells in the absence of Neurod1.
Neurod1 affects hair cell type differentiation through regulation of multiple genes
A role of Neurog1 in inner ear neurosensory lineage acquisition is well established along with Neurod1 as a downstream activator of Neurog1 [2], [15], [16]. We now provide evidence for a novel function of Neurod1 consistent with its early expression in the prosensory domain to suppress an alternate hair cell fate. To further understand the possible interactions we analyzed in a Neurod1 lacZ reporter the level of β-galactosidase expression in delaminating neuroblasts (Fig. 4A). We previously showed by in situ hybridization that Pax2-Cre results in complete and early deletion of Neurod1 in mutant mice [29]. We therefore needed to use the reporter as both wild-type neurons and neurons lacking Neurod1 will show the reporter. In contrast to wild-type mice, the expression of lacZ reporter was increased in the delaminating neurons of Neurod1 CKO mice (Fig. 4A’) while the expression in the otocyst was reduced. This early knock out of Neurod1 resulted in considerable changes in expression pattern of several genes analyzed in different embryonic stages with in situ hybridization (Fig. 4).
For example, Neurog1 persisted longer in its expression in the delaminating neurons in the absence of Neurod1 (Fig. 4B’,C’,D’), suggesting a negative regulation of Neurog1 by Neurod1. Sox2 is associated with progenitor and stem cell populations in the developing CNS tissues [42], which might also be true for the sensory progenitors of the cochlea [37], [43]. We therefore also investigated Sox2 expression in early embryos of Neurod1 mutant mice. In Neurod1 CKO mice, Sox2 expression was expanded most prominently in the dorsal vestibular but also in the ventral prosensory region of the cochlea (Fig. 4E’,F’,G’) compared to the wild-type littermates (Fig. 4E, F, G). Later expression of Sox2 was near the ectopic vesicles inside the ganglia (Fig. 3F’).
Among twenty-three different Fgfs and four FGF receptors, several are known to function in early otic induction and sensory specification [8], [44], [45], [46], [47]. We investigated Fgf8 expression due to its known early expression in delaminating neurons and later function in organ of Corti in supporting cell development [41], [48], [49]. We found Fgf8 expression in the early stage of development, which is substantially modified in the absence of Neurod1 (Fig. 4H–J’). Fgf8 was expressed in the prosensory domain and transiently in the delaminating neuroblasts (Fig. 4H,I,J) of wild-type mice. In Neurod1 CKO mice, Fgf8 was expressed in delaminating cells with moderate loss of expression in ganglia (Fig. 4H’,I’,J’) consistent with the neuronal loss in the absence of Neurod1 [29].
In summary, we suggested a compelling role of Neurod1 in specification of neurosensory precursors beyond its role in neuronal differentiation, possibly through an interaction with important genes like Neurog1, Sox2 and Fgf8. This suggestion is based on the fact that early deletion of Neurod1 results in substantial changes of expression of several genes that may directly or indirectly affect cell fate determination of the precursor population.
Inactivation of Neurod1 results in a shortened and disorganized organ of Corti
Having now established that Neurod1 affects cell fate acquisition in the ear ganglia we investigated the effect of loss of Neurod1 on hair cells. Previous work has shown a surprising overlap of apparent cell specific bHLH genes in inner ear development. For example, Neurog1 null mice not only lose all sensory neurons but also have truncated development of hair cells in several sensory epithelia [14], [17]. Recently, a lineage relationship between neurons and some hair cells was demonstrated [20]. Interestingly, despite absence of Neurog1, hair cells strongly express Neurod1 [17], suggesting that hair cell specific factors such as Atoh1 are also able to upregulate Neurod1 expression and can do so more effectively in the absence of Neurog1. Since Neurod1 is immediately downstream and directly regulated by Neurog1 [2], we investigated the effects of Neurod1 on hair cell development as they are affected by the absence of either Neurog1 or Neurod1 [14], [15], [16], [20]. Consistent with an expression of Neurod1 in hair cells [29] and previous suggestions about a shortened cochlea with disorganized hair cells in Neurod1 systemic null mice [15], [16], [50], we also found a truncation of the cochlea. When compared to wild-type, Neurod1 mutant cochlea was not as much shortened as Neurog1 null cochlea (Table 1). Size reduction in Neurod1 CKO mice was also apparent in other epithelia and canal cristae which were approximately 30% shorter than the control (Table 1; Fig. S1). These data suggest that Neurod1 exerts a comparable effect on hair cell formation as Neurog1 albeit at a reduced scale. The somewhat less severe effect could relate to the fact that Neurod1 is downstream to Neurog1 and some common sensory neuron/hair cell precursors may already have separated.
Table 1. Length of the Cochlea in wild-type, Neurod1 CKO and Neurog1 null mutations.
| Length of Cochlea (µm) | WT (n = 4) | Neurod1 CKO (n = 4) | Neurog1 null (n = 3) |
| Mean | 5907 | 2695 | 2449 |
| SD | 408 | 138 | 320 |
We next wanted to investigate whether the truncated cochlea has multiple rows of disorganized hair cells as previously reported in other mutants with shortened cochlear growth [9], [14]. There was a gradient of malformation of the hair cells in the base and apex of Neurod1 CKO mice (Fig. 5) as demonstrated with Myo VIIa immunofluorescence staining at different stages of development. We observed premature expression of Myo VIIa in the apex of Neurod1 mutant cochlea in comparison to wild-type mice (Fig. 5A,A”,B,B’). In contrast, the organization of the organ of Corti was normal in the basal half of the cochlea and comparable with the control mice (Fig. 5C–D’). However, the orientation of hair cells was severely disrupted in the apical half. In this location, multiple rows of inner and outer hair cells (IHCs, OHCs) were found (Fig. 5F). The misalignment of the hair cells in the apical half of mutant cochlea was more obvious in later stages (P7 shown here) and demonstrated two rows of IHCs and four to five rows of OHCs (Fig. 5F,F’). In addition, Myo VIIa was more prominently expressed in IHCs in the apex, as compared to the uniform expression in the base, and similar high levels of expression of Myo VIIa was found in scattered OHCs in the apex (Fig. 5F,F’). We therefore interpreted these cells as ‘ectopic IHC’s’. The apical tip consisted exclusively of IHCs in two disorganized rows without any OHCs (Fig. 5F”).
Figure 5. Neurod1 is necessary for development of an orderly patterned organ of Corti.
Myo VIIa immunocytochemistry shows upregulation of Myo VIIa in the wild-type starts around E16.5 from the mid-base and later progresses toward both base and apex with a regular organization of one row of inner and three rows of outer hair cells throughout the cochlea (A, A’, C, C’, E). In contrast, Myo VIIa is already expressed throughout the cochlea with disorganization of hair cells in the apical half (B, B’, D) in E16.5 Neurod1 CKO mutant littermates. In later stages, the basal half of the CKO mutant shows normal orientation of the hair cells (D, D’) whereas the apex of Neurod1 CKO mice shows multiple rows of both IHCs and OHCs with reduction of Myo VIIa intensity in most outer hair cells (F, F’). In addition, clusters of higher intensity of Myo VIIa positive cells are found in between outer hair cells with equivalent staining intensity to inner hair cells (arrows in F’). The apical tip of the mutant cochlea shows a partially duplicated row of inner hair cells with complete absence of outer hair cells (F”). Using espin immunocytochemistry we confirm the disorganization of the apical half of the mutant cochlea where two rows of inner hair stereocilia and four to five rows of outer hair stereocilia are observed (H–H’) along with some unusually displaced strongly stained inner hair stereocilia (arrow in I) in between faintly labeled outer hair stereocilia (I). IHC, inner hair cells; OHC, outer hair cells. Bar indicates 100 µm except F”; 10 µm in F”.
To evaluate the apical specialization of the organ of Corti of Neurod1 null mice, we performed the espin immunofluoresence labeling combined with the Myo VIIa. We found similar abnormality of stereocilia with espin as with Myo VIIa in hair cells in the apical half. The espin immunolabeling confirmed the disorganization of organ of Corti in the apex of Neurod1 CKO mice where multiple rows of IHCs and OHCs were found with two distinct types of stereocilia (Fig. 5H–H”). In the newborn mice, density of the stereocilia of both IHCs and OHCs were almost equal, but at later stages the density of stereocilia of OHCs was remarkably reduced with few displaced highly dense inner hair stereocilia in place of weakly labeled outer hair stereocilia (Fig. 5I). Near the apical tip only the inner type of stereocilia were found (data not shown).
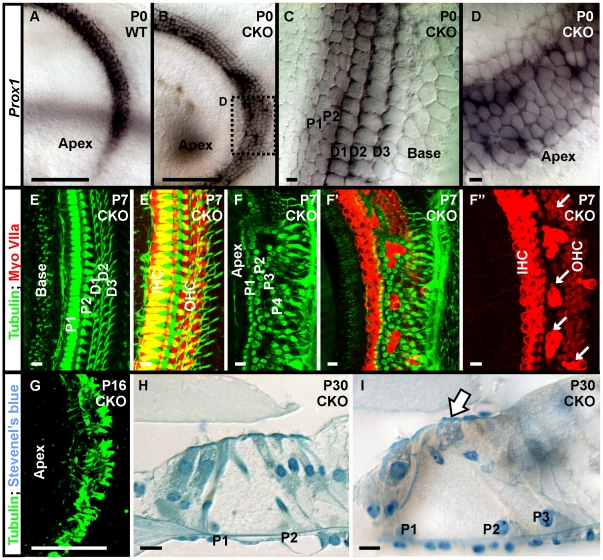
We next wanted to understand how the irregularity of hair cells affected the supporting cells. In situ hybridization with Prox1, a marker for Deiter's and Pillar cells [51], [52] revealed disorganized supporting cells (Fig. 6D). We also used β-tubulin immunofluoresecnce labeling in P7 and P16 Neurod1 CKO mutant mice (Fig. 6E–F’,G). We found the organization of supporting cells in the basal half of the cochlea was regular with thick processes of two Pillar cells and thin phalangeal processes of Deiter’s cells labeled with β-tubulin (Fig. 6E, E’). In the apical half, most Deiter’s cell processes were converted into thick Pillar cell processes (Fig. 6G) that surrounded the ectopic, strong Myo VIIa positive IHCs (Fig. 6F–F”).
Figure 6. Neurod1 regulates organization of supporting cells as well as hair cells.
A supporting cell marker, Prox1, shows uniform expression in two rows of pillar and three rows of Deiter’s cells in wild-type mice (A) and in the base of Neurod1 CKO mice (B,C). In contrast, the apex of Neurod1 CKO mice shows multiple disorganized rows of supporting cells with no clear distinction between Pillar and Deiter’s cells (D). At P7, combined tubulin and Myo VIIa immunocytochemistry shows normal organization of supporting cells in between hair cells in the base of the mutant cochlea (E, E’). This orientation is disrupted in the apex where Myo VIIa positive hair cells are surrounded by supporting cells with thick processes filled with tubulin. This is reminiscent of Pillar cells and not of the thinner phalangeal processes of Deiter’s cells (F, F”, G). Detailed histology in thin plastic sections of P30 mutant cochlea show not only the persistence of multiple rows of inner and outer hair cells in the apex (H) but also the formation of multiple rows of Pillar cells (P1, P2, P3) and inner hair cells in places of outer hair cells (arrow in I). These data suggest that Neurod1 is an important transcription factor that mediates the coordinated type specific hair cell and supporting cell development in the apical half of the organ of Corti which is necessary for the coordinated development of supporting cells. P1, P2, inner and outer Pillar cells; D1, D2, D3, 1st, 2nd, 3rd row of Deiter’s cells; IHC, inner hair cells; OHC, outer hair cells. Bar indicates 100 µm except C, D, H, I and 10 µm in C, D, H, I.
We further investigated this unusual phenotype in later stage of serially sectioned P30 cochlea. At this stage, while some normal OHCs near the apex had developed a cigar shape pattern, some abnormal OHCs resembled IHCs in their shape (Fig. 6H,I) as well as duplication of IHCs consistent with the finding with Myo VIIa staining (Fig. 6H). In addition, the supporting cells around these hair cells were markedly disorganized and had multiple rows of Pillar cells (Fig. 6I).
Apical disorganization may relate to premature expression of hair cell genes
We next investigated several genes relevant for neurosensory ear development to elucidate further the extent of defect at the level of gene expression. Atoh1 is a crucial factor for hair cell differentiation of the inner ear [1]. In the cochlea, Atoh1 is upregulated in a base to apex gradient, starting around E13.5 [17], [32]. We found a spatiotemporally altered Atoh1 expression in the Neurod1 mutant cochlea (Fig. 7B–B”). Atoh1 was expressed in the apex of Neurod1 mutant cochlea (Fig. 7B–B’) as early as E13.5 before it appeared in the wild-type littermate cochlea (Fig. 7A). At E14.5, Atoh1 was profoundly expressed in the entire cochlea of Neurod1 mutant mice (Fig. 7B’) whereas in the wild-type littermate, expression was weak in the apex with an obvious gradient suggesting a base to apex progression (Fig. 7A’). Therefore, Atoh1 expression gradient in the cochlea was not only altered in the absence of Neurod1 but also was expressed earlier than in the wild-type mice (compare Fig. 7A, B). A similar phenotype with premature expression of Atoh1 in the apex was reported in Neurog1 null mice [17]. This similarity could indicate a possibly similar mechanism as Neurod1 is downstream of Neurog1 [2] and that Neurog1 could directly or indirectly regulate Neurod1 expression in the apical hair cells.
Figure 7. Neurod1 controls hair cell specific gene expression in the apex.
Normally hair cell cycle exit and differentiation is delayed in the cochlea compared to vestibular epithelia and shows a pattern of apex-to-base progression of cell cycle exit. In contrast, there is a base-to-apex progression of differentiation, including upregulation of Atoh1 (A), Pou4f3 (C) and Nhlh1 (E) in wild-type mice. In Neurod1 CKO mice, the expression of these genes not only happens earlier but progresses from the apex to the base (compare A and B; C and D, D”; E, E’ and F, F’,F”). Atoh1 and Pou4f3 are essential for hair cell differentiation and maintenance, respectively. Their expression suggests a premature initiation of hair cell differentiation that normally is delayed in the apex compared to the base. Note that the apex in B is shown with 90° anti-clockwise rotation in B’. AC, anterior canal crista; HC, horizontal canal crista; PC, posterior canal crista; S, saccule; U, utricle. Bar indicates 100 µm.
Pou4f3 is responsible for cochlear and vestibular hair cell differentiation and survival. Deletion results not only in hair cell loss but also in delayed loss of ganglion neurons [33], [35], [53]. We have analyzed the expression of Pou4f3 in Neurod1 CKO mice with in situ hybridization. Pou4f3 expression starts shortly after the hair cell fate is committed [54]. We found a premature upregulation of Pou4f3 in the apex of the mutant cochlea much earlier than wild-type shown at E14.5 (Fig. 7D, D”) while in the wild-type littermate, Pou4f3 expression had just started near the base of the cochlea (Fig. 7C, C’).
Nhlh1 is later in development expressed in the hair cells of all sensory epithelia [36]. In wild-type mice, Nhlh1 was expressed and upregulated in the base of the cochlea at E16.5 with an apparent base to apex gradient (Fig. 7E’). In Neurod1 CKO mice, Nhlh1 expression was already evident in the apex of the cochlea at E14.5 (Fig. 7F). By E16.5, Nhlh1 was expressed in the entire cochlea of Neurod1 CKO mice, which is obviously more profound in comparison to weak expression near the base of the wild-type littermate (compare Fig. 7E’, F’, F”).
In summary, Myo VIIa, Atoh1, Pou4f3 and Nhlh1 showed an altered pattern of expression in Neurod1 CKO cochlea with premature expression in an apex-to-base instead of a base-to-apex progression as in wild-type littermates (Fig. 5A–B’, 7). This spatiotemporal aberration of the bHLH gene expression might alter the onset of differentiation of apical hair cells. The premature expression of these transcription factors in hair cells that show a delayed differentiation compared to their early cell cycle exit [17] may relate to the disorganization of apical hair cells in Neurod1 CKO mice.
Atoh1 expression was not only altered spatiotemporally but also persisted longer in the absence of Neurod1. For example, Atoh1 expression progressively reduced in the wild-type cochlea from base-to-apex started from P0 to onward with restricted expression only in the inner two rows of OHCs (Fig. S1A–A’; shown in P0). In contrast, Atoh1 expression persisted longer in all the hair cells throughout the cochlea of Neurod1 CKO mice (Fig. S1B–B”). Atoh1 was also aberrantly expressed in the non-sensory compartment which was most obvious in the cruciate eminence of the anterior cristae (AC) and also in striola region of the saccule and utricle (Fig. S1D–D”). Consistent with Atoh1 expression, we also found longer persisting Pou4f3 expression in the base and apex of the Neurod1 CKO cochlea in contrast to wild-type littermates (data not shown). We previously reported that loss of Neurod1 resulted in continued expression of Atoh1 in cerebellar granule cells [26]. Apparently, Neurod1 exerts a similar inhibitory influence on the expression of Atoh1 and its downstream genes Nhlh1 and Pou4f3.
Altered Fgf8 expression may relate to the cochlear histological changes
In mice, FGF3, FGF8 and FGF10 play a role in the early inductive events of the otic vesicle formation [8], [39], [46], [55], [56], [57], [58]. We observed Fgf8 expression in the delaminating sensory neuron in both wild-type and Neurod1 CKO mice as early as E10.5 (Fig. 4). Consistent with expression changes in Atoh1 and other downstream hair cell specific genes, Fgf8 was also expressed prematurely in the apex of the Neurod1 mutant mice (Fig. 8B’). Fgf8 was transiently expressed in the apex of wild-type mice but disappeared after E11.5 (compare Fig. 8A and B). However, absence of Neurod1 resulted in continued Fgf8 expression in the apex from E11.5 onward and thus resulted in premature and reversed expression pattern (Fig. 8A’, B’, C’). In contrast, in wild-type mice, Fgf8 expression started at E14.5 from the base of the cochlea progressing over time to the apex (Fig. 8C).
Figure 8. Fgf8 misexpression correlates with formation of ‘ectopic inner hair cells’.
In situ hybridization shows a persistent expression of Fgf8 in neurosensory precursors in Neurod1 mutant cochlea as early as E12.5 (arrow in B’) with premature expression in particular in the apex of the cochlea (C’) in comparison to wild-type (C). Fgf8 is expressed transiently in the prosensory domain in wild-type mice (arrow in A) and later is shown to be upregulated in the cochlea with a base-to-apex gradient (A,B,C). In newborn mice, Fgf8 is uniformly expressed in all inner hair cells almost along the entire length of the cochlea in wild-type mice (D, D’). In contrast, Neurod1 CKO mice display an increased expression level in the apex (E, F, H, I, J) and deviate from the single row labeling of only inner hair cells seen in the base (G). Two or more rows of inner hair cells are positive for Fgf8 and scattered single and multiple cells are interspersed among the multiple rows of outer hair cells which are also positive for Fgf8 (arrows in I). The apical tip shows up to three rows of Fgf8 positive cells (J). Radial sections through the apex of Fgf8 ISH reacted and Myo VIIa immunostained cochlea reveals co-localization of Fgf8 and Myo VIIa in inner as well as ‘ectopic inner hair cells’ scattered among outer hair cells (arrows in K–K”). SEM of P30 Neurod1 CKO mice reveals a normal organization of inner hair cells in a single row and three rows of outer hair cells in the base of Neurod1 CKO mice (L). In contrast, the apex shows inner hair cell sized stereocilia (arrows) interspersed among normal sized stereocilia bearing outer hair cells (arrowhead; M,N). Dotted line in H indicates border between normal and disorganized organ of Corti. IHC, inner hair cells; OHC, outer hair cells. Bar indicates 100 µm in A–F except D’; 10 µm in D’,G–J, K–K” and 1 µm in L–N.
In the cochlea, Fgf8 is expressed in IHCs from where it diffuses to bind to its receptor, Fgfr3, which leads to the development of Pillar cells instead of Deiter’s cells [41], [59], [60]. Our data on Myo VIIa expression in the Neurod1 CKO mutant suggested that some OHCs may achieve an inner hair cell-like phenotype (‘ectopic IHCs’; Fig. 5) and this may be due to an altered Fgf8 expression. We therefore examined Fgf8 expression to verify that these cells are ‘ectopic IHCs’ as Fgf8 is a marker of IHCs of the organ of Corti [44]. In wild-type mice, Fgf8 was expressed exclusively in the single row of IHCs (Fig. 8D,D’). In the Neurod1 CKO mutant, we found single rows of Fgf8 positive IHCs only in the basal half of the cochlea (Fig. 8E,G) whereas in the apical half, multiple rows of Fgf8 positive IHCs was observed as well as ectopic expression in some IHCs replacing OHCs (Fig. 8E,F,H,I) consistent with the observation of Myo VIIa expression (Fig. 5F,F’). Closer to the apical tip we found only Fgf8 expression in multiple rows of IHCs without OHCs (Fig. 8J). In conclusion, absence of Neurod1 altered Fgf8 expression in the apex of the mutant cochlea which directly or indirectly related to the change of stereotyped pattern and differentiation of apical hair cells in the organ of Corti.
To analyze more closely the effect of Fgf8 expression in the organ of Corti, we performed plastic sections of Fgf8 in situ reacted P0 ears sequentially immunolabeled with anti Myo VIIa antibody. Sections through the base showed the expected distribution of a single row of Fgf8 and Myo VIIa positive IHCs and three rows of OHCs (data not shown). While,radial sections through the apex confirmed both clusters of inner hair cells as well as single or multiple Fgf8 positive cells among the OHCs in Neurod1 CKO mice (Fig. 8K–K”) consistent with the whole mounted data.
We further investigated the consequences of Fgf8 misexpression and analyzed the differentiation of the stereocilia in P30 mice using SEM. Consistent with the patchy expression of Fgf8 in only some topographic OHCs we found a patchy aberration of stereocilia where some ‘OHCs’ had stereocilia twice as thick as others, resembling the diameter of inner hair cell stereocilia (Fig. 8M,N). The changes in stereocilia supported the evidence of ‘ectopic IHCs’, dispersed among OHCs.
In summary, absence of Neurod1 leads to premature upregulation of hair cell differentiation genes in the apex, severe disorganization of the apical hair cells and supporting cells, misexpression of Fgf8 in some ‘OHCs’, and development of inner hair cell-like stereocilia among OHCs. Consistent with its early expression in the inner ear prosensory region, Neurod1 plays a significant role in hair cell maturation through the suppression of several genes in the apex, most prominently Atoh1 and Fgf8. We tested whether a delayed knockout of Neurod1 using Tg(Atoh1-cre) could achieve these altered differentiation of hair cells. Despite a massive cerebellar phenotype of this CKO mouse [26], our data showed no effect in inner ear development or any alteration of phenotype of neurosensory cells (Fig. S2). Once the prosensory domain is specified in early embryonic stage, later deletion of Neurod1 has no effect in refinement of hair cell fate.
Discussion
Neurod1 is essential for neuronal differentiation in the cerebellum [26] and the ear [29] and can convert non-neuronal cells into neurons [61] through the regulation of over 500 downstream genes [62]. We analyzed the role of Neurod1 in inner ear neurosensory cell development using a newly generated Neurod1 conditional knockout mouse. We previously reported [15], [29] substantial loss of inner ear sensory neurons and disorganization of remaining afferent projections in Neurod1 systemic and conditional null mice. The expression of other bHLH genes such as Nhlh1 and Nhlh2 [36] may be responsible for partial rescue of those few sensory neurons that survive in the absence of Neurod1. We here identify two novel roles of Neurod1:
Neurod1 suppresses hair cell differentiation in sensory ganglia.
Neurod1 controls gene expression needed for outer hair cell maturation.
Neurod1 suppresses differentiation of ganglion cells into hair cells
A cascade of pro-neuronal bHLH genes transforms ectodermal cells into neurons and can do so by simply being misexpressed in the developing ectoderm [61] or the ear [43]. These bHLH proteins also determine cell fate in other tissues such as pancreas [16], gut [63] and Merkel cells [64]. bHLH genes are important for cell fate switch. Without expression of Atoh1, cells can change from a secretory to an absorptive phenotype [65].
In this study, we observed the formation of vesicles lined by hair cells in place of remaining ganglia in the Neurod1 CKO mice (Fig. 1). To further understand the molecular basis of formation of these ‘intraganglionic hair cells’, we analyzed expression of multiple hair cell specific genes. We found positive expression of Atoh1 and Pou4f3 and the hair cell marker Myo VIIa in these ‘intraganglionic hair cells’. Nhlh1 and Neurod1 are limited early on to neuronal expression but are found later in hair cells, including the ‘intraganglionic hair cells’ (shown with Nhlh1 in situ hybridization and Neurod1 lacZ expression). The overlapping function of Neurod1 and Nhlh1 may not only lead to the survival of some neurons but the absence of Neurod1 may allow premature and persistent expression of Atoh1 in these remaining ganglion neurons (Fig. 2D, D’). Consistent with the previous report of Atoh1 expression in some neurons [17], our in situ data also show a transient Atoh1 expression in the delaminating sensory neurons in the wild-type embryo. This limited expression of Atoh1 may normally be restrained by the expression of Neurod1 and absence of Neurod1 seems to allow continued expression. We suggest that this continued expression of Atoh1, combined with the absence of Neurod1 expression can result in the differentiation of ‘intraganglionic hair cells’ as well as the expression of other hair cell markers such as Myo VIIa, Pou4f3 and Nhlh1. In addition to hair cell markers, these or other cells within the ganglia show neurosensory markers such as Sox2, Nhlh2 and Fgf8.
Only three cell types with different embryonic origins are found in vestibular and cochlear ganglia: inner ear derived sensory neurons [66], mesoderm derived fibroblasts and neural crest derived Schwann cells [67]. A transformation of Schwann cells or fibroblasts, which never express Neurod1, is theoretically possible. However, the presence of multiple genes known to be expressed in neurosensory cells but not in Schwann cells or fibroblasts makes this a very improbable scenario. We therefore interpret our data to suggest that some inner ear derived ‘sensory neuron precursors’ adopt a hair cell fate in the absence of Neurod1. Such ‘hair cell’ formation from delaminated neurosensory precursors suggests a degree of flexibility of cell fate acquisition and is consistent with the emerging concept of lineage and possibly clonal neurosensory relationships in the ear [18], [19], [20]. In addition, these Myo VIIa positive ‘hair cells’ express marker genes otherwise only associated with hair cells in the ear, and reside around vesicles inside vestibular ganglion aggregations near the utricle and saccule. Since none of these markers ever appear in neural crest derived Schwann cells or in fibroblasts but at least Atoh1 is known to be expressed in sensory neurons [17], we suggest that of all three cell types found in wild-type ear ganglia it is the sensory neurons that are converted to hair cells. We name these hair cells as ‘intraganglionic hair cells’. Further work is needed to analyze in details the transformation of sensory neuron precursors into ‘intraganglionic hair cells’ and demonstrate the suppression of Atoh1 by Neurod1 at the molecular level either within a given cell or between cells via the delta/notch system.
Neurod1 helps to organize the organ of Corti by controlling spatiotemporal gene expression
Previous work showed that Neurod1 is expressed in hair cells [16], [17] in a shorter and disorganized cochlea [15], [50] of Neurod1 null mice. It has also been noted that some of the first row of OHCs obtain inner hair cell like appearance [16] and the IHCs may form multiple disorganized rows. We demonstrate for the first time the degree of disorganization of the cochlear apex with the formation of ectopic IHCs in place of OHCs in Neurod1 CKO mice and show the complete absence of OHC formation in the most apical part of the cochlea. We show expression of IHC marker genes in cells that topologically should be outer hair cells and the histological alteration of these cells such as diameter of stereocilia. Neurod1 mimics Neurog1 with respect to shortening, disorganization and gene expression alteration in the cochlea [14]. Like in Neurog1 null mice, Atoh1 is prematurely upregulated in the apex of Neurod1 CKO mice, suggesting a fate change of common precursors toward hair cells [17]. The most reduced sensory epithelia in Neurog1 null mice is the saccule [14], [31], now known to be affected because of lineage relationship of saccular neurons and hair cells [20].
In contrast, Neurod1 CKO mice show the most profound size reduction in canal cristae and cochlea (Table 1, Fig. S1B,D,D”). Effects of Neurod1 on overall growth are thus not simply a milder extension of Neurog1 effects. It is possible that simple premature expression of Atoh1 and its downstream genes disrupts convergent extension [68] and thus leads to the observed histological alteration of the cochlear apex. This is in agreement with the complete extension of the cochlea in the absence of Atoh1 expression and any differentiation of hair cells [31]. Other mutants with reduction in growth and multiple rows of hair cells show no mixing of inner and outer hair cells [9], [14]. Combined with the enhanced cell death as early as E9.5 in Neurod1 null mice [29], the early appearance of truncated growth in the cochlea suggests that common neuronal/hair cell precursors may die in the absence of Neurod1, reducing the growth of the organ of Corti and canal cristae. The expression of other bHLH genes such as Nhlh1 and Nhlh2 may rescue some common neurosensory precursors in the utricle and saccule resulting in near normal size growth. What additional gene(s) may mediate these differential sensory epithelia effects is unknown.
How can Neurod1 affect neuronal and hair cell differentiation?
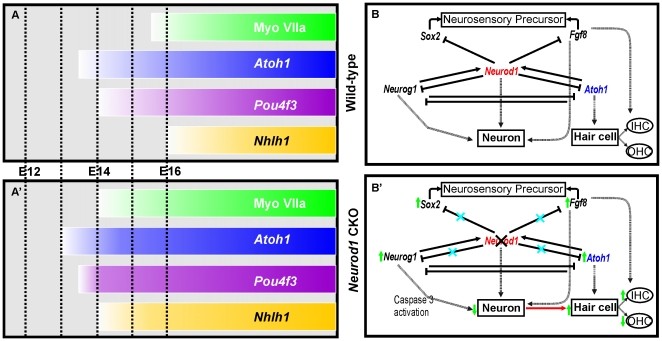
Neurod1 regulates several genes involved in hair cell differentiation. Atoh1, Pou4f3, Fgf8 and Nhlh1 are prematurely expressed in the apical half of the cochlea in Neurod1 CKO mutants (Fig. 9A,A’) and appear in hair cells within the sensory ganglia of the ear. There is also a transient change in Sox2 expression and in Neurog1 expression. Our results are best compatible with a suggestion that Neurod1 expression in neurosensory precursors suppresses specific downstream genes (Atoh1, Pou4f3, Nhlh1, Fgf8, Sox2) necessary for general neurosensory and specific hair cell differentiation (Fig. 9B,B’). For example, the upregulation of Fgf8 in some ‘outer hair cells’, which may change their fate to ‘inner hair cells’, suggest a more specific function of Neurod1 in regulation of Fgf8. The effect of Neurod1 on Neurog1 is likely due to a direct, intracellular feedback loop (Fig. 9B,B’) and is in line with previous reports of such a feedback loop in olfactory receptor cell development [69]. In contrast, the effect of Neurod1 on Atoh1 expression could be either directly in the same cell as in the cerebellum [26] or could be mediated through an intermediary such as Fgf8, Sox2 or an as yet to be determined factor within or between cells. Further analysis of other developing systems in which Neurod1 and Atoh1 are sequentially expressed or co-expressed, such as the dorsal cochlear nucleus [70] or the enteroendocrine intestine cells [71], are needed to establish generality of this feedback loop beyond the ear and the cerebellum.
Figure 9. Neurod1 regulates neuronal differentiation by suppression of premature hair cell differentiation of neurons possibly interacting with several target genes.
In the absence of Neurod1 several hair cell specific genes such as Myo VIIa, Atoh1, Pou4f3 and Nhlh1 are prematurely expressed with an inverse gradient of apex-to-base progression of hair cell differentiation instead of usual base-to-apex progression (cochlear expression shown with bars in A, A’). In addition, these genes are also expressed ectopically in the differentiating vestibular ganglia near the utricle and saccule. This substantial alteration of gene expression changes the organization of the apical part of the cochlea and results in the formation of ‘intragangliionic hair cells’. Our data and those of others suggest the following interaction of Neurod1 with Neurog1, Atoh1, Sox2 and Fgf8 to regulate inner ear cellular identity (B, B’). We propose that after early and transient activation of Neurod1 by Neurog1 and Atoh1 to differentiate neuron, Neurod1 suppresses Neurog1 to inhibit precursor proliferation and Atoh1 to inhibit hair cell differentiation in neurons. These three way interactions result in formation of neurons with delayed hair cell differentiation. Cross inhibition of Neurog1 and Atoh1 was previously suggested [17, 20] and we suggest that Neurod1 is a key intermediary player. Neurod1 also regulates other cell fate determining genes like Sox2 and Fgf8 which may more directly related to the observed cell fate switch. We suggest that Neurod1 deletion in early embryos disrupts this gene network and, as a consequence, the coordinated sequential neurosensory development of inner ear resulting in the transformation of some surviving neurons into ‘intraganglionic hair cells’ and alteration of the cell type specific differentiation of outer hair cells in the cochlea.
Cross-regulation of Neurog1 and Atoh1 have been proposed for the spinal cord [72] and the mammalian ear [20] in which hair cells are massively reduced in Neurog1 null mice [14]. However, in none of these cases has the interaction been directly demonstrated at the cellular or molecular level. Our data on the effect of Neurod1 CKO mutants suggests that Neurod1 is at least in the ear an intermediate factor that mediates such cross-inhibitory interactions between Neurog1 and Atoh1 (Fig. 9B, B’). The differences in effects of either loss of Neurog1 or Neurod1 on overall hair cell formation and specific hair cell developmental changes suggests that other downstream factors specific to Neurog1 or Neurod1 must exist that also mediate such cross-inhibitory interactions. Fgf8 and possibly Sox2 seem to be appropriate candidates to play this role. Further analysis of expression of these and other genes now identified as being changed in Neurod1 CKO mice are needed for Neurog1 null mice to fully understand the complexity of interaction of the genes in this developing system.
In summary, we propose that inner ear development resembles other developing systems insofar as sophisticated interactions of bHLH genes determine neuronal fate [3], [73]. In cooperation with other genes known to be expressed in the developing sensory neurons and hair cells [3], [4], [5], [74], [75], Neurod1 may achieve neuronal differentiation not only through upregulation of appropriate downstream target genes [62] but also through suppression of other bHLH genes that mediate other states of cellular differentiation such as Neurog1 [69] or Atoh1 [26]. In the absence of Neurod1, several cell fate determining genes that would normally be suppressed are prematurely or continuously expressed (Figs 4,9). These expression changes result most likely in a cell fate change of neurosensory precursors into topologically inappropriate cells such as ‘intraganglionic hair cells’ and ‘ectopic inner hair cells’ (Fig. 9B’). Through some of the 500 genes directly regulated by Neurod1 [62], Neurod1 may actually mediate the cross-regulation of Neurog1 and Atoh1 as recently suggested [20]. Our findings show a more refined action of Neurod1 in the developing ear than the previously suggested simple effects on neuronal survival and differentiation [15], [16], [29]. Neurod1 may interact with other genes expressed during neurosensory development [36], likely mimicking better described systems in the complexity of their interaction [76] particularly at the promoter level [77]. Fully understanding this interplay is necessary to allow, through regulation of the levels of expression of Neurod1, the generation of ‘intraganglionic hair cells’ in deaf patients. Such ‘intraganglionic hair cells’ could imitate regular hair cells and sustain long-term cochlear implant function by maintaining viable neurons.
Supporting Information
Atoh1 in situ hybridization shows a progressive base-to-apex reduction in wild-type cochlea (A, A’, A”) with faded expression in IHC and outermost OHCs. In contrast, Atoh1 remains uniformly expressed throughout the cochlea in Neurod1 CKO mice (B, B’, B”). Direct comparison of vestibular sensory epithelia shows that canal cristae are more reduced than utricle and saccule (C–F) which show mostly alterations in shape. Such qualitative changes are also apparent in anterior canal cristae where hair cells form in the non-sensory region of cruciate eminence in absence of Neurod1 (arrow in G,H). AC, anterior canal crista; HC, horizontal canal crista; S, saccule; U, utricle. Bar indicates 100 µm.
(0.63 MB TIF)
Our data suggest that Neurod1 specifies neurosensory precursors by refining cellular identity during early embryonic stage. We confirm this assumption studying the delayed knock out of Neurod1 using Tg (Atoh1cre), a CKO mutation that results in massive cerebellar defects (26). The in situ hybridizations of Atoh1 and Fgf8 show normal organization of inner ear with four rows of hair cells throughout the cochlea (A–B). We conclude that once Atoh1 has regulated its downstream target genes to specify the hair cell precursor's fate, later loss of Neurod1 cannot alter hair cell differentiation. IHC, inner hair cell; OHC, outer hair cell. Bar indicates 100 µm.
(0.52 MB TIF)
Acknowledgments
We like to thank Drs. K.-A. Nave and S. Goebbels, Max-Planck-Institute of Experimental Medicine, for providing the floxed Neurod1 mice used for this study and Drs. T. Ohyama and A. Groves for providing the Pax2-Cre line. The Leica TCS SP5 confocal microscope was purchased in part with a grant from the Roy. J. Carver foundation. We thank CLAS and the Office of the Vice President for Research (OVPR) for support. We wish to thank the following people for providing plasmids for in situ hybridization: Atoh1; Dr. Zoghbi; Neurog1, Dr. Ma; Fgf8, Dr. Pirvola; Pou4f3, Dr. Xiang; Nhlh1 and Nhlh2, Dr. Braun; Sox2, Dr. Cheah.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by a NIH grant (R01 DC 005590) to B.F. The support of CLAS and OVPR of the University of Iowa is acknowledged. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bermingham NA, Hassan BA, Price SD, Vollrath MA, Ben-Arie N, et al. Math1: an essential gene for the generation of inner ear hair cells. Science. 1999;284:1837–1841. doi: 10.1126/science.284.5421.1837. [DOI] [PubMed] [Google Scholar]
- 2.Ma Q, Chen Z, del Barco Barrantes I, de la Pompa JL, Anderson DJ. neurogenin1 is essential for the determination of neuronal precursors for proximal cranial sensory ganglia. Neuron. 1998;20:469–482. doi: 10.1016/s0896-6273(00)80988-5. [DOI] [PubMed] [Google Scholar]
- 3.Fritzsch B, Eberl DF, Beisel KW. The role of bHLH genes in ear development and evolution: revisiting a 10 year old hypothesis. 2010. CMLSE in press. [DOI] [PMC free article] [PubMed]
- 4.Abello G, Khatri S, Radosevic M, Scotting PJ, Giraldez F, et al. Independent regulation of Sox3 and Lmx1b by FGF and BMP signaling influences the neurogenic and non-neurogenic domains in the chick otic placode. Dev Biol. 2010;339:166–178. doi: 10.1016/j.ydbio.2009.12.027. [DOI] [PubMed] [Google Scholar]
- 5.Bell D, Streit A, Gorospe I, Varela-Nieto I, Alsina B, et al. Spatial and temporal segregation of auditory and vestibular neurons in the otic placode. Dev Biol. 2008;322:109–120. doi: 10.1016/j.ydbio.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 6.Fritzsch B, Beisel KW, Jones K, Farinas I, Maklad A, et al. Development and evolution of inner ear sensory epithelia and their innervation. J Neurobiol. 2002;53:143–156. doi: 10.1002/neu.10098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karis A, Pata I, van Doorninck JH, Grosveld F, de Zeeuw CI, et al. Transcription factor GATA-3 alters pathway selection of olivocochlear neurons and affects morphogenesis of the ear. J Comp Neurol. 2001;429:615–630. doi: 10.1002/1096-9861(20010122)429:4<615::aid-cne8>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 8.Pauley S, Wright TJ, Pirvola U, Ornitz D, Beisel K, et al. Expression and function of FGF10 in mammalian inner ear development. Dev Dyn. 2003;227:203–215. doi: 10.1002/dvdy.10297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pauley S, Lai E, Fritzsch B. Foxg1 is required for morphogenesis and histogenesis of the mammalian inner ear. Dev Dyn. 2006;235:2470–2482. doi: 10.1002/dvdy.20839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farinas I, Jones KR, Tessarollo L, Vigers AJ, Huang E, et al. Spatial shaping of cochlear innervation by temporally regulated neurotrophin expression. J Neurosci. 2001;21:6170–6180. doi: 10.1523/JNEUROSCI.21-16-06170.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beisel K, Hansen L, Soukup G, Fritzsch B. Regenerating cochlear hair cells: quo vadis stem cell. Cell Tissue Res. 2008;333:373–379. doi: 10.1007/s00441-008-0639-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oshima K, Shin K, Diensthuber M, Peng AW, Ricci AJ, et al. Mechanosensitive hair cell-like cells from embryonic and induced pluripotent stem cells. Cell. 141:704–716. doi: 10.1016/j.cell.2010.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Groves AK. The challenge of hair cell regeneration. Exp Biol Med (Maywood) 2010;235:434–446. doi: 10.1258/ebm.2009.009281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma Q, Anderson DJ, Fritzsch B. Neurogenin 1 null mutant ears develop fewer, morphologically normal hair cells in smaller sensory epithelia devoid of innervation. J Assoc Res Otolaryngol. 2000;1:129–143. doi: 10.1007/s101620010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim WY, Fritzsch B, Serls A, Bakel LA, Huang EJ, et al. NeuroD-null mice are deaf due to a severe loss of the inner ear sensory neurons during development. Development. 2001;128:417–426. doi: 10.1242/dev.128.3.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu M, Pereira FA, Price SD, Chu MJ, Shope C, et al. Essential role of BETA2/NeuroD1 in development of the vestibular and auditory systems. Genes Dev. 2000;14:2839–2854. doi: 10.1101/gad.840500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matei V, Pauley S, Kaing S, Rowitch D, Beisel KW, et al. Smaller inner ear sensory epithelia in Neurog 1 null mice are related to earlier hair cell cycle exit. Dev Dyn. 2005;234:633–650. doi: 10.1002/dvdy.20551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fritzsch B, Beisel KW, Bermingham NA. Developmental evolutionary biology of the vertebrate ear: conserving mechanoelectric transduction and developmental pathways in diverging morphologies. Neuroreport. 2000;11:R35–44. doi: 10.1097/00001756-200011270-00013. [DOI] [PubMed] [Google Scholar]
- 19.Fritzsch B, Beisel KW, Hansen LA. The molecular basis of neurosensory cell formation in ear development: a blueprint for hair cell and sensory neuron regeneration? Bioessays. 2006;28:1181–1193. doi: 10.1002/bies.20502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raft S, Koundakjian EJ, Quinones H, Jayasena CS, Goodrich LV, et al. Cross-regulation of Ngn1 and Math1 coordinates the production of neurons and sensory hair cells during inner ear development. Development. 2007;134:4405–4415. doi: 10.1242/dev.009118. [DOI] [PubMed] [Google Scholar]
- 21.Koundakjian EJ, Appler JL, Goodrich LV. Auditory neurons make stereotyped wiring decisions before maturation of their targets. J Neurosci. 2007;27:14078–14088. doi: 10.1523/JNEUROSCI.3765-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Satoh T, Fekete DM. Clonal analysis of the relationships between mechanosensory cells and the neurons that innervate them in the chicken ear. Development. 2005;132:1687–1697. doi: 10.1242/dev.01730. [DOI] [PubMed] [Google Scholar]
- 23.Kiernan AE, Xu J, Gridley T. The Notch ligand JAG1 is required for sensory progenitor development in the mammalian inner ear. PLoS Genet. 2006;2:e4. doi: 10.1371/journal.pgen.0020004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohyama T, Groves AK. Generation of Pax2-Cre mice by modification of a Pax2 bacterial artificial chromosome. Genesis. 2004;38:195–199. doi: 10.1002/gene.20017. [DOI] [PubMed] [Google Scholar]
- 25.Goebbels S, Bode U, Pieper A, Funfschilling U, Schwab MH, et al. Cre/loxP-mediated inactivation of the bHLH transcription factor gene NeuroD/BETA2. Genesis. 2005;42:247–252. doi: 10.1002/gene.20138. [DOI] [PubMed] [Google Scholar]
- 26.Pan N, Jahan I, Lee JE, Fritzsch B. Defects in the cerebella of conditional Neurod1 null mice correlate with effective Tg(Atoh1-cre) recombination and granule cell requirements for Neurod1 for differentiation. Cell Tissue Res. 2009;337:407–428. doi: 10.1007/s00441-009-0826-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matei VA, Feng F, Pauley S, Beisel KW, Nichols MG, et al. Near-infrared laser illumination transforms the fluorescence absorbing X-Gal reaction product BCI into a transparent, yet brightly fluorescent substance. Brain Res Bull. 2006;70:33–43. doi: 10.1016/j.brainresbull.2005.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.del Cerro M, Cogen J, del Cerro C. Stevenel's Blue, an excellent stain for optical microscopical study of plastic embedded tissues. Microsc Acta. 1980;83:117–121. [PubMed] [Google Scholar]
- 29.Jahan I, Kersigo J, Pan N, Fritzsch B. Neurod1 regulates survival and formation of connections in the mouse ear and brain. Cell, Tissue Res in press. 2010 doi: 10.1007/s00441-010-0984-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sekerkova G, Zheng L, Mugnaini E, Bartles JR. Espin actin-cytoskeletal proteins are in rat type I spiral ganglion neurons and include splice-isoforms with a functional nuclear localization signal. J Comp Neurol. 2008;509:661–676. doi: 10.1002/cne.21755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fritzsch B, Matei VA, Nichols DH, Bermingham N, Jones K, et al. Atoh1 null mice show directed afferent fiber growth to undifferentiated ear sensory epithelia followed by incomplete fiber retention. Dev Dyn. 2005;233:570–583. doi: 10.1002/dvdy.20370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen P, Johnson JE, Zoghbi HY, Segil N. The role of Math1 in inner ear development: Uncoupling the establishment of the sensory primordium from hair cell fate determination. Development. 2002;129:2495–2505. doi: 10.1242/dev.129.10.2495. [DOI] [PubMed] [Google Scholar]
- 33.Xiang M, Gan L, Li D, Chen ZY, Zhou L, et al. Essential role of POU-domain factor Brn-3c in auditory and vestibular hair cell development. Proc Natl Acad Sci U S A. 1997;94:9445–9450. doi: 10.1073/pnas.94.17.9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang EJ, Liu W, Fritzsch B, Bianchi LM, Reichardt LF, et al. Brn3a is a transcriptional regulator of soma size, target field innervation and axon pathfinding of inner ear sensory neurons. Development. 2001;128:2421–2432. doi: 10.1242/dev.128.13.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiang M, Maklad A, Pirvola U, Fritzsch B. Brn3c null mutant mice show long-term, incomplete retention of some afferent inner ear innervation. BMC Neurosci. 2003;4:2. doi: 10.1186/1471-2202-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kruger M, Schmid T, Kruger S, Bober E, Braun T. Functional redundancy of NSCL-1 and NeuroD during development of the petrosal and vestibulocochlear ganglia. Eur J Neurosci. 2006;24:1581–1590. doi: 10.1111/j.1460-9568.2006.05051.x. [DOI] [PubMed] [Google Scholar]
- 37.Kiernan AE, Pelling AL, Leung KK, Tang AS, Bell DM, et al. Sox2 is required for sensory organ development in the mammalian inner ear. Nature. 2005;434:1031–1035. doi: 10.1038/nature03487. [DOI] [PubMed] [Google Scholar]
- 38.Dabdoub A, Puligilla C, Jones JM, Fritzsch B, Cheah KS, et al. Sox2 signaling in prosensory domain specification and subsequent hair cell differentiation in the developing cochlea. Proc Natl Acad Sci U S A. 2008;105:18396–18401. doi: 10.1073/pnas.0808175105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pirvola U, Ylikoski J, Trokovic R, Hebert JM, McConnell SK, et al. FGFR1 is required for the development of the auditory sensory epithelium. Neuron. 2002;35:671–680. doi: 10.1016/s0896-6273(02)00824-3. [DOI] [PubMed] [Google Scholar]
- 40.Woods C, Montcouquiol M, Kelley MW. Math1 regulates development of the sensory epithelium in the mammalian cochlea. Nat Neurosci. 2004;7:1310–1318. doi: 10.1038/nn1349. [DOI] [PubMed] [Google Scholar]
- 41.Puligilla C, Feng F, Ishikawa K, Bertuzzi S, Dabdoub A, et al. Disruption of fibroblast growth factor receptor 3 signaling results in defects in cellular differentiation, neuronal patterning, and hearing impairment. Dev Dyn. 2007;236:1905–1917. doi: 10.1002/dvdy.21192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Graham V, Khudyakov J, Ellis P, Pevny L. SOX2 functions to maintain neural progenitor identity. Neuron. 2003;39:749–765. doi: 10.1016/s0896-6273(03)00497-5. [DOI] [PubMed] [Google Scholar]
- 43.Puligilla C, Dabdoub A, Brenowitz SD, Kelley MW. Sox2 induces neuronal formation in the developing mammalian cochlea. J Neurosci. 2010;30:714–722. doi: 10.1523/JNEUROSCI.3852-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pirvola U, Ylikoski J, Trokovic R, Hebert J, McConnell S, et al. FGFR1 Is Required for the Development of the Auditory Sensory Epithelium. Neuron. 2002;35:671–685. doi: 10.1016/s0896-6273(02)00824-3. [DOI] [PubMed] [Google Scholar]
- 45.Pirvola U, Spencer-Dene B, Xing-Qun L, Kettunen P, Thesleff I, et al. FGF/FGFR-2(IIIb) signaling is essential for inner ear morphogenesis. J Neurosci. 2000;20:6125–6134. doi: 10.1523/JNEUROSCI.20-16-06125.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wright TJ, Mansour SL. Fgf3 and Fgf10 are required for mouse otic placode induction. Development. 2003;130:3379–3390. doi: 10.1242/dev.00555. [DOI] [PubMed] [Google Scholar]
- 47.Hayashi T, Ray CA, Younkins C, Bermingham-McDonogh O. Expression patterns of FGF receptors in the developing mammalian cochlea. Dev Dyn. 2010;239:1019–1026. doi: 10.1002/dvdy.22236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Colvin JS, Bohne BA, Harding GW, McEwen DG, Ornitz DM. Skeletal overgrowth and deafness in mice lacking fibroblast growth factor receptor 3. Nat Genet. 1996;12:390–397. doi: 10.1038/ng0496-390. [DOI] [PubMed] [Google Scholar]
- 49.Shim K, Minowada G, Coling DE, Martin GR. Sprouty2, a mouse deafness gene, regulates cell fate decisions in the auditory sensory epithelium by antagonizing FGF signaling. Dev Cell. 2005;8:553–564. doi: 10.1016/j.devcel.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 50.Xia A, Visosky AM, Cho JH, Tsai MJ, Pereira FA, et al. Altered traveling wave propagation and reduced endocochlear potential associated with cochlear dysplasia in the BETA2/NeuroD1 null mouse. J Assoc Res Otolaryngol. 2007;8:447–463. doi: 10.1007/s10162-007-0092-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bermingham-McDonogh O, Oesterle EC, Stone JS, Hume CR, Huynh HM, et al. Expression of Prox1 during mouse cochlear development. J Comp Neurol. 2006;496:172–186. doi: 10.1002/cne.20944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fritzsch B, Dillard M, Lavado A, Harvey NL. Canal cristae growth and fiber extension to the outer hair cells require Prox1 activity. PLoS One. 2010;5:1–12. doi: 10.1371/journal.pone.0009377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Keithley EM, Erkman L, Bennett T, Lou L, Ryan AF. Effects of a hair cell transcription factor, Brn-3.1, gene deletion on homozygous and heterozygous mouse cochleas in adulthood and aging. Hear Res. 1999;134:71–76. doi: 10.1016/s0378-5955(99)00070-2. [DOI] [PubMed] [Google Scholar]
- 54.Ryan AF, Mullen LM, Doherty JK. Cellular targeting for cochlear gene therapy. Adv Otorhinolaryngol. 2009;66:99–115. doi: 10.1159/000218210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alvarez Y, Alonso MT, Vendrell V, Zelarayan LC, Chamero P, et al. Requirements for FGF3 and FGF10 during inner ear formation. Development. 2003;130:6329–6338. doi: 10.1242/dev.00881. [DOI] [PubMed] [Google Scholar]
- 56.Mansour SL, Goddard JM, Capecchi MR. Mice homozygous for a targeted disruption of the proto-oncogene int-2 have developmental defects in the tail and inner ear. Development. 1993;117:13–28. doi: 10.1242/dev.117.1.13. [DOI] [PubMed] [Google Scholar]
- 57.Martin K, Groves AK. Competence of cranial ectoderm to respond to Fgf signaling suggests a two-step model of otic placode induction. Development. 2006;133:877–887. doi: 10.1242/dev.02267. [DOI] [PubMed] [Google Scholar]
- 58.Ohyama T, Groves AK, Martin K. The first steps towards hearing: mechanisms of otic placode induction. Int J Dev Biol. 2007;51:463–472. doi: 10.1387/ijdb.072320to. [DOI] [PubMed] [Google Scholar]
- 59.Hayashi T, Ray CA, Bermingham-McDonogh O. Fgf20 is required for sensory epithelial specification in the developing cochlea. J Neurosci. 2008;28:5991–5999. doi: 10.1523/JNEUROSCI.1690-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kelley MW. Regulation of cell fate in the sensory epithelia of the inner ear. Nat Rev Neurosci. 2006;7:837–849. doi: 10.1038/nrn1987. [DOI] [PubMed] [Google Scholar]
- 61.Lee JE, Hollenberg SM, Snider L, Turner DL, Lipnick N, et al. Conversion of Xenopus ectoderm into neurons by NeuroD, a basic helix-loop-helix protein. Science. 1995;268:836–844. doi: 10.1126/science.7754368. [DOI] [PubMed] [Google Scholar]
- 62.Seo S, Lim JW, Yellajoshyula D, Chang LW, Kroll KL. Neurogenin and NeuroD direct transcriptional targets and their regulatory enhancers. Embo J. 2007;26:5093–5108. doi: 10.1038/sj.emboj.7601923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang Q, Bermingham NA, Finegold MJ, Zoghbi HY. Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science. 2001;294:2155–2158. doi: 10.1126/science.1065718. [DOI] [PubMed] [Google Scholar]
- 64.Bermingham NA, Hassan BA, Wang VY, Fernandez M, Banfi S, et al. Proprioceptor pathway development is dependent on Math1. Neuron. 2001;30:411–422. doi: 10.1016/s0896-6273(01)00305-1. [DOI] [PubMed] [Google Scholar]
- 65.Shroyer NF, Helmrath MA, Wang VY, Antalffy B, Henning SJ, et al. Intestine-specific ablation of mouse atonal homolog 1 (Math1) reveals a role in cellular homeostasis. Gastroenterology. 2007;132:2478–2488. doi: 10.1053/j.gastro.2007.03.047. [DOI] [PubMed] [Google Scholar]
- 66.Rubel EW, Fritzsch B. Auditory system development: primary auditory neurons and their targets. Annu Rev Neurosci. 2002;25:51–101. doi: 10.1146/annurev.neuro.25.112701.142849. [DOI] [PubMed] [Google Scholar]
- 67.Morris JK, Maklad A, Hansen LA, Feng F, Sorensen C, et al. A disorganized innervation of the inner ear persists in the absence of ErbB2. Brain Res. 2006;1091:186–199. doi: 10.1016/j.brainres.2006.02.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kelly M, Chen P. Shaping the mammalian auditory sensory organ by the planar cell polarity pathway. Int J Dev Biol. 2007;51:535–547. doi: 10.1387/ijdb.072344mk. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kawauchi S, Beites CL, Crocker CE, Wu HH, Bonnin A, et al. Molecular signals regulating proliferation of stem and progenitor cells in mouse olfactory epithelium. Dev Neurosci. 2004;26:166–180. doi: 10.1159/000082135. [DOI] [PubMed] [Google Scholar]
- 70.Fritzsch B, Pauley S, Feng F, Matei V, Nichols DH. The evolution of the vertebrate auditory system: transformations of vestibular mechanosensory cells for sound processing is combined with newly generated central processing neurons. International Journal of Comparative Psychology. 2006;19:1–24. [Google Scholar]
- 71.Ray SK, Leiter AB. The basic helix-loop-helix transcription factor NeuroD1 facilitates interaction of Sp1 with the secretin gene enhancer. Mol Cell Biol. 2007;27:7839–7847. doi: 10.1128/MCB.00438-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gowan K, Helms AW, Hunsaker TL, Collisson T, Ebert PJ, et al. Crossinhibitory activities of Ngn1 and Math1 allow specification of distinct dorsal interneurons. Neuron. 2001;31:219–232. doi: 10.1016/s0896-6273(01)00367-1. [DOI] [PubMed] [Google Scholar]
- 73.Ohsawa R, Kageyama R. Regulation of retinal cell fate specification by multiple transcription factors. Brain Res. 2008;1192:90–98. doi: 10.1016/j.brainres.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 74.Qian Y, Fritzsch B, Shirasawa S, Chen CL, Choi Y, et al. Formation of brainstem (nor)adrenergic centers and first-order relay visceral sensory neurons is dependent on homeodomain protein Rnx/Tlx3. Genes Dev. 2001;15:2533–2545. doi: 10.1101/gad.921501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ross SE, Mardinly AR, McCord AE, Zurawski J, Cohen S, et al. Loss of inhibitory interneurons in the dorsal spinal cord and elevated itch in Bhlhb5 mutant mice. Neuron. 65:886–898. doi: 10.1016/j.neuron.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Breitkreutz A, Choi H, Sharom JR, Boucher L, Neduva V, et al. A global protein kinase and phosphatase interaction network in yeast. Science. 2010;328:1043–1046. doi: 10.1126/science.1176495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schmidt D, Wilson MD, Ballester B, Schwalie PC, Brown GD, et al. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science. 2010;328:1036–1040. doi: 10.1126/science.1186176. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Atoh1 in situ hybridization shows a progressive base-to-apex reduction in wild-type cochlea (A, A’, A”) with faded expression in IHC and outermost OHCs. In contrast, Atoh1 remains uniformly expressed throughout the cochlea in Neurod1 CKO mice (B, B’, B”). Direct comparison of vestibular sensory epithelia shows that canal cristae are more reduced than utricle and saccule (C–F) which show mostly alterations in shape. Such qualitative changes are also apparent in anterior canal cristae where hair cells form in the non-sensory region of cruciate eminence in absence of Neurod1 (arrow in G,H). AC, anterior canal crista; HC, horizontal canal crista; S, saccule; U, utricle. Bar indicates 100 µm.
(0.63 MB TIF)
Our data suggest that Neurod1 specifies neurosensory precursors by refining cellular identity during early embryonic stage. We confirm this assumption studying the delayed knock out of Neurod1 using Tg (Atoh1cre), a CKO mutation that results in massive cerebellar defects (26). The in situ hybridizations of Atoh1 and Fgf8 show normal organization of inner ear with four rows of hair cells throughout the cochlea (A–B). We conclude that once Atoh1 has regulated its downstream target genes to specify the hair cell precursor's fate, later loss of Neurod1 cannot alter hair cell differentiation. IHC, inner hair cell; OHC, outer hair cell. Bar indicates 100 µm.
(0.52 MB TIF)