Abstract
Schizophrenia is a neurodevelopmental psychiatric disorder characterized by a variety of structural brain abnormalities that appear to progress across the course of illness. Schizophrenia also is highly heritable, and one gene that has emerged as a possible susceptibility factor is G72. G72 influences brain development and activity by an as-yet unclear mechanism, and multiple studies have reported associations between G72 and schizophrenia. We were interested in linking these domains of investigation by determining whether G72 also influences the rate of longitudinal structural brain changes in individuals with schizophrenia. As part of the Iowa Longitudinal Study of Recent Onset Psychoses, we genotyped four G72 polymorphisms previously associated with schizophrenia in 110 subjects with schizophrenia or schizoaffective disorder from whom we had obtained two brain MRI scans an average of three years apart. The four polymorphisms captured three haplotypes, one of which was strongly associated with an increased rate of frontal lobe volume decrement. This same haplotype was also associated with more severe psychotic symptoms at the time of the second scan. These data thus suggest that variation in G72 modulates the progressive brain changes that characterize schizophrenia.
Keywords: DAOA, MRI, schizophrenia, frontal lobe, psychosis
Introduction
Schizophrenia is a neuropsychiatric disorder that affects 1% of the population. The disease is characterized by positive symptoms such as delusions and hallucinations, negative symptoms such as social withdrawal and affective flattening, and disorganization of speech and behavior. The etiology of schizophrenia appears to be due primarily to genetic factors, as evidenced by a sibling relative risk of approximately 10 and an estimated heritability of 81% (Sullivan et al., 2003). Brain structure volumes are also affected by schizophrenia. Specifically, enlarged CSF volumes, reductions in temporal and frontal lobes, and abnormalities in subcortical structures have been found in cross-sectional studies of individuals with schizophrenia (Shenton et al., 2001). Longitudinal MRI studies show that individuals with schizophrenia experience changes in brain structure volumes over time (Gur et al., 1998), with a progressive reduction in frontal lobe white matter volume (Ho et al., 2003; Mathalon et al., 2001) shown to be modulated by antipsychotic administration (Styner et al., 2005).
The search for genetic contributors to these pathophysiological processes has recently focused attention on glutamatergic N-methyl-D-aspartic acid (NMDA) receptors (Ghaziuddin et al., 1995; Goff and Coyle 2001). Several studies in the late 1990’s showed linkage to schizophrenia on chromosome 13q32-34 (Christian et al., 2002). Two overlapping genes in this region, G72 and G30, were then found to be associated with schizophrenia (Chumakov et al., 2002). Initially, G72 was thought to interact with D-amino acid oxidase (DAO), an enzyme involved in attenuating NMDA receptor-mediated neurotransmission (Chumakov et al., 2002), but more recent data suggest that G72 regulates mitochondrial function without interacting with DAO (Kvajo et al., 2008a). Subsequent studies have evaluated the linkage between G72 and schizophrenia (Abecasis et al., 2004; Addington et al., 2004; Chen et al., 2004; Detera-Wadleigh and McMahon 2006; Fallin et al., 2005; Korostishevsky et al., 2004; Li and He 2007; Liu et al., 2006; Ma et al., 2006; Mulle et al., 2005; Schulze et al., 2005; Wang et al., 2004; Wood et al., 2006; Yue et al., 2006; Zou et al., 2005). Although some did not find association (Liu et al., 2006; Mulle et al., 2005), multiple others supported the contribution of these genes to schizophrenia susceptibility (Abecasis et al., 2004; Addington et al., 2004; Chen et al., 2004; Detera-Wadleigh and McMahon 2006; Fallin et al., 2005; Korostishevsky et al., 2004; Li and He 2007; Ma et al., 2006; Schulze et al., 2005; Shi et al., 2008; Wang et al., 2004; Wood et al., 2006; Yue et al., 2006; Zou et al., 2005).
Over the past ten years, we have collected DNA from research subjects in the Iowa Longitudinal Study of Recent Onset Psychoses who have undergone serial brain imaging studies. Using these data, we are examining the effect of glutamatergic genetic variation on quantitative traits in individuals with schizophrenia. For this study, we examined whether genetic variations in G72 affected brain volume change over time in schizophrenia. This represents a unique opportunity to investigate the longitudinal effect of genotype on brain volume in a disorder that shows progressive changes in brain structure.
Materials and Methods
Subjects
Subjects included 110 individuals with schizophrenia or schizoaffective disorder ascertained through the ongoing Iowa Longitudinal Study of Recent Onset Psychoses. The subjects were predominantly Caucasian (107/110) and male (78/110) reflecting the ethnic makeup of Iowa and the gender bias of schizophrenia. The age at the initial MRI scan was 26.7 ± 7.14 years. The longitudinal study design and methods have been previously described (Gupta et al., 1997). In brief, the subjects with schizophrenia or schizophrenia-spectrum disorders all entered the study within 5 years of their first hospitalization. At intake, the subjects were evaluated extensively with standardized clinical rating scales (Comprehensive Assessment of Symptoms and History (CASH)(Andreasen 1985)) and diagnoses were established by the best estimate method at a multi-disciplinary staffing. A neuropsychological battery and magnetic resonance imaging (MRI) of the brain were also obtained. These assessments have since been repeated at 2, 5, and 9 years, and every 3 years thereafter.
Of the 110 study subjects, 99 had DSM-IV schizophrenia while 11 had schizoaffective disorder. All analyses were performed with and without the schizoaffective individuals to determine whether they were associated with different results than the individuals with schizophrenia; in all instances they were not. Thus, given that it is unclear whether schizophrenia and schizoaffective disorder are genetically distinct entities, individuals with schizoaffective disorder were retained in the interest of sample size and power. All subjects had at least two brain MRI scans a minimum of 2 years apart.
Medications
Antipsychotic administration was recorded both prior to the first scan and in between scans. The data was converted to average chlorpromazine daily dose by calculating the total dose of antipsychotic during the time period, converting it to Chlorpromazine equivalents, and dividing by the total number of days. In order to decrease the effect of outliers, these data were then stratified into no antipsychotic, low dose antipsychotic (1–400mg chlorpromazine daily), medium dose antipsychotic (401–800mg chlorpromazine daily), and high dose antipsychotic (> 801 mg chlorpromazine daily). These data showed that study subjects had minimal prior treatment: 33 (30%) were neuroleptic-naïve, while 39 (35%) had received low doses of antipsychotics (<400mg chlorpromazine equivalents daily) across the course of their illnesses.
Genetic Analyses
DNA was prepared by high-salt extraction from whole blood. Four SNPs in the region of G72 (rs3696165, rs3696167, rs2391191, rs778294) were chosen for genotyping based on prior studies demonstrating positive association with schizophrenia (Abecasis et al., 2004; Addington et al., 2004; Chen et al., 2004; Detera-Wadleigh and McMahon 2006; Fallin et al., 2005; Korostishevsky et al., 2004; Li and He 2007; Ma et al., 2006; Schulze et al., 2005; Wang et al., 2004; Wood et al., 2006; Yue et al., 2006; Zou et al., 2005). Genotyping was performed with the fluorogenic 5’ nuclease method (TaqMan™, Applied Biosystems (ABI), Foster City, CA) using reagents obtained from ABI, including VIC and FAM labeled probes and TaqMan™ Universal PCR Master Mix. PCR and allele calling were performed on a StrataGene Mx3000P qPCR thermocycler. Replicate samples were included on all genotyping plates to ensure accurate allele calling.
MRI acquisition and image processing
Magnetic resonance images of the whole brain were obtained on a single 1.5-Tesla GE (General Electric Medical Systems, Milwaukee, Wisconsin) Signa MR scanner. Three MR sequences were acquired for each subject: T1-weighted spoiled grass, proton density (PD) and T2-weighted images. The images were processed using the locally developed BRAINS (Brain Research: Analysis of Images, Networks, and Systems) software package; detailed descriptions of image analysis methods and the imaging parameters have been previously described (Andreasen et al., 1996; Magnotta et al., 2002). In short, the T1-weighted images were spatially normalized and re-sampled so that the anterior-posterior axis of the brain was realigned parallel to the anterior-posterior commissure line, and the inter-hemispheric fissure was aligned on the other two axes. Using an automated image registration program, the T2 and PD weighted images were aligned to the spatially normalized T1 weighted image. These images were then linearly transformed into standardized stereotaxic Talairach atlas space, generating automated measurements of frontal, temporal, parietal, and occipital lobes, cerebellum, and subcortical regions (Andreasen et al., 1996). Tissue volumes were further classified into gray matter (GM), white matter (WM) and cerebrospinal fluid (CSF), by employing a discriminant analysis method of tissue segmentation (Harris et al., 1999). In this study, total volume, white matter volume, and gray matter volume were examined for each of the following regions: cerebrum, frontal lobe, temporal lobe, parietal lobe, and occipital lobe.
Symptoms
Because symptom severity not likely to be “progressive” in the same manner as brain deterioration, we examined genotype effects only on endpoint symptom severity (symptom severity at second MRI). We also tested only those haplotypes that produced significant effects on brain structure volume change.
Positive and negative symptoms were assessed using the Scale for Assessment of Positive Symptoms (SAPS) (Andreasen 1984) and Scale for Assessment of Negative Symptoms (SANS) (Andreasen 1983). All available sources of information were used to assess symptom severity, including patient reports, informant interviews and medical records. Items corresponding to the SANS/SAPS global items were rated using a score ranging from 0 (none) to 5 (severe), and then grouped into psychotic, disorganized, and negative symptom dimensions that have repeatedly been shown to cluster independently (Andreasen et al., 1995). The psychotic dimension summed SAPS ratings for hallucinations and delusions, the disorganized dimension included SAPS ratings for positive formal thought disorder, bizarre/disorganized behavior, and inappropriate affect, and the negative dimension included SANS ratings for attention, affective flattening, alogia, avolition/apathy, and anhedonia/asociality.
Statistical Analyses
Haploview was used to assess the haplotypes in the sample (Barrett et al., 2005). We first identified haplotypes for subjects who were homozygotes at all four SNPs, and found three haplotypes. Subjects who were heterozygous at one or more of the SNPs were evaluated and no further haplotypes were found. All subjects with genetic data were able to be assigned to the homozygous or heterozygous combinations of the three haplotypes.
Demographics and illness characteristics were stratified by genotype. ANOVA was used to test the difference between the group means for continuous variables, including age at onset, age at first MRI scan, and years between scans. Chi-square tests were used to test the differences between the distributions of the categorical variables, which included gender, neuroleptic use prior to first scan, neuroleptic use between scans, clozapine use prior to first scan, and clozapine use between scans.
We then performed three separate MANCOVAs in which we analyzed the effect of each haplotype jointly on brain structure volumes. Number of haplotype alleles (0, 1, 2), treated categorically, was the predictor variable, and the multivariate dependent variable was the change in brain volume regions between the two scans (including all regions of interest). Covariates included age at onset, age at first MRI scan, years between scans, gender, neuroleptic dose range prior to first scan, and neuroleptic dose range between scans. To follow-up on haplotypes with a statistically significant difference across all brain volumes, univariate ANCOVAs were used to find the regions showing the differences. To ensure that significant results were not due to non-normal distributions or outliers, we also performed non-parametric analogs of the ANCOVAs, which, in all cases, confirmed the parametric tests. Because MANCOVA automatically incorporates p-value correction for multiple testing, adjustment of the type-I error level is unnecessary. A p-value less than 0.05 was regarded as statistically significant.
To determine whether genotypes that influenced brain structure also influenced end-point symptomatology, we performed linear regression with dependent variables of total scores for the three symptom domains, adjusted for age, sex and medication use.
Results
We first tested the 4 SNPs for linkage disequilibrium (LD). The SNPs covered 38 kb, and D’ was 1 for all relationships. From these 4 SNPs, we identified three haplotypes which were all relatively common (Table 1).
Table 1.
G72 haplotype prevalences
| Prevalence |
||||||
|---|---|---|---|---|---|---|
| SNP |
Schizophrenia (n=110) |
Controls (n=120) |
||||
| Haplotype | rs3916965 | rs3916967 | rs2391191 | rs778294 | ||
| 1 | A | G | A | G | 0.40 | 0.35 |
| 2 | G | A | G | G | 0.33 | 0.35 |
| 3 | G | A | G | A | 0.27 | 0.30 |
The data were stratified by haplotype and evaluated. A comparison of the demographics and illness characteristics between the haplotype groups and all other subjects showed no significant demographic or medication use differences between the groups (Table 2).
Table 2.
Demographic and Illness Characteristics of Individuals with Schizophrenia
| Haplotype 1 alleles | Haplotype 2 alleles | Haplotype 3 alleles | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Total Sample | 0 | 1 | 2 | 0 | 1 | 2 | 0 | 1 | 2 | ||
| N | 110 | 37 | 57 | 16 | 45 | 58 | 7 | 60 | 41 | 9 | |
| Males | 71% | 78% | 65% | 75% | 78% | 66% | 71% | 65% | 76% | 89% | |
| Age at onset (years) | 22.0 ± 5.5 | 22.6 ± 5.8 | 21.6 ± 5.5 | 22.4 ± 5.0 | 20.7 ± 4.5 | 22.7 ± 6.1 | 25.1 ± 4.1 | 22.7 ± 5.6 | 21.6 ± 5.4 | 19.4 ± 4.6 | |
| Age at first MRI Scan (years) | 26.8 ± 6.9 | 26.9 ± 7.3 | 27.1 ± 5.6 | 25.4 ± 5.7 | 25.8 ± 6.2 | 27.6 ± 7.7 | 26.0 ± 3.5 | 26.4 ± 6.6 | 28.0 ± 7.3 | 23.3 ± 6.2 | |
| Years between scans ± sd | 3.0 ± 1.6 | 2.9 ± 1.8 | 3.1 ± 1.4 | 2.9 ± 1.8 | 2.9 ± 1.4 | 3.0 ± 1.6 | 3.3 ± 2.6 | 3.1 ± 1.8 | 2.9 ± 1.4 | 2.8 ± 1.4 | |
| Years between scans (min, max) | (1.1, 9.0) | (1.1, 9.0) | (1.8, 8.1) | (1.9, 8.2) | (1.9, 8.2) | (1.1, 9.0) | (2.0, 9.0) | (1.8, 9.0) | (1.1, 9.0) | (1.9, 6.4) | |
| Neuroleptic Naïve prior to study | 30% | 30% | 30% | 31% | 29% | 29% | 42% | 30% | 34% | 11% | |
| Clozapine use prior to study | 8% | 11% | 7% | 6% | 11% | 7% | 0% | 7% | 7% | 22% | |
| Interim Clozapine use | 18% | 19% | 17% | 19% | 27% | 14% | 0% | 13% | 22% | 33% | |
| Interim medication treatmenta | |||||||||||
| 0 mg | 6% | 3% | 9% | 6% | 11% | 3% | 0% | 5% | 7% | 11% | |
| 1 mg – 400 mg | 17% | 22% | 19% | 0% | 7% | 26% | 14% | 15% | 24% | 0% | |
| 401 mg – 800 mg | 24% | 24% | 23% | 25% | 24% | 23% | 29% | 23% | 24% | 22% | |
| > 800 mg | 53% | 51% | 49% | 69% | 58% | 48% | 57% | 57% | 44% | 67% | |
average chlorpromazine-equivalent daily dose
Relationships between changes in brain structure volumes over time and haplotypes were then investigated. Haplotypes 1 and 3 showed significant differences in brain structure volumes, with MANCOVA p-values of 0.01 and 0.04, respectively. Haplotype 1 produced a significant effect on frontal lobe tissue volume loss (Table 3: p = 0.008 for total frontal volume). This effect appeared, based on the allelic group means, to arise primarily from greater volume loss in the haplotype 1 homozygotes. An analysis that compared change in frontal tissue volume in haplotype 1 homozygotes versus all others was confirmatory (F = 9.75, p = 0.002). The haplotype 1 effect on frontal volume change did not appear to be tissue specific, with both gray and white matter showing patterns of association similar to that for overall frontal volume. The effects for gray and white matter, like that for overall volume, were also stronger when homozygotes were compared against all others (frontal gray: F = 4.83, p = 0.03; frontal white: F = 4.03, p = 0.05).
Table 3.
Changes in brain structure volumes during inter-scan interval (cc ± sd)
| Haplotype 1 alleles MANCOVA p=0.01 |
Haplotype 2 alleles MANCOVA p=0.59 |
Haplotype 3 alleles MANCOVA p=0.04 |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Region | Baseline Volume |
0 (n=37) |
1 (n=57) |
2 (n=16) |
p-value | 0 (n=45) |
1 (n=58) |
2 (n=7) |
p-value | 0 (n=60) |
1 (n=41) |
2 (n=9) |
p-value |
| Cerebral cortex | 1143 ± 130 | −11 ± 6.5 | 1.7 ± 5.2 | −27 ± 9.9 | 0.03 | −9.8 ± 6.0 | −7.8 ± 5.3 | 19 ± 15 | 0.21 | −2.9 ± 5.3 | −14 ± 6.4 | 0.8 ± 14 | 0.33 |
| Gray | 684 ± 79.5 | −12 ± 6.8 | 3.7 ± 5.5 | −16 ± 10 | 0.11 | −11 ± 6.2 | −1.9 ± 5.5 | 15 ± 16 | 0.26 | 0.1 ± 5.4 | −5.3 ± 6.6 | −30 ± 14 | 0.13 |
| White | 458 ± 64.7 | 0.3 ± 4.8 | −2.0 ± 3.9 | −11 ± 7.4 | 0.46 | 0.8 ± 4.3 | −5.8 ± 3.8 | 4.2 ± 11 | 0.44 | −3.0 ± 3.6 | −9.1 ± 4.4 | 31 ± 9.2 | 0.0008 |
| Frontal lobe | 438 ± 56.4 | −2.5 ± 3.4 | 0.8 ± 2.8 | −18 ± 5.3 | 0.008 | −7.3 ± 3.2 | −1.4 ± 2.8 | 10 ± 8.2 | 0.10 | −2.3 ± 2.86 | −5.0 ± 3.5 | 0.3 ± 7.4 | 0.75 |
| Gray | 264 ± 35.9 | −0.6 ± 3.1 | −0.1 ± 2.5 | −12 ± 4.8 | 0.10 | −6.3 ± 2.8 | 0.3 ± 2.5 | 7.0 ± 7.2 | 0.10 | −2.2 ± 2.5 | −0.1 ± 3.1 | −8.8 ± 6.5 | 0.48 |
| White | 175 ± 26.4 | −1.9 ± 2.0 | 0.9 ± 1.6 | −6.8 ± 3.0 | 0.07 | −1.1 ± 1.8 | −1.7 ± 1.6 | 3.1 ± 4.6 | 0.61 | −0.1 ± 1.5 | −4.9 ± 1.8 | 9.1 ± 3.9 | 0.005 |
| Temporal lobe | 226 ± 25.5 | −1.3 ± 2.5 | 1.5 ± 2.0 | −4.1 ± 3.8 | 0.38 | −1.2 ± 2.3 | 0.1 ± 2.0 | 2.4 ± 5.8 | 0.81 | 0.6 ± 20 | −1.7 ± 2.4 | 0.8 ± 5.1 | 0.75 |
| Gray | 154 ± 18.1 | −2.8 ± 2.2 | 1.2 ± 1.7 | −4.5 ± 3.3 | 0.20 | −2.5 ± 2.0 | −0.3 ± 1.7 | 3.0 ± 5.0 | 0.52 | 0.3 ± 1.7 | −2.0 ± 2.1 | −4.7 ± 4.5 | 0.49 |
| White | 372 ± 10.1 | 1.5 ± 1.0 | 0.3 ± 0.8 | 0.3 ± 1.5 | 0.64 | 1.2 ± .9 | 0.5 ± 0.8 | −0.7 ± 2.3 | 0.70 | 0.3 ± .8 | 0.3 ± 0.9 | 5.5 ± 2.0 | 0.05 |
| Parietal lobe | 251 ± 29.6 | −1.3 ± 2.2 | 3.4 ± 1.8 | −2.3 ± 3.3 | 0.14 | 2.6 ± 2.0 | −1.0 ± 1.8 | 7.2 ± 5.1 | 0.20 | 2.1 ± 1.7 | −2.0 ± 2.1 | 7.1 ± 4.5 | 0.13 |
| Gray | 137 ± 17.1 | −2.9 ± 1.6 | 2.3 ± 1.3 | −0.4 ± 2.5 | 0.05 | 0.6 ± 1.5 | −0.7 ± 1.3 | 4.0 ± 3.9 | 0.49 | 1.5 ± 1.3 | −1.0 ± 1.6 | −3.8 ± 3.4 | 0.25 |
| White | 114 ± 16.2 | 1.6 ± 1.4 | 1.1 ± 1.2 | −1.9 ± 2.2 | 0.40 | 1.9 ± 1.3 | −0.3 ± 1.1 | 3.2 ± 3.3 | 0.35 | 0.6 ± 1.1 | −1.0 ± 1.3 | 11 ± 2.8 | 0.0009 |
| Occipital lobe | 124 ± 18.1 | −1.0 ± 1.2 | −1.9 ± 1.0 | −0.8 ± 1.8 | 0.82 | −1.4 ± 1.1 | −1.7 ± 1.0 | 0.5 ± 2.8 | 0.76 | −1.2 ± 1.0 | −1.9 ± 1.2 | −0.6 ± 2.5 | 0.84 |
| Gray | 69 ± 11.2 | −1.4 ± 1.4 | 1.6 ± 1.1 | 2.4 ± 2.1 | 0.15 | 1.0 ± 1.3 | 0.3 ± 1.1 | 2.7 ± 3.2 | 0.75 | 2.0 ± 1.1 | −0.2 ± 1.3 | −3.5 ± 2.8 | 0.13 |
| White | 55 ± 11.4 | 0.4 ± 1.1 | −3.5 ± 0.9 | −3.3 ± 1.7 | 0.03 | −2.3 ± 1.1 | −2.0 ± 0.9 | −2.2 ± 2.7 | 0.97 | −3.2 ± 0.9 | −1.7 ± 1.1 | 2.9 ± 2.3 | 0.05 |
Table 3 shows average change in brain structure volumes between time 1 and time 2 MRI brain scans for various haplotype based groups of subjects. The effects of the three haplotypes on the brain structure volumes (taken together) was first tested with MANCOVAs. The independent predictor was number of haplotype alleles, treated as a categorical variable, and covariates included total intra-cranial volume, age, gender, inter-scan interval, anti-psychotic treatment prior to first scan, and anti-psychotic treatment during the inter-scan interval. The group-level p-value is given above the columns and the univariate p-values are given for each region and haplotype.
While haplotype 2 did not affect change in brain tissue volume, haplotype 3 produced a significant effect on cerebral cortical white matter volume change (p=0.0008). The pattern of effects was similar across all cerebral lobes, with individuals homozygous for haplotype 3 having experienced a significant increase in white matter volume. As with haplotype 1, this effect confirmed by a test comparing haplotype 3 homozygotes against all other subjects (cortical white matter: F = 14.20, p = 0.0003).
The robustness of these results was evaluated with the non-parametric Wilcoxen two-sample test. The results were unchanged, with p = 0.008 for the effect of haplotype 1 on total frontal volume change and p = 0.0027 for the effect of haplotype 3 on cerebral white matter volume change.
Given these results, we then tested whether homozygosity for haplotypes 1 and 3 influenced endpoint symptom severity (Table 4). Haplotype 1 homozygotes experienced more severe psychotic symptoms than other subjects (p=0.006), while haplotype 3 did not influence endpoint symptom severity. Given the influence of haplotype 1 on two quantitative traits, we tested whether frontal volume change was correlated with endpoint psychotic symptoms while controlling for age and gender. Although change in tissue volume was inversely correlated with increased severity of symptoms, the relationship was not statistically significant (partial r = −0.14, p = 0.17).
Table 4.
Symptom domain score at follow-up scan
| Haplotype 1 alleles | Haplotype 3 alleles | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Symptom Domain | All Subjects (n=110) |
0 (n=37) |
1 (n=57) |
2 (n=16) |
p-value | 0 (n=60) |
1 (n=41) |
2 (n=9) |
p-value |
| Psychotic | 3.0 | 2.8 | 2.5 | 4.5 | 0.006 | 2.9 | 2.8 | 3.0 | 0.90 |
| Disorganized | 1.9 | 1.8 | 1.8 | 2.0 | 0.71 | 2.0 | 1.8 | 1.0 | 0.21 |
| Negative | 10.2 | 9.7 | 9.6 | 11.1 | 0.22 | 10.6 | 9.2 | 8.0 | 0.17 |
Scores are adjusted for age, gender, inter-scan interval, and anti-psychotic usage. A higher score indicates increased symptom severity.
Discussion
Schizophrenia has long been known to be characterized by diffuse brain abnormalities. These manifest, in general, as increased ventricular size and diminished cerebral cortical tissue volume in cross-sectional comparisons with healthy controls. A question that cannot be answered by cross-sectional studies, however, is whether these abnormalities are progressive or static: are they fully expressed at diagnosis, remaining stable thereafter, or do they progress over the life span? And if the latter, is the progression due to medications, innate predisposition, or other factors? The first question has now been answered by longitudinal studies showing that individuals with schizophrenia experience, on average, a gradual loss of cortical tissue across the course of illness (Boos et al., 2007; Farrow et al., 2005; Whitford et al., 2006; Zipparo et al., 2008). We found, for example, that a cohort of individuals with schizophrenia who we have studied with serial MRI scans for up to 20 years have experienced on-going decrement in cortical gray matter volume, particularly in the frontal lobe (Ho et al., 2003). This has occurred at a more rapid rate than in a comparison group of healthy control individuals and during an age period—the twenties and thirties—when brain tissue volume typically remains stable.
The second question, that of causality, has not been answered as clearly. Evidence exists for effects of antipsychotic medication on some brain structure volumes, but the cortex appears to be either unaffected by such medications or protected by them from the on-going deteriorative disease process (Styner et al., 2005; Takahashi et al., 2007). In accord with this, some studies have shown that the degree of cortical volume loss is directly associated with duration of untreated psychosis (Perkins et al., 2005). In a study of twin pairs discordant for schizophrenia, progressive volume changes were found to be at least partially attributable to genetic factors (Brans et al., 2008). In a direct test of this hypothesis, we previously showed that variation in the brain derived neurotrophic factor gene (BDNF) influences longitudinal changes in frontal lobe tissue volume in individuals with schizophrenia (Ho et al., 2007).
With the hypothesis, therefore, that genetic factors modulate longitudinal brain changes, we investigated the role of a high interest gene, G72, in producing these changes. Variation in G72 has been associated with schizophrenia in multiple samples (Abecasis et al., 2004; Addington et al., 2004; Chen et al., 2004; Detera-Wadleigh and McMahon 2006; Fallin et al., 2005; Korostishevsky et al., 2004; Li and He 2007; Ma et al., 2006; Schulze et al., 2005; Shi et al., 2008; Wang et al., 2004; Wood et al., 2006; Yue et al., 2006; Zou et al., 2005). G72 has been shown to influence, in individuals with schizophrenia, symptom severity (Schulze et al., 2005; Yue et al., 2007) and cognitive processes that depend on frontal lobe function (Donohoe et al., 2007; Goldberg et al., 2006) . G72 has also been shown to influence hippocampal and prefrontal function in relatives of individuals with schizophrenia (Hall et al., 2008). The mechanisms through which G72 exerts these effects are unclear. The prevailing hypothesis, suggested by early studies, has been that G72 influences glutamate neurotransmission. G72 was thought to activate D-amino acid oxidase (DAO), which in turn oxidizes D-serine, an important co-agonist for the NMDA glutamate receptor. Individuals with schizophrenia have lower serum levels of D-serine than healthy controls, and use of D-serine as an adjunctive medication has been shown to improve the symptoms of schizophrenia (Boks et al., 2007). Recent data, however, suggests a potential alternative function for G72. Kvajo et al, reported that a specific isoform of G72 that stimulated dendritic arborization did not interact with DAO but rather influenced mitochondrial function (Kvajo et al., 2008b). They identified G72 as a primate specific, rapidly evolving gene, and hypothesized that it plays a role in supplying the increased energy needs of the primate brain. By either theory, G72 could plausibly influence brain morphology in individuals with schizophrenia.
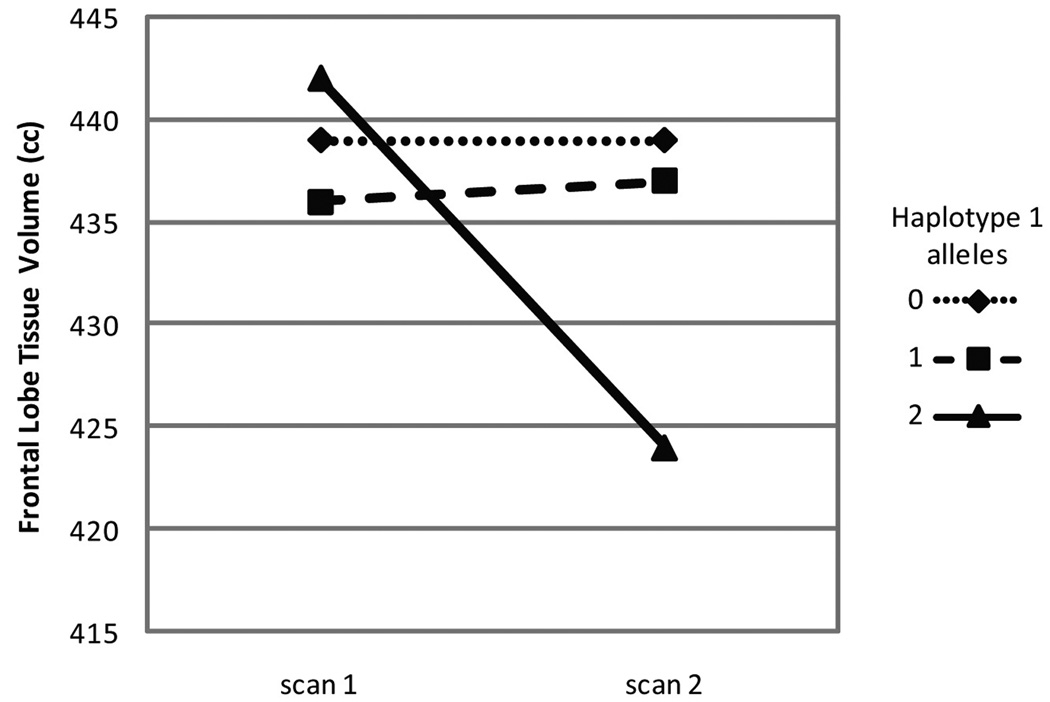
We therefore genotyped, in a sample of individuals with schizophrenia who have received serial brain imaging and symptom assessments, four G72 SNPs that have been associated individually and as haplotypes with both schizophrenia and bipolar disorder. Our analyses of these data showed that individuals with schizophrenia who were homozygous for a specific haplotype (haplotype 1) experienced an accelerated rate of frontal lobe tissue loss compared to subjects with other G72 haplotypes. When confounds such as age, interscan interval, and medication treatment were removed, the haplotype 1 homozygous state was associated with a loss of 4.1% of frontal lobe tissue volume, while non-homozygous individuals did not experience any change (Figure 1). Progressive decreases in frontal lobe volume have also been shown to be associated with greater symptom severity and impaired cognitive functioning (Ho et al., 2003). We therefore tested whether, at the time of the second scan, haplotype 1 was associated with symptomatology and found that psychotic symptoms were significantly more severe in the haplotype 1 homozygotes than in the rest of the sample (Table 4).
Average frontal lobe volumes at time 1 and time 2 MRI scans for haplotype 1 genotype groups.
Scans were obtained a minimum of 2 and an average of 3.0 ± 1.5 years apart. Linear regression was performed with haplotype as the independent variable and change in brain volume between the two scans as the dependent variable. The regression was adjusted for age at onset, age at first MRI scan, years between scans, gender, neuroleptic dose range prior to first scan, and neuroleptic dose range between scans. The decrease in frontal volume over time for haplotype 1 homozygotes is significantly different than for the other groups. In fact, the p-value for the estimated slope of the remaining subjects is 0.49, suggesting that there is no change in frontal lobe volumes for the subjects who are not haplotype 1 homozygotes.
We also found that individuals homozygous for haplotype 3 experienced a larger increase in white matter volume over time in comparison to the rest of the sample. This could indicate increased myelination and may represent a protective effect, consistent with one prior study that reported a protective variant within the G72 locus (Abecasis et al., 2004). The haplotype 3 homozygotes also experienced less severe positive and negative symptoms at the time of their second MRI, although the number of haplotype 3 homozygotes was small and the symptom effects were not statistically significant.
Relating these data to previous G72 studies is challenging because the markers examined and the findings are not consistent. Haplotype 1 has nonetheless been implicated in a number of reports. Of eight studies that evaluated G72 and schizophrenia and that reported the over-represented alleles (Addington et al., 2004; Chumakov et al., 2002; Hattori et al., 2003; Ma et al., 2006; Mulle et al., 2005; Schulze et al., 2005; Wang et al., 2004; Yue et al., 2006; Zou et al., 2005), six showed consistency with our haplotype 1 finding (Hattori et al., 2003; Mulle et al., 2005; Schulze et al., 2005; Wang et al., 2004; Yue et al., 2006; Zou et al., 2005). Yue et al found that a haplotype overlapping and consistent with our haplotype 1 increased the risk for both early-onset and male schizophrenia (Yue et al., 2006). Meta-analyses by Li & He and Detera-Wadleigh & McMahon found significant over-representation of the A allele in rs3916965 and the A allele in rs2391191, both of which are in haplotype 1 (Detera-Wadleigh and McMahon 2006; Li and He 2007). Thus, prior results found with the four SNPs used in our study implicate haplotype 1, thereby supporting our extension of these findings of a detrimental effect of haplotype 1 on brain structure volumes in schizophrenia
These data must be interpreted in the context of several limitations. First, because we only examined individuals with schizophrenia, we are unable to determine whether the identified associations are specific to schizophrenia or are more generalized. Also, the significant effects were found in the homozygote group samples, with small sample sizes. This diminishes our power to detect more subtle associations, and the actual genetic mechanisms at play are likely to be more complex than what we are modeling. In spite of these limitations, our study suggests that genetic variation in the gene G72 modulates symptom severity and progressive changes in brain structures that occur in individuals with schizophrenia.
Acknowledgements
This work was supported in part by grants MH68380, MH31593, MH40856, and MH43271 from the National Institute of Mental Health, and by a Nellie Ball Research Trust Fund Award and a NARSAD Young Investigator Award to THW.
References
- Abecasis GR, Burt RA, Hall D, Bochum S, Doheny KF, Lundy SL, Torrington M, Roos JL, Gogos JA, Karayiorgou M. Genomewide scan in families with schizophrenia from the founder population of Afrikaners reveals evidence for linkage and uniparental disomy on chromosome 1. Am J Hum Genet. 2004;74(3):403–417. doi: 10.1086/381713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addington AM, Gornick M, Sporn AL, Gogtay N, Greenstein D, Lenane M, Gochman P, Baker N, Balkissoon R, Vakkalanka RK, et al. Polymorphisms in the 13q33.2 gene G72/G30 are associated with childhood-onset schizophrenia and psychosis not otherwise specified. Biol Psychiatry. 2004;55(10):976–980. doi: 10.1016/j.biopsych.2004.01.024. [DOI] [PubMed] [Google Scholar]
- Andreasen NC. The Scale for the Assessment of Negative Symptoms (SANS) Iowa City: The University of Iowa; 1983. [PubMed] [Google Scholar]
- Andreasen NC. The Scale for the Assessment of Positive Symptoms (SAPS) Iowa City: The University of Iowa; 1984. [Google Scholar]
- Andreasen NC. Comprehensive Assessment of Symptoms and History (CASH) Iowa City: The University of Iowa; 1985. [DOI] [PubMed] [Google Scholar]
- Andreasen NC, Arndt S, Miller D, Flaum M, Nopoulos P. Correlational studies of the Scale for the Assessment of Negative Symptoms and the Scale for the Assessment of Positive Symptoms: an overview and update. [Review] [45 refs] Psychopathology. 1995;28(1):7–17. doi: 10.1159/000284894. [DOI] [PubMed] [Google Scholar]
- Andreasen NC, Rajarethinam R, Cizadlo T, Arndt S, Swayze VWn, Flashman LA, O'Leary DS, Ehrhardt JC, Yuh WT. Automatic atlas-based volume estimation of human brain regions from MR images. Journal of Computer Assisted Tomography. 1996;20(1):98–106. doi: 10.1097/00004728-199601000-00018. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Boks MP, Rietkerk T, van de Beek MH, Sommer IE, de Koning TJ, Kahn RS. Reviewing the role of the genes G72 and DAAO in glutamate neurotransmission in schizophrenia. Eur Neuropsychopharmacol. 2007;17(9):567–572. doi: 10.1016/j.euroneuro.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Boos HB, Aleman A, Cahn W, Pol HH, Kahn RS. Brain volumes in relatives of patients with schizophrenia: a meta-analysis. Arch Gen Psychiatry. 2007;64(3):297–304. doi: 10.1001/archpsyc.64.3.297. [DOI] [PubMed] [Google Scholar]
- Brans RG, van Haren NE, van Baal GC, Schnack HG, Kahn RS, Hulshoff Pol HE. Heritability of changes in brain volume over time in twin pairs discordant for schizophrenia. Arch Gen Psychiatry. 2008;65(11):1259–1268. doi: 10.1001/archpsyc.65.11.1259. [DOI] [PubMed] [Google Scholar]
- Chen YS, Akula N, Detera-Wadleigh SD, Schulze TG, Thomas J, Potash JB, DePaulo JR, McInnis MG, Cox NJ, McMahon FJ. Findings in an independent sample support an association between bipolar affective disorder and the G72/G30 locus on chromosome 13q33. Mol Psychiatry. 2004;9(1):87–92. doi: 10.1038/sj.mp.4001453. image 5. [DOI] [PubMed] [Google Scholar]
- Christian SL, McDonough J, Liu Cy CY, Shaikh S, Vlamakis V, Badner JA, Chakravarti A, Gershon ES. An evaluation of the assembly of an approximately 15-Mb region on human chromosome 13q32–q33 linked to bipolar disorder and schizophrenia. Genomics. 2002;79(5):635–656. doi: 10.1006/geno.2002.6765. [DOI] [PubMed] [Google Scholar]
- Chumakov I, Blumenfeld M, Guerassimenko O, Cavarec L, Palicio M, Abderrahim H, Bougueleret L, Barry C, Tanaka H, La Rosa P, et al. Genetic and physiological data implicating the new human gene G72 and the gene for D-amino acid oxidase in schizophrenia. Proc Natl Acad Sci U S A. 2002;99(21):13675–13680. doi: 10.1073/pnas.182412499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detera-Wadleigh SD, McMahon FJ. G72/G30 in schizophrenia and bipolar disorder: review and meta-analysis. Biol Psychiatry. 2006;60(2):106–114. doi: 10.1016/j.biopsych.2006.01.019. [DOI] [PubMed] [Google Scholar]
- Donohoe G, Morris DW, Robertson IH, McGhee KA, Murphy K, Kenny N, Clarke S, Gill M, Corvin AP. DAOA ARG30LYS and verbal memory function in schizophrenia. Mol Psychiatry. 2007;12(9):795–796. doi: 10.1038/sj.mp.4002026. [DOI] [PubMed] [Google Scholar]
- Fallin MD, Lasseter VK, Avramopoulos D, Nicodemus KK, Wolyniec PS, McGrath JA, Steel G, Nestadt G, Liang KY, Huganir RL, et al. Bipolar I disorder and schizophrenia: a 440-single-nucleotide polymorphism screen of 64 candidate genes among Ashkenazi Jewish case-parent trios. Am J Hum Genet. 2005;77(6):918–936. doi: 10.1086/497703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrow TF, Whitford TJ, Williams LM, Gomes L, Harris AW. Diagnosis-related regional gray matter loss over two years in first episode schizophrenia and bipolar disorder. Biol Psychiatry. 2005;58(9):713–723. doi: 10.1016/j.biopsych.2005.04.033. [DOI] [PubMed] [Google Scholar]
- Ghaziuddin M, Leininger L, Tsai L. Brief report: thought disorder in Asperger syndrome: comparison with high-functioning autism. Journal of Autism & Developmental Disorders. 1995;25(3):311–317. doi: 10.1007/BF02179292. [DOI] [PubMed] [Google Scholar]
- Goff DC, Coyle JT. The emerging role of glutamate in the pathophysiology and treatment of schizophrenia. Am J Psychiatry. 2001;158(9):1367–1377. doi: 10.1176/appi.ajp.158.9.1367. [DOI] [PubMed] [Google Scholar]
- Goldberg TE, Straub RE, Callicott JH, Hariri A, Mattay VS, Bigelow L, Coppola R, Egan MF, Weinberger DR. The G72/G30 Gene Complex and Cognitive Abnormalities in Schizophrenia. Neuropsychopharmacology. 2006 doi: 10.1038/sj.npp.1301049. [DOI] [PubMed] [Google Scholar]
- Gupta S, Andreasen NC, Arndt S, Flaum M, Hubbard WC, Ziebell S. The Iowa Longitudinal Study of Recent Onset Psychosis: one-year follow-up of first episode patients. Schizophrenia Research. 1997;23(1):1–13. doi: 10.1016/S0920-9964(96)00078-3. [DOI] [PubMed] [Google Scholar]
- Gur RE, Cowell P, Turetsky BI, Gallacher F, Cannon T, Bilker W, Gur RC. A follow-up magnetic resonance imaging study of schizophrenia. Relationship of neuroanatomical changes to clinical and neurobehavioral measures. Arch Gen Psychiatry. 1998;55(2):145–152. doi: 10.1001/archpsyc.55.2.145. [DOI] [PubMed] [Google Scholar]
- Hall J, Whalley HC, Moorhead TW, Baig BJ, McIntosh AM, Job DE, Owens DG, Lawrie SM, Johnstone EC. Genetic Variation in the DAOA (G72) Gene Modulates Hippocampal Function in Subjects at High Risk of Schizophrenia. Biol Psychiatry. 2008 doi: 10.1016/j.biopsych.2008.03.009. [DOI] [PubMed] [Google Scholar]
- Harris G, Andreasen NC, Cizadlo T, Bailey JM, Bockholt J, Magnotta V, Arndt S. Improving tissue classification in magnetic resonance imaging: A three-dimensional multispectral discriminant analysis method with automated training class selection. Journal of Computer Assisted Tomography. 1999;23(1):144–154. doi: 10.1097/00004728-199901000-00030. [DOI] [PubMed] [Google Scholar]
- Hattori E, Liu C, Badner JA, Bonner TI, Christian SL, Maheshwari M, Detera-Wadleigh SD, Gibbs RA, Gershon ES. Polymorphisms at the G72/G30 gene locus, on 13q33, are associated with bipolar disorder in two independent pedigree series. Am J Hum Genet. 2003;72(5):1131–1140. doi: 10.1086/374822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho BC, Andreasen NC, Dawson JD, Wassink TH. Association between brain-derived neurotrophic factor Val66Met gene polymorphism and progressive brain volume changes in schizophrenia. Am J Psychiatry. 2007;164(12):1890–1899. doi: 10.1176/appi.ajp.2007.05111903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho BC, Andreasen NC, Nopoulos P, Arndt S, Magnotta V, Flaum M. Progressive structural brain abnormalities and their relationship to clinical outcome: a longitudinal magnetic resonance imaging study early in schizophrenia. Arch Gen Psychiatry. 2003;60(6):585–594. doi: 10.1001/archpsyc.60.6.585. [DOI] [PubMed] [Google Scholar]
- Korostishevsky M, Kaganovich M, Cholostoy A, Ashkenazi M, Ratner Y, Dahary D, Bernstein J, Bening-Abu-Shach U, Ben-Asher E, Lancet D, et al. Is the G72/G30 locus associated with schizophrenia? single nucleotide polymorphisms, haplotypes, and gene expression analysis. Biol Psychiatry. 2004;56(3):169–176. doi: 10.1016/j.biopsych.2004.04.006. [DOI] [PubMed] [Google Scholar]
- Kvajo M, Dhilla A, Swor DE, Karayiorgou M, Gogos JA. Evidence implicating the candidate schizophrenia/bipolar disorder susceptibility gene G72 in mitochondrial function. Molecular psychiatry. 2008a;13(7):685–696. doi: 10.1038/sj.mp.4002052. [DOI] [PubMed] [Google Scholar]
- Kvajo M, Dhilla A, Swor DE, Karayiorgou M, Gogos JA. Evidence implicating the candidate schizophrenia/bipolar disorder susceptibility gene G72 in mitochondrial function. Mol Psychiatry. 2008b;13(7):685–696. doi: 10.1038/sj.mp.4002052. [DOI] [PubMed] [Google Scholar]
- Li D, He L. G72/G30 genes and schizophrenia: a systematic meta-analysis of association studies. Genetics. 2007;175(2):917–922. doi: 10.1534/genetics.106.061796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YL, Fann CS, Liu CM, Chang CC, Wu JY, Hung SI, Liu SK, Hsieh MH, Hwang TJ, Chan HY, et al. No association of G72 and D-amino acid oxidase genes with schizophrenia. Schizophr Res. 2006;87(1–3):15–20. doi: 10.1016/j.schres.2006.06.020. [DOI] [PubMed] [Google Scholar]
- Ma J, Qin W, Wang XY, Guo TW, Bian L, Duan SW, Li XW, Zou FG, Fang YR, Fang JX, et al. Further evidence for the association between G72/G30 genes and schizophrenia in two ethnically distinct populations. Mol Psychiatry. 2006;11(5):479–487. doi: 10.1038/sj.mp.4001788. [DOI] [PubMed] [Google Scholar]
- Magnotta VA, Harris G, Andreasen NC, O'Leary DS, Yuh WT, Heckel D. Structural MR image processing using the BRAINS2 toolbox. Comput Med Imaging Graph. 2002;26(4):251–264. doi: 10.1016/s0895-6111(02)00011-3. [DOI] [PubMed] [Google Scholar]
- Mathalon DH, Sullivan EV, Lim KO, Pfefferbaum A. Progressive brain volume changes and the clinical course of schizophrenia in men: a longitudinal magnetic resonance imaging study. Arch Gen Psychiatry. 2001;58(2):148–157. doi: 10.1001/archpsyc.58.2.148. [DOI] [PubMed] [Google Scholar]
- Mulle JG, Chowdari KV, Nimgaonkar V, Chakravarti A. No evidence for association to the G72/G30 locus in an independent sample of schizophrenia families. Mol Psychiatry. 2005;10(5):431–433. doi: 10.1038/sj.mp.4001619. [DOI] [PubMed] [Google Scholar]
- Perkins DO, Gu H, Boteva K, Lieberman JA. Relationship between duration of untreated psychosis and outcome in first-episode schizophrenia: a critical review and meta-analysis. Am J Psychiatry. 2005;162(10):1785–1804. doi: 10.1176/appi.ajp.162.10.1785. [DOI] [PubMed] [Google Scholar]
- Schulze TG, Ohlraun S, Czerski PM, Schumacher J, Kassem L, Deschner M, Gross M, Tullius M, Heidmann V, Kovalenko S, et al. Genotype-phenotype studies in bipolar disorder showing association between the DAOA/G30 locus and persecutory delusions: a first step toward a molecular genetic classification of psychiatric phenotypes. Am J Psychiatry. 2005;162(11):2101–2108. doi: 10.1176/appi.ajp.162.11.2101. [DOI] [PubMed] [Google Scholar]
- Shenton ME, Dickey CC, Frumin M, McCarley RW. A review of MRI findings in schizophrenia. Schizophr Res. 2001;49(1–2):1–52. doi: 10.1016/s0920-9964(01)00163-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Badner JA, Gershon ES, Liu C. Allelic association of G72/G30 with schizophrenia and bipolar disorder: A comprehensive meta-analysis. Schizophr Res. 2008;98(1–3):89–97. doi: 10.1016/j.schres.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styner M, Lieberman JA, McClure RK, Weinberger DR, Jones DW, Gerig G. Morphometric analysis of lateral ventricles in schizophrenia and healthy controls regarding genetic and disease-specific factors. Proc Natl Acad Sci U S A. 2005;102(13):4872–4877. doi: 10.1073/pnas.0501117102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch Gen Psychiatry. 2003;60(12):1187–1192. doi: 10.1001/archpsyc.60.12.1187. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Suzuki M, Tanino R, Zhou SY, Hagino H, Niu L, Kawasaki Y, Seto H, Kurachi M. Volume reduction of the left planum temporale gray matter associated with long duration of untreated psychosis in schizophrenia: a preliminary report. Psychiatry Res. 2007;154(3):209–219. doi: 10.1016/j.pscychresns.2006.10.001. [DOI] [PubMed] [Google Scholar]
- Wang X, He G, Gu N, Yang J, Tang J, Chen Q, Liu X, Shen Y, Qian X, Lin W, et al. Association of G72/G30 with schizophrenia in the Chinese population. Biochem Biophys Res Commun. 2004;319(4):1281–1286. doi: 10.1016/j.bbrc.2004.05.119. [DOI] [PubMed] [Google Scholar]
- Whitford TJ, Grieve SM, Farrow TF, Gomes L, Brennan J, Harris AW, Gordon E, Williams LM. Progressive grey matter atrophy over the first 2–3 years of illness in first-episode schizophrenia: a tensor-based morphometry study. Neuroimage. 2006;32(2):511–519. doi: 10.1016/j.neuroimage.2006.03.041. [DOI] [PubMed] [Google Scholar]
- Wood LS, Pickering EH, Dechairo BM. Significant Support for DAO as a Schizophrenia Susceptibility Locus: Examination of Five Genes Putatively Associated with Schizophrenia. Biol Psychiatry. 2006 doi: 10.1016/j.biopsych.2006.07.005. [DOI] [PubMed] [Google Scholar]
- Yue W, Kang G, Zhang Y, Qu M, Tang F, Han Y, Ruan Y, Lu T, Zhang J, Zhang D. Association of DAOA polymorphisms with schizophrenia and clinical symptoms or therapeutic effects. Neurosci Lett. 2007;416(1):96–100. doi: 10.1016/j.neulet.2007.01.056. [DOI] [PubMed] [Google Scholar]
- Yue W, Liu Z, Kang G, Yan J, Tang F, Ruan Y, Zhang J, Zhang D. Association of G72/G30 polymorphisms with early-onset and male schizophrenia. Neuroreport. 2006;17(18):1899–1902. doi: 10.1097/WNR.0b013e3280102ed4. [DOI] [PubMed] [Google Scholar]
- Zipparo L, Whitford TJ, Redoblado Hodge MA, Lucas S, Farrow TF, Brennan J, Gomes L, Williams LM, Harris AW. Investigating the neuropsychological and neuroanatomical changes that occur over the first 2–3 years of illness in patients with first-episode schizophrenia. Prog Neuropsychopharmacol Biol Psychiatry. 2008;32(2):531–538. doi: 10.1016/j.pnpbp.2007.10.011. [DOI] [PubMed] [Google Scholar]
- Zou F, Li C, Duan S, Zheng Y, Gu N, Feng G, Xing Y, Shi J, He L. A family-based study of the association between the G72/G30 genes and schizophrenia in the Chinese population. Schizophr Res. 2005;73(2–3):257–261. doi: 10.1016/j.schres.2004.01.015. [DOI] [PubMed] [Google Scholar]