Abstract
Background
B. subtilis is an important organism in the biotechnological application. The efficient expression system is desirable in production of recombinant gene products in B. subtilis. Recently, we developed a new inducible expression system in B. subtilis, which directed by B. subtilis maltose utilization operon promoter Pglv. The system demonstrated high-level expression for target proteins in B. subtilis when induced by maltose. However, the system was markedly repressed by glucose. This limited the application of the system as a high-expression tool in biotechnology field. The aim of this study was to further improve the Pglv promoter system and enhance its expression strength.
Results
Here, site-directed mutagenesis was facilitated to enhance the expression strength of Pglv. The transcription level from four mutants was increased. Production of β-Gal from the mutants reached the maximum 1.8 times as high as that of wildtype promoter. When induced by 5% maltose, the production of β-Gal from two mutants reached 14.3 U/ml and 13.8 U/ml, 63.5% and 57.5% higher than wildtype promoter (8.8 U/ml) respectively. Thus, site-directed mutagenesis alleviated the repression of glucose and improved the expression activity. To further improve the promoter system, the B. subtilis expression host was reconstructed, in which B. subtilis well-characterized constitutive promoter P43 replaced the promoter of the glv operon in B. subtilis chromosome through a double crossover event. The β-galactosidase production from the improved system (21.1 U/mL) increased compared to that from origin system. Meanwhile, the repression caused by glucose was further alleviated.
Conclusions
In this study, we obtained a mutated promoter Pglv-M1 through site-directed mutagenesis, which demonstrated high expression strength and alleviated the repression caused by glucose. Moreover, we alleviated the repression and enhanced the expression activity of the Pglv-M1 promoter system via reconstruction of the B. subtilis host. Thus, we provided a valuable expression system in B. subtilis.
Background
B. subtilis is an important organism in the biotechnological application, regarding its non-pathogenic and well-characterized biochemical and physiological property [1,2]. The genetic engineering of B. subtilis played a significant role in biotechnological application and industry [3,4]. A controllable expression system is desirable in efficient production of recombinant gene products in B. subtilis [5-7]. The most prominently and widely used induction systems in B. subtilis are mediated by promoters Pspac and Pxyl. The disadvantage of these systems is that the inducer is costly for industrial application [8-10].
Recently, we developed a new inducible expression system in B. subtilis [11], which directed by B. subtilis maltose utilization operon promoter Pglv [12,13]. The system demonstrated high-level expression for target proteins in B. subtilis when induced by maltose. Moreover, cheap and safe inducer makes the system a potential promoter system in industrial application. However, the system was markedly repressed by glucose, in which the glucose repressed the Pglv promoter via a catabolism repression element (cre) located downstream of the transcription origin site of the Pglv promoter [11-13]. This limited the application of the system as a high-expression tool in biotechnology field. To improve the Pglv promoter system, site-directed mutagenesis of several nucleotides downstream the transcription origin site of Pglv was facilitated via overlap polymerase chain reaction (PCR) in vitro in this study. To further alleviate the repression and enhance the expression strength of the Pglv promoter, the B. subtilis expression host was reconstructed, in which the constitutive promoter P43 replaced the promoter of the glv operon in the B. subtilis chromosome through a double crossover event.
Results and discussion
Site-direct mutagenesis of Pglv promoter and examination of the expression strength
In the mutant Pglv-M1, two bases located at the conservation sequence of catabolism repression element were mutated (GC→AT). According to the 3' sequence of 16 s RNA, the ribosome binding site was optimized in the mutants Pglv-M2 and Pglv-M3, based on Pglv-M1, Additionally, based on Pglv-M3, the two bases downstream Shine-Dalgarno sequence (SD sequence) of the Pglv promoter was mutated, resulting in Pglv-M4.
There is a typical conservation sequence of catabolism repression element (cre), overlapping with the ribosome binding site, downstream of the transcription origin site of the Pglv promoter [11,13]. In order to alleviate the repression of glucose and improve the expression strength of the Pglv promoter, the site-directed mutagenesis of cre sequence is performed. To further enhance the expression of gene downstream the promoter, we try to improve the ribosome-binding site sequence. According to the sequence of hydroxyl end of 16 S rRNA, the nucleotides downstream the SD sequence were select to modify in pJRINM4.
To examine the expression efficiency of the obtained four mutants, they were sub-cloned and engineered with synthetic ribosome binding site. The resultant recombinants pJRINM1, pJRINM2, pJRINM3 and pJRINM4, in which the bgaB was under the control of four mutants respectively, were transformed into B. subtilis 1A747 to investigate the expression of β-Gal. Real-time PCR assay (Figure 1A) showed that compared with the pLJ-7, the transcription amount from the pJRINM1, pJRINM2 and pJRINM3 were increased in different degrees, in which the mutant pJRINM1 is obviously prior to the pLJ-7. This suggested the site-directed mutagenesis of the Pglv promoter is efficient in these three mutants.
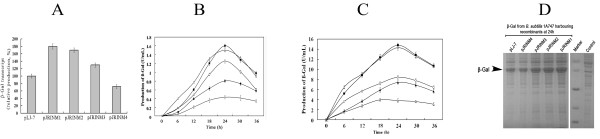
Figure 1.
RT-PCR analysis of the mutated promoters (A) and the production of β-Gal driven by these promoters (B, C and D). (A) Real-time PCR analysis of the transcription amount from pLJ-7, pJRINM1, pJRINM2, pJRINM3 and pJRINM4, respectively (16 s rDNA used as the control). (B) The production of β-Gal when cultured in LB. The black square, white triangle, white circle, black diamond and white diamond represents the production of β-Gal from B. subtilis 1A747 harboring pJRINM1, pJRINM2, pJRINM3, pLJ-7 and pJRINM4, respectively. (C) The production of β-Gal when cultured in LB supplemented with 5% maltose. The black square, white triangle, white circle, black diamond and white diamond represents the production of β-Gal from B. subtilis 1A747 harboring pJRINM1, pJRINM2, pJRINM3, pLJ-7 and pJRINM4; (D) SDS-PAGE analysis of β-Gal from the recombinants in B. subtilis 1A747 after 24 h cultured on a 12% SDS-polyacrylamide gel. β-Gal is indicated by the arrow. Molecular mass marker indicates (top to bottom): 116, 66, 45, 35 and 25 kDa.
To further verify the efficiency of mutated promoters, the β-Gal driven by these promoters was determined. The production of β-Gal from pJRINM1, pJRINM2 and pJRINM3 increased compared with that from pLJ-7. Amongst the production of β-Gal from pJRINM1, pJRINM2 was 1.8 fold (1.6 U/mL) and 1.7 fold (1.5 U/mL) of that from wildtype promoter (0.9 U/mL) after 24 h culture, respectively. Whereas, the production of β-Gal from pJRINM4 was obviously decreased, about 58.2% of that from wildtype promoter after 24 h culture (Figure 1B). This showed that site-directed mutagenesis in pJRINM4 has negative effect on the Pglv promoter. We speculated that the three mutated sites including the SD sequnence may change the space structure of the promoter, and the result reduced the transcript level.
While, the growth trend of B. subtilis 1A747 harbouring pJRINM1, pJRINM2, pJRINM3, pJRINM4 or pLJ-7 was approximately same (Additional file 1). It confirmed that the difference of β-Gal production was not caused by the cell amount.
To further assay these promoters, we tested the growth curves (Additional file 2) and the β-Gal activity (Figure 1C) under the induction conditions. When induced with 5% maltose, the production of β-Gal from pJRINM1 and pJRINM2 reached 14.3 U/mL and 13.8 U/ml, up by 63.5% and 57.5% compared with wildtype promoter (8.77U/mL) at 24 h, respectively. Meanwhile, the β-Gal production from pJRINM4 was 4.3 U/mL, only 48.8% of that from wildtype promoter.
SDS-PAGE assay (Figure 1D) demonstrated the β-Gal production from pJRINM1 and pJRINM2 was obviously higher than that from pLJ-7, further confirming that the two mutants enhanced the expression strength of Pglv promoter.
Effect of site-directed mutagenesis on the repression caused by glucose
To examine the effect of mutant on alleviating repression caused by glucose, the four recombinants were cultured in LB medium supplemented with 5% maltose and 5% glucose, and the growth curves (Additional file 3) and β-Gal production (Figure 2A) was measured, respectively. The β-Gal production from wildtype promoter was only 1/17 and 1/5 of that in medium without glucose supplement at 24 h and 30 h, respectively. Whereas the β-Gal production from pJRINM1 was 1/4 and 1/3 of that in medium without glucose supplement at 24 h and 30 h, respectively. While, the β-Gal production from pJRINM2, pJRINM3 and pJRINM4 was 1/6, 1/6 and 1/4 of that in medium without glucose supplement at 24 h, respectively. Thus, these mutants alleviated the repression caused by glucose, especially pJRINM1.
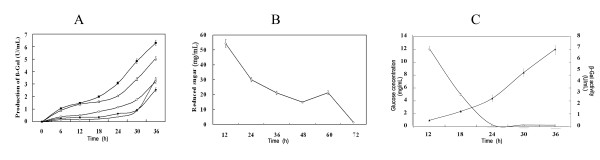
Figure 2.
Characterization of the mutated promoters. The production of β-Gal from B. subtilis 1A747 harboring mutants supplemented with 5% maltose plus 5% glucose. The black square, white triangle, white circle, black diamond and white diamond represents the production of β-Gal from B. subtilis 1A747 harboring pJRINM1, pJRINM2, pJRINM3, pLJ-7 and pJRINM4, respectively; (B) Reduced sugar concentration of culture of B. subtilis 1A747 harboring pJRINM1 in LB supplemented with 5% maltose; (C) Glucose actual concentration and β-Gal activity detection from B. subtilis 1A747 harboring pJRINM1 in LB supplemented with 5% maltose plus 5% glucose. The black triangle represents the β-Gal production. The white triangle represents the glucose actual concentration.
Taken together, since the pJRINM1 demonstrated advantage in improvement of expression strength and alleviation of repression caused by glucose, the mutated promoter Pglv-M1 is a good candidate as an expression element in B. subtilis biotechnology application. Subsequently, an effort was made to further probe the relation between glucose and β-Gal driven by the pJRINM1. When B. subtilis harbouring pJRINM1 was cultured in LB medium with 5% glucose, reducing sugar assay (Figure 2B) showed that the reducing sugar decreased from 12 h to 48 h. Additionally, we detected the actual concentration of glucose during the culture. Figure 2C showed a trend that with the decrease of the glucose concentration during cultivation, the β-Gal production from the recombinant sharply increased in the medium supplemented with 5% maltose plus 5% glucose.
Reconstruction of the B. subtilis strain to further improve the expression system
The abovementioned results suggested that the promoter Pglv-M1 alleviated the repression caused by glucose; however, the glucose still exerted repression on the system. We speculated that one of the possible causes of the residual repression was that the maltose utilization operon located in the chromosome of the B. subtilis was nevertheless negatively regulated by glucose, in which GlvR as a positive regulator of the Pglv-M1 promoter system was driven by the operon native promoter Pglv. To further improve the Pglv-M1 promoter system, we replaced maltose utilization operon promoter Pglv with B. subtilis constitutive promoter P43. Southern blot analysis (Figure 3A) and PCR detection indicated that the double crossover event occurred in the resultant strain B. subtilis BCYL as expected.
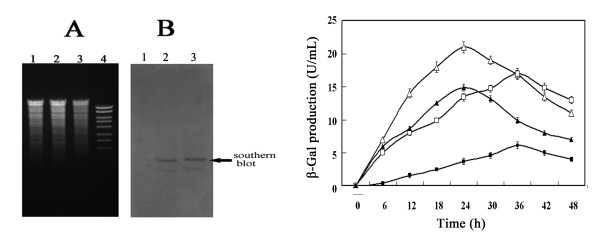
Figure 3.
Reconstruction of B. subtilis host. (A) Agarose-gel-electrophoresis of chromosome DNA digested with ApaI and EcoRI. Lane1, treated chromosome DNA from B. subtilis 1A747 [this served as the negative control]; lanes 2 and 3, treated chromosome DNA from B. subtilis BCYL; lane 4, molecular weigh marker. (B) Southern blot analysis of recombinant B. subtilis BCYL chromosome DNA. The lanes 1, 2 and 3 are that the same as in (A). (C) β-Gal production by the improved expression system. White triangle and white square represents the β-Gal production by B. subtilis BCYL harboring pJRINM1 cultures in LB with 5% maltose and LB with 5% maltose plus 5% glucose, respectively; black triangle and black square represents the β-Gal production by B. subtilis 1A747 harboring pJRINM1 when cultured in LB with 5% maltose and LB with 5% maltose plus 5% glucose, respectively.
To probe the efficiency of the improved expression system, the mutant pJRINM1 was transformed into BCYL and expression experiments were carried out. Figure 3B indicates that, with 5% maltose induction, β-Gal production reached a maximum (21.1 U/mL) at 24 h from B. subtilis BCYL harboring pJRINM1, and this was 50.3% higher than that from B. subtilis 1A747 harboring pJRINM1. For investigating the repression caused by glucose on the improved expression system, BCYL harboring pJRINM1 was cultivated in LB supplemented with 5% maltose plus 5% glucose, yielded about 4-fold higher production of β-Gal than that of B. subtilis 1A747 harboring pJRINM1 at 24 h (Figure 3B). Therefore, the reporter gene production driven by the improved expression system increased obviously when supplemented with 5% glucose or not.
Controllable and strong promoter is the essential element to achieve high-level expression of target gene in the B. subtilis genetic engineering. B. subtilis maltose utilization promoter is a potential control element in the biotechnological application; however, glucose as a repressor influenced the strength of Pglv. In the maltose-inducible system, maltose plays two roles as an inductor: on one hand, it positively regulates the transcription of the Pglv promoter; on the other hand, as a fermentation carbon source, its metabolism production-glucose repressed the activity of promoter. There is a dynamic balance between the two carbon sources during the fermentation. Maltose was a positive regulation factor and, meanwhile degraded as one of carbon sources during this process; in the later stage of fermentation, both the total carbon sources and glucose are at relatively low level. When induced by 5% maltose, the production of β-Gal reached the maximum at 24 h and then decreased. In the glucose repression experiment, expression strength of both the mutated and wildtype promoter had a drastic increase after cultured for 24 h. This may be the utilization of glucose, as an easy ferment carbon source, was prior to maltose and the amount of glucose in total carbon source declines to a relatively low level in the later stage of fermentation. As a result, the repression caused by glucose was relatively alleviated after 24 h.
Conclusions
In this study, we obtained a mutated promoter Pglv-M1 through site-directed mutagenesis, which demonstrated high expression strength and alleviated the repression caused by glucose. Moreover, we further alleviate the repression and enhance the expression activity of the Pglv-M1 promoter system via reconstruction of the B. subtilis host. Thus, we improved the promoter system and provided a valuable expression system in B. subtilis.
Methods
Bacterial strains, plasmids and growth conditions
B. subtilis 1A747 was a generous gift from the Bacillus Genetic Stock Center (BGSC). Escherichia coli DH5α was purchased from Novagen (Darmstadt, Germany). The bacterial strains were cultured in Luria-Bertani (LB) medium at 37°C. Maltose and glucose were added as required. The following concentrations of antibiotics were used for selection: 100 μg/mL ampicillin (Amp), 5 μg/mL chloramphenicol (Cm) and 50 μg/mL spectinomycin (Spec). The plasmids used in this study were listed in Additional file 4.
Primers and oligonucleotides
Polymerase chain reaction (PCR) primers and oligonucleotides used in this study were listed in Additional file 5.
General manipulation
General recombinant DNA technique was carried out using standard techniques [14]. The transformation of B. subtilis was performed by electroporation [15,16].
Site-directed mutagenesis of Pglv promoter via overlap PCR
The bases downstream the transcription original sites were site-directed mutated by using of overlap PCR. The sequences of primers that were used in this experiment are shown in Additional file 5, and the mechanism of the mutagenesis method and the positions of the primers are depicted in Figure 4.
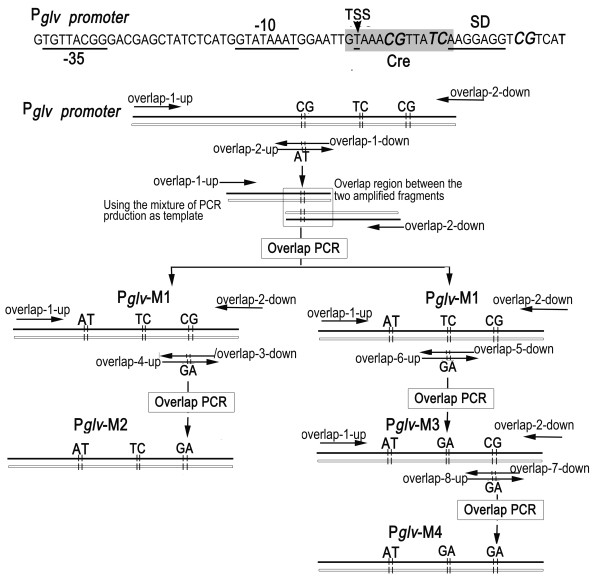
Figure 4.
Schematic diagram of the overlap PCR. -10, -35, TSS, SD, and Cre represent the -10 box, -35 box, transcription origin, SD box and catabolite repression element, respectively. Italic indicates the mutated nucleotides. Approximate positions of the primers are shown with arrows. Using primer pairs overlap-1-up/overlap-1-down and overlap-2-up/overlap-2-down, a 360 bp fragment upstream of the maltose promoter and a 300 bp fragment downstream of the maltose promoter were amplified from Bacillus subtilis 1A747 genome DNA, in which there are 30 bp overlap in the two amplified fragments and the two mutant sites were introduced by primers overlap-1-down and overlap-2-up. With overlap-1-up and overlap-2-down as primers, and the mixture of two obtained fragments as template, a 630 bp Pglv-M1 was overlap PCR amplified. Same mechanism was used to generate the other three mutants Pglv-M2, Pglv-M3 and Pglv-M4.
A 360 bp fragment upstream of the maltose promoter and a 300 bp fragment downstream of the maltose promoter waiting for site-directed mutagenesis were amplified from Bacillus subtilis 1A747 genome DNA as a template respectively, using primer pairs overlap-1-up/overlap-1-down and overlap-2-up/overlap-2-down, in which there are 30 bp overlap in the two amplified fragments and the two mutant sites were introduced by primers overlap-1-down and overlap-2-up. With overlap-1-up and overlap-2-down as primers, and the mixture of two fragments as template, a 630 bp Pglv-M1 was overlap PCR amplified.
According to the abovementioned protocol, using Pglv-M1 as template, the second mutant Pglv-M2 and third mutant Pglv-M3 were amplified via overlap PCR by using of primer pairs overlap-1-up/overlap-3-down and overlap-4-up/overlap-2-down, and overlap-1-up/overlap-5-down and overlap-6-up/overlap-2-down, respectively. The mutant was introduced by overlap section of overlap-3-down and overlap-4-up for Pglv-M2 and, overlap-5-down and overlap-6-up for Pglv-M3. Then, the fourth mutant Pglv-M4 was generated from Pglv-M3 using primer pairs overlap-1-up/overlap-7-down and overlap-8-up/overlap-2-down, in which the overlap section of overlap-7-down and overlap-8-up introduced the mutant.
Construction of plasmid vectors
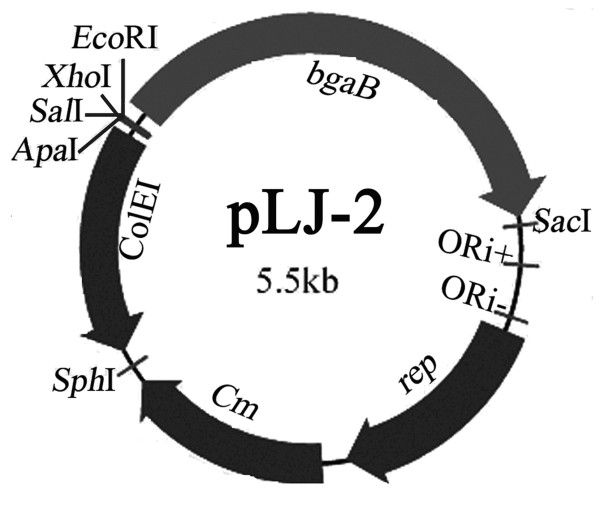
Using primer pair bga-up/bga-down, the bgaB coding for thermostable β-Gal was polymerase chain reaction (PCR) amplified from plasmid pDL. The obtained 2.0 kb fragment was digested with EcoRI and SacI, and cloned into pGJ103 digested with the same enzymes, resulting in pLJ-2 (Figure 5).
Figure 5.
Map of the promoter probe vector pLJ-2. ORI+, ORI- and rep represent the single-strand replication origin, the double strand origin and replication protein in B. subtilis, respectively. ColEI, bgaB and Cm represent E. coli ColEI replicon, chloramphenicol-resistance marker and coding gene of β-Gal. The unique restriction sites are marked on the outside of the map.
Using Pglv-1-up and Pglv-5-down as primers, PglvMa, PglvMb, PglvMc and PglvMd were PCR amplified from Pglv-M1, Pglv-M2, Pglv-M3 and Pglv-M4 (As shown in Figure 4), respectively. After digested with ApaI and BamHI, these amplified fragments were cloned into the corresponding sites of pBluskm, resulting in pJR1, pJR2, pJR3 and pJR4. A ribosome binding site (RBS) fragment INSD generated by annealing two artificially synthesized oligonucleotides INSID-1 and INSID-2, was cloned into the pJR1-4 digested by BamHI and SacI, respectively. Then, the four mutant promoters with engineered RBS were excised from the obtained four recombinants with ApaI and EcoRI, and cloned into the corresponding sites of pLJ-2, in which the bgaB was used as reporter, yielding pJRINM1, pJRINM2, pJRINM3 and pJRINM4, respectively.
Reconstruction of B. subtilis strain
With B. subtilis 1A747 chromosomal DNA as template, the two homogeneous arms GAf (using GlvA-fro-up and GlvA-fro-down as primers) and GAb (using GlvA-bac-up and GlvA-bac-down as primers) were amplified. A selection marker (spectinomycin resistance gene) was generated through PCR amplified from plasmid pDG1728, using primers Spec-I-up and Spec-I-down. And BamHI-SalI treated selection marker and KpnI-ApaI treated GAf was cloned into pBluskm, resulting in pYG34.
The expression cassette directed by P43 promoter was constructed as follows. First, the P43 promoter [3,17] was amplified from B. subtilis 1A747 chromosomal DNA, using primers P43-1-up and P43-1-down. After digestion with ApaI and EcoRI, the P43 promoter was cloned into the corresponding sites in pBluskm, yielding pB43. Then the GAb fragment, digested with EcoRI and SacI, was cloned into the corresponding sites in pB43, resulting in pCYL17. Finally, with primers P43-2-up and GlvA-bac-down, the P43 promoter and the GAb were amplified as a single product from pCYL17 and ligated with pYG34 after digestion with BamHI and SacI, resulting in pCYL25. The B. subtilis 1A747 was transformed with the linear fragment of pCYL25 obtained with ScaI to replace promoter Pglv with P43 via a double crossover event between the linearized pCYL25 and B. subtilis 1A747 chromosome. The spectinomycin resistance (SpecR) colonies were selected. And the resultant strain was named as BCYL.
Isolation of total RNA and Real-time PCR
The cultures was harvested at 24 h. Total RNA of bacteria was isolated by using SV total RNA isolation kit (Cat. Z3100, Promega). The cDNA chain was synthesized by using Reverse Transcription System (Cat. A3500, Promega).
And Real-time PCR was performed by using Real time PCR Kit (Cat. DRR041 S, TaKaRa). The bagB gene was amplified using bga-up and bga-down as primers B1-up/B1-down. With the primers 16s-up/16s-down, Bacillus subtilis 16 s rDNA was amplified as control, The PCR protocol was as follows: 2 min at 50°C, 10 min at 95°C, and then 35 cycles consisting of 45 s at 95°C, 1 min at 52°C, and 30 s at 72°C. Reactions were carried out in real-time PCR detection system (IQ5, Bio-RAD).
SDS-PAGE assay
Sodium dodecylsulfate polyacrylamide gel electrophoresis (SDS-PAGE) was performed as described previously [14].
β-Gal activity assay
Method has been described previously [[11,18] and [19]]. In brief, the culture was pelleted by centrifuge and resuspended in an equivalent volume of buffer Z [18,19]. Using buffer Z, 0.01 or 0.1 mL sample aliquots was diluted to 0.8 mL, and then added 0.01 mL of lysozyme stock (10 mg/ml). The mixture was incubated at 37°C for 30 min, and then, added 0.2 ml of 4 mg/ml o-nitrophenyl-b-Dgalactopyranoside (ONPG). After incubated at 55°C for 15 min, the reaction was stopped by adding 0.5 ml of 1 M Na2CO3. Absorbance was recorded at 420 nm with a spectrophotometer (HITACHI, U-3010). One unit of β-galactosidase activity was defined as the amount of enzyme necessary to release 1 μmol 2-nitrophenol from o-nitrophenylgalactopyranoside per minute at 55°C. β-galactosidase activity is expressed as units per mL sample.
Southern blot analysis
Southern blot analysis of chromosomal DNA digested with ApaI and EcoRI was carried out as described previously [14]. Probe labeling was performed with a DIG DNA labeling kit (Roche, Cat.No.1093657) according to the instructions, using the spectinomycin resistance gene as template.
Reducing sugar assay
Reducing sugar was measured by the dinitrosalicylic method (DNS) [20] using glucose as a standard.
Assay of glucose concentration
The concentration of glucose was determinate as previously described [21]
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
MMY initiated and coordinated the project. WWZ and MMY were responsible for site-directed mutagenesis and its analysis. WWZ and YLC performed the batch cultivation. MMY and Y-S Gong performed construction of expression system. All authors wrote the paper and approved the final version of the manuscript.
Supplementary Material
The growth curves from B. subtilis 1A747 harboring different plasmids when cultured in LB. (black diamond) represents OD595 from B. subtilis 1A747 harboring pLJ-7; (black square) represents OD595 from B. subtilis 1A747 harboring pJRINM1; (black triangle) represents OD595 from B. subtilis 1A747 harboring pJRINM2; cross (x) represents OD595 from B. subtilis 1A747 harboring pJRINM3; asterisk (*) represents OD595 from B. subtilis 1A747 harboring pJRINM4.
The growth curves from B. subtilis 1A747 harboring different plasmids when cultured in LB supplemented with 5% maltose. (black diamond) represents OD595 from B. subtilis 1A747 harboring pLJ-7; (black square) represents OD595 from B. subtilis 1A747 harboring pJRINM1; (black triangle) represents OD595 from B. subtilis 1A747 harboring pJRINM2; cross (×) represents OD595 from B. subtilis 1A747 harboring pJRINM3; asterisk (*) represents OD595 from B. subtilis 1A747 harboring pJRINM4.
The growth curves from B. subtilis 1A747 harboring different plasmids when cultured in LB supplemented with 5% maltose plus 5% glucose. (black diamond) represents OD595 from B. subtilis 1A747 harboring pLJ-7; (black square) represents OD595 from B. subtilis 1A747 harboring pJRINM1; (black triangle) represents OD595 from B. subtilis 1A747 harboring pJRINM2; cross (×) represents OD595 from B. subtilis 1A747 harboring pJRINM3; asterisk (*) represents OD595 from B. subtilis 1A747 harboring pJRINM4.
Plasmids used in this study.
Primers and oligonucletides used in this study.
Contributor Information
Yang M Ming, Email: yangjinxin@vip.163.com.
Zhang W Wei, Email: summertansy@163.com.
Chen Y Lin, Email: biodna@126.com.
Gong Y Sheng, Email: scimanuscript@126.com.
Acknowledgements
This work was supported by grants from Chinese National Natural Science Foundation (30871813). And we thank the Bacillus Genetic Stock Centre of Ohio State University for generously providing the study materials.
References
- Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, Azevedo V, Bertero MG, Bessieres P, Bolotin A, Borchert S. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- Schallmey M, Singh A, Ward OP. Developments in the use of Bacillus species for industrial production. Can J Microbiol. 2004;50:1–17. doi: 10.1139/w03-076. [DOI] [PubMed] [Google Scholar]
- Zhang XZ, Cui ZL, Hong Q, Li SP. High-level expression and secretion of methyl parathion hydrolase in Bacillus subtilis WB800. Appl Environ Microbiol. 2005;71:4101–4103. doi: 10.1128/AEM.71.7.4101-4103.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang AL, Liu H, Yang MM, Gong YS, Chen H. Assay and characterization of a strong promoter element from B. subtilis. Biochem Biophys Res Commun. 2007;354:90–95. doi: 10.1016/j.bbrc.2006.12.137. [DOI] [PubMed] [Google Scholar]
- Nguyen HD, Nguyen QA, Ferreira RC, Ferreira LCS, Tran LT, Schumann W. Construction of plasmid-based expression vectors for Bacillus subtilis exhibiting full structural stability. Plasmid. 2005;54:241–248. doi: 10.1016/j.plasmid.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Nguyen HD, Schumann W. Novel plasmid-based expression vectors for intra- and extracellular production of recombinant proteins in Bacillus subtilis. Prot Expr Purif. 2006;46:189–195. doi: 10.1016/j.pep.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Li W, Li HX, Ji SY, Li S, Yang MM, Chen YL. Characterization of two temperature-inducible promoters newly isolated from B. subtilis. Biochem Biophys Res Commun. 2007;358:1148–1153. doi: 10.1016/j.bbrc.2007.05.064. [DOI] [PubMed] [Google Scholar]
- Kim L, Mogk A, Schumann W. A xylose-inducible Bacillus subtilis integration vector and its application. Gene. 1996;87:53–61. doi: 10.1016/s0378-1119(96)00466-0. [DOI] [PubMed] [Google Scholar]
- Bhavsar AP, Zhao XM, Brown ED. Development and characterization of a xylose-dependent system for cloned gene in Bacillus subtilis: conditional complementation of a Teichoic acid mutant. Appl Environ Microbiol. 2001;67:403–410. doi: 10.1128/AEM.67.1.403-410.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl B, Wehrl W, Wiegert T, Homuth G, Schumann W. Development of a new integration site within the Bacillus subtilis chromosome and construction of compatible expression cassettes. J Bacteriol. 2001;183:2696–2699. doi: 10.1128/JB.183.8.2696-2699.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang MM, Zhang WW, Zhang XF, Cen PL. Construction and characterization of a novel maltose inducible expression vector in Bacillus subtilis. Biotechnol Lett. 2006;28:1713–1718. doi: 10.1007/s10529-006-9125-4. [DOI] [PubMed] [Google Scholar]
- Thompson J, Pikis A, Ruvinov SB, Henrissat B, Yamamoto H, Sekiguchi J. The gene glvA of Bacillus subtilis 168 encodes a metal-requiring NADH-dependent 6-phospho-glucosidase. J Bol Chem. 1998;273:27347–27356. doi: 10.1074/jbc.273.42.27347. [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Serizawa M, Thompson J, Sekiguchi J. Regulation of glvoperon in Bacillus subtilis : YfiA(GLVR) is a positive regulator of the operon that is repressed through CcpA and cre. J Bacteriol. 2001;183:5110–5121. doi: 10.1128/JB.183.17.5110-5121.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fitsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor: Cold Spring Harbor Press; 1989. [Google Scholar]
- Bron S. In: Molecular biological methods for Bacillus. Harwood CR, Cutting SM, editor. New York: John Wiley and Sons; 1990. Plasmids; pp. 75–174. [Google Scholar]
- Yang MM, Zhang WW, Bai XT, Li HX, Cen PL. Electroporation is a feasible method to introduce circularized or linearized DNA into B. subtilis chromosome. Mol Biol Rep. 2010. in press . [DOI] [PubMed]
- Wang PZ, Doi RH. Overlapping promoters transcribed by Bacillus subtilis σ55 and σ37 RNA polymerase holoenzymes during growth and stationary phase. J Biol Chem. 1984;259:8619–8625. [PubMed] [Google Scholar]
- Hirata H, Fukazawa T, Negoro S, Okada H. Structure of a β-galactosidase gene of Bacillus stearothermophilus. J Bacteriol. 1986;166:722–727. doi: 10.1128/jb.166.3.722-727.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning S, Leahy M, Sjheehan D. Nucleotide and deduced amino acidsequences of Rhizobium meliloti 102F34 lacZ gene: comparison with prokaryotic β-galactosidases and hunan β-glucuronidase. Gene. 1994;141:91–96. doi: 10.1016/0378-1119(94)90133-3. [DOI] [PubMed] [Google Scholar]
- Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem. 1959;31:426–428. doi: 10.1021/ac60147a030. [DOI] [Google Scholar]
- Cavari BZ, Phelps G. Sensitive Enzymatic Assay for Glucose Determination in Natural Waters. Appl Environ Microbiol. 1977;33:1237–1243. doi: 10.1128/aem.33.6.1237-1243.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short JM, Fernandez JM, Sorge JA, Huse WD. λ ZAP: A bacteriophage λ expression vector with in vivo excision properties. Nucleic Acids Res. 1988;16:7583–7600. doi: 10.1093/nar/16.15.7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerot-Fleury AM, Frandsen N, Stragier P. Plasmids for ectopic integration in Bacillus subtilis. Gene. 1996;180:57–61. doi: 10.1016/S0378-1119(96)00404-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The growth curves from B. subtilis 1A747 harboring different plasmids when cultured in LB. (black diamond) represents OD595 from B. subtilis 1A747 harboring pLJ-7; (black square) represents OD595 from B. subtilis 1A747 harboring pJRINM1; (black triangle) represents OD595 from B. subtilis 1A747 harboring pJRINM2; cross (x) represents OD595 from B. subtilis 1A747 harboring pJRINM3; asterisk (*) represents OD595 from B. subtilis 1A747 harboring pJRINM4.
The growth curves from B. subtilis 1A747 harboring different plasmids when cultured in LB supplemented with 5% maltose. (black diamond) represents OD595 from B. subtilis 1A747 harboring pLJ-7; (black square) represents OD595 from B. subtilis 1A747 harboring pJRINM1; (black triangle) represents OD595 from B. subtilis 1A747 harboring pJRINM2; cross (×) represents OD595 from B. subtilis 1A747 harboring pJRINM3; asterisk (*) represents OD595 from B. subtilis 1A747 harboring pJRINM4.
The growth curves from B. subtilis 1A747 harboring different plasmids when cultured in LB supplemented with 5% maltose plus 5% glucose. (black diamond) represents OD595 from B. subtilis 1A747 harboring pLJ-7; (black square) represents OD595 from B. subtilis 1A747 harboring pJRINM1; (black triangle) represents OD595 from B. subtilis 1A747 harboring pJRINM2; cross (×) represents OD595 from B. subtilis 1A747 harboring pJRINM3; asterisk (*) represents OD595 from B. subtilis 1A747 harboring pJRINM4.
Plasmids used in this study.
Primers and oligonucletides used in this study.