Abstract
Background
The anaplastic lymphoma kinase (ALK) gene is frequently involved in translocations that lead to gene fusions in a variety of human malignancies, including lymphoma and lung cancer. Fusion partners of ALK include NPM, EML4, TPM3, ATIC, TFG, CARS, and CLTC. Characterization of ALK fusion patterns and their resulting clinicopathological profiles could be of great benefit in better understanding the biology of lung cancer.
Results
RACE-coupled PCR sequencing was used to assess ALK fusions in a cohort of 103 non-small cell lung carcinoma (NSCLC) patients. Within this cohort, the EML4-ALK fusion gene was identified in 12 tumors (11.6%). Further analysis revealed that EML4-ALK was present at a frequency of 16.13% (10/62) in patients with adenocarcinomas, 19.23% (10/52) in never-smokers, and 42.80% (9/21) in patients with adenocarcinomas lacking EGFR and KRAS mutations. The EML4-ALK fusion was associated with non-smokers (P = 0.03), younger age of onset (P = 0.03), and adenocarcinomas without EGFR/KRAS mutations (P = 0.04). A trend towards improved survival was observed for patients with the EML4-ALK fusion, although it was not statistically significant (P = 0.20). Concurrent deletion in EGFR exon 19 and fusion of EML4-ALK was identified for the first time in a Chinese female patient with an adenocarcinoma. Analysis of ALK expression revealed that ALK mRNA levels were higher in tumors positive for the EML-ALK fusion than in negative tumors (normalized intensity of 21.99 vs. 0.45, respectively; P = 0.0018). However, expression of EML4 did not differ between the groups.
Conclusions
The EML4-ALK fusion gene was present at a high frequency in Chinese NSCLC patients, particularly in those with adenocarcinomas lacking EGFR/KRAS mutations. The EML4-ALK fusion appears to be tightly associated with ALK mRNA expression levels. RACE-coupled PCR sequencing is a highly sensitive method that could be used clinically for the identification of EML4-ALK-positive patients.
Background
The anaplastic lymphoma kinase (ALK) gene encodes a receptor tyrosine kinase (RTK) that has been discovered to be present in a number of fusion proteins consisting of the intracellular kinase domain of ALK and the amino-terminal portions of different genes [1,2]. Activated ALK is involved in the inhibition of apoptosis and the promotion of cellular proliferation through activation of downstream PI3K/Akt and MAPK signalling pathways [3]. Genetic alterations involving ALK, including gene fusions, amplification, and mutations, have been identified in anaplastic large cell lymphomas, inflammatory myofibroblastic tumors, and neuroblastoma, respectively [1,4-7]. To date, in studies from a variety of human cancer types, the reported fusion partners of ALK have included NPM [1], EML4 [8], MSN [9], TPM3 [10,11], ATIC [12-14], TFG [15], CARS [16], CLTC [17], and KIF5B [18]. In lung cancer, the primary ALK fusion detected was identified as EML4-ALK, followed by TFG-ALK and KIF5-ALK, although other unknown fusions may also exist which can not be detected due to limits of present technology [19].
The ALK-EML4 fusion attaches the ALK gene to a gene involved in microtubule formation and stabilization, "echinoderm microtubule associated protein-like 4" (EML4) [20,21]. This fusion generates a transforming fusion tyrosine kinase, several isoforms of which have been identified in lung cancers [8,22]. The frequency of the EML4-ALK fusion was first reported by Soda et al. to be approximately 6.7% (5/75) in non-small cell lung carcinomas (NSCLCs) in Japanese patients [8]. ALK and EML4 are both located on the short arm of chromosome 2, separated by 12 megabases (Mb) of sequence, and are oriented in opposite directions. To date, more than nine different variants of the EML4-ALK fusion have been identified. These variants consist of exons 20 to 29 of ALK fused to EML4 exon 13 (variant 1, V1), exon 20 (V2), exon 6 (V3a), exon 6 with an 11 amino acid (aa) insertion (V3b), exon 14 with an additional 11 nucleotide insertion of unknown origin at nucleotide 50 in exon 20 of ALK (V4), exon 2 (V5), exon 13 (V6), exon 14 with the fusion beginning at nucleotide 13 in exon 20 of ALK (V7), exon 15 (primarily also reported as "V4", in this article depicted as V8), and exon 18 ("V5", accordingly here as V9), as described in detail in Horn and Pao's review [23]. ALK gene fusions have been demonstrated to be oncogenic in 3T3 and Ba/F3 cellular models [24]. Although different variants of EML4-ALK fusion proteins may exhibit different enzymatic activities, EML4 retains the N-terminal coil-coiled domain (CC) in all EML4-ALK variants. This domain has been shown to be responsible for the dimerization and constitutive activation of EML4-ALK [24]. Importantly, in some cell lines harboring EML4-ALK fusions, targeting of ALK using specific inhibitors has shown promising efficacy for treatment of lung cancer through inhibition of Akt and induction of apoptosis. For this reason, ALK inhibitors have been developed and assessed in early clinical trials [25,26].
Published studies using reverse transcriptase-polymerase chain reaction (RT-PCR) analysis or fluorescent in situ hybridization (FISH) to characterize EML4-ALK fusions in lung cancer have revealed frequencies ranging from 0.5% to 7.5% in general lung cancer patients and a frequency of 13.5% in clinical factor-enriched cases [23], [27,28]. Here, we report the development of a technology based on rapid amplification of cDNA ends (RACE)-coupled PCR and sequencing to analyze expression of ALK fusion genes. This technology was designed to identify both known and novel fusion partners of ALK, followed by confirmation using qualitative specific RT-PCR. Using this method, we analysed the ALK fusion status in clinical lung cancer samples. Results from this study could, thus, provide a better understanding of ALK fusions with all potential partner genes in a clinical population. Furthermore, these results provide insight into the clinicopathological characteristics of ALK fusion-positive Chinese NSCLC patients.
Results
Identification of EML4-ALK fusions in 103 cases of NSCLC
RNA samples from a total of 103 NSCLC cases were reverse-transcribed to make cDNA, followed by oligo-dC tailing. Two successive rounds of PCR were used to identify potential fusion fragments (Fig. 1A, B). BigDye3.1-labeled products were then sequenced to identify fusions between ALK and potential partners (Fig. 1C). Based on a sequence alignment with the ALK reference sequence (NCBI accession number: NM_004304.3), in total, 12 samples were identified as ALK fusion-positive (Table 1).
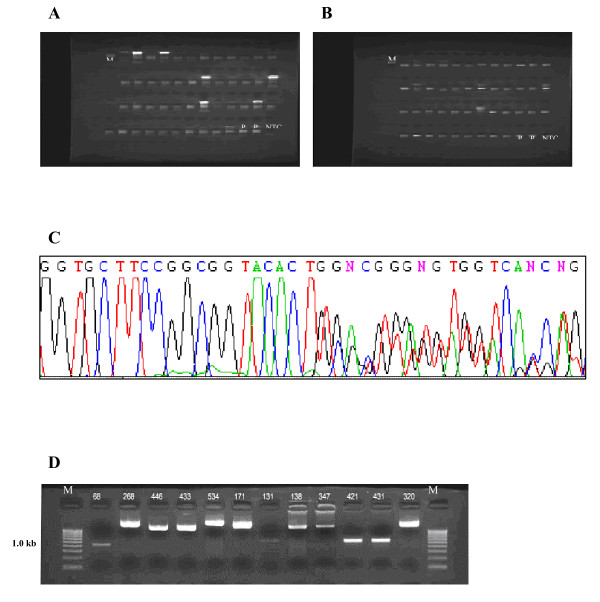
Figure 1.
Assessment of ALK fusion status by RACE-coupled PCR sequencing. (A) Representative image showing gel electrophoresis results for first-round PCR products. Lane M, marker (200 bp ladder); lanes P, positive controls (2 lanes); lane NTC, no template control; other lanes, samples 1 through 45. (B) Representative image showing gel electrophoresis results for second-round PCR products. Lane organization is identical to (A). (C) Representative sequencing chromatograph showing ALK fusion with EML4. (D) RT-PCR validation for samples positive for ALK fusion expression. RT-PCR was used to confirm EML4-ALK fusion transcript expression in 12 samples. Sample 68 is the V5 variant (E2;A20, 408 bp). Samples 268, 171, and 320 are V9 variants (E18;A20, 2256 bp). Samples 446, 433, 138, and 347 are V1 variants (E13;A20, 1689 bp). Sample 534 is the V2 variant (E20;A20, 2445 bp). Samples 131, 421, and 431 are V3a/b variants (E6;A20 or E6 ins33;A20, 867 and 900 bp, respectively). Lane M, marker (100 bp ladder).
Table 1.
EML4-ALK fusion variants identified in the 12 NSCLC cases.
| Variant | Published fusion type* | No. of positive samples | Frequency |
|---|---|---|---|
| V1 | EML4 E13-ALK E20 | 4 | 3.9% |
| V2 | EML4 E20-ALK E20 | 1 | 1.0% |
| V3a | EML4 E6-ALK E20 | 3 | 2.9% |
| V3b | EML4 E6-11 aa in INS6-ALK E20 | ||
| V4 | EML4 E15-ALK E20 | 0 | 0 |
| V5 | EML4 E2-ALK E20 | 1 | 1.0% |
| V6 | EML4 E18-ALK E20 | 3 | 2.9% |
*All fusion types have previously been published.
RT-PCR confirmation of expression of ALK fusion transcripts in positive samples
Fusion gene-specific primers were designed, and qualitative RT-PCR was conducted to confirm the presence of ALK fusions in positive samples identified by RACE-coupled PCR sequencing. Fusion RNAs from the 12 ALK fusion-positive samples were amplified by RT-PCR, and specific bands corresponding to the expected products were observed after gel electrophoresis (Fig. 1D).
Correlation of ALK fusion with ALK expression
Gene expression profiling was conducted on clinical samples using the Affymetrix GeneChip® Human Genome U133 plus 2.0 technology. Normalized expression intensities for the ALK and EML4 transcripts were extracted and plotted versus ALK fusion status (Fig. 2A). ALK was found to be significantly over-expressed in the 10 fusion-positive samples analyzed (2 EML4-ALK cases were not analyzed by microarray), but not in samples lacking ALK fusions or control adjacent tissues. ALK expression was significantly different in EML-ALK fusion-positive and -negative samples (normalized intensity values of 21.99 vs. 0.45, respectively; P = 0.0018). In contrast, EML4 expression did not differ significantly between the groups (Fig. 2B). These results indicate that presence of the ALK fusion was strongly associated with ALK mRNA expression levels, leading to a 50-fold increase in expression.
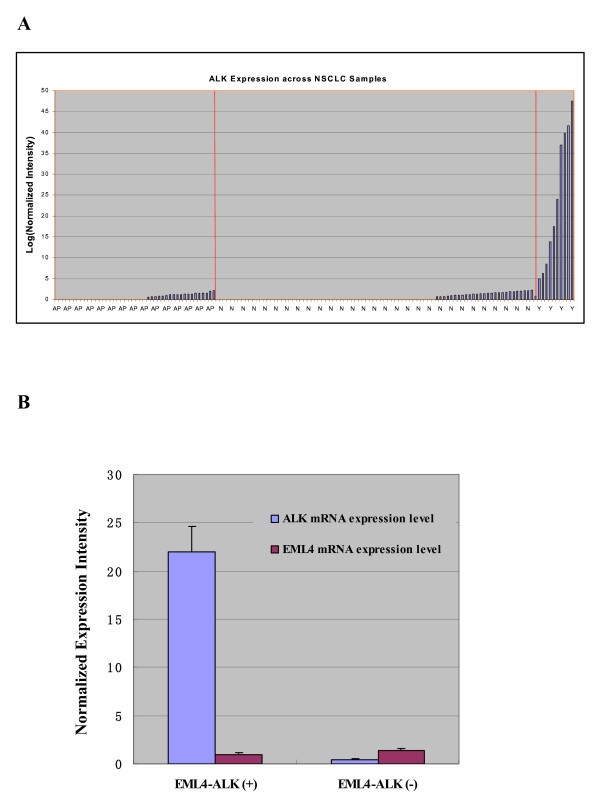
Figure 2.
ALK expression is associated with ALK fusion status. (A) ALK is overexpressed at the mRNA level in 10 fusion-positive cases analyzed using Affymetrix Genechip® U133 plus 2.0. AP, Adjacent normal tissues; N, Cancer samples with no fusion; Y, Cancer samples positive for fusion. (B) ALK is differentially expressed between EML-ALK fusion-positive and negative samples (normalized intensity of 21.99 vs. 0.45, respectively; P = 0.0018), but EML4 expression was not significantly different between the groups. Error bars indicate mean ± standard deviation.
Concurrent EGFR mutation and ALK fusion in one sample
For all samples analyzed for ALK fusion, DNA was obtained for sequencing of the EGFR gene to assess mutation status. In one sample, the patient was found to be heterozygous for 2235-2249 del15 in exon 19 of EGFR (Fig. 3A). In the same sample, EML4-ALK variant 3b was also present, in which exons 20 to 29 of ALK were connected to exon 6 of EML4, with an additional 33 bp insertion from intron 6 of EML4 (Fig. 3B, C). Histological adenocarcinoma was identified in this patient by morphological examination of hematoxylin and eosin-stained tissue samples (Fig. 3D).
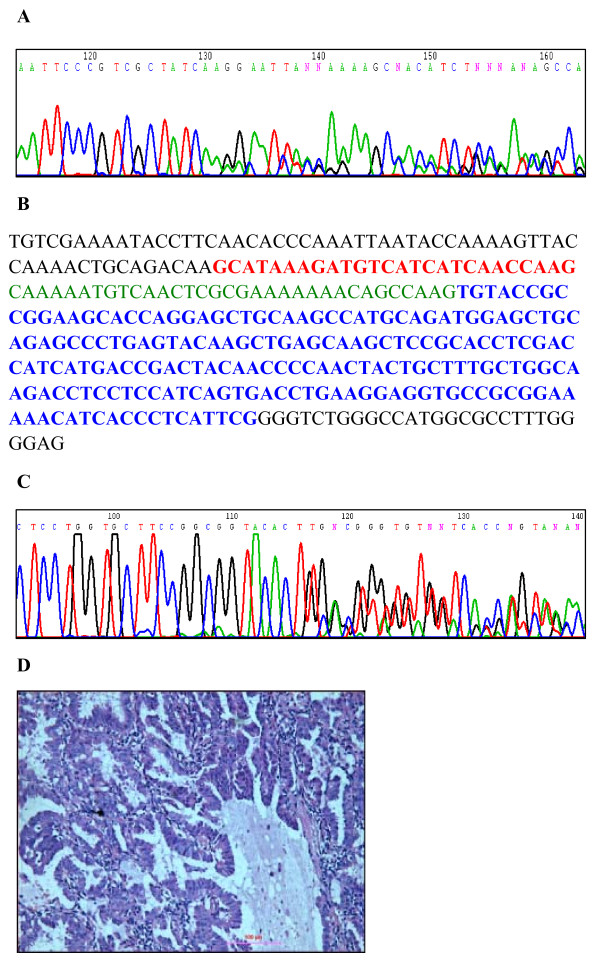
Figure 3.
Concurrent heterozygous ALK fusion and EGFR deletion in a single adenocarcinoma. (A) Chromatograph indicating a heterozygous deletion (2235-2249 del 15) in exon 19 of EGFR in a patient with EML4-ALK. (B) Sequence of EML4-ALK fusion variant 3b. Red sequence indicates exon 6 of EML4. Green sequence indicates an insertion of 33 bp from intron 6 of EML4 found in variant 3b. Blue sequence indicates exon 20 of ALK. (C) Chromatograph for EML4-ALK fusion variant 3b indicating the presence of a heterozygous fusion of ALK with exon 6 of EML4 with an additional 33 bp insertion. (D) Histological staining of the adenocarcinoma from the patient with both an EGFR mutation and EML4-ALK fusion. Scale bar = 100 μm.
Association of ALK fusion status with clinicopathological parameters
Of the 12 samples containing EML4-ALK fusions, 10 were identified as adenocarcinomas (one of which contained a mixed squamous carcinoma component), and two were identified as squamous cell carcinomas. In these patients, the presence of the ALK fusion was mutually exclusive with the presence of KRAS mutations. Notably, although the presence of ALK fusions was correlated with wild-type EGFR status (P = 0.04), we did identify one patient who had both the EML4-ALK fusion and an EGFR mutation.
To our knowledge, this is the first patient identified with a concurrent EGFR exon 19 deletion and the EML4-ALK fusion translocation. The concurrent mutations occurred in a female, non-smoking Chinese patient with a histological adenocarcinoma. EML4-ALK was significantly associated with non-smoking (P = 0.03). Patients with ALK fusions exhibited a significantly decreased number of smoking pack-years (5.0 vs. 18.5; P < 0.01) and were younger (53 vs. 61; P = 0.03) compared with patients without the fusion gene. No mutation in the kinase domain of ALK was detected by sequencing samples from the first group of 50 NSCLC cases collected consecutively (data not shown). Thus, mutation of ALK in NSCLC likely either does not occur or is rare. A trend towards improved survival was observed in the EML4-ALK cohort, though this was not statistically significant (P = 0.15; Fig. 4). ALK fusion-positive NSCLCs exhibited a hazard ratio of 0.54 (95% CI, 0.23-1.26) for overall survival.
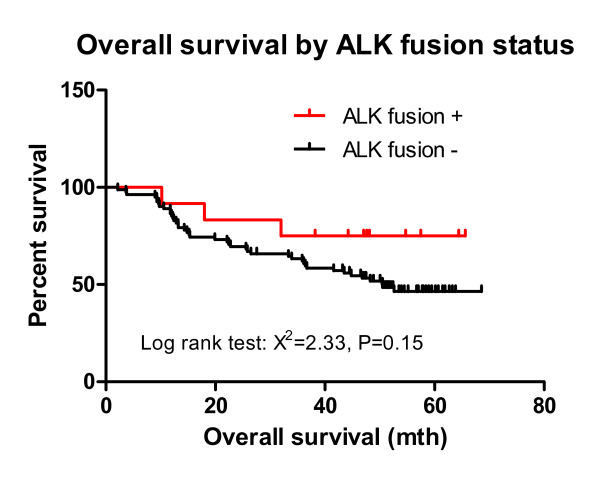
Figure 4.
Kaplan-Meier survival curve, stratified by ALK fusion status. Percent survival of patients with and without ALK fusions was plotted versus the number of months after surgery. Prior to the date of article submission, the ALK fusion-positive patients (n = 12) did not have enough events to determine the median overall survival, while the ALK fusion-negative patients (n = 101) had a median overall survival of 50.5 months. The hazard ratio for death in the ALK fusion-positive group was 0.54 (95% CI, 0.23-1.26). The log-rank (Mantel-Cox) test had a chi-squared value of 2.33 (P = 0.15).
Discussion
ALK belongs to the insulin receptor subfamily of receptor tyrosine kinases. Aberrant ALK activity has recently been shown to be present in anaplastic large cell lymphoma, as well as in solid tumors, including NSCLC [8,22]. Previous investigations have shown that translocation of ALK can result in fusion with the neighbouring gene, EML4, in cancer cells [19]. The fused genes then encode a fusion protein in which the intracellular tyrosine kinase domain of the ALK receptor is constitutively active. In all EML4-ALK fusion variants, the amino-terminal coiled-coil (CC) domain of EML4 has been shown to be retained in the fusion protein and is believed to be responsible for receptor dimerization and constitutive activation of the kinase domain (Fig. 5).
Figure 5.
Structure of the N-terminal exons of EML4. The coiled-coil domain (CC) is responsible for dimerization and constitutive activation of EML4-ALK; HELP, hydrophobic EMAP-like protein domain; WD, WD-repeat domain.
In the present study, the five transcript variants identified all possessed the CC domain and are, thus, likely to produce active, oncogenic EML4-ALK proteins in these NSCLC tumors. In future studies, we intend to investigate ALK kinase activity and activation of downstream signalling kinases in patient samples. These studies are particularly important, as data from Soda et al. [8] revealed that deletion of either the HELP or WD domains of EML4 can reduce kinase activity, by up to 50%. Thus, some variants may produce kinases with more activity than others.
To determine whether ALK fusions exhibit characteristic expression profiles at the mRNA level, we compared ALK expression based on microarray data (using the Affymetrix Genechip® human genome U133 plus 2.0 system) that corresponded to the same set of RNA samples. We found that ALK was expressed at higher levels in samples containing EML4-ALK, compared with samples that did not contain the fusion. Due to potential errors in microarray expression data arising from multiple correction procedures, real-time qRT-PCR should be performed to confirm ALK mRNA expression data. Additionally, use of immunohistochemistry could also confirm whether mRNA expression is correlated with EML4-ALK protein expression.
Careful review of the literature reveals that ALK fusion proteins are frequently present in lung cancer patients (Table 2). For example, Soda et al. found that the frequency of EML4-ALK fusion variants 1 and 2 was 6.7% (5/75) in a Japanese population using RT-PCR [20]. Taheuchi et al. [29] found a frequency of 4.35% (11/253) for EML4-ALK fusion variants 1-5 in Japanese patients using RT-PCR. Another RT-PCR-based study, by Koivunen et al. [30], showed that 3.6% (6/167) of Korean patients and 1.4% (2/136) of Caucasian patients with lung cancer possessed EML4-ALK variants 1, 3, and 4. Using a combination of FISH and RT-PCR, Perner et al. [31] found that 0.5% (3/603) of Caucasians with lung cancer have EML4-ALK variant 1. Wong et al. [32] found that EML4-ALK variants 1, 2, 3, and 9 (previously as "V5") were present at a frequency of 4.9% (13/266) in Chinese patients with lung cancer using RT-PCR. Also using RT-PCR, Martelli [33] found that 7.5% (9/120) of Italian and Spanish patients possessed EML4-ALK variants 1 and 3. In a study by Shaw and colleagues, NSCLC patients were selected for genetic screening on the basis of two or more of the following characteristics: female gender, Asian ethnicity, never/light smoking history, and adenocarcinoma histology. Analysis of EML4-ALK in these patients by FISH revealed a substantially higher frequency of the fusion (13%; 19/141) [34]. As is evident in these studies, RT-PCR and FISH represent the primary methods previously used to analyze the frequency of ALK fusions (Table 2).
Table 2.
ALK fusion variants previously reported in lung cancer.
| Citation | Freq. | Origin | Method | Number of Positives for each Fusion type | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| V1 | V2 | V3 | V4 | V5 | V6 | V7 | V8 | V9 | ||||
| Soda, et al. Nature 2007 | 6.7% (5/75) |
Japanese | RT-PCR | 3 | 2 | |||||||
|
Takeuchi et al. Clin Cancer Res. 2008 |
4.35% (11/253) |
Japanese | RT-PCR | 3 | 3 | 3 | 1 | 1 | ||||
|
Takeuchi et al. Clin Cancer Res. 2009 |
/ | Japanese | iAEP/RT-PCR | 1 | 1 | 3 | 1 | 1 | ||||
|
Koivunen et al. Clin Cancer Res. 2008 |
3.6% (6/167) | Korean | RT-PCR | 2 | 4 | 2 | ||||||
| 1.4% (2/136) | Caucasian | |||||||||||
| Inamura et al. J Thorac Oncol. 2008 | 2.3% (5/221) | Japanese | RT-PCR | 2 | 3 | |||||||
| Perner et al. Neoplasia 2008 | 0.5% (3/603) | Caucasian | FISH, RT-PCR | 3 | ||||||||
|
Wong et al. Cancer. 2009 |
4.9% (13/266) | Chinese | RT-PCR | 2 | 2 | 8 | 1 | |||||
|
Martelli et al. Am J Pathol. 2009 |
7.5% (9/120) | Italian and Spanish | RT-PCR | 7 | 2 | |||||||
| Present study |
11.6% (12/103) |
Chinese | RACE-coupled PCR sequenc-ing | 4 | 1 | 3 | 1 | 3 | ||||
Our data, which was based on non-selective hospital-obtained samples, revealed a frequency of 11.6% (12/103) for EML4-ALK fusion variants 1, 2, 3, 5, and 9 in Chinese NSCLC patients using RACE-coupled PCR sequencing technology. This rather high frequency of ALK fusion may be attributable the high sensitivity of RACE-PCR sequencing assays. Use of this technique has also confirmed that, in non-small cell lung cancer, the fusion of ALK appears to occur solely with EML4, as previously reported [35], [36], rather than with other genes.
Comparison of ALK fusion status with results characterizing the presence of EGFR and KRAS mutations in the same set of cancer samples revealed that ALK fusions occurred in the absence of KRAS mutations. Previous reports have indicated that ALK fusion can occur concurrently with EGFR mutations (1/305) and KRAS mutations (1/120), although these may be rare events [30,33]. Consistent with these observations, we also identified a patient with concurrent EGFR exon 19 deletion and EML4-ALK fusion. The patient, who was female, non-smoking, and Chinese with histological adenocarcinoma, exhibited post-surgery overall survival of more than 38 months. This observation was particularly interesting because of the abnormality of mutations in two potential "tumor-driving" receptor genes in a single tumor. Whether tumor progression is dependent on both kinases, or only one of the two receptors, is unclear in this case. Thus, clarification of the actual activation status of the two receptors is important in assessing the use of targeted therapies, including EGFR inhibitors and/or ALK inhibitors. Notably, the frequency of ALK fusions in patients with adenocarcinomas that did not exhibit EGFR/KRAS mutations reached 45.0% (9/21), strongly indicating that only a specific molecular subset of adenocarcinomas is characterized by EML4-ALK fusion. Thus, stratification of patients with ALK fusions may be useful in the development of personalized medical treatments for NSCLC.
Characterization of associations between ALK fusion status and clinicopathological variables revealed that EML4-ALK fusions were associated with adenocarcinoma, because fusions were identified in nine cases of adenocarcinoma, two cases of squamous carcinoma, and one case of adenocarcinoma with partial squamous carcinoma components. EML4-ALK fusion was also associated with non-smokers (P = 0.02), and patients who possessed the ALK fusion had significantly less smoking pack-years (6.9 vs. 19.1; P = 0.01). Grouping of patients revealed that the frequency of the EML4-ALK fusion was 16.13% (10/62) in patients with adenocarcinomas and 19.23% (10/52) in never-smokers. Patients with EML4-ALK fusions were also significantly younger (53 vs. 61; P = 0.03) in comparison to patients without the fusion gene. Results showing that ALK fusion is frequently associated with adenocarcinoma and younger onset of disease were consistent with previous reports [37].
No mutation in ALK was identified by sequencing samples from 50 NSCLC cases (data not shown). However, patients in the EML4-ALK cohort appeared to exhibit increased survival, compared to patients without the fusion, although this was not statistically significant (P = 0.15), with a hazard ratio of 0.54 (95% CI, 0.23-1.26) for overall survival. Considering that mutated EGFR is used as a prognostic indicator, better prognosis may be associated with the presence of EML4-ALK fusions in NSCLC, suggesting that EML4-ALK may represent an effective prognostic factor, similar to EGFR.
Recently, ALK fusion was reported to play a role in resistance to EGFR tyrosine kinase inhibitors (TKIs) [34]. However, cancers characterized by expression of ALK fusions are clinically-sensitive to specific ALK inhibitors [25,26]. Thus, ALK fusions may be useful as biomarkers for predicting the response to ALK TKI and resistance to EGFR TKI. Immunohistochemical analysis of ALK expression could serve as a screening tool and potential surrogate technique to assess ALK fusion status. Though the correlation between ALK gene rearrangement and EML4-ALK mRNA and protein levels has been disputed [33], [38], [39], our data suggests that ALK fusion did correlate with increased mRNA expression.
In the present study, patients with ALK fusions did not receive TKI treatment. However, our results, together with those from previous studies, indicate that correlations between ALK fusion status and the efficacy of EGFR TKI and ALK TKI require further investigation in a larger patient cohort. Concurrent EGFR mutation and EML4-ALK fusion in one patient in this study (1/12), together with data from previous reports [30,33], also suggests that treatment efficacy should also be assessed in this group of patients. Given the reported resistance of tumors with ALK fusions to EGFR TKI, in rare cases with concurrent ALK fusion and EGFR/KRAS mutation, the molecular heterogeneity of these altered oncogenes and potential cross-talk between their protein products requires further investigation. Results from such studies would be useful in the development of sequential or concurrent target therapies.
In summary, RACE-coupled PCR sequencing can serve as sensitive tool to identify ALK fusions with a variety of potential partners in patient samples. However, in the present study of NSCLC samples, only EML4 was identified as a partner protein. A high frequency (11.6%) of NSCLC samples exhibited expression of different EML4-ALK transcript variants. Patients with ALK fusions appeared to exhibit a trend towards improved survival in comparison to patients without fusions. Notably, the presence of ALK fusions was associated with histological adenocarcinomas and with wild-type EGFR and KRAS status, which may be of clinical relevance in targeted therapy using ALK inhibitors.
Conclusions
A high frequency of the EML4-ALK fusion gene was present in Chinese patients with NSCLC, particularly in patients with adenocarcinomas without EGFR/KRAS mutations. ALK fusion status was significantly associated with ALK mRNA expression levels. RACE-coupled PCR sequencing was highly sensitive and could serve as a method for identifying EML4-ALK fusion patients for targeted therapies in the clinic.
Methods
Lung cancer patients
In total, 103 NSCLC specimens were obtained from patients who underwent surgery for NSCLC from 2004 to 2006 at Guangdong General Hospital in the Guangdong Lung Cancer Institute of the Guangdong Academy of Medical Sciences. NSCLC samples included 62 adenocarcinomas, 29 squamous cell carcinomas, 11 large cell carcinomas, and 1 smooth muscle sarcoma.
Informed consent to use resected tissue for genetic analysis was obtained for all patients. The study was approved by the Institutional Review Boards (IRBs) of Guangdong General Hospital.
Demographic and clinicopathological profiles for the cases are shown in Table 3. Staging and histological classification were based on the World Health Organization (WHO) system. All 103 cases were analyzed for ALK fusion status, and both EGFR/KRAS mutation data and ALK fusion data were obtained for 96 of the 103 cases.
Table 3.
Summary of patient demographic and clinicopathological profiles.
| Characteristic | Number of Patients | ALK fusion | ||
|---|---|---|---|---|
| Positive | Negative | P-value | ||
| No. of patients | 103 | 12 (11.6%) | 91 | |
| Age (years; mean ± SD) | 52 (< 61 y) | 8 | 44 | 0.37 |
| 53 (≥ 61 y) | 4 | 39 | ||
| Gender | ||||
| Male | 74 (71.8%) | 7 | 67 | 0.27 |
| Female | 29 (28.2%) | 5 | 24 | |
| Smoking | ||||
| Smokers | 51 (49.5%) | 2 | 47 | 0.03 |
| Non-smokers | 52 (50.5%) | 10 | 44 | |
| Histology | ||||
| Adenocarcinoma | 62 (60.2%) | 10 | 52 | 0.08* |
| Squamous cell | 29 (28.1%) | 2 | 27 | |
| carcinoma | 11 (10.7%) | 0 | 11 | |
| Large-cell | 1 (1.0%) | 0 | 1 | |
| carcinoma | ||||
| Others | ||||
| Stage | ||||
| I | 63 (61.2%) | 8 | 55 | 0.72 |
| II | 18 (17.5%) | 1 | 17 | |
| III | 20 (19.4%) | 3 | 17 | |
| IV | 2 (1.9%) | 0 | 2 | |
* AC compared with combined SCC, LCC, and other histological types.
Nucleic acid extraction
Total RNA and DNA were extracted from lung tissue samples using the RNeasy kit (QIAGEN, Valencia, CA) and DNeasy kit (QIAGEN), respectively. All samples were pathologically assessed and trimmed at low temperature to ensure that more than 80% of the sample consisted of tumor tissue, while protecting the RNA from degradation. RNA quality was assessed by gel electrophoresis using an Agilent bio-analyzer 2100 (with RNA integrity number [RIN] > 6). DNA quality was assessed by routine gel electrophoresis.
ALK fusion analysis by RACE-coupled PCR sequencing
This methodology is based on the principles of 5'-RACE and RT-PCR. Briefly, an ALK gene-specific primer was used to reverse transcribe RNAs into cDNAs. The sequence of the primer was 5'-TTCAGGCAGCGTGTTCACAGCCA-3'. Reverse transcription was performed based on the manufacturer's recommended protocol for the TaKaRa RNA PCR (AMV) kit, Ver. 3.0 (Takara). Briefly, the RT reaction consisted of 2 μL MgCl2, 1 μL 10 × RT buffer, 3.75 μL H2O, 1 μL 10 mM dNTPs, 0.25 μL RNase inhibitor, 0.5 μL AMV, 0.5 μL 12.5 μM gene-specific primer, and 1 μL RNA (< 500 ng total RNA). The reaction was incubated at 42°C for 30 min, 99°C for 5 min, and 5°C for 5 min. After reverse transcription, cDNAs were purified using the High Pure PCR Product Purification Kit (Roche) according to the manufacturer's protocol. Purified cDNAs were further subjected to poly-cytidine (poly-C) tailing. Briefly, reactions containing 1.5 μL 1% BSA, 5.0 μL 5 × tailing buffer, 2.5 μL 2 mM dCTP, and 15.0 μL purified cDNA were gently mixed and incubated for 2-3 min at 94°C, followed by incubation for 2 min on ice. After the addition of 1 μL TdT, the reactions were incubated for an additional 30 min at 37°C. TdT was then inactivated by incubation at 65°C for 10 min, and the contents of the reaction were collected by brief centrifugation and placed on ice.
Two PCR reactions were performed to amplify target cDNA fragments spanning exon 20 of ALK and upstream sequences that may contain transcript sequences for any genes fused to ALK. Primers for the first and PCR reactions were as follows: forward primer 5'-GGCCA CGCGTCCACTAGTACGGGGGGGGGG-3' and reverse primer 5'-GGCACCTCCTTCAC GTCACTGATG-3' and forward primer 5'-GGCCAC GCGTCGA CTAGTAC-3' and reverse primer 5'-ACCAGGAAACAGCTAT GACCGGTCTTG CCAGCAAAGCAGTAG TTG -3', respectively. PCR reactions were performed according to the manufacturer's instructions for the HS PCR kit (Takara). PCR products were then purified and labelled using the BDT v3.1 Ready Reaction-5000 and SEQ Buffer 5000 Ready Reaction (Applied Biosystems) using the M13 sequencing primer, 5'-CAGGAAACAGCT ATGACC-3'. Sequencing was performed on a Genetic Analyzer 3730XL (Applied Biosystems). Target sequences of interest were aligned with the ALK reference sequence (NM_004304.3) to determine if a fusion with another gene was present.
EGFR mutation analysis by direct sequencing
Genomic DNA from each sample was used for sequence analysis of EGFR exons 18, 19, 20, and 21. These exons were amplified by PCR as previously reported [40], and the resulting PCR products were purified and labelled for sequencing according to the manufacturer's protocol for the BigDye 3.1 kit (Applied Biosystems).
Statistical analyses
Statistical analysis of associations between ALK fusion status and clinical factors was performed using the chi-squared test or Fisher's exact test. Student's t-test was used for comparisons between means. Pearson's correlation coefficient was calculated when assessing relationships between continuous variables. Kaplan-Meier survival curves were generated and the log-rank test was used to test the significance of differential survival. P values less than 0.05 were deemed to indicate statistical significance.
Competing interests
X. Zhang is the vice president and head of the AstraZeneca Innovation Center China (ICC). He is a member of the AstraZeneca global senior management team for the cancer and infection research areas. M. Zhu and L. Yin are senior scientists, and S. Zhang is a basic scientist at the AstraZeneca Innovation Center China. A patent application has been submitted for the RACE-coupled PCR sequencing method for ALK fusion detection.
Authors' contributions
XZ performed experiments and prepared the manuscript. SZ, LY, JA, ZX, JL, and SC performed experiments. XY, JY, and QZ helped to prepare tissue samples and obtained informed consent from the patients. XZ, MZ, and YW designed the experiments, supervised the project, and prepared the manuscript. All authors read and approved the final manuscript.
Note
The English in this document has been checked by at least two professional editors, both native speakers of English. For a certificate, please see:
Contributor Information
Xuchao Zhang, Email: zhxuchao3000@yahoo.com.cn.
Shirley Zhang, Email: Shirley.Zhang@astrazeneca.com.
Xuening Yang, Email: yangxuening@gmail.com.
Jinji Yang, Email: yangjinji2003@163.com.
Qing Zhou, Email: gzzhouqing@126.com.
Lucy Yin, Email: Lucy.Yin@astrazeneca.com.
Shejuan An, Email: asjuan@126.com.
Jiaying Lin, Email: gzlinjiaying3@yahoo.com.cn.
Shiliang Chen, Email: 295591612@qq.com.
Zhi Xie, Email: xiezi8045@sina.com.
Mike Zhu, Email: mike.zhu@astrazeneca.com.
Xiaolin Zhang, Email: xiaolin.zhang@astrazeneca.com.
Yi-long Wu, Email: syylwu@live.cn.
Acknowledgements
This work was supported by the Joint Research Program of the Guangdong General Hospital-AstraZeneca Innovation Center China Joint Laboratory. This work was also supported by the following grants: National Natural Science Foundation of China Grant No. 30772531 (to Y. Wu); Guangdong Science and Technology Department, GDSTD Industry Technology R&D Project No. 2007A032000002 (to Y. Wu); Key Science and Technology Project of Guangzhou Science and Technology Bureau No. 2007Z2-0081 (to Y. Wu); National "863" Division of Keystone Project No. 2006AA02A401 (to Y. Wu); and Key Science and Technology Project of Guangzhou Science and Technology Bureau No. 2006B60101010 (to Y. Wu).
References
- Morris SW, Kirstein MN, Valentine MB, Dittmer KG, Shapiro DN, Saltman DL, Look AT. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin's lymphoma. Science. 1994;263:1281–1284. doi: 10.1126/science.8122112. [DOI] [PubMed] [Google Scholar]
- Palmer RH, Vernersson E, Grabbe C, Hallberg B. Anaplastic lymphoma kinase: signalling in development and disease. Biochemical Journal. 2009;420:345–361. doi: 10.1042/BJ20090387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polgar D, Leisser C, Maier S, Strasser S, Ruger B, Dettke M, Khorchide M, Simonitsch I, Cerni C, Krupitza G. Truncated ALK derived from chromosomal translocation t(2;5)(p23;q35) binds to the SH3 domain of p85-PI3K. Mutation Research. 2005;570:9–15. doi: 10.1016/j.mrfmmm.2004.09.011. [DOI] [PubMed] [Google Scholar]
- Coffin CM, Hornick JL, Fletcher CD. Inflammatory myofibroblastic tumor: comparison of clinicopathologic, histologic, and immunohistochemical features including ALK expression in atypical and aggressive cases. American Journal of Surgical Pathology. 2007;31:509–520. doi: 10.1097/01.pas.0000213393.57322.c7. [DOI] [PubMed] [Google Scholar]
- Chen Y, Takita J, Choi YL, Kato M, Ohira M, Sanada M, Wang L, Soda M, Kikuchi A, Igarashi T. Oncogenic mutations of ALK kinase in neuroblastoma. Nature. 2008;455:971–974. doi: 10.1038/nature07399. [DOI] [PubMed] [Google Scholar]
- George RE, Sanda T, Hanna M, Frohling S, Luther W, Zhang J, Ahn Y, Zhou W, London WB, McGrady P. Activating mutations in ALK provide a therapeutic target in neuroblastoma. Nature. 2008;455:975–978. doi: 10.1038/nature07397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janoueix-Lerosey I, Lequin D, Brugieres L, Ribeiro A, de Pontual L, Combaret V, Raynal V, Puisieux A, Schleiermacher G, Pierron G. Somatic and germline activating mutations of the ALK kinase receptor in neuroblastoma.[see comment] Nature. 2008;455:967–970. doi: 10.1038/nature07398. [DOI] [PubMed] [Google Scholar]
- Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K, Hatanaka H. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer.[see comment] Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- Tort F, Campo E, Pohlman B, Hsi E. Heterogeneity of genomic breakpoints in MSN-ALK translocations in anaplastic large cell lymphoma. Human Pathology. 2004;35:1038–1041. doi: 10.1016/j.humpath.2004.05.006. [DOI] [PubMed] [Google Scholar]
- Kinoshita Y, Tajiri T, Ieiri S, Nagata K, Taguchi T, Suita S, Yamazaki K, Yoshino I, Maehara Y, Kohashi K. A case of an inflammatory myofibroblastic tumor in the lung which expressed TPM3-ALK gene fusion. Pediatric Surgery International. 2007;23:595–599. doi: 10.1007/s00383-006-1821-6. [DOI] [PubMed] [Google Scholar]
- Armstrong F, Lamant L, Hieblot C, Delsol G, Touriol C. TPM3-ALK expression induces changes in cytoskeleton organisation and confers higher metastatic capacities than other ALK fusion proteins. European Journal of Cancer. 2007;43:640–646. doi: 10.1016/j.ejca.2006.12.005. [DOI] [PubMed] [Google Scholar]
- Ma Z, Cools J, Marynen P, Cui X, Siebert R, Gesk S, Schlegelberger B, Peeters B, De Wolf-Peeters C, Wlodarska I, Morris SW. Inv(2)(p23q35) in anaplastic large-cell lymphoma induces constitutive anaplastic lymphoma kinase (ALK) tyrosine kinase activation by fusion to ATIC, an enzyme involved in purine nucleotide biosynthesis. Blood. 2000;95:2144–2149. [PubMed] [Google Scholar]
- Trinei M, Lanfrancone L, Campo E, Pulford K, Mason DY, Pelicci PG, Falini B. A new variant anaplastic lymphoma kinase (ALK)-fusion protein (ATIC-ALK) in a case of ALK-positive anaplastic large cell lymphoma. Cancer Research. 2000;60:793–798. [PubMed] [Google Scholar]
- Colleoni GW, Bridge JA, Garicochea B, Liu J, Filippa DA, Ladanyi M. ATIC-ALK: A novel variant ALK gene fusion in anaplastic large cell lymphoma resulting from the recurrent cryptic chromosomal inversion, inv(2)(p23q35) American Journal of Pathology. 2000;156:781–789. doi: 10.1016/S0002-9440(10)64945-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez L, Pinyol M, Hernandez S, Bea S, Pulford K, Rosenwald A, Lamant L, Falini B, Ott G, Mason DY. TRK-fused gene (TFG) is a new partner of ALK in anaplastic large cell lymphoma producing two structurally different TFG-ALK translocations. Blood. 1999;94:3265–3268. [PubMed] [Google Scholar]
- Cools J, Wlodarska I, Somers R, Mentens N, Pedeutour F, Maes B, De Wolf-Peeters C, Pauwels P, Hagemeijer A, Marynen P. Identification of novel fusion partners of ALK, the anaplastic lymphoma kinase, in anaplastic large-cell lymphoma and inflammatory myofibroblastic tumor. Genes, Chromosomes & Cancer. 2002;34:354–362. doi: 10.1002/gcc.10033. [DOI] [PubMed] [Google Scholar]
- Touriol C, Greenland C, Lamant L, Pulford K, Bernard F, Rousset T, Mason DY, Delsol G. Further demonstration of the diversity of chromosomal changes involving 2p23 in ALK-positive lymphoma: 2 cases expressing ALK kinase fused to CLTCL (clathrin chain polypeptide-like) Blood. 2000;95:3204–3207. [PubMed] [Google Scholar]
- Takeuchi K, Choi YL, Togashi Y, Soda M, Hatano S, Inamura K, Takada S, Ueno T, Yamashita Y, Satoh Y. KIF5B-ALK, a novel fusion oncokinase identified by an immunohistochemistry-based diagnostic system for ALK-positive lung cancer. Clinical Cancer Research. 2009;15:3143–3149. doi: 10.1158/1078-0432.CCR-08-3248. [DOI] [PubMed] [Google Scholar]
- Mano H. Non-solid oncogenes in solid tumors: EML4-ALK fusion genes in lung cancer. Cancer Science. 2008;99:2349–2355. doi: 10.1111/j.1349-7006.2008.00972.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollmann M, Parwaresch R, Adam-Klages S, Kruse ML, Buck F, Heidebrecht HJ. Human EML4, a novel member of the EMAP family, is essential for microtubule formation. Experimental Cell Research. 2006;312:3241–3251. doi: 10.1016/j.yexcr.2006.06.035. [DOI] [PubMed] [Google Scholar]
- Houtman SH, Rutteman M, De Zeeuw CI, French PJ. Echinoderm microtubule-associated protein like protein 4, a member of the echinoderm microtubule-associated protein family, stabilizes microtubules. Neuroscience. 2007;144:1373–1382. doi: 10.1016/j.neuroscience.2006.11.015. [DOI] [PubMed] [Google Scholar]
- Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, Nardone J, Lee K, Reeves C, Li Y. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- Horn L, Pao W. EML4-ALK: honing in on a new target in non-small-cell lung cancer.[comment] Journal of Clinical Oncology. 2009;27:4232–4235. doi: 10.1200/JCO.2009.23.6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soda M, Takada S, Takeuchi K, Choi YL, Enomoto M, Ueno T, Haruta H, Hamada T, Yamashita Y, Ishikawa Y. A mouse model for EML4-ALK-positive lung cancer. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:19893–19897. doi: 10.1073/pnas.0805381105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak EL, Camidge DR, Clark J, Shapiro GI, Maki RG, Ratain MJ, Solomon B, Bang Y, Ou S, Salgia R. Clinical activity observed in a phase I dose escalation trial of an oral c- met and ALK inhibitor, PF-02341066. Journal of Clinical Oncology. 2009;27:15s. doi: 10.1200/JCO.2008.21.7695. (suppl; abstr 3509):15s. [DOI] [Google Scholar]
- Bang Y, Kwak EL, Shaw AT, Camidge DR, Iafrate AJ, Maki RG, Solomon BJ, Ou SI, Salgia R, Clark JW. Clinical activity of the oral ALK inhibitor PF-02341066 in ALK-positive patients with non-small cell lung cancer (NSCLC) Journal of Clinical Oncology. 2010;28:7s. doi: 10.1200/JCO.2009.25.9937. (suppl; abstr 3):7s. [DOI] [Google Scholar]
- Solomon B, Varella-Garcia M, Camidge DR. ALK gene rearrangements: a new therapeutic target in a molecularly defined subset of non-small cell lung cancer. Journal of Thoracic Oncology: Official Publication of the International Association for the Study of Lung Cancer. 2009;4:1450–1454. doi: 10.1097/JTO.0b013e3181c4dedb. [DOI] [PubMed] [Google Scholar]
- Tan LH, Do E, Chong SM, Koay ES. Detection of ALK gene rearrangements in formalin-fixed, paraffin-embedded tissue using a fluorescence in situ hybridization (FISH) probe: a search for optimum conditions of tissue archiving and preparation for FISH. Molecular Diagnosis. 2003;7:27–33. doi: 10.2165/00066982-200307010-00005. [DOI] [PubMed] [Google Scholar]
- Takeuchi K, Choi YL, Soda M, Inamura K, Togashi Y, Hatano S, Enomoto M, Takada S, Yamashita Y, Satoh Y. Multiplex reverse transcription-PCR screening for EML4-ALK fusion transcripts. Clinical Cancer Research. 2008;14:6618–6624. doi: 10.1158/1078-0432.CCR-08-1018. [DOI] [PubMed] [Google Scholar]
- Koivunen JP, Mermel C, Zejnullahu K, Murphy C, Lifshits E, Holmes AJ, Choi HG, Kim J, Chiang D, Thomas R. EML4-ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer. Clinical Cancer Research. 2008;14:4275–4283. doi: 10.1158/1078-0432.CCR-08-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perner S, Wagner PL, Demichelis F, Mehra R, Lafargue CJ, Moss BJ, Arbogast S, Soltermann A, Weder W, Giordano TJ. EML4-ALK fusion lung cancer: a rare acquired event. Neoplasia (New York) 2008;10:298–302. doi: 10.1593/neo.07878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong DW, Leung EL, So KK, Tam IY, Sihoe AD, Cheng LC, Ho KK, Au JS, Chung LP, Pik Wong M. University of Hong Kong Lung Cancer Study G. The EML4-ALK fusion gene is involved in various histologic types of lung cancers from nonsmokers with wild-type EGFR and KRAS. Cancer. 2009;115:1723–1733. doi: 10.1002/cncr.24181. [DOI] [PubMed] [Google Scholar]
- Martelli MP, Sozzi G, Hernandez L, Pettirossi V, Navarro A, Conte D, Gasparini P, Perrone F, Modena P, Pastorino U. EML4-ALK rearrangement in non-small cell lung cancer and non-tumor lung tissues. American Journal of Pathology. 2009;174:661–670. doi: 10.2353/ajpath.2009.080755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw AT, Yeap BY, Mino-Kenudson M, Digumarthy SR, Costa DB, Heist RS, Solomon B, Stubbs H, Admane S, McDermott U. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK.[see comment] Journal of Clinical Oncology. 2009;27:4247–4253. doi: 10.1200/JCO.2009.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inamura K, Takeuchi K, Togashi Y, Hatano S, Ninomiya H, Motoi N, Mun MY, Sakao Y, Okumura S, Nakagawa K. EML4-ALK lung cancers are characterized by rare other mutations, a TTF-1 cell lineage, an acinar histology, and young onset. Modern Pathology. 2009;22:508–515. doi: 10.1038/modpathol.2009.2. [DOI] [PubMed] [Google Scholar]
- Shinmura K, Kageyama S, Tao H, Bunai T, Suzuki M, Kamo T, Takamochi K, Suzuki K, Tanahashi M, Niwa H. EML4-ALK fusion transcripts, but no NPM-, TPM3-, CLTC-, ATIC-, or TFG-ALK fusion transcripts, in non-small cell lung carcinomas. Lung Cancer. 2008;61:163–169. doi: 10.1016/j.lungcan.2007.12.013. [DOI] [PubMed] [Google Scholar]
- Inamura K, Takeuchi K, Togashi Y, Nomura K, Ninomiya H, Okui M, Satoh Y, Okumura S, Nakagawa K, Soda M. EML4-ALK fusion is linked to histological characteristics in a subset of lung cancers. Journal of Thoracic Oncology: Official Publication of the International Association for the Study of Lung Cancer. 2008;3:13–17. doi: 10.1097/JTO.0b013e31815e8b60. [DOI] [PubMed] [Google Scholar]
- Choi YL, Takeuchi K, Soda M, Inamura K, Togashi Y, Hatano S, Enomoto M, Hamada T, Haruta H, Watanabe H. Identification of novel isoforms of the EML4-ALK transforming gene in non-small cell lung cancer. Cancer Research. 2008;68:4971–4976. doi: 10.1158/0008-5472.CAN-07-6158. [DOI] [PubMed] [Google Scholar]
- Boland JM, Erdogan S, Vasmatzis G, Yang P, Tillmans LS, Johnson MR, Wang X, Peterson LM, Halling KC, Oliveira AM. Anaplastic lymphoma kinase immunoreactivity correlates with ALK gene rearrangement and transcriptional up-regulation in non-small cell lung carcinomas. Human Pathology. 2009;40:1152–1158. doi: 10.1016/j.humpath.2009.01.012. [DOI] [PubMed] [Google Scholar]
- Jiang SX, Yamashita K, Yamamoto M, Piao CJ, Umezawa A, Saegusa M, Yoshida T, Katagiri M, Masuda N, Hayakawa K, Okayasu I. EGFR genetic heterogeneity of nonsmall cell lung cancers contributing to acquired gefitinib resistance. International Journal of Cancer. 2008;123:2480–2486. doi: 10.1002/ijc.23868. [DOI] [PubMed] [Google Scholar]