Abstract
Objective
To consider recent findings from quantitative genetic research in the context of molecular genetic research, especially genome-wide association studies. We focus on findings that go beyond merely estimating heritability. We use learning abilities and disabilities as examples.
Method
Recent twin research in the area of learning abilities and disabilities was reviewed.
Results
Three findings from quantitative genetic research stand out for their far-reaching implications for child and adolescent psychiatry. First, common disorders such as learning difficulties are the quantitative extreme of the same genetic factors responsible for genetic influence throughout the normal distribution (the Common Disorders are Quantitative Traits Hypothesis). Second, the same set of genes is largely responsible for genetic influence across diverse learning and cognitive abilities and disabilities (the Generalist Genes Hypothesis). Third, experiences are just as influenced genetically as are behaviors and genetic factors mediate associations between widely used measures of the environment and behavioural outcomes (the Nature of Nurture Hypothesis).
Conclusions
Quantitative genetics can go far beyond the rudimentary ‘how much’ question about nature versus nurture, and can continue to provide important findings in the era of molecular genetics.
Keywords: Quantitative genetics, molecular genetics, twin studies, learning abilities, disabilities
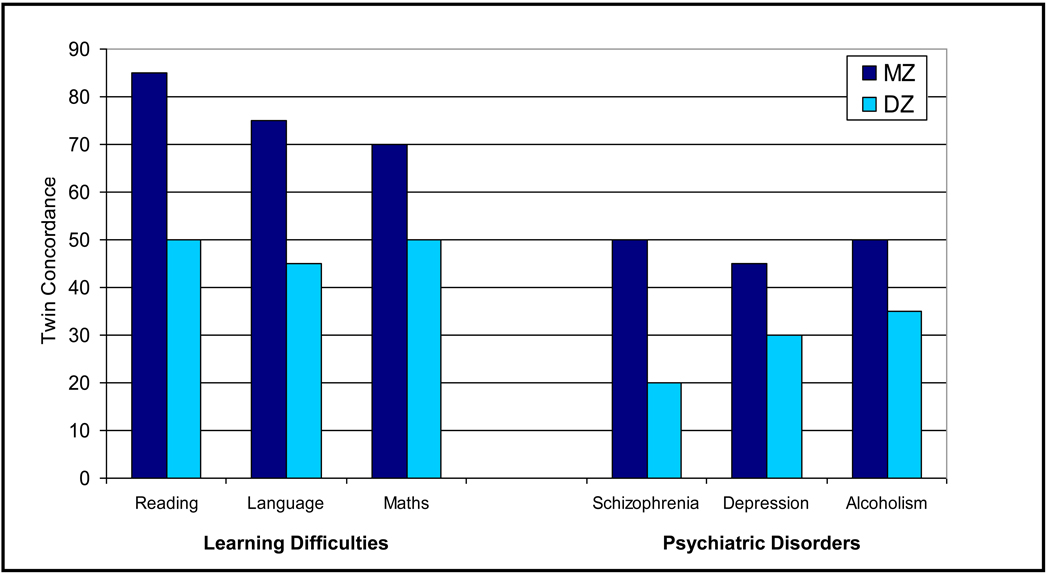
Quantitative genetic research--strain and selection studies in nonhuman animals and twin and adoption studies in our species--has demonstrated the ubiquitous importance of genetic influence on behavioral dimensions and disorders1. For learning disabilities, MZ and DZ twin concordances are about 85% and 50% respectively for reading; 75% and 45% for language; and 70% and 50% for mathematics2. These results indicate substantial genetic influence on learning difficulties, which is greater than for most other common psychiatric disorders. Figure 1 compares results for learning difficulties to those for three psychiatric disorders: schizophrenia (MZ=50%; DZ=20%); depression (45%; 30%); and alcoholism (50%; 35%)1. For the entire range of learning abilities rather than disabilities, heritability estimates are typically about 50%, meaning that about half of the variance in learning abilities can be attributed to genetic differences3. In terms of public acceptance of these findings, a large UK survey indicated that more than 90% of teachers and parents say that they believe genetics to be at least as important as the environment for learning abilities and disabilities4.
Figure 1.
MZ and DZ twin concordances of learning disabilities and for psychiatric disorders. Data extracted from review by Plomin et al.,1.
Note: DZ = Dizygotic; MZ = Monozygotic
Quantitative genetic methods estimate the cumulative effect of genetic influence regardless of the number of genes involved or the magnitude or complexity of their effects. If we could find the genes responsible for heritability there would be no more need for quantitative genetic designs because genetic influence could be assessed directly from each individual’s DNA rather than implied indirectly by genetic relatedness as in twin and adoption studies. However, although genome-wide association (GWA) research has had many successes5, it seems highly unlikely that most of the genes responsible for the heritability for any complex trait will be identified in the foreseeable future. The reason is that the largest effect sizes found in GWA efforts to date are very small, which means that many such genes of even smaller effect size will be needed to account for heritability6.
The largest effect sizes of replicable associations from GWA studies are odds-ratios of about 1.2 for case-control studies of disorders and less than 1% of the population variance for quantitative traits. These effect sizes are so small that samples in the thousands are needed to identify replicable associations, for example, in case-control studies of schizophrenia7, type 2 diabetes8 and obesity9, and in studies of quantitative traits such as lipids10 and height11. For this reason, it is not surprising that the first GWA study of reading ability that was powered to detect effect sizes of about 1% of the variance was unable to detect reliable associations of this magnitude with a sample of 400012. Similarly, GWA studies of cognitive abilities were unable to detect reliable associations in studies with 700 subjects13 and 3000 subjects14. Significant associations with cognition and memory reported in one GWA study with 350 subjects15,16 have not been replicated17.
If the largest effect sizes of replicable associations are so small, hundreds of genes of very small effect size will be needed to account for heritability which is typically about 50%. Moreover, finding the rest of the associations with even smaller effect sizes seems a daunting task--this has been called ‘the missing heritability’ problem18. For this reason, molecular genetics seems unlikely to replace quantitative genetics in the foreseeable future. Nonetheless, we hope that our prediction about GWA research is wrong and that it will be possible to identify most of the missing heritability, which coupled with decreasing genotyping costs, would put quantitative genetics out of business. This hope is not unrealistic in the long term: GWA research only began in 2007 and has only searched the genome for common single-nucleotide polymorphisms (SNPs). Hope for finding the missing heritability springs from the rapid pace of developments in GWA research which includes other types of polymorphisms such as copy-number variants (CNVs), other rare variants, non-coding RNA and the entire genome sequence which will capture variants of any type19. Moreover, finding any replicable associations between DNA variants and behavior is useful for research purposes as in the case of the FTO gene which is associated with body weight and obesity, and is a highly replicated finding across multiple studies20.
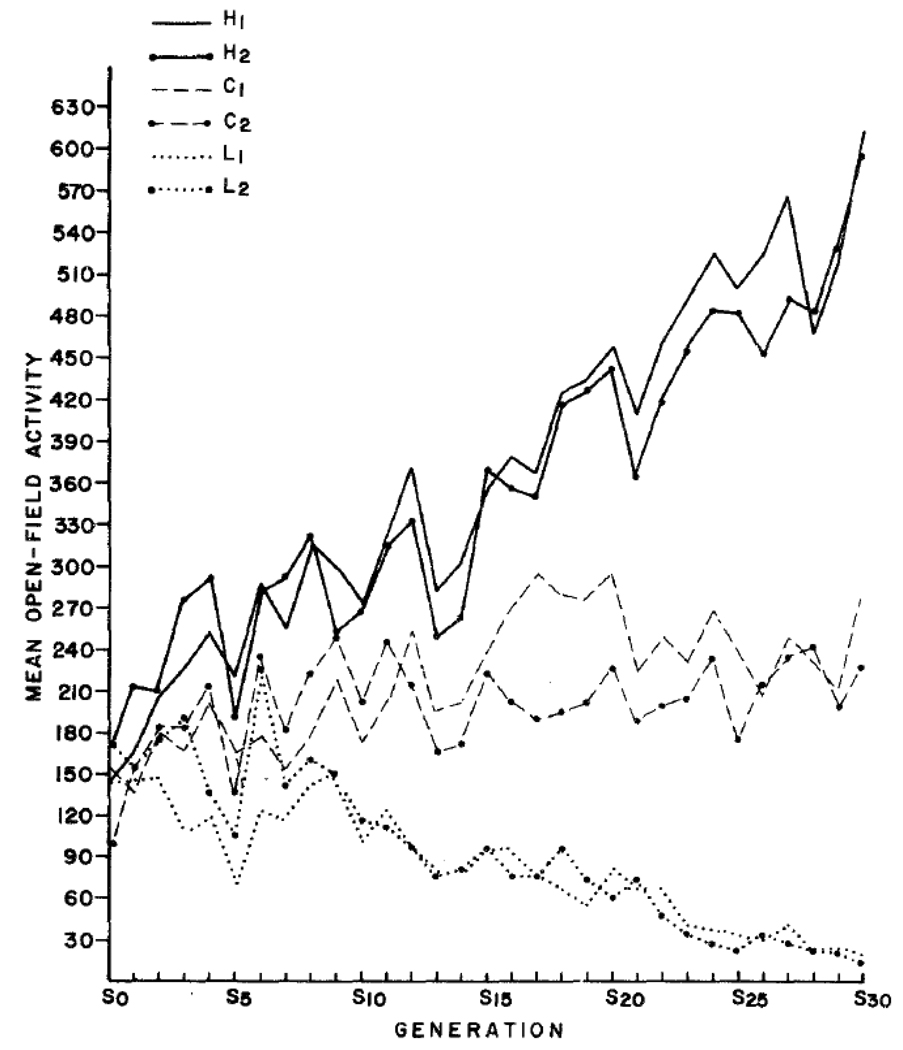
The GWA finding that many genes of small effect size are responsible for heritability should not have been a surprise because quantitative genetic research on complex traits in nonhuman animals using the selection design has for decades provided evidence that many genes of small effect are involved. If only a few genes were responsible for the heritability of a trait, selected lines would separate after a few generations and would not diverge any further in later generations. In contrast, selection studies of behavioral phenotypes as well as other complex traits show a linear response to selection even after dozens of generations of selection. For example, in one of the largest and longest selection studies of behavior, mice were selected for activity in a brightly lit box called an open field, where lower activity scores are presumed to index fearfulness21. As shown in Figure 2, strong evidence of genetic influence can be seen by the successful response to selection. After 30 generations of such selective breeding, a 30-fold average difference in activity was achieved–indeed, there was no overlap between the activity of the low and high lines. Moreover, the difference between the high and low lines steadily increases each generation, indicating that many genes contribute to variation in this behavior.
Figure 2.
Results of a selection study of open-field activity. Note: Beginning with an F3 cross between two inbred strains of mice, two lines were selected for high open-field activity (H1 and H2) in which the most active (lease fearful) mice were selected and mated with other high-active mice. Similarly, two lines were selected for low open-field activity (L1 and L2), and two lines were randomly mated within each line to sever as controls (C1 and C2). (With kind permission from Springer Science+Business Media: Behavior Genetics, Response to 30 generations of selection for open-field activity in laboratory mice, volume 8, 1978, page 3–13, J. C. DeFries, M. C. Gervais, & E. A. Thomas, figure 2, ©1978 by Plenum Publishing Corporation. All rights reserved.)
Our goal is not to denigrate GWA research–indeed, GWA is the focus of much of our own research12,14,22. Instead, our goal is to point to the bright future of quantitative genetic research, especially when its potential is exploited to go beyond merely estimating heritabilities. The main point of our paper is that quantitative genetic methods can go far beyond the rudimentary nature-nurture questions about whether and how much genes influence behavior to investigate how genes influence behavior. In this paper, we describe three examples of such quantitative genetic findings in relation to learning abilities and disabilities, before highlighting additional quantitative genetic methodologies that we predict will provide important biological and environmental findings as we enter the era of molecular genetics.
Common disorders are quantitative traits
Quantitative genetic research supports the conclusion that learning disabilities are the quantitative extremes of the genes responsible for the normal distribution of learning abilities3,23. DeFries-Fulker (DF) extremes analysis24 assesses genetic links between the extreme and the normal range by bringing together the dichotomous classification of learning disability and the quantitative trait of learning ability. Rather than assessing twin similarity in terms of concordance for a diagnostic cut-off (i.e. the disorder as qualitatively distinct from the normal range of controls), DF extremes analysis assesses twin similarity as the extent to which the mean standardized quantitative trait score of co-twins of the selected extreme probands is similar to the mean standardized score of those probands. This measure of twin similarity is called a group twin correlation (or transformed co-twin mean) in DF extremes analysis because it focuses on the mean quantitative trait score of co-twins rather than individual differences. Doubling the difference between MZ and DZ group twin correlations estimates the genetic contribution to the average phenotypic difference between the probands and the population. The ratio between this genetic estimate and the phenotypic difference between the probands and the population is called group heritability, the extent to which the phenotypic difference between the probands and the population can be explained by genetic differences. It should be noted that group heritability does not refer to individual differences among the probands—the question is not why one learning-disabled proband has slightly worse learning difficulties than another, but rather why probands as a group have more learning problems than the rest of the population. Finding group heritability implies that both learning disability and learning ability are heritable and, most importantly, that there are genetic links between learning disability and normal variation in learning ability. If a measure of extremes (or a diagnosis) were not linked genetically to a quantitative trait, group heritability would be zero. Research using this DF extremes method consistently show that group heritabilities are substantial for reading, language, mathematical disabilities as well as general learning disability2. These results suggest that common learning difficulties are the quantitative extreme of the same genetic factors responsible for the normal distribution of learning abilities.
This conclusion–that the polygenic distribution underlying behavioral traits, like learning disabilities, is normally distributed–has far-reaching conceptual and practical implications, especially for studies of qualitatively diagnosed common disorders, diseases and disabilities. Although there are a few GWA studies of quantitative traits, most notably height11, nearly all current GWA studies are based on qualitative diagnoses of cases and controls5,25. Fisher showed how quantitative traits can be explained by qualitative Mendelian inheritance if multiple genes are involved26, but what about the converse–common complex disorders that are diagnosed qualitatively? If, as GWA research indicates, multiple genes affect these disorders, their genetic liability is distributed quantitatively not qualitatively. Thus, there is a disconnect between qualitatively diagnosed disorders that are the focus of GWA studies and their quantitatively distributed polygenic liabilities. The resolution lies in recognizing that common disorders are the extremes of quantitative traits27. In other words, what we call common disorders such as learning disabilities are the quantitative extremes of continuous distributions of genetic risk. The obvious test of this hypothesis is that genes found for disorders in case-control studies will be associated, not just with differences between cases and controls, but with individual differences throughout the entire range of variation. For example, genes found to be associated with reading disability in case-control studies are predicted to be associated with reading ability for the entire range of variation, including good readers.
Several implications follow from thinking quantitatively about disorders, especially when some of the many genes are identified that are responsible for their heritability27. Independent of genetics, this trend towards thinking quantitatively can already be seen in the area of mental disorders28,29, although debates about diagnoses versus dimensions span the entire breadth of medicine30. The most novel implication of thinking quantitatively is that it leads to thinking positively. Thinking positively suggests that we should investigate mechanisms that push beyond normality; for example, not just fixing poor reading but promoting good reading. In the area of learning disabilities and abilities, quantitative genetic research has begun to address high cognitive abilities31.
Generalist genes
Quantitative genetic research has shown that the same genes affect different learning abilities and disabilities2,32,33. In other words, when genes are found that predict reading disability or ability, the same genes are also highly likely to predict mathematics disability or ability. Because these results suggest that a single set of genes has general effects across diverse learning abilities and disabilities, this is called the Generalist Genes Hypothesis.
These surprising findings derive from multivariate genetic analysis which investigates not only the variance of traits considered one at a time but also the covariance among traits1,34. Multivariate genetic analysis estimates the extent to which genetic and environmental factors that affect one trait also affect another trait. It yields a statistic called the genetic correlation which indexes the correlation between genetic effects on the two traits independent of the heritabilities of the two traits. That is, the genetic correlation between two traits can be 1.0 even when the heritabilities of the two traits are modest. The genetic correlation can be roughly interpreted as the likelihood that genes found to be associated with one trait will also be associated with the other trait.
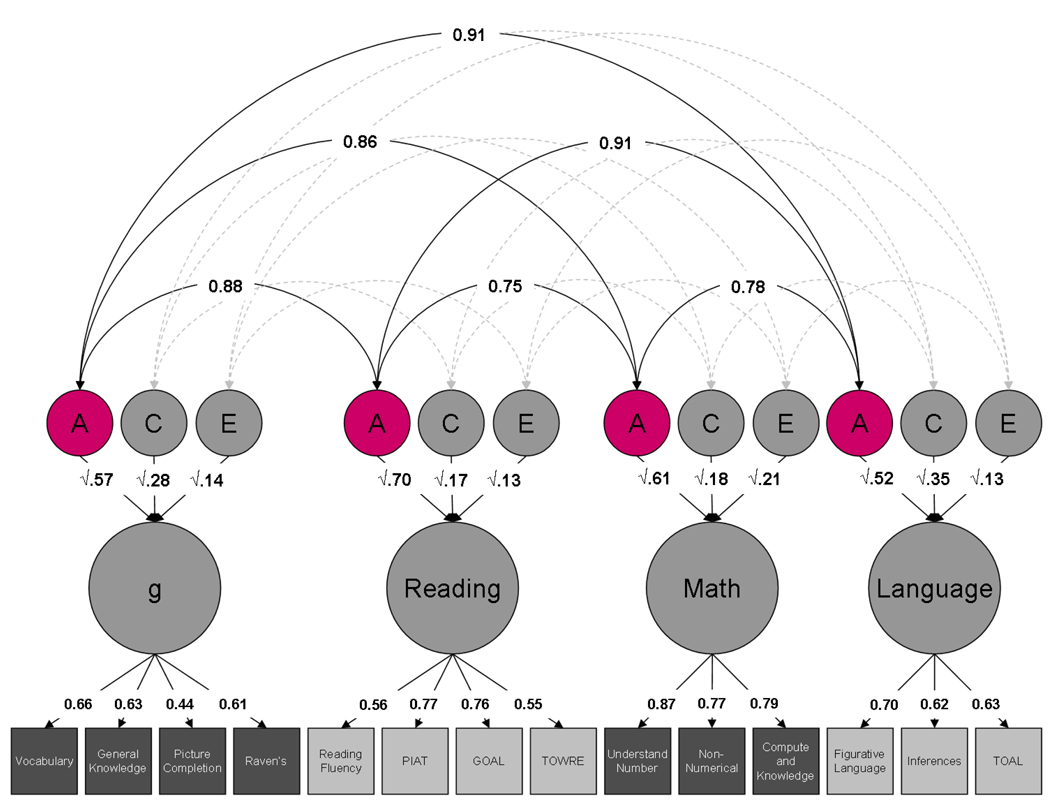
In a review of a dozen multivariate genetic studies of learning abilities and difficulties, the average genetic correlation was 0.70 between reading and language, reading and mathematics, and language and mathematics2. Similar results emerge from more recent research in middle childhood35,36 and early adolescence37. Figure 3 shows the results of the latest multivariate genetic test of the General Genes Hypothesis37. The genetic correlations among the latent variables range from 0.75 to 0.91. Results similarly supportive of the General Genes Hypothesis have been found using this same dataset for low ability38 and high ability39.
Figure 3.
Multivariate genetic common pathway model for 14 cognitive tests for more than 5000 pairs of twins at 12 years of age. Note: Squares represent measured traits; circles represent latent factors. The lower tier of arrows represents factor loadings; the middle tier represents genetic and environmental path coefficients; the curved arrows at the top represent correlations between genetic latent factors. Estimates of cross-trait additive genetic effects (A) are highlighted. Reprinted by permission of the publisher (Taylor & Francis Ltd, http://www.tandf.co.uk/journals) from Davis, Haworth & Plomin. Learning abilities and disabilities: generalist genes in early adolescence. Cognitive Neuropsychiatry 2009;14(4):312-31.) A=Additive genetic effects; C=Shared (common) environmental effects; E=non-shared environmental effects; g = general cognitive ability; GOAL = Global Online Assessment for Learning, Formative Assessment in Literacy for Key Stage 3; PIAT = Peabody Individual Achievement Test (reading comprehension); TOAL = Test of Adolescent and Adult Language; TOWRE = Test of Word Reading Efficiency.
The good news is that if the same set of genes is largely associated with most learning disabilities it should be easier to find these genes. However, because the genetic correlations are less than 1.0, genes also contribute to making children better at some abilities than others. That is, when genes are identified that are responsible for genetic influence on reading ability, most of these genes will also be associated with mathematics ability, but some will not40.
If genetic correlations are so high between learning abilities, it makes sense to expect that components within each learning domain are even more highly correlated genetically, and that is the case. Genetic correlations range between 0.60 and 0.90 within the domains of language, reading, and mathematics2. For example, in a recent study, five components of mathematics including computation, interpretation, and non-numerical processes were assessed via the Internet in a study of more than 1000 10-year-old twin pairs41. The average genetic correlation between the five components of mathematics was 0.91.
Moreover, the general effects of genes appear to extend beyond specific learning abilities such as reading and mathematics to other cognitive abilities such as verbal abilities (e.g. vocabulary and word fluency) and non-verbal abilities (e.g. spatial and memory). Multivariate genetic research on diverse cognitive abilities consistently finds genetic correlations greater than 0.50 and often near 1.0 across diverse cognitive abilities42. Similar results suggesting substantial genetic overlap have been found for more basic information processing measures such as speed of processing as well as measures of brain volume42.
Phenotypic correlations among diverse tests of cognitive abilities led Charles Spearman in 1904 to call this general factor g in order to avoid the many connotations of the word intelligence43. To what extent do generalist genes for g overlap with generalist genes for learning abilities? A review of about a dozen such studies concludes that genetic correlations between g and learning abilities are substantial but somewhat lower than the genetic correlations among learning abilities2. This result suggests that most (but not all) generalist genes that affect learning abilities are even more general in that they also affect other sorts of cognitive abilities included in the g factor.
The Generalist Genes Hypothesis suggests that genetic nosology differs from current diagnoses which are based on symptoms rather than etiology. Because genetic effects are general, they dissolve distinctions between diverse learning difficulties that ostensibly differ so much in terms of the cognitive processes involved. When these generalist genes are identified, they will greatly accelerate research on general mechanisms at all levels of analysis from genes to brain to behavior44.
Multivariate genetic research also has an interesting story to tell about environmental influences on learning abilities. Genetic research distinguishes two types of environmental influences. Environmental influences that make family members similar are called shared environment. And environmental influences that do not contribute to resemblance among family members are called non-shared environment, which also includes error of measurement. Multivariate genetic analyses indicate that shared environmental influences are generalists: Shared environmental correlations among learning and cognitive abilities are as high as genetic correlations. For example, in two recent studies, the shared environmental correlation was 0.74 between reading and mathematics at 7 years45 and the average shared environmental correlation was 0.86 between five components of mathematics at 10 years41. An obvious hypothesis that has not yet been rigorously tested is that some monolithic factors such as the socioeconomic status of the family or school quality might be responsible for these generalist shared environmental effects.
In contrast to these generalist effects of shared environment, non-shared environmental effects are specialists: Non-shared environmental correlations are low. For example, in the same two studies, the non-shared environmental correlation was 0.39 between reading and mathematics at 7 years45 and the average non-shared environmental correlation was 0.24 between five components of mathematics at 10 years41. Little is known about specific non-shared environmental influences that are the source of specialist environments largely because most environmental research focuses on shared environmental factors such as family background. Further investigation is needed to identify these specialist non-shared environmental influences which are specific to each trait and each age. This is particularly important, as quantitative genetics has highlighted that non-shared environmental influences are the main source of environmental influences on traits, and that the effect of shared environmental factors is low and decreases with age.
The nature of nurture
The great strength of quantitative genetic methods is that they investigate the net effect of genetic and environmental influences simultaneously which means that quantitative genetic studies are as much studies of the environment controlling for genetic effects as they are genetic studies controlling for environmental effects, as illustrated in the previous section. It is also possible to use quantitative genetic designs to explore the interplay between nature and nurture, especially when measures of the environment are included46,47. Quantitative genetic theory includes two concepts at the genotype-environment (GE) interface-GE interaction and GE correlation-although there are other ways to address the interplay between nature and nurture48,49. GE interaction refers to genetic sensitivity to environments in the sense that the effects of the environment can depend on genetics and the effects of genetics can depend on the environment48. In contrast, GE correlation refers to genetic exposure to the environment in that experiences can be correlated with genotype; for this reason GE correlation has been called the nature of nurture50. In other words, genetic effects on behavior do not stop at the skin; genetic effects need to be considered in relation to an ‘extended phenotype’ that includes effects on individuals’ environments51,52. Although GE interaction is currently the focus of much molecular genetic research53–55, our reading of the quantitative genetic literature suggests that GE correlation is a more widespread phenomenon47.
Investigating GE correlation in quantitative genetic research involves treating environmental measures as dependent measures to assess the extent to which these measures, which ostensibly assess the environment, in fact show genetic influence. Beginning with the pioneering work of Rowe56,57, dozens of twin and adoption studies have shown ubiquitous genetic influence on widely used measures of the environment58. A recent review of 55 independent genetic studies that included measures of the environment reported an average heritability of 0.27 across 35 different environmental measures, including not just measures of the family environment, such as parenting, but also measures outside the family such as peer groups, classroom environments, and life events59. For example, one recent developmental study of 1800 twin pairs interviewed retrospectively about peer group deviance showed heritabilities from 0.40 to 0.50 from childhood to young adulthood60.
Evidence for the heritability of environmental measures led to the first GWA study of an environmental measure61, a measure called CHAOS62 which assesses ‘environmental confusion’ in the home and which is more strongly associated with cognitive development in childhood than any other proximal measure of the environment63,64. Similar to other GWA studies described earlier, no replicable associations were found, but the point is that heritable variation in environmental measures implies that variation in DNA sequence is ultimately responsible for their heritability. Other molecular genetic studies have begun to focus on GE correlation, rather than just considering GE correlation as a confounding factor in research on GE interaction65.
Of course, finding genetic influence on environmental measures does not mean that environments are inherited any more than the heritability of reading means that words are inherited. What these findings mean is that genetic factors affect children’s experiences, mediated for example by genetic influences on the children’s personality. Three types of GE correlation have been described66,67. Passive GE correlation occurs because genetically related family members provide an environment correlated with a child’s genetic propensities. Evocative GE correlation happens because children evoke reactions from others based on the child’s genetic propensities. Active GE correlation involves children’s selection, modification and creation of environments correlated with their genetic propensities. A developmental theory of GE correlation proposes that during childhood, the influence of passive GE correlation declines and the importance of the active kind increases as children begin to make their way in the world beyond their families67.
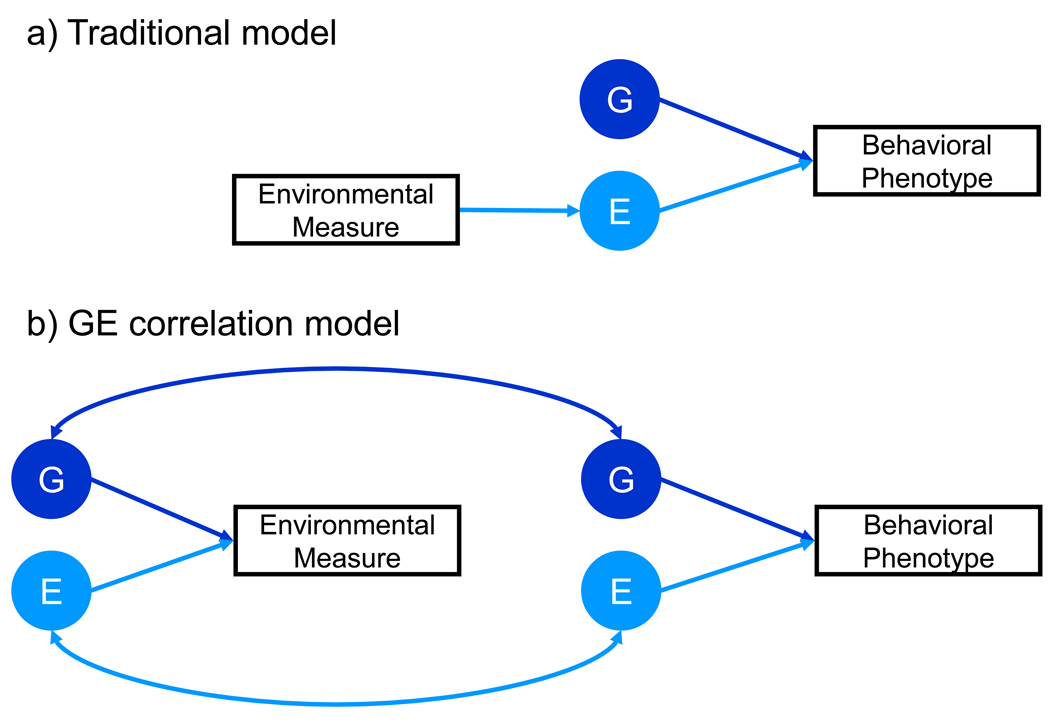
Genetic influence on experience suggests a new perspective on the environment, as illustrated in Figure 4. The traditional model of the environment makes the reasonable assumption that an environmental measure indexes the environmental contribution to behavior. In contrast, the GE correlation model takes into account the interface with genetics in two ways: (1) an environmental measure can be influenced genetically and (2) the association between an environmental measure and a behavioral phenotype can be mediated genetically. Multivariate genetic analysis, described in the previous section, can be applied to investigate the genetic and environmental etiology of the association between an environmental measure and a behavioral phenotype. The first such multivariate genetic analysis found that the phenotypic correlation of 0.61 between maternal negativity and adolescent antisocial behavior was more than half mediated by genetic factors46. Subsequent multivariate genetic studies have reported similar results–for example, between measures of the environment such as parenting, peer deviance, and life events and measures of behavior such as psychopathology, academic achievement and cognitive ability1. A general rule of thumb is that the size of the genetic influence on an environmental measure is a good indication of the extent of the genetic links between that environment and the behavioral outcome measure.
Figure 4.
The relationship between environmental measures and behavioral phenotypes: a) Traditional model and b) genotype-environment (GE) correlation model.
The GE correlation model of the environment predicts not only that DNA variants can be found that are associated with measures of the environment but also that these DNA variants will mediate associations between environmental measures and behavioral measures. This will be the definitive test of the GE correlation model. As is the case with all quantitative genetic research, more precise questions can be asked when we are able to include specific genes in addition to specific measures of the environment. For example, it will be possible to test the extent to which GE correlation arises for passive, evocative or active reasons and to test the hypothesis that genetic effects become less passive and more active during childhood and adolescence. An early study using a composite of SNPs associated with general cognitive ability in a sample of 7000 7-year-old children found GE correlations with preschool proximal measures of the family environment (chaos and discipline) rather than distal measures (maternal education and father's occupational class), suggesting evocative rather than passive GE correlation68.
The GE correlation model points to a radically different view of the way the environment works. Instead of a child being a passive recipient of environmental events, which is a holdover from the days of stimulus-response learning theory, the GE correlation model supports an active view of experience in which children make their own environments in part on the basis of their genetic proclivities58. That is, children select, modify, construct and even re-construct experiences for genetic reasons, creating correlations between their genotypes and their environments. Where do parents and policy makers fit in this GE correlation model? Sandra Scarr, who has done much of the seminal research in this area, concluded that “Parents’ most important job is to provide support and opportunities, not to try to shape children’s enduring characteristics. Policy makers’ most important role is to help parents provide support and opportunities for their children” 69,p.204.
Future directions for quantitative genetics
As summarized above, quantitative genetic research continues to contribute clinically relevant findings, as well as charting the direction for future molecular genetic research. In addition to the three findings outlined above, there are several other quantitative genetic methodologies that we predict will continue to be informative during, and beyond the era of molecular genetics, and which we highlight here.
The first and most notable contribution that quantitative genetics makes over molecular genetics is that quantitative genetics is as much about the environment, as it is about genetics. Although some molecular genetic studies have begun to include environmental measures54,55,61,70, the majority of these do so at a candidate gene level and not genome-wide. In addition, just as genetic influences are typically of small effect size, the same is likely true for environmental influences, making environmental influences as difficult to detect as DNA variants. The benefit of quantitative genetics is that we do not need to know specifically what these environmental influences are, just as we do not need to know which specific genes are involved. This means that quantitative genetics can investigate how environmental influences function, even before specific factors are identified, and can also help to chart the direction for future environmental research that focuses on identifying these environmental measures. The ideal situation would be to know what the specific genetic and environmental influences are, and quantitative genetics will be influential in identifying these. Quantitative genetic research has provided some of the best evidence for the role of the environment in complex traits, most notably indicating that environmental influences typically operate on an individual-by-individual basis and not generally on a family-by-family basis71. Quantitative genetics also provides a valuable method for identifying specific environmental factors while controlling for genetic influence: the MZ differences design72. The MZ differences design can also be extended to investigate longitudinal and multivariate pathways73.
Molecular genetic research currently focuses on univariate analyses, although some multivariate GWA studies are underway, the methods for multivariate and longitudinal GWA studies are in their infancy, whereas such models for quantitative genetic research have been extensively developed and are widely used34. In addition, twin studies are typically cohort studies that have collected a variety of phenotypes during development. In contrast, most GWA studies are focused on one particular disorder, so few can be extended to investigate multivariate and longitudinal research questions. Molecular genetic research in complex traits will really come into its own when we have GWA data on large population-representative cohort studies with multiple phenotypes and environmental data, and once we have determined optimal statistical techniques for analyzing the millions of data points that would come out of such molecular genetic studies. But for the time-being, quantitative genetics provides the only validated method for assessing multivariate and longitudinal etiological questions.
Finally, just as quantitative genetic methodologies can be used to study environmental measures as the dependent variable, they can also be applied to biological data. Recent studies have highlighted the utility of applying the twin design to gene expression, methylation (epigenetics), and copy-number variant data74,75,76, especially in relation to understanding the genetic and environmental origins of these biological traits77
Conclusion
Genome-wide association studies are struggling to identify a few of the many genes responsible for the ubiquitous heritability of common disorders and complex dimensions such as learning disabilities and abilities because the largest effect sizes are so small. In contrast, we hope that these three examples of findings from quantitative genetics, as well as the future uses of quantitative genetic research highlighted here, illustrate the potential of quantitative genetics to continue to make discoveries with far-reaching ramifications for child and adolescent psychiatry.
Acknowledgments
Preparation of this paper was supported in part by grants from the U.K. Medical Research Council (G050079), the Wellcome Trust (WT084728) and the U.S. National Institute of Child Health and Human Development (HD44454). CMAH is supported by an MRC/ESRC Interdisciplinary Fellowship (G0802681).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
This article represents one of several articles published in the xxx issue of the Journal of the American Academy of Child and Adolescent Psychiatry that explores the intersection of genetics and mental health disorders in children and adolescents. The editors invite the reader to investigate the additional articles on this burgeoning area of developmental psychopathology.
References
- 1.Plomin R, DeFries JC, McClearn GE, McGuffin P. Behavioral Genetics. 5th Edition. New York: Worth; 2008. [Google Scholar]
- 2.Plomin R, Kovas Y. Generalist genes and learning disabilities. Psychol.Bull. 2005:592–617. doi: 10.1037/0033-2909.131.4.592. [DOI] [PubMed] [Google Scholar]
- 3.Kovas Y, Haworth CMA, Dale PS, Plomin R. The genetic and environmental origins of learning abilities and disabilities in the early school years. Monogr.Soc.Res.Child.Dev. 2007;72(3):vii–160. doi: 10.1111/j.1540-5834.2007.00439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walker SO, Plomin R. The nature - nurture question: Teachers' perceptions of how genes and the environment influence educationally relevant behavior. Educ.Psychol. 2005;25:509–515. [Google Scholar]
- 5.Donnelly P. Progress and challenges in genome-wide association studies in humans. Nature. 2008;456(7223):728–731. doi: 10.1038/nature07631. [DOI] [PubMed] [Google Scholar]
- 6.McCarthy MI, Abecasis GR, Cardon LR, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat.Rev.Genet. 2008;9(5):356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 7.Stefansson H, Ophoff RA, Steinberg S, et al. Common variants conferring risk of schizophrenia. Nature. 2009;460:744–747. doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zeggini E, Scott LJ, Saxena R, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat.Genet. 2008;40:638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frayling TM, Timpson NJ, Weedon MN, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316(5826):889. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manolio TA. Cohort studies and the genetics of complex disease. Nat.Genet. 2009;41(1):5–6. doi: 10.1038/ng0109-5. [DOI] [PubMed] [Google Scholar]
- 11.Weedon MN, Lango H, Lindgren CM, et al. Genome-wide association analysis identifies 20 loci that influence adult height. Nat.Genet. 2008;40(5):575–583. doi: 10.1038/ng.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meaburn EL, Harlaar N, Craig IW, Schalkwyk LC, Plomin R. QTL association scan of early reading disability and ability using pooled DNA and 100K SNP microarrays in a sample of 5,500 children. Mol.Psychiatry. 2008;13:729–740. doi: 10.1038/sj.mp.4002063. [DOI] [PubMed] [Google Scholar]
- 13.Seshadri S, DeStefano A, Au R, et al. Genetic correlates of brain aging on MRI and cognitive test measures: a genome-wide association and linkage analysis in the Framingham Study. BMC.Med.Genet. 2007;8:S15. doi: 10.1186/1471-2350-8-S1-S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Butcher LM, Davis OSP, Craig IW, Plomin R. Genome-wide quantitative trait locus association scan of general cognitive ability using pooled DNA and 500K single nucleotide polymorphism microarrays. Genes.Brain.Behav. 2008;7:435–446. doi: 10.1111/j.1601-183X.2007.00368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huentelman MJ, Papassotiropoulos A, Craig DW, et al. Calmodulin-binding transcription activator 1 (CAMTA1) alleles predispose human episodic memory performance. Hum.Mol.Genet. 2007;16(12):1469. doi: 10.1093/hmg/ddm097. [DOI] [PubMed] [Google Scholar]
- 16.Papassotiropoulos A, Stephan DA, Huentelman MJ, et al. Common Kibra alleles are associated with human memory performance. Science. 2006;314(5798):475–478. doi: 10.1126/science.1129837. [DOI] [PubMed] [Google Scholar]
- 17.Need AC, Attix DK, McEvoy JM, et al. Failure to replicate effect of Kibra on human memory in two large cohorts of European origin. Am.J.Med.Genet.B.Neuropsychiatr.Genet. 2008:667–668. doi: 10.1002/ajmg.b.30658. [DOI] [PubMed] [Google Scholar]
- 18.Maher B. Personal genomes: The case of the missing heritability. Nature. 2008;456(7218):18. doi: 10.1038/456018a. [DOI] [PubMed] [Google Scholar]
- 19.Plomin R, Davis OSP. The future of genetics in psychology and psychiatry: microarrays, genome-wide association, and non-coding RNA. J.Child.Psychol.Psychiat. 2009;50(1–2):63–71. doi: 10.1111/j.1469-7610.2008.01978.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walley AJ, Asher JE, Froguel P. The genetic contribution to non-syndromic human obesity. Nat.Rev.Genet. 2009;10(7):431–442. doi: 10.1038/nrg2594. [DOI] [PubMed] [Google Scholar]
- 21.DeFries JC, Gervais MC, Thomas EA. Response to 30 generations of selection for open-field activity in laboratory mice. Behav.Genet. 1978;8:3–13. doi: 10.1007/BF01067700. [DOI] [PubMed] [Google Scholar]
- 22.Ronald A, Butcher LM, Docherty S, et al. A genome-wide association study of social and non-social autistic-like traits in the general population using pooled DNA, 500K SNP microarrays and both community and diagnosed autism replication samples. Behav.Genet. doi: 10.1007/s10519-009-9308-6. In press. DOI 10.1007/s10519-009-9308-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haworth CMA, Kovas Y, Petrill SA, Plomin R. Developmental origins of low mathematics performance and normal variation in twins from 7 to 9 years. Twin.Res.Hum.Genet. 2007;10(1):106–117. doi: 10.1375/twin.10.1.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeFries JC, Fulker DW. Multiple regression analysis of twin data. Behav.Genet. 1985;15:467–473. doi: 10.1007/BF01066239. [DOI] [PubMed] [Google Scholar]
- 25.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fisher RA. The correlation between relatives on the supposition of Mendelian inheritance. T.Roy.Soc.Edin. 1918;52:399–433. [Google Scholar]
- 27.Plomin R, Haworth CMA, Davis OSP. Common disorders are quantitative traits. Nat.Rev.Genet. 2009;10:872–878. doi: 10.1038/nrg2670. [DOI] [PubMed] [Google Scholar]
- 28.First MB, Pincus HA, Levine JB, Williams JBW, Ustun B, Peele R. Clinical utility as a criterion for revising psychiatric diagnoses. Am.J.Psychiatry. 2004;161(6):946–954. doi: 10.1176/appi.ajp.161.6.946. [DOI] [PubMed] [Google Scholar]
- 29.The Psychiatric GWAS Consortium Steering Committee. A framework for interpreting genome-wide association studies of psychiatric disorders. Mol.Psychiatry. 2009;14:10–17. doi: 10.1038/mp.2008.126. [DOI] [PubMed] [Google Scholar]
- 30.Sackett DL, Rosenberg WMC, Gray JAM, Haynes RB, Richardson WS. Evidence based medicine: what it is and what it isn't. Brit.Med.J. 1996;312(7023):71–72. doi: 10.1136/bmj.312.7023.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Plomin R, Haworth CMA. Genetics of High Cognitive Abilities. Behav.Genet. 2009;39(4):347–349. doi: 10.1007/s10519-009-9277-9. [DOI] [PubMed] [Google Scholar]
- 32.Bartels M, Rietveld MJ, van Baal GC, Boomsma DI. Heritability of educational achievement in 12-year-olds and the overlap with cognitive ability. Twin.Res. 2002 December;5(6):544–553. doi: 10.1375/136905202762342017. [DOI] [PubMed] [Google Scholar]
- 33.Wainwright MA, Wright MJ, Luciano M, Geffen GM, Martin NG. Multivariate genetic analysis of academic skills of the Queensland core skills test and IQ highlight the importance of genetic g. Twin.Res.Hum.Genet. 2005;8(6):602–608. doi: 10.1375/183242705774860259. [DOI] [PubMed] [Google Scholar]
- 34.Neale MC, Maes HHM. Methodology for genetic studies of twins and families. Dordrecht, The Netherlands: Kluwer Academic Publishers B.V.; 2003. [Google Scholar]
- 35.Davis OSP, Kovas Y, Harlaar N, et al. Generalist genes and the Internet generation: etiology of learning abilities by web testing at age 10. Genes.Brain.Behav. 2008;7:455–462. doi: 10.1111/j.1601-183X.2007.00370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haworth CMA, Kovas Y, Dale PS, Plomin R. Science in Elementary School: Generalist Genes and School Environments. Intelligence. 2008;36:694–701. [Google Scholar]
- 37.Davis OSP, Haworth CMA, Plomin R. Learning abilities and disabilities: generalist genes in early adolescence. Cognit.Neuropsychiatry. 2009;14(4):312–331. doi: 10.1080/13546800902797106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haworth CMA, Kovas Y, Harlaar N, et al. Generalist genes and learning disabilities: a multivariate genetic analysis of low performance in reading, mathematics, language and general cognitive ability in a sample of 8000 12-year-old twins. J.Child.Psychol.Psychiat. 2009;50(10):1318–1325. doi: 10.1111/j.1469-7610.2009.02114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haworth CMA, Dale PS, Plomin R. Generalist Genes and High Cognitive Abilities. Behav.Genet. 2009;39(4):437–445. doi: 10.1007/s10519-009-9271-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haworth CMA, Meaburn EL, Harlaar N, Plomin R. Reading and Generalist Genes. Mind.Brain.Educ. 2007;1(4):173–180. doi: 10.1111/j.1751-228X.2007.00018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kovas Y, Petrill SA, Plomin R. The origins of diverse domains of mathematics: Generalist genes but specialist environments. J.Educ.Psychol. 2007;99(1):128. doi: 10.1037/0022-0663.99.1.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deary IJ, Spinath FM, Bates TC. Genetics of intelligence. Eur.J.Hum.Genet. 2006;14(6):690–700. doi: 10.1038/sj.ejhg.5201588. [DOI] [PubMed] [Google Scholar]
- 43.Spearman C. "General intelligence," objectively determined and measured. Am.J.Psychol. 1904;15:201–292. [Google Scholar]
- 44.Kovas Y, Plomin R. Generalist genes: Implications for cognitive sciences. Trends.Cogn.Sci. 2006;10:198–203. doi: 10.1016/j.tics.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 45.Kovas Y, Harlaar N, Petrill SA, Plomin R. 'Generalist genes' and mathematics in 7-year-old twins. Intelligence. 2005;5:473–489. doi: 10.1016/j.intell.2005.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Plomin R, Reiss D, Hetherington EM, Howe GW. Nature and nurture: Genetic contributions to measures of the family environment. Dev.Psychol. 1994;30:32–43. [Google Scholar]
- 47.Plomin R, Davis OSP. Gene-environment interactions and correlations in the development of cognitive abilities and disabilities. In: MacCabe J, O'Daly O, Murray RM, McGuffin P, Wright P, editors. Beyond Nature and Nurture: Genes, Environment and their Interplay in Psychiatry. Andover, UK: Thomson Publishing Services; 2006. pp. 35–45. [Google Scholar]
- 48.Kendler KS, Eaves LJ. Models for the joint effects of genotype and environment on liability to psychiatric illness. Am.J.Psychiatry. 1986;143:279–289. doi: 10.1176/ajp.143.3.279. [DOI] [PubMed] [Google Scholar]
- 49.Rutter M. Genes and behavior: Nature-nurture interplay explained. Blackwell Publ.; 2007. [Google Scholar]
- 50.Plomin R, Bergeman CS. The nature of nurture: Genetic influences on "environmental" measures. Behav.Brain.Sci. 1991;14:373–427. [Google Scholar]
- 51.Dawkins R. The Extended Phenotype: The Long Reach of the Gene. Oxford: Oxford University Press; 1983. [Google Scholar]
- 52.Dawkins R. Extended phenotype-But not too extended. A reply to Laland, Turner and Jjablonka. Biol.Philos. 2004;19(3):377–396. [Google Scholar]
- 53.Caspi A, Moffitt TE. Gene-environment interactions in psychiatry: joining forces with neuroscience. Nat.Rev.Neurosci. 2006;7(7):583–590. doi: 10.1038/nrn1925. [DOI] [PubMed] [Google Scholar]
- 54.Risch N, Herrell R, Lehner T, et al. Interaction Between the Serotonin Transporter Gene(5-HTTLPR), Stressful Life Events, and Risk of Depression: A Meta-analysis. JAMA. 2009;301(23):2462. doi: 10.1001/jama.2009.878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Uher R, McGuffin P. The moderation by the serotonin transporter gene of environmental adversity in the aetiology of mental illness: review and methodological analysis. Mol.Psychiatry. 2007;13(2):131–146. doi: 10.1038/sj.mp.4002067. [DOI] [PubMed] [Google Scholar]
- 56.Rowe DC. Environmental and genetic influences on dimensions of perceived parenting: A twin study. Dev.Psychol. 1981;17:203–208. [Google Scholar]
- 57.Rowe DC. A biometrical analysis of perceptions of family environment: A study of twin and singleton sibling relationships. Child.Dev. 1983;54:416–423. [PubMed] [Google Scholar]
- 58.Plomin R. Genetics and experience:The interplay between nature and nurture. Thousand Oaks, California: Sage Publications Inc.; 1994. [Google Scholar]
- 59.Kendler KS, Baker JH. Genetic influences on measures of the environment: a systematic review. Psychol.Med. 2007;37(05):615–626. doi: 10.1017/S0033291706009524. [DOI] [PubMed] [Google Scholar]
- 60.Kendler KS, Jacobson KC, Gardner CO, Gillespie N, Aggen SA, Prescott CA. Creating a social world: a developmental twin study of peer-group deviance. Arch.Gen.Psychiatry. 2007;64(8):958. doi: 10.1001/archpsyc.64.8.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Butcher LM, Plomin R. The Nature of Nurture: A Genomewide Association Scan for Family Chaos. Behav.Genet. 2008;38(4):361–371. doi: 10.1007/s10519-008-9198-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Matheny AP, Wachs TD, Ludwig JL, Phillips K. Bringing order out of chaos: Psychometric characteristics of the Confusion, Hubbub and Order Scale. J.Appl.Dev.Psychol. 1995;16:429–444. [Google Scholar]
- 63.Deater-Deckard K, Mullineaux PY, Beekman C, Petrill SA, Schatschneider C, Thompson LA. Conduct problems, IQ, and household chaos: a longitudinal multi-informant study. J.Child.Psychol.Psychiat. 2009;50(10):1301–1308. doi: 10.1111/j.1469-7610.2009.02108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Petrill SA, Pike A, Price TS, Plomin R. Chaos in the home and socioeconomic status are associated with cognitive development in early childhood: Environmental mediators identified in a genetic design. Child.Dev. 2004;32(5):445–460. [Google Scholar]
- 65.Jaffee SR, Price TS. Gene-environment correlations: a review of the evidence and implications for prevention of mental illness. Mol.Psychiatry. 2007;12(5):432–442. doi: 10.1038/sj.mp.4001950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Plomin R, DeFries JC, Loehlin JC. Genotype-environment interaction and correlation in the analysis of human behaviour. Psychol.Bull. 1977;85:309–322. [PubMed] [Google Scholar]
- 67.Scarr S, McCartney K. How people make their own environments: A theory of genotype-->environmental effects. Child.Dev. 1983;54:424–435. doi: 10.1111/j.1467-8624.1983.tb03884.x. [DOI] [PubMed] [Google Scholar]
- 68.Harlaar N, Butcher L, Meaburn E, Craig IW, Plomin R. A behavioural genomic analysis of DNA markers associated with general cognitive ability in 7-year-olds. J.Child.Psychol.Psychiat. 2005;46:1097–1107. doi: 10.1111/j.1469-7610.2005.01515.x. [DOI] [PubMed] [Google Scholar]
- 69.Scarr S. How people make their own environments: Implications for parents and policy makers. Psychol.Public.Pol.L. 1996;2(2):204–228. [Google Scholar]
- 70.van Os J, Rutten BPF. Gene-Environment-Wide Interaction Studies in Psychiatry. Am.J.Psychiatry. 2009;166(9):964–966. doi: 10.1176/appi.ajp.2008.09060904. [DOI] [PubMed] [Google Scholar]
- 71.Plomin R, Asbury K, Dunn J. Why are children in the same family so different? Nonshared environment a decade later. Can.J.Psychiat. 2001;46(3):225–233. doi: 10.1177/070674370104600302. [DOI] [PubMed] [Google Scholar]
- 72.Pike A, Reiss D, Hetherington EM, Plomin R. Using MZ differences in the search for nonshared environmental effects. J.Child.Psychol.Psychiat. 1996;37:695–704. doi: 10.1111/j.1469-7610.1996.tb01461.x. [DOI] [PubMed] [Google Scholar]
- 73.Burt SA, McGue M, Iacono WG, Krueger RF. Differential parent-child relationships and adolescent externalizing symptoms:Cross-lagged analyses within a monozygotic twin differences design. Dev.Psychol. 2006;42(6):1289–1298. doi: 10.1037/0012-1649.42.6.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bruder CEG, Piotrowski A, Gijsbers AACJ, et al. Phenotypically concordant and discordant monozygotic twins display different DNA copy-number-variation profiles. Am.J.Hum.Genet. 2008;82(3):763–771. doi: 10.1016/j.ajhg.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hasler R, Begun A, Freitag-Wolf S, et al. Genetic control of global gene expression levels in the intestinal mucosa: a human twin study. Physiol.Genomics. 2009;38(1):73–79. doi: 10.1152/physiolgenomics.00010.2009. [DOI] [PubMed] [Google Scholar]
- 76.Heijmans BT, Kremer D, Tobi EW, Boomsma DI, Slagboom PE. Heritable rather than age-related environmental and stochastic factors dominate variation in DNA methylation of the human IGF2/H19 locus. Hum.Mol.Genet. 2007;16(5):547. doi: 10.1093/hmg/ddm010. [DOI] [PubMed] [Google Scholar]
- 77.Kan KJ, Ploeger A, Raijmakers MEJ, Dolan CV, van der Maas HLJ. Nonlinear epigenetic variance: review and simulations. Dev.Sci. 2009;13(1):11–27. doi: 10.1111/j.1467-7687.2009.00858.x. [DOI] [PubMed] [Google Scholar]