Abstract
In meiosis, programmed DNA breaks repaired by homologous recombination (HR) can be processed into inter-homolog crossovers that promote the accurate segregation of chromosomes. In general, more programmed DNA double-strand breaks (DSBs) are formed than the number of inter-homolog crossovers, and the excess DSBs must be repaired to maintain genomic stability. Sister-chromatid (inter-sister) recombination is postulated to be important for the completion of meiotic DSB repair. However, this hypothesis is difficult to test because of limited experimental means to disrupt inter-sister and not inter-homolog HR in meiosis. We find that the conserved Structural Maintenance of Chromosomes (SMC) 5 and 6 proteins in Caenorhabditis elegans are required for the successful completion of meiotic homologous recombination repair, yet they appeared to be dispensable for accurate chromosome segregation in meiosis. Mutations in the smc-5 and smc-6 genes induced chromosome fragments and dismorphology. Chromosome fragments associated with HR defects have only been reported in mutants, which have disrupted inter-homolog crossover. Surprisingly, the smc-5 and smc-6 mutations did not disrupt the formation of chiasmata, the cytologically visible linkages between homologous chromosomes formed from meiotic inter-homolog crossovers. The mutant fragmentation defect appeared to be preferentially enhanced by the disruptions of inter-homolog recombination but not by the disruptions of inter-sister recombination. Based on these findings, we propose that the C. elegans SMC-5/6 proteins are required in meiosis for the processing of homolog-independent, presumably sister-chromatid-mediated, recombination repair. Together, these results demonstrate that the successful completion of homolog-independent recombination is crucial for germ cell genomic stability.
Author Summary
Sperm and oocytes are essential for the faithful transmission of genetic information during sexual reproduction. As germ cells mature into sperm and oocytes, DNA double-strand breaks (DSBs) are deliberately created on each chromosome and a subset of DSBs is repaired to form meiotic crossovers between homologous chromosomes. Because germ cells must undergo this programmed process of deliberate DNA damage and repair, identifying repair factors active in germ cells and determining the requirement of their functions in meiotic DSB repair are important first steps in understanding infertility and developmental disorders caused by defective sperm and oocytes. In this manuscript, we find that the evolutionarily conserved SMC-5 and SMC-6 proteins fulfill a critical role in preserving genomic stability in germ cells in C. elegans. Our findings further describe the genetic mechanisms by which the C. elegans SMC-5/6 proteins function in meiotic DSB repair. These data reveal that inter-sister homologous recombination, a repair mechanism thought to function as a back-up repair method in meiosis, serves a more significant role in normal meiosis than was previously appreciated.
Introduction
Homologous recombination (HR) utilizes an undamaged homologous DNA template to repair DNA double-strand breaks (DSBs). The use of template-mediated repair minimizes the likelihood of DNA sequence alterations arising during the repair process. For mitotic cells following DNA replication, the sister chromatid is the predominant repair template because of the close proximity of sister chromatids maintained by sister-chromatid cohesion [1], [2], and inter-sister recombination provides an important high fidelity pathway for DSB repair.
Meiosis is a specialized cell cycle in which diploid progenitor cells divide to produce haploid gametes [3], . The chromosome copy number is reduced in meiosis during the reductional division, in which homologous chromosomes are bioriented at metaphase to ensure that each daughter cell receives a haploid complement of chromosomes. The correct biorientation of homologous chromosomes generally requires the formation of physical linkages between homologous chromosomes called chiasmata, which are formed by reciprocal chromatid exchanges between homologous chromosomes that can occur through inter-homolog recombination. To promote chiasmata formation in meiosis I, a germ cell will purposely create up to hundreds of programmed DSBs which are repaired by HR [5]. While a small subset of DSB is repaired to form chiasmata, it is generally thought that the remaining DSBs must be efficiently repaired to preserve the genomic stability of the germ cell.
Even though homologous recombination between sister chromatids will not contribute to chiasmata formation, sister-chromatid recombination is responsible for a portion of meiotic DSB repair in a variety of species [6]–[11], and is thought to promote genomic stability in germ cells especially when inter-homolog recombination is compromised or unavailable [7], [10], [12]–[14]. However, whether meiotic sister-chromatid recombination is crucial for germ cell genomic stability, when inter-homolog repair is functional, has not been determined.
The C. elegans homolog of the mammalian breast and ovarian cancer susceptibility gene brc-1 was implicated specifically for meiotic sister-chromatid recombination [7]. The brc-1(tm1145) mutant showed a delayed progression in HR repair, but there were no significant changes in chiasmata formation [7]. The brc-1 mutation appeared to impede homolog-independent repair, because it caused the appearance of chromosome fragments when combined with mutations that disrupted inter-homolog repair [7]. However, the brc-1 mutant by itself only exhibited a mild chromosome fragmentation defect [7]. These results suggest that brc-1 and by extension sister-chromatid recombination are not required to complete meiotic DSB repair. The degree to which sister-chromatid recombination was disrupted by the brc-1 mutation is unclear.
If sister-chromatid recombination in meiosis were necessary for the proper repair of meiotic DSB, then mutations that disrupted meiotic sister-chromatid recombination could result in chromosome anomalies (e.g. dicentric chromosomes), which could lead to mis-segregation and aneuploidy [15], [16]. Because mis-segregation and aneuploidy in meiosis also are the expected outcomes from the loss of inter-homolog recombination, it would be difficult to distinguish an inter-sister recombination defect from an inter-homolog recombination defect. The study of sister-chromatid recombination in meiosis would be simpler in an experimental organism, in which the presence of chromosome fragments and rearrangements would not necessarily lead to mis-segregation. One such model organism is the nematode Caenorhabditis elegans that can segregate partial chromosome duplications and fusion chromosomes with surprisingly high fidelity [17], [18]. We hypothesized that severe DSB repair defects in sister-chromatid recombination could be decoupled from chromosome mis-segregation when studied in C. elegans.
For a candidate meiotic sister-chromatid recombination factor, we chose the Structural Maintenance of Chromosomes (SMC) 5 and 6 protein complex, because the human and yeast Smc5/6 complexes have previously been implicated in sister-chromatid recombination in mitosis [19]–[21]. The mouse and fission yeast Smc5/6 protein complexes also are expressed during meiosis and the loss of the fission yeast complex leads to aneuploid spore formation [22], [23]. However, the requirement for the Smc5/6 complex specifically for meiotic sister-chromatid recombination has not been addressed.
The main objective of this research is to address the requirement and the function of the C. elegans SMC-5 and SMC-6 proteins for DSB repair in meiotic germ cells. The C. elegans smc-5 and smc-6 mutants exhibited defects in the processing of RAD-51 HR intermediates in meiosis. Similar to the brc-1 mutant, chiasmata formation and meiotic chromosome segregation were apparently unaffected in the smc-5 and smc-6 mutants. In this study, we demonstrate that the RAD-51 defect is due to homolog-independent repair. More importantly, we find that the severe loss-of-function mutation in smc-5 or smc-6 is sufficient to elicit a chromosome fragmentation defect in meiotic germ cells. The fragments appeared to be associated with homolog-independent repair of programmed meiotic DSBs. Consistent with a loss in sister-chromatid recombination, the smc-5 and smc-6 mutant fragmentation defect was enhanced by inter-homolog recombination mutations, but not by mutations that reduced sister-chromatid recombination or cohesion. While the smc-5 and smc-6 mutants were initially viable, the mutant strains would gradually lose fecundity and exhibit other germ cell defects. These results reveal that the SMC-5/6 proteins function in homolog-independent, likely sister-chromatid-mediated, recombination in meiosis, and that homolog-independent recombination is required for germ cell genomic stability.
Results
The identification of the C. elegans smc-5 and smc-6 deletion mutants and the production of specific antibodies to the SMC-5 and SMC-6 proteins
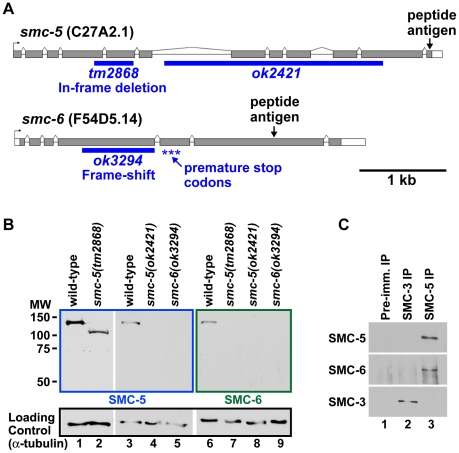
The C. elegans SMC-5 homolog C27A2.1 and the SMC-6 homolog F54D5.14 were identified previously based on protein sequence homology [24]. We generated antibodies to the SMC-5 and SMC-6 homologs that detected proteins of the predicted size from wild-type worm lysates (Figure 1B, lanes 1, 3 and 6). Western blot analyses of the mutant worm lysates confirmed the specificities of the antibodies, because the detected bands were absent in the smc-5(ok2421) and smc-6(ok3294) mutants (Figure 1B, lanes 4 and 9) that were predicted to have severe disruptions in protein function (Figure 1A). The SMC-5 antibodies also detected a smaller protein band in the smc-5(tm2868) mutant (Figure 1B, lane 2), in agreement with the predicted in-frame deletion in the smc-5(tm2868) encoded protein (Figure 1A). Immunoprecipitation analysis revealed that the C. elegans SMC-5 and SMC-6 proteins specifically co-precipitated from whole worm lysates (Figure 1C), as expected from the known association between the Smc5 and Smc6 proteins in yeast and in human [23], [25]–[27].
Figure 1. Identification and detection of the SMC-5 and SMC-6 homologs in C. elegans.
(A) Two diagrams illustrate the predicted exon-intron structures of the smc-5 and smc-6 genes and the locations of the tm2868, ok2421 and ok3294 deletion mutations (blue bars). The exons are represented by the grey boxes and the introns by the black lines. The tm2868 deletion removes 444 basepairs from exon 4 to exon 5, and it is predicted to generate an in-frame deletion of 133 codons. The ok2421 deletion removes 2,420 basepairs from intron 6 to exon 11. The ok3294 lesion deletes the last 830 basepairs from exon 4, and should result in a frame-shift at codon 192 and the introduction of premature termination codons (asterisks). The corresponding peptide regions used for antibody production are indicated. (B) Western blot analysis of whole worm lysates using antibodies raised to SMC-5 (lanes 1–5), antibodies to SMC-6 (lanes 6–9), and antibodies to alpha-tubulin as a loading control (lanes 1–9). The genotypes of the worms used are indicated above the western blot images. Two-fold more lysates were loaded in lanes 1–2 to detect the smaller SMC-5 product from the smc-5(tm2868) mutant lysates. (C) Western blot analysis of immunoprecipitates from wild-type embryo lysates using pre-immune (lane 1), SMC-3 (lane 2) and SMC-5 antibodies (lane 3). Antibodies for western blot detection are indicated to the left of the western blot images.
The SMC-5 and SMC-6 proteins are enriched in the adult germline
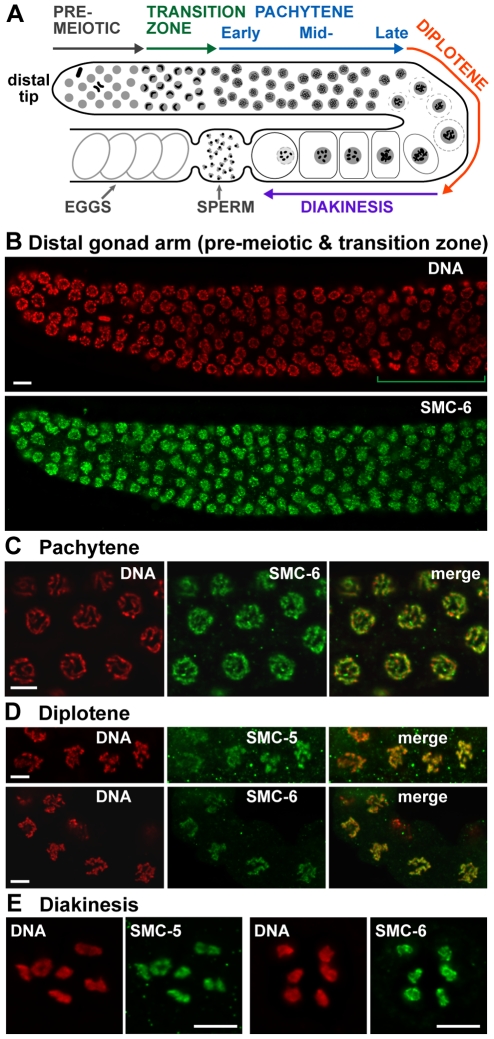
The SMC-6 protein was detected by indirect immunofluorescence microscopy in the nuclei of germ cells throughout the adult hermaphrodite gonad (Figure 2). Beginning at the distal tip region of the gonad (Figure 2A), SMC-6 staining is detected in the nucleus of germ cells in mitotic proliferation, pre-meiotic S phase and in the early stages of meiosis (the transition zone), which are equivalent to leptotene and zygotene (Figure 2B). Interestingly, SMC-6 staining became more enriched on chromosomes at pachytene (Figure 2C), which coincided with the timing of meiotic DSB repair [14]. The SMC-6 immunofluorescence became more intense as the germ cells exited pachytene and progressed through the diplotene and diakinesis stages of prophase (Figures 2D and 2E). The pachytene and diakinesis staining of SMC-6 was specifically disrupted by the smc-6(ok3294) mutation (Figures S1C and S1E). Even though we could not detect SMC-5 immunostaining on pachytene chromosomes, we found that the smc-5(tm2868) and smc-5(ok2421) mutations reduced SMC-6 staining on pachytene chromosomes (Figures S1A and S1B), indicating a possible dependency on SMC-5 for the localization of SMC-6 at pachytene. The immunostaining for SMC-5 was detected on diplotene and diakinesis chromosomes specifically in wild-type (Figures 2D and 2E), but not in the smc-5(tm2868) and smc-5(ok2421) mutant oocytes (Figures S1D and S1H). The SMC-5 and SMC-6 chromosomal staining in diplotene (data not shown) and diakinesis oocytes (Figures S1F, S1G and S1I) also appeared to be interdependent. In addition to germ cell staining, SMC-5 and SMC-6 immunostaining were detected in somatic cells during early embryogenesis (data not shown). The three smc-5 and smc-6 mutations caused frequent chromatin-bridges to appear in the intestine, even though immunostaining was significantly weaker in the intestine than the germline (Bickel and Chan, unpublished observations). These results suggest that the SMC-5/6 proteins accumulate in the soma and the germline, with greater enrichment seen in the germ cells.
Figure 2. SMC-5 and SMC-6 accumulate in germ cells and on meiotic chromosomes.
(A) A drawing representation of an adult hermaphrodite gonad arm. The progression of germ cell proliferation and meiosis are indicated by the arrows starting from the distal tip region of the gonad arm. (B) SMC-6 staining was detected in the nucleus of germ cells in the distal gonad arm. Mitotic germ nuclei are located in the pre-meiotic region proximal to the distal tip. Pre-meiotic S phase nuclei should be located proximal to the transition zone nuclei [66]. The green bracket marks the transition zone region. (C) SMC-6 staining in pachytene nuclei appeared to accumulate on chromosomes, which differed from the more diffuse nucleoplasmic staining in the distal gonad arm in (B). In the diplotene (D) and diakinesis (E) stages of meiotic prophase I, SMC-5 and SMC-6 staining were detected exclusively on chromosomes. Scale bars = 5 µm.
Mutants of smc-5 and smc-6 exhibited reduced fecundity
The homozygous smc-5 and smc-6 mutants (F1) produced by heterozygous mutant parents and the F2 offspring produced by the homozygous smc-5 and smc-6 F1 mutants exhibited near wild-type embryonic viability (98% to 99% viable with n>2,000 per genotype). Since the F2 mutants lacked maternal and zygotic expression of the SMC-5 or the SMC-6 protein, we conclude that the C. elegans SMC-5 and SMC-6 proteins are dispensable for viability. However, the smc-5 and smc-6 mutants were difficult to maintain as homozygous mutant strains and were prone to becoming sterile (Figure S2A). The transgenerational sterility phenotype is typically associated with genomic instability in the germ cells [28]–[30]. By contrast, the homozygous brc-1(tm1145) mutant strains remained fecund (Figure S2A), which suggests the smc-5 and smc-6 mutations may be more disruptive to normal germ cell functions.
In agreement with the observed enrichment for the SMC-5 and SMC-6 proteins in germ cells, the smc-5 and smc-6 mutants had significantly smaller gonads containing fewer germ cells (Figure S3). In comparison to wild-type, there was on average a 30% to 50% reduction in fertilized eggs produced by the smc-5 and smc-6 F1 mutants and in smc-5 RNAi-treated worms (Figure S2B). Figure S2C shows that the smc-5(tm2868) mutant and the wild-type worms both began to lay eggs at approximately 1 day after L4 development. However, fertilized egg production declined in the smc-5(tm2868) mutant at an earlier age than wild-type (Figure S2C) and the mutant hermaphrodites began to lay unfertilized oocytes (data not shown). Similar phenotypes were also observed for the smc-5(ok2421) and the smc-6(ok3294) mutants (data not shown). The C. elegans hermaphrodite produces and stores a supply of sperm during late larval development, which will be used to fertilize oocytes produced in adulthood. The appearance of unfertilized oocytes could be caused by a premature depletion of hermaphrodite-produced sperm. The smc-5(tm2868) mutant hermaphrodites indeed showed an absence of sperm at an earlier age in adulthood (Figure S2D). However, male sperm supplied by mating failed to restore egg production (Figure S2E), which indicates that the loss of fecundity is not simply due to an insufficient amount of sperm.
Germ cells from the smc-5 and smc-6 mutants exhibited phenotypic defects in DNA damage repair–hypersensitivity to ionizing radiation and increased germ cell apoptosis
In order to determine if the smc-5 and smc-6 mutants have defects in the maintenance of germ cell genomic stability, we tested whether germ cells lacking SMC-5/6 functions are hypersensitive to ionizing radiation (IR). We followed a published protocol that examines the viability of eggs produced from radiation-damaged germ cells [31]. For each dose of gamma radiation exposure examined, the smc-5 and smc-6 mutants exhibited drastically reduced viability in comparison to wild type, consistent with the mutant germ cells being hypersensitive to DNA damage (Figure 3A).
Figure 3. Loss of function mutations in smc-5 and smc-6 conferred hypersensitivity to ionizing radiation (IR) and increased DNA-damage responses in germ cells.
(A) Graph of the viability of the eggs produced from mock and radiation-exposed germ cells in mutant F1 and wild-type L4-stage larvae as described [31]. 297 eggs or more were counted for each genotype and at each dose of IR. (B) Relative mRNA abundance for egl-1 after normalization to gamma tubulin tbg-1 as measured by quantitative RT-PCR [32]. Error bars represent standard errors. The asterisks (*) indicate significant changes from the wild-type control (p<0.001, t-Test). Gel analysis confirmed the size of each RT-PCR product and is shown underneath the graph. (C) Micrographs of germ cell corpses (white arrows) detected by CED-1::GFP fluorescence showed a greater number of corpses in the smc-5(tm2868) mutant germline. The average number of corpses per gonad and the standard error are indicated. Scale bars = 10 µm.
Because the smc-5 and smc-6 F1 mutants showed the aforementioned gonadal defects (Figures S2 and S3) even before IR treatment, we examined whether DNA damage may already be present in the germline of unchallenged smc-5 and smc-6 mutants. A well-characterized DNA damage response in the C. elegans hermaphrodite germline is the induction of apoptosis by CEP-1/p53, via the transactivation of the pro-apoptotic gene egl-1 [32]. Quantitative RT-PCR analysis showed a 5- to 6-fold increase in egl-1 transcript levels in the smc-5 and smc-6 mutant worms in comparison to wild type (Figure 3B). There was also an approximate two-fold increase in germ cell corpses in all three smc-5 and smc-6 mutant strains as detected by acridine orange staining (Table S1). This staining was specifically suppressed in the smc-5(tm2868);ced-3(n717) double mutant, which had an impaired ability to implement apoptosis (Table S1). The germ cell corpses also were confirmed in the smc-5(tm2868) mutant using a CED-1::GFP reporter that marked cell corpses during endocytosis (Figure 3C and Table S2). The RNAi knockdowns of three core pro-apoptotic genes ced-3, ced-4, and egl-1 suppressed the appearance of the GFP-positive germ cells in the smc-5(tm2868) mutant, indicating that they were apoptotic germ cell corpses (Table S2). In summary, the molecular, cytological and functional evidence demonstrated that the smc-5 and smc-6 mutations resulted in increased DNA damage response.
The smc-5 and smc-6 mutants showed an abnormal accumulation of SPO-11-dependent homologous recombination intermediates
We next examined whether meiotic DSB repair was compromised in the smc-5 and smc-6 mutant germ cells, by monitoring the appearance and disappearance of the HR strand exchange protein RAD-51 on meiotic chromosomes [33]. DNA DSBs are formed by the SPO-11 topoisomerase-like proteins at meiosis entry [34]. Following 5′-to-3′ single-strand end resection, the RAD-51 proteins localize to the single-stranded DNA to promote the invasion of intact homologous DNA template [14], [33]. RAD-51 focal formation usually reaches a maximum density (foci per nucleus) at early-pachytene and RAD-51 foci begin to disappear at mid-pachytene [14], [35], [36]. By late pachytene, there are few RAD-51 foci as HR repair is completed.
For comparison of equivalent gonadal regions between the wild-type and the mutant strains, we limited our analysis of RAD-51 focal staining to mutant gonad arms that were close in size to wild type. RAD-51 staining appeared in the distal region of the smc-5 and smc-6 mutant gonads, where it is rarely seen in wild-type (Figures S4A–S4C); the quantification of the results is shown in Figure S4F. The germ cells at the distal region are in mitosis and pre-meiotic DNA replication, and therefore prior to meiotic entry. We suspected that these RAD-51 foci were independent of meiotic DSB. In agreement with this prediction, we found that the RAD-51 foci in the distal gonads of both smc-5 mutant alleles (tm2868 and ok2421) persisted even though meiotic DSB formation was blocked by the spo-11(ok79) null mutation [34] (Figures S4D and S4E). The budding and fission yeast Smc5/6 mutants are known to exhibit Rad51-dependent DNA replication defects [20], [37]–[42]. The abnormal RAD-51 staining seen here in the pre-meiotic germline in C. elegans may represent a DNA replication defect.
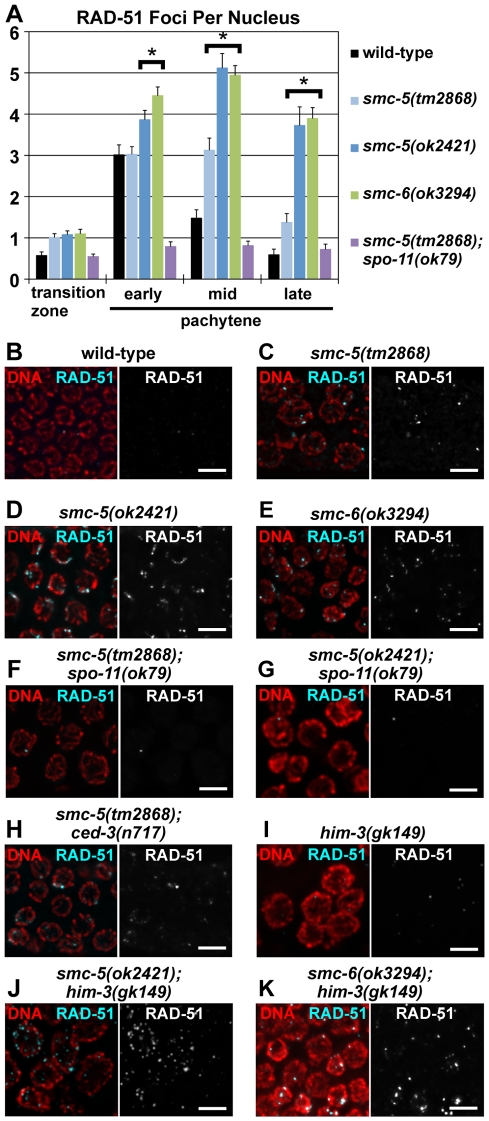
In the transition zone and at early pachytene, the meiotic germ cells exhibited slightly more RAD-51 foci in the smc-5 and smc-6 mutants than in wild-type (Figure 4A). The difference in the number of RAD-51 foci between the mutants and the wild-type was significantly more pronounced at mid- and late-pachytene (Figures 4A–4E). In contrast to the pre-meiotic RAD-51 staining, the aberrant pachytene RAD-51 staining in the smc-5(tm2868) and the smc-5(ok2421) mutants was significantly reduced by the spo-11(ok79) mutation (Figures 4F and 4G), which suggests that the RAD-51 staining defect at pachytene resulted primarily from a defect in meiotic DSB repair rather than from prior DNA damage in the pre-meiotic region.
Figure 4. Aberrant RAD-51 accumulation in meiotic germ cells from the smc-5 and smc-6 mutants.
(A) The average numbers of RAD-51 foci per nucleus in the transition zone and pachytene regions of the gonad arms are represented in the bar graph; the error bars represent standard errors. The pachytene region was divided into three equal length areas (early, mid and late) and each quantified separately. The smc-5 and smc-6 mutants showed significant increases in RAD-51 foci in comparison to wild-type at the mid- to late-pachytene regions (p≤0.01; two-tailed t-Test). The numerical values and sample sizes are summarized in Table S3. (B–K) Micrographs of RAD-51 immunofluorescence and DAPI-DNA fluorescence in late pachytene nuclei from the wild-type and the mutant germlines. Scale bars = 5 µm.
Because apoptosis in germ cells could generate corpses that have elevated RAD-51 staining, we also examined the smc-5(tm2868);ced-3(n717) double mutant that was deficient for germ cell apoptosis. The ced-3 mutation failed to suppress the aberrant RAD-51 staining at late pachytene in the smc-5(tm2868) mutant, thus ruling out the possibility that the RAD-51 defect was induced by apoptosis (Figure 4H).
Homologous recombination appears to be the predominant pathway for meiotic double-strand break repair in C. elegans [43], [44]. The lig-4(ok716) mutant in the canonical non-homologous end-joining (NHEJ) pathway has no measurable effects on RAD-51 accumulation in the germline [10]. Therefore, the aberrant RAD-51 staining in the smc-5 and smc-6 mutants likely reflects a defect in homologous recombination repair rather than NHEJ.
SMC-5 and SMC-6 are dispensable for inter-homolog crossover formation
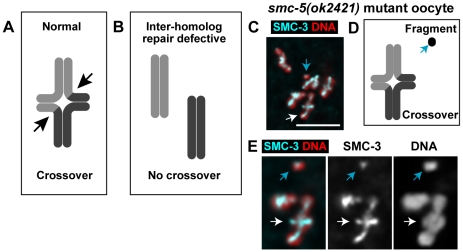
The failure to efficiently form inter-homolog crossover and the subsequent chromosome mis-segregation in meiosis would produce two expected phenotypes: low embryonic viability due to aneuploidy and higher than normal frequency of XO males due to X chromosome mis-segregation. The smc-5 and smc-6 F2 mutants exhibited normal embryonic viability (98% to 99%) and wild-type frequencies of male self-progeny (0.1% to 0.2%). Chiasmata formation appeared to be normal in the smc-5 and smc-6 mutant oocytes, because the mutant oocytes had an average of six DNA figures per oocyte consistent with normal inter-homolog linkage from the six pairs of homologous chromosomes (Table S4). We also observed the restructuring of meiotic chromosomes at late prophase consistent with chiasmata formation (Figure 5), as confirmed by cohesin immunostaining and DNA-DAPI fluorescence of the chromatid axes (Figures 5C–5E). The ZHP-3 proteins are proposed to couple synaptonemal complex morphogenesis and crossover formation and to mark precursor sites for inter-homolog crossover formation in late pachytene [45], [46]. The average number of ZHP-3 foci remained unchanged at the wild-type level of six foci per nucleus in the smc-5(ok2421) and smc-6(ok3294) mutant germ cells (Figure S5; [45]). Thus, the combination of phenotypic and cytological evidence confirmed inter-homolog crossover formation in the smc-5 and smc-6 mutant germlines.
Figure 5. Chromosome fragmentation defects in smc-5 and smc-6 mutant diakinesis oocytes that successfully formed crossovers.
Cartoon representations (A and B) of a pair of homologous chromosomes at diakinesis. (A) In wild-type, inter-homolog association is maintained by cohesion between the recombinant chromatids. The location of the chromatid exchange is indicated by the two black arrows, which provides the focal point for the restructuring of chromatid axes that results in a “cross-shape” or cruciform structure of the diakinesis chromosomes in C. elegans [67]. (B) If inter-homolog crossover failed to form, the homologous chromosomes would prematurely separate and the chromosome axes will not have the cruciform shape. (C) Micrograph of an smc-5(ok2421) mutant oocyte containing six linked homolog pairs with the wild-type cruciform shape, indicating successful inter-homolog crossover formation. Projection depth and scale bar = 5 µm. (D) A cartoon representation of the linked homolog and DNA fragment shown in (C and E). (E) Magnified view of a linked homolog pair (white arrow) and a chromosome fragment (light blue arrow) from (C) for which cohesin SMC-3 staining also was detected on the fragment. Projection depth = 2 µm.
The smc-5 and smc-6 mutations appeared to disrupt homolog-independent homologous recombination in meiosis
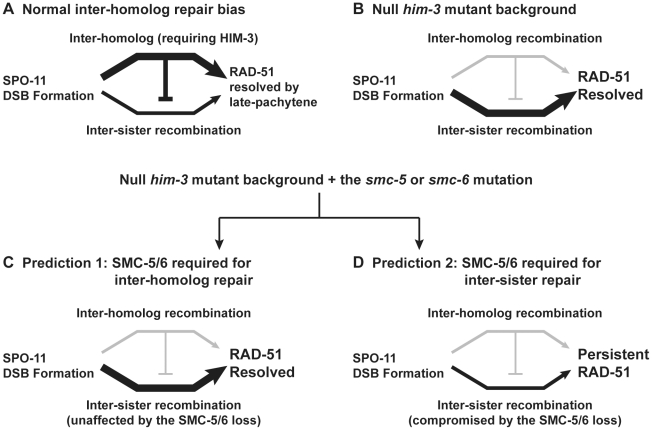
Based on our findings, the SMC-5/6 proteins appear to be required for homologous recombination that does not affect inter-homolog crossover formation, which possibly involves inter-homolog non-crossover repair or homolog-independent sister-chromatid recombination. To address the two possibilities, we tested whether the smc-5(ok2421) and the smc-6(ok3294) mutations will confer the RAD-51 defect in the homolog synapsis mutant him-3(gk149). HIM-3 is an axial element of the synaptonemal complex that promotes inter-homolog recombination by stabilizing the close association of homologous chromosomes in wild-type (Figure 6A; [47]). In the absence of HIM-3 (Figure 6B), inter-homolog non-crossover repair should be disrupted. If the smc-5 or the smc-6 mutation also disrupted inter-homolog non-crossover repair, then the him-3 double mutant with smc-5 or smc-6 should be similar to the him-3 single mutant (Figure 6C). Conversely, if the SMC-5/6 proteins promote homolog-independent recombination, which should be functional in the him-3 mutant [48], then the RAD-51 staining in the him-3 double mutant should be more persistent than the him-3 single mutant (Figure 6D).
Figure 6. Possible outcomes for the combined genetic disruptions of smc-5 or smc-6 and him-3.
(A) Schematic diagram depicts the biased repair of SPO-11 DSBs via inter-homolog recombination repair in meiosis. (B) In the absence of HIM-3, SPO-11 DSBs are repaired via homolog-independent (i.e. sister-chromatid) recombination [48]. (C–D) Two predicted repair outcomes for the double mutants. (C) If the SMC-5/6 proteins function mainly in inter-homolog repair which is already disrupted by the him-3 mutation, then the overall repair efficiency as monitored by the removal of RAD-51 staining should be the same in the double mutants as compared to the smc-5 and smc-6 single mutants. (D) If the SMC-5/6 proteins function mainly in inter-sister repair, then the double mutant should exacerbate the repair defects and the RAD-51 intermediates should persist at late pachytene.
The RAD-51 focal staining at late pachytene in the him-3(gk149) null mutant was similar to wild-type (Figure 4I; [48]). The smc-5(ok2421) or the smc-6(ok3294) mutation in the him-3 mutant genetic background drastically increased RAD-51 focal staining at late pachytene (Figures 4J and 4K), indicating that the smc-5 and smc-6 mutations impeded homolog-independent homologous recombination repair in meiosis.
The smc-5(ok2421) and the smc-6(ok3294) mutants exhibited chromosome fragmentation and dismorphology at diakinesis
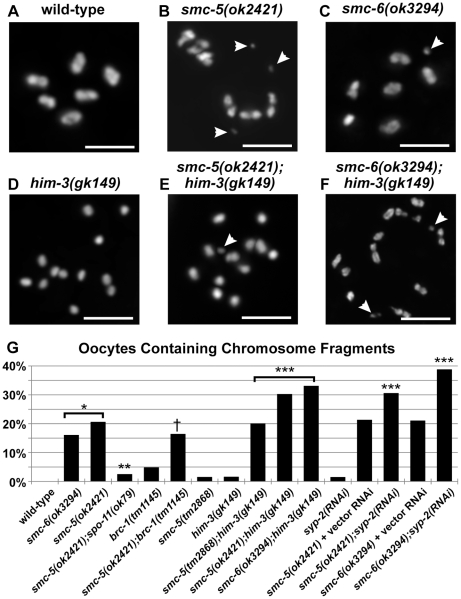
Approximately 20% of the diakinesis oocytes from the smc-5(ok2421) and smc-6(ok3294) mutants contained chromosome fragments (blue arrows in Figure 5, and white arrowheads in Figure 7). These fragments were disproportionately smaller than the linked homologs at diakinesis, and also showed staining for the cohesin SMC-3 protein indicating they were derived from chromosomes (Figure 5E). By contrast, no fragments were observed in wild-type oocytes (Figures 7A and 7G). Intriguingly, the fragmentation defect was rarely seen in the smc-5(tm2868) mutant oocytes and appeared to correlate with the severity of SMC-5 and SMC-6 protein disruption because the smc-5(tm2868) mutants still produced a truncated SMC-5 protein (Figure 1B). The fragmentation defect also correlated with the more severe RAD-51 staining defect at late pachytene that was found in the smc-5(ok2421) and smc-6(ok3294) mutants (Figure 4A). Similar to the RAD-51 defect, the spo-11(ok79) null mutation potently reduced fragmentation from 20.5% in smc-5(ok2421) mutant oocytes to 2.4% in the smc-5;spo-11 double mutant oocytes (Figure 7G).
Figure 7. Chromosome fragmentation defects linked to homolog-independent repair of meiotic DSB.
(A–F) Micrographs of DAPI-stained diakinesis chromosomes are shown at the same magnification. Chromosomal fragments are indicated by white arrowheads, and scale bars = 5 µm. (G) A graph of the percentages of diakinesis oocytes containing chromosome fragments. The smc-5(ok2421) and the smc-6(ok3294) mutant oocytes showed significantly higher frequency of chromosome fragmentation over wild-type (*p<0.001, Fisher's Exact Test). The smc-5(ok2421) fragmentation defect was drastically suppressed by the spo-11(ok79) mutation (**p = 0.006). The disruption of inter-homolog recombination by destabilizing homolog-synapsis consistently enhanced the fragmentation defects in all three smc-5 and smc-6 mutant strains (***). By contrast, the brc-1 mutation affecting sister-chromatid recombination [7] failed to enhance the fragmentation defect of the smc-5(ok2421) mutation (†). The numerical values, sample sizes, and statistical comparisons are summarized in Table S5.
The brc-1(tm1145) mutant reportedly exhibited a low penetrance fragmentation defect [7], which prompted us to compare the fragmentation defect in the brc-1(tm1145) mutant oocytes to the smc-5 and smc-6 mutants. There was an approximate three- to four-fold lower incidence of the fragmentation defect in the brc-1(tm1145) mutant oocytes in comparison to the smc-6(ok3294) and the smc-5(ok2421) mutants, respectively (Figure 7G). The smc-5 and smc-6 mutant oocytes also exhibited an unresolved diakinesis chromosome defect that was absent in the brc-1(tm1145) mutant and the wild-type oocytes (Figures S6A and S6B). Unresolved diakinesis chromosomes often appeared with a severe loss of homologous recombination function as found in the rad-51(lg8701) and other HR repair mutants (Figure S6A; [14],[33],[44],[49]). These results together indicate that the smc-5(ok2421) and the smc-6(ok3294) mutants may be more severely compromised for meiotic homologous recombination repair than the brc-1(tm1145) mutant.
We utilized chromosome fragmentation at diakinesis as a quantitative readout to determine the genetic interactions of smc-5 and smc-6 with inter-homolog recombination mutations or sister-chromatid recombination and cohesion mutations. If the fragmentation defect of the smc-5 and smc-6 mutants was due to disruptions in sister-chromatid recombination, then additional mutations that disrupted inter-homolog recombination (e.g. homolog-synapsis mutants) should enhance the fragmentation defect. Moreover, additional mutations that disrupted sister-chromatid recombination or cohesion would be expected to not enhance the fragmentation defect. We examined two homolog-synapsis defective conditions, using the him-3(gk149) null mutation and RNAi knockdown of syp-2, which encodes a synaptonemal complex central element protein [14]. While the loss of function for him-3 or syp-2 in the wild-type genetic background caused a modest chromosome fragmentation defect (1.4% to 1.5%), the homolog synapsis deficiencies consistently enhanced the penetrance of the fragmentation defects in the smc-5(ok2421) and the smc-6(ok3294) mutants. There were synergistic increases from an average of 20% fragmentation for the smc-5(ok2421) and smc-6(ok3294) single mutants to 30% to 39% for the double mutants (Figure 7G). This enhancement was more pronounced in the sensitized smc-5(tm2868) hypomorphic mutant. The smc-5(tm2868);him-3(gk149) double mutant oocytes exhibited chromosome fragments in 20% of the oocytes, which was a 10-fold increase in fragmentation compared to the smc-5(tm2868) and the him-3(gk149) single mutants (Figure 7G). By contrast, double mutants with brc-1(tm1145) (Figure 7G) or the cohesin Smc1 mutant, him-1(e879), failed to enhance the fragmentation defect (Figure S6C). The brc-1(tm1145) and the him-1(e879) mutations disrupt sister-chromatid recombination and cohesin function, respectively [7], [50]. Thus, the specific enhancements of the fragmentation defect by the loss of homolog-synapsis and not by further disruptions to sister-chromatid recombination support the hypothesis that the SMC-5/6 proteins function in meiotic sister-chromatid recombination.
Discussion
The successful completion of homolog-independent homologous recombination is crucial for genomic stability in germ cells
The requirement for homolog-independent recombination under normal meiotic conditions, when inter-homolog repair is functional, has not been tested. Challenges in addressing this question are the lack of experimental means to robustly disrupt homolog-independent recombination in meiosis without also perturbing inter-homolog recombination.
This study provides evidence that the SMC-5/6 proteins in C. elegans are required for the successful completion of homologous recombination repair of meiotic DSBs, consistent with similar observations in fission yeast [22]. Our findings further pinpoint this requirement for the SMC-5/6 proteins in C. elegans to homolog-independent homologous recombination. The smc-5 and smc-6 mutations disrupted the normal progression of homolog-independent homologous recombination intermediates in the him-3(gk149) mutants (Figures 4J and 4K). Although we have not directly assessed sister-chromatid recombination in the meiotic germ cells, the genetic interactions between smc-5(ok2421) and brc-1(tm1145) (Figure 7G) and between smc-5(ok2421) or smc-6(ok3294) with the him-1(e879) mutation (Figure S6C) are consistent with sister-chromatid recombination being disrupted in the smc-5(ok2421) and smc-6(ok3294) mutants. Additionally, the smc-5(ok2421) and smc-6(ok3294) mutants each showed a gradual transgenerational sterility defect, consistent with the loss of germ cell genomic stability. Together, the cytological and phenotypic evidence from the smc-5 and smc-6 mutants provides the first experimental support that the proper execution of homolog-independent recombination in meiosis is crucial for genomic stability in germ cells.
Chromosome fragmentation at diakinesis
A chromosome fragment arising simply from incomplete DSB repair in a chromatid should remain bound to its sister chromatid by cohesion and should not appear as a free fragment at diakinesis. Sister-chromatid cohesion is presumably maintained in the smc-5 and smc-6 mutants as evident by the intact chiasma and the cohesin SMC-3 staining on diakinesis chromosomes (Figure 5E and Table S4). Therefore, we hypothesize that the appearance of fragments may require the localized loss of chromatid cohesion near the site of homolog-independent repair in addition to incomplete or aberrant DSB repair. In budding yeast, cohesin-mediated repair in G2 phase of the mitotic cell cycle can establish de novo damage-induced cohesion [51], [52], and cohesin-mediated repair in both budding yeast and human requires the Smc5/6 complexes [19]–[21], [53]. Although evidence suggests that damage-induced cohesion may not form during meiosis in budding yeast [54], whether damage-induced cohesion exists in meiosis in other species remains to be determined. Intriguingly, the human Rec8 cohesin subunit that functions in meiosis was able to rescue damage-induced cohesion in the budding yeast scc1(mcd1) mutant [54].
As another possibility, the loss of Smc5/6 function could produce abnormal chromosome structures and fragments through ectopic recombination. The fission and budding yeast Smc5/6 complexes are thought to function in a late step in recombination repair, perhaps in the regulation of HR intermediates [20], [37], [55], [56]. Aside from DNA damage response, the yeast Smc5/6 complexes also are required for genomic stability during DNA replication. Budding and fission yeast mutants deficient for the Smc5/6 complexes were found to accumulate Rad51-dependent joint molecules near collapsed replication forks [37], [38]. These aberrant structures are thought to result from ectopic template switching events in the resuscitation of collapsed replication forks [38], [57]. A similar DNA replication defect in the C. elegans smc-5 and smc-6 mutant germ cells may account for the abnormal SPO-11-independent RAD-51 staining seen in the pre-meiotic region (Figure S4). This function may be analogous to restraining ectopic strand invasion events during homologous recombination repair. Such a defect during homologous recombination repair could explain the increased staining intensity for RAD-51 in the smc-5 and smc-6 mutant germ cells at pachytene. The proposed roles for the SMC-5/6 complex in damage-induced cohesion and the regulation of ectopic recombination may underlie meiotic defects not only in programmed DSB repair but also inappropriate cohesion loss near the DNA lesion, and the combination of both defects may account for the increased frequency for chromosome fragments at diakinesis.
An experimental model to examine homolog-independent homologous recombination repair
We showed that chromosome fragmentation and dismorphology defects were present in the smc-5 and smc-6 mutant oocytes just prior to fertilization, but they had no discernable effects on offspring viability and chromosome segregation. This finding supports our hypothesis that C. elegans, as an experimental model, could decouple the successful completion of meiotic DSB repair from the potentially lethal effects of chromosome fragments and other abnormal chromosome configurations. C. elegans may be uniquely useful to study the molecular mechanisms involved in homolog-independent repair, which may be utilized in many species to maintain genomic integrity during meiosis [6]–[11].
Materials and Methods
Genetics and phenotypic analyses
The C. elegans Bristol (N2) strain served as the wild-type control and all strains were maintained at 20°C under standard growth conditions [58]. Genetic mutations, rearrangements and transgenic reporters used in this study are as follows:
LGI: him-1(e879) [50]
LGII: smc-5(tm2868), smc-5(ok2421), smc-6(ok3294) [In this study], mIn1[dpy-10(e128) mIs14[myo-2::gpf; pes-10::gfp]] [59]
LGIII: brc-1(tm1145) [7]
LGIV: him-3(gk149) [48], spo-11(ok79) [34], rad-51(lg8701) [33], nT1[qIs51] (IV:V), nT1[unc-?(n754) let-?] (IV;V) [60]
LGV: bcIs39[lim-7::ced-1-gfp] [32],[61]
RNAi knockdown for smc-5 was induced by microinjection of 1–2 mg/mL of double-stranded RNA specific to the smc-5 gene in phosphate-buffered saline (PBS) in adult hermaphrodites at 1 day after L4 development, and mock control worms were injected with PBS containing no RNA. The smc-5 RNA was transcribed in vitro using T7 RNA polymerase (Promega) and a double-stranded DNA template containing T7 promoter sequences (underlined in lower case below) amplified from wild-type N2 genomic DNA using the following primers:
5′ gcgtaatacgactcactatagggTCCGTGTGCATTTCTTGCTC 3′ and
5′ gcgtaatacgactcactatagggATAGGCTTTCGAGGCATCAC 3′
The RNAi knockdown of syp-2 was performed as described [45].
For the germline apoptosis analysis, worm strains containing the CED-1::GFP reporter construct [32] were grown at 20°C until the L4 larval stage and were then shifted to 25°C for 16 hours prior to analysis for GFP-positive germ corpses.
For ionizing radiation hypersensitivity, L4 hermaphrodites from wild-type and F1 mutant strains were irradiated in a Philips RT250 orthovoltage unit at 30, 60 and 90 Gy of ionizing radiation. The worms recovered for 24 hours before eggs were collected and counted for embryo lethality as described [31]. 297 to 545 embryos were counted for each genotype per dose of radiation.
Quantitative RT-PCR analysis
Approximately 500 embryos were collected and grown for 72 hours at 20°C to young adulthood (approximately 1 day after L4 larval development), at which time they were harvested for RNA extraction using TRIzol reagent (Invitrogen). 250 ng of extracted RNA were used for oligo(dT)-primed cDNA synthesis using Superscript III First Strand Synthesis Kit (Invitrogen). Quantitative PCR were performed using the ABI 7500 Fast Real Time PCR system and the SYBR Green PCR kit (5 Prime) following manufacturer's protocol but at reduced 15 µL reaction volumes. PCR primers for egl-1 and gamma-tubulin tbg-1 are described in [32]. All RT-PCR products were confirmed by melt curve analysis and by gel analysis to verify that the amplification depended on reverse transcription and the cDNA products were the expected size (Figure 3B). The amounts of template cDNA were titrated to validate the measurements. The mRNA abundance was calculated based on threshold cycle numbers (Ct) and normalized to tbg-1 using the equation: relative abundance = 2−[(Ct,gene-of-interest)−(Ct,normalization control)]
Antibody preparation, western blotting and immunoprecipitation
Rabbit antibodies were raised to peptides containing an amino terminal cysteine-glycine (CG) linker and the following sequences:
SMC-5 carboxyl terminus: TNSHGKHYDTSAKIDATFAKMGISA
SMC-6 internal residues 849–868: DAMEMVENDKKNHPMPPGET
Antibodies were purified on peptide affinity columns prior to use. Rat antibodies to C. elegans SMC-3 used for immunofluorescence were described in [50]. Commercial rabbit anti-human Smc3 antibodies (Bethyl Laboratories, Inc., cat. no. A300-060A) were used for the immunoprecipitation study. The cross-reactivity of anti-human Smc3 antibodies to the C. elegans SMC-3 protein was confirmed by western blot and mass spectrometry analysis (data not shown) of precipitated proteins. Other antibodies used in the study include mouse anti-tubulin DM1alpha (Sigma), mouse anti-p-granule OIC1D4 (Developmental Studies Hybridoma Bank, [62]) and guinea pig anti-ZHP-3 [45]. Rabbit anti-RAD-51 antibodies obtained from two different sources had nearly identical staining (Strategic Diagnostics, Inc., cat. no. 2948.00.02; and [33])
Worm lysate preparation, western blot and immunoprecipitation were performed as described [50], [63]. The detection of the SMC-5 and SMC-6 proteins in whole worm lysates required 50 to 100 wild-type worms, and 150 to 200 smc-5(tm2868) mutant worms for the truncated SMC-5 protein.
Fluorescence microscopy
All micrographs were captured on an Olympus BX61 epifluorescence compound microscope with a Hamamatsu ORCA ER camera. Images of whole gonad arms were captured with a 10× Plan Fluorite (NA 0.3) dry objective. All other images were captured with a 60× Plan Apochromat (NA 1.35) oil objective at 0.25 to 0.28 µm z-sections and deconvolved using Huygens Essential software version 3.4 (Scientific Volume Imaging). Images were processed using ImageJ and Photoshop CS2 software packages. Slide preparation was performed as described [64] with the exception of ZHP-3 and RAD-51 (Strategic Diagnostics, Inc.) that were processed as described by [45]. Quantification of RAD-51 foci was performed on germline stained with the RAD-51 antibodies described by [33]. The intensity of RAD-51 fluorescence for individual gonads was normalized to the background fluorescence found in the rachis of the gonad. RAD-51 counts for the pre-meiotic nuclei were limited to the first eight rows of germ nuclei from the distal tip. The meiotic region of the germline was divided into four equal length sections from the beginning of transition zone (polarized chromosome morphology) to the exit from pachytene (onset of cellularization). The four sections were classified as transition zone, early-, mid- and late-pachytene.
Statistical evaluations were performed using the two-tailed t-Test using Microsoft Excel and the two-tailed Fisher's Exact test calculated as described [65].
Supporting Information
Immunostaining detection of the SMC-5 and SMC-6 proteins in the C. elegans germline is specific. Micrographs of pachytene and diakinesis germ cells from the smc-5 and smc-6 mutants co-stained with antibodies to the cohesin SMC-3 protein (A–E) and the SMC-6 protein (A–C, E, F and I) or the SMC-5 protein (D, G and H). The smc-5 and smc-6 mutations specifically reduced the immunostaining for their cognate proteins (A–E, and H). The enrichment of the SMC-5 and SMC-6 proteins on diakinesis chromosomes appeared to be inter-dependent (F, G and I). The smc-5(tm2868) mutation may retain some biological function, therefore we cannot rule out the possibility that some residual SMC-5/6 proteins are still present, but are below the limit of detection by immunostaining. Scale bars = 5 µm.
(1.41 MB TIF)
The smc-5 and smc-6 mutants exhibited compromised germline functions. (A) Transgenerational sterility appeared in the smc-5(ok2421) and smc-6(ok3294) homozygous mutant strains but was absent in the wild-type and the brc-1(tm1145) mutant strains. Individual L4 hermaphrodites were isolated to establish independent colonies (n = 19 to 20 colonies per genotype). For the mutant strains, the L4 hermaphrodites used to establish the colonies were homozygous F1 mutants produced by heterozygous mutant parents. Each colony was then maintained for 12 generations, during which time five L4 hermaphrodites from each plate were transferred to a new plate every four to five days to allow the next generation of offspring to develop into L4 larvae. If a plate representing an independent colony has less than five viable L4 progeny, then the line is considered to be “sterile”. The line-graph presents the percentages of the starting independent colonies that are sterile at each generation. (B) The smc-5 and smc-6 mutants produced fewer fertilized eggs compared to the wild-type strain. Fertilized eggs produced by individual hermaphrodites were counted every six to 12 hours for four days starting from the late L4 larval stage. Ten or more hermaphrodites were analyzed per genotype, generation and RNAi treatment condition. The bar graph represents the average of the total eggs produced by an individual hermaphrodite per genotype/RNAi condition, and the error bars represent the SEMs. (C) A comparison of the average number of eggs produced per day between age-matched wild-type (n = 6) and the smc-5(tm2868) homozygous F1 mutant strain (n = 17). The color key shown in (C) also applies to (D and E). (D) Gonads from the wild-type and the smc-5(tm2868) mutant hermaphrodites at the specified ages were dissected, DAPI-DNA stained and visualized on a compound epifluorescence microscope for the presence or absence of sperm in the spermatheca. 12 to 21 hermaphrodites were examined for each genotype. (E) The smc-5(tm2868) mutant hermaphrodites were allowed to mate with males marked by a Pmyo-2::GFP reporter for 24 hours. Following the mating procedure, individual hermaphrodites were then transferred to separate plates and the number of larval offspring produced was counted. Mated hermaphrodites were identified based on the presence of male and GFP-positive larval offspring, and vice versa for unmated hermaphrodites. Hermaphrodites that did not produce any offspring were excluded from the analysis. The bar graph represents the average number of offspring produced by mated (n = 24) and unmated (n = 12) smc-5(tm2868) F1 mutant hermaphrodites, and the error bars represent the SEMs.
(0.51 MB TIF)
The smc-5 and smc-6 F1 mutants have smaller gonads with few germ cells in comparison to the wild-type. (A–D) Micrographs of dissected DAPI-stained gonads from age-matched wild-type and smc-5 and smc-6 mutants are shown at the same magnification. The white bracket indicates the approximate length of the gonad arm from the distal tip to the end of pachytene.
(0.41 MB TIF)
Aberrant RAD-51 focal staining is found in the pre-meiotic region in the smc-5 and smc-6 mutants. (A–E) Micrographs of DAPI and RAD-51 antibody staining in dissected gonads. The genotypes are indicated at the top of each set of micrographs. The white dashed lines mark the pre-meiotic regions. Scale bars = 5 µm. (F) The average numbers of RAD-51 foci per nucleus are presented in the bar graph. The error bars represent the SEMs.
(2.48 MB TIF)
ZHP-3 localization appeared normal in the smc-5 and smc-6 mutants. Micrographs of DAPI and ZHP-3 antibody stained germ cells at the late pachytene stage. The genotypes are indicated at the top of each set of micrographs. The average numbers of ZHP-3 foci per late pachytene germ cell (± SEM) are indicated for the wild-type, the smc-5(ok2421) and the smc-6(ok3294) mutants. Scale bars = 5 µm.
(0.78 MB TIF)
The smc-5 and smc-6 mutant oocytes exhibit chromosome dismorphology resembling defects seen in the rad-51(lg8701) mutant. (A) Micrographs of diakinesis chromosomes visualized by DAPI-DNA fluorescence in which the chromosomes failed to resolve properly in the rad-51(lg8701), smc-5(ok2421) and smc-6(ok3294) mutants. (B) The bar graph represents the percentages of oocytes at the “−1” to “−3” positions of the gonad with less than 4 resolved DNA bodies (n = 30 oocytes per genotype). (C) The fragmentation defect of the smc-5(ok2421) and smc-6(ok3294) mutants were not enhanced by the cohesin him-1(e879) mutation. For each genotype, embryos were harvested and grown at the permissive temperature of 15-degree C for the him-1(e879) mutation until the worms had developed into late-stage L4 larvae. The worms were then shifted to the restrictive temperature of 25-degree C for 16 hours to disrupt cohesin function [1], before they were dissected and analyzed for the presence of DAPI-stained chromosome fragments. The difference in growth temperature had no obvious effects on the frequency of chromosome fragmentation in the smc-5(ok2421) and smc-6(ok3294) single mutants. More importantly, the him-1(e879) mutation did not enhance the fragmentation defect in the double mutants with either the smc-5(ok2421) and the smc-6(ok3294) mutant (Fisher's Exact Test, p values >0.8). The measurement counts and statistical comparisons are summarized in Table S5. [1] Chan RC, Chan A, Jeon M, Wu TF, Pasqualone D, et al. (2003) Chromosome cohesion is regulated by a clock gene paralogue TIM-1. Nature 423:1002–1009.
(0.29 MB TIF)
Average germ corpses per gonad measured by acridine-orange staining. Acridine-orange staining was carried out by incubating the worms in a M9 solution containing 50 µg/mL of acridine-orange (Anaspec) for 3 hours followed by washes and destaining with additional M9 solution as described [2]. The RNAi inactivation of pro-apoptotic genes was performed by the feeding RNAi method [3], for which the worms were fed on the RNAi vector containing bacteria for two successive generations. [2] Gartner A, MacQueen AJ, Villeneuve AM (2004) Methods for analyzing checkpoint responses in Caenorhabditis elegans. Methods Mol Biol 280:257–274. [3] Kamath RS, Ahringer J (2003) Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30:313–321.
(0.03 MB DOC)
Average germ corpses per gonad measured by CED-1::GFP.
(0.03 MB DOC)
Average number of RAD-51 foci per nucleus ± SEM in different regions of the germline from wild-type and mutant hermaphrodites. The sample size number (n) indicates the number of germ nuclei examined for each region per genotype.
(0.03 MB DOC)
Average number of similar size DAPI-stained DNA figures in diakinesis oocytes.
(0.03 MB DOC)
Proportion of oocytes with one or more chromosome fragments at diakinesis. Statistical comparisons were performed using the Fisher's Exact Test [4]. [4] Agresti A (1992) A survey of exact inference for contingency tables. Statistical Science 7:131–153.
(0.07 MB DOC)
Acknowledgments
We thank the Caenorhabditis Genetics Center, the C. elegans Gene Knockout Consortium and the Japanese National Bioresource Project for worm strains, A. Gartner for RAD-51 antibodies and helpful suggestions, N. Bhalla for ZHP-3 antibodies, B. Meyer for SMC-5 and SMC-6 peptides, and J. Li for the use of the real-time PCR machine. We also thank D. Mets, J. Moran, T. Glover, T. Wilson, K. Nabeshima, G. Csankovzski and reviewers for critical comments.
Footnotes
The authors have declared that no competing interests exist.
This work was supported by internal institutional funds and the Biological Sciences Scholars Program at the University of Michigan. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kadyk LC, Hartwell LH. Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics. 1992;132:387–402. doi: 10.1093/genetics/132.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sjogren C, Nasmyth K. Sister chromatid cohesion is required for postreplicative double-strand break repair in Saccharomyces cerevisiae. Curr Biol. 2001;11:991–995. doi: 10.1016/s0960-9822(01)00271-8. [DOI] [PubMed] [Google Scholar]
- 3.Page SL, Hawley RS. Chromosome choreography: the meiotic ballet. Science. 2003;301:785–789. doi: 10.1126/science.1086605. [DOI] [PubMed] [Google Scholar]
- 4.Zickler D, Kleckner N. Meiotic chromosomes: integrating structure and function. Annu Rev Genet. 1999;33:603–754. doi: 10.1146/annurev.genet.33.1.603. [DOI] [PubMed] [Google Scholar]
- 5.Martinez-Perez E, Colaiacovo MP. Distribution of meiotic recombination events: talking to your neighbors. Curr Opin Genet Dev. 2009;19:105–112. doi: 10.1016/j.gde.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwacha A, Kleckner N. Identification of joint molecules that form frequently between homologs but rarely between sister chromatids during yeast meiosis. Cell. 1994;76:51–63. doi: 10.1016/0092-8674(94)90172-4. [DOI] [PubMed] [Google Scholar]
- 7.Adamo A, Montemauri P, Silva N, Ward JD, Boulton SJ, et al. BRC-1 acts in the inter-sister pathway of meiotic double-strand break repair. EMBO Rep. 2008;9:287–292. doi: 10.1038/sj.embor.7401167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cromie GA, Hyppa RW, Taylor AF, Zakharyevich K, Hunter N, et al. Single Holliday junctions are intermediates of meiotic recombination. Cell. 2006;127:1167–1178. doi: 10.1016/j.cell.2006.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lange J, Skaletsky H, van Daalen SK, Embry SL, Korver CM, et al. Isodicentric Y chromosomes and sex disorders as byproducts of homologous recombination that maintains palindromes. Cell. 2009;138:855–869. doi: 10.1016/j.cell.2009.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smolikov S, Eizinger A, Hurlburt A, Rogers E, Villeneuve AM, et al. Synapsis-defective mutants reveal a correlation between chromosome conformation and the mode of double-strand break repair during C. elegans meiosis. Genetics. 2007;176:2027–2033. doi: 10.1534/genetics.107.076968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Webber HA, Howard L, Bickel SE. The cohesion protein ORD is required for homologue bias during meiotic recombination. J Cell Biol. 2004;164:819–829. doi: 10.1083/jcb.200310077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayashi M, Chin GM, Villeneuve AM. C. elegans germ cells switch between distinct modes of double-strand break repair during meiotic prophase progression. PLoS Genet. 2007;3:e191. doi: 10.1371/journal.pgen.0030191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mahadevaiah SK, Turner JM, Baudat F, Rogakou EP, de Boer P, et al. Recombinational DNA double-strand breaks in mice precede synapsis. Nat Genet. 2001;27:271–276. doi: 10.1038/85830. [DOI] [PubMed] [Google Scholar]
- 14.Colaiacovo MP, MacQueen AJ, Martinez-Perez E, McDonald K, Adamo A, et al. Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev Cell. 2003;5:463–474. doi: 10.1016/s1534-5807(03)00232-6. [DOI] [PubMed] [Google Scholar]
- 15.Haber JE, Thorburn PC, Rogers D. Meiotic and mitotic behavior of dicentric chromosomes in Saccharomyces cerevisiae. Genetics. 1984;106:185–205. doi: 10.1093/genetics/106.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murnane JP. Telomeres and chromosome instability. DNA Repair (Amst) 2006;5:1082–1092. doi: 10.1016/j.dnarep.2006.05.030. [DOI] [PubMed] [Google Scholar]
- 17.Dernburg AF. Here, there, and everywhere: kinetochore function on holocentric chromosomes. J Cell Biol. 2001;153:F33–38. doi: 10.1083/jcb.153.6.f33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hillers KJ, Villeneuve AM. Chromosome-wide control of meiotic crossing over in C. elegans. Curr Biol. 2003;13:1641–1647. doi: 10.1016/j.cub.2003.08.026. [DOI] [PubMed] [Google Scholar]
- 19.Potts PR, Porteus MH, Yu H. Human SMC5/6 complex promotes sister chromatid homologous recombination by recruiting the SMC1/3 cohesin complex to double-strand breaks. Embo J. 2006;25:3377–3388. doi: 10.1038/sj.emboj.7601218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Torres-Rosell J, Machin F, Farmer S, Jarmuz A, Eydmann T, et al. SMC5 and SMC6 genes are required for the segregation of repetitive chromosome regions. Nat Cell Biol. 2005;7:412–419. doi: 10.1038/ncb1239. [DOI] [PubMed] [Google Scholar]
- 21.De Piccoli G, Cortes-Ledesma F, Ira G, Torres-Rosell J, Uhle S, et al. Smc5-Smc6 mediate DNA double-strand-break repair by promoting sister-chromatid recombination. Nat Cell Biol. 2006;8:1032–1034. doi: 10.1038/ncb1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pebernard S, McDonald WH, Pavlova Y, Yates JR, 3rd, Boddy MN. Nse1, Nse2, and a novel subunit of the Smc5-Smc6 complex, Nse3, play a crucial role in meiosis. Mol Biol Cell. 2004;15:4866–4876. doi: 10.1091/mbc.E04-05-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taylor EM, Moghraby JS, Lees JH, Smit B, Moens PB, et al. Characterization of a novel human SMC heterodimer homologous to the Schizosaccharomyces pombe Rad18/Spr18 complex. Mol Biol Cell. 2001;12:1583–1594. doi: 10.1091/mbc.12.6.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cobbe N, Heck MM. The evolution of SMC proteins: phylogenetic analysis and structural implications. Mol Biol Evol. 2004;21:332–347. doi: 10.1093/molbev/msh023. [DOI] [PubMed] [Google Scholar]
- 25.McDonald WH, Pavlova Y, Yates JR, 3rd, Boddy MN. Novel essential DNA repair proteins Nse1 and Nse2 are subunits of the fission yeast Smc5-Smc6 complex. J Biol Chem. 2003;278:45460–45467. doi: 10.1074/jbc.M308828200. [DOI] [PubMed] [Google Scholar]
- 26.Zhao X, Blobel G. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc Natl Acad Sci U S A. 2005;102:4777–4782. doi: 10.1073/pnas.0500537102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sergeant J, Taylor E, Palecek J, Fousteri M, Andrews EA, et al. Composition and architecture of the Schizosaccharomyces pombe Rad18 (Smc5-6) complex. Mol Cell Biol. 2005;25:172–184. doi: 10.1128/MCB.25.1.172-184.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ahmed S, Hodgkin J. MRT-2 checkpoint protein is required for germline immortality and telomere replication in C. elegans. Nature. 2000;403:159–164. doi: 10.1038/35003120. [DOI] [PubMed] [Google Scholar]
- 29.Chin GM, Villeneuve AM. C. elegans mre-11 is required for meiotic recombination and DNA repair but is dispensable for the meiotic G(2) DNA damage checkpoint. Genes Dev. 2001;15:522–534. doi: 10.1101/gad.864101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smelick C, Ahmed S. Achieving immortality in the C. elegans germline. Ageing Res Rev. 2005;4:67–82. doi: 10.1016/j.arr.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 31.Gartner A, Milstein S, Ahmed S, Hodgkin J, Hengartner MO. A conserved checkpoint pathway mediates DNA damage–induced apoptosis and cell cycle arrest in C. elegans. Mol Cell. 2000;5:435–443. doi: 10.1016/s1097-2765(00)80438-4. [DOI] [PubMed] [Google Scholar]
- 32.Schumacher B, Schertel C, Wittenburg N, Tuck S, Mitani S, et al. C. elegans ced-13 can promote apoptosis and is induced in response to DNA damage. Cell Death Differ. 2005;12:153–161. doi: 10.1038/sj.cdd.4401539. [DOI] [PubMed] [Google Scholar]
- 33.Alpi A, Pasierbek P, Gartner A, Loidl J. Genetic and cytological characterization of the recombination protein RAD-51 in Caenorhabditis elegans. Chromosoma. 2003;112:6–16. doi: 10.1007/s00412-003-0237-5. [DOI] [PubMed] [Google Scholar]
- 34.Dernburg AF, McDonald K, Moulder G, Barstead R, Dresser M, et al. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell. 1998;94:387–398. doi: 10.1016/s0092-8674(00)81481-6. [DOI] [PubMed] [Google Scholar]
- 35.Martinez-Perez E, Villeneuve AM. HTP-1-dependent constraints coordinate homolog pairing and synapsis and promote chiasma formation during C. elegans meiosis. Genes Dev. 2005;19:2727–2743. doi: 10.1101/gad.1338505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mets DG, Meyer BJ. Condensins regulate meiotic DNA break distribution, thus crossover frequency, by controlling chromosome structure. Cell. 2009;139:73–86. doi: 10.1016/j.cell.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ampatzidou E, Irmisch A, O'Connell MJ, Murray JM. Smc5/6 is required for repair at collapsed replication forks. Mol Cell Biol. 2006;26:9387–9401. doi: 10.1128/MCB.01335-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Branzei D, Sollier J, Liberi G, Zhao X, Maeda D, et al. Ubc9- and mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell. 2006;127:509–522. doi: 10.1016/j.cell.2006.08.050. [DOI] [PubMed] [Google Scholar]
- 39.Branzei D, Vanoli F, Foiani M. SUMOylation regulates Rad18-mediated template switch. Nature. 2008;456:915–920. doi: 10.1038/nature07587. [DOI] [PubMed] [Google Scholar]
- 40.Irmisch A, Ampatzidou E, Mizuno K, O'Connell MJ, Murray JM. Smc5/6 maintains stalled replication forks in a recombination-competent conformation. Embo J. 2009;28:144–155. doi: 10.1038/emboj.2008.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Torres-Rosell J, De Piccoli G, Cordon-Preciado V, Farmer S, Jarmuz A, et al. Anaphase onset before complete DNA replication with intact checkpoint responses. Science. 2007;315:1411–1415. doi: 10.1126/science.1134025. [DOI] [PubMed] [Google Scholar]
- 42.Torres-Rosell J, Machin F, Aragon L. Smc5-Smc6 complex preserves nucleolar integrity in S. cerevisiae. Cell Cycle. 2005;4:868–872. doi: 10.4161/cc.4.7.1825. [DOI] [PubMed] [Google Scholar]
- 43.Clejan I, Boerckel J, Ahmed S. Developmental modulation of nonhomologous end joining in Caenorhabditis elegans. Genetics. 2006;173:1301–1317. doi: 10.1534/genetics.106.058628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin JS, Winkelmann N, Petalcorin MI, McIlwraith MJ, Boulton SJ. RAD-51-dependent and -independent roles of a Caenorhabditis elegans BRCA2-related protein during DNA double-strand break repair. Mol Cell Biol. 2005;25:3127–3139. doi: 10.1128/MCB.25.8.3127-3139.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bhalla N, Wynne DJ, Jantsch V, Dernburg AF. ZHP-3 acts at crossovers to couple meiotic recombination with synaptonemal complex disassembly and bivalent formation in C. elegans. PLoS Genet. 2008;4:e1000235. doi: 10.1371/journal.pgen.1000235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saito TT, Youds JL, Boulton SJ, Colaiacovo MP. Caenorhabditis elegans HIM-18/SLX-4 interacts with SLX-1 and XPF-1 and maintains genomic integrity in the germline by processing recombination intermediates. PLoS Genet. 2009;5:e1000735. doi: 10.1371/journal.pgen.1000735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zetka MC, Kawasaki I, Strome S, Mueller F. Synapsis and chiasma formation in Caenorhabditis elegans require HIM-3, a meiotic chromosome core component that functions in chromosome segregation. Genes Dev. 1999;13:2258–2270. doi: 10.1101/gad.13.17.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Couteau F, Nabeshima K, Villeneuve A, Zetka M. A component of C. elegans meiotic chromosome axes at the interface of homolog alignment, synapsis, nuclear reorganization, and recombination. Curr Biol. 2004;14:585–592. doi: 10.1016/j.cub.2004.03.033. [DOI] [PubMed] [Google Scholar]
- 49.Rinaldo C, Bazzicalupo P, Ederle S, Hilliard M, La Volpe A. Roles for Caenorhabditis elegans rad-51 in meiosis and in resistance to ionizing radiation during development. Genetics. 2002;160:471–479. doi: 10.1093/genetics/160.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chan RC, Chan A, Jeon M, Wu TF, Pasqualone D, et al. Chromosome cohesion is regulated by a clock gene paralogue TIM-1. Nature. 2003;423:1002–1009. doi: 10.1038/nature01697. [DOI] [PubMed] [Google Scholar]
- 51.Strom L, Karlsson C, Lindroos HB, Wedahl S, Katou Y, et al. Postreplicative formation of cohesion is required for repair and induced by a single DNA break. Science. 2007;317:242–245. doi: 10.1126/science.1140649. [DOI] [PubMed] [Google Scholar]
- 52.Unal E, Heidinger-Pauli JM, Koshland D. DNA double-strand breaks trigger genome-wide sister-chromatid cohesion through Eco1 (Ctf7). Science. 2007;317:245–248. doi: 10.1126/science.1140637. [DOI] [PubMed] [Google Scholar]
- 53.Lindroos HB, Strom L, Itoh T, Katou Y, Shirahige K, et al. Chromosomal association of the Smc5/6 complex reveals that it functions in differently regulated pathways. Mol Cell. 2006;22:755–767. doi: 10.1016/j.molcel.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 54.Heidinger-Pauli JM, Unal E, Guacci V, Koshland D. The kleisin subunit of cohesin dictates damage-induced cohesion. Mol Cell. 2008;31:47–56. doi: 10.1016/j.molcel.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 55.Cost GJ, Cozzarelli NR. Smc5p promotes faithful chromosome transmission and DNA repair in Saccharomyces cerevisiae. Genetics. 2006;172:2185–2200. doi: 10.1534/genetics.105.053876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pebernard S, Wohlschlegel J, McDonald WH, Yates JR, 3rd, Boddy MN. The Nse5-Nse6 dimer mediates DNA repair roles of the Smc5-Smc6 complex. Mol Cell Biol. 2006;26:1617–1630. doi: 10.1128/MCB.26.5.1617-1630.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Murray JM, Carr AM. Smc5/6: a link between DNA repair and unidirectional replication? Nat Rev Mol Cell Biol. 2008;9:177–182. doi: 10.1038/nrm2309. [DOI] [PubMed] [Google Scholar]
- 58.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Edgley ML, Riddle DL. LG II balancer chromosomes in Caenorhabditis elegans: mT1(II;III) and the mIn1 set of dominantly and recessively marked inversions. Mol Genet Genomics. 2001;266:385–395. doi: 10.1007/s004380100523. [DOI] [PubMed] [Google Scholar]
- 60.Ferguson EL, Horvitz HR. Identification and characterization of 22 genes that affect the vulval cell lineages of the nematode Caenorhabditis elegans. Genetics. 1985;110:17–72. doi: 10.1093/genetics/110.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou Z, Hartwieg E, Horvitz HR. CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans. Cell. 2001;104:43–56. doi: 10.1016/s0092-8674(01)00190-8. [DOI] [PubMed] [Google Scholar]
- 62.Strome S. Asymmetric movements of cytoplasmic components in Caenorhabditis elegans zygotes. J Embryol Exp Morphol. 1986;97(Suppl):15–29. [PubMed] [Google Scholar]
- 63.Chan RC, Severson AF, Meyer BJ. Condensin restructures chromosomes in preparation for meiotic divisions. J Cell Biol. 2004;167:613–625. doi: 10.1083/jcb.200408061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Howe M, McDonald KL, Albertson DG, Meyer BJ. HIM-10 is required for kinetochore structure and function on Caenorhabditis elegans holocentric chromosomes. J Cell Biol. 2001;153:1227–1238. doi: 10.1083/jcb.153.6.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Agresti A. A survey of exact inference for contingency tables. Statistical Science. 1992;7:131–153. [Google Scholar]
- 66.Jaramillo-Lambert A, Ellefson M, Villeneuve AM, Engebrecht J. Differential timing of S phases, X chromosome replication, and meiotic prophase in the C. elegans germ line. Dev Biol. 2007;308:206–221. doi: 10.1016/j.ydbio.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 67.Nabeshima K, Villeneuve AM, Hillers KJ. Chromosome-wide regulation of meiotic crossover formation in Caenorhabditis elegans requires properly assembled chromosome axes. Genetics. 2004;168:1275–1292. doi: 10.1534/genetics.104.030700. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Immunostaining detection of the SMC-5 and SMC-6 proteins in the C. elegans germline is specific. Micrographs of pachytene and diakinesis germ cells from the smc-5 and smc-6 mutants co-stained with antibodies to the cohesin SMC-3 protein (A–E) and the SMC-6 protein (A–C, E, F and I) or the SMC-5 protein (D, G and H). The smc-5 and smc-6 mutations specifically reduced the immunostaining for their cognate proteins (A–E, and H). The enrichment of the SMC-5 and SMC-6 proteins on diakinesis chromosomes appeared to be inter-dependent (F, G and I). The smc-5(tm2868) mutation may retain some biological function, therefore we cannot rule out the possibility that some residual SMC-5/6 proteins are still present, but are below the limit of detection by immunostaining. Scale bars = 5 µm.
(1.41 MB TIF)
The smc-5 and smc-6 mutants exhibited compromised germline functions. (A) Transgenerational sterility appeared in the smc-5(ok2421) and smc-6(ok3294) homozygous mutant strains but was absent in the wild-type and the brc-1(tm1145) mutant strains. Individual L4 hermaphrodites were isolated to establish independent colonies (n = 19 to 20 colonies per genotype). For the mutant strains, the L4 hermaphrodites used to establish the colonies were homozygous F1 mutants produced by heterozygous mutant parents. Each colony was then maintained for 12 generations, during which time five L4 hermaphrodites from each plate were transferred to a new plate every four to five days to allow the next generation of offspring to develop into L4 larvae. If a plate representing an independent colony has less than five viable L4 progeny, then the line is considered to be “sterile”. The line-graph presents the percentages of the starting independent colonies that are sterile at each generation. (B) The smc-5 and smc-6 mutants produced fewer fertilized eggs compared to the wild-type strain. Fertilized eggs produced by individual hermaphrodites were counted every six to 12 hours for four days starting from the late L4 larval stage. Ten or more hermaphrodites were analyzed per genotype, generation and RNAi treatment condition. The bar graph represents the average of the total eggs produced by an individual hermaphrodite per genotype/RNAi condition, and the error bars represent the SEMs. (C) A comparison of the average number of eggs produced per day between age-matched wild-type (n = 6) and the smc-5(tm2868) homozygous F1 mutant strain (n = 17). The color key shown in (C) also applies to (D and E). (D) Gonads from the wild-type and the smc-5(tm2868) mutant hermaphrodites at the specified ages were dissected, DAPI-DNA stained and visualized on a compound epifluorescence microscope for the presence or absence of sperm in the spermatheca. 12 to 21 hermaphrodites were examined for each genotype. (E) The smc-5(tm2868) mutant hermaphrodites were allowed to mate with males marked by a Pmyo-2::GFP reporter for 24 hours. Following the mating procedure, individual hermaphrodites were then transferred to separate plates and the number of larval offspring produced was counted. Mated hermaphrodites were identified based on the presence of male and GFP-positive larval offspring, and vice versa for unmated hermaphrodites. Hermaphrodites that did not produce any offspring were excluded from the analysis. The bar graph represents the average number of offspring produced by mated (n = 24) and unmated (n = 12) smc-5(tm2868) F1 mutant hermaphrodites, and the error bars represent the SEMs.
(0.51 MB TIF)
The smc-5 and smc-6 F1 mutants have smaller gonads with few germ cells in comparison to the wild-type. (A–D) Micrographs of dissected DAPI-stained gonads from age-matched wild-type and smc-5 and smc-6 mutants are shown at the same magnification. The white bracket indicates the approximate length of the gonad arm from the distal tip to the end of pachytene.
(0.41 MB TIF)
Aberrant RAD-51 focal staining is found in the pre-meiotic region in the smc-5 and smc-6 mutants. (A–E) Micrographs of DAPI and RAD-51 antibody staining in dissected gonads. The genotypes are indicated at the top of each set of micrographs. The white dashed lines mark the pre-meiotic regions. Scale bars = 5 µm. (F) The average numbers of RAD-51 foci per nucleus are presented in the bar graph. The error bars represent the SEMs.
(2.48 MB TIF)
ZHP-3 localization appeared normal in the smc-5 and smc-6 mutants. Micrographs of DAPI and ZHP-3 antibody stained germ cells at the late pachytene stage. The genotypes are indicated at the top of each set of micrographs. The average numbers of ZHP-3 foci per late pachytene germ cell (± SEM) are indicated for the wild-type, the smc-5(ok2421) and the smc-6(ok3294) mutants. Scale bars = 5 µm.
(0.78 MB TIF)
The smc-5 and smc-6 mutant oocytes exhibit chromosome dismorphology resembling defects seen in the rad-51(lg8701) mutant. (A) Micrographs of diakinesis chromosomes visualized by DAPI-DNA fluorescence in which the chromosomes failed to resolve properly in the rad-51(lg8701), smc-5(ok2421) and smc-6(ok3294) mutants. (B) The bar graph represents the percentages of oocytes at the “−1” to “−3” positions of the gonad with less than 4 resolved DNA bodies (n = 30 oocytes per genotype). (C) The fragmentation defect of the smc-5(ok2421) and smc-6(ok3294) mutants were not enhanced by the cohesin him-1(e879) mutation. For each genotype, embryos were harvested and grown at the permissive temperature of 15-degree C for the him-1(e879) mutation until the worms had developed into late-stage L4 larvae. The worms were then shifted to the restrictive temperature of 25-degree C for 16 hours to disrupt cohesin function [1], before they were dissected and analyzed for the presence of DAPI-stained chromosome fragments. The difference in growth temperature had no obvious effects on the frequency of chromosome fragmentation in the smc-5(ok2421) and smc-6(ok3294) single mutants. More importantly, the him-1(e879) mutation did not enhance the fragmentation defect in the double mutants with either the smc-5(ok2421) and the smc-6(ok3294) mutant (Fisher's Exact Test, p values >0.8). The measurement counts and statistical comparisons are summarized in Table S5. [1] Chan RC, Chan A, Jeon M, Wu TF, Pasqualone D, et al. (2003) Chromosome cohesion is regulated by a clock gene paralogue TIM-1. Nature 423:1002–1009.
(0.29 MB TIF)
Average germ corpses per gonad measured by acridine-orange staining. Acridine-orange staining was carried out by incubating the worms in a M9 solution containing 50 µg/mL of acridine-orange (Anaspec) for 3 hours followed by washes and destaining with additional M9 solution as described [2]. The RNAi inactivation of pro-apoptotic genes was performed by the feeding RNAi method [3], for which the worms were fed on the RNAi vector containing bacteria for two successive generations. [2] Gartner A, MacQueen AJ, Villeneuve AM (2004) Methods for analyzing checkpoint responses in Caenorhabditis elegans. Methods Mol Biol 280:257–274. [3] Kamath RS, Ahringer J (2003) Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30:313–321.
(0.03 MB DOC)
Average germ corpses per gonad measured by CED-1::GFP.
(0.03 MB DOC)
Average number of RAD-51 foci per nucleus ± SEM in different regions of the germline from wild-type and mutant hermaphrodites. The sample size number (n) indicates the number of germ nuclei examined for each region per genotype.
(0.03 MB DOC)
Average number of similar size DAPI-stained DNA figures in diakinesis oocytes.
(0.03 MB DOC)
Proportion of oocytes with one or more chromosome fragments at diakinesis. Statistical comparisons were performed using the Fisher's Exact Test [4]. [4] Agresti A (1992) A survey of exact inference for contingency tables. Statistical Science 7:131–153.
(0.07 MB DOC)