Abstract
Calcium has a pivotal role in biological functions, and serum calcium levels have been associated with numerous disorders of bone and mineral metabolism, as well as with cardiovascular mortality. Here we report results from a genome-wide association study of serum calcium, integrating data from four independent cohorts including a total of 12,865 individuals of European and Indian Asian descent. Our meta-analysis shows that serum calcium is associated with SNPs in or near the calcium-sensing receptor (CASR) gene on 3q13. The top hit with a p-value of 6.3×10-37 is rs1801725, a missense variant, explaining 1.26% of the variance in serum calcium. This SNP had the strongest association in individuals of European descent, while for individuals of Indian Asian descent the top hit was rs17251221 (p = 1.1×10-21), a SNP in strong linkage disequilibrium with rs1801725. The strongest locus in CASR was shown to replicate in an independent Icelandic cohort of 4,126 individuals (p = 1.02×10-4). This genome-wide meta-analysis shows that common CASR variants modulate serum calcium levels in the adult general population, which confirms previous results in some candidate gene studies of the CASR locus. This study highlights the key role of CASR in calcium regulation.
Author Summary
Calcium levels in blood serum play an important role in many biological processes. The regulation of serum calcium is under strong genetic control. This study describes the first meta-analysis of a genome-wide association study from four cohorts totaling 12,865 participants of European and Indian Asian descent. Confirming previous results in some candidate gene studies, we find that common polymorphisms at the calcium-sensing receptor (CASR) gene locus are associated with serum calcium concentrations. We show that CASR variants give rise to the strongest signals associated with serum calcium levels in both European and Indian Asian populations, while no other locus reaches genome-wide significance. Our results show that CASR is a key player in genetic regulation of serum calcium in the adult general population.
Introduction
Calcium is the most abundant mineral in the human body contributing approximately one kilogram to the average adult human body mass. Whereas 99% of calcium is stored in the skeleton and teeth, the remaining 1% circulates in the bloodstream and is involved in many physiological processes including its function as a universal cellular signaling molecule [1]–[2]. Calcium plays a key role in membrane potential, which is important for muscle contraction, heart rate regulation and generation of nerve impulses. Calcium also influences bone metabolism, ion transport and many other cellular processes [3]. Approximately 2/5 of calcium in the extracellular fluid is found in blood serum. The level of serum calcium is under tight hormonal control with a normal concentration of 2.15–2.55 mmol/L. Serum calcium is under strong genetic control, with twin studies showing that the variance in total calcium due to genetic effects is between 50% and 78% [4]–[5].
While skeletal calcium is important in numerous clinical disorders, in particular bone and mineral disorders, the clinical role of serum calcium is less clear. Several [6]–[7] (but not all [8]) studies indicated that elevated serum calcium levels are associated with an increased risk of cardiovascular disease. Patients with hyperparathyroidism, who suffer from chronic hypercalcemia, have a high prevalence of hypertension and increased cardiovascular mortality [9]–[11]. However, the mechanisms underlying the putative association of serum calcium with increased cardiovascular morbidity and mortality remain unclear.
Rare monogenic forms of hypo- or hypercalcemia have been described, including disorders involving the calcium-sensing receptor (CASR, locus 3q13) gene. Heterozygous and homozygous CASR mutations that inactivate CASR are responsible, respectively, for familial hypocalciuric hypercalcemia, type 1 (also known as familial benign hypercalcemia) (OMIM #145980) [12]–[13] and neonatal severe hyperparathyroidism (OMIM #239200) [13]. On the other hand, mutations that result in CASR activation lead to autosomal dominant hypocalcemia (OMIM #146200) [14]. Mutations in many other genes have also been found to lead to disturbances of serum calcium levels (Table 1).
Table 1. Serum calcium candidate genes.
| Gene | Gene region | Disease | OMIM | # SNPs* | Top SNP† | Top SNP p-value† |
| AIRE | chr21: 44,530,191-44,542,530 | Autoimmune polyendocrine syndrome, type I | 240300 | 870 | rs2838473, rs13052277, rs717177 | 2.48E-04, 2.78E-05, 4.55E-03 |
| ALPL | chr1: 21,581,175-21,650,208 | Hypophosphatasia, infantile | 241500 | 786 | rs6426723, rs1256348, rs4654973 | 6.88E-03, 1.14E-03, 7.30E-04 |
| BSND | chr1: 55,176,638-55,186,485 | Bartter syndrome, type 4 | 602522 | 1007 | rs17111592, rs11584093, rs6588528 | 4.61E-03, 8.40E-05, 1.70E-02 |
| CASR | chr3:123,385,220-123,488,032 | Familial hypocalciuric hypercalcemia, type 1; neonatal severe hyperparathyroidism; autosomal dominant hypocalcemia | 145980; 239200; 146200; 241200 | 921 | rs1801725, rs1801725, rs17251221 | 6.29E-37, 2.58E-18, 1.07E-21 |
| CDKN1B | chr12: 12,761,576-12,766,569 | Bartter syndrome, type 4 | 131100 | 855 | rs3825271, rs888200, rs11055225 | 7.05E-04, 3.56E-03, 8.66E-04 |
| CLCNKA | chr1: 16,093,672-16,105,850 | Bartter syndrome, type 4 | 602522 | 545 | rs12405694, rs16852052, rs6661012 | 1.34E-02, 2.51E-02, 1.00E-02 |
| CLCNKB | chr1: 16,115,658-16,128,782 | Bartter syndrome, type 3; Bartter syndrome, type 4 | 607364; 602522 | 520 | rs12405694, rs16852052, rs6661012 | 1.34E-02, 2.51E-02, 1.00E-02 |
| CLDN16 | chr3: 191,588,543-191,611,035 | Hypomagnesemia 3, renal | 248250 | 1381 | rs11714779, rs11714779, rs9682599 | 3.15E-04, 2.86E-04, 3.18E-02 |
| CYP27B1 | chr12: 56,442,384-56,447,145 | Vitamin D-dependant rickets type I | 264700 | 567 | rs11172284, rs810204, rs715930 | 2.18E-03, 2.36E-03, 3.26E-02 |
| GATA3 | chr10: 8,136,673-8,157,170 | Hypoparathyroidism; sensorineural deafness; renal disease | 146255 | 1492 | rs11812109, rs12359361, rs2765399 | 1.30E-03, 2.86E-03, 4.35E-03 |
| GCM2 | chr6: 10,981,450-10,990,084 | Familial hyperparathyroidism | 146200 | 908 | rs16870899, rs16870899, rs6457160 | 4.88E-03, 4.59E-03, 3.36E-03 |
| GNAS | chr20: 56,900,130-56,919,640 | Pseudohypoparathyroidism, type IA; Pseudohypoparathyroidism, type IB | 103580; 603233 | 949 | rs2145477, rs911297, rs6015375 | 2.38E-04, 3.95E-03, 1.12E-02 |
| HRPT2 | chr1: 189,822,81-189,952,713 | Hyperparathyroidism (familial isolated hyperparathyroidism); parathyroid carcinoma | 145000; 608266 | 836 | rs10737627, rs913478, rs2887613 | 1.15E-02, 6.50E-03, 4.71E-02 |
| KCNJ1 | chr11: 128,213,12-128,242,478 | Bartter syndrome, antenatal, type 2 | 241200 | 1059 | rs948215, rs3897566, rs7116606 | 1.08E-03, 1.50E-03, 3.46E-05 |
| MEN1 | chr11: 64,327,572-64,335,342 | Hyperparathyroidism (familial isolated hyperparathyroidism); multiple endocrine neoplasia, type I | 145000; 131100 | 498 | rs7947143, rs7947143, rs11820322 | 1.49E-02, 6.83E-03, 2.92E-03 |
| PHEX | chrX: 21,810,216-22,025,985 | Hypophosphatemic rickets, X-linked dominant | 307800 | NA | NA | NA |
| PTH | chr11: 13,470,178-13,474,143 | Familial hyperparathyroidism | 146200 | 1292 | rs10832087, rs10500780, rs1502242 | 9.61E-04, 1.71E-03, 1.96E-03 |
| PTH1R | chr3: 46,894,240-46,920,291 | Jansen's metaphyseal chondrodysplasia | 156400 | 511 | rs1402151, rs883739, rs6442037 | 1.03E-02, 7.54E-03, 2.18E-02 |
| RET | chr10: 42,892,533-42,944,955 | Multiple endocrine neoplasia, type I | 131100 | 856 | rs3026762, rs3026762, rs12265792 | 2.10E-04, 1.90E-04, 4.10E-02 |
| SLC12A1 | chr15: 46,285,790-46,383,568 | Bartter syndrome, antenatal, type I | 601678 | 925 | rs1025759, rs596942, rs919129 | 2.25E-03, 1.29E-02, 2.30E-04 |
| SLC4A1 | chr17: 39,682,566-39,700,993 | Renal tubular acidosis, distal, autosomal dominant | 179800 | 494 | rs12602991, rs12602991, rs708384 | 2.18E-03, 7.20E-03, 4.05E-03 |
| TBCE | chr1: 231,856,81-231,938,321 | Hypoparathyroidism-retardation-dysmorphism syndrome | 241410 | 591 | rs12133603, rs12133603, rs291353 | 2.34E-03, 2.18E-03, 5.38E-03 |
| TRPM6 | chr9: 74,566,965-74,732,564 | Hypomagnesemia with secondary hypocalcemia | 602014 | 1123 | rs877809, rs877809, rs12550903 | 9.31E-04, 1.20E-03, 6.14E-03 |
| VDR | chr12: 46,521,589-46,585,081 | Vitamin D-resistant rickets type II; vitamin D-dependent rickets, type II | 277440; 259700 | 1066 | rs1859441, rs1859441, rs11168354 | 1.22E-03, 9.59E-04, 3.34E-04 |
Genes which have been shown to lead to disturbances of serum calcium levels. For each gene, we report the top SNP for the meta-analyses of all cohorts, European cohorts only and Indian Asian cohorts only.
*500 kb upstream and downstream the gene region.
†Combined, European, and Indian Asian.
In the present study, we report results obtained from meta-analysis of genome-wide associations of serum calcium levels from four cohorts with a total of 12,865 participants. We first describe the design of the study and its main finding, that variants in CASR give rise to the strongest signals associated with serum calcium levels in both European and Indian Asian populations. Our results confirm previous studies showing that mutations in CASR are associated with serum calcium levels in young healthy women [15]–[16] and extend this observation to men and women across a large spectrum of age. We show that CASR is a key player in the genetic regulation of serum calcium in men and women from the general adult population.
Results
We performed a meta-analysis for genome-wide associations of serum calcium, determined by subtracting the estimated amount of calcium bound to albumin from the total serum calcium, to infer the amount of ionized calcium (see Materials and Methods). Our study included four cohorts: (i) 5404 European individuals from the Cohorte Lausanne (CoLaus) [17]–[18], (ii) 5548 European and Indian Asian individuals from the London Life Sciences Population (LOLIPOP) Study from West London UK [19]–[20], (iii) 1196 European individuals from the InCHIANTI Study (Tuscany, Italy) [21], and (iv) 717 individuals of European descent from the Baltimore Longitudinal Study of Aging (BLSA) study based in the Baltimore-Washington DC area [22], totaling 12,865 participants (see Table 2 for more detailed characteristics of each cohort).
Table 2. Characteristics of participants, by study.
| CoLaus | LOLIPOP European Whites | LOLIPOP Indian Asians | InCHIANTI | BLSA | deCODE | |
| Sample size | 5404 | 1601 | 3947 | 1196 | 717 | 4126 |
| Gender (males/females) | 2542/2862 | 1397/204 | 3832/115 | 533/663 | 390/327 | 1313/2813 |
| Age(years)* | 53.43(34.9,75.4) | 54.5(22.6,75.0) | 50.7(35.0,74.9) | 68.22(21,102) | 70.4(22,98) | 60.1(7,103) |
| Pre/Post menopause | 1210/1652 | 100/104 | 56/59 | 79/584 | 38/289 | 872/1836 (105 with pre and post measurements) |
| Serum calcium(mmol/L)* | 2.29(0.094) | 2.41(0.12) | 2.37(0.11) | 2.36(0.10) | 2.31(0.11) | 2.38(0.14) |
| Corrected serumCalcium (mmol/L)* | 2.18(0.09) | 2.31(0.09) | 2.29(0.09) | 2.30(0.09) | 2.30(0.1) | 2.31(0.14) |
| Serum albumin[g/L]* | 44.2(2.5) | 43.7(2.9) | 43.4(2.9) | 42.3(3.1) | 40.4(3.5) | 42.9(3.9) |
Characteristics are shown for CoLaus, LOLIPOP European, LOLIPOP Indian Asian, InCHIANTI, BLSA and deCODE. Corrected serum calcium, designed to estimate the amount of biologically active serum calcium, is defined as Ca_corrected = total serum calcium [mmol/L] + (40 - albumin [g/L])/40.
*Values represent mean (range) or mean (sd).
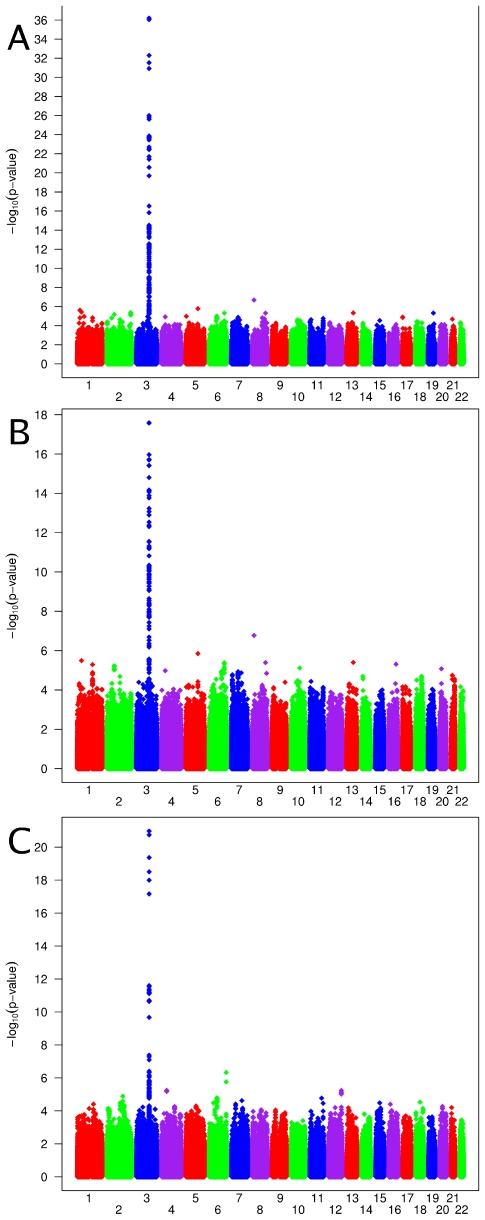
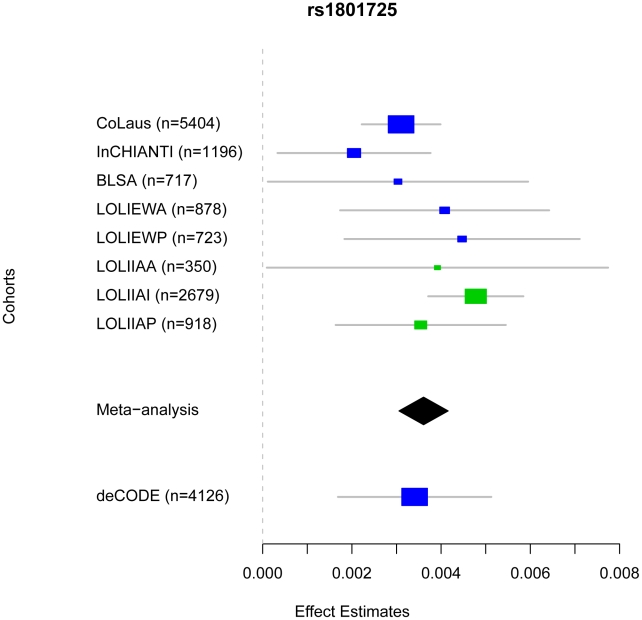
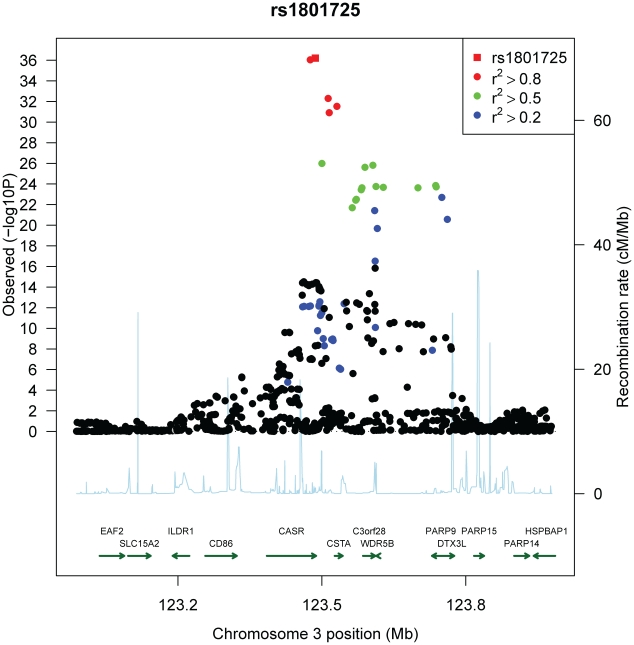
Genome-wide association scans were performed first independently for each cohort using linear regression and then the effect sizes from each cohort were meta-analyzed (see Materials and Methods). Due to the possibility of population substructure obscuring effects of genetic variants, meta-analysis was performed separately for (i) combined European and Indian Asian cohorts (N = 12,865) and restricted to cohorts of (ii) European (N = 8,919), and (iii) Indian Asian descent (N = 3,947). The meta-analyses yielded 100 SNPs from the combined cohorts, 70 SNPs when restricting to European cohorts and 22 SNPs restricting to Indian Asian cohorts that exceeded the genome-wide significance threshold of 5×10-8 (Figure 1A–1C) (the full list is provided in Table S2A, S2B, S2C). All SNPs reaching statistical significance clustered around the CASR locus at 3q13. The most significant SNP in the (i) combined and (ii) European meta-analyses was rs1801725 (p = 6.29×10-37, p = 2.58×10-18, respectively) and in the (iii) Indian Asian meta-analysis was rs17251221 (p = 1.07×10-21). These two SNPs are less than 11 kb apart and are in high linkage disequilibrium with each other (r2 = 0.946, 0.494, 1.0, 1.0 in HapMap CEU, CHB, JPT, YRI, respectively), and therefore most likely derive from the same association signal. We find that rs1801725 explains 1.26% of the variance in serum calcium, with the effect sizes and standard errors of the serum calcium increasing T allele in individual cohorts shown in Figure 2 and Table S3. According to our additive model, each rs1801725 T allele increases log10 serum calcium (in units mmol/L) by 3.61×10-3, equivalent to a multiplicative effect of 1.008 on serum calcium (see also Table S2). At an average serum calcium level of 2.25 mmol/L, each rs1801725 T allele yields an increase of 0.01874 mmol/L, or 21% of one standard deviation of serum calcium levels in a normal population. The regional pattern of association of SNPs around the CASR locus, and their linkage disequilibrium with rs1801725, are shown in Figure 3. Of note, rs1042636, which has been associated with decreased serum calcium [23], also achieved genome-wide significance with the G minor allele associated with decreased serum calcuim (p = 4.96×10-9). However, conditional on the rs1801725 locus, located 12 bps upstream, the rs1042636 p-value became 3.32×10−4, indicating that the two loci share contributions to serum calcium levels.
Figure 1. Genome-wide association results.
Manhattan plots showing significance of association of all SNPs in the meta-analysis for (A) combined European and Indian Asian cohorts, (B) European cohorts only and (C) Indian Asian cohorts only. SNPs are plotted on the x-axis according to their position on each chromosome against association with serum calcium concentrations on the y-axis (shown as −log10 p-values).
Figure 2. Comparison of rs1801725 significance across cohorts.
The effect size and 95% confidence intervals of the serum calcium increasing T allele of SNP rs1801725 are shown separately for each cohort (CoLaus, LOLIPOP_EWA, LOLIPOP_EWP, LOLIPOP_IAA, LOLIPOP_IAI, LOLIPOP_IAP, BLSA, InCHIANTI) and for the replication cohort deCODE. European cohorts are shown in blue and Indian Asian cohorts are drawn in green. The size of the box is proportional to the precision 1/se2 and the meta-analysis estimate and 95% confidence interval across all cohorts is given by a diamond.
Figure 3. Regional association plot of the CASR locus.
Plots show genomic position on the x-axis and −log10 p-values on the y-axis for SNPs in the CASR locus on chromosome 3. The sentinel hit is shown as a red square. Patterns of linkage disequilibrium between the sentinel SNP and all other SNPs are color-coded. Red circles indicate high correlation (r2>0.8), green circles indicate moderate correlation (r2>0.5), blue circles indicate low correlation (r2>0.2) and black circles indicate no correlation (r2<0.2). The fine-scale recombination rates from [72]–[73] are plotted in light blue.
To confirm the rs1801725 signal, we analyzed the association pattern with serum calcium in a separate cohort. We used a subset of 4,126 Icelandic individuals from the deCODE study [24]–[25] with serum calcium measurements. We found the rs1801725 T allele to be strongly associated with increased serum calcium (p = 1.02×10-4), replicating the key meta-analysis result.
While only the CASR locus reached nominal genome-wide significance for association with serum calcium, the top regions with p<10-5 are shown in Table 3. These SNPs cover 12 regions, the significance of which is displayed across cohorts in Figure S3. There were no SNPs in other candidate genes (which have previously been shown to be involved in disorders associated with disturbed serum calcium levels) that were associated with serum calcium at genome-wide significance. The most significant SNPs within 500 kb of the gene transcripts are shown in Table 1. Considering the set of 18,611 distinct SNPs mapping to the set of serum calcium candidate genes excluding CASR, we find no significant association (at significance level 0.05 and applying the Bonferroni correction for multiple testing, giving a cut-off p-value of 2.69×10-6, see also Figure S4). Indeed, fixing the sample size and genome-wide significance threshold our study is well-powered (≥0.80) to detect SNPs explaining at least 0.31% of the variance. Therefore the common SNPs within the candidate genes (excluding CASR) likely play at best a small role in serum calcium regulation.
Table 3. Significance of top SNPs.
| Combined European and Indian Asian Cohorts | |||||||||||
| db SNP | Chr | Position (Build 35) | Coded Allele | Non- Coded Allele | Frequency Coded Allele | Beta | Se | Lower 95% CI | Upper 95% CI | P-value | R2 |
| rs1801725 | 3 | 123486447 | T | G | 17.75 | 3.61E-03 | 2.81E-04 | 3.06E-03 | 4.16E-03 | 6.29E-37 | 0.0126 |
| rs17120351 | 8 | 14731368 | T | C | 96.24 | 4.03E-03 | 7.67E-04 | 2.52E-03 | 5.53E-03 | 2.06E-07 | 0.0021 |
| rs7448017 | 5 | 117800594 | T | G | 17.83 | -1.73E-03 | 3.57E-04 | -2.43E-03 | -1.03E-03 | 1.65E-06 | 0.0018 |
| rs742393 | 1 | 25271187 | G | A | 78.86 | 2.11E-03 | 4.44E-04 | 1.24E-03 | 2.98E-03 | 2.41E-06 | 0.0018 |
| rs16827695 | 1 | 40956147 | G | A | 94.52 | 3.46E-03 | 7.41E-04 | 2.01E-03 | 4.92E-03 | 3.74E-06 | 0.0017 |
| rs1550532 | 2 | 234046848 | G | C | 74.24 | -1.15E-03 | 2.48E-04 | -1.64E-03 | -6.66E-04 | 4.28E-06 | 0.0017 |
| rs261503 | 13 | 81305121 | G | A | 91.35 | 2.45E-03 | 5.29E-04 | 1.41E-03 | 3.49E-03 | 4.60E-06 | 0.0017 |
| rs10119 | 19 | 50098513 | G | A | 73.82 | -1.63E-03 | 3.53E-04 | -2.33E-03 | -9.41E-04 | 4.76E-06 | 0.0017 |
| rs17666460 | 6 | 149298132 | G | A | 11.16 | 1.93E-03 | 4.18E-04 | 1.11E-03 | 2.75E-03 | 4.81E-06 | 0.0017 |
| rs16902486 | 8 | 129024247 | G | C | 4.26 | -2.66E-03 | 5.75E-04 | -3.78E-03 | -1.53E-03 | 4.83E-06 | 0.0017 |
| rs17005914 | 2 | 70721826 | T | C | 21.4 | -1.47E-03 | 3.24E-04 | -2.11E-03 | -8.38E-04 | 6.76E-06 | 0.0016 |
| rs10455097 | 6 | 74550153 | C | A | 47.75 | -9.58E-04 | 2.15E-04 | -1.38E-03 | -5.37E-04 | 9.94E-06 | 0.0015 |
We report SNPs with p-values <1E-05 filtered by distinct regions, determined by merging SNPs within 1 Mb of each other. Results are given for different cohort subsets of combined European and Indian Asian cohorts, European cohorts, and Indian Asian cohorts. The Beta and SE values represent the effect of the coded allele on log10 corrected serum calcium levels; R2 represents the fraction of variation explained by the SNP.
We analyzed the association of the top SNP with several calcium-related outcomes (coronary heart disease, myocardial infarction, hypertension, stroke, osteoarthritis, osteoporosis and kidney stones). The number of cases and controls for each outcome and each cohort is given in Table S4. Logistic regression including age and pseudosex (see Materials and Methods) as covariates did not find any significant association between rs1801725 and the calcium-related outcomes, after correcting for multiple testing (effect sizes and standard errors for the T allele are listed in Table S5). Power calculations show that given the sample sizes for the clinical traits above, our study has good power (≥0.80) to detect odds ratios of 1.20, 1.13, 1.77, 1.27, 1.27, 1.24 and 1.75, respectively. As the smallest p-values from calcium-related traits were for osteoarthritis and osteoporosis (bonferroni-corrected p = 0.21, 0.44, respectively), we further investigated bone density traits. None of deCODE hip bone mineral density or spine bone mineral density (N = 6657 and 6838, respectively) nor InCHIANTI total bone density, trabecular bone density, cortical bone density, cortical bone thickness or cortical bone area (N = 1196) bonferroni-adjusted p-values for eight traits were significant.
Discussion
This genome-wide scan of 12,865 individuals revealed CASR as the most significant (and only genome-wide significant) locus influencing serum calcium levels. Specifically, we found evidence for a strong association of SNPs located in the CASR locus with serum calcium levels in both Europeans and Indian Asians. The strongest locus in CASR was further shown to replicate in an independent Icelandic cohort of 4,126 individuals.
The top signal (rs1801725, 2956G>T) explains 1.26% of the variance in serum calcium. Indeed, this is similar to results from other GWAS of human height [26]–[29], body mass index [30]–[31], serum urate [32]–[34] and serum lipid concentrations [34]–[36], for which the genome-wide significant loci uncovered thus far collectively explain only a small fraction of the phenotypic variance (usually at least one order of magnitude less than the total additive genetic variance estimated from heritability studies [37]–[38]). The rs1801725 T allele (A986S) was associated with higher serum calcium, consistent with previous findings (see Table S6). The rs1801725 polymorphism (with T allele frequencies of 16.76%, 19.98% in European and Indian Asian cohorts, respectively) affects serum calcium levels of a substantial proportion of the population.
The rs1801725 polymorphism encodes a missense variant in exon 7 of the CASR gene, which leads to a non-conservative amino-acid change (serine substitution for alanine-986, A986S corresponding to nucleotides 2956G>T) in the cytoplasmic tail of CASR. In vitro studies showed that mutations within the C-terminal tail may influence several aspects of CASR function, such as signal transduction, intracellular trafficking and cell surface expression [39]–[41]. However, PolyPhen predicts rs1801725 to be a benign substitution. It is presently unclear whether this substitution gives rise to functional variants, as functional studies have yielded conflicting results [42]–[43]. Deep sequencing of this region may help identifying the causal variants. While it is still not possible to infer a direct causal role, it is of interest to note that the SNP gives rise to an amino acid change in the C-terminal tail of CASR, a domain which plays a key role in the receptor function and may potentially influence intracellular trafficking following CASR activation by extracellular calcium.
Several studies have reported associations of A986S and nearby CASR mutations with various phenotypes. The A986S CASR polymorphism has been associated with variations in circulating calcium levels in healthy adults in some studies [15], [23], [44]–[45], but not in others [46]–[47]. The fact that the latter studies were underpowered (sample size ranging from 148 to 1252) to detect a small effect size likely explains these inconsistent results. The rs1042636 (R990G) polymorphism has been associated with the magnitude of parathyroid hormone (PTH) secretion in patients with primary hyperparathyroidism [48], and preliminary results suggest that it could influence response to cinacalcet, a calcimimetic used to treat secondary hyperparathyroidism in patients with end-stage renal disease [49]. In a meta-analysis, 986S was associated with a 49% increased risk (P = 0.002) of primary hyperparathyroidism [47], [50]–[51]. Among patients with primary hyperparathyroidism, the AGQ haplotype (i.e. 986A, 990G, 1011Q, which is associated with lower serum calcium levels and hypercalciuria [52]) was associated with increased risk, and the SRQ haplotype with decreased risk, of kidney stones [50].
CASR has been previously considered as a candidate gene for osteoporosis [53] and coronary heart disease as well as increased total and cardiovascular mortality [54]. In our meta-analysis, we found no significant association of rs1801725 with these calcium-related phenotypes. A recent meta-analysis focusing on effects of candidate genes on osteoporosis also reports negative results for CASR [55]. Furthermore, results on the association of elevated serum calcium with increased cardiovascular risk in the general population are controversial [6]–[8]. It is therefore not clear to what extent serum calcium might predict cardiovascular risk. The SNPs identified in this meta-analysis could serve as genetic instruments in future studies, such as Mendelian randomization analysis in longitudinal cohorts, to further investigate the causal effect of serum calcium on osteoporosis and on cardiovascular disease risk (see Table S5 for rs1801725 effects and standard errors).
Our meta-analysis suffers from some limitations. First, we used corrected serum calcium and not directly measured ionized serum calcium. The correlation between corrected serum calcium and ionized serum calcium varies between 0.66 and 0.87 [56]–[58]. We can hypothesize that the association of ionized serum calcium with CASR variants would be stronger than the one with corrected serum calcium because ionized calcium is the form physiologically active on CASR. Second, data on serum phosphate, PTH or vitamin D are not available, so that we cannot explore further these relationships. Third, sample sizes for calcium-related clinical traits were limited, many clinical traits in CoLaus were self-reported instead of clinically diagnosed, and we incur a multiple testing penalty due to the number of clinical traits posited to be associated with serum calcium. However, the major strengths of the study are the hypothesis-free nature of GWAS studies, the large sample meta-analysis and the inclusion of multiple populations.
Materials and Methods
Cohorts
CoLaus is a population-based sample from Lausanne, Switzerland, consisting of 5435 individuals between 35 and 75 years old (after QC) of which a subset of 5404 had available serum calcium measurements. The study design and protocols have been described previously [17]–[18]. The CoLaus study was approved by the Institutional Ethic's Committee of the University of Lausanne. The London Life Sciences Prospective Population Study (LOLIPOP) is an ongoing population-based cohort study of ∼30,000 Indian Asian and European white men and women, aged 35–75 years living in West London, United Kingdom [59]. All study participants gave written consent including for genetic studies. The LOLIPOP study is approved by the local Research Ethics Committee. The participants included in the present study are a subset of 3947 Indian Asians and 1601 Europeans from the LOLIPOP cohort study. LOLIPOP individuals are separated by origin as well as the genotyping platform, with IAA, IAI or IAP denoting Indian Asians genotyped on Affymetrix, Illumina or Perlegen platforms, respectively, and EWA and EWI denoting Europeans genotyped on Affymetrix or Illumina platforms, respectively (see Table S1). The InCHIANTI study is a population-based epidemiological study aimed at evaluating the factors that influence mobility in the older population living in the Chianti region in Tuscany, Italy. The details of the study have been previously reported [60]. Overnight fasted blood samples were taken for genomic DNA extraction, and measurement of serum calcium. For this study, 1196 subjects with serum calcium and GWAS data were analyzed. The study protocol was approved by the Italian National Institute of Research and Care of Aging Institutional Review and Medstar Research Institute (Baltimore, MD). The Baltimore longitudinal study on Aging (BLSA) study is a population-based study aimed to evaluate contributors of healthy aging in the older population residing predominantly in the Baltimore-Washington DC area [61]. Starting in 1958, participants are examined every one to four years depending on their age. Blood samples were collected for DNA extraction. This analysis focused on a subset of the participants (N = 717) of European ancestry. The BLSA has continuing approval from the Institutional Review Board (IRB) of Medstar Research Institute. Approval was obtained from local ethic committees for all studies and all participants signed informed written consent. The deCODE study consists of individuals who visited a private outpatient laboratory, the Laboratory in Mjodd, Reykjavik, Iceland between 1997 and 2008. The main referral center for this laboratory is a multispecialty medical clinic in Reykjavik (Laeknasetrid). For the serum calcium analysis we used information on 4,126 individuals with both genome-wide SNP data and measured serum calcium and serum albumin. The samples for bone density analysis have previously been described in detail [24]–[25]. For this study 6,657 individuals with total hip bone mineral density (BMD) and 6,838 individuals with lumbar spine BMD and SNP data were available for analysis. All participants gave informed consent and the study was approved by the Data Protection Commission of Iceland (DPC) and the National Bioethics Committee of Iceland.
Clinical data
For each CoLaus participant a venous blood sample was collected under fasting conditions. Measurements were conducted using a Modular P apparatus (Roche Diagnostics, Switzerland). Total serum calcium was measured by O-cresolphtalein (2.1% – 1.5% maximum inter and intra-batch CVs); albumin was measured by bromocresol green (2.5% – 0.4%). To further characterize the identified genetic variants, we analyzed the association with several outcomes postulated to be correlated with serum calcium. Within the CoLaus study, we have questionnaire responses to queries about personal histories of osteoporosis, osteoarthritis, myocardial infarction and stroke in addition to clinical data determining hypertension status, defined as previously described [17]. The assessment of LOLIPOP study participants was carried out by a trained research nurse, during a 45 minute appointment according to a standardized protocol and with regular QC audits. An interviewer-administered questionnaire was used to collect data on medical history, family history, current prescribed medication, and cardiovascular risk factors. Physical assessment included anthropometric measurements (height, weight, waist, hip) and blood pressure. Blood was collected after an 8 hour fast for biochemical analysis, including glucose, insulin, total and HDL cholesterol and triglycerides, and whole blood was taken for DNA extraction [59]. InCHIANTI serum albumin concentrations were determined as percentage of total protein using agarose electrophoretic technique (Hydragel Protein (E) 15/30, Sebia, Issy-les-Moulineaux, France). Serum calcium was measured using calorimetric assay (Roch Diagnostic, GmbH, Mannheim, Germany) by a Roche-Hitachi autoanalyzer (The intra-assay CV and 0.9% and the inter-assay CV was 1.5%). Measures of bone density, bone dimensions and osteoporosis diagnosis were assessed by peripheral quantitative computed tomography (pQCT) using the XCT 2000 device (Stratec Medizintechnik, Pforzheim, Germany) [62]. BLSA albumin concentrations were measured by a calorimetric assay using bromocresol green (Ortho-Clinical Diagnostics). Calcium concentrations were measured by a calorimetric assay (Vitros 5,1,FS).
Genome-wide genotyping and imputation
CoLaus participants were genotyped using Affymetrix Human Mapping 500 K Array. For the genome-wide association stage, genotyping in LOLIPOP participants was carried out using the Illumina 317 K mapping array, Affymetrix Human Mapping 500 K array, and Perlegen, 284 K platforms (Table S1). Participants of the InCHIANTI and BLSA studies were genotyped using Illumina Infinium HumanHap 550 K SNP arrays were used for genotyping [21]. Imputation of allele dosage of SNPs was performed using either MACH [63] or IMPUTE [64] with parameters and quality control filters as described in . All European cohorts imputed SNPs typed in the HapMap CEU population; LOLIPOP Indian Asian cohort imputed SNPs using mixed HapMap populations, given that this showed greater concordance with real genotypes compared with use of any one HapMap population. SNPs were excluded if cohort-specific imputation quality as assessed by r2.hat (MACH) or .info (IMPUTE) metrics were <0.30. In total, 2,557,252 genotyped or imputed SNPs had data from one or more cohorts and were analyzed. Genotypes in deCODE were measured using either humanHap300, humanHap300-duo or humanCNV370.
Statistical analysis
Individual genome-wide association analysis
Biologically active serum calcium is estimated by the correction, Ca_corrected = total serum calcium [mmol/L] + (40 - albumin [g/L])/40. Individuals with values <1.9 or >3 were removed as these were extreme outliers. Linear-regression analyses were carried out using an additive genetic model on log10-transformed corrected calcium levels adjusted for age and pseudosex (a factor variable with three values: males, pre-menopausal females and post-menopausal females). BLSA also included the first two and LOLIPOP included the first four ancestry principal components in the regression, respectively. Regression analyses were performed with QUICKTEST [65] (CoLaus), MACH2qtl (LOLIPOP) [63] or MERLIN [66] (InCHIANTI, BLSA).
Meta-analysis
The results from all cohorts were combined into a fixed-effects meta-analysis using inverse variance weighting. Tests for heterogeneity were assessed using Cochran's Q statistic and the log of the related H statistic [67] after grouping LOLIPOP subsets into European and Indian subsets. For rs1801725 and rs1042636 the p-values were (0.07657, 0.1432) and (0.3450, 0.8876), respectively, indicating limited between-study variability. The analysis was implemented in R and run on a quad-core Linux machine. SNPs were reported provided they had effect size estimates in at least 2 of the 5 European cohorts, in at least 2 of the 3 Indian Asian cohorts, or in at least 3 of the 8 total cohorts. For the overall meta-analysis, residual inflation of the test statistic was corrected using genomic control [68]. The inflation factor was 1.0207 for the all combined cohorts, 1.0068 for European cohorts and 1.0286 for Indian Asian cohorts. Where reported, individual study p-values are corrected for inflation using genomic control methods for genotyped and imputed SNPs combined (inflation factors for individual studies were 1.0139 (CoLaus), 0.9891 (LOLIPOP EWA), 0.9994 (LOLIPOP EWP), 0.9967 (LOLIPOP IAA), 1.0131 (LOLIPOP IAI), 0.9985 (LOLIPOP IAP), 0.9842 (InCHIANTI), 1.0019 (BLSA)). The regional association plot (Figure 3) was created modifying a publically available R script [69]. The map of fine-scale recombination rates was downloaded from the HapMap website http://www.hapmap.org/downloads/recombination/ using Phase II HapMap data (release 21). Quantile-quantile plots of the association results are shown in Figure S1A, S1B, S1C, study-specific quantile-quantile plots are shown in Figure S2. Associations below p = 5×10−8 were considered genome-wide significant, which corresponds to a Bonferroni correction for the estimated one million independent common variant tests in the human genome of European individuals [70]. The analysis of osteoporosis status in CoLaus and InCHIANTI was performed using logistic regression including age and pseudosex as covariates in QUICKTEST [65]. Linkage disequilibrium was estimated from HapMap CEU (2007-01, build 35 non-redundant) genotypes. LD r2 statistics were estimated for SNPs within 500 kb using Haploview [71].
Association of rs1801725 with calcium-related outcomes
For each related trait (coronary heart disease, hypertension, kidney stones, myocardial infarction, osteoarthritis, osteoporosis and stroke) we performed a fixed-effects meta-analysis of the logistic regression coefficients. We applied the bonferroni correction to adjust for multiple testing. We performed Wald-based power calculations using a type I error of 0.05/7 and meta-analysis coefficient estimates and standard errors to estimate the sample size for each trait giving power 0.80.
Supporting Information
Quantile-Quantile plots of genome-wide association results. Observed -log10 p-values on the y-axis are plotted against theoretical -log10 p-values on the x-axis resulting from each meta-analysis. Results are color-coded by chromosome. The top results largely derive from the CASR locus on chromosome 3. Results are shown separately for (A) all cohorts, (B) European cohorts and (C) Indian Asian cohorts.
(6.48 MB TIF)
Study-specific quantile-quantile plots. Shown are observed -log10 p-values plotted against expected -log10 p-values resulting from each single study after applying genomic control correction. The study-specific λ-values were λ = 1.0139 (CoLaus), λ = 0.9891 (LOLIPOP_EWA), λ = 0.9994 (LOLIPOP_EWP), λ = 0.9967 (LOLIPOP_IAA), λ = 1.0131 (LOLIPOP_IAI), λ = 0.9985 (LOLIPOP_IAP), λ = 0.9842 (InCHIANTI), λ = 1.0019 (BLSA). For the combined European and Indian Asian, European only and Indian Asian only meta-analyses the inflation factors were 1.0207, 1.0068, and 1.0286, respectively.
(4.36 MB TIF)
Comparison of significance across cohorts. The effect size and 95% confidence intervals of SNPs which do not reach genome-wide significance in the combined European and Indian Asian meta-analysis are shown separately for each cohort (CoLaus, LOLIPOP_EWA, LOLIPOP_EWP, LOLIPOP_IAA, LOLIPOP_IAI, LOLIPOP_IAP, BLSA, InCHIANTI). European cohorts are drawn in blue and Indian Asian cohorts are drawn in green. The size of the box is proportional to the precision 1/se2 and the meta-analysis estimate and 95% confidence interval across all cohorts is given by a diamond.
(0.38 MB TIF)
Candidate gene QQ-plots. For 18611 SNPs mapping to candidate genes (excluding CASR), we compare observed -log10 p-values to the mean quantiles of the uniform distribution. As a comparison, we randomly choose a set of genes from which we select the same number of SNPs. From 1,000 random draws we calculate the 95th percentile of -log10 p-values (in blue). Results are shown separately for all cohorts, European only and Indian Asian only (A–C). CoLaus permuted phenotype results comparing observed p-values to the 95th percentile of -log10 p-values from 100 permutations are shown in (D).
(0.41 MB TIF)
Genotyping, imputation, and analysis procedures by study. The genotyping platforms, quality control (QC) filters applied before imputation, imputation software, number of SNPs, and genotype-phenotype association software are shown for each study.
(0.05 MB DOC)
Complete list of genome-wide significant SNPs. Below is the list of all SNPs that exceeded the threshold of genome-wide significance (p<5×10e-8). Position is given for NCBI Build 35. Meta-analysis is performed by inverse variance weighted fixed effect regression. The coded allele is the allele to which the beta (effect) estimate refers. Results are shown separately for (A) European and Indian Asian cohorts, (B) European cohorts only, and (C) Indian Asian cohorts only.
(0.24 MB DOC)
Significance of top SNPs by cohort. Shown are study-specific results of the SNPs with genomic control (GC) p-values <1E-05 filtered by distinct regions, determined by merging SNPs within 1 Mb of each other. Results are shown separately for (A) European and Indian Asian cohorts, (B) European cohorts, and (C) Indian Asian cohorts.
(0.08 MB DOC)
Number of cases and controls for calcium-related outcomes. For several related phenotypes, we test the association of rs1801725 with these binary responses. Shown here are the number of cases and controls for each phenotype in each cohort and the total across cohorts.
(0.04 MB DOC)
Logistic regression of clinical phenotypes on rs1801725. We report the effect size and standard error of the rs1801725 T allele from logistic regressions of each clinical phenotype.
(0.04 MB DOC)
Studies of CASR mutations and serum calcium. A survey of previous studies which investigate the relationship between CASR mutations and levels of serum calcium.
(0.05 MB DOC)
Acknowledgments
The authors would like to thank those who agreed to participate in the studies and the many colleagues who contributed to collection and phenotypic characterization of the clinical samples, to genotyping, and to statistical analyses. The computations for CoLaus imputation were performed in part at the Vital-IT center for high performance computing of the Swiss Institute of Bioinformatics.
Footnotes
Vincent Mooser and Dawn Waterworth are full-time employees of GlaxoSmithKline, a pharmaceutical company. Peter Vollenweider and Gerard Waeber received financial support from GlaxoSmithKline to build the CoLaus study. Daniel F Gudbjarsson, Unnur Styrkarsdottir, Hilma Holm, Unnur Thorsteinsdottir and Kari Stefansson are employees of deCODE genetics, a biotechnology company.
The CoLaus study is supported by GlaxoSmithKline, by the Faculty of Biology and Medicine of the University of Lausanne, Switzerland, and by a grant from the Swiss National Science Foundation: 33CSCO-122661. JS Beckman is supported by a grant form the Swiss National Foundation (310000-112552). M Bochud is supported by grants from the Swiss National Foundation (PROSPER 3200BO-111362/1, 3233BO-111361/1) and by the Swiss School of Public Health Plus (SSPH+). S Bergmann is grateful for financial support from the Giorgi-Cavaglieri Foundation, the Swiss National Science Foundation (Grant #3100AO-116323/1), the Swiss Institute of Bioinformatics and the European Framework Project 6 (through the AnEuploidy and EuroDia projects). The BLSA was supported in part by the Intramural Research Program of the NIH, National Institute on Aging. A portion of that support was through a R&D contract with Medstar Research Institute. The InCHIANTI study baseline (1998–2000) was supported as a “targeted project” (ICS110.1/RF97.71) by the Italian Ministry of Health and in part by the US National Institute on Aging (Contracts: 263 MD 9164 and 263 MD 821336). Y Li is partially supported by DK078150-03 and DK056350 (to the University of North Carolina Nutrition Obesity Research Center). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Carafoli E. Calcium-mediated cellular signals: a story of failures. Trends Biochem Sci. 2004;29:371–379. doi: 10.1016/j.tibs.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 2.Carafoli E. Calcium–a universal carrier of biological signals. Delivered on 3 July 2003 at the Special FEBS Meeting in Brussels. FEBS J. 2005;272:1073–1089. doi: 10.1111/j.1742-4658.2005.04546.x. [DOI] [PubMed] [Google Scholar]
- 3.Carafoli E. The ambivalent nature of the calcium signal. J Endocrinol Invest. 2004;27:134–136. [PubMed] [Google Scholar]
- 4.Whitfield JB, Martin NG. The effects of inheritance on constituents of plasma: a twin study on some biochemical variables. Ann Clin Biochem. 1984;21 (Pt 3):176–183. doi: 10.1177/000456328402100303. [DOI] [PubMed] [Google Scholar]
- 5.Williams PD, Puddey IB, Martin NG, Beilin LJ. Platelet cytosolic free calcium concentration, total plasma calcium concentration and blood pressure in human twins: a genetic analysis. Clin Sci (Lond) 1992;82:493–504. doi: 10.1042/cs0820493. [DOI] [PubMed] [Google Scholar]
- 6.Leifsson BG, Ahren B. Serum calcium and survival in a large health screening program. J Clin Endocrinol Metab. 1996;81:2149–2153. doi: 10.1210/jcem.81.6.8964843. [DOI] [PubMed] [Google Scholar]
- 7.Lind L, Skarfors E, Berglund L, Lithell H, Ljunghall S. Serum calcium: a new, independent, prospective risk factor for myocardial infarction in middle-aged men followed for 18 years. J Clin Epidemiol. 1997;50:967–973. doi: 10.1016/s0895-4356(97)00104-2. [DOI] [PubMed] [Google Scholar]
- 8.Dhingra R, Sullivan LM, Fox CS, Wang TJ, D'Agostino RB, Sr, et al. Relations of serum phosphorus and calcium levels to the incidence of cardiovascular disease in the community. Arch Intern Med. 2007;167:879–885. doi: 10.1001/archinte.167.9.879. [DOI] [PubMed] [Google Scholar]
- 9.Palmer M, Adami HO, Bergstrom R, Jakobsson S, Akerstrom G, et al. Survival and renal function in untreated hypercalcaemia. Population-based cohort study with 14 years of follow-up. Lancet. 1987;1:59–62. doi: 10.1016/s0140-6736(87)91906-4. [DOI] [PubMed] [Google Scholar]
- 10.Lundgren E, Lind L, Palmer M, Jakobsson S, Ljunghall S, et al. Increased cardiovascular mortality and normalized serum calcium in patients with mild hypercalcemia followed up for 25 years. Surgery. 2001;130:978–985. doi: 10.1067/msy.2001.118377. [DOI] [PubMed] [Google Scholar]
- 11.Wermers RA, Khosla S, Atkinson EJ, Grant CS, Hodgson SF, et al. Survival after the diagnosis of hyperparathyroidism: a population-based study. Am J Med. 1998;104:115–122. doi: 10.1016/s0002-9343(97)00270-2. [DOI] [PubMed] [Google Scholar]
- 12.Heath H, III, Odelberg S, Jackson CE, Teh BT, Hayward N, et al. Clustered inactivating mutations and benign polymorphisms of the calcium receptor gene in familial benign hypocalciuric hypercalcemia suggest receptor functional domains. J Clin Endocrinol Metab. 1996;81:1312–1317. doi: 10.1210/jcem.81.4.8636323. [DOI] [PubMed] [Google Scholar]
- 13.Pollak MR, Brown EM, Chou YH, Hebert SC, Marx SJ, et al. Mutations in the human Ca(2+)-sensing receptor gene cause familial hypocalciuric hypercalcemia and neonatal severe hyperparathyroidism. Cell. 1993;75:1297–1303. doi: 10.1016/0092-8674(93)90617-y. [DOI] [PubMed] [Google Scholar]
- 14.Pollak MR, Brown EM, Estep HL, McLaine PN, Kifor O, et al. Autosomal dominant hypocalcaemia caused by a Ca(2+)-sensing receptor gene mutation. Nat Genet. 1994;8:303–307. doi: 10.1038/ng1194-303. [DOI] [PubMed] [Google Scholar]
- 15.Cole DE, Peltekova VD, Rubin LA, Hawker GA, Vieth R, et al. A986S polymorphism of the calcium-sensing receptor and circulating calcium concentrations. Lancet. 1999;353:112–115. doi: 10.1016/s0140-6736(98)06434-4. [DOI] [PubMed] [Google Scholar]
- 16.Cole DE, Vieth R, Trang HM, Wong BY, Hendy GN, et al. Association between total serum calcium and the A986S polymorphism of the calcium-sensing receptor gene. Mol Genet Metab. 2001;72:168–174. doi: 10.1006/mgme.2000.3126. [DOI] [PubMed] [Google Scholar]
- 17.Firmann M, Mayor V, Marques-Vidal PM, Bochud M, Pecoud A, et al. The CoLaus study: a population-based study to investigate the epidemiology and genetic determinants of cardiovascular risk factors and metabolic syndrome. BMC Cardiovasc Disord. 2008. [DOI] [PMC free article] [PubMed]
- 18.Rodondi N, Cornuz J, Marques-Vidal P, Butler J, Hayoz D, et al. Aspirin use for the primary prevention of coronary heart disease: A population-based study in Switzerland. Prev Med. 2008;46:137–144. doi: 10.1016/j.ypmed.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 19.Chambers JC, Elliott P, Zabaneh D, Zhang W, Li Y, et al. Common genetic variation near MC4R is associated with waist circumference and insulin resistance. Nat Genet. 2008;40:716–718. doi: 10.1038/ng.156. [DOI] [PubMed] [Google Scholar]
- 20.Kooner JS, Chambers JC, Aguilar-Salinas CA, Hinds DA, Hyde CL, et al. Genome-wide scan identifies variation in MLXIPL associated with plasma triglycerides. Nat Genet. 2008;40:149–151. doi: 10.1038/ng.2007.61. [DOI] [PubMed] [Google Scholar]
- 21.Melzer D, Perry JR, Hernandez D, Corsi AM, Stevens K, et al. A genome-wide association study identifies protein quantitative trait loci (pQTLs). PLoS Genet. 2008;4:e1000072. doi: 10.1371/journal.pgen.1000072. doi: 10.1371/journal.pgen.1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schock NW, Greulich RC, Andres R, Arenberg D, Costa PT, et al. Washington D.C: 1984. Normal Human Aging: The Baltimore Longitudinal Study of Aging. [Google Scholar]
- 23.Scillitani A, Guarnieri V, De GS, Muscarella LA, Battista C, et al. Blood ionized calcium is associated with clustered polymorphisms in the carboxyl-terminal tail of the calcium-sensing receptor. J Clin Endocrinol Metab. 2004;89:5634–5638. doi: 10.1210/jc.2004-0129. [DOI] [PubMed] [Google Scholar]
- 24.Styrkarsdottir U, Halldorsson BV, Gretarsdottir S, Gudbjartsson DF, Walters GB, et al. New sequence variants associated with bone mineral density. Nat Genet. 2009;41:15–17. doi: 10.1038/ng.284. [DOI] [PubMed] [Google Scholar]
- 25.Styrkarsdottir U, Halldorsson BV, Gretarsdottir S, Gudbjartsson DF, Walters GB, et al. Multiple genetic loci for bone mineral density and fractures. N Engl J Med. 2008;358:2355–2365. doi: 10.1056/NEJMoa0801197. [DOI] [PubMed] [Google Scholar]
- 26.Gudbjartsson DF, Walters GB, Thorleifsson G, Stefansson H, Halldorsson BV, et al. Many sequence variants affecting diversity of adult human height. Nat Genet. 2008;40:609–615. doi: 10.1038/ng.122. [DOI] [PubMed] [Google Scholar]
- 27.Lettre G, Jackson AU, Gieger C, Schumacher FR, Berndt SI, et al. Identification of ten loci associated with height highlights new biological pathways in human growth. Nat Genet. 2008;40:584–591. doi: 10.1038/ng.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weedon MN, Lettre G, Freathy RM, Lindgren CM, Voight BF, et al. A common variant of HMGA2 is associated with adult and childhood height in the general population. Nat Genet. 2007;39:1245–1250. doi: 10.1038/ng2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weedon MN, Lango H, Lindgren CM, Wallace C, Evans DM, et al. Genome-wide association analysis identifies 20 loci that influence adult height. Nat Genet. 2008;40:575–583. doi: 10.1038/ng.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loos RJ, Lindgren CM, Li S, Wheeler E, Zhao JH, et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet. 2008;40:768–775. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doring A, Gieger C, Mehta D, Gohlke H, Prokisch H, et al. SLC2A9 influences uric acid concentrations with pronounced sex-specific effects. Nat Genet. 2008;40:430–436. doi: 10.1038/ng.107. [DOI] [PubMed] [Google Scholar]
- 33.Li S, Sanna S, Maschio A, Busonero F, Usala G, et al. The GLUT9 gene is associated with serum uric acid levels in Sardinia and Chianti cohorts. PLoS Genet. 2007;3:e194. doi: 10.1371/journal.pgen.0030194. doi: 10.1371/journal.pgen.0030194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wallace C, Newhouse SJ, Braund P, Zhang F, Tobin M, et al. Genome-wide association study identifies genes for biomarkers of cardiovascular disease: serum urate and dyslipidemia. Am J Hum Genet. 2008;82:139–149. doi: 10.1016/j.ajhg.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sandhu MS, Waterworth DM, Debenham SL, Wheeler E, Papadakis K, et al. LDL-cholesterol concentrations: a genome-wide association study. Lancet. 2008;371:483–491. doi: 10.1016/S0140-6736(08)60208-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goldstein DB. Common genetic variation and human traits. N Engl J Med. 2009;360:1696–1698. doi: 10.1056/NEJMp0806284. [DOI] [PubMed] [Google Scholar]
- 38.Maher B. Personal genomes: The case of the missing heritability. Nature. 2008;456:18–21. doi: 10.1038/456018a. [DOI] [PubMed] [Google Scholar]
- 39.Bai M, Trivedi S, Brown EM. Dimerization of the extracellular calcium-sensing receptor (CaR) on the cell surface of CaR-transfected HEK293 cells. J Biol Chem. 1998;273:23605–23610. doi: 10.1074/jbc.273.36.23605. [DOI] [PubMed] [Google Scholar]
- 40.Bai M, Trivedi S, Lane CR, Yang Y, Quinn SJ, et al. Protein kinase C phosphorylation of threonine at position 888 in Ca2+o-sensing receptor (CaR) inhibits coupling to Ca2+ store release. J Biol Chem. 1998;273:21267–21275. doi: 10.1074/jbc.273.33.21267. [DOI] [PubMed] [Google Scholar]
- 41.Gama L, Breitwieser GE. A carboxyl-terminal domain controls the cooperativity for extracellular Ca2+ activation of the human calcium sensing receptor. A study with receptor-green fluorescent protein fusions. J Biol Chem. 1998;273:29712–29718. doi: 10.1074/jbc.273.45.29712. [DOI] [PubMed] [Google Scholar]
- 42.Vezzoli G, Terranegra A, Arcidiacono T, Biasion R, Coviello D, et al. R990G polymorphism of calcium-sensing receptor does produce a gain-of-function and predispose to primary hypercalciuria. Kidney Int. 2007;71:1155–1162. doi: 10.1038/sj.ki.5002156. [DOI] [PubMed] [Google Scholar]
- 43.Harding B, Curley AJ, Hannan FM, Christie PT, Bowl MR, et al. Functional characterization of calcium sensing receptor polymorphisms and absence of association with indices of calcium homeostasis and bone mineral density. Clin Endocrinol (Oxf) 2006;65:598–605. doi: 10.1111/j.1365-2265.2006.02634.x. [DOI] [PubMed] [Google Scholar]
- 44.Kelly C, Gunn IR, Gaffney D, Devgun MS. Serum calcium, urine calcium and polymorphisms of the calcium sensing receptor gene. Ann Clin Biochem. 2006;43:503–506. doi: 10.1258/000456306778904632. [DOI] [PubMed] [Google Scholar]
- 45.Laaksonen MM, Outila TA, Karkkainen MU, Kemi VE, Rita HJ, et al. Associations of vitamin D receptor, calcium-sensing receptor and parathyroid hormone gene polymorphisms with calcium homeostasis and peripheral bone density in adult Finns. J NutrigenetNutrigenomics. 2009;2:55–63. doi: 10.1159/000204960. [DOI] [PubMed] [Google Scholar]
- 46.Bollerslev J, Wilson SG, Dick IM, Devine A, Dhaliwal SS, et al. Calcium-sensing receptor gene polymorphism A986S does not predict serum calcium level, bone mineral density, calcaneal ultrasound indices, or fracture rate in a large cohort of elderly women. Calcif Tissue Int. 2004;74:12–17. doi: 10.1007/s00223-002-0066-1. [DOI] [PubMed] [Google Scholar]
- 47.Cetani F, Borsari S, Vignali E, Pardi E, Picone A, et al. Calcium-sensing receptor gene polymorphisms in primary hyperparathyroidism. J Endocrinol Invest. 2002;25:614–619. doi: 10.1007/BF03345085. [DOI] [PubMed] [Google Scholar]
- 48.Yamauchi M, Sugimoto T, Yamaguchi T, Yano S, Kanzawa M, et al. Association of polymorphic alleles of the calcium-sensing receptor gene with the clinical severity of primary hyperparathyroidism. Clin Endocrinol (Oxf) 2001;55:373–379. doi: 10.1046/j.1365-2265.2001.01318.x. [DOI] [PubMed] [Google Scholar]
- 49.Rothe HM, Shapiro WB, Sun WY, Chou SY. Calcium-sensing receptor gene polymorphism Arg990Gly and its possible effect on response to cinacalcet HCl. Pharmacogenet Genomics. 2005;15:29–34. doi: 10.1097/01213011-200501000-00005. [DOI] [PubMed] [Google Scholar]
- 50.Scillitani A, Guarnieri V, Battista C, De GS, Muscarella LA, et al. Primary hyperparathyroidism and the presence of kidney stones are associated with different haplotypes of the calcium-sensing receptor. J Clin Endocrinol Metab. 2007;92:277–283. doi: 10.1210/jc.2006-0857. [DOI] [PubMed] [Google Scholar]
- 51.Miedlich S, Lamesch P, Mueller A, Paschke R. Frequency of the calcium-sensing receptor variant A986S in patients with primary hyperparathyroidism. Eur J Endocrinol. 2001;145:421–427. doi: 10.1530/eje.0.1450421. [DOI] [PubMed] [Google Scholar]
- 52.Vezzoli G, Tanini A, Ferrucci L, Soldati L, Bianchin C, et al. Influence of calcium-sensing receptor gene on urinary calcium excretion in stone-forming patients. J Am Soc Nephrol. 2002;13:2517–2523. doi: 10.1097/01.asn.0000030077.72157.d2. [DOI] [PubMed] [Google Scholar]
- 53.Kim KS, Kim GS, Hwang JY, Lee HJ, Park MH, et al. Single nucleotide polymorphisms in bone turnover-related genes in Koreans: ethnic differences in linkage disequilibrium and haplotype. BMC Med Genet. 2007;8:70. doi: 10.1186/1471-2350-8-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marz W, Seelhorst U, Wellnitz B, Tiran B, Obermayer-Pietsch B, et al. Alanine to serine polymorphism at position 986 of the calcium-sensing receptor associated with coronary heart disease, myocardial infarction, all-cause, and cardiovascular mortality. J Clin Endocrinol Metab. 2007;92:2363–2369. doi: 10.1210/jc.2006-0071. [DOI] [PubMed] [Google Scholar]
- 55.Richards JB, Kavvoura FK, Rivadeneira F, Styrkarsdottir U, Estrada K, et al. Collaborative meta-analysis: associations of 150 candidate genes with osteoporosis and osteoporotic fracture. Ann Intern Med. 2009;151:528–537. doi: 10.7326/0003-4819-151-8-200910200-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Björkman MP, Sorva AJ, Tilvis RS. Calculated serum calcium is an insufficient surrogate for measured ionized calcium. Archives of Gerontology and Geriatrics. 1979;49:348–350. doi: 10.1016/j.archger.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 57.Robertson WG, Marshall RW. Calcium measurements in serum and plasma–total and ionized. CRC Crit Rev Clin Lab Sci. 1979;11:271–304. doi: 10.3109/10408367909105859. [DOI] [PubMed] [Google Scholar]
- 58.Ladenson JH, Lewis JW, Boyd JC. Failure of total calcium corrected for protein, albumin, and pH to correctly assess free calcium status. J Clin Endocrinol Metab. 1978;46:986–993. doi: 10.1210/jcem-46-6-986. [DOI] [PubMed] [Google Scholar]
- 59.Chambers JC, Zhang W, Li Y, Sehmi J, Wass MN, et al. Genome-wide association study identifies variants in TMPRSS6 associated with hemoglobin levels. Nature Genetics. 2009. [DOI] [PMC free article] [PubMed]
- 60.Ferrucci L, Bandinelli S, Benvenuti E, Di Iorio A, Macchi C, et al. Subsystems contributing to the decline in ability to walk: bridging the gap between epidemiology and geriatric practice in the InCHIANTI study. J Am Geriatr Soc. 2000;48:1618–1625. doi: 10.1111/j.1532-5415.2000.tb03873.x. [DOI] [PubMed] [Google Scholar]
- 61.Shock NW, Greulich RC, Arenberg D, Costa PT, Lakatta EG, et al. Washington, D.C.: National Institutes of Health; 1984. Normal Human Aging: The Baltimore Longitudinal Study of Aging. [Google Scholar]
- 62.Russo CR, Lauretani F, Bandinelli S, Bartali B, Di Iorio A, et al. Aging bone in men and women: beyond changes in bone mineral density. Osteoporos Int. 2003;14:531–538. doi: 10.1007/s00198-002-1322-y. [DOI] [PubMed] [Google Scholar]
- 63.Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu Rev Genomics Hum Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 65.Johnson T, Kutalik Z. QUICKTEST. 2008.
- 66.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin–rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 67.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 68.Bacanu SA, Devlin B, Roeder K. The power of genomic control. Am J Hum Genet. 2000;66:1933–1944. doi: 10.1086/302929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 70.Dudbridge F, Gusnanto A. Estimation of significance thresholds for genomewide association scans. Genet Epidemiol. 2008;32:227–234. doi: 10.1002/gepi.20297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 72.McVean GA, Myers SR, Hunt S, Deloukas P, Bentley DR, et al. The fine-scale structure of recombination rate variation in the human genome. Science. 2004;304:581–584. doi: 10.1126/science.1092500. [DOI] [PubMed] [Google Scholar]
- 73.Winckler W, Myers SR, Richter DJ, Onofrio RC, McDonald GJ, et al. Comparison of fine-scale recombination rates in humans and chimpanzees. Science. 2005;308:107–111. doi: 10.1126/science.1105322. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Quantile-Quantile plots of genome-wide association results. Observed -log10 p-values on the y-axis are plotted against theoretical -log10 p-values on the x-axis resulting from each meta-analysis. Results are color-coded by chromosome. The top results largely derive from the CASR locus on chromosome 3. Results are shown separately for (A) all cohorts, (B) European cohorts and (C) Indian Asian cohorts.
(6.48 MB TIF)
Study-specific quantile-quantile plots. Shown are observed -log10 p-values plotted against expected -log10 p-values resulting from each single study after applying genomic control correction. The study-specific λ-values were λ = 1.0139 (CoLaus), λ = 0.9891 (LOLIPOP_EWA), λ = 0.9994 (LOLIPOP_EWP), λ = 0.9967 (LOLIPOP_IAA), λ = 1.0131 (LOLIPOP_IAI), λ = 0.9985 (LOLIPOP_IAP), λ = 0.9842 (InCHIANTI), λ = 1.0019 (BLSA). For the combined European and Indian Asian, European only and Indian Asian only meta-analyses the inflation factors were 1.0207, 1.0068, and 1.0286, respectively.
(4.36 MB TIF)
Comparison of significance across cohorts. The effect size and 95% confidence intervals of SNPs which do not reach genome-wide significance in the combined European and Indian Asian meta-analysis are shown separately for each cohort (CoLaus, LOLIPOP_EWA, LOLIPOP_EWP, LOLIPOP_IAA, LOLIPOP_IAI, LOLIPOP_IAP, BLSA, InCHIANTI). European cohorts are drawn in blue and Indian Asian cohorts are drawn in green. The size of the box is proportional to the precision 1/se2 and the meta-analysis estimate and 95% confidence interval across all cohorts is given by a diamond.
(0.38 MB TIF)
Candidate gene QQ-plots. For 18611 SNPs mapping to candidate genes (excluding CASR), we compare observed -log10 p-values to the mean quantiles of the uniform distribution. As a comparison, we randomly choose a set of genes from which we select the same number of SNPs. From 1,000 random draws we calculate the 95th percentile of -log10 p-values (in blue). Results are shown separately for all cohorts, European only and Indian Asian only (A–C). CoLaus permuted phenotype results comparing observed p-values to the 95th percentile of -log10 p-values from 100 permutations are shown in (D).
(0.41 MB TIF)
Genotyping, imputation, and analysis procedures by study. The genotyping platforms, quality control (QC) filters applied before imputation, imputation software, number of SNPs, and genotype-phenotype association software are shown for each study.
(0.05 MB DOC)
Complete list of genome-wide significant SNPs. Below is the list of all SNPs that exceeded the threshold of genome-wide significance (p<5×10e-8). Position is given for NCBI Build 35. Meta-analysis is performed by inverse variance weighted fixed effect regression. The coded allele is the allele to which the beta (effect) estimate refers. Results are shown separately for (A) European and Indian Asian cohorts, (B) European cohorts only, and (C) Indian Asian cohorts only.
(0.24 MB DOC)
Significance of top SNPs by cohort. Shown are study-specific results of the SNPs with genomic control (GC) p-values <1E-05 filtered by distinct regions, determined by merging SNPs within 1 Mb of each other. Results are shown separately for (A) European and Indian Asian cohorts, (B) European cohorts, and (C) Indian Asian cohorts.
(0.08 MB DOC)
Number of cases and controls for calcium-related outcomes. For several related phenotypes, we test the association of rs1801725 with these binary responses. Shown here are the number of cases and controls for each phenotype in each cohort and the total across cohorts.
(0.04 MB DOC)
Logistic regression of clinical phenotypes on rs1801725. We report the effect size and standard error of the rs1801725 T allele from logistic regressions of each clinical phenotype.
(0.04 MB DOC)
Studies of CASR mutations and serum calcium. A survey of previous studies which investigate the relationship between CASR mutations and levels of serum calcium.
(0.05 MB DOC)