Abstract
The evolution of duplicate genes has been a topic of broad interest. Here, we propose that the conservation of gene family size is a good indicator of the rate of sequence evolution and some other biological properties. By comparing the human–chimpanzee–macaque orthologous gene families with and without family size conservation, we demonstrate that genes with family size conservation evolve more slowly than those without family size conservation. Our results further demonstrate that both family expansion and contraction events may accelerate gene evolution, resulting in elevated evolutionary rates in the genes without family size conservation. In addition, we show that the duplicate genes with family size conservation evolve significantly more slowly than those without family size conservation. Interestingly, the median evolutionary rate of singletons falls in between those of the above two types of duplicate gene families. Our results thus suggest that the controversy on whether duplicate genes evolve more slowly than singletons can be resolved when family size conservation is taken into consideration. Furthermore, we also observe that duplicate genes with family size conservation have the highest level of gene expression/expression breadth, the highest proportion of essential genes, and the lowest gene compactness, followed by singletons and then by duplicate genes without family size conservation. Such a trend accords well with our observations of evolutionary rates. Our results thus point to the importance of family size conservation in the evolution of duplicate genes.
Keywords: gene duplication, gene essentiality, gene family size conservation, singleton, primate evolution
Introduction
Gene duplication has been a focus of molecular evolution for decades for its importance in functional innovation and its influences on the evolutionary rates of genes. The evolution of duplicate genes, however, is an issue of debate. Traditionally, duplicate genes are considered as functionally redundant and thus are subject to positive selection or relaxed purifying selection (Ohno 1970; Johnson et al. 2001; Fortna et al. 2004; Nei and Rooney 2005; Bailey and Eichler 2006). Therefore, duplicate genes are expected to evolve faster than singletons, a view that is supported by a wide spectrum of studies (Garczarek et al. 2000; Johnson et al. 2001; Kondrashov et al. 2002; Nembaware et al. 2002; Ranson et al. 2002; McLysaght et al. 2003; Han et al. 2009; Jaillon et al. 2009). However, it has also been reported that duplicate genes tend to evolve at a slower pace than singletons (Yang et al. 2003; Davis and Petrov 2004; Jordan et al. 2004). It was suggested that the evolutionary rates of duplicate genes reflect the functionality of the ancestral singletons (Jordan et al. 2004) and that structural constraints may have more influence than dispensability on the evolutionary rate of a protein (Yang et al. 2003). These observations imply that factors other than duplication per se are involved in the evolution of duplicate genes.
Generally, duplicate genes are destined to one of the three evolutionary fates: subfunctionalization, neofunctionalization, and pseudogenization (Ohno 1970). In the former two cases, the functions of the duplicated genes diverge from each other. The function of each copy is retained for phenotypic stability or functional innovation. Therefore, the duplicated genes will be subject to purifying selection after the subfunctionalization or neofunctionalization (where positive selection can be involved; Han et al. 2009). The size of such a gene family is also expected to stabilize afterward within reasonable time, for the subsequent deduction of genes will be detrimental, whereas addition of gene copies may lead to dosage imbalance or genetic perturbation (He and Zhang 2006). In comparison, in the case of pseudogenization, the numbers of duplicate genes are instable because of the gene birth-and-death process (Nei and Rooney 2005). The evolution of such genes is thus expected to be rapid. Nevertheless, it is noteworthy that the three processes (i.e., subfunctionalization, neofunctionalization, and pseudogenization) can alternatively occur during the evolution of gene families. In other words, the numbers of genes in the current gene families may be the mixed results of these processes. By comparing the genomes of closely related species, we may be able to find differences in evolutionary rates between functionally stable families (of which the family sizes are conserved across species) and the families where functional divergence is occurring (where the birth-and-death process keeps going on). Therefore, we reason that the conservation of gene family size in multiple species may reflect certain functional constraints (e.g., dosage balance), which in turn could influence the evolutionary rates of duplicate genes. To test this hypothesis, we conducted an extensive analysis to compare the evolutionary rates of duplicate genes of families with and without size conservation in three closely related primates—human, chimpanzee, and rhesus macaque, whose genomes are completely (or nearly completely) sequenced and well annotated. Furthermore, the relatively short divergence times among these species and the large numbers of gene families that each genome possesses enable us to systematically analyze the dynamics of duplicate genes in recent primate evolution. Because evolutionary rates are correlated with other biological factors, such as gene essentiality (Gu et al. 2003; He and Zhang 2006; Li et al. 2006), expression level (Pal et al. 2001; Drummond et al. 2005; Liao et al. 2006), expression breadth (Duret and Mouchiroud 2000; Winter et al. 2004; Zhang and Li 2004; Liao et al. 2006), and gene compactness (Liao et al. 2006), we also studied the relationships between gene family size conservation and these biological features.
Materials and Method
Data Retrieval
The protein-coding genes in human, chimpanzee, and macaque; gene families; orthology assignments; single-nucleotide polymorphism (SNP) data; and human–chimpanzee and human–macaque evolutionary rates (including the Ka, Ks, and Ka/Ks values) were downloaded from the Ensembl Genome Browser at http://www.ensembl.org/ (version 49). Note that the gene families and orthology assignments were determined using the Markov Cluster algorithm (Enright et al. 2002) with Ensembl parameter settings. The longest isoform was selected in cases of alternative splicing. For the analysis of gene essentiality, two data sets were used to avoid potential data bias: human essential genes (Liao and Zhang 2008) and human orthologues of mouse lethal genes (Liao and Zhang 2007). The former set includes 120 human genes that are associated with lethality before puberty or infertility (Jimenez-Sanchez et al. 2001). The latter includes the human orthologues of 2,301 mouse genes, for which the homozygous null mutations were annotated as lethal before reproduction or as sterile (the knockout phenotypes were downloaded from the Mouse Genome Informatics at http://www.informatics.jax.org/). The retroposed genes with introns were downloaded from the study by Fablet et al. (2009). The gene expression levels were determined with reference to the data set downloaded from http://biogibbs.stanford.edu/∼yxing/MBE/. This data set was generated by examining the transcriptomes of six human tissues (heart, kidney, liver, muscle, spleen, and testis) using a high-density exon array platform (Xing et al. 2007). The expression level of a gene was defined as the average signal intensity across these six examined tissues. The expression breadths of human genes were derived from Gene Atlas V2 data set (http://symatlas.gnf.org/), of which 73 nonpathogenic tissues were selected for the analysis of the study. We used an average difference value of 200 as the threshold for a gene to be considered as expressed in a given tissue (Su et al. 2002; Yang et al. 2005). To assign the probe set to the Ensembl-annotated genes, we aligned the sequences of each probe set against the Ensembl coding sequences (CDSs) using the BLAST (basic local alignment search tool) package. Only the probe sets that were 100% identical to the coding sequences were considered. To avoid ambiguity, we removed any probe set that matched to more than one gene.
Extraction of Human–Chimpanzee–Macaque Orthologous Families
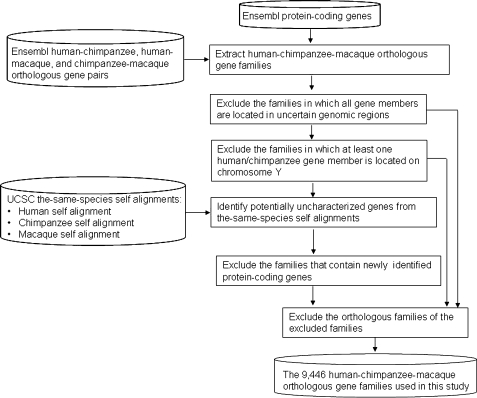
In this study, only the families that are present in all the three primate genomes (designated as “H-C-M” homologous families) are considered. To minimize potential errors in family size assignments, we excluded three types of families (also see fig. 1): 1) the families of which all the member genes are located in uncertain genomic regions, 2) the families that include at least one human or chimpanzee member gene that is located on chromosome Y (because the sequences of Y chromosome are unavailable for rhesus macaque), and 3) the families that include potentially uncharacterized protein-coding genes (which will be described in the next paragraph). Consequently, a total of 9,446 H-C-M families were extracted, which contained 17,211 human genes, 16,580 chimpanzee genes, and 17,402 macaque genes (table 1). The H-C-M families were further divided into two types of families: families whose sizes remain the same (i.e., H=C=M families) and those whose sizes vary among the three species (i.e., non-H=C=M families). To eliminate the potential confounding factor of gene copy number variations (CNVs), the families that included at least one human member gene that overlaps with experimentally validated genomic CNVs (downloaded from the Database of Genomic Variants [Iafrate et al. 2004] at http://projects.tcag.ca/variation/) were excluded from the H=C=M families (table 1). Throughout this study, “H=C=M families” indicate the H=C=M families in which the human genes do not overlap with any CNVs. The list of the human, chimpanzee, and macaque genes analyzed in this study is available at http://idv.sinica.edu.tw/trees/Duplication/Duplication.html.
FIG. 1.
The data collation processes.
Table 1.
The Human–Chimpanzee–Macaque Orthologous Gene Families Analyzed in this Study.
| Types of Gene Families | Number of Families | Number of Genes |
||
| H | C | M | ||
| H-C-M families (H, C, M>0) | 9,446 | 17,211 | 16,580 | 17,402 |
| H=C=M | 5,318 | 6,750 | 6,750 | 6,750 |
| H=C=M=1 | 4,343 | 4,343 | 4,343 | 4,343 |
| H=C=M>1 | 975 | 2,407 | 2,407 | 2,407 |
| Non-H=C=M | 1,639 | 5,985 | 5,354 | 6,176 |
| Dup-H≠C≠M (or H≠C≠M; H,C,M>1) | 143 | 1,359 | 1,102 | 1,251 |
Identification of Potentially Uncharacterized Protein-Coding Genes
To identify potential protein-coding genes that have not been annotated by Ensembl, we downloaded all the genomic regions that were alignable with Ensembl-annotated protein-coding genes in the same-species self-chained alignments from CNVVdb (Chen et al. 2009) at http://cnvvdb.genomics.sinica.edu.tw/. Subsequently, all the protein isoforms of each Ensembl gene were aligned against these sequences using the BLAT protein-to-DNA alignment package (Kent 2002). A potentially uncharacterized protein-coding gene was defined as such a BLAT match that satisfies all the following criteria: 1) it was not an annotated functional gene or pseudogene (annotated by Ensembl or PseudoPipe; Zhang et al. 2006), 2) it was not shorter than 80% of the length of the query protein, 3) it was not disrupted by any premature stop codons, 4) its “internal exons” were flanked by legal splicing sites (i.e., the GT-AG/GC-AG rule), and 5) it had a start codon. Accordingly, we identified 146, 117, and 101 potentially uncharacterized genes in human, chimpanzee, and rhesus macaque, respectively. The gene families that contained these potentially uncharacterized genes were excluded from our analyses because the sizes of such families were uncertain.
Measurement of Tissue Specificity
The tissue specificity (τ) is defined as 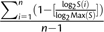 , where n is the number of human tissues examined in this study (i.e., n = 73), S(i) indicates the signal intensity (Hubbell et al. 2002) of the gene of interest in tissue i, and Max(S) is the highest expression signal of the gene across all examined tissues (Yanai et al. 2005). A large τ value indicates high tissue specificity. Note that S(i) is arbitrarily set as 100 if it is smaller than 100. This practice can minimize the influence of noises caused by low signal intensities of the expression data (Liao and Zhang 2006; Liao et al. 2006).
, where n is the number of human tissues examined in this study (i.e., n = 73), S(i) indicates the signal intensity (Hubbell et al. 2002) of the gene of interest in tissue i, and Max(S) is the highest expression signal of the gene across all examined tissues (Yanai et al. 2005). A large τ value indicates high tissue specificity. Note that S(i) is arbitrarily set as 100 if it is smaller than 100. This practice can minimize the influence of noises caused by low signal intensities of the expression data (Liao and Zhang 2006; Liao et al. 2006).
Results and Discussion
Genes with Family Size Conservation Evolve More Slowly than Those without Family Size Conservation
We first compare the evolutionary rates of the H=C=M families and the non-H=C=M families. As shown in table 2, the median Ka, Ks, and Ka/Ks values of human–chimpanzee and human–macaque 1:1 orthologues of the H=C=M families are all significantly smaller than those of the non-H=C=M families (all P values < 0.05 by the two-tailed Wilcoxon rank sum test). This result suggests that in primates, the genes of size-conserved families evolve more slowly than those of non-size–conserved families at both the RNA and protein levels.
Table 2.
The Evolutionary Rates (Ka, Ks, and the Ka/Ks ratio) of the Human–Chimpanzee and Human–Macaque 1:1 Orthologous Gene Pairs in H=C=M and non-H=C=M families.
| Human vs. Chimpanzee (median value) |
Human vs. Macaque (median value) |
|||||
| Types of Gene Families | Ka | Ks | Ka/Ks | Ka | Ks | Ka/Ks |
| H=C=M | 0.0027 | 0.0108 | 0.2516 | 0.0140 | 0.0662 | 0.2093 |
| Non-H=C=M | 0.0030 | 0.0119 | 0.2715 | 0.0169 | 0.0762 | 0.2301 |
| P valuea | <10−4 | <10−6 | <0.05 | <10−11 | <10−15 | <0.001 |
P values were estimated by using the two-tailed Wilcoxon rank sum test.
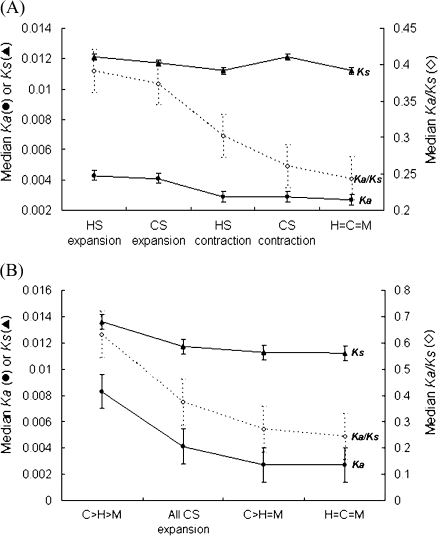
The next question to ask is whether family size expansion and contraction have different effects on the evolutionary rates of the affected families. Using rhesus macaque as the outgroup species, we can extract from non-H=C=M families the families that have potentially undergone expansion or contraction in human and chimpanzee and classify them into four subgroups (table 3): human-specific (HS) expansion (281 families), HS contraction (91 families), chimpanzee-specific (CS) expansion (87 families), and CS contraction (289 families). The HS/CS expansion and contraction families may have, respectively, experienced net gene gain and loss events in terms of maximum parsimony of evolution. Figure 2A illustrates the median Ka, Ks, and Ka/Ks values between human genes and their chimpanzee counterparts in these four subgroups compared with those of the H=C=M families. We have four observations. First, the median Ka and Ka/Ks values are significantly higher in the HS expansion families than in the CS contraction families, both of which have a larger number of genes in human than in chimpanzee (fig. 2A, both P values < 0.001). A similar trend is also observed in the comparison between the CS expansion and HS contraction families, both of which have a larger family size in chimpanzee than in human (fig. 2A, both P values < 0.05).
Table 3.
Potential Family Expansion/Contraction in the Human and Chimpanzee Genomes with Reference to the Family Sizes of the Corresponding Rhesus Macaque Families.
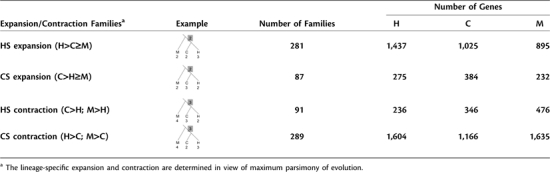 |
FIG. 2.
Comparisons of median Ka (filled circles), Ks (filled triangles), and Ka/Ks (open diamonds) values of human genes and their closest counterparts in chimpanzee in (A) HS/CS expansion, HS/CS contraction, and H=C=M families; and (B) the C>H>M, all CS expansion (C>H≥M), C>H=M, and H=C=M families. Error bars represent the standard errors.
Second, the median Ka and Ka/Ks values are higher in HS/CS expansion families than in HS/CS contraction families. Previous studies have indicated that amino acid substitutions accelerate after gene duplication (Garczarek et al. 2000; Johnson et al. 2001; Ranson et al. 2002; McLysaght et al. 2003) and that gene family size changes may accelerate gene evolution (Demuth et al. 2006). Our results suggest that the previous view can be further refined in that genes of the expanded families tend to evolve faster than those of the contracted families.
Third, the median Ka and Ka/Ks values are both significantly higher in the CS/HS expansion families than in the H=C=M families (all P value < 0.001; fig. 2A). For a CS expansion family (C>H≥M), the “extra” gene copies in chimpanzee may be functionally redundant and thus be subject to weak selection pressure. Because we only consider the human genes and their closest counterparts in chimpanzee, the extra chimpanzee gene copies are not included in the calculation of evolutionary rates. We thus expect the evolutionary rates in this group to be close to those in the H=C=M group. Surprisingly, however, we find the opposite to be true. To see why, we divide the CS expansion families (including 275 human genes) into two subgroups: the families of which the sizes are the same in human and macaque (C>H=M, including 168 human genes) and the families of which the sizes differ between human and macaque (C>H>M, including 107 human genes). We find that the median Ka, Ks, and Ka/Ks values of the C>H=M families are very close to those of the H=C=M families (the difference is statistically insignificant, see fig. 2B). The result implies that the genes in the C>H=M and those in the H=C=M families are under similar levels of selection pressure. In contrast, the other subgroup (C>H>M) of families have significantly higher median Ka, Ks, and Ka/Ks values than the CS expansion families (all P values < 0.001) and, of course, the H=C=M families (fig. 2B). The result indicates that the elevated Ka and Ka/Ks values in the CS expansion families are dominated by the C>H>M families, of which the sizes are not conserved between human and rhesus macaque. To see whether this observation is generally true, we performed a similar study for the HS expansion families (H>C≥M). We find that the trend that the elevated Ka and Ka/Ks values in the HS expansion families are dominated by the families without the family size constraint remains the same (see supplementary fig. S1, Supplementary Material online). Furthermore, to investigate whether the “extra” human gene copies (which have no orthologs in chimpanzee) are indeed subject to relaxed purifying selection, we compared the coding SNP density of these extra genes and that of the genes that have orthologues in chimpanzee. We find that the former genes tend to have a higher average coding SNP density than the latter (6.9 vs. 5.8 SNPs per kb, P value < 0.05 by the two-tailed t-test), implying that these extra gene copies tend to be functionally redundant. With the above results, there are two possible reasons (which are not mutually exclusive) for the difference in evolutionary rates between size-conserved and non-size–conserved gene families. Take the CS expansion families as an example. The first reason is that, as stated above, selection pressure at the nucleotide level is associated with family size preservation. In C>H=M families, the family size constraint between human and rhesus macaque may imply stronger selective constraint at the nucleotide-level changes on these families than the C>H>M families. The second reason is that, in C>H>M families, human and chimpanzee may have experienced family expansion events or combinations of family expansion and contraction events after the human–chimpanzee divergence. These gene gain/loss events may have resulted in increased evolutionary rates in this subgroup.
Fourth, figure 2A also shows that the HS and CS contraction families both tend to have slightly higher median Ka and Ka/Ks values than the H=C=M groups (though the differences are insignificant, also see supplementary fig. S2, Supplementary Material online). It has been suggested that relaxed selection may be responsible for gene family size contraction (Demuth et al. 2006). A well-known example is the loss of olfactory receptors in primates (also found in the HS contraction families), which may have resulted from relaxed selection on odorant perception (Gilad et al. 2004).
In short, our results demonstrate that both gene family expansion and contraction events may accelerate gene evolution, resulting in elevated evolutionary rates in the non-H=C=M families. We also demonstrate that the direction of family size change is meaningful in this regard, with family size expansion accelerating gene evolution more than family size contraction.
Singletons Evolve Faster than Duplicate Genes with Family Size Conservation but More Slowly than Those without Family Size Conservation
We have shown that size-conserved gene families tend to evolve more slowly than non-size–conserved families. Since there has been a controversy on whether duplicate genes evolve faster than singleton genes, we are interested in exploring whether the factor of family size conservation plays an important role in the difference in evolutionary rates between these two types of genes. First, we compared the evolutionary rates of singletons and duplicate genes (designated as H=C=M=1 and H=C=M>1, respectively) while controlling the factor of family size conservation. As shown in table 4, genes of the H=C=M>1 families have much lower median Ka and Ka/Ks values (all P values < 10−11) than those of the H=C=M=1 families for either human–chimpanzee or human–macaque orthologues. In addition, we also find that the evolutionary rates of different H=C=M>1 subfamilies (e.g., H=C=M=2, H=C=M=3, and H=C=M>3) have similar evolutionary rates despite the variations in family size and that all these families have significantly lower Ka and Ka/Ks values than singletons (supplementary fig. S3, Supplementary Material online).
Table 4.
The Evolutionary Rates (Ka, Ks, and the Ka/Ks ratio) of the Human–Chimpanzee and Human–Macaque 1:1 Orthologous Gene Pairs in H=C=M=1, H=C=M>1, and Dup-H≠C≠M Families.
| Human vs. Chimpanzee (median value) |
Human vs. Macaque (median value) |
|||||
| Types of Gene Families | Ka | Ks | Ka/Ks | Ka | Ks | Ka/Ks |
| H=C=M=1 | 0.0030 | 0.0107 | 0.2727 | 0.0152 | 0.0658 | 0.2318 |
| H=C=M>1 | 0.0022 | 0.0109 | 0.2127 | 0.0115 | 0.0668 | 0.1728 |
| P value (H=C=M=1 vs. H=C=M>1)a | <10−11 | NS | <10−12 | <10−14 | NS | <10−15 |
| Dup-H≠C≠M (H≠C≠M; H,C,M>1) | 0.0050 | 0.0121 | 0.4237 | 0.0267 | 0.0859 | 0.3350 |
| P value (H=C=M>1 vs. Dup-H≠C≠M)a | <10−15 | <0.01 | <10−15 | <10−15 | <10−15 | <10−15 |
| P value (H=C=M=1 vs. Dup-H≠C≠M)a | <10−15 | <10−4 | <10−12 | <10−15 | <10−15 | <10−11 |
NOTE.—NS, not significant.
P values are estimated by using the two-tailed Wilcoxon rank sum test.
Second, we ask whether the evolutionary rates differ between the duplicate genes of size-conserved (i.e., H=C=M>1) and non-size–conserved families. Accordingly, we extract the non-size–conserved duplicate gene families from non-H-C-M families (i.e., H≠C≠M and H, C, M>1; designated as “dup-H≠C≠M” families) and compare the evolutionary rates of these families with those of the H=C=M>1 families. Note that the factor of gene duplicability is controlled in this comparison. Also note that the gene duplicability is unambiguous here because all the orthologous primate families include multiple genes. Table 4 shows that the genes of dup-H≠C≠M families have significantly larger median Ka and Ka/Ks values than those of both H=C=M>1 families and H=C=M=1 families for either human–chimpanzee or human–macaque orthologues (all P values < 10−11).
Taken together, duplicate genes with family size conservation tend to evolve more slowly than singletons, whereas the reverse is true for the duplicate genes without family size conservation. One possible reason is that family size preservation maintains the genetic stability important for the survival of organisms (e.g., the stability of central cellular and developmental processes) (He and Zhang 2006). Gene duplication and deletion may cause dosage imbalance or other genetic disturbances (Papp et al. 2003). Consequently, the size conservation of multigene families across species implies strong selection pressure on the size of these families, consistent with our previous observation that gene family expansion and contraction events may accelerate evolution. Our result therefore modifies the previous view of different evolutionary forces between singletons and duplicate genes and demonstrates that gene family size conservation is an informative indicator for the evolutionary rates of duplicate genes.
Considering that the differentiation between singleton and duplicate gene families may change with family clustering criteria, we examine whether different clustering criteria have affected our results. Accordingly, we BLAST-align the singleton genes against all the Ensembl protein-coding genes and reassign the singletons to the gene families whose member(s) matches the query singletons using two different cutoff thresholds at E = 0.001 and E = 0.1. This practice effectively reduces the number of singleton genes and thus can minimize the effects of the potential confounding factor that fast-evolving genes tend to be classified as singletons. In fact, the overall tendency that singletons evolve faster than H=C=M>1 families but more slowly than dup-H≠C≠M families at the protein level holds well in both of the reassigned data sets (supplementary table S1, Supplementary Material online). Therefore, gene family clustering criteria do not seem to affect our conclusions.
Another potential confounding factor in our analysis is CNV, which may result in ambiguities of H=C=M family assignments. To address this issue, we analyze the evolutionary rates of two sets of H=C=M families: those that include (supplementary table S2, Supplementary Material online) and those that do not include (tables 1, 2, and 4) the families with their member genes overlapping with human CNVs (see Material and Methods). The evolutionary rates are then compared with singletons and duplicate gene families as stated above. Again, the overall tendencies hold well, suggesting that CNVs have no significant impacts on our study (supplementary table S2, Supplementary Material online).
We then investigate whether the GC content and codon usage bias of the compared genes are associated with the observed differences in evolutionary rates. Our result shows that dup-H≠C≠M families have higher GC content and codon usage bias than those of H=C=M=1 and H=C=M>1 families (supplementary table S3, Supplementary Material online). The difference in GC content is especially conspicuous at the 4-fold degenerate sites. However, no significant differences at the 0-fold sites are observed. Therefore, GC content and codon usage bias cannot account for the increased Ka values in dup-H≠C≠M families. Furthermore, if these two sequence features indeed have caused an increase in Ks values in the dup-H≠C≠M families, the elevated Ka/Ks ratios in these families (compared with the H=C=M families) will lend even stronger support for our claim that family size conservation is an important determinant of evolutionary rates of multigene families.
Next, we are interested to know whether H=C=M=1, H=C=M>1, and dup-H≠C≠M families differ from each other in terms of biological functions, which could account for the observed differences in evolutionary rates among these families. We thus compare the distributions of gene ontology (GO) functional categories of these three types of families. Our result shows that the differences in the distribution of GO categories are not consistent with the differences in evolutionary rates of the three types of gene families (see supplementary fig. S4 and table 4). Therefore, the differences in biological functions among these gene families do not seem to correlate with the differences in evolutionary rates.
Families with Size Conservation Have a High Proportion of Essential Genes
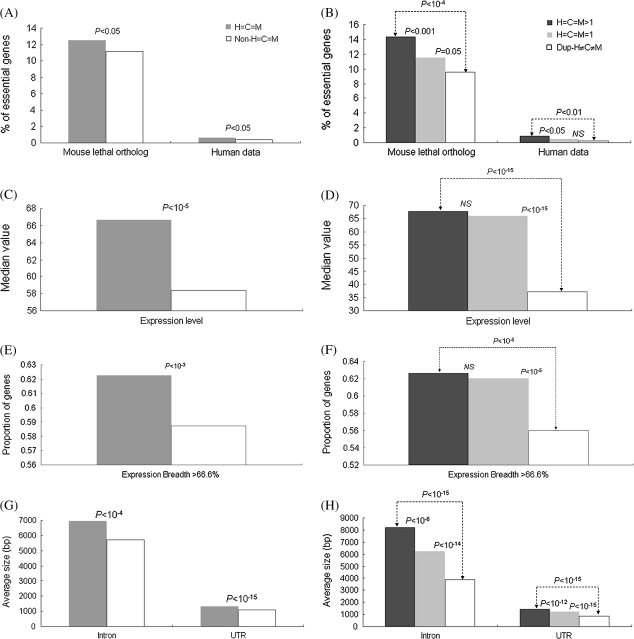
Because essential genes are known to evolve slowly (Wall et al. 2005; Zhang and He 2005; Liao et al. 2006; Larracuente et al. 2008), we then ask whether the member genes of size-conserved families tend to be essential. Accordingly, we compare the proportion of essential genes (including human essential genes [Liao and Zhang 2008] and human orthologues of mouse lethal genes [Liao and Zhang 2007]) between the H=C=M families and the non-H=C=M families. Figure 3A shows that the genes of H=C=M families have a significantly higher proportion of essential genes than those of non-H=C=M families for both data sets (both P values < 0.05). Subsequently, we probe the relationship between gene essentiality and gene duplicability. Figure 3B shows that the genes of H=C=M>1 families have a significantly higher proportion of essential genes (for both essential gene data sets) than those of singletons (both P values < 0.05). If the factor of family size conservation is not considered, the difference in the proportion of essential genes between singletons and duplicate genes becomes statistically insignificant (P values > 0.5 for both data sets by the two-tailed Fisher’s exact test). Interestingly, this observation is consistent with the previous reports that gene essentiality and gene duplicability are uncorrelated in mammals (Liang and Li 2007; Liao and Zhang 2007). Our result thus suggests that the apparent lack of correlation between gene essentiality and duplicability may not be true, supporting a recent claim that gene duplicability and essentiality are correlated after controlling for confounding factors (Liang and Li 2009). Furthermore, our analysis shows that families with size conservation tend to be more functionally important than those without size conservation. In fact, the H=C=M>1 families have the highest percentage of essential genes, followed by the H=C=M=1 families, and then by dup-H≠C≠M families (fig. 3B). The overall trend accords well with what we observe in the analysis of evolutionary rates (table 4).
FIG. 3.
The left panel compares (A) the proportions of essential genes, (C) the expression levels, (E) the expression breadth, and (G) the gene compactness (average intron/UTR length) between gene families with and without size conservation (“H=C=M” and “non-H=C=M,” respectively). The right panel (B, D, F, and H) compares the four same features in the same order between singleton gene families with size conservation (“H=C=M=1”) and multigene families with or without size conservation (“H=C=M>1” and “dup-H≠C≠M,” respectively). The P values were estimated by using the two-tailed Fisher’s exact test (A, B, E, and F), the two-tailed Wilcoxon rank sum test (C and D), and the two-tailed t-test (G and H). NS, not significant.
Families with Size Conservation Have a Higher Level of Gene Expression and Expression Breadth and a Lower Level of Gene Compactness than Those without Size Conservation
Having demonstrated that the families with size conservation tend to have low evolutionary rates and a high proportion of gene essentiality, we then ask whether family size conservation is correlated with other biological properties, such as expression level, expression breadth, and gene compactness (measured by the average lengths of introns and untranslated regions [UTRs]), all of which are known to be associated with evolutionary rates. As shown in figure 3C, E, and G, the genes of H=C=M families have a higher expression level (P value < 10−5 by the two-tailed Wilcoxon rank sum test), and a higher proportion of broadly expressed genes (P value < 10−3 by the two-tailed Fisher’s exact test), and a longer average intron/UTR length (both P values < 10−4 by the two-tailed t-test) than those of non-H=C=M families. Because evolutionary rate has been shown to be negatively correlated with expression level (Pal et al. 2001; Drummond et al. 2005; Liao et al. 2006) and expression breadth (Duret and Mouchiroud 2000; Winter et al. 2004; Zhang and Li 2004; Liao et al. 2006) but positively correlated with gene compactness (Liao et al. 2006), this observation is consistent with our results of evolutionary rate analyses. Again, for the above three biological features, the measurements of singleton genes fall in between those of H=C=M>1 families and dup-H≠C≠M families (fig. 3D, F, and H). Note that a similar trend is observed if we use the tissue specificity index τ (see Materials and Method) to reexamine expression breadth of the data (supplementary fig. S5, Supplementary Material online). Also note that because retroposed genes could contribute to short intron lengths and cause biases in the average intron size, we exclude retroposed genes and reanalyze the data. We find that the overall tendency still holds well (supplementary fig. S6, Supplementary Material online).
Concluding Remarks
In this study, we demonstrate that family size conservation is a good indicator of the biological/evolutionary features of duplicate genes. The duplicate genes of size-conserved families have lower evolutionary rates, a higher proportion of essential genes, higher expression levels, a higher proportion of broadly expressed genes, and lower gene compactness than those of non-size–conserved families. Therefore, this study points out the importance to distinguish between the two types of duplicate genes when comparing the biological features of singleton and duplicate genes. We give two examples of how this differentiation may affect evolutionary studies. First, we show that duplicate genes do not necessarily evolve faster than singleton genes. In fact, our result indicates that the duplicate genes of non-size–conserved families evolve faster than singletons, which in turn evolve faster than the duplicate genes of size-conserved families. Second, by taking into consideration the factor of family size conservation, we find a correlation between gene duplicability and essentiality. In sum, our study indicates that family size conservation is an important indicator of the evolution of duplicate genes and should be included in future studies of duplicate genes.
Supplementary Material
Supplementary tables S1–S3 and Supplementary figures S1–S6 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Supplementary Material
Acknowledgments
We especially thank Dr Ben-Yang Liao and Mr Meng-Pin Weng for technical assistance in processing the gene essentiality and expression data. This work is supported by the Genomics Research Center and Biodiversity Research Center, Academia Sinica, Taiwan; the National Health Research Institutes (NHRI), Taiwan, to T.J.C. (contract NHRI-EX98-9408PC); National Science Council, Taiwan, to T.J.C. (contract NSC 96-2628-B-001-005-MY3); NHRI intramural funding to F.C.C.; National Institute of Health (grant GM30998) to W.H.L.
References
- Bailey JA, Eichler EE. Primate segmental duplications: crucibles of evolution, diversity and disease. Nat Rev Genet. 2006;7:552–564. doi: 10.1038/nrg1895. [DOI] [PubMed] [Google Scholar]
- Chen FC, Chen YZ, Chuang TJ. CNVVdb: a database of copy number variations across vertebrate genomes. Bioinformatics. 2009;25:1419–1421. doi: 10.1093/bioinformatics/btp166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JC, Petrov DA. Preferential duplication of conserved proteins in eukaryotic genomes. PLoS Biol. 2004;2:E55. doi: 10.1371/journal.pbio.0020055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demuth JP, De Bie T, Stajich JE, Cristianini N, Hahn MW. The evolution of mammalian gene families. PLoS ONE. 2006;1:e85. doi: 10.1371/journal.pone.0000085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DA, Bloom JD, Adami C, Wilke CO, Arnold FH. Why highly expressed proteins evolve slowly. Proc Natl Acad Sci U S A. 2005;102:14338–14343. doi: 10.1073/pnas.0504070102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Mouchiroud D. Determinants of substitution rates in mammalian genes: expression pattern affects selection intensity but not mutation rate. Mol Biol Evol. 2000;17:68–74. doi: 10.1093/oxfordjournals.molbev.a026239. [DOI] [PubMed] [Google Scholar]
- Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fablet M, Bueno M, Potrzebowski L, Kaessmann H. Evolutionary origin and functions of retrogene introns. Mol Biol Evol. 2009;26:2147–2156. doi: 10.1093/molbev/msp125. [DOI] [PubMed] [Google Scholar]
- Fortna A, Kim Y, MacLaren E, et al. (16 co-authors) Lineage-specific gene duplication and loss in human and great ape evolution. PLoS Biol. 2004;2:E207. doi: 10.1371/journal.pbio.0020207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garczarek L, Hess WR, Holtzendorff J, van der Staay GW, Partensky F. Multiplication of antenna genes as a major adaptation to low light in a marine prokaryote. Proc Natl Acad Sci U S A. 2000;97:4098–4101. doi: 10.1073/pnas.070040897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilad Y, Przeworski M, Lancet D. Loss of olfactory receptor genes coincides with the acquisition of full trichromatic vision in primates. PLoS Biol. 2004;2:E5. doi: 10.1371/journal.pbio.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Steinmetz LM, Gu X, Scharfe C, Davis RW, Li WH. Role of duplicate genes in genetic robustness against null mutations. Nature. 2003;421:63–66. doi: 10.1038/nature01198. [DOI] [PubMed] [Google Scholar]
- Han MV, Demuth JP, McGrath CL, Casola C, Hahn MW. Adaptive evolution of young gene duplicates in mammals. Genome Res. 2009;19:859–867. doi: 10.1101/gr.085951.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Zhang J. Higher duplicability of less important genes in yeast genomes. Mol Biol Evol. 2006;23:144–151. doi: 10.1093/molbev/msj015. [DOI] [PubMed] [Google Scholar]
- Hubbell E, Liu WM, Mei R. Robust estimators for expression analysis. Bioinformatics. 2002;18:1585–1592. doi: 10.1093/bioinformatics/18.12.1585. [DOI] [PubMed] [Google Scholar]
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Wincker P. “Changing by doubling”, the impact of Whole Genome Duplications in the evolution of eukaryotes. C R Biol. 2009;332:241–253. doi: 10.1016/j.crvi.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Jimenez-Sanchez G, Childs B, Valle D. Human disease genes. Nature. 2001;409:853–855. doi: 10.1038/35057050. [DOI] [PubMed] [Google Scholar]
- Johnson ME, Viggiano L, Bailey JA, Abdul-Rauf M, Goodwin G, Rocchi M, Eichler EE. Positive selection of a gene family during the emergence of humans and African apes. Nature. 2001;413:514–519. doi: 10.1038/35097067. [DOI] [PubMed] [Google Scholar]
- Jordan IK, Wolf YI, Koonin EV. Duplicated genes evolve slower than singletons despite the initial rate increase. BMC Evol Biol. 2004;4:22. doi: 10.1186/1471-2148-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. BLAT–the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov FA, Rogozin IB, Wolf YI, Koonin EV. Selection in the evolution of gene duplications. Genome Biol. 2002;3:RESEARCH0008. doi: 10.1186/gb-2002-3-2-research0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larracuente AM, Sackton TB, Greenberg AJ, Wong A, Singh ND, Sturgill D, Zhang Y, Oliver B, Clark AG. Evolution of protein-coding genes in Drosophila. Trends Genet. 2008;24:114–123. doi: 10.1016/j.tig.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Li L, Huang Y, Xia X, Sun Z. Preferential duplication in the sparse part of yeast protein interaction network. Mol Biol Evol. 2006;23:2467–2473. doi: 10.1093/molbev/msl121. [DOI] [PubMed] [Google Scholar]
- Liang H, Li WH. Gene essentiality, gene duplicability and protein connectivity in human and mouse. Trends Genet. 2007;23:375–378. doi: 10.1016/j.tig.2007.04.005. [DOI] [PubMed] [Google Scholar]
- Liang H, Li WH. Functional compensation by duplicated genes in mouse. Trends Genet. 2009;25:441–442. doi: 10.1016/j.tig.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao BY, Scott NM, Zhang J. Impacts of gene essentiality, expression pattern, and gene compactness on the evolutionary rate of mammalian proteins. Mol Biol Evol. 2006;23:2072–2080. doi: 10.1093/molbev/msl076. [DOI] [PubMed] [Google Scholar]
- Liao BY, Zhang J. Low rates of expression profile divergence in highly expressed genes and tissue-specific genes during mammalian evolution. Mol Biol Evol. 2006;23:1119–1128. doi: 10.1093/molbev/msj119. [DOI] [PubMed] [Google Scholar]
- Liao BY, Zhang J. Mouse duplicate genes are as essential as singletons. Trends Genet. 2007;23:378–381. doi: 10.1016/j.tig.2007.05.006. [DOI] [PubMed] [Google Scholar]
- Liao BY, Zhang J. Null mutations in human and mouse orthologs frequently result in different phenotypes. Proc Natl Acad Sci U S A. 2008;105:6987–6992. doi: 10.1073/pnas.0800387105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLysaght A, Baldi PF, Gaut BS. Extensive gene gain associated with adaptive evolution of poxviruses. Proc Natl Acad Sci U S A. 2003;100:15655–15660. doi: 10.1073/pnas.2136653100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Rooney AP. Concerted and birth-and-death evolution of multigene families. Annu Rev Genet. 2005;39:121–152. doi: 10.1146/annurev.genet.39.073003.112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nembaware V, Crum K, Kelso J, Seoighe C. Impact of the presence of paralogs on sequence divergence in a set of mouse-human orthologs. Genome Res. 2002;12:1370–1376. doi: 10.1101/gr.270902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. Evolution by gene and genome duplication. Berlin: Springer; 1970. [Google Scholar]
- Pal C, Papp B, Hurst LD. Highly expressed genes in yeast evolve slowly. Genetics. 2001;158:927–931. doi: 10.1093/genetics/158.2.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp B, Pal C, Hurst LD. Dosage sensitivity and the evolution of gene families in yeast. Nature. 2003;424:194–197. doi: 10.1038/nature01771. [DOI] [PubMed] [Google Scholar]
- Ranson H, Claudianos C, Ortelli F, Abgrall C, Hemingway J, Sharakhova MV, Unger MF, Collins FH, Feyereisen R. Evolution of supergene families associated with insecticide resistance. Science. 2002;298:179–181. doi: 10.1126/science.1076781. [DOI] [PubMed] [Google Scholar]
- Su AI, Cooke MP, Ching KA, et al. (14 co-authors) Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci U S A. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall DP, Hirsh AE, Fraser HB, Kumm J, Giaever G, Eisen MB, Feldman MW. Functional genomic analysis of the rates of protein evolution. Proc Natl Acad Sci U S A. 2005;102:5483–5488. doi: 10.1073/pnas.0501761102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter EE, Goodstadt L, Ponting CP. Elevated rates of protein secretion, evolution, and disease among tissue-specific genes. Genome Res. 2004;14:54–61. doi: 10.1101/gr.1924004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Ouyang Z, Kapur K, Scott MP, Wong WH. Assessing the conservation of mammalian gene expression using high-density exon arrays. Mol Biol Evol. 2007;24:1283–1285. doi: 10.1093/molbev/msm061. [DOI] [PubMed] [Google Scholar]
- Yanai I, Benjamin H, Shmoish M, et al. (12 co-authors) Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics. 2005;21:650–659. doi: 10.1093/bioinformatics/bti042. [DOI] [PubMed] [Google Scholar]
- Yang J, Gu Z, Li WH. Rate of protein evolution versus fitness effect of gene deletion. Mol Biol Evol. 2003;20:772–774. doi: 10.1093/molbev/msg078. [DOI] [PubMed] [Google Scholar]
- Yang J, Su AI, Li WH. Gene expression evolves faster in narrowly than in broadly expressed mammalian genes. Mol Biol Evol. 2005;22:2113–2118. doi: 10.1093/molbev/msi206. [DOI] [PubMed] [Google Scholar]
- Zhang J, He X. Significant impact of protein dispensability on the instantaneous rate of protein evolution. Mol Biol Evol. 2005;22:1147–1155. doi: 10.1093/molbev/msi101. [DOI] [PubMed] [Google Scholar]
- Zhang L, Li WH. Mammalian housekeeping genes evolve more slowly than tissue-specific genes. Mol Biol Evol. 2004;21:236–239. doi: 10.1093/molbev/msh010. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Carriero N, Zheng D, Karro J, Harrison PM, Gerstein M. PseudoPipe: an automated pseudogene identification pipeline. Bioinformatics. 2006;22:1437–1439. doi: 10.1093/bioinformatics/btl116. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.