Abstract
Comparison of expression levels and breadth and evolutionary rates of intronless and intron-containing mammalian genes shows that intronless genes are expressed at lower levels, tend to be tissue specific, and evolve significantly faster than spliced genes. By contrast, monomorphic spliced genes that are not subject to detectable alternative splicing and polymorphic alternatively spliced genes show similar statistically indistinguishable patterns of expression and evolution. Alternative splicing is most common in ancient genes, whereas intronless genes appear to have relatively recent origins. These results imply tight coupling between different stages of gene expression, in particular, transcription, splicing, and nucleocytosolic transport of transcripts, and suggest that formation of intronless genes is an important route of evolution of novel tissue-specific functions in animals.
Keywords: alternative splicing, intronless genes, monomorphic genes, polymorphic genes, mammalian gene evolution
Introduction
In all eukaryotes, at least some genes contain introns, and in multicellular organisms, genes with multiple introns constitute a substantial majority (Roy and Gilbert 2006). Moreover, alternative splicing, with additional contributions from alternative transcription initiation and termination, is the basis for the functional diversity of the transcriptomes in multicellular eukaryotes, at least, in vertebrates (Blencowe 2006; Kim et al. 2008). The extent of alternative splicing in multicellular organisms has been repeatedly revised upward (Mironov et al. 1999; Modrek et al. 2001; Lareau et al. 2004). The latest estimates using deep sequencing of the human transcriptome suggest that over 90% of human intron-containing genes are alternatively spliced at least in some tissues and under some conditions (Wang et al. 2008).
Introns enhance the efficiency of transcription initiation and elongation in spliced genes. Moreover, due to interactions between spliceosomal proteins and the polyadenylation machinery, messenger RNA (mRNA) nuclear export receptors, and RNA-binding proteins, splicing can actively promote 3′-end formation, polyadenylation, and mRNA export (Le Hir et al. 2003) and enhance transcript stability (Wang et al. 2007). It has been suggested that expression profiles of monomorphic genes from which only a single transcript is produced substantially differ from those of polymorphic genes whose transcripts are diversified via alternative splicing as well as alternative transcription (Wang et al. 2008; Wegmann et al. 2008). Here, we analyze expression and evolution of different architectural classes of human genes and reveal dramatic differences between intronless and spliced genes but not between monomorphic and polymorphic genes.
Gene Architectures, Expression Patterns, and Evolutionary Rates Dramatically Differ between Spliced and Intronless Genes
We analyzed the architectures, expression profiles, and rates of evolution of annotated human transcripts deposited in the major sequence databases. The majority of transcripts in the University of California–Santa Cruz (UCSC) and Ensembl databases were assigned to alternatively spliced and/or alternatively transcribed genomic loci, in agreement with the notion that alternative events occur in most human genes. This dominance of alternatively expressed genes notwithstanding a considerable fraction of genes possess only one annotated transcript (ca. 38%) or even contain no introns at all (ca. 5%) (table 1), according to UCSC database (the discrepancy between the latest estimates of the extent of alternative splicing obtained through deep sequencing of the human transcriptome [Wang et al. 2008] and the fraction of genes that are annotated as being alternatively spliced in the current databases most likely stems from the lack of annotation of isoforms produced at low levels). Given that we analyzed only intact genes with readily detectable levels of transcription, we assumed that all these were bona fide functional genes, rather than pseudogenes.
Table 1.
Intronless, monomorphic, and polymorphic genes in different age classes of human genes.
| Number of Genes | Evolutionary Age of Genes |
||||||
| Gene Type | Cellular | Eukaryota | Metazoa | Chordata | Mammalia | Primates | |
| Polymorphic | 11,082 | 3,111 (0.609) | 3,310 (0.617) | 2,474 (0.560) | 1,623 (0.481) | 505 (0.389) | 59 (0.249) |
| Monomorphic | 7,651 | 1,903 (0.372) | 1,874 (0.349) | 1,745 (0.395) | 1,325 (0.393) | 645 (0.497) | 159 (0.671) |
| Intronless | 1,072 | 96 (0.019) | 184 (0.034) | 199 (0.045) | 426 (0.126) | 148 (0.114) | 19 (0.080) |
| Mono/Poly | 0.883 | 0.826 | 1.022 | 1.185 | 1.826 | 4.2 | |
| Intronless/Poly | 0.317 | 0.541 | 0.834 | 2.719 | 3 | 3.4 | |
Protein CDSs of 22,849 human genes were classified according to the ancestral taxa where the proteins first appeared. Ancestral homologues were identified in RefSeq sequences from organisms of different taxonomic levels by BLASTP search with an expectation cutoff value of 10−6 using a procedure described in Supplementary Material online. Frequencies of gene types in evolutionary age classes are shown in parentheses. Mono/poly and intronless/poly ratios were normalized taking into account numbers of genes in the groups.
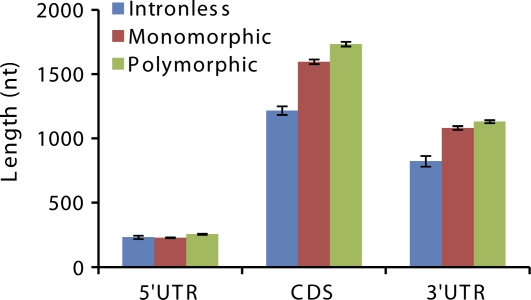
In an attempt to gain insight into the relationships between the complexity of gene architecture and expression, on the one hand, and gene evolution, on the other hand, we classified mammalian gene loci into three classes: 1) intronless genes, 2) monomorphic genes with one annotated isoform, and 3) polymorphic genes producing several alternative transcripts. Among intronless genes, coding sequences (CDSs) and 3′ untranslated regions (3′UTRs) are on average substantially shorter than the respective domains of intron-containing genes (P < 10−30 for CDS, P < 10−8 for 3′UTR; hereinafter, all P values were calculated using the Mann–Whitney U test) (fig. 1), although for 5′UTRs this effect is marginal. The relationship between monomorphic and polymorphic genes is also complex: the CDS and 3′UTRs of polymorphic genes tend to be somewhat longer than those of monomorphic genes (P < 10−6, P < 10−8), whereas the 5′UTRs are on average not dramatically different from those in monomoprhic genes (P < 0.005). As expected, polymorphic genes on average have a greater number of introns than monomorphic genes (P < 10−5); the difference in intron density is not so pronounced but significant as well (P < 0.01; supplementary table S1, Supplementary Material online). Similarly, CDS and 3′UTRs of intronless genes in mouse are significantly shorter than those in intron-containing genes (data not shown).
FIG. 1.
Means of functional domain lengths in mature transcripts of intronless, monomorphic, and polymorphic genes. Gene structure and lengths of functional domains were determined from genomic coordinates of the human genome (March 2006 assembly) that were downloaded from the UCSC genome server (http://genome.ucsc.edu). Monomorphic or polymorphic gene status was defined from the number of annotated transcript isoforms, as described in Supplementary Material online.
Intronless genes typically are expressed at a significantly lower level and in a narrower range of tissues than monomorphic or polymorphic genes (fig. 2; P < 10−65 and P < 10−82 for expressed sequence tag [EST], P < 10−13 and P < 10−12 for the Genomics Institute of the Novartis Research Foundation [GNF] Atlas 2, respectively). The same trends were observed when mammal-specific and primate-specific intronless genes were excluded from the analysis in order to eliminate any possibility of contamination of the set of intronless genes with pseudogenes (data not shown). By contrast, there was no dramatic difference in the expression of monomorphic as compared with polymorphic genes, and among the polymorphic genes, no strong dependence of expression on the number of isoforms was observed (fig. 2). The same trends were observed for mouse intronless, monomorphic, and polymorphic genes, as inferred from the analysis of the mouse GNF Atlas 2 expression data (supplementary fig. S1A, Supplementary Material online). Notably, when monomorphic and intronless genes were pooled together, as it was done in a previous study (Wegmann et al. 2008), expression of the pooled group significantly and consistently differed from the expression of polymorphic genes (supplementary fig. S1B, Supplementary Material online), in agreement with the observations of Wegmann et al. (2008). Taking into account that retroposed genes have a characteristic property to acquire introns in 5′UTRs after retroposition (Brosius and Gould 1992; Brosius 1999), we also analyzed separately the group of genes with completely intronless CDSs and with intron-containing 5′UTRs. These genes are few in numbers and show intermediate values of expression level and breadth between intronless and monomorphic genes (supplementary table S2, Supplementary Material online). The expression breadth for this group of genes (with intronless CDS and intron-containing 5′UTRs) was significantly different from the expression levels of both intronless and monomorphic genes (P < 5 × 10−3 and P < 10−9 from EST data; P < 10−2 and P < 5 × 10−5 from GNF Atlas 2 data; supplementary table S2, Supplementary Material online). Similar relationships were observed for expression level in these three groups of genes.
FIG. 2.
Means of expression levels and breadth in human intronless (Int0), monomorphic (Mono), and polymorphic (Poly) genes with different numbers of isoforms (from 2 to 4 and more). Gene expression levels and breadths were evaluated by tallying the numbers of gene-specific EST sequences from normal human tissues in GenBank (Ogurtsov et al. 2008) and from GenAtlas expression data, as described in Supplementary Material online.
We further found that the rates of evolution of the CDS among approximately 9,000 pairs of orthologous genes from human and macaque were significantly higher for intronless genes, as compared with spliced genes, in both non-synonymous and synonymous positions (P < 0.0001 for Kn and P < 10−7 for Ks); by contrast, the difference between the evolutionary rates of monomorphic and polymorphic genes was not significant (fig. 3). It has been shown previously that mammalian and primate-specific human and mouse genes including intronless ones evolve faster than genes of more ancient origin (Agarwal 2005; Wolf et al. 2009); however, we observed the exact same trends among “old,” evolutionarily conserved intronless genes (i.e., when mammal-specific and primate-specific genes were excluded from the analysis; see supplementary fig. S1C, Supplementary Material online). Of course, it has to be kept in mind that genes obviously are highly dynamic units, so the divide between “old” and “new” intronless genes is to some extent conditional given that some evolutionary conserved intronless could evolve by retroposition of spliced genes. For the evolutionary rates of the UTRs (K5 and K3), a different trend was observed; these domains evolve at approximately the same rates in human intronless and monomorphic genes (fig. 3). Evolutionary rates of CDSs in the group of genes with intronless CDSs and intron-containing 5′UTRs are close to those of intronless genes and the differences for both Kn and Ks are marginal between these two groups.
FIG. 3.
Means of evolutionary rates in intronless, monomorphic, and polymorphic (Poly) genes with different numbers of isoforms (from 2 to 4 and more). Rates of synonymous (dS) and non-synonymous (dN) substitutions in the protein CDSs and evolutionary rates in 5′UTRs (K5) and 3′UTRs (K3) were estimated from sequence alignments of approximately 9,000 human and macaque orthologous genes, as described previously (Ogurtsov et al. 2008).
The rate of evolution of the CDS shows significant inverse correlation with expression level in all studied model organisms (Pal et al. 2001; Drummond and Wilke 2009), and a similar trend has been reported for 3′UTRs but not for 5′UTRs (Jordan et al. 2004). In the current data set, we observed significant inverse correlations of both Kn and K3 with expression breadth among both monomorphic and polymorphic genes (P < 0.001) as well as intronless genes (P < 10−6) (supplementary fig. S2, Supplementary Material online). Given the connection between expression level and evolution rate of protein-coding genes, we performed a multiple regression analysis and found that the use of evolutionary variables alone (Kn, Kn/Ks, K5, and K3) allowed prediction of expression breadth and level independent of gene structural features, namely, lengths of introns and CDSs, numbers of introns, and number of transcribed isoforms (R = 0.238; supplementary fig. S3A, Supplementary Material online). A model that used structural parameters alone yielded R = 0.178 on the validation set (R = 0.227 on the training set; supplementary fig. S3B, Supplementary Material online). The two groups of variables had orthogonal (independent) predictive power, that is, R2 values for cumulative structural and evolutionary predictions were close to the sum of R2 values for independent structural and evolutionary predictions (see Supplementary Material online for details). The plot of predicted versus actual expression breadth for the validation set using combined parameters is shown in supplementary figure S3C (see Supplementary Material online for details). Thus, the evolutionary and structural variables independently predict the gene expression pattern. In other words, their predictions are nonredundant so that the combined model explains the maximum fraction of the variation in expression (the sum of R2 values for the two groups of variables).
Finally, we grouped human genes into several classes according to their apparent evolutionary age, that is, the phylogenetic depth at which reliable homologs are detectable for the gene in question ([Wolf et al. 2009], and see Supplementary Material online for details). Counterintuitively but in line with a previous report (Irimia et al. 2007), we found that ancient classes (i.e., genes with homologues in bacteria, archaea, or unicellular eukaryotes) were enriched for polymorphic genes as compared with the “younger” genes (table 1). Intronless genes were found to be mostly eukaryote specific, and for the majority, no homologues were detectable outside Chordata (table 1). The ratio between monomorphic and polymorphic genes was dramatically increased only in mammal-specific and primate-specific classes, whereas the ratio between intronless and alternative spliced genes was 3-fold greater in the chordate-specific group than in Metazoa-specific group (table 1). These observations are in agreement both with the findings presented above, namely, that intronless genes are expressed at a lower level and evolve faster than intron-containing genes, and with the previous report on a similar pattern of differences between “younger” and “older” genes (Wolf et al. 2009).
Concluding Remarks
The main result of the present analysis is that the most pronounced differences in expression profiles and evolutionary rates, as well as in the size of genome loci and transcribed domains are observed between intronless and spliced genes. By contrast, there was no dramatic difference between the two classes of spliced genes, monomorphic and polymorphic, in either the evolutionary or expression characteristics (with the exception of the evolutionary rates of the UTRs). When intronless and monomorphic genes were lumped together and compared with polymorphic genes, significant differences were found for all analyzed variables, creating an illusion of a major distinction between genes that undergo alternative splicing and those that do not. However, when intronless genes are isolated in a separate class, it becomes clear that splicing per se is a critical correlate of gene expression and evolutionary rates (at least that of the CDS). Most likely, this connection goes beyond correlation, that is, splicing actually is an important determinant of expression and, through expression, of gene evolutionary rates (Drummond and Wilke 2009). Indeed, several experimental studies indicate that intron-containing genes are more efficiently expressed than the same genes after removal of introns (Le Hir et al. 2003; Nott et al. 2004) and in particular that splicing enhances mRNA export from the nucleus (Reed and Hurt 2002; Valencia et al. 2008). In a more general context, these findings are compatible with the concept of extensive coupling between eukaryotic cellular machineries for transcription, splicing, nucleo-cytoplasmic transport, nonsense-mediated decay, and translation (Maniatis and Reed 2002; Maciag et al. 2006; Komili and Silver 2008). Of course, these findings reveal general trends and do not imply that there are no mechanisms for high-level expression of intronless genes as seen, in particular, for histones (Marzluff 2005).
Considering the dramatic differences in expression profile and protein evolutionary rates between intronless and spliced genes, but not between monomorphic and polymorphic genes, we submit that the very concept of a “monomorphic gene” might not be robust because any gene that carries at least one intron and hence interacts with the spliceosomal machinery has the potential of being alternatively spliced under specific conditions. This conclusion is in agreement with the recent estimate indicating that the overwhelming majority of genomic loci in mammals are subject to alternative splicing (Wang et al. 2008). Previous analyses suggest that much of this alternative splicing is conserved in evolution and by inference is functional, but a substantial fraction is aberrant and non-functional (Sorek et al. 2004; Yeo et al. 2005).
The current results highlight distinct features of intronless genes in vertebrates. It appears that many intronless genes are evolutionary innovations, so their formation, at least in part, via reverse transcription–mediated mechanisms, could be an important route of evolution of tissue-specific functions of animals (Brosius and Gould 1992). In line with their recent evolutionary origin, intronless genes mostly encode regulatory proteins and components of signal transduction pathways (Hill and Sorscher 2006). However, we would like to mention that this is a generalization, and there are examples that contradict the general rules, such as intronless histone genes, which are abundantly and ubiquitously expressed. Expression pathways and regulation of intronless genes are interesting subjects for experimental study.
Supplementary Material
Supplementary figs. S1 and S3, supplementary tables S1 and S2, and Supplementary Material are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Supplementary Material
Acknowledgments
The authors’ research is supported by the intramural funds of the Department of Health and Human Services (National Library of Medicine, National Institutes of Health).
References
- Agarwal SM. Evolutionary rate variation in eukaryotic lineage specific human intronless proteins. Biochem Biophys Res Commun. 2005;337:1192–1197. doi: 10.1016/j.bbrc.2005.09.172. [DOI] [PubMed] [Google Scholar]
- Blencowe BJ. Alternative splicing: new insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- Brosius J. Many G-protein-coupled receptors are encoded by retrogenes. Trends Genet. 1999;15:304–305. doi: 10.1016/s0168-9525(99)01783-7. [DOI] [PubMed] [Google Scholar]
- Brosius J, Gould SJ. On “genomenclature”: a comprehensive (and respectful) taxonomy for pseudogenes and other “junk DNA”. Proc Natl Acad Sci U S A. 1992;89:10706–10710. doi: 10.1073/pnas.89.22.10706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DA, Wilke CO. The evolutionary consequences of erroneous protein synthesis. Nat Rev Genet. 2009;10:715–724. doi: 10.1038/nrg2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill AE, Sorscher EJ. The non-random distribution of intronless human genes across molecular function categories. FEBS Lett. 2006;580:4303–4305. doi: 10.1016/j.febslet.2006.06.051. [DOI] [PubMed] [Google Scholar]
- Irimia M, Rukov JL, Penny D, Roy SW. Functional and evolutionary analysis of alternatively spliced genes is consistent with an early eukaryotic origin of alternative splicing. BMC Evol Biol. 2007;7:188. doi: 10.1186/1471-2148-7-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan IK, Marino-Ramirez L, Wolf YI, Koonin EV. Conservation and coevolution in the scale-free human gene coexpression network. Mol Biol Evol. 2004;21:2058–2070. doi: 10.1093/molbev/msh222. [DOI] [PubMed] [Google Scholar]
- Kim E, Goren A, Ast G. Alternative splicing: current perspectives. Bioessays. 2008;30:38–47. doi: 10.1002/bies.20692. [DOI] [PubMed] [Google Scholar]
- Komili S, Silver PA. Coupling and coordination in gene expression processes: a systems biology view. Nat Rev Genet. 2008;9:38–48. doi: 10.1038/nrg2223. [DOI] [PubMed] [Google Scholar]
- Lareau LF, Green RE, Bhatnagar RS, Brenner SE. The evolving roles of alternative splicing. Curr Opin Struct Biol. 2004;14:273–282. doi: 10.1016/j.sbi.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Le Hir H, Nott A, Moore MJ. How introns influence and enhance eukaryotic gene expression. Trends Biochem Sci. 2003;28:215–220. doi: 10.1016/S0968-0004(03)00052-5. [DOI] [PubMed] [Google Scholar]
- Maciag K, Altschuler SJ, Slack MD, Krogan NJ, Emili A, Greenblatt JF, Maniatis T, Wu LF. Systems-level analyses identify extensive coupling among gene expression machines. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100045. 2006.0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- Marzluff WF. Metazoan replication-dependent histone mRNAs: a distinct set of RNA polymerase II transcripts. Curr Opin Cell Biol. 2005;17:274–280. doi: 10.1016/j.ceb.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Mironov AA, Fickett JW, Gelfand MS. Frequent alternative splicing of human genes. Genome Res. 1999;9:1288–1293. doi: 10.1101/gr.9.12.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrek B, Resch A, Grasso C, Lee C. Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res. 2001;29:2850–2859. doi: 10.1093/nar/29.13.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott A, Le Hir H, Moore MJ. Splicing enhances translation in mammalian cells: an additional function of the exon junction complex. Genes Dev. 2004;18:210–222. doi: 10.1101/gad.1163204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogurtsov AY, Marino-Ramirez L, Johnson GR, Landsman D, Shabalina SA, Spiridonov NA. Expression patterns of protein kinases correlate with gene architecture and evolutionary rates. PLoS One. 2008;3:e3599. doi: 10.1371/journal.pone.0003599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal C, Papp B, Hurst LD. Highly expressed genes in yeast evolve slowly. Genetics. 2001;158:927–931. doi: 10.1093/genetics/158.2.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R, Hurt E. A conserved mRNA export machinery coupled to pre-mRNA splicing. Cell. 2002;108:523–531. doi: 10.1016/s0092-8674(02)00627-x. [DOI] [PubMed] [Google Scholar]
- Roy SW, Gilbert W. The evolution of spliceosomal introns: patterns, puzzles and progress. Nat Rev Genet. 2006;7:211–221. doi: 10.1038/nrg1807. [DOI] [PubMed] [Google Scholar]
- Sorek R, Shamir R, Ast G. How prevalent is functional alternative splicing in the human genome? Trends Genet. 2004;20:68–71. doi: 10.1016/j.tig.2003.12.004. [DOI] [PubMed] [Google Scholar]
- Valencia P, Dias AP, Reed R. Splicing promotes rapid and efficient mRNA export in mammalian cells. Proc Natl Acad Sci U S A. 2008;105:3386–3391. doi: 10.1073/pnas.0800250105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HF, Feng L, Niu DK. Relationship between mRNA stability and intron presence. Biochem Biophys Res Commun. 2007;354:203–208. doi: 10.1016/j.bbrc.2006.12.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegmann D, Dupanloup I, Excoffier L. Width of gene expression profile drives alternative splicing. PLoS One. 2008;3:e3587. doi: 10.1371/journal.pone.0003587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf YI, Novichkov PS, Karev GP, Koonin EV, Lipman DJ. Inaugural article: the universal distribution of evolutionary rates of genes and distinct characteristics of eukaryotic genes of different apparent ages. Proc Natl Acad Sci U S A. 2009;106:7273–7280. doi: 10.1073/pnas.0901808106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo GW, Van Nostrand E, Holste D, Poggio T, Burge CB. Identification and analysis of alternative splicing events conserved in human and mouse. Proc Natl Acad Sci U S A. 2005;102:2850–2855. doi: 10.1073/pnas.0409742102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.